Abstract
Abnormal differentiation of muscle is closely associated with aging (sarcopenia) and diseases such as cancer and type II diabetes. Thus, understanding the mechanisms that regulate muscle differentiation will be useful in the treatment and prevention of these conditions. Protein lysine acetylation and methylation are major post-translational modification mechanisms that regulate key cellular processes. In this study, to elucidate the relationship between myogenic differentiation and protein lysine acetylation/methylation, we performed a PCR array of enzymes related to protein lysine acetylation/methylation during C2C12 myoblast differentiation. Our results indicated that the expression pattern of HDAC11 was substantially increased during myoblast differentiation. Furthermore, ectopic expression of HDAC11 completely inhibited myoblast differentiation, concomitant with reduced expression of key myogenic transcription factors. However, the catalytically inactive mutant of HDAC11 (H142/143A) did not impede myoblast differentiation. In addition, wild-type HDAC11, but not the inactive HDAC11 mutant, suppressed MyoD-induced promoter activities of MEF2C and MYOG (Myogenin), and reduced histone acetylation near the E-boxes, the MyoD binding site, of the MEF2C and MYOG promoters. Collectively, our results indicate that HDAC11 would suppress myoblast differentiation via regulation of MyoD-dependent transcription. These findings suggest that HDAC11 is a novel critical target for controlling myoblast differentiation.
Keywords: HDAC11, lysine acetylation, myoblast differentiation, MyoD-dependent transcription
INTRODUCTION
The differentiation of myoblasts into myotubes is important for the development of muscle-specific structures and contractile function (Berendse et al., 2003; Burattini et al., 2004). Myoblast differentiation is regulated by two families of transcription factors (Molkentin et al., 1995; Sabourin and Rudnicki, 2000), namely, the basic helix-loop-helix (bHLH) myogenic regulatory factors (MRFs) that consist of MyoD (Myf-3) (Davis et al., 1987), Myogenin (Myf-1) (Edmondson and Olson, 1989), Myf5 (Braun et al., 1989) and Mrf4 (Myf-6) (Braun et al., 1990), and myocyte enhancer factor 2 (MEF2) factors that contain MEF2A, B, C and D (Molkentin et al., 1995). All MRFs contain a conserved basic domain for DNA binding and a bHLH domain necessary for heterodimerization with ubiquitous bHLH E-proteins (Sabourin and Rudnicki, 2000). Among the MRFs, Myf5 and MyoD are required for myogenic lineage determination and myoblast proliferation (Lassar and Skapek, 1994; Olson and Klein, 1994; Rudnicki et al., 1992; 1993; Weintraub et al., 1991), whereas Myogenin and Mrf4 play critical roles in the fusion of myoblasts into myotubes and terminal differentiation (Hasty et al., 1993, Nabeshima et al., 1993). The ability of MyoD to determine myogenic lineage depends on its functions in chromatin remodeling at silent loci and in the transcriptional activation of muscle genes (Bergstrom et al., 2002; Gerber et al., 1997). The MEF2 family, which consists of four vertebrate MEF2 family members, has been found to have muscle-specific DNA-binding activity when myoblasts differentiate into myotubes (Gossett et al., 1989; Olson et al., 1995; Rhodes and Konieczny, 1989). MEF2A, B, and D are ubiquitously expressed, whereas MEF2C is selectively expressed in skeletal muscle, brain and spleen (Martin et al., 1993; McDermott et al., 1993; Pollock and Treisman, 1991; Yu et al., 1992). The role of MEF2C during myogenic differentiation has been well studied because of its abundant expression in skeletal muscle (Martin et al., 1993; McDermott et al., 1993). MyoD is able to bind and target promoters of MEF2C and MYOG (Myogenin) in order to promote muscle specific gene expression, resulting in myotube formation (Buchberger et al., 1994; Dodou et al., 2003; Faralli and Dilworth, 2012). On the other hand, the interactions between MRFs/MEF2 and transcriptional co-repressors/co-activators may also determine muscle differentiation. Representative chromatin remodeling enzymes include lysine acetyltransferase and lysine methyltransferase (De la Serna et al., 2006; Guasconi and Puri, 2009; Puri and Sartorelli, 2000). Among the histone modifications these enzymes initiate, histone deacetylation, H3-K9 demethylation and H3-K27 trimethylation are closely associated with transcriptional suppression (heterochromatin) (Caretti et al., 2004; De la Serna et al., 2006; Guasconi and Puri, 2009; Pasini et al., 2008). Elimination of theses transcriptional corepressors from MRFs and MEF2 induces the activation of muscle specific promoters (Ait-Si-Ali et al., 2004; McKinsey et al., 2002). In contrast, H3-R8 demethylation, H3-R17 demethylation, and H3-K4 trimethylation are related to transcriptional activation (Guasconi and Puri, 2009). In addition, H3-K9 and H3-K14 histone acetylation is associated with the augmented expression of genes (Karmodiya et al., 2012; Srivastava et al., 2014).
As mentioned above, the lysine modification of histones is one of the major mechanisms that regulates gene expression. In the present study, we searched for factors related to histone acetylation and methylation that may regulate muscle differentiation. PCR-based screening analysis showed that histone deacetylase 11 (HDAC11) would be a highly influential histone modification factor involved in the regulation of myoblast differentiation. HDAC enzymes remove acetyl groups from lysine residues within histones H3 and H4 of the nucleosome. In addition, HDAC enzymes can directly bind to MyoD and MEF2, and target muscle-specific gene promoters as well as their own promoters (Iezzi et al., 2004; Lu et al., 2000; Mal, 2006; Mal and Harter, 2003; Mal et al., 2001). HDAC11 has recently been identified as a unique member of the class IV HDAC, consisting of only a catalytic domain (Gao et al., 2002; Haberland et al., 2009; Voelter et al., 2005). However, the relationship between HDAC11 and myoblast differentiation has not been established. Therefore, in this study, we investigated the effect of HDAC11 on myoblast differentiation using wild-type HDAC11 and its catalytically inactive mutant (H142/143A) (Rajendran et al., 2011; Thangapandian et al., 2011; 2012). Our novel findings of the role of HDAC11 in myoblast differentiation provide an improved understanding of the factors regulating myoblast differentiation.
MATERIALS AND METHODS
Cell culture
Murine C2C12 myoblasts were obtained from the American Type Culture Collection (ATCC). Cells were cultured in a DMEM high-glucose medium (Gibco-Invitrogen) supplemented with 15% heat-inactivated fetal bovine serum (FBS) and 1% antibiotic-antimycotic solution, as a growth medium (GM), at 37°C in a humidified atmosphere of 5% CO2. To initiate myogenic differentiation, C2C12 myoblasts with 50–70% confluence were grown in a DMEM culture medium containing 2% horse serum and 1% antibiotic-antimycotic solution as a differentiation medium (DM). The differentiation medium was changed every day until the indicated times.
Quantitative PCR (qPCR) analysis
Total RNA was isolated from undifferentiated or differentiated C2C12 cell lines using the RNeasy® Lipid Tissue Mini Kit (Qiagen, USA). We used 2 μg of total RNA to generate cDNA with the MMLV Reverse Transcriptase and random primers (Promega). The cDNA was analyzed by quantitative PCR using a SYBR green PCR kit and a C1000 Touch™ Thermal Cycler (Bio-Rad). All data were normalized to ribosomal L32 expression.
Western blot analysis
Cells were lysed in a protein lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 50 mM NaF, 5 mM β-glycero-phosphate, 1 mM sodium orthovanadate, 1 mM DTT, and a protease inhibitor cocktail (Roche). Western blot analyses was performed on 5–30 μg whole-cell extracts. Proteins were separated by electrophoresis on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes (Pall Corporation). Membranes were blocked with 5% skim milk and probed with primary antibodies. Anti-HDAC11 was obtained from Abcam. Antibodies against myosin heavy chain (MHC) and M2-Flag were purchased from Sigma-Aldrich. Antibodies against HSP90, MyoD1, MEF2C, and Myogenin were obtained from Santa Cruz Biotechnology. Antisera against phospho-AMPK, AMPK, phospho-AKT and AKT were from Cell Signaling Technology. The specific signals were amplified by horseradish peroxidase-conjugated secondary anti-rabbit IgG or anti-mouse IgG (Santa Cruz Biotechnology) and were visualized using an enhanced chemiluminescence system (BD Biosciences).
Generation of stable cell lines
The coding sequence of HDAC11 was amplified from mouse cDNA using PCR and cloned into the pRetroX-IRES-ZsGreen1 vector with an N-terminal FLAG tag (Clontech). The catalytically inactive HDAC11 mutant (H142/143A) was generated using QuikChange site-directed mutagenesis kit (Agilent). To generate retroviruses expressing GFP only, HDAC11 and HDAC11 (H142/143A) mutant constructs were individually co-transfected into GP2-293 cells with pVSV-G (Clontech) using Lipofectamine 2000 (Gibco-Invitrogen). At 48–72 h after transfection, the media with retrovirus-containing supernatant were collected and passed through a 0.45 μm filter. C2C12 myoblasts were infected with retroviruses expressing GFP control, HDAC11 or its mutant in the presence of polybrene (8μg/ml). Infected C2C12 myoblasts were enriched by determining the degree of GFP expression using a FACSAria cell sorter (BD Biosciences). At differentiation day 5, stable cell lines were confirmed by Western blot assay and qPCR analysis.
Immunocytochemistry (ICC)
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were incubated in 0.1% Triton X-100 for 15 min at room temperature for permeabilization. Cells were then incubated with blocking buffer (Roche) in a 0.1% Triton X-100 for 1 h at room temperature, followed by overnight incubation at 4°C with the primary antibody against MHC. Cells were then incubated with the secondary antibody (Santa Cruz). Cellular nuclei were stained with DAPI solution (Invitrogen), and images were captured with a Leica DM IL LED microscope with a Leica DFC 450C.
Luciferase reporter assay
C2C12 stable cell lines were maintained with DMEM growth media (GM). For transfection, cells were seeded in triplicate in 24-well plates at 1 × 105 cells per well. Trans IT®-LT1 transfection reagents (Mirus Bio) were used in accordance with the manufacturer’s instructions. Each transfection was performed with 300 ng of reporter constructs fused with the luciferase gene (pGL4.12-MEF2C and pGL4.12-4RTK promoter), 50 ng of a CMV-β-galactosidase plasmid, and 5 ng of expression plasmid for pcDNA3-flag-MyoD. Total cell lysates were prepared 48 h after transfection, and promoter activities were assessed using the Dual-Luciferase reporter assay system (Promega) (Lee et al., 2016).
Immunoprecipitation (IP)
C2C12 stable cells were washed twice with ice-cold PBS and lysed in a protein lysis buffer. The resulting cell lysates were incubated with anti-acetyl-lysine agarose (EMD Millipore) overnight at 4°C. The beads were subsequently washed three times each with protein lysis buffer, followed by suspension in the sample buffer. The eluted proteins were resolved by 10%SDS-polyacrylamide (PAGE) gel electrophoresis and analyzed by Western blotting with anti-annexinA1 (Abcam).
Chromatin immunoprecipitation (ChIP)
Cross-linking of protein complexes, nuclear isolation, and immunoprecipitation assays in C2C12 cells expressing wild-type or mutant HDAC11 were performed as previously described (Oh et al., 2012). Cells were treated with 1% formaldehyde for 20 min to cross-link histones to DNA, and were washed twice in PBS. Crosslinked cells were then lysed with cell lysis buffer (0.1% NP40, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 25 mM HEPES pH 7.8, and a protease inhibitor cocktail). Subsequently, cell lysates were centrifuged at 5,000 rpm for 2 min at 4°C, and the supernatants were removed. The pellets were lysed with a nuclear lysis buffer (1% Triton X-100, 140 mM NaCl, 1 mM EDTA, 0.1% Na-deoxycholate, 0.1% SDS, 50 mM HEPES pH7.9 and a protease inhibitor cocktail). The nuclear lysates were sonicated at 20% output (Sonics VC505) and then centrifuged at 13,000 rpm for 10 min at 4°C. The supernatants were collected as chromatin samples. For each immunoprecipitation reaction, the supernatant proteins were immunoprecipitated with anti-histone H3K9 antibodies (Santa Cruz Biotechnology). Anti-rabbit IgG was used as a negative control. Precipitated DNA fragments were analyzed by PCR using primers against relevant mouse promoters.
Statistical analysis
Experimental data are shown as the mean ± standard deviation (SD) or ± standard error of the mean (SEM), as indicated in the figure legends. Comparisons of different groups were performed using two-tailed unpaired Student’s t-tests. In all statistical comparisons, P values < 0.05 were considered statistically significant.
RESULTS
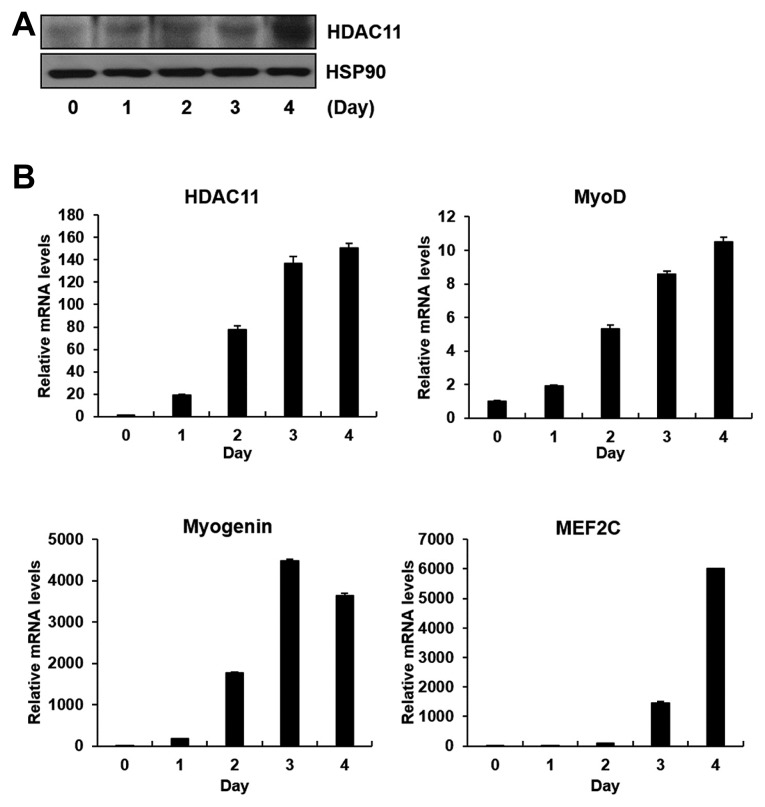
HDAC11 expression is markedly and consistently increased during mouse skeletal myoblast differentiation
Recently, it has been reported that the PTM of histone proteins affect the expression levels of myogenic genes during myoblast differentiation (De la Serna et al., 2006; Guasconi and Puri, 2009; Puri and Sartorelli, 2000). To access the relationship between histone modification and myoblast differentiation, we focused on enzymes involved in histone acetylation and methylation, two major PTMs of histones for regulating gene transcription. We observed changes in mRNA expression levels after differentiation compared with before differentiation in C2C12 myoblasts using qPCR analysis. Among the enzymes that demonstrated P values < 0.01, those whose correlation with myogenic differentiation is not well known and are novel enough to investigate are shown in Table 1. While several genes satisfy both P value (<0.01) and fold-change (2-fold) criteria, the change in mRNA levels of HDAC11 was strikingly increased more than 100 times after differentiation compared with before differentiation of C2C12 myoblasts (Table 1). Changes in mRNA and protein levels of HDAC11 were further confirmed (Figs. 1A and 1B). As shown in Fig. 1B, HDAC11 mRNA expression gradually increased with the induction of myogenic differentiation markers (MyoD, Myogenin, and MEF2C) during myoblast differentiation.
Table 1.
The list of enzymes showing significant changes in gene expression levels during myoblast differentiation
| Modification | Name of gene | Function | Fold change (4 day/0 day) | p-value |
|---|---|---|---|---|
| Acetylation | PCAF | Acetyltransferase | 3.02 | ** |
| SRC-1 | Acetyltransferase | 8.42 | ** | |
| HDAC11 | Deacetylase | 107.57 | ** | |
| Methylation | MLL2 | Methyltransferase | 2.69 | * |
| MLL3 | Methyltransferase | 2.45 | * | |
| PRDM9 | Methyltransferase | 2.33 | * | |
| ASH1 | Methyltransferase | 2.04 | ** | |
| SETD7 | Methyltransferase | 2.32 | * | |
| JHDM1D | Demethylase | 8.83 | * | |
| KDM5B | Demethyalse | 5.77 | * |
P < 0.01,
P < 0.005
Fig. 1.
HDAC11 is up-regulated during myogenic differentiation.
C2C12 myoblasts were differentiated for the indicated periods. (A) Western blot analysis showing protein expression level of HDAC11 during myoblast differentiation. (B) qPCR analysis showing mRNA levels of HDAC11 and myogenic differentiation markers (MyoD, Myogenin, and MEF2C) during myoblast differentiation.
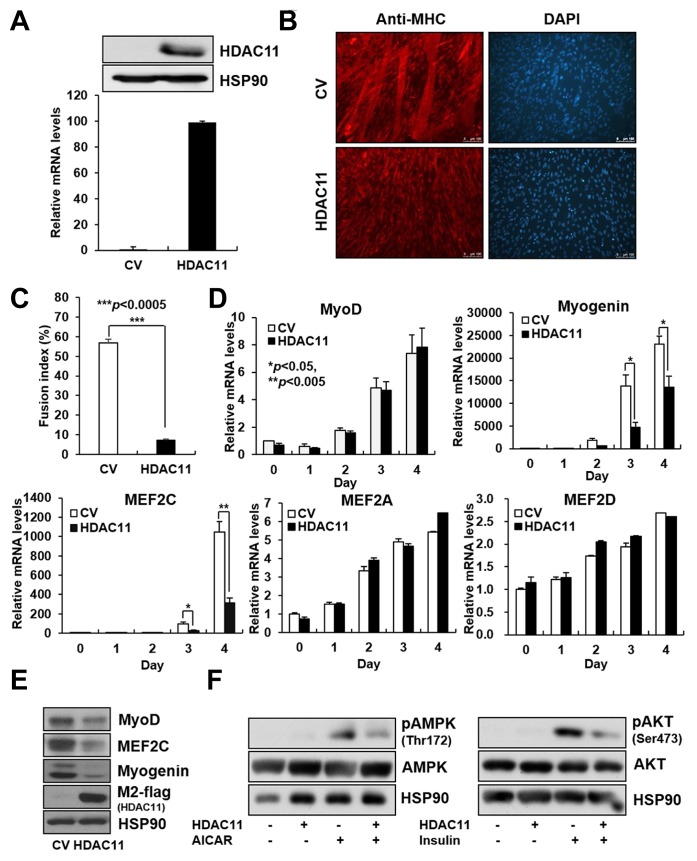
Stable expression of HDAC11 impedes C2C12 myoblast differentiation
Our results obtained from screening analysis indicated the possibility of HDAC11 as a highly influential histone modification factor involved in the regulation of myoblast differentiation. To investigate the effect of HDAC11 on myoblast differentiation, we generated C2C12 stable cell lines expressing flag-tagged HDAC11 or retrovirus control. Expression levels of HDAC11 in C2C12 stable cell lines were confirmed both at mRNA and protein levels (Fig. 2A). The expression of HDAC11 suppressed C2C12 myoblast differentiation, as assessed by MHC staining (Fig. 2B), and markedly reduced the fusion index of myocytes (Fig. 2C). This was concomitant with a decrease in mRNA and protein levels of the muscle-specific transcription factors, MEF2C and Myogenin (Figs. 2D and 2E). MyoD protein levels were slightly decreased by HDAC11 expression, whereas its mRNA levels did not change (Figs. 2D and 2E). Among other MEF2 family members, expression levels of MEF2A and D were not influenced by HDAC11 expression (Fig. 2D), and MEF2B gene expression was not detected (data not shown). Further, to test whether abnormal differentiation of skeletal muscle cells by HDAC11 can affect its function, we evaluated the phosphorylation levels of AKT and AMPK, two main regulators in the glucose uptake signaling pathway. As shown in Fig. 2F, HDAC11 expression reduced the active phosphorylated forms of AKT and AMPK in skeletal muscle cells.
Fig. 2.
The ectopic expression of HDAC11 inhibits myogenic differentiation.
C2C12 myoblasts were stably infected with retroviruses expressing control vector (CV) or HDAC11. C2C12 stable cells were differentiated for 4 days. (A) The expression levels of HDAC11 in stable cell lines were determined by western blot and qPCR analysis. (B) Myotube formations were determined by ICC using an anti-myosin heavy chain (MHC). (C) The fusion index was calculated as the ratio of the nuclei in MHC-positive tubes verses the total number of nuclei. Data represent the mean from three different spots ± SEM (***P < 0.005). (D, E) The mRNA and protein levels of myogenic differentiation markers in C2C12 cells with stable expression of HDAC11 were determined by qPCR analysis and western blot analysis. (F) Differentiated myotubes were treated with 500 μM AICAR (an analog of AMP that is capable of stimulating AMPK activity) for 2 h or insulin for 15 min. Phosphorylation levels of AMPK and AKT were detected by western blot analysis. Data in (D) represent the mean ± SD (*P < 0.05, **P < 0.005).
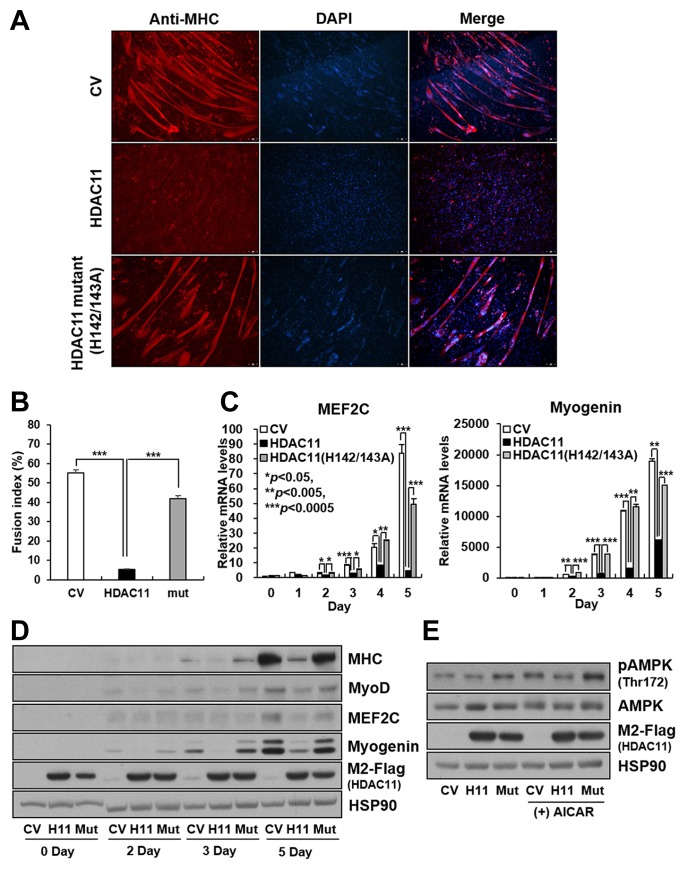
The deacetylase activity of HDAC11 is involved in the suppression of myoblast differentiation
To further investigate if the enzymatic activity of HDAC11 is associated with myoblast differentiation, we generated C2C12 stable cell lines expressing flag-tagged wild-type and mutant (H142A/H143A) HDAC11. It has been previously reported that substitution of histidine to alanine on amino acids 142 and 143 of HDACs can block their deacetylase activity (Rajendran et al., 2011; Thangapandian et al., 2011; 2012). In contrast to HDAC11 expression, expression of the catalytically inactive mutant of HDAC11 did not suppress myogenic differentiation (Fig. 3A), and it was confirmed by the measurement of the fusion myogenic index (Fig. 3B). Consistent with these results, mRNA and protein expression of transcription factors associated with myoblast differentiation such as MEF2C and Myogenin were also suppressed by wild-type HDAC11 expression, but not that of the catalytically inactive mutant (Figs. 3C and 3D). In addition, HDAC11 expression reduced the AMPK phosphorylation levels, whereas its deacetylase inactive mutant did not influence the phosphorylation state of AMPK (Fig. 3E).
Fig. 3.
The enzymatic activity of HDAC11 is involved in suppression of myogenic differentiation.
C2C12 stable cells expressing control vector (CV), HDAC11, or HDAC11 (H142/143A) mutant were differentiated for 5 days. (A) Multinucleated myotubes were characterized by immunofluorescence using anti-myosin heavy chain (MHC). (B) The fusion index was calculated as the ratio of the nuclei in MHC-positive tubes verses the total number of nuclei. Data represent the mean from three different spots ± SEM (***P < 0.005). (C, D) Cells were differentiated for the indicated times. The mRNA and protein levels of myogenic markers were determined by qPCR analysis and western blot analysis. (E) Differentiated myotubes were treated with 500 μM AICAR for 2 h. The phosphorylation level of AMPK was detected by western blot analysis. Data in (C) represent the mean ± SD (*P < 0.05, **P < 0.005, ***P < 0.0005). (CV, control vector; H11, HDAC11; Mut, HDAC11 (H142/143A) mutant)
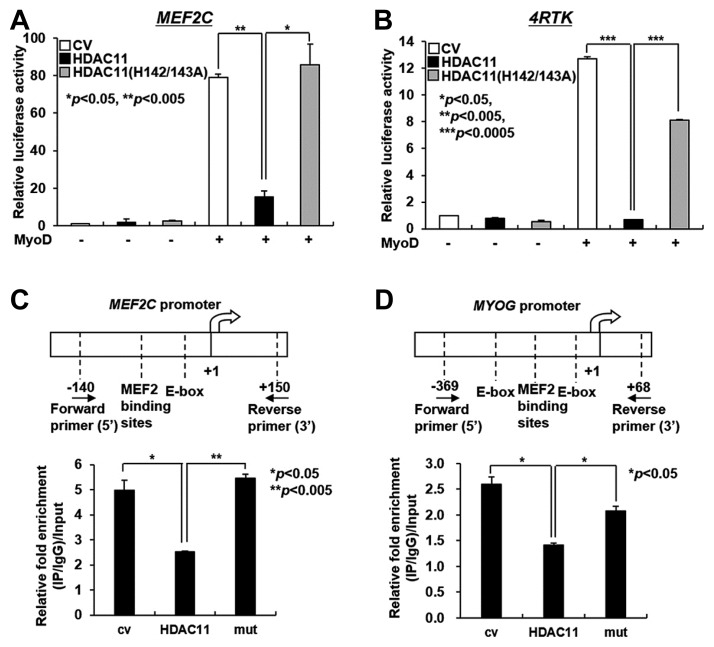
HDAC11 suppresses myoblast differentiation in a MyoD-dependent manner
MyoD is one of the transcription factors that regulates the transcription levels of MEF2C and Myogenin by directly binding to their promoters. This prompted us to investigate the effects of HDAC11 as a histone deacetylase on the MyoD-dependent promoter activities of MEF2C and Myogenin. MyoD-induced MEF2C promoter activity was decreased in the C2C12 cell line expressing wild-type HDAC11, but not in its enzymatically inactive mutant (H142A/H143A) cell line (Fig. 4A). Next, to investigate the effect of HDAC11 on My-oD-dependent Myogenin transcription, we used a MyoD-dependent promoter 4RTK luciferase construct, which contains four tandem E-box transcription factor binding sites from the muscle creatine kinase (MCK) promoter (Weintraub et al., 1990). MyoD regulates the expression of Myogenin by binding to the E-box region in the MYOG (Myogenin) promoter. As shown in Fig. 4B, MyoD-induced luciferase activity of 4RTK was decreased in the C2C12-HDAC11 cell line, but not in its enzymatically inactive mutant cell line. These data suggest that HDAC11 suppresses transcription of the MEF2C and Myogenin in a MyoD-dependent manner in order to inhibit myogenic differentiation. To further examine whether HDAC11 influences the acetylation state near the E-boxes, the MyoD binding site, of the MEF2C and MYOG promoters (Mal and Hater, 2003; Wang et al., 2001), we performed chromatin immunoprecipitation (ChIP) assays in C2C12 cells expressing wild-type or catalytically inactive HDAC11 using an anti-histone H3K9 antibody (Liu et al., 2009). Histone acetylation near the E-boxes of the MEF2C and MYOG promoters was reduced by HDAC11, but not by the catalytically inactive HDAC11 mutant (Figs. 4C and 4D). As previously reported, the binding of HDAC to MyoD may inhibit MyoD-dependent myogenic gene transcription (Mal et al., 2003). Therefore, to further elucidate the relationship between HDAC11 and MyoD during myogenic differentiation, we measured the interaction between the two molecules. As shown in Fig. 5A, HDAC11 physically interacted with MyoD, and wild type HDAC11, but not the inactive HDAC11 mutant, strongly bound to endogenous MyoD in a time-dependent manner during myoblast differentiation (Fig. 5B). These data suggest that the deacetylase activity of HDAC11 is essential for MyoD binding, followed by the regulation of MyoD-dependent transcription.
Fig. 4.
HDAC11 decreases histone acetylation at the promoter regions of MEF2C and MYOG and impedes transcription of MEF2C and MYOG in a MyoD-dependent manner.
(A, B) C2C12 myoblasts expressing HDAC11 or the HDAC11 (H142/143A) mutant were co-transfected with 300 ng of pGL4-luciferase constructs (MEF2C or 4RTK) and 5 ng of vector expressing MyoD for 48 h. (C, D) The chromatin immunoprecipitation experiment at day 3 after differentiation was performed to check the effect of HDAC11 on the acetylated histone H3K9 at the promoter region of MEF2C and MYOG. Data in (A–D) represent the mean ± SD (*P < 0.05, **P < 0.005, ***P < 0.0005). (cv, control vector; mut, HDAC11 (H142/143A) mutant)
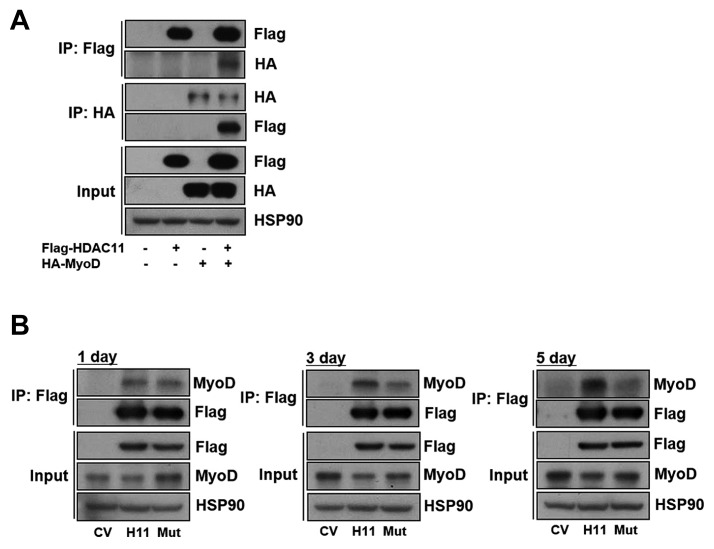
Fig. 5.
HDAC11 directly interacts with MyoD.
(A) 293T cells were co-transfected with Flag-HDAC11 and HA-MyoD for 48 h. Extracts from transfected cells were immunoprecipitated with anti-Flag or anti-HA, followed by western blot using anti-Flag and anti-HA. (B) C2C12 cells expressing CV control, HDAC11 or HDAC11 (H142/143A) mutant were differentiated for the indicated times. Cell lysates were immunoprecipitated with anti-Flag, followed by Western blot using anti-Flag and anti-MyoD. (CV, control vector; H11, HDAC11; Mut, HDAC11 (H142/143A) mutant)
DISCUSSION
HDAC11 was discovered in 2002, and classified as a class IV HDAC, where it was the only member. However, the functional and physiological role of HDAC11 has remained largely unknown until now. Here we show for the first time that HDAC11 can regulate myogenic differentiation. Our findings demonstrated that HDAC11 was significantly up-regulated during myoblast differentiation. Ectopic expression of wild-type HDAC11 induced the suppression of myoblast differentiation, whereas the catalytically inactive mutant did not affect myoblast differentiation. Although the effect of HDAC11 depletion in C2C12 cells was not shown in this study, it was thought that disruption of proper differentiation by HDAC11 depletion in C2C12 cells would excessively promote myogenic differentiation. Further, we investigated whether HDAC11-suppressed myoblast differentiation could affect muscle function by observing phosphorylation levels of AMPK and AKT (Figs. 2F and 3E). Actually, it has been reported that AMPK and AKT could affect transcriptional activity of MyoD (Fu et al., 2013; Xu and Wu, 2000). Therefore, it was possible that reduced phosphorylation levels of AMPK and AKT in C2C12 stable cell line expressing HDAC11 might influence transcriptional activity of MyoD. However, the interaction of HDAC11 with MyoD increased in a time-dependent manner during myogenic differentiation, although the ectopic expression of HDAC11 slightly decreased the expression of endogenous MyoD protein (Fig. 5). These data strongly supported that inhibition of MyoD-dependent transcription was most directly driven by HDAC11 at least in this model.
MyoD controls myoblast proliferation and myogenic lineage progression. It is intimately involved in muscle differentiation by regulating the transcription of muscle-specific genes such as MEF2C and Myogenin (Bergstrom et al., 2002; Buchberger et al., 1994; Dodou et al., 2003; Faralli and Dilworth, 2012; Gerber et al., 1997). MyoD can directly bind to the promoter regions of MEF2C and MYOG, and MyoD-induced muscle gene expression can be mediated by chromatin remodeling (Bergstrom et al., 2002; Gerber et al., 1997). Therefore, we hypothesized that inhibition of myogenic differentiation by HDAC11 would be associated with histone deacetylation at the promoter regions of MyoD target genes such as MEF2C and MYOG. As expected, HDAC11 lowered the promoter activities of MEF2C and MYOG, and reduced histone acetylation levels near the MyoD binding sites of the MEF2C and MYOG promoters (Fig. 4). Under physiological conditions, it was thought that HDAC11 would function as a negative regulator, finely controlling muscle differentiation (Fig. 6).
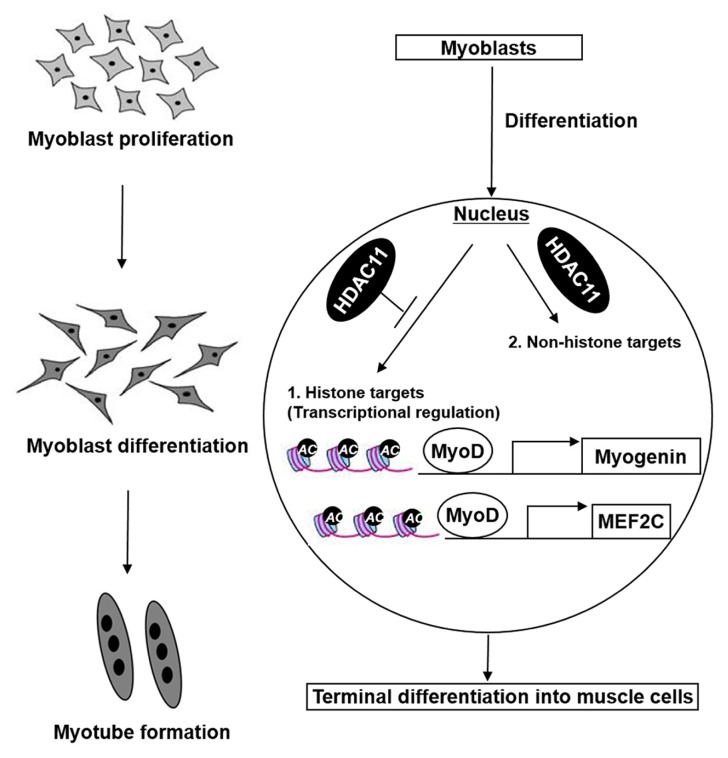
Fig. 6.
A proposed model for HDAC11-mediated suppression of myoblast differentiation.
The expression level of HDAC11 was gradually increased during the process of myogenic differentiation. Elevated expression of HDAC11 down-regulated the levels of histone acetylation at promoter regions of MEF2C and MYOG, and consequently suppressed myoblast differentiation in a MyoD-dependent manner. HDAC11 could also target non-histone factors as substrates for deacetylation during myoblast differentiation. In conclusion, HDAC11 would be essential for proper myogenic differentiation, and its regulatory mechanism would provide important clues for fine regulation of myogenic differentiation.
The mechanism for HDACs that underlie the regulation of gene expression is not limited to histone modification. Recently, it was reported that HDACs can regulate target gene expression by modulating mRNA stability (Krishnan et al., 2010). Thus, we also investigated whether HDAC11 can affect the stability of MyoD, MEF2C, and Myogenin mRNA. However, no effects of HDAC11 on mRNA stability were observed (data not shown). To gain further insight into the mechanism of HDAC11-induced inhibition of myoblast differentiation, we attempted to find non-histone targets of HDAC11 by Western blot analysis using an anti-acetyl-lysine antibody after 2-dimesional gel electrophoresis. We obtained several candidates such as annexin A1, voltage-dependent anion-selective channel protein 1, enolase 1B, and guanine nucleotide-binding protein subunit β-1 (Supplementary Fig. 1A). In the case of annexin A1, validation using Western blot analysis showed that our data were reliable (Supplementary Fig. 1B). These efforts strongly indicated that HDAC11 may also target non-histone proteins as substrates for deacetylation. However, more detailed studies are necessary to clarify the effects of HDAC11 on non-histone targets during myoblast differentiation.
In addition to HDAC11, our screening analysis detected several undescribed enzymes, including SRC-1, PCAF, JHDM1D, and KDM5B that may be associated with myoblast differentiation (Table 1). Further investigation of these enzymes is warranted to better understand the regulation of myogenic differentiation by histone modification.
Taken together, our novel findings revealed that HDAC11 impedes myoblast differentiation in a MyoD-dependent manner via modulation of histone acetylation in the MEF2C and MYOG promoters. In addition, other enzymes demonstrating expression changes during myoblast differentiation, and several non-histone candidate target proteins of HDAC11, were also discovered. These data should provide further understanding of the regulation of myoblast differentiation.
Supplementary data
ACKNOWLEDGMENTS
This work was supported by a grant from the KRIBB and by the National Research Foundation of Korea (NRF) Grants (2016R1C1B2010257, 2015M3A9D7029882 and 2017M3 A9C4065954).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ait-Si-Ali S., Guasconi V., Fritsch L., Yahi H., Sekhri R., Naguibneva I., Robin P., Cabon F., Polesskaya A., Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendse M., Grounds M.D., Lloyd C.M. Myoblast structure affects subsequent skeletal myotube morphology and sarcomere assembly. Exp Cell Res. 2003;291:435–450. doi: 10.1016/j.yexcr.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Bergstrom D.A., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Braun T., Buschhausen-Denker G., Bober E., Tannich E., Arnold H.H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989;8:701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Bober E., Winter B., Rosenthal N., Arnold H.H. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J. 1990;9:821–831. doi: 10.1002/j.1460-2075.1990.tb08179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A., Ragge K., Arnold H.H. The myogenin gene is activated during myocyte differentiation by pre-existing, not newly synthesized transcription factor MEF–2. J Biol Chem. 1994;269:17289–17296. [PubMed] [Google Scholar]
- Burattini S., Ferri P., Battistelli M., Curci R., Luchetti F., Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur J Histochem. 2004;48:223–233. [PubMed] [Google Scholar]
- Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- De la Serna I.L., Ohkawa Y., Imbalzano A.N. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- DeFronzo R.A., Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009;32:S157–S163. doi: 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E., Xu S.M., Black B.L. Mef2c is activated directly by myogenicbasic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Olson E.N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- Faralli H., Dilworth F.J. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp Funct Genomics. 2012;2012:836374. doi: 10.1155/2012/836374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Zhao J.X., Liang J., Zhu M.J., Foretz M., Viollet B., Du M. AMP-activated protein kinase mediates myogenin expression and myogenesis via histone deacetylase 5. Am J Physiol Cell Physiol. 2013;305:C887–C895. doi: 10.1152/ajpcell.00124.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Cueto M.A., Asselbergs F., Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- Gerber A.N., Klesert T.R., Bergstrom D.A., Tapscott S.J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Gossett L.A., Kelvin D.J., Sternberg E.A., Olson E.N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasconi V., Puri P.L. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R.L., Olson E.N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P., Bradley A., Morris J.H., Edmondson D.G., Venuti J.M., Olson E.N., Klein W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Iezzi S., Di Padova M., Serra C., Caretti G., Simone C., Maklan E., Minetti G., Zhao P., Hoffman E.P., Puri P.L., et al. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan M., Singh A.B., Smith J.J., Sharma A., Chen X., Eschrich S., Yeatman T.J., Beauchamp R.D., Dhawan P. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene. 2010;29:305–312. doi: 10.1038/onc.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A.B., Skapek S.X., Novitch B. Regulatory mechanisms that coordinate skeletal muscle differentiation and cell cycle withdrawal. Curr Opin Cell Biol. 1994;6:788–794. doi: 10.1016/0955-0674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Lee D.S., Choi H., Han B.S., Kim W.K., Lee S.C., Oh K-J., Bae K-H. c-Jun regulates adipocyte differentiation via the KLF15-mediated mode. Biochem Biophys Res Commun. 2016;469:552–558. doi: 10.1016/j.bbrc.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Liu H., Hu Q., D’Ercole J., Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57:1–12. doi: 10.1002/glia.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., McKinsey T.A., Zhang C.L., Olson E.N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- Mal A.K. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A., Harter M.L. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci USA. 2003;100:1735–1739. doi: 10.1073/pnas.0437843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A., Sturniolo M., Schiltz R.L., Ghosh M.K., Harter M.L. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.F., Schwarz J.J., Olson E.N. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J.C., Cardoso M.C., Yu Y.T., Andres V., Leifer D., Krainc D., Lipton S.A., Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., Olson E.N. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Black B.L., Martin J.F., Olson E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Oh K.J., Park J., Kim S.S., Oh H., Choi C.S., Koo S.H. TCF7L2 modulates glucose homeostasis by regulating CREB- and FoxO1-dependent transcriptional pathway in the liver. PLOS Genet. 2012;8:e1002986. doi: 10.1371/journal.pgen.1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., Klein W.H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- Olson E.N., Perry M., Schulz R.A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- Pasini D., Hansen K.H., Christensen J., Agger K., Cloos P.A., Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and polycomb-repressive complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R., Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Puri P.L., Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Rajendran P., Williams D.E., Ho E., Dashwood R.H. Metabolism as a key to histone deacetylase inhibition. Crit Rev Biochem Mol Biol. 2011;46:181–199. doi: 10.3109/10409238.2011.557713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes S.J., Konieczny S.F. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Braun T., Hinuma S., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Schnegelsberg P.N., Stead R.H., Braun T., Arnold H.H., Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sabourin L.A., Rudnicki M.A. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Bhowmick K., Chatterjee S., Basha J., Kundu T.K., Dhar S.K. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014;281:5265–5278. doi: 10.1111/febs.13067. [DOI] [PubMed] [Google Scholar]
- Thangapandian S., John S., Lee Y., Kim S., Lee K.W. Dynamic structure-based pharmacophore model development: a new and effective addition in the histone deacetylase 8 (HDAC8) inhibitor discovery. Int J Mol Sci. 2011;12:9440–9462. doi: 10.3390/ijms12129440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangapandian S., John S., Lee K.W. Molecular dynamics simulation study explaining inhibitor selectivity in different class of histone deacetylases. J Biomol Struct Dyn. 2012;29:677–698. doi: 10.1080/07391102.2012.10507409. [DOI] [PubMed] [Google Scholar]
- Voelter-Mahlknecht S., Ho A.D., Mahlknecht U. Chromosomal organization and localization of the novel class IV human histone deacetylase 11 gene. Int J Mol Med. 2005;16:589–598. [PubMed] [Google Scholar]
- Wang D.Z., Valdez M.R., McAnally J., Richardson J., Olson E.N. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Lockshon D., Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T.K., Turner D., Rupp R., Hollenberg S., et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Xu Q., Wu Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J Biol Chem. 2000;275:36750–36757. doi: 10.1074/jbc.M005030200. [DOI] [PubMed] [Google Scholar]
- Yu Y.T., Breitbart R.E., Smoot L.B., Lee Y., Mahdavi V., Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.