ABSTRACT
In a concept study the ability to induce heterogeneous ice formation by Bacterial Ghosts (BGs) from Escherichia coli carrying ice nucleation protein InaZ from Pseudomonas syringae in their outer membrane was investigated by a droplet-freezing assay of ultra-pure water. As determined by the median freezing temperature and cumulative ice nucleation spectra it could be demonstrated that both the living recombinant E. coli and their corresponding BGs functionally display InaZ on their surface. Under the production conditions chosen both samples belong to type II ice-nucleation particles inducing ice formation at a temperature range of between −5.6 °C and −6.7 °C, respectively. One advantage for the application of such BGs over their living recombinant mother bacteria is that they are non-living native cell envelopes retaining the biophysical properties of ice nucleation and do no longer represent genetically modified organisms (GMOs).
KEYWORDS: bacterial ghosts, ice nucleation, ice nucleation protein
Introduction
Ice nucleation proteins (INPs) of ice nucleation active (ina+) bacteria have the property to catalyze heterogeneous ice formation of supercooled water by orienting water molecules into an ice-like structure.1-3 INPs are integrated to the outer cell membrane of Gram-negative bacteria where they form large multimers,4-6 or can be released into the surrounding environment known as extracellular ice nucleating material.7,8 Generally, ina+ bacteria are Gram-negative, epiphytic and pathogenic as Pseudomonas syringae, Pseudomonas putidia, Erwinia herbicola, Erwinia ananas, Xanthomonas campestris, or Pantoea ananatis among others.7-17 The well characterized plant pathogen Pseudomonas syringae represents one of the most efficient INA bacterial ice nucleus known, initiating plant damaging ice formation at temperatures of −2°C.11,18 Aside from the habitat of an epiphytic plant pathogen, Pseudomonas syringae, as well as other INA bacteria were found in clouds, rain, snow and streams indicating that they are disseminated with the earth hydrological cycle.19-22 Airborne ice-active bacteria are involved in cloud condensation – acting as cloud condensation nuclei (CCN) – and precipitation.23,24
Recombinant bacterial INPs were already functionally expressed in Gram-negative non-ice nucleation active Escherichia coli, as well as in plants.25,26 INPs are also used for molecular biologic applications, using ice-nucleation activity as a reporter gene system.27 In addition, the N-terminal domain of INPs, which link the protein to the outer cell membrane is used as cell surface display system.28-30
In this concept study the ice nucleation protein Z (InaZ) of Pseudomonas syringae was expressed in E. coli which were further processed to Bacterial Ghosts (BGs). BGs are empty cell envelopes of Gram-negative bacteria produced by the expression of cloned gene E of bacteriophage Phix17431-35 The conversion of a single living bacterium into a BG is a rapid process in the range of milliseconds35 and can be characterized by puncturing the bacterium cell envelope from the inside to the outside expelling its cytoplasmic content to the surrounding medium. Electron micrographs of this process can be depicted from the original description in 199033 and a more recent one using this E-mediated lysis of E. coli for cryo-electron tomography of membrane protein complexes within the native cell envelope complex remaining intact after E-mediated lysis.34
Any protein, lipid, polysaccharide and complex structures derived from the molecular building blocks of the cell envelope complex of a living bacterium (either natural or recombinant expressed) is retained in BGs.35,36 The formation of the E-specific transmembrane tunnel structure seals the periplasmic space retaining all constituents of this compartment with a minimal loss of less than 10% as compared with 90% of constituents from the cytoplasmic space measured by marker enzyme release and electron microscopic studies during the E-lysis process.33,37,38 The loss of cytoplasm including its nucleic acids content, ribosomes, soluble proteins and other solutes is compensated by an influx of water through the envelope shell which is freely permeable for water.35 The advantage of BGs over their living mother bacteria is that they are non-living but contain a native cell envelope with their biophysical and biochemical characteristics.34,35
BGs represent a versatile alternative to bacteria when properties of the cell envelope are in the focus of interest as they represent the state and composition of the cell envelope at the time point of their production. In this communication it was investigated whether recombinant proteins introduced into the envelope complex of E. coli such as INP also retain their functional properties in the BG envelope. So far surface changes in BGs have only been tested immunologically by inserting foreign antigens sequences into a loop of OmpA facing the outside of the outer membrane of E. coli but not functionally for the biophysical integrity of structural components.39 Inserting INP into the cell envelope of E. coli and measuring its ice nucleation activities is the first example reporting the structural and functional stability of a recombinant protein being inserted into the outer membrane of BGs.
Results
E-mediated lysis for production of ice nucleation-active bacterial ghosts
To produce BGs carrying INPs genes inaZ and E, carried on different plasmids were transformed into E. coli C41. Briefly, 2 E-lysis plasmids have been used for this study (Fig. 1), pGLysivb which carries lysis gene E fused with an in vivo biotinylation sequence under transcriptional control of a cI857 repressor gene and mutated - λpRmut- operator system40,41 and pGLMivb which represents a modification of pGLysivb where E-mediated cell lysis is controlled by LacIq / PTAC repressor / promoter system. To express ice nucleation protein InaZ, in the follow called INP, its full length sequence was fused to a C-terminal His-tag sequence located in plasmid pBINP downstream of the arabinose inducible PBAD promoter (Fig. 1). Induction of INP was constitutive for the overnight culture of E. coli C41 (pBINP) clones used for E-lysis studies with plasmids pGLysivb and pGLMivb. The chemical and temperature inducible E-lysis plasmids were tested for lysis efficiency in combination with the INP expressed from pBINP in E. coli strain C41. With lysis plasmid pGLMivb expression of gene E is driven by the IPTG inducible PTAC promotor, whereas with plasmid pGLysivb E-mediated lysis is induced by a temperature upshift of the culture from 23°C to 42°C under control of temperature-sensitive λ PRmut – cI857 promoter – repressor system. E. coli C41 was selected to produce INP-BGs as this strain is able to maintain cellular vitality despite recombinant membrane protein overexpression. 42
Figure 1.
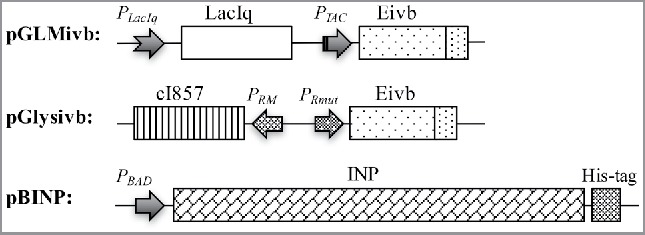
Partial map of plasmids used for production of BGs carrying INP.
In E. coli C41 carrying pBINP, expression of INP was induced by the addition of 0.2% arabinose to the culture medium. The culture was grown at 23°C as it has been shown that temperatures below 25°C have beneficial effects on ice nucleation activity.,25,43 When the culture reached an optical density at 600 nm (OD600) of 0.55–0.6 E-specific lysis was induced either by a temperature up-shift from 23 to 42°C (pGlysivb), or by addition of 0.5 mM IPTG (pGLMivb) and is referred to time point (Tp) 0 min in the corresponding growth curves (Fig. 2). The growth and lysis of the bacteria was monitored by measuring the OD600, flow cytometry, light microscopy and by determination of colony forming units (cfu) analysis (Figs. 2–4).
Figure 2.
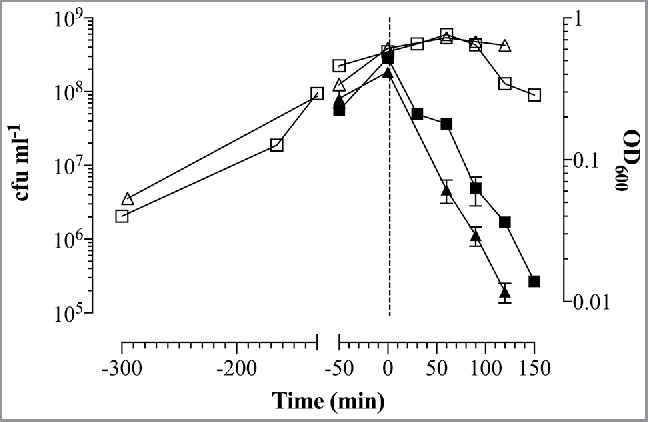
Growth and E-lysis of E. coli C41 carrying plasmids for expression of INP and BG production. E. coli C41 (pBINP, pGLMivb) (quad) and E. coli C41 (pBINP, pGLysivb) (triangle) cultures were grown in LBv supplemented with 0.2% arabinose to induce INP expression at 23°C from plasmid pBINP. At time point zero (illustrated by vertical sketched line) E-mediated lysis was induced from pGLysivb by temperature shift from 23°C to 42°C and from pGLMivb by addition of 0.5 mM IPTG. Open symbols indicate OD600 values of growth and lysis phase. During E-lysis colony forming units (closed symbols) were determined to calculate lysis efficiency. After 120 min (E. coli C41 [pBINP, pGLysivb]) and 150 min (E. coli C41 [pBINP, pGLMivb]) BG production was completed. Data were obtained from 3 independent experiments. Error bars indicate standard errors.
Figure 3.
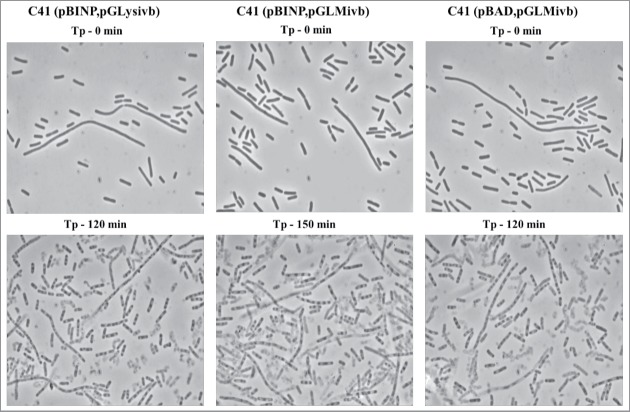
Light microscopy pictures of E. coli C41 (pBINP, pGLysivb), E. coli C41 (pBINP, pGLMivb), and E. coli C41 (pBAD, pGLysivb) strains at time point of lysis induction (Tp - 0min) and end point of E-mediated lysis after 120 min and 150 min, respectively (Tp - 120 min, Tp - 150 min).
Figure 4.
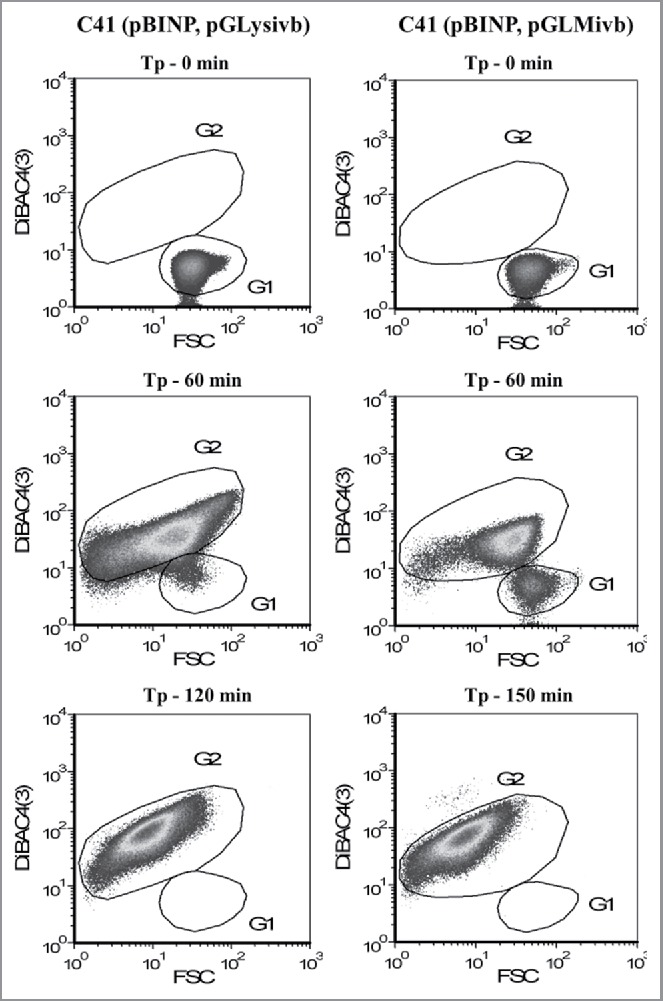
Flow cytometry density dot plots showing E-lysis process of E. coli C41 (pBINP, pGLysivb) and E. coli C41 (pBINP, pGLMivb) strains. Illustrated populations are discriminated from non-cellular background via Rh414 staining. Dot plots illustrate fluorescence intensity with Dibac4(3) vs. FSC–forward scatter; G1: living cells, G2: BGs. Tp - 0 min: indicates time point of lysis induction; Tp - 60 min: 60 min after lysis induction; Tp - 120 min: end of lysis phase for E. coli C41 (pBINP, pGLysivb); Tp - 150 min: end of lysis phase for E. coli C41 (pBINP, pGLMivb).
During the growth phase of E. coli C41 carrying pBINP elongation of the cell body was observed in some of the bacteria. This elongation was unspecific in regard of INP expression as E. coli C41 cells carrying plasmid pBAD24 (the backbone plasmid of pBINP) and the lysis plasmid pGLysivb exhibit the same phenotype (Fig. 4).
The E-lysis process of E. coli C41 (pBINP, pGLysivb) and E. coli C41 (pBINP, pGLMivb) was also monitored online via fluorescence-based flow cytometry (FCM) (Fig. 3), which allows an almost online process control to discriminate the ratio of full non-lysed bacteria and BGs already formed.44 In a FCM sample of bacterial culture the use of the phospholipids staining dye RH414 enables the visualization of bacteria and the discrimination of non-cellular background. DiBAC4(3), which enters depolarized cell membranes and binds to intracellular proteins or membranes, provides an assessment of bacterial cell viability.45 During E-lysis process of E. coli C41 (pBINP, pGLysivb) and E. coli C41 (pBINP, pGLMivb) BGs exhibit a strong DiBAC4(3) fluorescence signal and a complete shift of DiBAC-negative living cells (G1) to DiBAC-positive BGs (G2), can be detected at the end of lysis (Fig. 3). The lysis of the temperature inducible C41(pBINP,pGLysivb) culture and the chemically induced C41 (pBINP, pGLMivb) culture reached their time point of 99.9% lysis efficiency (LE) at 120 min and 150 min after lysis induction, respectively (Fig. 2). The presence of INP in C41 bacteria did not affect E-mediated lysis when compared with C41 bacteria carrying plasmid pBAD24 and lysis plasmid pGLysivb (Data not shown).
To inactivate E-lysis escape mutants and non-lysed bacteria at time of harvest, the cultures were further incubated for 2h at 23°C with 0.17% β-propiolactone. This standard procedure for inactivation of any nucleic acids remaining in BG preparations45 ensures that BGs carrying INP and BGs without INP which were used in the following for the droplet-freezing assay did not contain any viable cells.
The attempts to trace pBINP encoded INP by its C-terminal His-tag were not successful and made it necessary to raise antibodies against INP. For this purpose, a N-terminal truncated version of INP (-NINP) was used and purified. -NINP was injected into rabbits to produce INP specific antibodies (see Materials and Methods) and with this antiserum Western blot analysis was performed to detect the presence of expressed INP at time point of E-lysis induction (0min) and after β-propiolactone treatment of harvested INP-BG samples (Fig. 5). The calculated molecular weight (Mr) of the 1200 aa long INP is 118,6 kDa and the band visible at 120 kDa in samples of full E. coli C41 bacteria with expressed INP and BGs derived from them (Fig. 5, lane 1 to 4) corresponds to the full length INP. Furthermore, an additional protein band at 87 kDa was detected in the INP bearing samples of E. coli C41 (pBINP, pGLMivb) and E. coli C41 (pBINP, pGLysivb). These 2 prominent bands were not detected in E. coli C41 carrying the backbone plasmid pBAD24 for the INP expression plasmid pBINP (Fig. 5, lane 5 to 6). As ATG and corresponding positional Shine-Dalgarno (SD) sequences for a truncated 87kDa INP could not be detected in the nucleotide sequence of inaZ it is most likely that the lower migrating protein band represents an INP degradation product. Such a INP degradation product has also been reported for InaV 46
Figure 5.
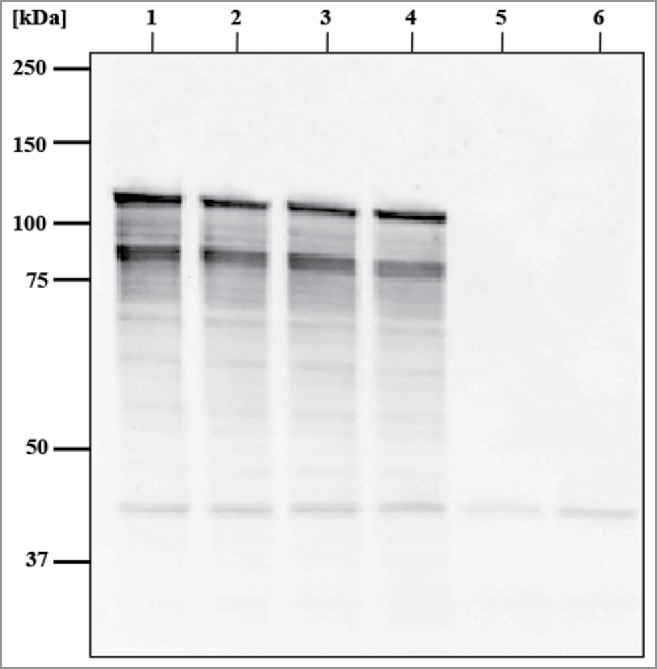
Western blot analysis of E. coli C41 expressing INP and harboring E-lysis plasmid pGLysivb or pGLMivb, respectively, and their derived BGs. Samples were taken at time point of E-lysis induction (Tp - 0 min) and after β-propiolactone treatment of BG samples. Western blotting was performed with rabbit anti-H-NINP serum and anti-rabbit IgG horseradish peroxidase conjugated antibodies. Lane1: E. coli C41 (pBINP, pGLMivb); lane 2: INP-BG-pLM, BG derived form of E. coli C41 (pBINP, pGLMivb); lane 3: E. coli C41 (pBH-NINPL, pGLysivb); lane 4: INP-BG-pLy, BG derived form of E. coli C41 (pBINP, pGLysivb); lane 5: E. coli C41 (pBAD, pGLysivb); lane 6: BG, BG derived form of E. coli C41 (pBAD, pGLysivb). Positions of molecular size marker proteins are indicated in kilodaltons (kDa).
Ice nucleation activity of BGs carrying INP (INP-BGs)
Ice-nucleating activity of full bacteria and BGs carrying INP, produced from E. coli C41 with lysis plasmid pGLysivb (INP-BG-pLy) or pGLMivb (INP-BG-pLM) respectively, was determined by a droplet-freezing assay (for details see Material and Methods). Living E. coli C41 cells carrying INP and lysis plasmid pGLysivb (C41-INP) not induced for lysis were used as a positive control. Living E. coli C41 cells carrying pBAD24 and pGLysivb (C41–24-pLy) as well as their BG derived form (BG), carrying no INP were included as a negative control.
As the heterogeneous freezing of a droplet of ultra-pure water at a given temperature is determined by the most active ice nucleus inside the droplet (meaning that only these nuclei is detectable 4,11) a series of dilutions from each suspension was tested. The particular freezing temperatures of the tested droplets plotted against the temperature, was used to determine the median freezing temperature T50 (Fig. 6B) and cumulative ice nucleation spectra (Fig. 7) of the samples. T50 is the temperature at which 50% of the droplets placed on the cooling device are frozen and was calculated out of the freezing spectra of each tested suspension containing 5 × 108 cells ml−1 (Fig. 6A).
Figure 6.
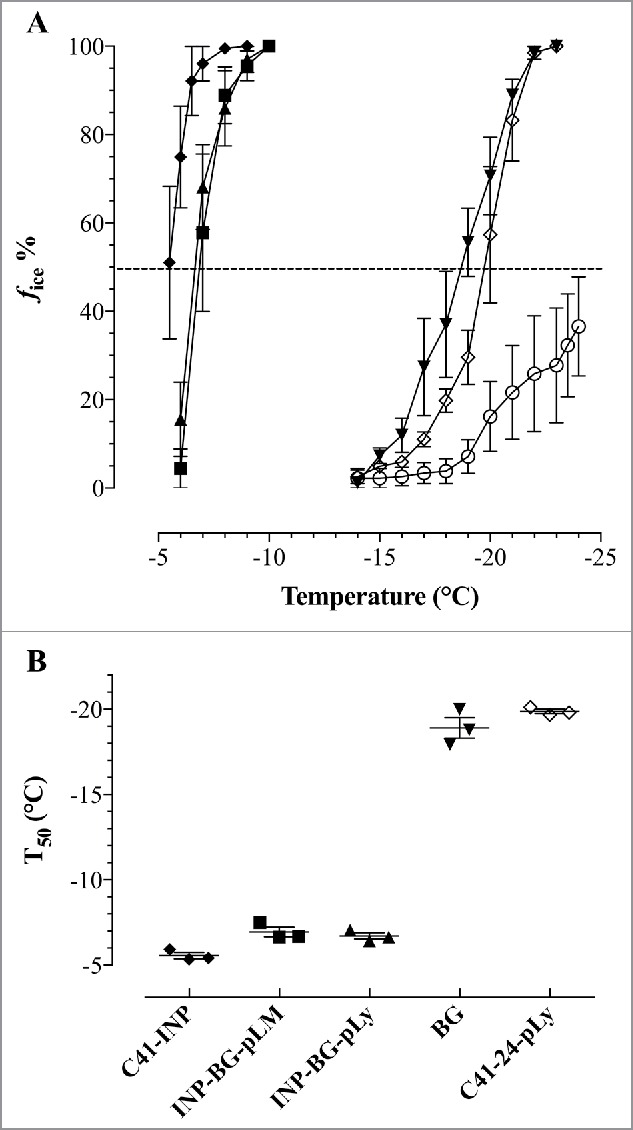
Average number of freezing spectra (A) and median freezing temperatures (T50) (B) of INP-BGs living E. coli C41 and E. coli C41 BGs. E.coli C41 (pBINP, pGLMivb) (INP-BG-pLM) (▪) C41 (pBINP, pGLysivb) (INP-BG-pLy) (▴), E.coli C41(pBINP, pGLysivb) cells carrying INP (C41-INP) (♦), E.coli C41(pBAD24, pGLysivb) ghosts (BGs) (▾) and C41 (pBAD24, pGLysivb) cells (C41–24-pLy) (◊). 210×10 µl droplets of ROTISOLV® water (○) were included as unspecific control. (A) Ice nucleation curve plotted as number fraction of frozen droplets in percent (fice %) at any temperature. (B) T50, temperature where 50% of all droplets are frozen. Forty-five 10 µl droplets of each suspension containing 5 × 108 cells ml−1 were tested by droplet freezing. Average numbers are obtained from 3 independent experiments. Error bars represent the standard errors.
Figure 7.
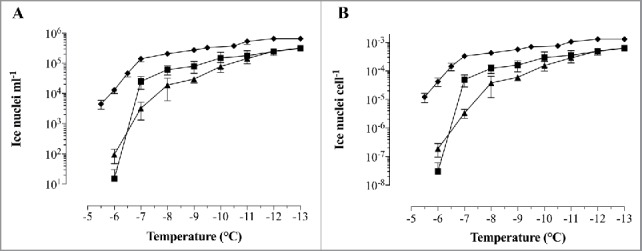
Cumulative concentration of ice nucleation activity for INP-BGs and E. coli C41-INP. E. coli C41 (pBINP, pGLysivb) (▴) and E. coli C41 (pBINP, pGLMivb) (▪) - BGs. Not lysed E. coli C41 (pBINP, pGLysivb) (♦) harvested before lysis induction. (A) Average Number of ice nuclei ml−1 at a given temperature N(T). (B) Average of Nucleation frequency (NF) expressed as ice nuclei cell−1. Data obtained from 3 independent experiments. Error bars represent the standard errors.
Both independent prepared INP-BG samples (INP-BG-pLM and INP-BG-pLy) showed a very close T50 values of −6.9°C and −6.7°C respectively, with first freezing events starting at −6.0°C. With living of E. coli C41 carrying INP (C41-INP) the first freezing event was noted at −5.5°C. The T50 value for this group was calculated to be at −5.6°C. The T50 for E. coli C41 cells (C41–24-pLy) and corresponding BGs carrying no INP was at −20.1°C and −18.9°C, respectively. The first frozen droplets detected for C41–24-pLy and C41 BGs were at −14°C, therefore the ice nucleation activity of INP-BGs was monitored up to a temperature decrease of −13°C not to detect INP unspecific induction of ice formation. For further characterization of the ice nucleation activity of INP-BGs the cumulative number N(T), of ice nuclei ml−1 active at a given temperature (Fig. 7A) and the nucleation frequency (NF) (Fig. 7B) were determined. The NF, obtained by normalizing the number of ice nuclei ml−1 for the number of cells in the original suspension, describes the fraction of cells in the suspension enfolding active ice nucleation sites at a given temperature.25 For both INP-BGs, ice nucleation was detected at −6°C, with a nucleation frequency of 3 × 10−8 for INP-BG-pLM and 1,9 × 10−7 for INP-BG-pLy. As the temperature decreased from −6°C to −7°C, the total number of ice nucleation active sites per cell at −7°C for INP-BG-pLM compared with INP-BG-pLy varies significantly 5 × 10−5 and 3.3 × 10−6, respectively. At a temperature of −10°C both types of INP-BGs exhibit closer NF activity values of 1.6 × 10−4 for INP-BG-pLy and 3 × 10−4 for INP-BG-pLM. With further decrease in temperature down to −13°C, NF of INP-BG-pLM and INP-BG-pLy was 6.3 × 10−4 and 6.4 × 10−4. Ice nucleation activity of living E. coli C41 carrying INP (C41-INP) was detected at higher temperature (−5.5°C) with an obviously higher NF of 1.2 × 10−5 which increased to 3.3 × 10−4 at −7°C. When temperature decreased to-13°C, an increase in NF up to 1.3 × 10−3 for C41-INP was detected.
Discussion
This communication is the first report of E. coli BGs carrying INP on their cell surface (INP-BGs). INP-BGs are able to lower the freezing point of ultra-pure water to a considerable content. The efficiency of bacteria nucleating ice-formation at a certain temperature can be subdivided into 3 distinct classes, type I acts at −5°C or warmer, type II from −5 to −7°C and type III below −7°C.47,48 According to this classification model the T50 value of INP-BGs determined in this study are belonging to type II (Fig. 6B) and with additional type III ice nucleation BGs in the samples (Fig. 7). Negative control BGs without INP and live E. coli C41 bacteria alone exhibit a T50 value of −18.9°C and −20.1°C, respectively (Fig 7).
Recombinant live bacteria E. coli C41 expressing INP on their surface also belong with their T50 value to type II ina+ particles, however, they possess slightly more ice catalyzing sites active at temperature of −6 to −7°C than the corresponding INP-BGs (Fig. 7). This difference might be explained by the fact that although the rod shaped bacterial cell envelope in BGs is retained it lacks the inner cytoplasmic turgor which makes the live bacteria chock-full which could provide a better positioning of neighboring INPs for ice nucleation. The crucial question, what makes a good ice nucleation center seems to depend on the coupling of surface crystallinity and hydrophilicity.49 Recently, the idea of ice templating by INPs has been proven experimentally showing that hydrogen bonding at the water-bacteria contact induce structural order and drive phase transitions of water in bacterial hydration shell.3
Former findings that a growth temperature of 30°C and higher of bacteria expressing INP had significant negative effects on the number of active ice nuclei were also investigated.25,43 INP-BGs were produced with 2 different lysis plasmids (pGLMivb and pGLysivb) controlling E-lysis by a chemical and temperature-sensitive promotor system, respectively. INP-BGs produced with pGLMivb were kept at 23°C for the whole production process whereas INP-BGs produced with pGLysivb were shifted to 42°C for 2h for the E-induced lysis. When the 2 different INP-BGs were compared for their ice-nucleation activity INP-BGs-pGLy (2h at 42°C) showed nearly the same ice nucleation frequency at −6°C. However, at −7°C INP-BGs-pGLM kept at 23°C exhibit a significant difference in the number of ice nucleation active sites per cell compared with INP-BGs-pGLy with NF of 5 × 10−5 versus 3.3 × 10−6 (Fig. 7). This observation indicates a slight negative effect of higher temperature on nucleation frequency of INP-BGs. A bit more the differences were detectable with the living INP bacteria (C41-INPC) with respect to the T50 value at lower temperature of −5.6°C (Fig. 6) and the (higher) total number of ice nuclei per cell at the first droplet freezing temperature (Fig. 7). Posttranslational modifications of INP are supposed to modify the stability of INP accumulations.6,50 Independent from potential temperature effect on INP formation and the firm cell-envelope of living cells mentioned above host cell phospholipase is induced by protein E.51 This enzymatic activity makes the envelope membranes more fluid at the E-mediated lysis tunnel area and could account for the −1°C difference detected in the T50 value of INP-BGs and live C41-INP parent bacteria.
Full length INP and a major INP degradation product have been detected with the rabbit antibodies raised against purified InaZ protein truncated with its N-terminal anchoring sequence (Fig. 5). The comparison of INP in E. coli C41 bacteria before and after E- mediated lysis show the same intensity of the INP specific bands in the live bacteria and their corresponding BGs. The chemical induced BGs seem to have a little bit better expression of INP and this could also explain the difference of number of ice nuclei at −7°C discussed above. Other studies observed ice nucleation activity for recombinant ice nucleation active E. coli similar to the P. syringae wild type strain acting as typ I ice nuclei (below −5°C).25,52 In the latter case INP of ina+ E. coli was expressed during logarithmic and stationary growth phase of the bacteria46,53 whereas E-protein mediated lysis requires active growth and cell division of the host bacteria limiting the INP expression time considerably.54-56
It should be mentioned that the experiments described in this communication did not have the aim to optimize the expression rates nor the quantity of INP per BG. Fed-Batch production of BGs could significantly increase the level of INP per BG and the total yield of INP-BG.57
The investigations performed were rather concept studies to answer the question whether BGs have the ability to induce ice nucleation at all. BGs from a recombinant Gram-negative bacterium as in our case E. coli C41 carrying INP could have the advantage over live parent bacteria that they are non-living and most important that they are no longer genetically modified organisms (GMOs) but only products thereof. In this respect they do not represent any biohazard in modifying microbial communities and escape arguments against GMOs in all different aspects. In addition β-propiolactone (BPL) treatment (which alkylates nucleic acids) of the produced BGs inactivate E-lysis escape mutants assuring a total safe BG preparation.45 The ability of INA BGs to lower the freezing time and temperature of goods could give an edge over its commercial competitors improving the economics and safety of snow making.
In the past BGs have been used for various purposes such as vaccines, carrier and targeting vehicles, miniature bioreactors and have been produced from a range of Gram-negative bacteria, as for example Salmonella tryphimurium, Actinobacillus pleuropneumoniae, Klebsiella pneumoniae, Vibrio cholera, Mannheimia haemolytica, Bordetella bronchiseptica, Shigella flexneri, Ralstonia eutropha, Pectobacterium cypripedii and various pathogenic and non-pathogenic E. coli strains including probiotic E.coli Nissle 1917.58-60 The use of INP-BGs from non-pathogenic E. coli strains or other harmless bacterial strains could find applications for advanced freezing processes and texturing of frozen food, water solubility of BG adsorbed compounds, BG ice nucleation in the atmosphere, cloud formation or artificial rain. One particular advantage for the latter 2 applications could be that BGs represent a light version of bacteria having lost almost ¾ of their weight when compared with the live parent bacteria. Therefore, INP-BGs should be able to use atmospheric transport ways more efficiently than their heavy brothers and sisters. Even if biologic ice nucleation in clouds on a global scale is in the uppermost estimate of 0.6%61 INP-BGs could find their niche in local applications competing with the use of toxic chemicals for cloud seeding or as both cloud condensing nuclei and heterogeneous ice nuclei for the production of clouds and precipitation.23
Beside various commercial applications of INP-BGs mentioned above it is anticipated that INP-BGs could also be a valuable tool to further investigate the molecular mechanisms governing biologic ice growth. In a recent study BGs have been used for cryo-electron tomography (cryoET) of native E. coli envelope membranes.34 CryoET has become a powerful tool for direct visualization of 3D structures of membrane protein complexes smaller than 300 nm at molecular resolution within native cell membranes. This application of cryoET for structural and functional studies of membrane protein complexes could become applied to INP. Furthermore, genetic engineered E. coli BGs as carrier of INP could extend the tool box for systematic studies in basic science manipulating various cell envelope structures such as lipids, polysaccharides and other which influence the efficiency of the biologic ice nucleation process.50,62
Materials and methods
Strains, plasmids and media
The E. coli strain K-12 5-α (New England BioLabs, NEB) has been used for routine cloning and E. coli C41(DE3) (Lucigen) for INP-BG production. Bacterial cultures were grown in animal protein-free, vegetable variant of Luria-Bertani (LBv: 10.0 g/l soy peptone (Carl Roth), 5.0 g/l yeast extract (Carl Roth), and 5.0 g/l NaCl). Appropriate antibiotics were supplemented to maintain the respective plasmids. The final concentration of antibiotics was as follows: ampicillin, 100 μg ml−1; gentamycin, 20 μg ml−1. The plasmids pBAD2463 and pGLysivb40,41 used in this study have been described previously. Lysis plasmid pGLMivb was obtained from Bird-C plasmid collection and represents a modification of lysis plasmid pGLysivb, where the temperature inducible λpR-cI857 promoter repressor system is exchanged by a chemical inducible LaqIq-repressor-Ptac-promoter system. Plasmid pEX-A2INP (Eurofins Genomics) harbors a chemically synthesized 3600 bp inaZ gene encoding INP of Pseudomonas syringae.64
Plasmid construction
To express, the INP under control of PBAD promoter the inaZ gene was cloned into the plasmid pBAD24. The sequence of inaZ without the termination codon was amplified by PCR where plasmid pEX-A2INP was used as template and primers inaZ-fwd and inaZ-his-rev to introduce terminal restriction sites EcoRI (5´- end) and HindIII (3´-end), as well as a 6x-His tag coding sequence and 2 termination codons at the 3´-end. Sense: (inaZ-fwd) 5´-GTACCGGAATTCAAT GAATCTCGACAAGGC- 3´; anti-sense: (inaZ-his-rev) 5´- CTGGGAAGCTTTTA TTAATGGTGATGGTGATGGTGAGAGCCGGATCCCTTTACCTCTATCCAGTCATC -3´. The amplified PCR fragment was cloned into the corresponding sites of the bacterial expression vector pBAD24, resulting in plasmid pBINP.
To construct pBH-NINP, expressing a N-terminal His-tagged truncated INP (H-NINP) lacking 485 bp of the 525 bp long N-terminal domain sequence, a 3174 bp PCR product was produced by PCR amplification using plasmid pEX-A2INP as template. Following primers were used: his-inaZ-fwd (5´-GTACCGGAATTCA ATGCACCATCACCATCACCATGGATCCGGCTCTATGAATCTCGACAAGGC-3´) to introduce terminal 6x-His tag coding sequence and EcoRI- restriction site at 5´-end; inaZ-rev: (5´-GACCCAAGCTTCTACTTTACCTCTATCCAGTCATC-3´) for a 3´-end HindIII site. The amplified PCR product and plasmid pBAD24 were digested with appropriate restriction enzymes and ligated into their corresponding sites to get pBH-NINP.
Growth, E-lysis and inactivation of bacteria
E. coli C41 harboring pBINP and E-lysis plasmid were grown in LBv supplemented with suitable antibiotics and 0.2% L-arabinose at 23°C to induce constitutive expression of the ice nucleation protein sequence under control of PBAD. When the culture reached an optical density at 600 nm (OD600) of 0.6 E-mediated lysis was initiated. Expression of gene E in lysis plasmid pGLysivb was induced by a temperature up-shift of the growing cultures from 23°C to 42°C. Whereas the expression of gene E from lysis plasmid pGLMivb in cultures kept at 23°C was induced with 0.5 mM isopropyl-D-1-thiogalactopyranoside (IPTG). After the completion of E-lysis process, the cell harvest was washed 4 times with 1x Vol. of sterile de-ionized water (dH2O) by centrifugation and suspended in 1x Vol of sterile dH2O. To inactivate any surviving E-lysis escape mutants from BG production, 0.17% (v/v) of the DNA-alkylating agent β-propiolactone (BPL, 98.5%, Ferak) was added to the washed harvest and kept for 120 min at 23°C with agitation. This mixture was washed twice with 1x Vol. of sterile dH2O and once with ROTISOLV® water (Carl Roth) and finally resuspended in 1/10x Vol. ROTISOLV®.
Monitoring and determination of E-lysis efficiency
The BG production was monitored by light-microscopy (Leica DM R microscope, Leica Microsystems), by checking the optical density at 600nm (OD600) and fluorescence-activated flow cytometry according to Langemann et al.44,45 Flow cytometry (FCM) was performed using a CyFlow® SL flow cytometer (Partec) with a 488-nm blue solid-state laser. The membrane potential-sensitive dye DiBAC4(3) (abs./em.: 493/516 nm, FL1,) was used for the evaluation of cell-viability. Fluorescent dye RH414 (abs./em. 532/760 nm) was used for staining phospholipid membranes and discriminating non-cellular background. 1 ml samples were diluted to provide appropriate cell counts and stained with 1.5 μl DiBAC4(3) (0.5mM) and 1.5 μl RH414 (2 mM), both from AnaSpec. Collected data was analyzed using FloMax V 2.52 (CyFlow SL; Quantum Analysis) and presented as 2D density dot plots illustrating forward scatter (FSC) against DiBAC fluorescence signal (FL1, DiBAC). To determine the colony forming units (cfu) during growth and lysis of E. coli C41 expressing INP bacterial samples were collected at different time points, diluted with saline and 50 μl samples were plated as triplicates on Plate Count Agar (Carl Roth) using a WASP spiral plater (Don Whitley Scientific). The plates were incubated at 36°C overnight and analyzed the next day using ProtoCOL SR 92000 colony counter (Synoptics Ltd). Lysis efficiency (LE) is defined as ratio of BGs after complete lysis to total cell counts and can be calculated by using following equation:
| (1) |
where, t0 is the time point of lysis induction (LI) and t is any time after LI.
Production of INP antibodies
N-terminal His-tagged INP lacking the N-terminal domain (H-NINP) was expressed from pBH-NINP in E. coli C41and H-NINP affinity purification of the protein was performed under denaturing conditions by using a nickel- agarose column (QIAGEN) according to the manufacturer's instructions. With this protein fraction custom made INP specific polyclonal serum was produced by Moravian Biotechnology in rabbits after 4 rounds of immunization.
Western blot analysis
Pellets (5 × 10−8 cells or BGs ml−1) were resuspended in 1x SDS gel-loading buffer, boiled for 5 min and electrophoretically separated on Bolt™ 8% Bis-Tris Plus gel by using XCell SureLock™ Mini-Cell electrophoresis system (Thermo Fisher Scientific). The proteins were then transferred to nitrocellulose membrane (GE Healthcare) with transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) using the XCell II™ Blot Module (Thermo Fisher Scientific). The membrane was placed in TBST (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.05% Tween-20) with 5% milk powder (Carl Roth) overnight at 4°C. Immunodetection was performed using rabbit polyclonal α-H-INP antibody serum followed by α-rabbit IgG-HRP (GE Healthcare). Detection was performed with an Amersham ECL Western blot detection kit (GE Healthcare) and developed with ChemiDoc™ XRS (Bio-Rad).
Ice-nucleating activity measurements
The ability to nucleate ice formation was measured by droplet freezing assay using a modified device based on a method of Vali.65 The cooling unit was constructed with the help of instruction notes by Quick-Ohm (Quick-Ohm Küpper). Hence, to detect ice nuclei in the suspensions to be tested (each suspension was adjusted to a concentration of 5 × 108 cells ml−1 determined by FCM) active at lower temperatures, a series of tenfold dilutions from each suspension (ranging from 5 × 108 cells ml−1 to 5 × 104 cells ml−1) was tested resulting in respective ice nucleus spectrum. Forty-five 10 μl droplets if not indicated otherwise of each dilution were distributed on a sterile aluminum plate coated with a hydrophobic film. The plate surface was washed with acetone before coating and flushed with a stream of filtered air before the sample droplets were placed. The plate was surrounded by styrofoam and covered by a plexiglas plate for isolation. The temperature of the working plate was decreased by 2 in series circuited 2-stage Peltier elements of the type TEC2–127–63–04, controlled by a QC-PC-C01C temperature controller (Quick-Ohm Küpper). The temperature controller was energized by a 10 kΩ potentiometer (M22S-R10K, Eaton). To measure the surface temperature of the plate a small precision temperature sensor TS-NTC-103A (B+B Thermo – Technik) was affixed, which is connected to the controller and the controller-display QC-PC-D-100 (Quick-Ohm Küpper). The plate surface can be cooled by the Peltier elements to a maximum temperature of −24.5°C. The temperature variation of the plate surface determined by a Voltcraft PL-120 T1 thermometer and a type K temperature sensor (1xK NL 1000, B+B Thermo – Technik) was ± 0.4°C. Samples were tested for its ice nucleation activity from −2 to −13°C at a constant rate of 0.5 or 1°C decrements. After a 30 sec dwell time at each temperature the Plexiglas plate was removed and the number of frozen droplets was recorded by a Panasonic Luminex DMC-FS3 digital camera.
The different samples were compared by the median freezing temperature (T50), which represents the temperature where 50% of all droplets are frozen. The T50 was calculated with the equation reported by Kishimoto et al.66
| (2) |
where F1 and F2 are the number of frozen drops at temperature T1 and adjacent temperature T2, and are just below and above 50% of the total number of tested drops (n).
The cumulative number N(T), of ice nuclei ml−1 active at a given temperature was calculated by an analogical variant of the equation of Vali 65 reported by Govindarajan and Lindow:4
| (3) |
where f = fraction of droplets unfrozen at temperature T, described as total number of droplets used divided by unfrozen droplets (N0/ NU), V = volume of each droplet used (10 µl), D = the number of 1:10serial dilutions of the original suspension. N(T) was normalized for the number of cells present in each suspension to obtain the nucleation frequency (NF) per cell by dividing ice nuclei −ml through cell density (cell−ml).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Abbas Muhammad for fruitful discussions and reviewing of the manuscript.
Funding
This work was funded by BIRD-C.
References
- [1].Gurian-Sherman D, Lindow SE. Bacterial ice nucleation: significance and molecular basis. Faseb J 1993; 7(14):1338-43; PMID:8224607 [DOI] [PubMed] [Google Scholar]
- [2].Warren G, Wolber P. Molecular aspects of microbial ice nucleation. Mol Microbiol 1991; 5(2):239-43; PMID:2041468 [DOI] [PubMed] [Google Scholar]
- [3].Pandey R, Usui K, Livingstone RA, Fischer SA, Pfaendtner J, Backus EH, Nagata Y, Fröhlich-Nowoisky J, Schmüser L, Mauri S, et al.. Ice-nucleating bacteria control the order and dynamics of interfacial water. Sci Adv 2016; 2(4):e1501630; PMID:27152346; http//dx.doi.org/ 10.1126/sciadv.1501630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Govindarajan AG, Lindow SE. Size of bacterial ice-nucleation sites measured in situ by radiation inactivation analysis. Proc Natl Acad Sci U S A 1988; 85(5):1334-8; PMID:16593912; http//dx.doi.org/ 10.1073/pnas.85.5.1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lindow SE, Lahue E, Govindarajan AG, Panopoulos NJ, Gies D. Localization of ice nucleation activity and the iceC gene product in Pseudomonas syringae and Escherichia coli. Mol Plant Microbe Interact 1989; 2(5):262-72; PMID:2520825 [DOI] [PubMed] [Google Scholar]
- [6].Kozloff LM, Turner MA, Arellano F. Formation of bacterial membrane ice-nucleating lipoglycoprotein complexes. J Bacteriol 1991; 173(20):6528-36; PMID:1917877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Phelps P, Giddings TH, Prochoda M, Fall R. Release of cell-free ice nuclei by Erwinia herbicola. J Bacteriol 1986; 167(2):496-502; PMID:3525514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Muryoi N, Matsukawa K, Yamade K, Kawahara H, Obata H. Purification and properties of an ice-nucleating protein from an ice-nucleating bacterium, Pantoea ananatis KUIN-3. J Biosci Bioeng 2003; 95(2):157-63; PMID:16233385 [DOI] [PubMed] [Google Scholar]
- [9].Lorv JS, Rose DR, Glick BR. Bacterial ice crystal controlling proteins. Scientifica (Cairo) 2014; 2014:976895; PMID:24579057; http//dx.doi.org/ 10.1155/2014/976895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Maki LR, Galyan EL, Chang-Chien MM, Caldwell DR. Ice nucleation induced by pseudomonas syringae. Appl Microbiol 1974; 28(3):456-9; PMID:4371331; http//dx.doi.org/ 10.1111/1462-2920.12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lindow SE, Arny DC, Upper CD. Bacterial ice nucleation: a factor in frost injury to plants. Plant Physiol 1982; 70(4):1084-9; PMID:16662618; http//dx.doi.org/ 10.1104/pp.70.4.1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun X, Griffith M, Pasternak JJ, Glick BR. Low temperature growth, freezing survival, and production of antifreeze protein by the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Can J Microbiol 1995; 41(9):776-84; PMID:7585354; http//dx.doi.org/ 10.1139/m95-107 [DOI] [PubMed] [Google Scholar]
- [13].Lindow SE, Arny DC, Upper CD. Erwinia herbicola: A Bacterial Ice Nucleus Active in Increasing Frost Injury to Corn. Phytopathology. 1978;68:523-7; http//dx.doi.org/ 10.1094/Phyto-68-523. [DOI] [Google Scholar]
- [14].Kawahara H. Cryoprotectants and Ice-Binding Proteins In: Margesin R, Schinner F, Marx JC, Gerday C, editors. Psychrophiles: from Biodiversity to Biotechnology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008. p. 229-46 [Google Scholar]
- [15].Abe K, Watabe S, Emori Y, Watanabe M, Arai S. An ice nucleation active gene of Erwinia ananas. Sequence similarity to those of Pseudomonas species and regions required for ice nucleation activity. FEBS Lett 1989; 258(2):297-300; PMID:2599095; http//dx.doi.org/ 10.1016/0014-5793(89)81678-3 [DOI] [PubMed] [Google Scholar]
- [16].Zhao JL, Orser CS. Conserved repetition in the ice nucleation gene inaX from Xanthomonas campestris pv. translucens. Mol Gen Genet 1990; 223(1):163-6; PMID:2259339; http//dx.doi.org/ 10.1007/BF00315811 [DOI] [PubMed] [Google Scholar]
- [17].Miller AM, Figueiredo JE, Linde GA, Colauto NB, Paccola-Meirelles LD. Characterization of the inaA gene and expression of ice nucleation phenotype in Pantoea ananatis isolates from Maize White Spot disease. Genet Mol Res 2016; 15(1):15017863; PMID:26985943; http//dx.doi.org/ 10.4238/gmr.15017863 [DOI] [PubMed] [Google Scholar]
- [18].Lindow SE, Hirano SS, Barchet WR, Arny DC, Upper CD. Relationship between ice nucleation frequency of bacteria and frost injury. Plant Physiol 1982; 70(4):1090-3; PMID:16662619; http//dx.doi.org/ 10.1104/pp.70.4.1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Christner BC, Morris CE, Foreman CM, Cai R, Sands DC. Ubiquity of biological ice nucleators in snowfall. Science 2008; 319(5867):1214; PMID:18309078; http//dx.doi.org/ 10.1126/science.1149757 [DOI] [PubMed] [Google Scholar]
- [20].Huffman JA, Prenni AJ, DeMott PJ, Pöhlker C, Mason RH, Robinson NH, et al.. High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmos Chem Phys 2013; 13(13):6151-64; http//dx.doi.org/ 10.5194/acp-13-6151-2013 [DOI] [Google Scholar]
- [21].Morris CE, Conen F, Alex Huffman J, Phillips V, Poschl U, Sands DC. Bioprecipitation: a feedback cycle linking earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob Chang Biol 2014; 20(2):341-51; PMID:24399753; http//dx.doi.org/ 10.1111/gcb.12447 [DOI] [PubMed] [Google Scholar]
- [22].Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffiere A, Yan S, Dominguez H, Thompson BM. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2008; 2(3):321-34; PMID:18185595; http//dx.doi.org/ 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- [23].Möhler O, DeMott PJ, Vali G, Levin Z. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences 2007; 4(6):1059-71; http//dx.doi.org/ 10.5194/bg-4-1059-2007 [DOI] [Google Scholar]
- [24].Möhler O, Georgakopoulos DG, Morris CE, Benz S, Ebert V, Hunsmann S, et al.. Heterogeneous ice nucleation activity of bacteria: new laboratory experiments at simulated cloud conditions. Biogeosciences 2008; 5(5):1425-35; http//dx.doi.org/ 10.5194/bg-5-1425-2008 [DOI] [Google Scholar]
- [25].Orser C, Staskawicz BJ, Panopoulos NJ, Dahlbeck D, Lindow SE. Cloning and expression of bacterial ice nucleation genes in Escherichia coli. J Bacteriol 1985; 164(1):359-66; PMID:3900043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baertlein DA, Lindow SE, Panopoulos NJ, Lee SP, Mindrinos MN, Chen TH. Expression of a bacterial ice nucleation gene in plants. Plant Physiol 1992; 100(4):1730-6; PMID:16653190; http//dx.doi.org/ 10.1104/pp.100.4.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lindgren PB, Frederick R, Govindarajan AG, Panopoulos NJ, Staskawicz BJ, Lindow SE. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J 1989; 8(5):1291-301; PMID:2548841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van Bloois E, Winter RT, Kolmar H, Fraaije MW. Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol 2011; 29(2):79-86; PMID:21146237; http//dx.doi.org/ 10.1016/j.tibtech.2010.11.003 [DOI] [PubMed] [Google Scholar]
- [29].Gao F, Ding H, Feng Z, Liu D, Zhao Y. Functional display of triphenylmethane reductase for dye removal on the surface of Escherichia coli using N-terminal domain of ice nucleation protein. Bioresour Technol 2014; 169:181-7; PMID:25058292; http//dx.doi.org/ 10.1016/j.biortech.2014.06.093 [DOI] [PubMed] [Google Scholar]
- [30].Fan LH, Liu N, Yu MR, Yang ST, Chen HL. Cell surface display of carbonic anhydrase on Escherichia coli using ice nucleation protein for CO(2) sequestration. Biotechnol Bioeng 2011; 108(12):2853-64; PMID:21732326; http//dx.doi.org/ 10.1002/bit.23251 [DOI] [PubMed] [Google Scholar]
- [31].Henrich B, Lubitz W, Plapp R. Lysis of Escherichia coli by induction of cloned phi X174 genes. Mol Gen Genet 1982; 185(3):493-7; PMID:6285147 [DOI] [PubMed] [Google Scholar]
- [32].Schon P, Schrot G, Wanner G, Lubitz W, Witte A. Two-stage model for integration of the lysis protein E of phi X174 into the cell envelope of Escherichia coli. FEMS Microbiol Rev 1995; 17(1–2):207-12; PMID:7669347 [DOI] [PubMed] [Google Scholar]
- [33].Witte A, Wanner G, Blasi U, Halfmann G, Szostak M, Lubitz W. Endogenous transmembrane tunnel formation mediated by phi X174 lysis protein E. J Bacteriol 1990; 172(7):4109-14; PMID:2141836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fu X, Himes BA, Ke D, Rice WJ, Ning J, Zhang P. Controlled bacterial lysis for electron tomography of native cell membranes. Structure 2014; 22(12):1875-82; PMID:25456413; http//dx.doi.org/ 10.1016/j.str.2014.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Witte A, Wanner G, Sulzner M, Lubitz W. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch Microbiol 1992; 157(4):381-8; PMID:1534215 [DOI] [PubMed] [Google Scholar]
- [36].Witte A, Wanner G, Lubitz W, Holtje JV. Effect of phi X174 protein E-mediated lysis on murein composition of Escherichia coli. FEMS Microbiol Lett 1998; 164(1):149-57; PMID:9675861; http//dx.doi.org/ 10.1016/S0378-1097(98)00210-9 [DOI] [PubMed] [Google Scholar]
- [37].Witte A, Lubitz W. Biochemical characterization of phi X174-protein-E-mediated lysis of Escherichia coli. Eur J Biochem 1989; 180(2):393-8; PMID:2522390; http//dx.doi.org/ 10.1111/j.1432-1033.1989.tb14661.x [DOI] [PubMed] [Google Scholar]
- [38].Witte A, Blasi U, Halfmann G, Szostak M, Wanner G, Lubitz W. Phi X174 protein E-mediated lysis of Escherichia coli. Biochimie 1990; 72(2–3):191-200; PMID:2143087; http//dx.doi.org/ 10.1016/0300-9084(90)90145-7 [DOI] [PubMed] [Google Scholar]
- [39].Jechlinger W, Haller C, Resch S, Hofmann A, Szostak MP, Lubitz W. Comparative immunogenicity of the hepatitis B virus core 149 antigen displayed on the inner and outer membrane of bacterial ghosts. Vaccine 2005; 23(27):3609-17; PMID:15855021; http//dx.doi.org/ 10.1016/j.vaccine.2004.11.078 [DOI] [PubMed] [Google Scholar]
- [40].Jechlinger W, Szostak MP, Witte A, Lubitz W. Altered temperature induction sensitivity of the lambda pR/cI857 system for controlled gene E expression in Escherichia coli. FEMS Microbiol Lett 1999; 173:347-52; http//dx.doi.org/ 10.1111/j.1574-6968.1999.tb13524.x [DOI] [PubMed] [Google Scholar]
- [41].Haidinger W, Mayr UB, Szostak MP, Resch S, Lubitz W. Escherichia coli Ghost Production by Expression of Lysis Gene E and Staphylococcal Nuclease. Appl Environ Microbiol 2003; 69(10):6106-13; PMID:14532068; http//dx.doi.org/ 10.1128/AEM.69.10.6106-6113.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Högbom M, van Wijk KJ, Slotboom DJ, Persson JO, et al.. Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci U S A 2008; 105(38):14371-6; PMID:18796603; http//dx.doi.org/ 10.1073/pnas.0804090105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gurian-Sherman D, Lindow SE. Differential effects of growth temperature on ice nuclei active at different temperatures that are produced by cells of Pseudomonas syringae. Cryobiology 1995; 32(2):129-38; PMID:7743815; http//dx.doi.org/ 10.1006/cryo.1995.1012 [DOI] [PubMed] [Google Scholar]
- [44].Langemann T, Mayr UB, Meitz A, Lubitz W, Herwig C. Multi-parameter flow cytometry as a process analytical technology (PAT) approach for the assessment of bacterial ghost production. Appl Microbiol Biotechnol 2016; 100(1):409-18; PMID:26521248; http//dx.doi.org/ 10.1007/s00253-015-7089-9 [DOI] [PubMed] [Google Scholar]
- [45].Langemann T, Koller VJ, Muhammad A, Kudela P, Mayr UB, Lubitz W. The Bacterial Ghost platform system: production and applications. Bioeng Bugs 2010; 1(5):326-36; PMID:21326832; http//dx.doi.org/ 10.4161/bbug.1.5.12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schmid D, Pridmore D, Capitani G, Battistutta R, Neeser J-R, Jann A. Molecular organisation of the ice nucleation protein InaV fromPseudomonas syringae. FEBS Letters 1997; 414(3):590-4; http//dx.doi.org/ 10.1016/s0014-5793(97)01079-x [DOI] [PubMed] [Google Scholar]
- [47].Turner MA, Arellano F, Kozloff LM. Three separate classes of bacterial ice nucleation structures. J Bacteriol 1990; 172(5):2521-6; PMID:2158972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ruggles JA, Nemecek-Marshall M, Fall R. Kinetics of appearance and disappearance of classes of bacterial ice nuclei support an aggregation model for ice nucleus assembly. J Bacteriol 1993; 175(22):7216-21; PMID:8226668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bi Y, Cabriolu R, Li T. Heterogeneous ice nucleation controlled by the coupling of surface crystallinity and surface hydrophilicity. J Phys Chem C 2016; 120(3):1507-14; http//dx.doi.org/ 10.1021/acs.jpcc.5b09740 [DOI] [Google Scholar]
- [50].Turner MA, Arellano F, Kozloff LM. Components of ice nucleation structures of bacteria. J Bacteriol 1991; 173(20):6515-27; PMID:1917876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lubitz W, Pugsley AP. Changes in host cell phospholipid composition of φX174 gene E product. FEMS Microbiol Lett 1985; 30(1–2):171-5; http//dx.doi.org/ 10.1111/j.1574-6968.1985.tb01006.x [DOI] [Google Scholar]
- [52].Hew CL, Yang DS. Protein interaction with ice. Eur J Biochem 1992; 203(1–2):33-42; PMID:1730239 [DOI] [PubMed] [Google Scholar]
- [53].Bassi AS, Ding DN, Gloor GB, Margaritis A. Expression of single chain antibodies (ScFvs) for c-myc oncoprotein in recombinant Escherichia coli membranes by using the ice-nucleation protein of Pseudomonas syringae. Biotechnol Prog 2000; 16(4):557-63; PMID:10933828; http//dx.doi.org/ 10.1021/bp000053k [DOI] [PubMed] [Google Scholar]
- [54].Blasi U, Henrich B, Lubitz W. Lysis of Escherichia coli by cloned phi X174 gene E depends on its expression. J Gen Microbiol 1985; 131(5):1107-14; PMID:3160821; http//dx.doi.org/ 10.1099/00221287-131-5-1107 [DOI] [PubMed] [Google Scholar]
- [55].Witte A, Brand E, Mayrhofer P, Narendja F, Lubitz W. Mutations in cell division proteins FtsZ and FtsA inhibit phiX174 protein-E-mediated lysis of Escherichia coli. Arch Microbiol 1998; 170(4):259-68; PMID:9732440 [DOI] [PubMed] [Google Scholar]
- [56].Witte A, Lubitz W, Bakker EP. Proton-motive-force-dependent step in the pathway to lysis of Escherichia coli induced by bacteriophage phi X174 gene E product. J Bacteriol 1987; 169(4):1750-2; PMID:2951368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Meitz A, Sagmeister P, Lubitz W, Herwig C, Langemann T. Fed-Batch Production of Bacterial Ghosts Using Dielectric Spectroscopy for Dynamic Process Control. Microorganisms 2016; 4(2):E18; PMID:27681912; http//dx.doi.org/ 10.3390/microorganisms4020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jalava K, Hensel A, Szostak M, Resch S, Lubitz W. Bacterial ghosts as vaccine candidates for veterinary applications. J Control Release 2002; 85(1–3):17-25; PMID:12480307; http//dx.doi.org/ 10.1016/S0168-3659(02)00267-5 [DOI] [PubMed] [Google Scholar]
- [59].Mayr UB, Walcher P, Azimpour C, Riedmann E, Haller C, Lubitz W. Bacterial ghosts as antigen delivery vehicles. Adv Drug Deliv Rev 2005; 57(9):1381-91; PMID:15878634; http//dx.doi.org/ 10.1016/j.addr.2005.01.027 [DOI] [PubMed] [Google Scholar]
- [60].Sührer I, Langemann T, Lubitz W, Weuster-Botz D, Castiglione K. A novel one-step expression and immobilization method for the production of biocatalytic preparations. Microb Cell Fact 2015; 14:180; PMID:26577293; http//dx.doi.org/ 10.1186/s12934-015-0371-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hoose C, Kristjánsson JE, Burrows SM. How important is biological ice nucleation in clouds on a global scale? Environ Res Lett 2010; 5(2):024009 [Google Scholar]
- [62].Kozloff LM, Turner MA, Arellano F, Lute M. Phosphatidylinositol, a phospholipid of ice-nucleating bacteria. J Bacteriol 1991; 173(6):2053-60; PMID:1848220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 1995; 177(14):4121-30; PMID:7608087; http//dx.doi.org/0021-9193/95/$04.00+0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Green RL, Warren GJ. Physical and functional repetition in a bacterial ice nucleation gene. Nature 1985; 317(6038):645-8; http//dx.doi.org/ 10.1038/317645a0 [DOI] [Google Scholar]
- [65].Vali G. Quantitative Evaluation of Experimental Results an the Heterogeneous Freezing Nucleation of Supercooled Liquids. J Atmospheric Sci 1971; 28(3):402-9; http//dx.doi.org/doi: [DOI] [Google Scholar]
- [66].Kishimoto T, Yamazaki H, Saruwatari A, Murakawa H, Sekozawa Y, Kuchitsu K, Price WS, Ishikawa M. High ice nucleation activity located in blueberry stem bark is linked to primary freeze initiation and adaptive freezing behaviour of the bark. AoB Plants 2014; 6:plu044; PMID:25082142; http//dx.doi.org/ 10.1093/aobpla/plu044 [DOI] [PMC free article] [PubMed] [Google Scholar]