ABSTRACT
Three DNA polymerases, polymerases α, δ, and ε (Pol α, Pol δ, and Pol ε), are responsible for eukaryotic genome duplication. When DNA replication stress is encountered, DNA synthesis stalls until the stress is ameliorated. However, it is not known whether there is a difference in the association of each polymerase with active and stalled replication forks. Here, we show that each DNA polymerase has a distinct pattern of association with active and stalled replication forks. Pol α is enriched at extending Okazaki fragments of active and stalled forks. In contrast, although Pol δ contacts the nascent lagging strands of active and stalled forks, it binds to only the matured (and not elongating) Okazaki fragments of stalled forks. Pol ε has greater contact with the nascent single-stranded DNA (ssDNA) of the leading strand on active forks than on stalled forks. We propose that the configuration of DNA polymerases at stalled forks facilitates the resumption of DNA synthesis after stress removal.
KEYWORDS: DNA replication, replication stress, DNA polymerase, ChIP-ssSeq, strand-specific sequencing
INTRODUCTION
During eukaryotic genome duplication, replication stress is known to cause DNA synthesis to stall until the stress is alleviated. Replication stress includes lesions induced by endogenous and exogenous DNA-damaging agents, ribonucleotide misincorporation, and the formation of secondary structures or DNA-RNA hybrids (1, 2). To better understand how genome integrity is maintained throughout replication stress, it is critical to determine how replisomes associate with DNA in active and stalled replication forks.
In budding yeast (Saccharomyces cerevisiae), DNA replication initiates at multiple sites, termed autonomously replicating sequences (ARSs) or replication origins. These origins are regulated temporally, with some origins firing early and others firing late in S phase of the cell cycle (3). In budding yeast, one of the primary responses to DNA replication stress is activation of the Mec1 and Rad53 kinase signaling cascade, a process equivalent to ataxia telangiectasia mutated- and Rad3-related (ATR) activation in human cells. Activated checkpoint kinases inhibit the firing of late replication origins, maintain the stability of stalled replication forks, and help restart DNA synthesis.
DNA polymerases α, ε, and δ (Pol α, Pol ε, and Pol δ) are the main replicative DNA polymerases for the eukaryotic nuclear genome, but other proteins are also involved in the process of DNA replication, including the origin recognition complex (ORC) and replicative helicase minichromosome maintenance (MCM) proteins. In G1/S transition, MCM is activated through the formation of the CMG (Cdc45, Mcm2-7, and GINS) complex and phosphorylation by two kinases, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) (4). Activated CMG helicase unwinds double-stranded DNA (dsDNA) at origins and recruits the single-stranded DNA (ssDNA)-binding protein RPA; RPA facilitates the recruitment of Pol α, which synthesizes RNA primers followed by short DNA chains to initiate leading strands and Okazaki fragment synthesis. Pol ε and Pol δ extend Pol α's products. In budding yeast, based on mutation biases introduced by Pol ε and δ mutants, it was proposed that Pol ε and δ are responsible for the synthesis of the leading and lagging strands, respectively (5–7). Recently, by mapping ribonucleotides introduced by DNA Pol ε and Pol δ mutants genome-wide in both budding yeast and fission yeast (Schizosaccharomyces pombe) (8–11), it was deduced by that Pol ε and Pol δ are involved in the synthesis of the leading and lagging strands, respectively. We have shown that Pol ε and Pol δ are enriched at nascent leading and lagging strands, respectively (12). However, a recent study of budding yeast suggests that Pol δ is involved in the synthesis of both leading and lagging strands and Pol ε is involved in DNA repair (13).
We and others have shown that proliferating cell nuclear antigen (PCNA), a processivity factor for Pol ε and Pol δ, is unloaded from lagging strands of stalled DNA replication forks and that this unloading is regulated by checkpoint kinases (12). Therefore, while replication proteins still associate with replisomes under stress so that DNA synthesis can resume once DNA replication stress is terminated, their contacts with DNA at stalled forks may differ from those with active forks. To analyze the interactions of DNA polymerases with replication forks in budding yeast, we evaluated the association of Pol α, Pol ε, and Pol δ with DNA in active and hydroxyurea (HU)-stalled replication forks (HU is an inhibitor of ribonucleotide reductase) by using chromatin immunoprecipitation plus strand-specific next-generation DNA sequencing (ChIP-ssSeq). Here, we report an in-depth analysis of protein ChIP-ssSeq data sets, which reveals distinct patterns of interaction of Pol δ and Pol ε with DNA at active and stalled replication forks. We suggest that these changes in contacts with DNA directly or indirectly help maintain the stability of stalled forks and facilitate the resumption of DNA synthesis after amelioration of replication stress.
RESULTS
Rationale for analyzing replication proteins using ChIP-ssSeq.
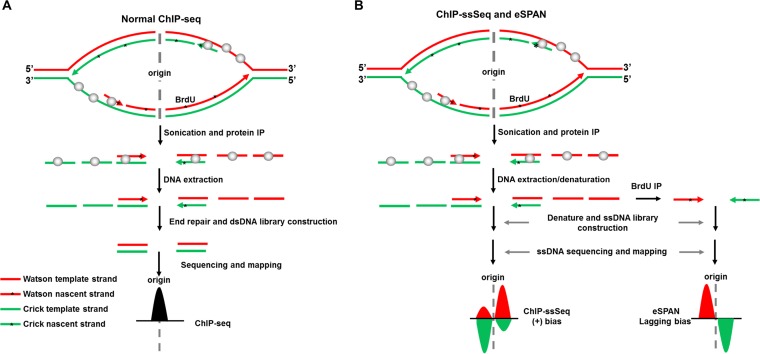
Chromatin immunoprecipitation-DNA sequencing (ChIP-seq) has been widely used to study the patterns of association of proteins of interest with chromatin (14). Most ChIP-seq libraries are prepared using protocols that involve ligation of dsDNA, which often leads to loss of ssDNA and strand-specific information (Fig. 1A). During DNA replication, dsDNA is unwound to generate ssDNA, which serves as the template for DNA synthesis. In the process, replication proteins, including ssDNA-binding proteins RPA and DNA polymerases, may partially interact with the ssDNA or DNA-RNA hybrids. In addition, DNA replication forks consist of the leading and lagging strands where DNA synthesis proceeds continuously and discontinuously, respectively and strand-specific information helps elucidate how a protein interacts with the forks.
FIG 1.
Schematic of the ChIP-ssSeq method used to detect the association of a protein with ssDNA. (A) The standard ChIP-seq process does not detect protein-ssDNA interactions. Here, we use a protein that binds to both dsDNA and ssDNA as an example to elucidate the process. The upper part of the schematic shows a replication bubble. The first step is the protein ChIP. To prepare a library for sequencing, ChIP DNA is extracted. After DNA end repair, only dsDNA is ligated with adaptors. Therefore, the sequence reads contain location information for protein-dsDNA interactions. The target protein is shown as gray balls. The black peak represents the DNA location. (B) The ChIP-ssSeq procedure preserves strand-specific information. The replication protein ChIP process is the same as the standard ChIP-seq. ChIP-ssSeq library preparation utilizes an ssDNA ligase to ligate denatured ChIP ssDNA to a 3′ adaptor, which marks the same end of each ssDNA molecule. The second DNA strand is synthesized and extended with a 3′ complementary oligonucleotide. After end repair, the 5′ end is ligated to a 5′ end adaptor with T4 DNA ligase. After sequencing, the reads are mapped to the Watson and Crick strands of the yeast genome to determine the location of the target protein and strand-specific information. ChIP-seq, chromatin immunoprecipitation and sequencing; ChIP-ssSeq, chromatin immunoprecipitation and strand-specific sequencing; dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; *, nucleotide analog BrdU, which is incorporated into nascent DNA during DNA replication. As a comparison, the outline for eSPAN procedures is also shown in panel B. The eSPAN procedure involves immunoprecipitation of protein-associated newly synthesized DNA marked with BrdU using antibodies against BrdU. Therefore, eSPAN detects the association of a protein with nascent DNA at DNA replication forks.
We previously reported the development of the enrichment and sequencing of protein-associated nascent DNA (eSPAN) method, which detects the association of a replication protein with nascent leading-/lagging-strand DNA (Fig. 1B, bottom right) (12). However, this method loses the information on how a protein interacts with ssDNA, which is prevalent at DNA replication forks. We also generated ChIP-ssSeq data sets during the process of obtaining eSPAN data sets. Briefly, protein ChIP DNA was denatured and ligated to the 3′ end of an adaptor (oligonucleotide) (Illumina) with an ssDNA ligase; ssDNA was then converted into dsDNA and ligated to a second adaptor (15, 16). The sequence reads were mapped to the Watson and Crick strands of the S. cerevisiae yeast genome (Fig. 1B). Since DNA for a protein ChIP-ssSeq likely contains both template and nascent DNA (Fig. 1B, left), ChIP-ssSeq will allow us to deduce how a DNA replication protein associates with single-stranded template DNA. As discussed and shown below, ChIP-ssSeq and eSPAN are two complementary methods, each of which reveals unique information on the association of a protein at DNA replication forks.
RPA ChIP-ssSeq shows that RPA is enriched at the lagging-strand template.
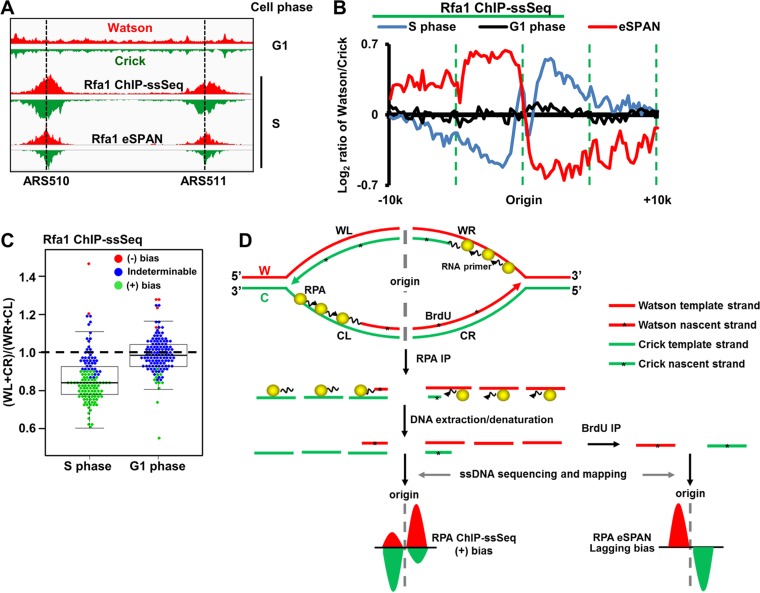
We first analyzed Rfa1 (the large subunit of the RPA complex) ChIP-ssSeq data sets to gain insight into how RPA associates with DNA replication forks. Briefly, yeast cells were arrested at G1 and then released into early S phase in the presence of HU for 45 min. Rfa1 ChIP was performed with G1 cells and early-S-phase cells. Rfa1 was barely detectable at the replication origin (ARS607) or at a distal site (ARS607 plus 8 kb, unreplicated region) at G1 (see Fig. S1A and B in the supplemental material). In contrast, Rfa1 was enriched 10-fold at ARS607 compared with its occurrence at the distal site (ARS607 plus 8 kb) in the presence of HU (Fig. S1B), indicating that RPA is recruited to DNA replication forks during S phase. Under these conditions, replication checkpoint kinase Rad53 is activated, as shown by Western blotting of Rad53 (Fig. S1C). In addition, the fact that late origins were not fired under these conditions also reflects the activation of Rad53 checkpoint kinase. The Rfa1 ChIP-ssSeq peaks surrounding each origin at ARS510 and ARS511 were asymmetric (Fig. 2A), consistent with RPA binding to ssDNA and not to dsDNA. We note that a previous study shows that RPA binds asymmetrically to resected ssDNA in a double-strand break site (17).
FIG 2.
RPA binds preferentially to ssDNA on the lagging-strand template. (A) Snapshot of RPA ChIP-ssSeq peaks at 2 early replication origins, ARS510 and ARS511. Log-phase cells were synchronized to G1 with α-factor and then released into 0.2 M hydroxyurea (HU) for 45 min. G1- and early-S-phase cells were collected for RPA ChIP-ssSeq. The RPA ChIP was performed using antibodies against the FLAG epitope, which was fused to the C terminus of Rfa1 (the large subunit of the RPA complex). Similar results were obtained by using an Rfa1-specific antibody (data not shown). The red and green regions represent the normalized sequence read densities of the Watson and Crick strands, respectively. Rfa1 eSPAN peaks are included for comparison (12). (B) The average pattern of Rfa1 ChIP-ssSeq peaks shows positive bias. The average log2 ratios of sequence reads of Watson and Crick strands at 134 early replication origins were calculated using a 200-bp sliding window and then normalized against input values to obtain the average bias pattern (blue). The lagging strand bias pattern of Rfa1 eSPAN peaks (red) (12) was used for comparison. (C) The dot-and-box plot shows the bias patterns of Rfa1 ChIP-ssSeq peaks at individual origins. The ratio of sequence reads of lagging to leading strands was calculated at each of 134 early replication origins; each dot represents the result for one origin. (+), positive; (−), negative. (D) Schematic of a mechanism that accounts for the positive bias pattern of RPA based on ChIP-ssSeq analysis and the lagging-strand bias pattern based on eSPAN analysis. The yellow balls represent the RPA complex. *, nucleotide analog BrdU, which is incorporated into nascent DNA during DNA replication. Note that the eSPAN peak bias reflects whether a protein binds to nascent leading and lagging strands, whereas ChIP-ssSeq peak bias indicates whether a protein binds to ssDNA or dsDNA.
To analyze Rfa1 ChIP-ssSeq results quantitatively on a genome-wide scale, we first calculated the average bias pattern, which is the average log2 ratio of sequencing reads of Watson strand over Crick strand, using a 200-bp sliding window surrounding 134 early replication origins. The average bias pattern of Rfa1 ChIP-ssSeq peaks indicated that on the right side of the origin, RPA bound more to the Watson strand, whereas on the left side of the origin, it bound more to the Crick strand (Fig. 2B). We categorized this finding as a positive bias pattern to differentiate it from the leading-strand bias pattern revealed by the eSPAN method, which detects the association of a protein with newly synthesized DNA (12). As a control, the Rfa1 ChIP-ssSeq using G1 cells did not show any bias (Fig. 2B), suggesting that bias seen in early S phase reflects how RPA associates with DNA replication forks in the presence of HU. We also analyzed the bias pattern of Rfa1 ChIP-ssSeq peaks at each of the 134 individual replication origins (Fig. 2C). Rfa1 ChIP-ssSeq peaks showed positive bias for most origins (n = 89 [66%]), whereas the Rfa1 ChIP-ssSeq using G1-phase cells showed no bias for the majority of origins (n = 119 [89%]). These results support the idea that RPA binds ssDNA of DNA replication forks stalled by HU.
While RPA is known to bind single-stranded template DNA, it may also contact nascent DNA at replication forks indirectly through protein-protein interactions. Indeed, RPA eSPAN reveals that RPA binds more to nascent lagging strands (12). Rfa1 ChIP-DNA contains the template-strand and nascent-strand DNA. Two potential mechanisms account for the positive bias pattern of Rfa1 ChIP-ssSeq peaks. First, a positive bias may indicate that more RPA binds to the lagging-strand template DNA than to the corresponding leading-strand template DNA (Fig. 2D). Second, RPA may bind more to nascent leading strands than to the corresponding nascent lagging strands. However, the latter explanation contradicts the RPA eSPAN results outlined above (Fig. 2D) (12). Based on our Rfa1 ChIP-ssSeq and Rfa1 eSPAN results, we suggest that more RPA binds to lagging-strand template than to leading-strand template of HU-stalled forks (Fig. 2D). The above-described RPA ChIP experiment was performed under HU conditions. We also performed the RPA ChIP-ssSeq under normal conditions. The results showed the same positive bias pattern (Fig. S1D and E), suggesting that RPA is more enriched at lagging-strand template DNA than at leading-strand template DNA at both active and HU-stalled forks. This explanation is consistent with the proposed model of RPA preferentially binding the lagging template strand to protect gaps between Okazaki fragments (18). To our knowledge, this result is the first experimental demonstration that more RPA binds to lagging-strand template than to leading-strand template. In addition to DNA replication, RPA is also involved in the DNA repair process and activation of DNA replication checkpoints (19, 20).
PCNA ChIP-ssSeq shows no strand bias at replication forks.
We analyzed PCNA ChIP-ssSeq data sets obtained from cells cultured with or without HU. No obvious strand bias was observed from the analysis of the average bias pattern of all early replication origins or the analysis of the bias patterns of individual origins (Fig. S2A to C). These results indicate that PCNA, a processivity factor of DNA polymerases that is loaded onto primer-template junctions, contacts dsDNA, including both template and nascent DNA, at active and HU-stalled replication forks.
MCM ChIP-ssSeq shows no strand bias at stalled replication forks.
Analysis of Mcm6 ChIP-ssSeq showed no significant strand bias of HU-stalled forks (Fig. S2D to F), suggesting that the MCM helicase binds to dsDNA. Similar results were obtained for Mcm4 ChIP-ssSeq (Fig. S2D to F). This observation seems to contradict the idea that the MCM complex travels along the leading strand (12, 21) and our eSPAN results showing that MCM associates preferentially with nascent leading-strand DNA compared to its association with nascent lagging-strand DNA. One likely explanation for our ChIP-ssSeq results is that the MCM helicase, while encircling one leading template DNA strand, still makes indirect contact with another lagging template strand of an HU-stalled fork. Indeed, it has been shown that the MCM protein complex interacts with both Pol ε, which is enriched at leading strands, and Pol α, which is enriched at lagging strands (22–25). For the rest of our studies, we focused on analysis of how three DNA polymerases associate with active and HU-stalled replication forks.
Pol α ChIP-ssSeq indicates that Pol α preferentially binds to DNA-RNA hybrids at lagging strands of active and HU-stalled replication forks.
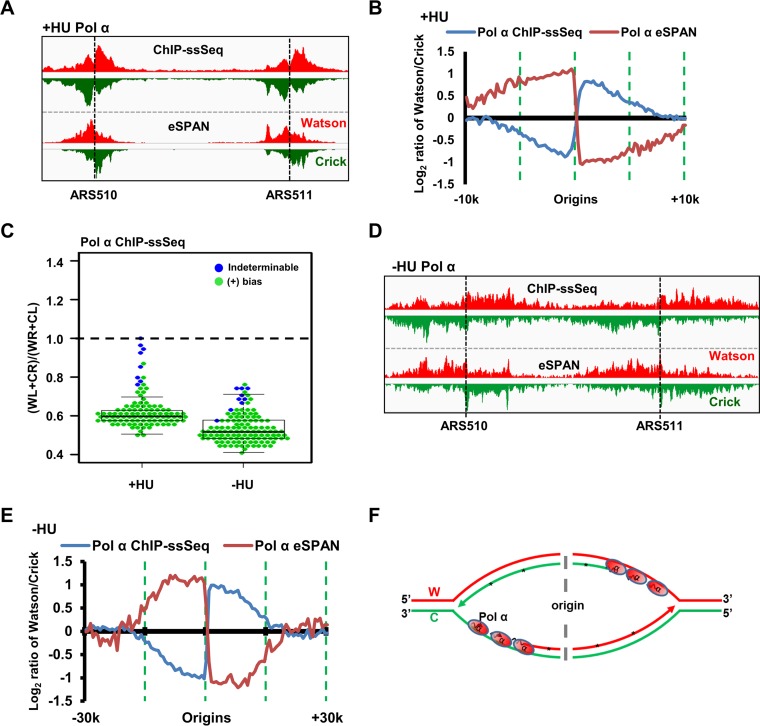
Pol α was enriched at the early replication origin (ARS607) compared to its occurrence at the distal site (ARS607 plus 8 kb) when cells were released from G1 to early S phase in the presence of HU (Fig. S3A and B), consistent with the results showing that Pol α associates with replicating DNA even in the presence of HU (26, 27). Inspection of Pol α ChIP-ssSeq results for replication origins ARS510 and ARS511 showed that Pol α ChIP-ssSeq peaks showed strong positive bias patterns (Fig. 3A and B). Analysis of the average bias pattern of 134 early replication origins confirmed that the positive bias pattern of Pol α ChIP-ssSeq peaks at individual forks occurs on a genome-wide scale (Fig. 3B), with 126 of 134 peaks (94%) showing positive bias (Fig. 3C). We also determined how Pol α bound to active replication forks by performing ChIP-ssSeq using cells released into S phase without HU at a lower temperature (Fig. 3C and S3C). The Pol α ChIP-ssSeq peaks also showed positive bias based on the analysis of the average bias pattern of early replication origins, as well as the analysis of individual origins (Fig. 3C to E).
FIG 3.
Pol α preferentially binds the single-stranded lagging-strand template DNA of both active and HU-stalled replication forks. (A to C) Pol α ChIP-ssSeq peaks at HU-stalled forks exhibit a positive (+) bias pattern. (A) Snapshot of Pol α ChIP-ssSeq and eSPAN peaks at ARS510 and ARS511. The signals represent normalized sequence read densities. Red and green represent the Watson and Crick strands, respectively. (B) Analysis of the average bias pattern of Pol α ChIP-ssSeq. Pol α eSPAN is used for comparison. (C) Analysis of bias patterns of Pol α ChIP-ssSeq of HU-stalled and active replication forks at individual origins (early replication origins only). (D, E) Pol α ChIP-ssSeq peaks at active forks show positive-strand bias. G1-synchronized yeast cells were released into fresh medium at 16°C in the presence of BrdU for 72 min. (D) Snapshot of Pol α ChIP-ssSeq and eSPAN peaks at ARS510 and ARS511. (E) Analysis of the average bias of Pol α ChIP-ssSeq peaks at active forks. (F) Schematic of a mechanism that shows why Pol α ChIP-ssSeq has a positive-strand bias pattern. W, Watson strand; C, Crick strand.
Pol α ChIP DNA consists of the lagging-strand template and newly synthesized RNA-DNA primer. The positive bias pattern of Pol α ChIP-ssSeq peaks indicates that more Pol α binds to lagging-strand template DNA than to leading-strand template DNA of active and HU-stalled replication forks. Supporting this idea, the published Pol α eSPAN peaks indicate that Pol α physically binds more to nascent lagging strands than to leading strands at active and HU-stalled replication forks (Fig. 3B, E, and F) (12). Because Pol α is involved in the synthesis of RNA and DNA primers, the positive bias indicates that Pol α binds to initiating Okazaki fragments at HU-stalled forks and to the elongating Okazaki fragments of active replication forks (Fig. 3F).
Pol δ is enriched at elongating Okazaki fragments of lagging-strand template DNA only of active replication forks.
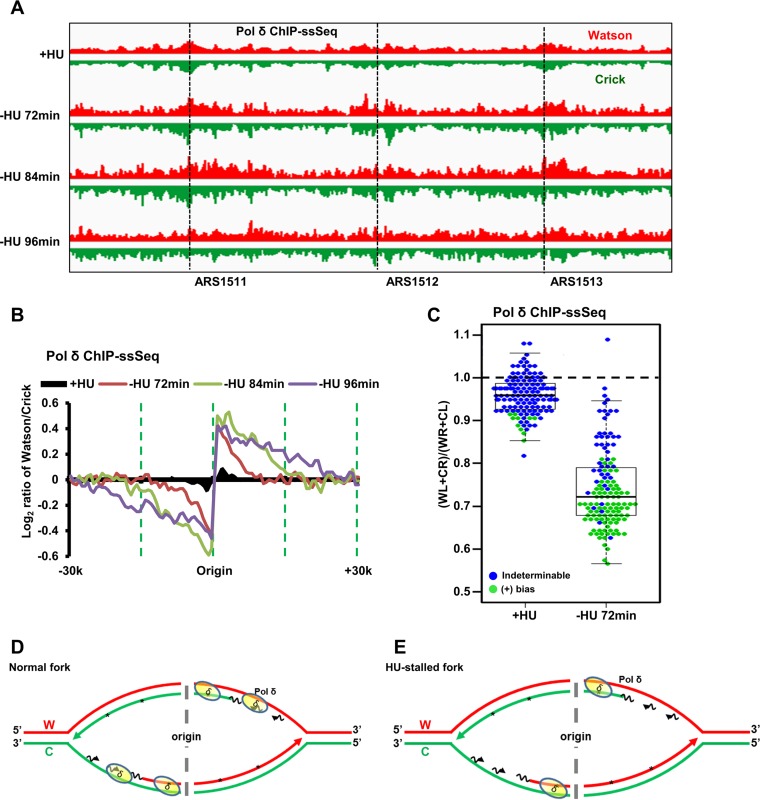
We next analyzed Pol δ (catalytic subunit) ChIP-ssSeq data obtained using cells released from G1 arrest into early S phase in the presence of HU. Pol δ ChIP-PCR analysis showed that Pol δ was enriched at replication forks originating from ARS607 compared to its occurrence at the unreplicated distal site (ARS607 plus 8 kb) (Fig. S4A and B). Pol δ ChIP-ssSeq peaks at HU-stalled forks did not reveal any bias pattern based on analysis of the average bias of 134 forks from early replication origins or on analysis of individual origins (Fig. 4A to C), suggesting that Pol δ binds equally to Watson and Crick strands of HU-stalled replication forks. In contrast, Pol δ ChIP-ssSeq peaks at active replication forks without HU showed low but consistent levels of positive bias in the analyses of the average bias pattern and of individual forks at all three time points (Fig. 4B and C). The difference in Pol δ ChIP-ssSeq peak biases between active and HU-stalled forks is unlikely to be due to any difference in input samples (Fig. S4C). Thus, Pol δ associates with DNA differentially at active and HU-stalled replication forks.
FIG 4.
Pol δ binds dsDNA at HU-stalled replication forks and preferentially binds lagging-strand template ssDNA of active forks. (A) Snapshot of Pol δ ChIP-ssSeq at three origins (ARS1511, ARS1512, and ARS1513) with HU or three time points without HU. G1-synchronized yeast cells were released into fresh medium at 16°C without HU. Samples were collected at the indicated time points (72 min, 84 min, and 96 min) for Pol δ ChIP-ssSeq of active forks. Samples were collected at 45 min after G1 release into HU at 30°C for Pol δ ChIP-ssSeq at HU-stalled forks. Note that Pol δ ChIP efficiency was relatively low compared to that of Pol ε and Pol α, despite repeated attempts. (B) Pol δ ChIP-ssSeq peaks show positive bias at active forks and no bias at HU-stalled forks. The average bias patterns of ChIP-ssSeq peaks at 134 early replication origins are shown. The bias patterns indicate that Pol δ associates with dsDNA but that it associates with ssDNA more frequently at active forks. (C) Dot-and-box plot shows the bias patterns of Pol δ ChIP-ssSeq peaks at 134 individual early replication origins of HU-stalled and active forks. Each dot represents one origin. (D, E) Schematics show the Pol δ-DNA interactions at active forks (D) and HU-stalled forks (E).
In principle, Pol δ binds both template and nascent DNA. Therefore, Pol δ ChIP-ssSeq peaks should show no bias at both active and HU-stalled forks. The eSPAN analysis of Pol δ indicates that Pol δ preferentially binds nascent lagging strands of HU-stalled and active replication forks (Fig. 4B) (12). We therefore deduce from the positive bias pattern of Pol δ ChIP-ssSeq peaks that Pol δ associates more with lagging-strand template DNA of active forks, which most likely reflects that Pol δ can associate with newly initiated Okazaki fragments with only very short sequences of nascent RNA of active forks (Fig. 4D and E). In contrast, this mode of association of Pol δ is lost at HU-stalled forks, which provides an explanation for the lack of bias of Pol δ ChIP-ssSeq peaks of HU-stalled forks. We have shown recently that the DNA polymerase clamp, PCNA, is unloaded from lagging strands of HU-stalled forks (12). PCNA is important for the activity of Pol δ, likely because it is important for tethering Pol δ at DNA replication forks. Therefore, the unloading of PCNA from lagging strands of HU-stalled forks may contribute to the loss of the association of Pol δ with newly initiated Okazaki fragments at HU-stalled forks, whereas Pol α still binds to them.
Pol ε-DNA interaction is different for active and HU-stalled replication forks.
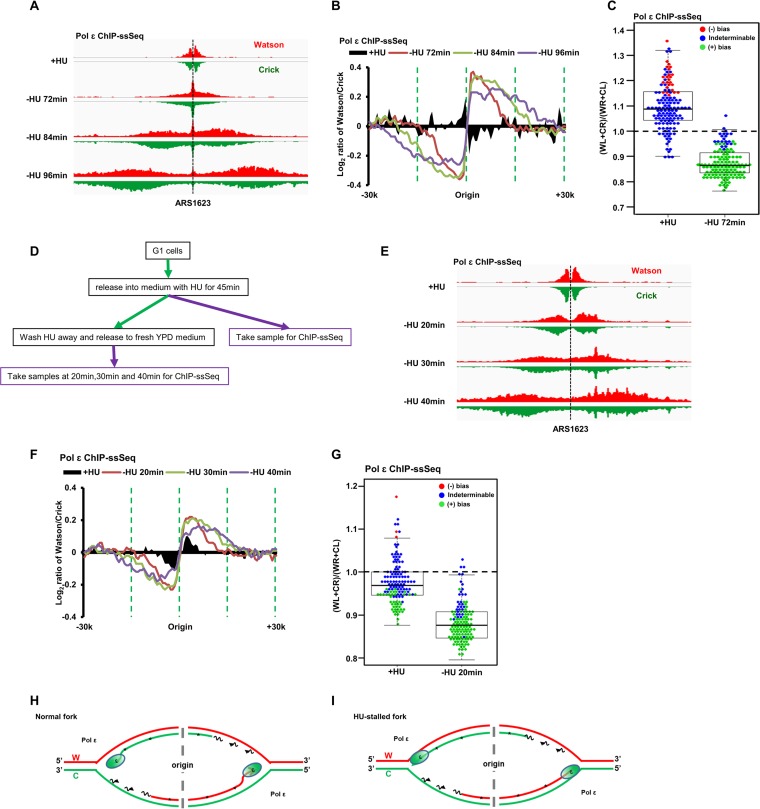
After determining the association of Pol α and Pol δ with DNA, we next used ChIP-ssSeq to examine how Pol ε interacts with DNA. Pol ε ChIP-PCR analysis indicated that Pol ε binds to replicating DNA at HU-stalled replication forks (Fig. S4A and B). Similar to the results for Pol δ, Pol ε ChIP-ssSeq demonstrated that Pol ε did not show significant bias at HU-stalled replication forks from early replication origins (Fig. 5A to C), indicating that Pol ε is cross-linked to dsDNA, including the template strand and nascent leading strand of HU-stalled replication forks. Remarkably, Pol ε ChIP-ssSeq showed positive bias at actively replicating forks at all time points considered (72, 84, and 96 min after release from G1) (Fig. 5B). The bias pattern was detected at the majority of individual origins (Fig. 5C), suggesting that the Pol ε-DNA interaction at active forks differs from that at stalled forks.
FIG 5.
Pol ε binds dsDNA at HU-stalled replication forks and preferentially binds leading nascent ssDNA of active forks. (A to C) Pol ε ChIP-ssSeq peaks show a positive bias pattern at active forks and no bias at HU-stalled forks. The experiments were performed as described in the legend to Fig. 4, except that Pol ε ChIP-ssSeq was performed. (A) Snapshot of Pol ε ChIP-ssSeq peaks at ARS1623. (B) Average bias patterns of Pol ε ChIP-ssSeq peaks using a 200-bp sliding window (early replication origins only). (C) Dot-and-box plot shows the bias patterns of Pol ε ChIP-ssSeq peaks at 134 individual early origins. (D to G) The bias patterns of Pol ε ChIP-ssSeq for HU-stalled and active forks are different. The bias patterns indicate that Pol ε associates with dsDNA but that it associates with ssDNA more frequently at active forks. (D) Flowchart of the experimental procedure. Yeast cells arrested in G1 were released into HU for 45 min. A fraction of cells were collected for Pol ε ChIP-ssSeq. The remaining cells were released into fresh medium after removal of HU. Samples were collected at the indicated time points after release for Pol ε ChIP-ssSeq. (E) Snapshot of Pol ε ChIP-ssSeq peaks at ARS1623 obtained using HU-arrested or released cells. (F) Analysis of the bias patterns of Pol ε ChIP-ssSeq peaks at HU-stalled and active forks (3 time points after release). (G) Analysis of Pol ε ChIP-ssSeq peaks at individual origins at active and HU-stalled forks. (H, I). Schematics showing Pol ε at active (H) and HU-stalled (I) forks. We propose that Pol ε binds to both template and nascent DNA of active forks but with a higher frequency to nascent DNA than to template DNA, which leads to the generation of Pol ε ChIP-ssSeq peak bias at active forks.
The above-described Pol ε ChIP-ssSeq data for HU-stalled and active replication forks were obtained from independent experiments, and the Pol ε ChIP-ssSeq bias was small. Therefore, we performed additional experiments to confirm that the different association patterns of Pol ε with DNA changed in stalled versus active forks. Briefly, yeast cells were arrested in G1 with α-factor and then released into HU-containing medium for 45 min. A fraction of cells were collected for Pol ε ChIP-ssSeq, and the remaining cells were released into fresh medium without HU. Samples were used to perform Pol ε ChIP-ssSeq at 3 time points after release from HU (20, 30, and 40 min) (Fig. 5D). Analysis of Pol ε ChIP-ssSeq data sets showed no bias pattern for peaks obtained using cells treated with HU, whereas peaks from cells after HU removal showed positive bias (Fig. 5E to G). We noticed that the results of Pol ε ChIP-ssSeq under HU conditions, as shown in Fig. 5B and F, appear to show the opposite trend. This is likely due to the facts that the Pol ε ChIP-ssSeq peaks at most origins showed indeterminable bias (no bias) and variations at a small number of origins contributed to the apparent changes in the insignificant bias pattern (compare Fig. 5C and G). Nonetheless, we observed very consistent results of Pol ε ChIP-ssSeq at active forks from each of the 3 time points of two independent experiments, supporting the idea that the association of Pol ε with DNA is altered when active forks become stalled by HU treatment.
Once again, two potential models explain the positive bias pattern of Pol ε ChIP-ssSeq peaks (Fig. 5H). Based on the Pol ε eSPAN results (Fig. 5B) (12), Pol ε binds preferentially to the leading strand. Therefore, it is possible that in addition to contact with leading-strand DNA, Pol ε also directly contacts the lagging-strand template DNA during normal replication. This mechanism is unlikely because it is hard to put the Cdc45-MCM-GINS complex, which is known to associate with Pol ε on the leading strand (23), in front of Pol ε. Second, Pol ε may not contact the leading-strand template DNA tightly, binding only to nascent DNA on the leading strands of active forks (Fig. 5H). We suggest that this mode of interaction with leading nascent DNA facilitates its ability to proofread or repair misincorporated nucleotides by using its 3′-to-5′ exonuclease activity (28). At a stalled fork, Pol ε may backtrack and associate with dsDNA, including both template and nascent strands (Fig. 5I).
DISCUSSION
Our present study reveals several novel insights into the contacts of proteins with active and HU-stalled DNA replication forks. First, we provide experimental evidence that RPA is enriched at lagging-strand template DNA compared to its occurrence at the corresponding leading-strand template DNA, consistent with the replication model of the role of RPA in DNA replication. Second, we show that Pol α associates with the lagging-strand template DNA of both active and HU-stalled forks. And third, we show that both Pol δ and Pol ε bind to HU-stalled forks differently than to active forks. Specifically, Pol δ binds to both initiating and elongating Okazaki fragments at active forks and is likely lost/removed from initiating Okazaki fragments at HU-stalled forks, where Pol α remains present. Pol ε likely backtracks and associates with dsDNA at HU-stalled forks. These results provide insight into how DNA synthesis can resume soon after the removal of HU-induced replication stress.
Advantages and limitations of the ChIP-ssSeq method and its comparison with the eSPAN method.
The library preparation of traditional ChIP-seq includes steps for dsDNA repair and dsDNA ligation. During the sample preparation process, protein-bound ssDNA and strand-specific information are lost (Fig. 1A). Generally, this loss is not an issue because most proteins bind dsDNA. However, during DNA replication, dsDNA is transiently unwound into ssDNA. Therefore, determining whether a DNA replication protein binds to ssDNA will help elucidate its mode of action in DNA synthesis. Here, we used ChIP-ssSeq to gain insights into how DNA replication proteins bind to active and stalled replication forks. Since DNA for protein ChIP-ssSeq potentially contains both template and nascent DNA, we analyzed ChIP-ssSeq peaks by calculating the average log2 ratio of sequence reads of Watson over Crick strands. If a protein contacts dsDNA, including both template and nascent DNA, the ratio should be zero, without any bias toward the Watson or Crick strand. If a protein binds ssDNA, the average log2 ratio of sequence reads of Watson over Crick strands of ChIP-ssSeq peaks is not zero. Indeed, ssDNA-binding protein RPA ChIP-ssSeq peaks at both active and HU-stalled forks exhibit a positive bias, indicating that more RPA is present at lagging-strand templates than at leading-strand templates, consistent with replication models of RPA in DNA replication.
We reported previously the development of the eSPAN method, which detects how a protein associates with newly synthesized DNA at DNA replication forks (12). Using this method, we detect the association of different DNA replication proteins with nascent leading or lagging strands of DNA replication forks. For instance, we observed that Pol ε and Pol δ are enriched at nascent leading and lagging strands, respectively, consistent with their division of labor during DNA synthesis. Interestingly, we also show that RPA is enriched at lagging strands using eSPAN. This result appears to contradict the idea that RPA binds and stabilizes single-stranded template DNA. Using ChIP-ssSeq, we show that RPA is enriched at lagging-strand templates compared to its occurrence at leading-strand templates. One explanation for the apparent discrepancy between the RPA ChIP-ssSeq and eSPAN results is that RPA binds preferentially to lagging-strand templates but contacts nascent DNA indirectly, likely through other proteins. In this way, RPA eSPAN peaks show a lagging-strand bias. Similarly, the PCNA eSPAN results show that PCNA is enriched at nascent lagging-strand DNA of active forks and nascent leading-strand DNA of HU-stalled forks, suggesting that PCNA is unloaded from lagging strands of HU-stalled forks (12). However, PCNA ChIP-ssSeq peaks at HU-stalled forks show no bias pattern. One explanation is that PCNA contacts both template DNA and nascent DNA at HU-stalled forks, which gives rise to the no-bias pattern of PCNA ChIP-ssSeq peaks. Therefore, the bias patterns of eSPAN peaks and ChIP-ssSeq peaks have different meanings: the eSPAN peak bias indicates how a protein associates with nascent leading and lagging strands of DNA replication forks, whereas the ChIP-ssSeq peak bias reflects how a protein binds to ssDNA and dsDNA. Together, these results comparing ChIP-ssSeq and eSPAN analyses of different proteins, including RPA, PCNA and MCM, and PCNA and DNA polymerases (see below), indicate that the ChIP-ssSeq and eSPAN methods provide complementary information on how a protein associates with DNA replication forks.
In theory, ChIP-ssSeq is suitable for studying DNA repair and RNA transcription when strand-specific information is needed. In fact, some reports indicate that similar ChIP-ssSeq approaches can study the DNA repair process (17, 29). These studies used either sticky-end dsDNA adaptor ligation or intramolecular microhomology to generate libraries. We adopted the ssDNA library preparation method developed by Meyer et al. (15), which was used to analyze highly damaged DNA from ancient human samples. The advantage of this method, compared with the two published ssDNA library preparation methods, is high efficiency. Meyer's method can generate libraries from very small quantities of DNA (16), and therefore, it is very suitable for constructing libraries from the small amounts of DNA isolated by ChIP experiments. We expect that ChIP-ssSeq may also yield useful information for other processes besides DNA replication and repair. For instance, allele-specific DNA methylation is known to occur frequently in mammalian cells (30). A combination of suitable library preparation methods with immunoprecipitation of methylated DNA will, in principle, be able to differentiate between methylated and unmethylated alleles. Future studies are needed to test this idea.
Association of Pol α with active and HU-stalled replication forks.
The Pol α-primase complex synthesizes primers for subsequent DNA synthesis by Pol δ and Pol ε, likely at the lagging and leading strands, respectively, of DNA replication forks. Previously, we used the eSPAN method to show that Pol α-primase is enriched at the nascent lagging strand of the DNA replication fork; this finding was consistent with the classical replication models that require a Pol α-primase complex for each Okazaki fragment (31, 32). In this study, we used the Pol α ChIP-ssSeq method to show that Pol α also binds more to the lagging-strand template DNA during DNA replication, further supporting the role of Pol α-primase in the classical DNA replication model. Interestingly, we observed that Pol α-primase remains bound to the lagging-strand template DNA at HU-stalled replication forks. We suggest that the association of the Pol α-primase complex with the template strand under this condition may facilitate the resumption of the replication process soon after amelioration of DNA replication stress. In addition, this association may serve as the target of cell cycle checkpoint kinases that regulate the arrest of DNA replication forks during stress. Consistent with this idea, previous work has shown that primase connects DNA replication to the DNA damage response (33).
Altered association of Pol δ and Pol ε with DNA replication forks stalled by replication stress.
Pol δ replicates both leading and lagging strands in the simian virus 40 (SV40) in vitro replication system (34–36). However, genetic evidence from the past decade supports Pol ε as the leading-strand replication enzyme in yeast (5, 6, 8, 37, 38), and Pol δ is responsible for replicating lagging-strand DNA. Recently, the division of labor between Pol ε and Pol δ in DNA synthesis has come into question with genetic analyses of mismatch repair-deficient DNA polymerase mutants (13). Therefore, several eukaryotic DNA replication models have been proposed (39). In every model, Pol ε is always physically linked with MCM helicase on the leading strand, regardless of whether it is the major active leading-strand DNA polymerase or just a repair enzyme. The result is fully compatible with our eSPAN data, indicating that Pol ε is enriched at the replicating leading strands. In contrast, Pol δ is enriched at the nascent lagging strands of DNA replication forks. We show here that Pol δ and Pol ε bind asymmetrically to DNA of active replication forks, suggesting that these two polymerases also bind to ssDNA and not solely to dsDNA at active replication forks. In contrast, at HU-stalled forks, Pol δ and Pol ε were bound predominantly to dsDNA. We suggest that at HU-stalled forks, Pol ε may backtrack to contact dsDNA.
A previous study has shown that MCM localization can be displaced several hundred base pairs from the origin by transcription regulation (40). While it is possible that transcriptional alteration during HU block contributes to the lack of bias in Pol ε and Pol δ ChIP-ssSeq peaks at HU-stalled forks, it is unlikely for the following reasons. First, we show that the Pol ε ChIP-ssSeq peak bias pattern reappears after we release cells from HU block to fresh medium, suggesting that Pol ε bias is associated with active replication forks. Second, it is known that HU has no apparent effect on the initiation of DNA replication at early replication origins, based on studies from many laboratories. Moreover, the observation that transcription can shift MCM localization was made in rat1 mutant cells in which transcription termination was reduced, whereas at HU-stalled forks, we did not observe such dramatic alterations in MCM distribution (12).
We noticed that the bias of Pol δ and Pol ε ChIP-ssSeq peaks at active forks is small compared to that of Rfa1 or Pol α. This small bias is not likely an artifact of calculation, because we analyzed ChIP-ssSeq data sets using two different methods. First, we used a 200-bp sliding window to calculate bias from either 10 or 30 kb surrounding each replication origin of HU-stalled and active forks, respectively. The trend of each data point of Pol δ and Pol ε ChIP-ssSeq shows that the bias, while small, is not random. Second, we also analyzed whether there is bias of Pol δ and Pol ε ChIP-ssSeq peaks at individual replication forks (Fig. 4C and 5C and G) and found that the bias, while small, is statistically significant. Unlike RPA, which binds ssDNA, and Pol α, which synthesizes primers for Pol δ and Pol ε, most Pol δ and Pol ε likely still contacts dsDNA, including template DNA and newly synthesized DNA. Therefore, it is not surprising that the Pol δ and Pol ε ChIP-ssSeq bias is smaller than that of RPA or Pol α. In conclusion, the bias changes at HU-stalled forks and active forks of Pol δ and Pol ε ChIP-ssSeq, while small, reflect the polymerase-DNA spatial contacts at active forks.
MATERIALS AND METHODS
Yeast strains.
The S. cerevisiae yeast strains used in this study were derived from strain W303 (leu2-3,112 ura3-1 his3-11 trp1-1 ade2-1 can1-100). Genotypes are listed in Table S1 in the supplemental material.
ChIP-ssSeq procedure.
ChIP-ssSeq experiments were performed as described previously (12). Briefly, α-factor was used to synchronize yeast cells at G1 (5 μg/ml and 50 ng/ml for wild-type BAR and bar1 mutant strains, respectively). To analyze the association of proteins with active forks, G1-arrested cells were released into chilled (16°C) yeast extract-peptone-dextrose (YPD) medium containing 400 mg/liter bromodeoxyuridine (BrdU), and samples were collected at different time points. We treated cells with HU, an inhibitor of ribonucleotide reductase, to deplete cells of deoxynucleoside triphosphate (dNTP). Thus, HU stalls the progression of DNA replication forks and inhibits the firing of late replication origins (41, 42). To analyze protein association with HU-stalled forks, cells were released into fresh medium containing 400 mg/liter BrdU and 0.2 M HU for 45 min. To perform ChIP, samples were incubated with 1% paraformaldehyde at 25°C for 20 min and then quenched with 0.125 M glycine for 5 min. Cells were lysed with glass beads, and the chromatin pellet was washed and sonicated to shear DNA to an average fragment size of about 200 to 400 bp. Sheared chromatin was immunoprecipitated with anti-FLAG antibody F1804 or anti-RPA antibody (gift of Steven Brill). After extensive washing, cross-links of the immunoprecipitated chromatin were reversed. DNA was recovered with the Chelex-100 protocol (43). Recovered DNA was purified with a PCR purification kit (Qiagen). ChIP DNA was used for quantitative PCR (Q-PCR) analysis (Table S2) and was treated at 95°C for 5 min before being used for library preparation of ssDNA in accordance with previously published procedures (15).
ChIP-ssSeq and analysis.
The ssDNA libraries were sequenced using paired-end sequencing on Illumina Hi-Seq 2000 or 2500 machines. Reads were first mapped to the yeast genome (sacCer3) using Bowtie2 software (44). Consistent paired-end reads were chosen for subsequent analysis. We noted that after removal of duplicated reads, paired-end reads with the same ends were rarely detected in our samples even for Mcm6 ChIP-seq using G1 cells. This is likely due to the fact that chromatin was sheared by sonication and the ends were processed during library preparation. The genome-wide read coverage of Watson and Crick strands was calculated by using BEDTools (45). The reads of the Watson and Crick strands were merged for peak calling by using MACS software (46).
We used our previously mapped DNA origin data set for analysis (12). To calculate the average bias pattern, the average log2 ratios of sequencing reads of Watson strand over Crick strand surrounding 134 early replication origins (10 and 30 kb up- and downstream from HU-stalled and active forks, respectively) were calculated using a sliding window of 200 bp. Duplicate reads were excluded from the calculations. These ratios were then normalized against the corresponding input to obtain the average bias pattern of ChIP-ssSeq. To analyze bias at individual origins, each peak region was separated into 4 quadrants: the Watson strand at the left (WL) and right (WR) of an origin and the Crick strand at the left (CL) and right (CR) of an origin. The sequence reads in each quadrant were counted. Binomial distribution was used to calculate the P value to determine whether sequence reads at the leading strand (WL+CR) were different from those of the lagging strand (WR+CL) at each replication fork. The log2 ratio {log2 [(WL+CR)/(WR+CL)]} at each replication origin was calculated and used to determine whether a ChIP-ssSeq peak had a positive- or negative-strand bias.
The published Gene Expression Omnibus (GEO) data set used in this study is available under accession number GSE52614.
Supplementary Material
ACKNOWLEDGMENT
We thank Oscar Aparicio for yeast strains and plasmids, Steven Brill for the anti-RPA antibody, and June Oshiro for editing our manuscript.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00190-17.
REFERENCES
- 1.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazouzi A, Velimezi G, Loizou JI. 2014. DNA replication stress: causes, resolution and disease. Exp Cell Res 329:85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio OM. 2013. Location, location, location: it's all in the timing for replication origins. Genes Dev 27:117–128. doi: 10.1101/gad.209999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell SD, Botchan MR. 2013. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 5:a012807. doi: 10.1101/cshperspect.a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. 2007. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. 2008. Division of labor at the eukaryotic replication fork. Mol Cell 30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stillman B. 2008. DNA polymerases at the replication fork in eukaryotes. Mol Cell 30:259–260. doi: 10.1016/j.molcel.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ, Kunkel TA. 2015. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol 22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daigaku Y, Keszthelyi A, Muller CA, Miyabe I, Brooks T, Retkute R, Hubank M, Nieduszynski CA, Carr AM. 2015. A global profile of replicative polymerase usage. Nat Struct Mol Biol 22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reijns MA, Kemp H, Ding J, de Proce SM, Jackson AP, Taylor MS. 2015. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh KD, Balachander S, Hesselberth JR, Storici F. 2015. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat Methods 12:251–257. doi: 10.1038/nmeth.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. 2014. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell 56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson RE, Klassen R, Prakash L, Prakash S. 2015. A major role of DNA polymerase delta in replication of both the leading and lagging DNA strands. Mol Cell 59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andres AM, Eichler EE, Slatkin M, Reich D, Kelso J, Paabo S. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gansauge MT, Meyer M. 2013. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc 8:737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 17.Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC, Casellas R. 2013. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Rep 3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell M, Langston L, Stillman B. 2013. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marechal A, Zou L. 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 21.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME. 2014. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A 111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta S, van Deursen F, de Piccoli G, Labib K. 2013. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol 23:543–552. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 25.Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. 2009. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J 28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K. 2012. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45:696–704. doi: 10.1016/j.molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Tran HT, Gordenin DA, Resnick MA. 1999. The 3′–>5′ exonucleases of DNA polymerases delta and epsilon and the 5′–>3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol 19:2000–2007. doi: 10.1128/MCB.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou ZX, Zhang MJ, Peng X, Takayama Y, Xu XY, Huang LZ, Du LL. 2013. Mapping genomic hotspots of DNA damage by a single-strand-DNA-compatible and strand-specific ChIP-seq method. Genome Res 23:705–715. doi: 10.1101/gr.146357.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 31.Waga S, Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu Rev Biochem 67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 32.Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 33.Marini F, Pellicioli A, Paciotti V, Lucchini G, Plevani P, Stern DF, Foiani M. 1997. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J 16:639–650. doi: 10.1093/emboj/16.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsurimoto T, Stillman B. 1991. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem 266:1961–1968. [PubMed] [Google Scholar]
- 35.Tsurimoto T, Stillman B. 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem 266:1950–1960. [PubMed] [Google Scholar]
- 36.Waga S, Stillman B. 1994. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 37.Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. 2014. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol 21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams JS, Clausen AR, Lujan SA, Marjavaara L, Clark AB, Burgers PM, Chabes A, Kunkel TA. 2015. Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nat Struct Mol Biol 22:291–297. doi: 10.1038/nsmb.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stillman B. 2015. Reconsidering DNA polymerases at the replication fork in eukaryotes. Mol Cell 59:139–141. doi: 10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gros J, Kumar C, Lynch G, Yadav T, Whitehouse I, Remus D. 2015. Post-licensing specification of eukaryotic replication origins by facilitated Mcm2-7 sliding along DNA. Mol Cell 60:797–807. doi: 10.1016/j.molcel.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santocanale C, Diffley JF. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 42.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 43.Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.