ABSTRACT
Alphaherpesviruses that establish persistent infections rely partly on their ability to evade host antiviral responses, notably the type I interferon (IFN) response. However, the mechanisms employed by alphaherpesviruses to avoid this response are not well understood. Pseudorabies virus (PRV) is an economically important pathogen and a useful model system for studying alphaherpesvirus biology. To identify PRV proteins that antagonize type I IFN signaling, we performed a screen by using an IFN-stimulated response element reporter in the swine cell line CRL. Unexpectedly, we identified the dUTPase UL50 as a strong inhibitor. We confirmed that UL50 has the ability to inhibit type I IFN signaling by performing ectopic expression of UL50 in cells and deletion of UL50 in PRV. Mechanistically, UL50 impeded type I IFN-induced STAT1 phosphorylation, likely by accelerating lysosomal degradation of IFN receptor 1 (IFNAR1). In addition, this UL50 activity was independent of its dUTPase activity and required amino acids 225 to 253 in the C-terminal region. The UL50 encoded by herpes simplex virus 1 (HSV-1) also possessed similar activity. Moreover, UL50-deleted PRV was more susceptible to IFN than UL50-proficient PRV. Our results suggest that in addition to its dUTPase activity, the UL50 protein of alphaherpesviruses possesses the ability to suppress type I IFN signaling by promoting lysosomal degradation of IFNAR1, thereby contributing to immune evasion. This finding reveals UL50 as a potential antiviral target.
IMPORTANCE Alphaherpesviruses can establish lifelong infections and cause many diseases in humans and animals. Pseudorabies virus (PRV) is a swine alphaherpesvirus that threatens pig production. Using PRV as a model, we found that this alphaherpesvirus could utilize its encoded dUTPase UL50 to induce IFNAR1 degradation and inhibit type I IFN signaling in an enzymatic activity-independent manner. Our finding reveals a mechanism employed by an alphaherpesvirus to evade the immune response and indicates that UL50 is an important viral protein in pathogenesis and is a potential target for antiviral drug development.
KEYWORDS: alphaherpesviruses, interferon signaling, type I interferon receptor 1, dUTPase UL50, pseudorabies virus
INTRODUCTION
The family Herpesviridae consists of large DNA viruses capable of establishing lifelong infection in hosts. Members of this family are causative agents of a variety of human and animal diseases and are further categorized into three subfamilies: alpha-, beta-, and gammaherpesviruses (1). Alphaherpesviruses are neurotropic and include herpes simplex virus 1 and 2 (HSV-1 and HSV-2), varicella-zoster virus (VZV), simian varicella virus (SVV), equine herpesvirus 1 (EHV1), bovine herpesvirus 1 (BHV1), and pseudorabies virus (PRV) (1–3). PRV infection can cause Aujeszky's disease in pigs, which can result in substantial economic losses. PRV resembles HSV-1 in many ways, despite substantial divergence at the sequence level, and is an important model virus for studying alphaherpesvirus biology in cell culture and in natural hosts (2, 4).
Viral infections trigger rapid host innate immune responses to combat infection. Among these, the type I interferon (IFN) response is prominent and plays a critical role in viral suppression, immunomodulation, and the regulation of the adaptive immune response (5, 6). Type I IFNs, represented by IFN-α and IFN-β, exert functions by binding to their shared receptor, comprising two subunits: IFNAR1 and IFNAR2. Because they lack an intrinsic protein kinase domain, IFNAR1 and IFNAR2 rely on preassociated members of the Janus protein tyrosine kinase family (JAKs) for signal transduction (7). Engagement of IFNs induces dimerization of IFNAR1 and IFNAR2, resulting in JAK transphosphorylation and activation. Activated JAKs subsequently phosphorylate STAT1 and STAT2, resulting in the formation of a heterodimer of pSTAT1/pSTAT2, which complexes with IFN regulatory factor 9 (IRF9). This trimeric complex, referred to as IFN-stimulated gene factor 3 (ISGF3), then rapidly shuttles to the nucleus, where it binds to a specific DNA sequence known as the IFN-stimulated response element (ISRE) and stimulates transcription of a number of IFN-stimulated genes (ISGs). Many of the gene products have potent antiviral functions (6, 8, 9).
Viruses in turn have evolved various strategies to counteract the interferon response, including inhibiting IFN production, suppressing IFN signaling, and neutralizing the functions of various ISGs. Evading the interferon response is particularly important for herpesviruses so they can establish persistent infections in hosts. Recent studies have significantly advanced our knowledge on the strategies employed by alphaherpesviruses, particularly HSV-1, to inhibit IFN production (8, 10–17).
Several studies have indicated that type I IFN signaling is impaired in alphaherpesvirus-infected cells. Human alphaherpesvirus infections dramatically suppress the expression of IFN-induced genes and render the infected cells less responsive to interferon than control cells in terms of IFN-triggered early signaling events, including the phosphorylation of STAT1 and STAT2 (17–19). In HSV-1-infected cells, the phosphorylation of JAKs and STATs is severely impaired at the early stage of infection and is accompanied by a reduction of JAK1 and STAT2 levels. The loss of JAK1 is mediated partly by the virion host shutoff protein UL41, which induces upregulation of suppressor of cytokine signaling 3 (SOCS3) (19, 20). The viral early protein UL54 (ICP27) plays a role in inhibiting STAT1 phosphorylation (21). In VZV-infected cells, IRF9 is degraded by the expression of the immediate early viral protein ORF63, and STAT2 phosphorylation is inhibited via other viral proteins (3). In HSV-2-infected cells, both STAT2 protein and mRNA are degraded (22). It appears that the key components of the type I IFN pathway are often the targets of viral proteins, either directly or indirectly, and that multiple viral components are involved in this process.
PRV infection suppresses IFN-induced upregulation of a subset of ISGs and STAT1 phosphorylation, indicating an impairment of IFN signaling in PRV-infected cells (23). However, the interaction between PRV infection and the interferon signaling pathway at the molecular level is largely unknown. In this study, to identify the PRV proteins that have potential to inhibit IFN signaling, we performed an unbiased screen by using an IFN-stimulated response element reporter in the swine cell line CRL. Unexpectedly, we identified the dUTPase UL50 as the strongest inhibitor among the 40 viral proteins screened. dUTPase catalyzes the hydrolysis of dUTP into dUMP and inorganic pyrophosphate and provides the dUMP precursor for dTTP biosynthesis (24). Furthermore, dUTPase maintains a low cellular dUTP/dTTP ratio to prevent the incorporation of uracil moieties into DNA; thus, this protein is critically important for DNA replication and genomic stability (24). Interestingly, several studies with gammaherpesviruses have shown that the dUTPase ORF54, a homologue of UL50, is capable of immune modulation that is independent of its dUTPase enzymatic activity (25–27). Here we report that independent of their dUTPase activity, the UL50 proteins of the alphaherpesviruses PRV and HSV-1 possess the ability to suppress interferon signaling by promoting lysosomal degradation of IFNAR1.
RESULTS
PRV UL50 inhibits the IFN-α signaling pathway.
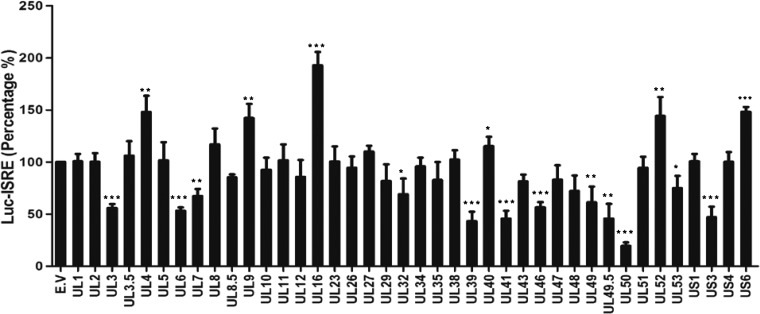
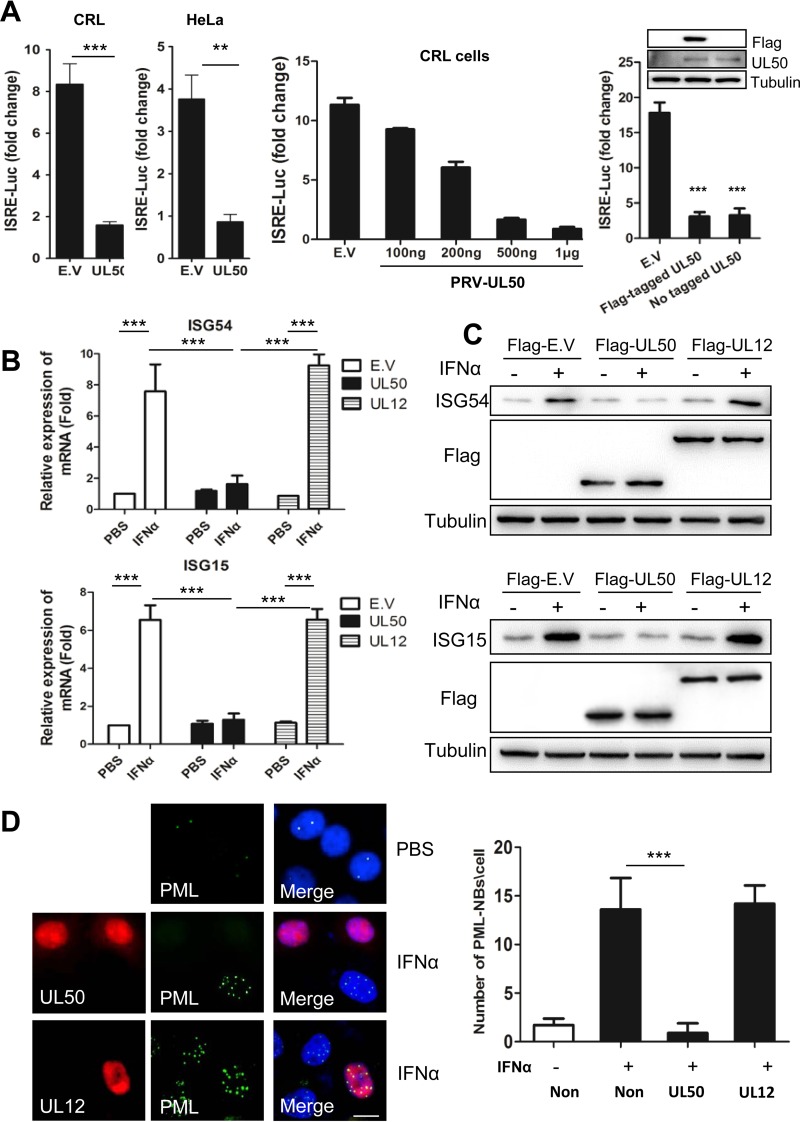
To screen for PRV proteins that antagonize type I IFN signaling, we individually transfected a plasmid encoding each viral protein or a vector control together with a firefly luciferase reporter driven by the IFN-stimulated response element (pGL3-ISRE-Luc) and a transfection control plasmid constitutively expressing renilla luciferase (pRL-TK) into the porcine macrophage cell line CRL. At 12 h posttransfection, the cells were treated with or without porcine IFN-α, and the induction of the ISRE reporter was measured by a dual-luciferase assay. Among the 40 PRV proteins we screened, UL50, a dUTPase, showed the strongest inhibition of IFN-α induction, and UL12, among many other PRV proteins, did not show much effect (Fig. 1). PRV UL50 diminished the activation of ISRE by IFN-α to only 19% and 23% of the control levels in porcine and human cells, respectively (Fig. 2A, left panel), and in a dose-dependent manner (Fig. 2A, middle panel). Both Flag-tagged and untagged UL50 proteins showed similar levels of inhibition of IFN-α induction (Fig. 2A, right panel). To verify the inhibitory activity of UL50 on IFN-α signaling, we examined the effect of UL50 expression on the induction of ISGs by IFN-α. Quantitative reverse transcription-PCR (qRT-PCR) and Western blot analysis demonstrated that the IFN-α-induced upregulation of ISG15 and ISG54, two classical ISGs, was substantially inhibited by Flag-tagged UL50 compared to that with the empty vector (E.V) and Flag-tagged UL12-transfected controls at both the mRNA (Fig. 2B) and protein (Fig. 2C) levels. In addition, immunofluorescence analysis revealed that the IFN-α-induced upregulation of promyelocytic leukemia protein (PML), another important ISG protein that forms nuclear bodies, was suppressed in nearly all the cells expressing UL50, while no inhibition was observed in UL12-expressing cells (Fig. 2D). Taken together, these data demonstrate that UL50 antagonizes interferon signaling.
FIG 1.
Inhibition of IFN-α-induced activation of ISRE by PRV proteins. Porcine CRL cells were cotransfected with 500 ng of plasmid expressing a PRV protein or with empty vector (E.V) together with 100 ng of pGL3-ISRE-Luc and 10 ng of pRL-TK. pRL-TK was used as an internal control for transfection efficiency. Twelve hours after transfection, cells were incubated in medium containing porcine IFN-α (1,000 U/ml) for 24 h and then harvested and analyzed for luciferase activity. The values represent percentages of IFN-α-induced ISRE activity in the cells expressing viral proteins compared to that in cells with empty vector. Data are means and standard deviations (SD) for three independent experiments. Statistical analyses were performed by Student's t test, using GraphPad Prism software. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 2.
PRV UL50 blocks IFN-α signaling. (A) PRV UL50 suppressed the IFN-α-induced activation of ISRE. (Left) Porcine CRL cells and human HeLa cells were cotransfected with 500 ng (or a different dose [middle panel]) of plasmid expressing PRV UL50 or with empty vector (E.V) together with 100 ng of pGL3-ISRE-Luc and 10 ng of pRL-TK. Twelve hours after transfection, the cells were incubated in medium containing porcine IFN-α (1,000 U/ml) for 24 h and then harvested and analyzed for luciferase activity. (Right) The Flag tag did not interfere with the suppressive effect of PRV UL50 on IFN-α-induced activation of ISRE. Porcine CRL cells were cotransfected with 500 ng of plasmid expressing either Flag-tagged PRV UL50 or untagged PRV UL50 or with empty vector (E.V) together with luciferase-expressing plasmids and then analyzed for luciferase activity. The Flag and UL50 proteins were verified by Western blotting. The values represented fold changes of ISRE activity between IFN-α-treated and untreated cells. Data are means and SD for three independent experiments. (B) PRV UL50 inhibited IFN-α-mediated induction of ISG15 and ISG54 at the mRNA level. HeLa cells were transfected with 2 μg of plasmid, as indicated. Twenty-four hours after transfection, the cells were incubated in medium containing human IFN-α (1,000 U/ml) for an additional 4 h. The mRNA levels of ISG15 and ISG54 were then detected by qRT-PCR. The results were obtained from three independent experiments and are means and SD. Statistical analyses of the above-described experiments were performed using GraphPad Prism software to perform Student's t test. **, P < 0.01; ***, P < 0.001. (C) PRV UL50 inhibited IFN-α-induced expression of ISG15 and ISG54 at the protein level. CRL cells (upper panels) and HeLa cells (lower panels) were transfected with 1 μg of plasmid, as indicated. Twenty-four hours after transfection, the cells were incubated in medium containing porcine or human IFN-α (1,000 U/ml) for 24 h. The expression of ISG54, ISG15, and Flag was then determined by Western blotting. Tubulin was used as a reference control. (D) PRV UL50 disrupted the endogenous PML nuclear bodies (PML-NBs) induced by IFN-α. PK15 cells were transfected with 1 μg of Flag-tagged UL50 or UL12. (Left) Twelve hours after transfection, the cells were treated with porcine IFN-α (1,000 U/ml) for 24 h, and PML, UL50, and UL12 were visualized by immunofluorescence assay with specific antisera. The nucleus was stained with DAPI. Bar = 10 μm. (Right) The number of PML-NBs in 50 cells was quantified, and means and SD are shown. Statistical analyses were performed by ANOVA, using GraphPad Prism software. ***, P < 0.001.
PRV UL50 inhibits IFN-α-induced STAT1 phosphorylation and translocation.
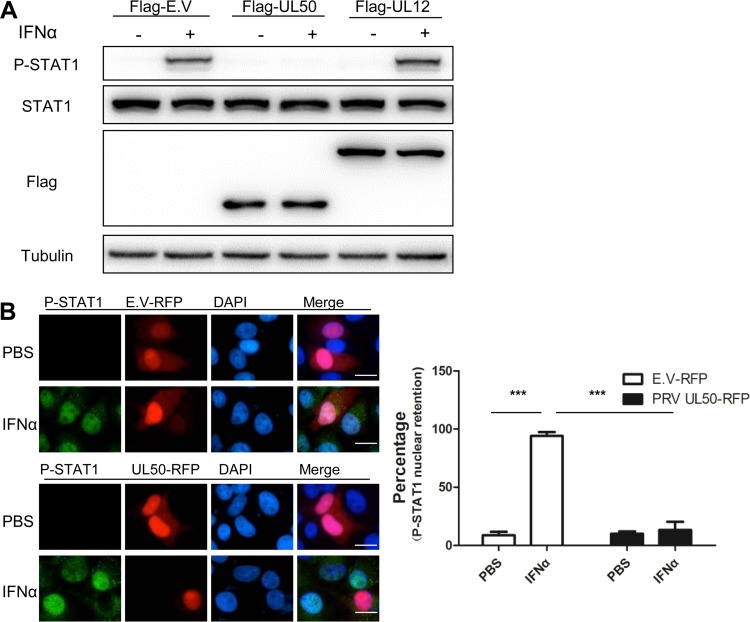
We then examined whether IFN-α-induced STAT1 phosphorylation was affected by UL50, as it is a critical signaling event in the type I IFN response and was shown to be suppressed by PRV infection (23). Western blot analysis showed that the ectopic expression of UL50, not UL12, in CRL cells abrogated IFN-α-induced STAT1 phosphorylation without affecting STAT1 protein expression (Fig. 3A). Moreover, immunofluorescence analysis of PK15 cells revealed that the IFN-α-induced nuclear accumulation of P-STAT1 was severely impaired in cells expressing UL50 (Fig. 3B). Collectively, these data demonstrate that UL50 expression in cells blocks IFN-α-induced phosphorylation and subsequent nuclear translocation of STAT1.
FIG 3.
PRV UL50 blocks the IFN-α-induced phosphorylation and nuclear accumulation of STAT1. (A) CRL cells were transfected with E.V or with 1 μg of plasmid expressing Flag-tagged PRV UL50 or PRV UL12 (viral negative control). Twenty-four hours after transfection, the cells were treated or not with porcine IFN-α (1,000 U/ml) for 30 min. STAT1, P-STAT, Flag, and tubulin were detected by Western blotting. (B) UL50 prevents the IFN-α-induced nuclear retention of P-STAT1. PK15 cells were transfected with 1 μg of plasmid expressing either control red fluorescent protein (RFP) or RFP-tagged PRV UL50. Twenty-four hours after transfection, the cells were incubated in medium containing either PBS or porcine IFN-α (1,000 U/ml) for 90 min. (Left) Immunofluorescence analysis of P-STAT1 and UL50 was performed using antibodies against P-STAT1 and RFP. The nucleus was stained with DAPI. Bars = 10 μm. (Right) The number of cells with P-STAT1 nuclear retention in 50 RFP- or UL50-expressing cells was quantified and expressed as a percentage of the total number of counted cells. Data are means and SD for three independent experiments. Statistical analyses were performed by ANOVA, using GraphPad Prism software. ***, P < 0.001.
PRV UL50 induces IFNAR1 degradation in lysosomes.
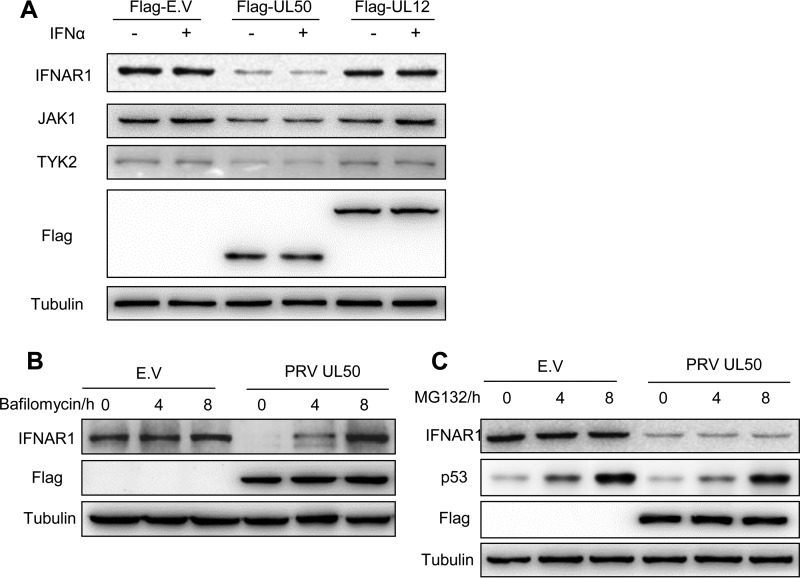
To unravel the mechanism underlying UL50 inhibition of STAT1 phosphorylation, we compared the expression levels of the components upstream of STAT1 phosphorylation in type I IFN signaling between CRL cells transfected with UL50 and those transfected with the empty vector or UL12 by Western blotting. We found that UL50 expression remarkably reduced IFNAR1 expression regardless of IFN-α treatment and modestly decreased the expression of JAK1 and TYK2, particularly in the cells treated with IFN-α (Fig. 4A). The reduced IFNAR1 in UL50-transfected cells was restored to the control level by treating the cells with the lysosome inhibitor bafilomycin for 8 h (Fig. 4B), but it was not restored by treatment with the proteasome inhibitor MG132, which effectively prevents p53 from degradation (Fig. 4C). This result indicates that UL50 likely induces lysosomal degradation of IFNAR1.
FIG 4.
UL50 inhibits IFNAR1 through lysosomal degradation. CRL cells were transfected with E.V or with 1 μg of plasmid expressing Flag-tagged PRV UL50 or PRV UL12. (A) Twenty-four hours after transfection, the cells were treated or not with porcine IFN-α (1,000 U/ml) for 30 min. IFNAR1, JAK1, TYK2, Flag, and tubulin were detected by Western blotting. Twenty-four hours after transfection, the cells were incubated in medium containing the lysosomal inhibitor bafilomycin (10 μM) (B) or the proteasome inhibitor MG132 (5 μM) (C) for 0, 4, and 8 h. The IFNAR1, p53, Flag, and tubulin proteins were detected by Western blotting. Equivalent volumes of dimethyl sulfoxide (DMSO) were added to the untreated control.
The UL50 proteins of PRV and HSV-1 inhibit type I IFN signaling independent of its dUTPase catalytic activity.
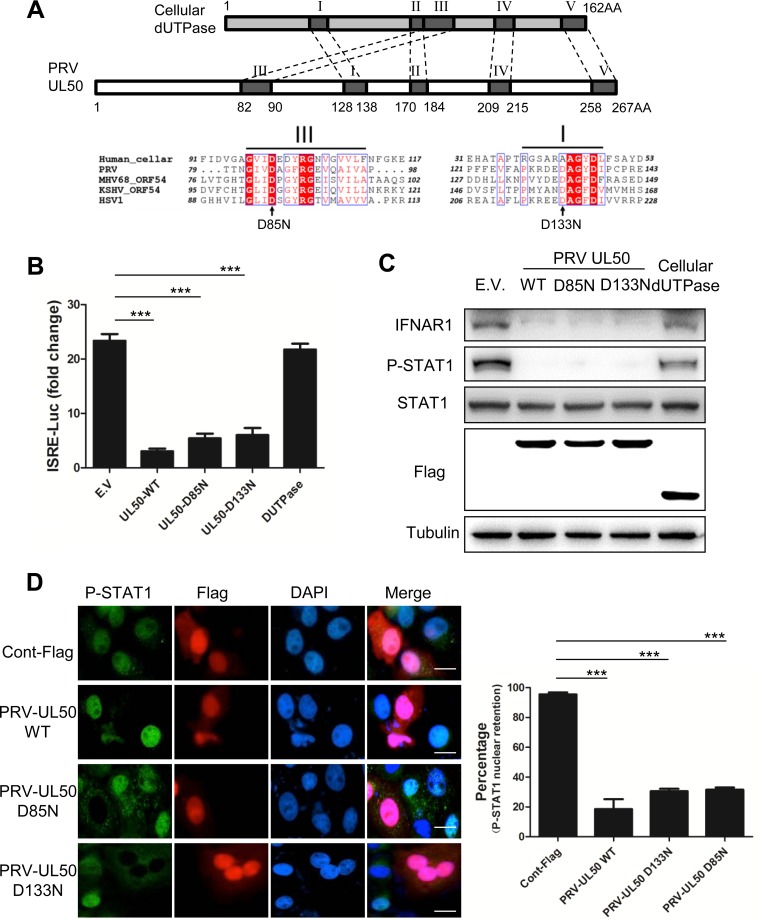
Since UL50 is a dUTPase, we then examined whether the dUTPase activity was required for its anti-IFN function. The catalytic center of dUTPases is formed by five conserved amino acid (aa) motifs (24, 26). A conserved aspartic acid residue in motif III has been found to be critical for catalytic activity for several herpesvirus dUTPases, including that of HSV-1 (27). Another Asp residue important for catalytic activity of herpesvirus dUTPases lies in motif 1 and is conserved in herpesvirus dUTPases (27). Mutation of the corresponding Asp in the Epstein-Barr virus (EBV) dUTPase ORF54 (Asp131) resulted in a large loss of catalytic activity (28). Sequence alignments reveal that these two Asp residues correspond to Asp85 and Asp133, respectively, in PRV (Fig. 5A). We individually mutated these two aspartic acids to asparagines. The generated mutants, referred to as UL50-D85N and UL50-D133N, are presumed to be dUTPase deficient. The ISRE-luciferase reporter assay showed that these two UL50 mutants still retained the ability to suppress IFN-α induction, exhibiting 23% (D85N) and 26% (D133N) of the control induction level, whereas the human cellular dUTPase did not show any inhibition (Fig. 5B). In addition, Western blot and immunofluorescence analyses further showed that, similar to WT UL50, the two mutants were also able to degrade IFNAR1 (Fig. 5C) and to block the IFN-α-induced phosphorylation (Fig. 5C) and nuclear accumulation (Fig. 5D) of STAT1, whereas the human cellular dUTPase did not (Fig. 5C). Collectively, these results demonstrated that PRV UL50 inhibited the IFN-α signaling pathway independent of its dUTPase activity.
FIG 5.
PRV UL50 inhibits IFN-α signaling independent of its dUTPase activity. (A) Amino acid sequence alignments of dUTPases from different herpesviruses and humans. The schematic shows the structures of PRV UL50 and human cellular dUTPase. The five functional motifs of PRV UL50 (I to V) were marked by comparing the deduced amino acid sequences between PRV UL50 and its corresponding human cellular dUTPase. Deduced amino acid sequence alignments of the C-terminal ends of functional dUTPase derived from humans, PRV UL50, HSV-1 UL50, KSHV ORF54, and MHV68 ORF54 are shown. Two conserved aspartic acid residues, at positions 85 and 133 of PRV UL50, were predicted to be critical for dUTPase activity and are marked by black arrows. (B) CRL cells were cotransfected with E.V or with 500 ng of plasmid expressing PRV wild-type UL50 (WT), a PRV UL50 mutant (UL50-D133N or UL50-D85N), or cellular dUTPase, along with 100 ng of pGL3-ISRE-Luc and 10 ng of pRL-TK. pRL-TK was used as an internal control for transfection efficiency. Twelve hours after transfection, the cells were incubated in medium containing porcine IFN-α (1,000 U/ml) for 24 h and then harvested and analyzed for luciferase activity. The values represent fold changes of ISRE activity between IFN-α-treated and untreated cells. (C) CRL cells were transfected with empty vector (E.V) or with 1 μg of plasmid expressing Flag-tagged PRV wild-type UL50 (WT), a PRV UL50 mutant (D85N or D133N), or human cellular dUTPase. Twenty-four hours after transfection, the cells were treated or not with porcine IFN-α (1,000 U/ml) for 30 min. The cells were harvested, and the protein levels of IFN-α receptor 1 (IFNAR1), STAT1, P-STAT, Flag, and tubulin were determined by Western blotting. (D) PK15 cells were transfected with 1 μg of plasmid expressing either control Flag, Flag-tagged PRV-UL50 WT, or a mutant (D85N or D133N). Twenty-four hours after transfection, the cells were incubated in medium containing porcine IFN-α (1,000 U/ml) for 90 min. (Left) P-STAT1 and Flag-tagged UL50 were visualized by immunofluorescence assay with antibodies against P-STAT1 or Flag. The nuclei were detected by DAPI staining. Bars = 10 μm. (Right) The number of cells with P-STAT1 nuclear retention in 50 Flag- or UL50-expressing cells was quantified and expressed as a percentage of the total number of counted cells. Data are presented as means and SD for three independent experiments. Statistical analyses for panels B and D were performed by ANOVA, using GraphPad Prism software. ***, P < 0.001.
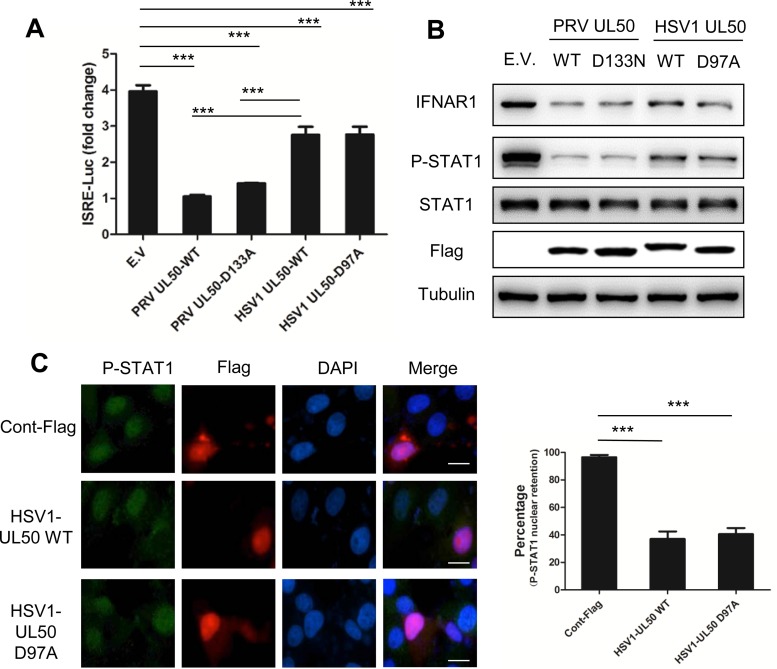
We further determined whether the UL50 dUTPase of human HSV-1 had a similar activity. An HSV-1 UL50 mutant (UL50-D97A) was constructed in which the conserved Asp97 residue in motif III was replaced with Ala. This mutant has been shown to be dUTPase activity deficient (29). We found that the induction of reporter activity (Fig. 6A), STAT1 phosphorylation (Fig. 6B), and nuclear translocation (Fig. 6C) by IFN-α in human HeLa cells was suppressed by the HSV-1 UL50 wild type (WT) and the UL50-D97A mutant. IFNAR1 expression was also reduced by HSV-1 UL50 WT and the UL50-D97A mutant (Fig. 6B). Similar results were also observed in CRL cells (data not shown). Note that compared to that of PRV UL50, HSV-1 UL50 showed a weaker inhibitory activity in these assays (Fig. 6A and B). Altogether, these data suggest that HSV-1 UL50 can inhibit IFN signaling and induce IFNAR1 reduction independent of its dUTPase activity, even though the activity is milder than that of PRV UL50.
FIG 6.
HSV-1 UL50 also inhibits IFN-α signaling in a dUTPase-independent manner. (A) HeLa cells were cotransfected with empty vector (E.V) or with 500 ng of plasmid expressing PRV wild-type UL50 (PRV UL50-WT) or mutant UL50 (UL50-D133A) or HSV-1 wild-type UL50 (WT) or mutant UL50 (UL50-D97A), along with 100 ng of pGL3-ISRE-Luc and 10 ng of pRL-TK. pRL-TK was used as an internal control for transfection efficiency. Twelve hours after transfection, the cells were incubated in medium containing human IFN-α (1,000 U/ml) for an additional 24 h and then harvested and analyzed for luciferase activity. Data are presented as means and SD for three independent experiments. Statistical analyses were performed by ANOVA, using GraphPad Prism software. ***, P < 0.001. (B) HeLa cells were transfected with empty vector (E.V) or with 1 μg of plasmid expressing Flag-tagged PRV wild-type UL50 (WT) or mutant UL50 (D133A) or HSV-1 wild-type UL50 (WT) or mutant UL50 (D97A). Twenty-four hours after transfection, the cells were treated or not with human IFN-α (1,000 U/ml) for 30 min. The cells were harvested, and the protein levels of IFN-α receptor 1 (IFNAR1), STAT1, P-STAT, Flag, and tubulin were detected by Western blotting. (C) HeLa cells were transfected with 1 μg of plasmid expressing either control Flag or Flag-tagged HSV-1 UL50 WT or mutant UL50 (D97A). Twenty-four hours after transfection, the cells were incubated in medium containing human IFN-α (1,000 U/ml) for 90 min. (Left) P-STAT1 and Flag-tagged UL50 were visualized by immunofluorescence assay with antibodies against P-STAT1 and Flag. The nuclei were detected by DAPI staining. Bars = 10 μm. (Right) The number of cells with P-STAT1 nuclear retention in 50 Flag- or UL50-expressing cells was quantified and expressed as a percentage of the total number of counted cells. Data are presented as means and SD for three independent experiments. Statistical analyses were performed by ANOVA, using GraphPad Prism software. ***, P < 0.001.
The region corresponding to aa 225 to 253 in the C terminus of PRV UL50 is required for UL50-mediated inhibition of IFN-α signaling.
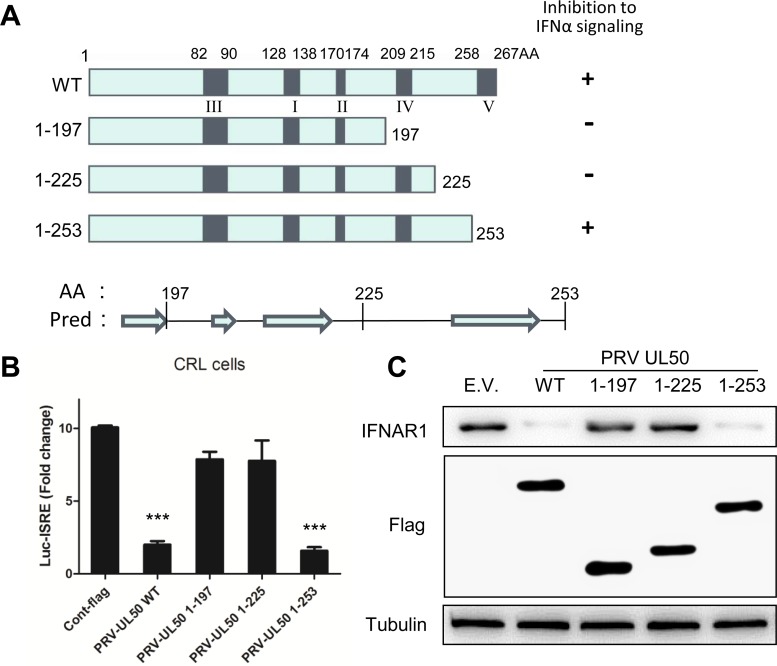
All dUTPases contain five conserved sequence motifs, but the motif order is different between cellular and herpesviral dUTPases due to gene duplication and divergence of the latter (28, 30, 31). To examine whether any of these conserved sequences are involved in the inhibition of IFN-α signaling, we generated a series of deletion mutants (Fig. 7A) and measured their ability to inhibit IFN-α signaling in the ISRE reporter assay. A C-terminal deletion of UL50 to aa 253, which lacks the V motif, did not affect the anti-IFN activity of UL50, but further deletion to aa 225 and other deletions completely abolished this activity (Fig. 7B and data not shown). Accordingly, only the 1-253 deletion mutant retained the ability to reduce IFNAR1 (Fig. 7C). These results indicate that the aa sequence from positions 225 to 253, which lies between motifs IV and V of UL50, is critical for UL50 inhibition of IFN-α signaling.
FIG 7.
Amino acids 225 to 253 in the C-terminal region of PRV UL50 are necessary for UL50-mediated inhibition of IFN signaling. (A) Schematic of PRV UL50 structures and deletion mutants. The five conserved functional motifs of PRV UL50 are marked I to V from the N to the C terminus. Three deletion mutants were constructed based on the predicted secondary protein structures by using the PSIPRED Protein Sequence Analysis Workbench. Pred, predicted secondary structures at the C terminus of UL50; AA, amino acid. Arrows show the predicted β-strands. (B) CRL cells were cotransfected with empty vector (E.V) or with 500 ng of plasmid expressing PRV wild-type UL50 (WT) or a deletion mutant (1-197, 1-225, or 1-253), along with 100 ng of pGL3-ISRE-Luc and 10 ng of pRL-TK. pRL-TK was used as an internal control for transfection efficiency. Twelve hours after transfection, the cells were incubated in medium containing porcine IFN-α (1,000 U/ml) for an additional 24 h and then harvested and analyzed for luciferase activity. (C) CRL cells were transfected with empty vector (E.V) or with 1 μg of plasmid expressing Flag-tagged PRV wild-type UL50 (WT) or a deletion mutant (1-197, 1-225, or 1-253). Twenty-four hours later, the cells were harvested, and the protein levels of IFN-α receptor 1 (IFNAR1), Flag, and tubulin were detected by Western blotting. The data in panel B are presented as means and SD for three independent experiments. Statistical analyses were performed by Student's t test, using GraphPad Prism software. ***, P < 0.001.
PRV UL50 interferes with IFN-α signaling during virus infection.
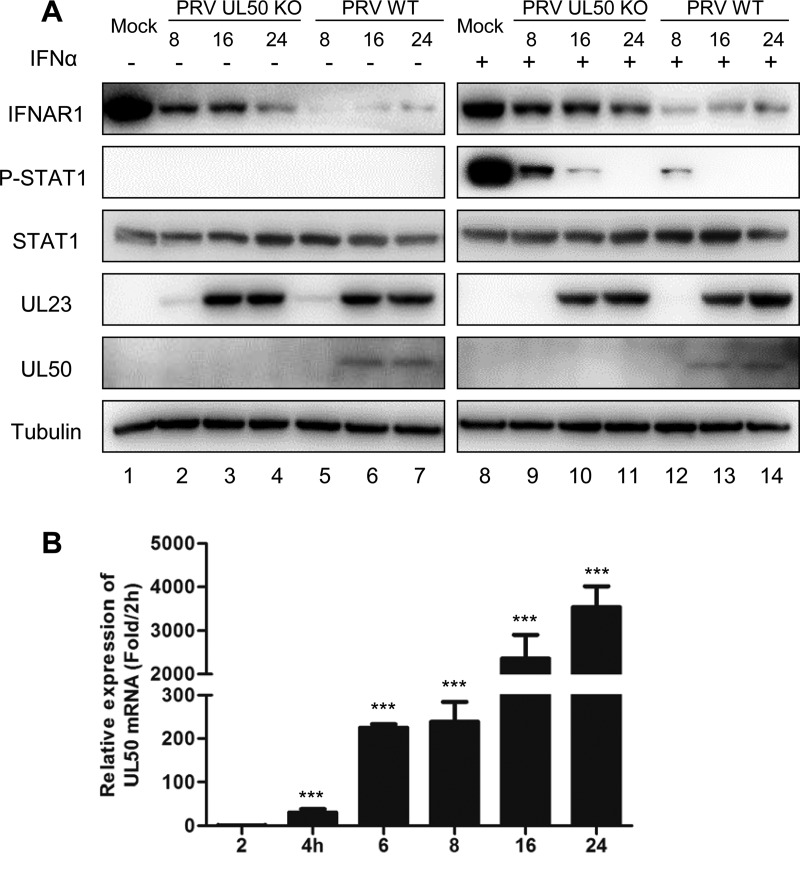
To evaluate the contribution of UL50 to the inhibition of type I IFN signaling during PRV infection, we infected porcine PK15 cells with wild-type (PRV-WT) or UL50-deleted (PRV-UL50 KO) PRV of strain BarthaK61, which we generated previously (25), at a multiplicity of infection (MOI) of 1. At different time points postinfection, the cells were treated with or without porcine IFN-α for 30 min. At this time, the cells were subjected to Western blot analysis of IFNAR1, P-STAT1, STAT1, and PRV UL23 (to show the viral loads). Compared with the mock treatment condition, both PRV-WT and PRV-UL50 KO infections reduced the IFNAR1 level (Fig. 8A, lanes 2 to 7 and lanes 9 to 14) and blocked IFN-α-induced STAT1 phosphorylation (Fig. 8A, lanes 9 to 14) in a time-dependent manner. However, these effects were much weaker in PRV-UL50 KO-infected cells. qRT-PCR analysis showed a significant accumulation of UL50 transcripts in the early stage of PRV infection (Fig. 8B), and a substantial amount of UL50 protein was detected at 16 h postinfection in PRV-WT-transfected cells (Fig. 8A). These results indicate that UL50 indeed plays a role in antagonizing IFN-α signaling during viral infection and that other mechanisms are also involved.
FIG 8.
PRV UL50 interferes with IFN-α signaling during virus infection. (A) PK15 cells were infected with either wild-type PRV (PRV WT) or a recombinant PRV UL50-knockout virus (PRV UL50 KO) (MOI = 1). At 8, 16, or 24 h postinfection, the cells were incubated in medium containing porcine IFN-α (1,000 U/ml) for an additional 30 min. The cells were then harvested, and the protein levels of IFNAR1, P-STAT1, STAT1, UL23, UL50, and tubulin were analyzed by Western blotting. (B) Levels of UL50 mRNA over the early stages of the PRV-WT infection time course. PK15 cells were infected with PRV-WT (MOI = 1), and at 2, 4, 6, 8, 16, and 24 h postinfection, the cells were harvested and the mRNA levels of UL50 detected by qRT-PCR. The results were obtained from three independent experiments and are means and SD. Statistical analyses of the above-described experiments were performed by Student's t test, using GraphPad Prism software. ***, P < 0.001.
PRV deficient in UL50 is more sensitive to IFN-α-mediated viral inhibition.
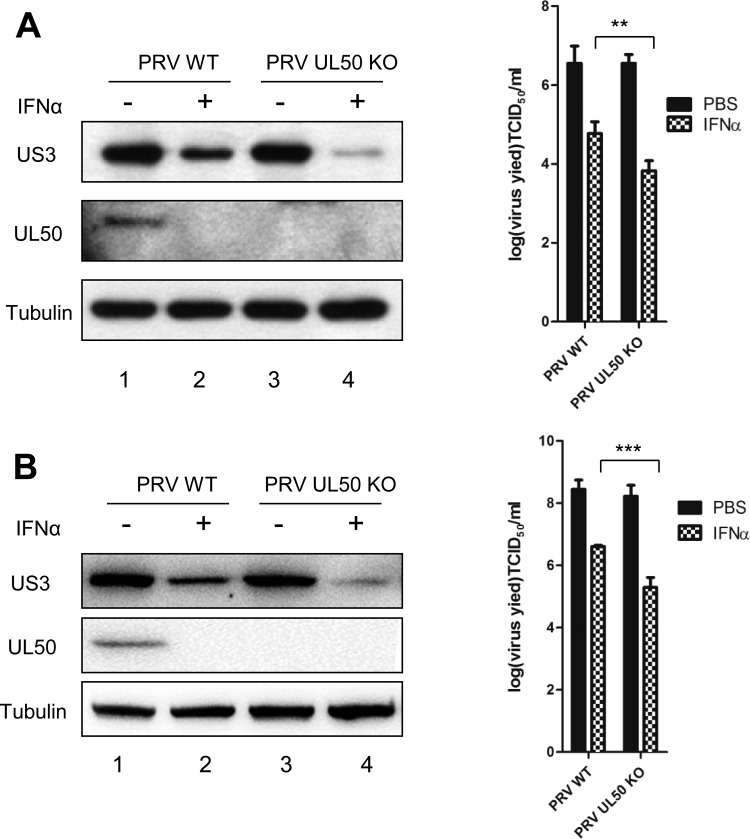
To further examine whether UL50 antagonizes the antiviral function of IFN-α during viral infection, we pretreated PK15 cells (Fig. 9A) and CRL cells (Fig. 9B) with IFN-α and then infected the cells with PRV-WT or PRV-UL50 KO, followed by an additional 24 h of IFN-α treatment. The viral protein US3 in the infected cells was examined by Western blotting, and the infectious viral particles in the culture medium were measured by determining the 50% tissue culture infective dose (TCID50). Although deletion of UL50 from PRV barely affected viral replication in the absence of IFN-α treatment compared to that with PRV-WT, it rendered the virus more sensitive to IFN-α. In PRV-UL50 KO-infected PK15 and CRL cells treated with IFN-α, lower levels of US3 (Fig. 9A and B, left panels) and approximately 8.7- to 10-fold decreases in the average infectious titer produced (Fig. 9A and B, right panels) were observed compared to those with PRV-WT-infected cells treated with IFN-α. These results indicate that UL50 exerts an anti-IFN-α function during viral infection.
FIG 9.
PRV deficient in UL50 is more sensitive to IFN-α-mediated viral inhibition. (A) PK15 cells were treated with porcine IFN-α (1,000 U/ml). Twenty-four hours later, the cells were infected with either wild-type PRV or recombinant PRV-UL50 KO for 1 h (MOI = 0.1). The infected cells were then incubated in medium containing porcine IFN-α (1,000 U/ml) for an additional 24 h. The cells were harvested and lysed for Western blotting (left) and TCID50 assay (right). (B) CRL cells were treated with porcine IFN-α (500 U/ml). Twelve hours later, the cells were infected with either PRV-WT or recombinant PRV-UL50 KO for 1 h (MOI = 1). The infected cells were then incubated in medium containing porcine IFN-α (500 U/ml) for an additional 24 h. Cells were harvested and lysed for Western blotting (left) and TCID50 assay (right). The virus protein (US3 and UL50) levels were examined by Western blotting, and tubulin was included as a loading control. The results were obtained from three independent experiments and are means and SD. Statistical analyses were performed by ANOVA, using GraphPad Prism software. **, P < 0.01; ***, P < 0.001.
DISCUSSION
The central finding of this study is that PRV UL50 possesses the ability to antagonize type I IFN signaling and facilitates PRV replication in the presence of IFN-α. Evading the type I IFN response is important for alphaherpesviruses to establish persistent infections in hosts. We found that UL50 can promote IFNAR1 degradation and inhibit IFN-α-induced STAT1 phosphorylation upon ectopic UL50 expression and PRV infection. This mechanism is likely shared by HSV-1. Our results highlight the importance of alphaherpesvirus UL50 in immune evasion and implicate it as a potential antiviral target.
PRV UL50 was identified as a strong inhibitor of type I IFN signaling through an unbiased screen, which was a surprising result because the primary function of dUTPase is associated with dUTP metabolism, but it was not unprecedented. It has been demonstrated that the dUTPase ORF54 in gammaherpesviruses possesses an immune modulation function (32–34). Interestingly, this function of ORF54 is independent of its dUTPase catalytic activity. For instance, the mouse herpesvirus 68 (MHV68) and Kaposi's sarcoma-associated herpesvirus (KSHV) ORF54 proteins are able to downregulate the immune response independent of their dUTPase activity (32). Similar to that of ORF54, the ability of UL50 to inhibit the IFN response is likely to be dUTPase activity independent, as both the WT and the dUTPase-deficient mutant (D97A) of HSV-1 UL50 showed similar levels of inhibition of the IFN response. In addition, the strong ability of PRV UL50 to antagonize the IFN response was not affected by point mutations in the catalytic sites (D85 and D133) or by deletion of motif V, which is essential for the catalytic activity of a dUTPase, from PRV UL50. In contrast, overexpression of the human cellular dUTPase did not interfere with IFN signaling. Our study provides the first evidence that alphaherpesvirus UL50 also plays a role in immune modulation, supporting the notion that viral dUTPases may have acquired additional functions as immune modulators (32–34).
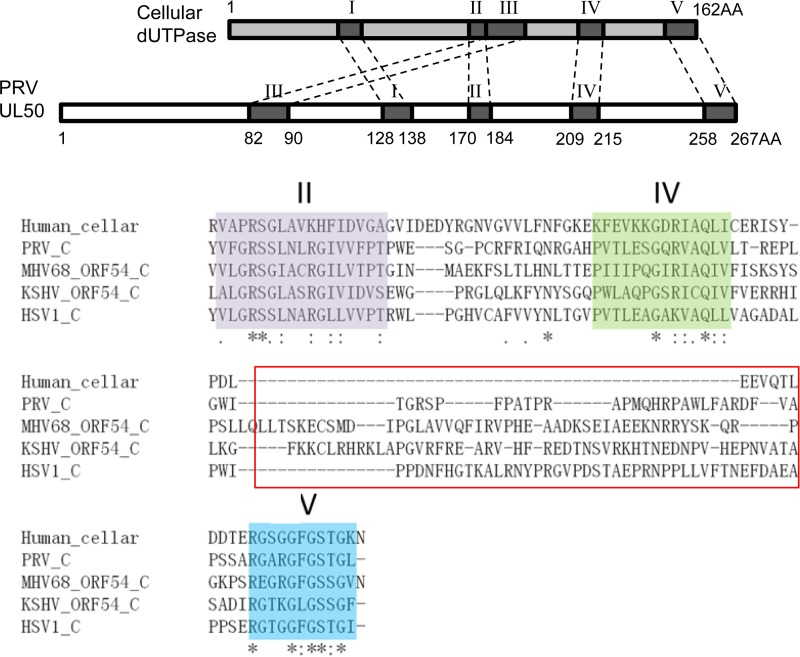
The immune modulation functions performed by herpesviral dUTPases may be mediated by a motif(s) and/or structures that are of viral origin. dUTPases are ubiquitously expressed in most organisms, including prokaryotes, eukaryotes, and some viruses (26, 35, 36). Most dUTPases contain five conserved aa motifs. These motifs are critical for the formation of the catalytic center of the enzyme (26, 30). Cellular dUTPases exist as homotrimers, with each unit contributing one or two of the five motifs (24). The dUTPases encoded by alpha- and gammaherpesviruses are twice as long in aa sequence as their cellular counterparts, which is proposed to be a result of gene duplication. Over the course of evolution, motifs I, II, IV, and V in the N-terminal half and motif III in the C-terminal half of herpesviral dUTPase were lost. However, the overall structures of monomeric herpesviral dUTPase and the trimeric cellular dUTPase are extremely similar (27). Based on secondary structure predictions, McGeehan et al. speculated that a unique domain that occupies the position of motif III in the C-terminal region of herpesviral dUTPase, between motifs II and IV, might contribute to the newly defined functions of this protein (27, 36); however, the biological evidence to support this is lacking. Through a series of deletion analysis studies based on the conserved motifs and secondary structures, we found that the region consisting of aa 225 to 253, between motifs IV and V, is required for PRV UL50 to inhibit IFN-α signaling. Interestingly, this region is nearly absent in the cellular dUTPase but is present in other herpesviral dUTPases shown to inhibit IFN-α signaling, including HSV-1 UL50 as well as KSHV and MHV68 ORF54, with varied aa lengths and compositions (Fig. 10). Thus, the addition of a stretch of aa between motifs IV and V may contribute to the anti-IFN function of herpesviral dUTPases. Sequence analysis of this region in PRV revealed a possible β-strand, which also exists in the HSV-1, MHV68, and KSHV dUTPases. More comparative studies and mutagenesis-based analyses are required to further elucidate whether this β-strand or other residues are critical in supporting the IFN-α-inhibitory function of these herpesvirus dUTPases.
FIG 10.
Amino acid sequence alignments of dUTPases between different herpesviruses and humans. (Top) Schematic drawing of the structures of PRV UL50 and human cellular dUTPase. (Bottom) Deduced amino acid sequence alignments of the C-terminal ends of functional dUTPase derived from humans, PRV UL50, HSV-1 UL50, KSHV ORF54, and MHV68 ORF54. The functional region (aa 225 to 253) of PRV UL50 responsible for anti-IFN activity is marked by a red rectangle.
Our data suggest that PRV UL50-induced IFNAR1 degradation may be the core mechanism behind its anti-IFN function. Engagement of IFNAR1 and -2 is crucial in initiating the IFN-α signaling cascade, and the concentrations of the receptors determine the sensitivity and strength of the pathway. IFN-induced downregulation of IFNAR1 is a key mechanism for turning off IFN signaling. Loss of IFNAR1 is often observed in herpesvirus-infected cells (32, 33, 37). PRV infection nearly abolished the expression of IFNAR1 (Fig. 7). Several studies with gammaherpesviruses have connected downregulation of IFNAR1 with ORF54. For instance, MHV68 ORF54 downregulates IFNAR1 (32), and KSHV ORF54 has the ability to decrease the levels of several cytokine receptors, including IFNAR1 (33). Our findings extended this ability of ORF54 to PRV UL50 and further provided some insights into the mechanism by suggesting that UL50 may accelerate lysosomal degradation of IFNAR1.
PRV infection-induced loss of IFNAR1 is very robust and is only partially restored upon deletion of UL50 from PRV, indicating that other mechanisms are involved. It has been shown that the HSV-1 infection-induced innate immune response and endoplasmic reticulum (ER) stress can result in IFNAR1 degradation by engaging PERK and p38, respectively (37, 38). We suspect that this might also be the case for PRV infection. Both viral infection-induced cellular responses and UL50 expression may contribute to the loss of IFNAR1. Moreover, we also cannot rule out the possibility that IFNAR1 is targeted by other viral proteins for degradation. Studies with other viruses have indicated that viral proteins can reduce IFNAR1 expression by either accelerating IFNAR1 degradation (39) or blocking its maturation (40). Our data clearly suggest that UL50 induces lysosomal degradation of IFNAR1, which is consistent with the nature of IFNAR1 as a membrane protein. IFNAR1 processes a degradation sequence which can be phosphorylated under certain cellular stresses independent of IFN treatment, resulting in the phosphorylation-dependent ubiquitination, endocytosis, and degradation of IFNAR1 (37, 38). Roupelieva et al. showed that recombinant ORF54 can stimulate certain pattern recognition receptors extracellularly, suggesting that these proteins may adopt some patterns that are foreign to hosts (41). It would be interesting to examine whether these herpesvirus dUTPases can activate certain intracellular stress response pathways, which in turn may induce IFNAR1 phosphorylation and degradation.
From our screening results, several other PRV proteins in addition to UL50 also bear the ability to inhibit IFN-induced reporter activity, though to a much weaker extent than that with UL50. Although more studies are required to confirm the inhibitory effect of these proteins on IFN signaling, nevertheless these results suggest that PRV may dispatch multiple proteins to inhibit IFN signaling. It has been suggested that alphaherpesviruses may use various strategies to evade the antiviral effect of IFN. For instance, UL41 and UL54 of HSV-1 have been found to be responsible for the loss of JAK1 and inhibition of STAT1 phosphorylation in HSV-1-infected cells (19, 21). Moreover, viral infection per se may also contribute to IFN evasion. Thus, PRV infection may also induce multiple mechanisms to inhibit IFN signaling, to which UL50 only partly contributes.
UL50 contributes to the neuropathogenesis caused by PRV infection. Previous studies suggested that UL50 dUTPase activity may compensate for the low cellular dUTPase levels in differentiated neuronal cells, thereby supporting rapid viral replication in these cells (42). Our results indicate that the immune evasion activity of UL50 may also play a role in the pathogenicity of PRV infection. Thus, UL50 may be a good target for anti-PRV drug development.
In conclusion, we identified UL50 as a novel interferon signaling inhibitor that acts by targeting IFNAR1 for lysosomal degradation independent of its dUTPase activity, thus contributing to the immune evasion ability of PRV. This finding may help to explain the role of UL50 in the neuropathogenesis of PRV infection and implicates it as a potential target for antiviral drug design.
MATERIALS AND METHODS
Cell culture and viruses.
PK15 cells (porcine kidney cells) and HeLa cells were cultured in Dulbecco's minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin (100 U/ml)-streptomycin (100 μg/ml). CRL cells, a porcine alveolar macrophage cell line, were grown in the above-described DMEM containing 20% FBS. The cell culture medium, serum, and antibiotics were purchased from Invitrogen. All cells were maintained at 37°C in 5% CO2.
The PRV BarthaK61 strain vaccine (lot number 2012002) was purchased from Weike Biotech Co., Harbin, China. The recombinant PRV UL50-knockout virus (PRV-UL50 KO) was described previously (25). The viruses were propagated and titrated on PK15 cells. The viral titer was determined and expressed as the 50% tissue culture infective dose (TCID50)/ml. Briefly, 5 × 104 PK15 cells were infected with 100 μl of 10-fold serially diluted PRV in a 96-well plate. Then, at 3 to 6 days postinfection, the cytopathic effect (CPE) in each well was observed and scored. The TCID50 was calculated by the Reed-Muench method. Viral samples were stored at −80°C until use.
Plasmids.
The PRV UL50 gene was amplified from the BarthaK61 genome. The PRV UL50 (D85N and D133N) and HSV-1 (D97A) kinase-dead mutants were constructed by overlap extension PCR. The gene encoding human cellular dUTPase was amplified from HCT116 cells. The HSV-1 UL50-expressing plasmid was obtained from Chunfu Zheng (Soochow University, Jiangsu, China). All of the cloned wild-type and mutant constructs were inserted into the Flag-tagged pRK5 vector between BamHI and HindIII sites and confirmed by sequencing. The primers used for UL50-related gene amplification are listed in Table 1. Other primer sequences for PRV gene cloning are available upon request.
TABLE 1.
Primers used for PRV, HSV-1, and human dUTPase cloning
| Primer (restriction enzyme site) | Sequence (5′–3′) |
|---|---|
| PRV-UL50-F (BglII) | TATAGATCTGGCCCCTCGGTGGAGACGATG |
| PRV-UL50-R (HindIII) | TATAAGCTTCGGGGTTGTCTGTGCCCAAT |
| PRV-UL50 1-197-R (HindIII) | TTAAAGCTTTCACTGGATCCGGAAGCGGCA |
| PRV-UL50 1-225-R (HindIII) | TTAAAGCTTTCAGATCCAGCCCAGCGGCTC |
| PRV-UL50 1-253-R (HindIII) | TTAAAGCTTTCAGGCGACAAAGTCCCGCGC |
| PRV-UL50 D85N-F | AACGGGATCGTGAACGCGGGCTTTCGC |
| PRV-UL50 D85N-R | GCGAAAGCCCGCGTTCACGATCCCGTT |
| PRV-UL50 D133N-F | AAGCGCGACGAGAACGCCGGATACGAC |
| PRV-UL50 D133N-R | GTCGTATCCGGCGTTCTCGTCGCGCTT |
| HSV1-UL50-F (BamHI) | ACGACGATGACAAGGGATCCATGAGTCAGTGGGGATCCGG |
| HSV1-UL50-R (HindIII) | GGGCCATGGCGGCCAAGCTTCTAAATACCGGTAGAGCCAA |
| HSV1-UL50 D97A-F | CTGGGTCTTATCGCATCGGGGTACCGC |
| HSV1-UL50 D97A-R | GCGGTACCCCGATGCGATAAGACCCAG |
| Human-DUTPase-F (BamHI) | CGCGGATCCATGCCCTGCTCTGAAGA |
| Human-DUTPase-R (HindIII) | CCCAAGCTTTTAATTCTTTCCAGTGGA |
Antibodies and reagents.
The production of an antibody against PRV US3 has been described previously (25). UL50 and UL23 antibodies against glutathione S-transferase (GST)-fused full-length proteins were raised in mice following a standard procedure as described previously (43). Rabbit anti-IFNAR1, rabbit anti-phosphorylated STAT1 (P-STAT1), and rabbit anti-STAT1 antibodies were purchased from Cell Signaling Technology. Mouse anti-Flag antibodies were purchased from Sigma. Rabbit anti-tubulin primary and horseradish peroxidase (HRP)-conjugated goat anti-mouse and anti-rabbit secondary antibodies were purchased from Santa Cruz Biotechnology. Tetramethyl rhodamine isocyanate (TRITC)- and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody and DAPI (4′,6-diamidino-2-phenylindole) were purchased from Beijing Ding Guo Chang Sheng Biotech. Co. Ltd.
Recombinant porcine IFN-α was a gift from Wenjun Liu (Chinese Academy of Sciences, Beijing, China). Recombinant human IFN-α was purchased from Peprotech. The proteasome inhibitor MG132 and the lysosome inhibitor bafilomycin were purchased from Sigma.
Immunofluorescence microscopy.
Cells grown on glass coverslips were fixed in 4% paraformaldehyde for 30 min at room temperature and then permeabilized with 0.2% Triton X-100 for 15 min on ice. After washing and blocking in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) for 30 min, the cells were incubated with specific primary antibodies for 1 h at room temperature followed by secondary antibodies for 30 min. Nuclei were stained with DAPI for 3 to 5 min. Images were captured using a Nikon Eclipse Ni-E microscope or a Leica Wetzlar GmbH microscope. The captured images were processed and analyzed using SPOT software (Nikon).
Western blot analysis.
Whole-cell extracts were prepared in lysis buffer supplemented with phosphatase inhibitors. The proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk in PBST (PBS containing 0.5% Tween 20) for 2 h at room temperature and were then incubated with specific primary antibodies overnight at 4°C followed by secondary antibodies for 45 min at room temperature. The reactive protein bands were visualized using an enhanced chemiluminescence (ECL) reagent.
RNA isolation and qRT-PCR.
Total RNA was extracted from cells by use of TRIzol (Invitrogen) following the manufacturer's instructions and then reverse transcribed into cDNA by use of a Quantitect reverse transcription kit (Qiagen). Quantitative RT-PCR (qRT-PCR) reactions were performed using a FastSYBR mixture (CWBiotech) and a ViiATM7 real-time PCR system (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene to normalize the target genes. Gene-specific primers for qPCR included ISG54 forward (5′-GACACGGTTAAAGTGTGGAG-3′) and reverse (5′-GGTACTGGTTGTCAGGATTC-3′), ISG15 forward (5′-CAGATCACCCAGAAGATCG-3′) and (5′-CCCTTGTTATTCCTCACCAG-3′), PRV-UL50 forward (5′-CTTCTTCGAGGTCTTTGCGC-3′) and reverse (5′-ATGTCGTATCCGGCGTCCT-3′), GAPDH-Human forward (5′-CCTTCCGTGTCCCTACTGCCAAC-3′) and reverse (5′-GACGCCTGCTTCACCACCTTCT-3′), and GAPDH-Porcine forward (5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′) and reverse (5′-CATGTGGGCCATGAGGTCCACCAC-3′) primers.
Transfection and luciferase reporter assays.
Cells were transfected with an empty vector (E.V) or with plasmids expressing viral proteins together with 100 ng of the reporter plasmid pGL3-ISRE-luc (firefly) and 10 ng of pRL-TK by using the Lipofectamine 3000 reagent (Invitrogen) or JetPRIME DNA transfection reagent (Polyplus-Transfection SA) as described in the manufacturer's instructions. The total amount of DNA was held constant by adding vector control plasmid. Twelve hours after transfection, cells were treated with or without 1,000 U/ml of IFN-α for 24 h. Cell lysates were measured for firefly and renilla luciferase activities by using a dual-luciferase reporter assay kit (Promega) according to the manufacturer's instructions. The firefly luciferase activities were normalized to the renilla luciferase activity. The ISRE activity fold changes between IFN-α-treated and untreated cells were calculated.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism software to perform Student's t test or analysis of variance (ANOVA) on at least three independent replicates. P values of <0.05 were considered to be statistically significant for each test.
ACKNOWLEDGMENTS
We thank Chunfu Zheng (Soochow University) for the gift of the HSV-1 UL50 plasmid, Wenjun Liu (Chinese Academy of Sciences) for recombinant porcine IFN-α, and Meili Li (Guangxi Medical University) for reagents.
This work was supported by the National Key Research and Development Program of China (grant 2016YFD0500100), the National Natural Science Foundation of China (grant 31500703), the Project for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (grant 2015SKLAB6-12), and the Chinese Universities Scientific Fund (grant 2017QC042).
REFERENCES
- 1.Steiner I, Benninger F. 2013. Update on herpes virus infections of the nervous system. Curr Neurol Neurosci Rep 13:414. doi: 10.1007/s11910-013-0414-8. [DOI] [PubMed] [Google Scholar]
- 2.Pomeranz LE, Reynolds AE, Hengartner CJ. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev 69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verweij MC, Wellish M, Whitmer T, Malouli D, Lapel M, Jonjic S, Haas JG, DeFilippis VR, Mahalingam R, Fruh K. 2015. Varicella viruses inhibit interferon-stimulated JAK-STAT signaling through multiple mechanisms. PLoS Pathog 11:e1004901. doi: 10.1371/journal.ppat.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brittle EE, Reynolds AE, Enquist LW. 2004. Two modes of pseudorabies virus neuroinvasion and lethality in mice. J Virol 78:12951–12963. doi: 10.1128/JVI.78.23.12951-12963.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katze MG, He Y, Gale M Jr. 2002. Viruses and interferon: a fight for supremacy. Nat Rev Immunol 2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 7.de Weerd NA, Samarajiwa SA, Hertzog PJ. 2007. Type I interferon receptors: biochemistry and biological functions. J Biol Chem 282:20053–20057. doi: 10.1074/jbc.R700006200. [DOI] [PubMed] [Google Scholar]
- 8.Schulz KS, Mossman KL. 2016. Viral evasion strategies in type I IFN signaling—a summary of recent developments. Front Immunol 7:498. doi: 10.3389/fimmu.2016.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Wang K, Li J, Zheng C. 2013. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol 87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Jin H, Valyi-Nagy T, Cao Y, Yan Z, He B. 2012. Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J Virol 86:2188–2196. doi: 10.1128/JVI.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verpooten D, Ma Y, Hou S, Yan Z, He B. 2009. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J Biol Chem 284:1097–1105. doi: 10.1074/jbc.M805905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 87:12814–12827. doi: 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Wang K, Wang S, Zheng C. 2013. Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. J Virol 87:12935–12948. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. 2011. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 85:11079–11089. doi: 10.1128/JVI.05098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su C, Zhan G, Zheng C. 2016. Evasion of host antiviral innate immunity by HSV-1, an update. Virol J 13:38. doi: 10.1186/s12985-016-0495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel MA, James SF, Traktinskiy I, Wyborny A, Choe A, Rempel A, Baird NL, Gilden D. 2014. Inhibition of phosphorylated-STAT1 nuclear translocation and antiviral protein expression in human brain vascular adventitial fibroblasts infected with varicella-zoster virus. J Virol 88:11634–11637. doi: 10.1128/JVI.01945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chee AV, Roizman B. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J Virol 78:4185–4196. doi: 10.1128/JVI.78.8.4185-4196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokota S, Yokosawa N, Okabayashi T, Suzutani T, Miura S, Jimbow K, Fujii N. 2004. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J Virol 78:6282–6286. doi: 10.1128/JVI.78.12.6282-6286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson KE, Song B, Knipe DM. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487–494. doi: 10.1016/j.virol.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadeppagari RK, Sanchez RL, Foster TP. 2012. HSV-2 inhibits type-I interferon signaling via multiple complementary and compensatory STAT2-associated mechanisms. Virus Res 167:273–284. doi: 10.1016/j.virusres.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brukman A, Enquist LW. 2006. Suppression of the interferon-mediated innate immune response by pseudorabies virus. J Virol 80:6345–6356. doi: 10.1128/JVI.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris JM, McIntosh EM, Muscat GE. 1999. Structure/function analysis of a dUTPase: catalytic mechanism of a potential chemotherapeutic target. J Mol Biol 288:275–287. doi: 10.1006/jmbi.1999.2680. [DOI] [PubMed] [Google Scholar]
- 25.Xu A, Qin C, Lang Y, Wang M, Lin M, Li C, Zhang R, Tang J. 2015. A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol Lett 37:1265–1272. doi: 10.1007/s10529-015-1796-2. [DOI] [PubMed] [Google Scholar]
- 26.Baldo AM, McClure MA. 1999. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J Virol 73:7710–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davison AJ, Stow ND. 2005. New genes from old: redeployment of dUTPase by herpesviruses. J Virol 79:12880–12892. doi: 10.1128/JVI.79.20.12880-12892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freeman L, Buisson M, Tarbouriech N, Van der Heyden A, Labbe P, Burmeister WP. 2009. The flexible motif V of Epstein-Barr virus deoxyuridine 5′-triphosphate pyrophosphatase is essential for catalysis. J Biol Chem 284:25280–25289. doi: 10.1074/jbc.M109.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato A, Tsuda S, Liu Z, Kozuka-Hata H, Oyama M, Kawaguchi Y. 2014. Herpes simplex virus 1 protein kinase US3 phosphorylates viral dUTPase and regulates its catalytic activity in infected cells. J Virol 88:655–666. doi: 10.1128/JVI.02710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cedergren-Zeppezauer ES, Larsson G, Nyman PO, Dauter Z, Wilson KS. 1992. Crystal structure of a dUTPase. Nature 355:740–743. doi: 10.1038/355740a0. [DOI] [PubMed] [Google Scholar]
- 31.Jons A, Mettenleiter TC. 1996. Identification and characterization of pseudorabies virus dUTPase. J Virol 70:1242–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leang RS, Wu TT, Hwang S, Liang LT, Tong L, Truong JT, Sun R. 2011. The anti-interferon activity of conserved viral dUTPase ORF54 is essential for an effective MHV-68 infection. PLoS Pathog 7:e1002292. doi: 10.1371/journal.ppat.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madrid AS, Ganem D. 2012. Kaposi's sarcoma-associated herpesvirus ORF54/dUTPase downregulates a ligand for the NK activating receptor NKp44. J Virol 86:8693–8704. doi: 10.1128/JVI.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariza ME, Glaser R, Kaumaya PT, Jones C, Williams MV. 2009. The EBV-encoded dUTPase activates NF-kappa B through the TLR2 and MyD88-dependent signaling pathway. J Immunol 182:851–859. doi: 10.4049/jimmunol.182.2.851. [DOI] [PubMed] [Google Scholar]
- 35.McClure MA. 2001. Evolution of the DUT gene: horizontal transfer between host and pathogen in all three domains of life. Curr Protein Pept Sci 2:313–324. doi: 10.2174/1389203013381062. [DOI] [PubMed] [Google Scholar]
- 36.McGeehan JE, Depledge NW, McGeoch DJ. 2001. Evolution of the dUTPase gene of mammalian and avian herpesviruses. Curr Protein Pept Sci 2:325–333. doi: 10.2174/1389203013380964. [DOI] [PubMed] [Google Scholar]
- 37.Qian J, Zheng H, Huangfu WC, Liu J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. 2011. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog 7:e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharya S, Qian J, Tzimas C, Baker DP, Koumenis C, Diehl JA, Fuchs SY. 2011. Role of p38 protein kinase in the ligand-independent ubiquitination and down-regulation of the IFNAR1 chain of type I interferon receptor. J Biol Chem 286:22069–22076. doi: 10.1074/jbc.M111.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia C, Vijayan M, Pritzl CJ, Fuchs SY, McDermott AB, Hahm B. 2015. Hemagglutinin of influenza A virus antagonizes type I interferon (IFN) responses by inducing degradation of type I IFN receptor 1. J Virol 90:2403–2417. doi: 10.1128/JVI.02749-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lubick KJ, Robertson SJ, McNally KL, Freedman BA, Rasmussen AL, Taylor RT, Walts AD, Tsuruda S, Sakai M, Ishizuka M, Boer EF, Foster EC, Chiramel AI, Addison CB, Green R, Kastner DL, Katze MG, Holland SM, Forlino A, Freeman AF, Boehm M, Yoshii K, Best SM. 2015. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe 18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roupelieva M, Griffiths SJ, Kremmer E, Meisterernst M, Viejo-Borbolla A, Schulz T, Haas J. 2010. Kaposi's sarcoma-associated herpesvirus Lana-1 is a major activator of the serum response element and mitogen-activated protein kinase pathways via interactions with the Mediator complex. J Gen Virol 91:1138–1149. doi: 10.1099/vir.0.017715-0. [DOI] [PubMed] [Google Scholar]
- 42.Pyles RB, Sawtell NM, Thompson RL. 1992. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol 66:6706–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 2 Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]