ABSTRACT
Zika virus (ZIKV) has caused significant outbreaks and epidemics in the Americas recently, raising global concern due to its ability to cause microcephaly and other neurological complications. A stable and efficient infectious clone of ZIKV is urgently needed. However, the instability and toxicity of flavivirus cDNA clones in Escherichia coli hosts has hindered the development of ZIKV infectious clones. Here, using a novel self-splicing ribozyme-based strategy, we generated a stable infectious cDNA clone of a contemporary ZIKV strain imported from Venezuela to China in 2016. The constructed clone contained a modified version of the group II self-splicing intron P.li.LSUI2 near the junction between the E and NS1 genes, which were removed from the RNA transcripts by an easy-to-establish in vitro splicing reaction. Transfection of the spliced RNAs into BHK-21 cells led to the production of infectious progeny virus that resembled the parental virus. Finally, potential cis-acting RNA elements in ZIKV genomic RNA were identified based on this novel reverse genetics system, and the critical role of 5′-SLA promoter and 5′-3′ cyclization sequences were characterized by a combination of different assays. Our results provide another stable and reliable reverse genetics system for ZIKV that will help study ZIKV infection and pathogenesis, and the novel self-splicing intron-based strategy could be further expanded for the construction of infectious clones from other emerging and reemerging flaviviruses.
IMPORTANCE The ongoing Zika virus (ZIKV) outbreaks have drawn global concern due to the unexpected causal link to fetus microcephaly and other severe neurological complications. The infectious cDNA clones of ZIKV are critical for the research community to study the virus, understand the disease, and inform vaccine design and antiviral screening. A panel of existing technologies have been utilized to develop ZIKV infectious clones. Here, we successfully generated a stable infectious clone of a 2016 ZIKV strain using a novel self-splicing ribozyme-based technology that abolished the potential toxicity of ZIKV cDNA clones to the E. coli host. Moreover, two crucial cis-acting replication elements (5′-SLA and 5′-CS) of ZIKV were first identified using this novel reverse genetics system. This novel self-splicing ribozyme-based reverse genetics platform will be widely utilized in future ZIKV studies and provide insight for the development of infectious clones of other emerging viruses.
KEYWORDS: Zika virus, self-splicing intron, infectious cDNA clone, cis-acting RNA elements, reverse genetics
INTRODUCTION
Zika virus (ZIKV) belongs to the flavivirus genus within the family of Flaviviridae, together with many other pathogens with global impacts, such as dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), and yellow fever virus (YFV). Originally discovered in the Zika forest of Uganda in 1947 (1), ZIKV has been neglected for the past several decades. Since 2007, it has successfully expanded into the Pacific (2–5) and the Americas (6–8), resulting in significant outbreaks and epidemics. More importantly, ZIKV infection was unexpectedly linked to congenital defects and severe neurological complications, including microcephaly (9–12) and Guillain-Barré syndrome (3, 13). Despite the timely and cooperative efforts of the scientific and public health community in the past years, the mechanisms of ZIKV emergence and pathogenicity remains largely unknown, and no vaccine or specific antiviral is commercially available.
Reverse genetics systems represent one of the most powerful tools for the study of viral replication and pathogenicity and have now been widely utilized in vaccine design and antiviral screening. Development of infectious clones of ZIKV have been well regarded as a priority in response to the ZIKV crisis (14, 15). Similar to other flaviviruses, the ZIKV genome is an approximately 11-kb, positive-sense, single-stranded RNA molecule, which contains a single open reading frame (ORF) flanked by 5′ and 3′ untranslated regions. The ORF encodes the polyprotein precursor, which is further cleaved into three structural (capsid, premembrane/membrane, and envelope) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins by viral and host proteases. In general, cloning of the full-length cDNA of flavivirus downstream of either a T7/SP6 promoter or a eukaryotic RNA polymerase II promoter is sufficient to enable the recovery of progeny virus from the corresponding in vitro transcribed RNA or plasmid DNA. However, the cDNAs of flavivirus genomes are known to be toxic and unstable in common Escherichia coli hosts (16–20), which can be primarily attributed to the leaky expression of toxic viral proteins from the cryptic prokaryotic promoters in viral cDNA (19, 20). This phenomenon has significantly hindered the development of flavivirus infectious clones (16–18, 21). To bypass this obstacle, different approaches have been tested. Early reverse genetics systems of flaviviruses were usually based on the in vitro ligation of separately cloned viral 5′-half and 3′-half cDNA fragments (16, 22), and similar strategies are still being utilized (23–25). For the generation of a full-length flavivirus infectious clone, the screening of different E. coli strains is usually needed (17), and the optimization of the assembly sequence of viral cDNA fragments is often required (18). Some infectious clones were generated by introducing silent mutations in the viral ORF to eliminate the cryptic prokaryotic promoters in viral cDNA (19), whereas another study showed that introducing a short spacer sequence, which contains multiple stop codons in both directions into the end of the envelope protein coding region, can effectively stabilize the cDNA clone of DENV3 (26). Short eukaryotic introns, which were in vivo spliced after transfection into susceptive cell lines, have also been used to stabilize flavivirus cDNA clones (27), including two recently reported infectious clones of ZIKV (15, 28).
The P.li.LSUI2 group II intron was originally discovered in the gene of the rRNA large subunit in the mitochondrial genome of the brown algae Pylaiella littoralis (29). Like many other group II introns, the P.li.LSUI2 intron encodes an intron-encoded protein (IEP) with RNA maturase and reverse transcriptase activities (30), which are required for in vivo self-splicing and retro-homing (31). The P.li.LSUI2 intron was shown to be highly efficient in self-splicing under in vitro reaction conditions, and its catalytic mechanisms have been investigated thoroughly using biochemical and structural approaches (32, 33). These advantages of P.li.LSUI2 promote the exploration of its potential applications in the stabilization of the infectious clones of ZIKV.
In the present study, we cloned the cDNA fragments of a contemporary ZIKV strain isolated in 2016 (34) and successfully assembled the full-length cDNA using the self-splicing group II intron-based strategy. The transcribed RNA was subjected to self-splicing in vitro, and transfection of the spliced RNAs into BHK-21 cells resulted in the production of infectious progeny virus. The parental ZIKV and the recovered one have similar growth profiles, plaque morphologies, and mouse neurovirulence phenotypes, suggesting that the recovered ZIKV retains the biological properties of the parental virus well. Moreover, using this newly established reverse genetics system, we have revealed the critical role of a panel of RNA elements, including the 5′ stem-loop A (SLA) (35–37) and cyclization sequences (CS) (38) during ZIKV viral RNA (vRNA) replication, highlighting the reliability and potential of this new self-splicing intron-based strategy in the generation of flavivirus infectious clones.
RESULTS
Recovery of ZIKV by in vitro self-splicing from the intron-containing cDNA clone.
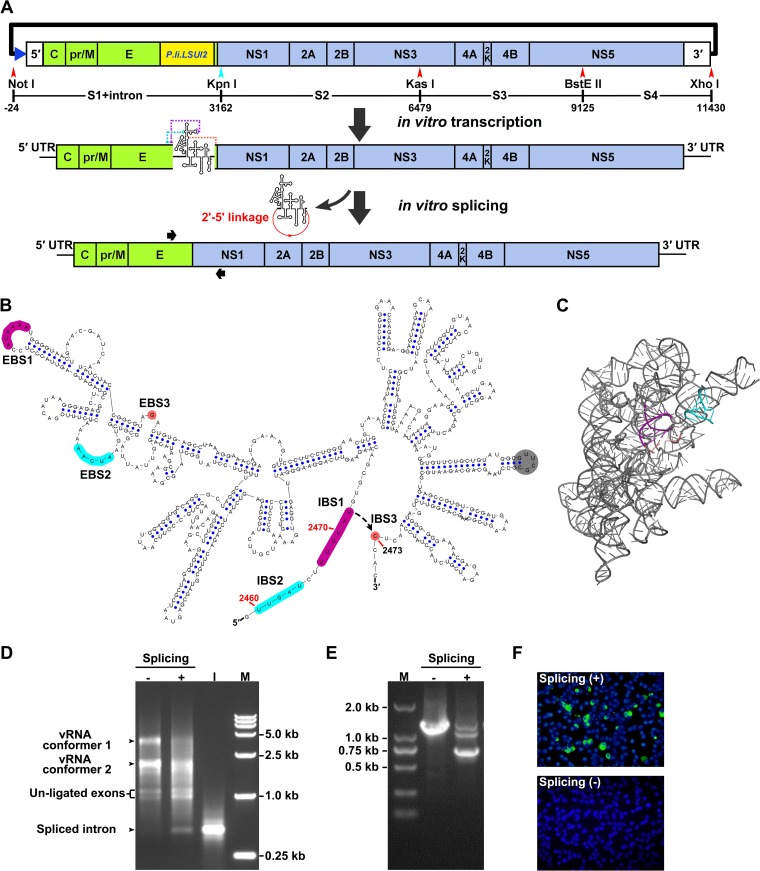
During our initial attempts, we found that direct assembly of the full-length cDNA of ZIKV GZ01 strain was unsuccessful, likely due to its toxicity to the E. coli host (19, 20). We rationalized that introducing an intron sequence rich in stop codons would efficiently terminate the translations of toxic peptides from viral cDNA and thus stabilize the corresponding cDNA clone of ZIKV. We chose the well-characterized P.li.LSUI2 group II intron (32, 33) as the spacer sequence, and its IEP-coding sequence was removed to prevent the unwanted splicing of the viral cDNA-derived transcripts in E. coli cells. The full-length genome of ZIKV was amplified by four primer pairs to generate the fragments S1 to S4, and the P.li.LSUI2 intron sequence was inserted between positions 2472 and 2473 in the viral cDNA (Fig. 1A and B). To ensure correct splicing, the exon binding sequences (EBSs) of P.li.LSUI2 were modified to recognize the flanking ZIKV-originated intron binding sequences (IBSs). The secondary structure of the engineered P.li.LSUI2 intron is shown in Fig. 1B and a three-dimensional (3-D) structural model of the P.li.LSUI2 intron is shown in Fig. 1C, with the EBS-IBS regions highlighted. As expected, the intron-containing S1 fragment was successfully assembled with the S2-S4 region to generate a full-length cDNA clone, the pACNR-GZ01-Intron-IC.
FIG 1.
Design and generation of the group II intron-based infectious clone of ZIKV. (A) Construction strategy of the infectious clone of ZIKV isolate GZ01. The SP6 promoter (blue triangle) was placed upstream of the 5′ end of ZIKV genome. The positions of the S1 to S4 fragments and endonucleases used for fragment assembly are labeled, and the numbering was calculated by setting the first nucleotide of ZIKV genome as “+1.” Note that the length of the intron sequence was calculated for the numbering. The artificially introduced KpnI site is shown in cyan. The cDNA sequence of P.li.LSUI2 group II intron was inserted near the border of the E and NS1 genes in the ZIKV genome. The EBS sequences of the inserted intron were modified to recognize the flanking ZIKV sequences. The intron-containing viral RNA transcripts were purified and subjected to in vitro splicing, which leads to the self-splicing of the intron from the viral genome and the generation of an intact viral genome. The binding locations of the primer pair used for RT-PCR detection in panel E and Fig. 2A are indicated by black arrows. (B) Secondary structure of the modified P.li.LSUI2 intron, which was inserted between position 2472 and 2473 in ZIKV genome. The EBS and IBS regions are highlighted in different colors. The EBS1 to -3 of the inserted intron was engineered to become complementary with positions 2467 to 2472, 2460 to 2464, and 2473, respectively, in the ZIKV genome. The gray circle indicates the original position of the IEP-coding region. The secondary structure was generated by the VARNA software (63). (C) 3-D structural model based on the original intron (PDB 4R0D) was generated by PyMol (pymol.org). The EBS and IBS regions were highlighted as in panel B. (D) Characterization of the in vitro splicing reaction. The electrophoresis patterns of the unspliced and spliced RNA transcripts were analyzed using a 1% agarose–TAE gel, and RNA, which was in vitro transcribed from the intron cDNA (lane “I”) and ran in parallel. Four hundred nanograms of each sample was loaded per well. The two slowly migrating bands correspond to different conformers of full-length RNA. Lane M, DNA marker DL 15,000. (E) RT-PCR was performed using the unspliced or spliced RNA transcripts as templates. Portions (5 μl) of PCR products were loaded onto a 1% agarose–TAE gel. Lane M, DNA marker DL 2,000. (F) Viral protein expression in transfected BHK-21 cells. A total of 500 ng of unspliced and spliced in vitro-transcribed RNA was transfected into BHK-21 cells, and viral E protein expression was detected by IFA at 72 h posttransfection.
The RNA preparation transcribed from the linearized pACNR-GZ01-Intron-IC was subjected to in vitro splicing reaction to remove the intron sequence. After an incubation at 45°C for 1 h, we first compared the electrophoresis patterns of the unspliced and spliced RNA. It was unambiguously shown that a band with the same size as the intron RNA was generated after the 45°C treatment (Fig. 1D, the bands labeled as spliced intron), and the bands corresponding to ZIKV genomic RNA from the in vitro splicing reaction migrated slightly faster than the corresponding bands in the unspliced control lane, suggesting the occurrence of the splicing reactions (Fig. 1D). To further confirm that the self-splicing process is successfully performed, the spliced and unspliced samples were subjected to reverse transcription-PCR (RT-PCR) and DNA sequencing. The size of major RT-PCR product from the spliced sample was consistent with the size of the corresponding ZIKV genome fragments, whereas the control product from the unspliced sample, which should be ca. 0.6 kb larger, was also in agreement with its predicted size (Fig. 1E). Sequencing of the RT-PCR product from the spliced sample confirmed that the ZIKV genome was correctly spliced (data not shown). These results suggested that full-length viral RNA could be efficiently generated after in vitro self-splicing.
Transfection of the in vitro-spliced RNA into BHK-21 cells resulted in robust viral protein expression in the transfected cells (Fig. 1F), whereas no or only minimal levels of viral protein expression were detected in the BHK-21 cells transfected with the unspliced RNA (Fig. 1F). Moreover, infectious virus can be detected in the supernatants of the transfected cells, whereas the unspliced, intron-containing RNA failed to produce infectious ZIKV. Thus, we developed a novel group II intron-based ZIKV infectious clone, pACNR-GZ01-Intron-IC, and the efficient recovery of infectious ZIKV was dependent on the self-splicing activity of the modified P.li.LSUI2 intron.
Characterization of the recovered ZIKV and its parental strain.
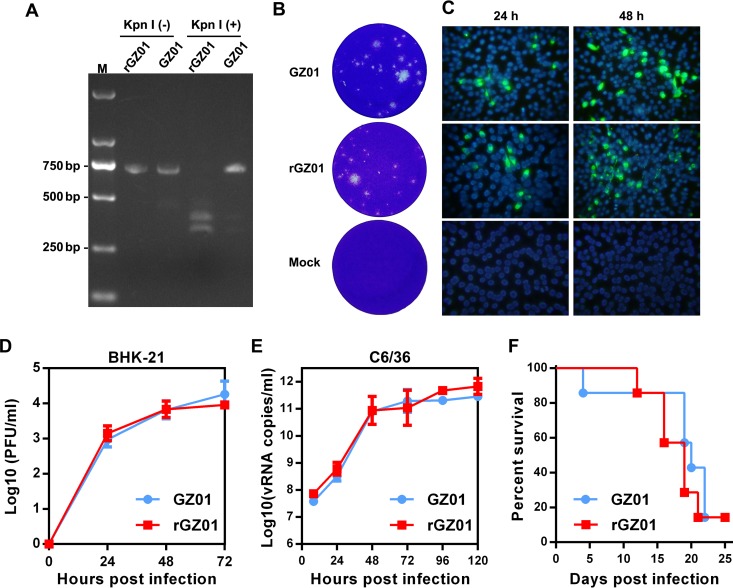
The recovered ZIKV (rGZ01) was passaged in C6/36 cells once to obtain a virus stock. First, the presence of the KpnI marker in the rGZ01 genome was confirmed by restriction endonuclease digestion (Fig. 2A), as well as DNA sequencing (data not shown) of the RT-PCR products from viral RNA of rGZ01. Then, a series of different assays were performed to characterize whether the rGZ01 retains its original biological characteristics like the parental GZ01. First, plaque formation assays showed the recovered rGZ01 and the parental GZ01 shared similar plaque morphologies in BHK-21 cells (Fig. 2B). Moreover, indirect immunofluorescence assay (IFA) demonstrated that ZIKV-positive cells increased at similar pattern in rGZ01 or GZ01-infected BHK-21 cells (Fig. 2C). Growth curve analysis in BHK-21 and mosquito C6/36 cells showed that the rGZ01 replicated with the same efficiency as its parental virus GZ01 (Fig. 2D and E).
FIG 2.
Comparison of biological characteristics between recovered GZ01 and the parental strain. (A) Identification of the KpnI-marker in recovered ZIKV. Viral RNA was isolated from supernatants of GZ01 and rGZ01 and ZIKV cDNA fragments were amplified by RT-PCR and digested by the endonuclease KpnI. The KpnI-digested PCR products [KpnI (+)] and undigested controls [KpnI (−)] were analyzed by electrophoresis in a 1.5% agarose–TAE gel. Lane M, DNA marker DL 2,000. (B) Plaque morphologies of rGZ01 and parental GZ01. Mock, uninfected BHK-21 cells. (C) BHK-21 cells were infected with an MOI of 0.1 of rGZ01 and GZ01, respectively, and viral E protein expression was monitored by IFA at different time points after infection. Uninfected BHK-21 cells were subjected to IFA and the results were shown in parallel (mock). (D and E) Comparison of the growth kinetics of rGZ01 and GZ01. (D) Infectious viruses in the supernatants of MOI 0.1-infected BHK-21 cells were determined by plaque assay. (E) C6/36 cells were infected with rGZ01 and GZ01 at an MOI of 0.1, culture supernatants were collected at different time points postinfection, and vRNA levels were determined by qRT-PCR. (F) Neurovirulence of rGZ01 and GZ01 in neonatal BALB/c mice. One-day-old BALB/c suckling mice (n = 7) were incubated with 10 PFU of rGZ01 or GZ01, respectively, and the survival statistics were monitored daily.
The mouse neurovirulence model of ZIKV has been well established previously (39, 40). We further tested the neurovirulence of rGZ01 and compared it with the parental GZ01. Upon intracerebral injection, both rGZ01 and GZ01 caused similar clinical symptoms in suckling mice, including inactivity, motor weakness, and bilateral hind limb paralysis, and most animals died within 22 days. No significant difference was detected between the survival curves of rGZ01 and GZ01 (Fig. 2F).
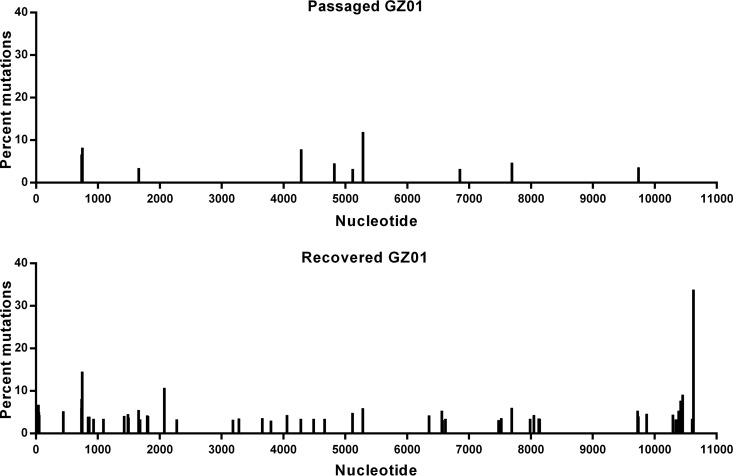
In addition, to profile the genetic variations of ZIKV during recovery and passaging in cells, high-throughput sequencing of rGZ01 and the passaged GZ01 were performed using an Illumina MiSeq sequencing machine. No amino acid substitutions were identified in either virus, and a panel of intra-host single nucleotide variants (iSNVs) distributed within the full genome were identified in individual samples (Fig. 3). Albeit there are more substitution sites in the recovered GZ01 than that in the passaged one, the substitution frequencies for both viruses were quite low, and few consensus sequence changes were detected. Taken together, these results showed that rGZ01 maintains similar biological characteristics to the parental strain GZ01, demonstrating that the catalytic intron-based infectious clone can be a good tool for the investigation of ZIKV pathogenesis.
FIG 3.
Mutational analysis of rGZ01 and its parental strain. Viral samples were analyzed by Illumina MiSeq high-throughput sequencing, and the mutational profiles of rGZ01 and the parental GZ01 strain were shown. Intrahost single nucleotide variant (iSNV) detection was performed using the sCLC genomic workbench v9.0 (CLCbio), and the site with substitution frequency above 5% was considered to be an iSNV.
Identification of RNA elements crucial for ZIKV vRNA replication.
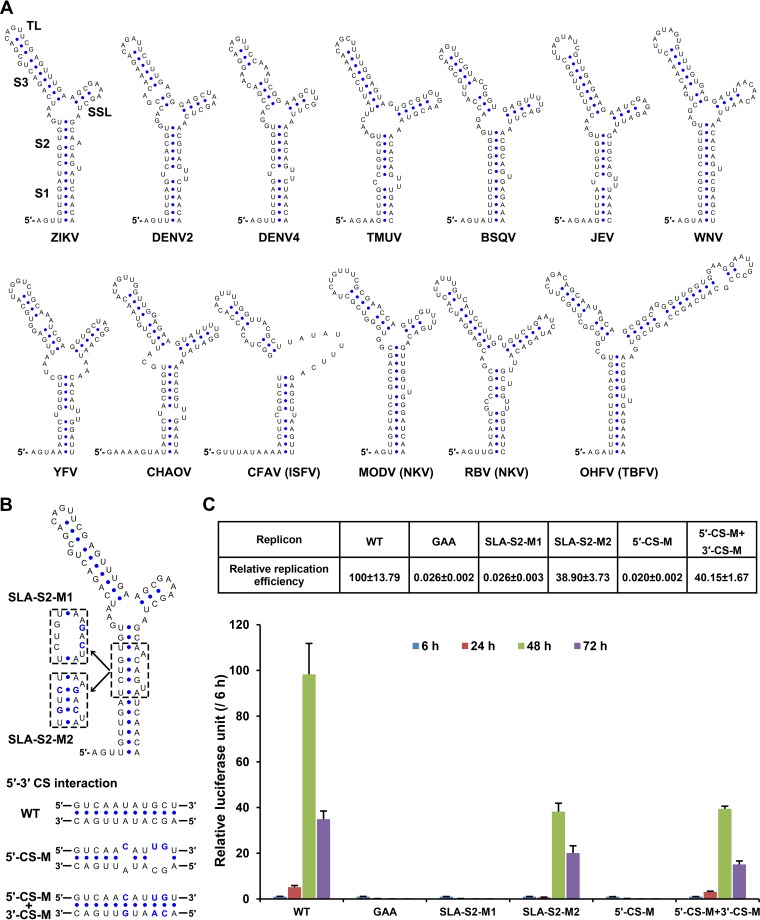
Flavivirus vRNA replication requires the participation of various cis-acting RNA elements in genomic 5′ and 3′ ends. The 5′ stem-loop A (SLA) structure binds to viral NS5, and the latter is translocated to the 3′ end to initiate minus-strand RNA synthesis though 5′-3′ terminal interactions (35), which include the 5′-3′ upstream AUG region (UAR), 5′-3′ downstream AUG region (DAR) and 5′-3′ cyclization sequence (CS) base-pairing interactions (38, 41–44). The dynamic modulation of viral NS5 recruitment to the 5′ end by the 5′-UAR-flanking stem (UFS) is also critical for efficient vRNA replication (45). The core sequence (5′, UCAAUAUGCU; 3′, AGCAUAUUGA) of the CS element is in consensus with the mosquito-borne flaviviruses (38, 46), whereas the secondary structure of the SLA element is highly conserved in all known species of flaviviruses as shown by Mfold (47, 48) predictions (Fig. 4A). However, the functional roles of potential cis-acting RNA elements in ZIKV genomic RNA have not been investigated yet.
FIG 4.
Functional analysis of 5′ SLA and CS using ZIKV reporter replicon. (A) Secondary structures of the SLA elements from different flaviviruses. The substructures were labeled on the SLA structure of ZIKV (GZ01). The top loop (TL), side stem-loop (SSL), and stem 1 to stem 3 (S1-S3) are indicated. DENV2, dengue virus type 2 (GenBank U87411); DENV4, dengue virus type 4 (GenBank AF326573); TMUV, Tembusu virus (GenBank JF895923); BSQV, Bussuquqara virus (GenBank AY632536); JEV, Japanese encephalitis virus (GenBank M18370); WNV, West Nile virus (GenBank DQ211652); YFV, yellow fever virus (GenBank X03700); CHAOV, Chaoyang virus (GenBank NC_017086); CFAV, cell fusing agent virus (GenBank M91671); MODV, modoc virus (GenBank AJ242984); RBV, rio bravo virus (GenBank JQ582840); OHFV, Omsk hemorrhagic fever virus (GenBank AY193805); ISFV, insect-specific flavivirus; NKV, flavivirus with non-known vector, TBFV, tick-borne flavivirus. (B) Mutations targeting the SLA and 5′-CS. The mutated nucleotides were shown in a blue font. (C) Replication kinetics of the wild-type and mutated ZIKV replicons in transfected BHK-21 cells. The ratios of raw luciferase units measured at different time points after transfection to the value measured at 6 h posttransfection (relative luciferase units) are shown. The percentages of relative luciferase units of various mutants at 48 h posttransfection to the corresponding value of the wild-type replicon are listed above. Data are expressed as means ± the standard deviations. The GAA mutant contains a GDD-to-GAA mutation in the catalytic motif of the NS5 RdRp domain.
Here, we first explored the roles of SLA and CS elements with the replicon system of ZIKV (49), in which the structural genes were replaced by the Renilla luciferase reporter gene. Replicon RNAs were transfected into BHK-21 cells, and Renilla luciferase reporter activities were monitored at 6, 24, 48, and 72 h posttransfection (p.t.; Fig. 4B and C). For the wild-type (WT) replicon, the luciferase activity at 48 h p.t. was ∼100-fold higher than that measured at 6 h p.t., indicating robust vRNA replication, whereas the replicons containing a GAA mutation in the catalytic GDD motif of the NS5 RNA-dependent RNA polymerase (RdRp) domain exhibited no vRNA replication (Fig. 4C), as expected. We also showed that disrupting the stem 2 of the SLA structure (mutant SLA-S2-M1) or mutating the 5′-CS element (mutant 5′-CS-M) abolished vRNA replication. In contrast, the reconstitution of the SLA stem 2 (SLA-S2-M2) or 5′-3′ CS interaction (5′-CS-M + 3′-CS-M) restored vRNA replication of the corresponding replicons, although these two mutants still replicated moderately less efficiently than the WT replicon. These results suggested that the SLA element and 5′-3′ CS interaction were crucial for ZIKV vRNA replication.
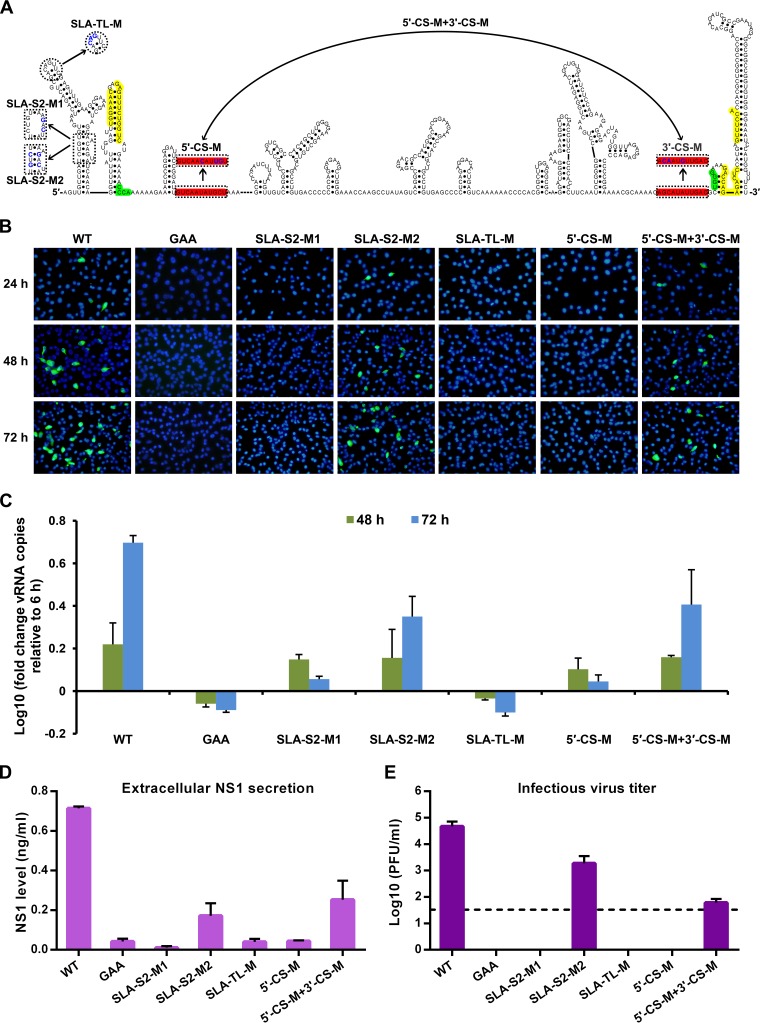
We then introduced the corresponding mutations into the newly developed infectious clone of ZIKV strain GZ01, and an additional mutation targeting the SLA top loop (SLA-TL-M) was also generated accordingly (Fig. 5). All mutant viruses, together with the WT ZIKV, were recovered in BHK-21 cells as described above. IFA results showed that mutations into the semiconserved top-loop sequence of SLA (SLA-TL-M) or the 5′-CS sequence (5′-CS-M) completely abolished viral protein expression in BHK-21 cells (Fig. 5B). Disruption of the base pairing of the SLA stem 2 region (SLA-S2-M1) was also deleterious for viral replication. In contrast, restoration of the SLA stem 2 (SLA-S2-M2) or reconstitution of the 5′-3′ CS interaction by introducing complementary mutations into the 3′-CS sequence (5′-CS-M + 3′-CS-M) enabled viral protein expression of the corresponding mutants (Fig. 5B). The vRNA levels in the transfected supernatants were then analyzed by quantitative RT-PCR (qRT-PCR) (Fig. 5C), and the accumulation of vRNA in the supernatant of cells transfected with the WT, the SLA-S2-M2, or the 5′-CS-M + 3′-CS-M RNA was comparable, whereas no vRNA replication was detected for the GAA or the SLA-S2-M1, the SLA-TL-M, and the 5′-CS-M groups. Moreover, viral NS1 secretion determined by enzyme-linked immunosorbent assay (ELISA) showed that NS1 was detected in the supernatants of the WT, the SLA-S2-M2 or the 5′-CS-M + 3′-CS-M groups, whereas in the other groups, only the background level of NS1 secretion could be detected (Fig. 5D). In agreement with these results, infectious virus release was able to detected for the WT, the SLA-S2-M2, or the 5′-CS-M + 3′-CS-M groups (Fig. 5E), highlighting the crucial roles of the SLA element and 5′-3′ CS interaction in viral replication. Combined the consistent results from the mutagenesis of replicon and infectious clone (Fig. 4 and 5), we conclude that the 5′-SLA element and 5′-3′ CS interaction were crucial for ZIKV replication, similar to other flaviviruses.
FIG 5.
Mutagenesis analysis of crucial RNA elements of ZIKV based on the GZ01 infectious clone. (A) Terminal secondary structure of ZIKV GZ01 strain. The demonstrated structure was based on the linear conformation of the viral genome. The three pairs of 5′-3′ complementary sequences, which were required for genome cyclization, were highlighted using different colors as follows: yellow, 5′-3′ UAR; green, 5′-3′ DAR; and red, 5′-3′ CS. The designed mutations were demonstrated and the mutated nucleotides are shown in blue font. The SLA-TL-M mutant contains mutations in the conserved terminal loop of the 5′-SLA element, and the SLA-S2-M1/M2 mutants disrupted or reconstituted the stem 2 in the 5′-SLA element. Mutations were also introduced into the 5′-CS to generate 5′-CS-M, whereas the 5′-CS-M + 3′-CS-M mutant combined the complementary mutations in the 5′- and 3′-CS to restore their base-pairing interaction. The 3′-CS-M mutant, which was not included in the analysis, was labeled in gray. (B) Equal amounts (400 ng per well) of in vitro-spliced wild-type and mutated viral RNA preparations were transfected into BHK-21 cells in a 24-well format, and viral E protein expression was detected by IFA at different time points after transfection. (C) qRT-PCR detection of vRNA accumulation in the supernatants of the transfected cells. The results are expressed as fold changes of the vRNA copies relative to the values measured at 6 h posttransfection. (D) Detection of the viral NS1 secretion in the culture supernatants at 72 h posttransfection by ELISA. (E) In total, equal portions (100 μl) of supernatants collected at 72 h posttransfection were used to infect C6/36 cells in 24-well plates. After 6 days, the infectious viruses in the culture supernatants were detected by plaque assay. The experiments shown in panels C to E were performed in triplicates and were shown as the means ± standard errors. The data were analyzed by one-way (D and E) or two-way (C) ANOVA, and biologically significant differences were also confirmed by multiple comparisons.
DISCUSSION
A series of approaches (14, 15, 24, 28, 50–52) have been applied for the generation of ZIKV reverse genetics platforms. Since direct assembly of the native ZIKV full genome in E. coli had been successful only in a few cases (14, 50), most established reverse genetics systems of ZIKV were based on either in vitro assembly of viral cDNA fragments (24, 52) or eukaryotic intron-based and HCMV promoter-driven, “infectious DNA” clones (15, 28). In the present study, we successfully utilized the group II intron P.li.LSUI2 to stabilize the cDNA clone of ZIKV in a commonly used E. coli host strain. The lack of an IEP-coding region in the intron sequence keeps it inactive in host cells; however, by changing the concentrations of monovalent and divalent ions in vitro, the intron becomes active and splices itself from RNA transcripts, resulting in an intact viral genome. Compared with conventional infectious clones of ZIKV, although one additional step of in vitro splicing is required in our system, the easy-to-setup in vitro splicing reaction is much simpler to perform than most of the in vitro assembly based systems. In addition, our in vitro splicing system has advantages over the previous described in vivo splicing system (15, 28). The efficiency of eukaryotic intron-based “infectious DNA” clones depends largely on the activity of the spliceosome complex of the transfected cells, and the in vivo transcription/splicing process varies in different cells. Thus, these eukaryotic intron-based infectious clones are not applicable for viral replication studies. In addition, the intron-based reverse genetics system retained the potential to convert into in vivo splicing-based systems since the self-splicing reaction can be facilitated by the RNA maturase activity of IEP in vivo (30, 31, 53), which can be supplied in trans.
cis-Acting RNA elements are involved not only in viral replication (35, 36, 41, 44–46, 54) and translation (55, 56) of flaviviruses but also in host adaption (57), neurovirulence (58), and pathogenicity (59–61). Using the ZIKV replicon and the newly developed infectious clone of ZIKV, we have shown that 5′-SLA and 5′-3′ CS interaction are indispensable for ZIKV replication. It was also showed that the 5′-SLA and 5′-3′ CS base-pairing restoration mutants consistently replicated moderately lower than the WT, suggesting that tertiary or quaternary interactions may be involved in the functionalization of these cis-acting RNA elements. Previously, studies suggested that the conserved UFS element is required for ZIKV replication (45) and that the SL-I/xrRNA1 structure is responsible for the production of ZIKV sfRNA (62). Thus, the RNA-based regulation mechanism of ZIKV should be highly similar to other well-studied mosquito-borne flaviviruses. These results suggest that the reverse genetics system described herein can also be used for the investigation of ZIKV replication and the selection and evaluation of anti-ZIKV medicines.
Flaviviruses are likely to continue to emerge and re-emerge around the world in the future. The group II intron-based strategy described here is ready to be applied to other flaviviruses. Thus, we believe it has the potential to be developed into a powerful tool for the rapid establishment of conventional reverse genetics systems of emerging flaviviruses in the future.
MATERIALS AND METHODS
Cells and virus.
The baby hamster kidney fibroblast cell line BHK-21 (ATCC CCL10) was cultured in Dulbecco modified Eagle medium (DMEM; Gibco, Thermo Fisher Scientific) containing 5% fetal bovine serum (FBS; Biowest) and 1% penicillin-streptomycin (PS; Thermo Fisher Scientific). The Aedes albopictus cell line C6/36 (ATCC CRL-1660) were cultured using RPMI 1640 (Gibco, Thermo Fisher Scientific) containing 10% FBS and 1% PS (Thermo Fisher Scientific).
The contemporary Asian lineage ZIKV isolate GZ01 (KU820898) was isolated from a Chinese patient that had returned from Venezuela in 2016 (34). Viral RNA for the amplification of genomic fragments was extracted from the culture supernatants of GZ01-infected BHK-21 cells.
DNA molecular manipulation.
Four cDNA fragments (S1 to S4) covering the full-length genome of the ZIKV GZ01 strain were amplified by RT-PCR and cloned into pGEM-T Easy (Promega), except for the S2 fragment, which was cloned into the pACNR plasmid with a preintroduced multiple cloning site using the KpnI/KasI restriction sites. This pACNR-S2 plasmid served as the basis for fragment assembly and the S3 and S4 fragments were subsequently assembled with pACNR-S2. The modified P.li.LSUI2 intron sequence was chemically synthesized and cloned by Sangon Biotech, China. The ZIKV S1 fragment and the intron sequence were fused using overlapping PCR, and the SP6 promoter sequence was placed upstream of the S1 fragment. Finally, the S1-intron sequence was cloned into pACNR-S234 to generate the corresponding pACNR-GZ01-Intron-IC. The schematic construction of the infectious clone is shown in Fig. 1A.
To generate SLA and 5′-CS mutants, site-directed mutagenesis was performed using the S1-intron plasmid as a template, and for the introduction of mutations into 3′-CS, the cloned S4 in pGEM-T Easy (Promega) vector was used as the site-directed mutagenesis template. The fragments containing the corresponding mutations were subcloned back into the corresponding full-length constructs to achieve the desired mutants. The GAA mutant was generated by site-directed mutagenesis together with subcloning. Primers used for site-directed mutagenesis are listed in Table 1.
TABLE 1.
Primers utilized to introduce mutations in the infectious clone of ZIKV isolate GZ01
| Primer | Sequence (5′-3′)a | Application |
|---|---|---|
| SLA-TL-M-F | CAGACTGCGAgtcTTCGAGTTTGAAGCG | To introduce mutations into SLA top loop |
| SLA-TL-M-R | ATTCACACAGATCAACAAC | |
| SLA-S2-M1-F | AAGCTAGCAAgacTATCAACAGGTTTTATTTTG | To introduce mutations to disrupt the stem 2 of SLA |
| SLA-S2-M1-R | TCGCTTCAAACTCGAACTG | |
| SLA-S2-M2-F | AGTTGTTGATgtcTGTGAATCAGACTG | To introduce mutations to restore the stem 2 of SLAb |
| SLA-S2-M2-R | CTATAGTGTCACCTAAATGC | |
| 5′-CS-M-F | AAGCTAGCAAcattgTATCAACAGG | To introduce mutations into 5′ cyclization sequence |
| 5′-CS-M-R | TCGCTTCAAACTCGAACTG | |
| 3′-CS-M-F | ACGCAAAACAcaatgTTGACGCTGGGAAAGACCAG | To introduce mutations into 3′ cyclization sequence |
| 3′-CS-M-R | TTTCCGGGGGGTCTCCTC | |
| GAA-F | AGTCAGTGGAgcagcaTGCGTTGTGAAGC | To introduce mutations into the GDD motif of NS5 |
| GAA-R | GCCATTCGTTTGAGCCTA |
Mutations are indicated in boldface letters.
This mutant was achieved based on pregenerated SLA-S2-M1 mutant.
In vitro RNA preparation.
In vitro transcription was performed using the Ribomax SP6 large-scale RNA production kit (Promega) as previously described (46). The RNA products were cleaned-up using the Purelink RNA minikit (Thermo Fisher Scientific), mixed with equal amounts of 2× splicing buffer (80 mM Tris-HCl [pH 7.4], 2 M NH4Cl, 20 mM MgCl2, and 0.04% sodium dodecyl sulfate) (32, 33), and incubated at 45°C for 1 h. Then, a secondary RNA cleanup protocol was performed to remove the salts from RNA preparations. The final RNA stocks were quantified using spectrophotometry and stored at −80°C until use.
Transfection and virus stock preparation.
A total of 5 × 104 BHK-21 cells were seeded onto 24-well plates with or without sterilized coverslips to grow to ca. 50% confluence at the time of transfection, which was performed using Lipofectamine 2000 (Thermo Fisher Scientific). At the indicated time points p.t., coverslips were fixed with acetone-methanol (3/7) at −20°C, and ZIKV E protein expression was detected by IFA as previously described (45). At day 3 p.t., culture supernatants were collected and used to infect C6/36 cells maintained in RPMI 1640 (Thermo Fisher Scientific) containing 2% FBS (Biowest). The C6/36 cells were cultured at 28°C for 6 days and then were freeze-thawed once. Cell debris was removed by centrifugation at 6,000 × g, 4°C for 15 min, and the supernatants were dispensed into single-use aliquots, which were stored at −80°C.
RT-PCR.
RT-PCR was performed using SuperScript III One-Step RT-PCR System with Platinum Taq (Thermo Fisher Scientific) following the user protocols to monitor the efficiency of the in vitro splicing reactions. For the identification of the genetic marker in the recovered virus, viral RNA was extracted from virus-containing culture supernatants using the PureLink RNA minikit (Thermo Fisher Scientific). RT-PCR was performed as described above, and the PCR products were subjected to KpnI digestion and DNA Sanger sequencing. DNA sequencing reactions were performed by SinoGenoMax, China.
Plaque formation assay.
Plaque assays were performed on the BHK-21 cell line using virus stocks prepared in C6/36 cells. A total of 300 μl of 10-fold-serial-diluted virus samples was added onto cell monolayers in 12-well plates. After a 1.5-h incubation at 37°C under 5% CO2, the virus dilutes in DMEM (Thermo Fisher Scientific) with 2% FBS were aspirated. Then, 1 ml of DMEM with 2% FBS and 1% low melting point agarose (Promega) was coated on the infected cells, which were cultured for another 4 days. The infected cells were then fixed with 4% formaldehyde, followed by staining with 1% crystal violet dissolved in 20% ethanol. Photos of plaques were captured using a digital camera or a BioSpectrum imaging system (UVP).
Viral propagation analyses.
BHK-21 and C6/36 cells were infected with rGZ01 or GZ01 at a multiplicity of infection (MOI) of 0.1. At the indicated time points after infection, culture supernatants were collected, and infectious virus was detected by plaque assay. Alternately, viral RNA was isolated using a PureLink RNA minikit, and vRNA copies were determined using qRT-PCR methods described previously (34). Lastly, in vitro-transcribed viral RNA was serial diluted to generate standards for absolute quantification. For the evaluation of protein expression, BHK-21 cells were seeded onto sterilized coverslips in 24-well plates, and were cultured overnight to reach 95% confluence. The cells were infected at an MOI of 0.1 with rGZ01 or GZ01. At the indicated time points, the coverslips were fixed, and IFAs were performed as described above.
Mice experiments.
The experimental procedures were approved by the Animal Experiment Committee of the Laboratory Animal Center, AMMS, China, and the experiments were performed according to the guidelines of the Chinese Regulations of Laboratory Animals (Ministry of Science and Technology of People's Republic of China) and Laboratory Animal-Requirements of Environment and Housing Facilities (GB 14925-2010; National Laboratory Animal Standardization Technical Committee). For the neurovirulence test, 1-day-old BALB/c mice received an intracerebral injection with 10 PFU of ZIKV, and the mortality and clinical symptoms were monitored for 25 days. The survival curve was analyzed by the log-rank test.
Replicon assay.
The ZIKV replicon containing the Renilla luciferase reporter gene was used for mutagenesis analysis (49). The mutants were generated using similar strategies for mutagenesis based on pACNR-GZ01-Intron-IC. One day prior to transfection, 2 × 104 BHK-21 cells were seeded into each well of 48-well plates, followed by incubation at 37°C in 5% CO2. Triplicate transfections were performed using Lipofectamine 2000 reagent (Thermo Fisher Scientific). For each well of BHK-21 cells, 500 ng of replicon RNA was transfected. The cell lysates were collected at 6, 24, 48, and 72 h after transfection. The Renilla luciferase activity was measured using a Renilla luciferase assay system (Promega) with the GloMax Discover system (Promega).
Detection of vRNA and NS1 in transfected supernatants.
Transfections were performed in the 48-well format as described under “Replicon assay” above. At 6 h posttransfection, the culture media containing the Lipofectamine 2000/RNA complexes were discarded, and the transfected cells were washed three times with 0.5 ml of FBS-free DMEM. Then, 0.25 ml of fresh DMEM containing 5% FBS was added to the cells, which were cultured at 37°C under 5% CO2. At 6, 48, and 72 h, the culture medium was collected. vRNA was isolated, and qRT-PCR was performed as described above. Detection of the NS1 protein was performed using a Zika virus NS1 ELISA kit (Biofront) according to instructions provided by the manufacturer.
High-throughput sequencing.
Viral RNA was extracted from the virus stocks of the passaged, parental GZ01 and rGZ01 prepared in C6/36 cells. Deep-sequencing was performed using Illumina MiSeq (Illumina). Intrahost single nucleotide variant (iSNV) detection was performed using CLC genomic workbench v9.0 (CLCbio) as previously described (34). The sites with a substitution frequency above 5% were considered to be an iSNV.
Statistical analysis.
The one-way analysis of variance (ANOVA) and Dunnett's multiple-comparison tests were performed to analyze the replicon data in Fig. 4. The log rank test was performed for the survival analysis. The two-way ANOVA was performed for the analysis of the vRNA levels in Fig. 5, and the one-way ANOVA and Fisher LSD tests were performed to analyze the NS1 secretion and infectious virus release results. Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software).
ACKNOWLEDGMENTS
This study was supported by the National Science and Technology Major Project of China (grants 2017ZX09101005 and 2017ZX10304402), the National Key Research and Development Project of China (grant 2016YFD0500304), the National Natural Science Foundation of China (NSFC; grants 31770190, 81661148054 and 81661130162). C.-F.Q. was supported by the Excellent Young Scientist (grant 81522025), Innovative Research Group (grant 81621005), and the Newton Advanced Fellowship from the UK Academy of Medical Sciences (NAF003\1003).
REFERENCES
- 1.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus I isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 3.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. 2014. Zika virus infection complicated by Guillain-Barre syndrome: case report, French Polynesia, December 2013. Euro Surveill 19:20720. [DOI] [PubMed] [Google Scholar]
- 4.Cao-Lormeau VM, Roche C, Teissier A, Robin E, Berry AL, Mallet HP, Sall AA, Musso D. 2014. Zika virus, French Polynesia, South pacific, 2013. Emerg Infect Dis 20:1085–1086. doi: 10.3201/eid2006.131413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tognarelli J, Ulloa S, Villagra E, Lagos J, Aguayo C, Fasce R, Parra B, Mora J, Becerra N, Lagos N, Vera L, Olivares B, Vilches M, Fernandez J. 2016. A report on the outbreak of Zika virus on Easter Island, South Pacific, 2014. Arch Virol 161:665–668. doi: 10.1007/s00705-015-2695-5. [DOI] [PubMed] [Google Scholar]
- 6.Brasil P, Pereira JP Jr, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baiao AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerbino-Neto J, Mesquita EC, Souza TM, Parreira V, Wittlin BB, Durovni B, Lemos MC, Vizzoni A, Bispo de Filippis AM, Sampaio SA, Goncalves Bde S, Bozza FA. 2016. Clinical manifestations of Zika virus infection, Rio de Janeiro, Brazil, 2015. Emerg Infect Dis 22:1318–1320. doi: 10.3201/eid2207.160375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Quinonez JA, Escobar-Escamilla N, Wong-Arambula C, Vazquez-Pichardo M, Torres-Longoria B, Lopez-Martinez I, Ruiz-Matus C, Kuri-Morales P, Ramirez-Gonzalez JE. 2016. Asian genotype Zika virus detected in traveler returning to Mexico from Colombia, October 2015. Emerg Infect Dis 22:937–939. doi: 10.3201/eid2205.160190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, Vizjak A, Pizem J, Petrovec M, Avsic Zupanc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z. 2016. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 11.Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, Mysorekar IU, Diamond MS. 2016. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165:1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, Faria Dde P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrao-Braga PC. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabrizius RG, Anderson K, Hendel-Paterson B, Kaiser RM, Maalim S, Walker PF. 2016. Guillain-Barre syndrome associated with Zika virus infection in a traveler returning from Guyana. Am J Trop Med Hyg 95:1161–1165. doi: 10.4269/ajtmh.16-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett AD, Vasilakis N, Weaver SC, Shi PY. 2016. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsetsarkin KA, Kenney H, Chen R, Liu G, Manukyan H, Whitehead SS, Laassri M, Chumakov K, Pletnev AG. 2016. A full-length infectious cDNA clone of Zika virus from the 2015 epidemic in Brazil as a genetic platform for studies of virus-host interactions and vaccine development. mBio 7:e01114-16. doi: 10.1128/mBio.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice CM, Grakoui A, Galler R, Chambers TJ. 1989. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biologist 1:285–296. [PubMed] [Google Scholar]
- 17.Lai CJ, Zhao BT, Hori H, Bray M. 1991. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc Natl Acad Sci U S A 88:5139–5143. doi: 10.1073/pnas.88.12.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamshchikov VF, Wengler G, Perelygin AA, Brinton MA, Compans RW. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294–304. doi: 10.1006/viro.2000.0795. [DOI] [PubMed] [Google Scholar]
- 19.Pu SY, Wu RH, Yang CC, Jao TM, Tsai MH, Wang JC, Lin HM, Chao YS, Yueh A. 2011. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J Virol 85:2927–2941. doi: 10.1128/JVI.01986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Aaskov J, Lott WB. 2011. Identification of a cryptic prokaryotic promoter within the cDNA encoding the 5′ end of dengue virus RNA genome. PLoS One 6:e18197. doi: 10.1371/journal.pone.0018197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J Gen Virol 84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- 22.Sumiyoshi H, Hoke CH, Trent DW. 1992. Infectious Japanese encephalitis virus RNA can be synthesized from in vitro-ligated cDNA templates. J Virol 66:5425–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos JJ, Cordeiro MT, Bertani GR, Marques ET, Gil LH. 2014. A two-plasmid strategy for engineering a dengue virus type 3 infectious clone from primary Brazilian isolate. An Acad Bras Cienc 86:1749–1759. doi: 10.1590/0001-3765201420130332. [DOI] [PubMed] [Google Scholar]
- 24.Weger-Lucarelli J, Duggal NK, Bullard-Feibelman K, Veselinovic M, Romo H, Nguyen C, Ruckert C, Brault AC, Bowen RA, Stenglein M, Geiss BJ, Ebel GD. 2017. Development and characterization of recombinant virus generated from a New World Zika virus infectious clone. J Virol 91:e01765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edmonds J, van Grinsven E, Prow N, Bosco-Lauth A, Brault AC, Bowen RA, Hall RA, Khromykh AA. 2013. A novel bacterium-free method for generation of flavivirus infectious DNA by circular polymerase extension reaction allows accurate recapitulation of viral heterogeneity. J Virol 87:2367–2372. doi: 10.1128/JVI.03162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaney JE Jr, Hanson CT, Firestone CY, Hanley KA, Murphy BR, Whitehead SS. 2004. Genetically modified, live attenuated dengue virus type 3 vaccine candidates. Am J Trop Med Hyg 71:811–821. [PubMed] [Google Scholar]
- 27.Yamshchikov V, Mishin V, Cominelli F. 2001. A new strategy in design of +RNA virus infectious clones enabling their stable propagation in Escherichia coli. Virology 281:272–280. doi: 10.1006/viro.2000.0793. [DOI] [PubMed] [Google Scholar]
- 28.Schwarz MC, Sourisseau M, Espino MM, Gray ES, Chambers MT, Tortorella D, Evans MJ. 2016. Rescue of the 1947 Zika virus prototype strain with a cytomegalovirus promoter-driven cDNA clone. mSphere 1:e00246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontaine JM, Rousvoal S, Leblanc C, Kloareg B, Loiseaux-de Goer S. 1995. The mitochondrial LSU rDNA of the brown alga Pylaiella littoralis reveals alpha-proteobacterial features and is split by four group IIB introns with an atypical phylogeny. J Mol Biol 251:378–389. doi: 10.1006/jmbi.1995.0441. [DOI] [PubMed] [Google Scholar]
- 30.Zerbato M, Holic N, Moniot-Frin S, Ingrao D, Galy A, Perea J. 2013. The brown algae Pl.LSU/2 group II intron-encoded protein has functional reverse transcriptase and maturase activities. PLoS One 8:e58263. doi: 10.1371/journal.pone.0058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enyeart PJ, Mohr G, Ellington AD, Lambowitz AM. 2014. Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and noncoding RNA analysis. Mobile DNA 5:2. doi: 10.1186/1759-8753-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa M, Fontaine JM, Loiseaux-de Goer S, Michel F. 1997. A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J Mol Biol 274:353–364. doi: 10.1006/jmbi.1997.1416. [DOI] [PubMed] [Google Scholar]
- 33.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. 2014. Crystal structure of a eukaryotic group II intron lariat. Nature 514:193–197. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li XF, Dong HL, Huang XY, Qiu YF, Wang HJ, Deng YQ, Zhang NN, Ye Q, Zhao H, Liu ZY, Fan H, An XP, Sun SH, Gao B, Fa YZ, Tong YG, Zhang FC, Gao GF, Cao WC, Shi PY, Qin CF. 2016. Characterization of a 2016 clinical isolate of Zika virus in non-human primates. EBioMedicine 12:170–177. doi: 10.1016/j.ebiom.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lodeiro MF, Filomatori CV, Gamarnik AV. 2009. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol 83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong H, Zhang B, Shi PY. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381:123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 38.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng YQ, Zhao H, Li XF, Zhang NN, Liu ZY, Jiang T, Gu DY, Shi L, He JA, Wang HJ, Sun ZZ, Ye Q, Xie DY, Cao WC, Qin CF. 2016. Isolation, identification and genomic characterization of the Asian lineage Zika virus imported to China. Sci China Life Sci 59:428–430. doi: 10.1007/s11427-016-5043-4. [DOI] [PubMed] [Google Scholar]
- 40.Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M Jr, Virgin HW, Klein RS, Sen GC, Diamond MS. 2012. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog 8:e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 373:1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Friebe P, Harris E. 2010. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J Virol 84:6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friebe P, Shi PY, Harris E. 2011. The 5′ and 3′ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J Virol 85:1900–1905. doi: 10.1128/JVI.02037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, Zhao H, Yu JY, Qin CF. 2016. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. eLife 5:e17636. doi: 10.7554/eLife.17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu ZY, Li XF, Jiang T, Deng YQ, Zhao H, Wang HJ, Ye Q, Zhu SY, Qiu Y, Zhou X, Qin ED, Qin C. 2013. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J Virol 87:6804–6818. doi: 10.1128/JVI.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M. 2003. Mfold web server for Nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuker M, Jacobson AB. 1998. Using reliability information to annotate RNA secondary structures. RNA 4:669–679. doi: 10.1017/S1355838298980116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Deng YQ, Wang S, Ma F, Aliyari R, Huang XY, Zhang NN, Watanabe M, Dong HL, Liu P, Li XF, Ye Q, Tian M, Hong S, Fan J, Zhao H, Li L, Vishlaghi N, Buth JE, Au C, Liu Y, Lu N, Du P, Qin FX, Zhang B, Gong D, Dai X, Sun R, Novitch BG, Xu Z, Qin CF, Cheng G. 2017. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity 46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Shan C, Zou J, Muruato AE, Bruno DN, de Almeida Medeiros Daniele B, Vasconcelos PF, Rossi SL, Weaver SC, Xie X, Shi PY. 2017. A cDNA clone-launched platform for high-yield production of inactivated Zika vaccine. EBioMedicine 17:145–156. doi: 10.1016/j.ebiom.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atieh T, Baronti C, de Lamballerie X, Nougairede A. 2016. Simple reverse genetics systems for Asian and African Zika viruses. Sci Rep 6:39384. doi: 10.1038/srep39384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Widman DG, Young E, Yount BL, Plante KS, Gallichotte EN, Carbaugh DL, Peck KM, Plante J, Swanstrom J, Heise MT, Lazear HM, Baric RS. 2017. A reverse genetics platform that spans the Zika virus family tree. mBio 8:e02014-16. doi: 10.1128/mBio.02014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuura M, Noah JW, Lambowitz AM. 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clyde K, Harris E. 2006. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol 80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiu WW, Kinney RM, Dreher TW. 2005. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J Virol 79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villordo SM, Filomatori CV, Sanchez-Vargas I, Blair CD, Gamarnik AV. 2015. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog 11:e1004604. doi: 10.1371/journal.ppat.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye Q, Li XF, Zhao H, Li SH, Deng YQ, Cao RY, Song KY, Wang HJ, Hua RH, Yu YX, Zhou X, Qin ED, Qin CF. 2012. A single nucleotide mutation in NS2A of Japanese encephalitis-live vaccine virus (SA14-14-2) ablates NS1′ formation and contributes to attenuation. J Gen Virol 93:1959–1964. doi: 10.1099/vir.0.043844-0. [DOI] [PubMed] [Google Scholar]
- 59.Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. eLife 3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roby JA, Pijlman GP, Wilusz J, Khromykh AA. 2014. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses 6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi PY, Nix JC, Beckham JD, Kieft JS. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354:1148–1152. doi: 10.1126/science.aah3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darty K, Denise A, Ponty Y. 2009. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]