ABSTRACT
Epstein-Barr virus latent membrane protein 1 (LMP1) is expressed in multiple human malignancies, including nasopharyngeal carcinoma and Hodgkin and immunosuppression-associated lymphomas. LMP1 mimics CD40 signaling to activate multiple growth and survival pathways, in particular, NF-κB. LMP1 has critical roles in Epstein-Barr virus (EBV)-driven B-cell transformation, and its expression causes fatal lymphoproliferative disease in immunosuppressed mice. Here, we review recent developments in studies of LMP1 signaling, LMP1-induced host dependency factors, mouse models of LMP1 lymphomagenesis, and anti-LMP1 immunotherapy approaches.
KEYWORDS: gamma herpesvirus, NF-κB, CD40, super-enhancer, oncogene, LMP1
INTRODUCTION
Epstein-Barr virus (EBV) was the first human tumor virus identified (1) and is associated with 200,000 human cancers annually (2). EBV's potent B-cell growth-transforming properties became apparent shortly after its discovery, providing an important insight into the association between EBV and human cancers (3, 4). Subsequent studies identified that EBV transforms primary B cells through expression of its latency III program, comprising nine viral proteins, two small RNAs, and miRNAs.
Elegant reverse genetic analyses revealed that only five EBV oncoproteins and viral miRNAs are necessary for the conversion of primary B cells into continuously proliferating lymphoblastoid cell lines (LCLs) (5, 6). Of these, only latent membrane protein 1 (LMP1) transforms rodent fibroblasts (7) and is sufficient to cause B-cell lymphoproliferative disease in mice (8, 9). LMP1 is highly expressed in immune suppression-associated lymphomas, including posttransplant lymphoproliferative disease and Hodgkin lymphoma (5).
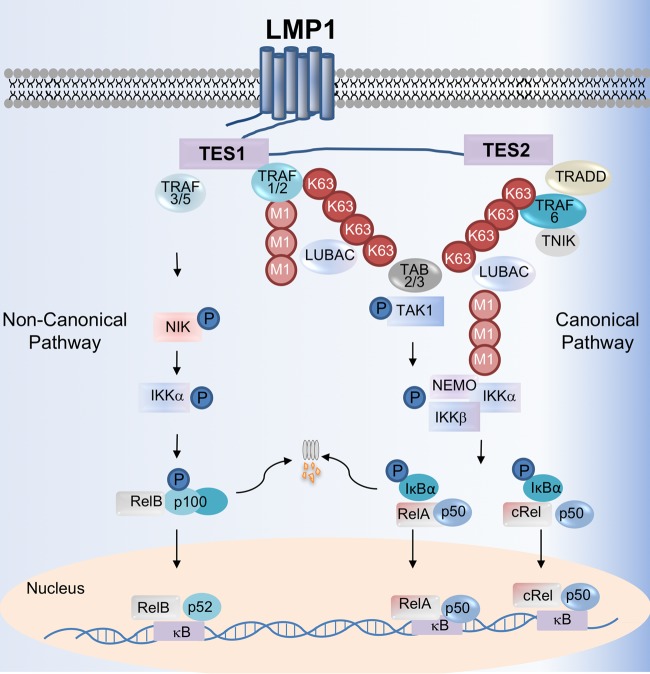
LMP1 signals from lipid rafts to induce proliferation, survival, migration, and immune evasion pathways (10, 11). Transmembrane domains induce LMP1 clustering to activate signaling from a 200-residue cytoplasmic tail. A reverse genetic analysis identified two LMP1 signaling domains (Fig. 1) critical for EBV-mediated B-cell growth transformation (12, 38, 87, 88). Together, these transformation effector site (TES)/C-terminal activation region (CTAR) 1 and 2 mimic CD40, a member of the tumor necrosis factor (TNF) receptor family and key B-cell costimulatory receptor. LMP1 constitutively activates NF-κB, mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase, and interferon regulatory factor pathways. The LMP1 CTAR3 domain also activates SUMOylation pathways to control migration and promote latency (13–15).
FIG 1.
LMP1-medated canonical and noncanonical NF-κB activation pathways. The LMP1 TES2 domain recruits TRADD to activate TRAF6, which assembles K63-linked pUb chains. K63 pUb chains recruit and activate the kinase TAK1 and the LUBAC complex, which attaches methionine 1-linked (M1) pUb chains to NEMO. TAK1 activates IKKβ, which then phosphorylates IκBα to enable RelA:p50 and cRel:p50 nuclear translocation. Canonical NF-κB is further activated by TES1, which recruits TRAF1 and TRAF2, likely as a heterotrimer. TES1 triggers K63 pUb chain attachment to TRAF2, which together with TRAF1, recruits and activates LUBAC. K63 and M1-linked pUb chains facilitate IKKβ activation by TAK1. LMP1 TES1 also stimulates noncanonical NF-κB activation, perhaps by sequestering TRAF3 to enable accumulation of the kinase NIK. IKKα is then activated by NIK and phosphorylates p100 to trigger its proteasomal processing into the mature p52 NF-κB subunit. p52 complexes translocate into the nucleus to regulate target gene expression. Not shown here, concurrent LMP1-mediated canonical and noncanonical pathway activation causes nuclear translocation of up to 13 NF-κB transcription factor dimers.
In this GEM, we focus on recently identified aspects of LMP1-mediated NF-κB activation, which is critical for lymphoblastoid B-cell transformation, growth, and survival, and is therefore a potential therapeutic target. Yet, genomic approaches identified thousands of LMP1/NF-κB target genes, raising the question of which are critical for LMP1-mediated oncogenicity. We highlight the use of chromatin immunoprecipitation sequencing (ChIP-seq) and clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 genetic approaches to identify key LMP1 and viral super-enhancer-induced host dependency factors. We also review recent advances in mouse models of LMP1 lymphomagenesis and adoptive immunotherapy approaches to eradicate LMP1+ cells. Readers are referred to a recent LMP1 review for aspects not covered here due to space constraints (16).
LMP1-MEDIATED CANONICAL NF-κB PATHWAY
LMP1-mediated canonical NF-κB activation remodels the host cell transcriptional program (2) and is critical for LCL survival (17, 18). Canonical NF-κB blockade decreases the mitochondrial membrane potential and rapidly induces apoptosis (18, 19). Hence, it is of significant interest to understand how LMP1 TES2 activates NF-κB.
TES2 initiates NF-κB activity by recruiting the adaptor protein TNF receptor (TNFR) associated death domain (TRADD). Yet, in contrast to TNFR, TES2 binds the TRADD death domain to prevent caspase activation (Fig. 1) (20, 21). Also, in contrast to the TNFR pathway, TRAF6 is apparently the first ubiquitin ligase activated by TES2 and is essential for downstream NF-κB and MAPK signaling (22–25). While CD40 and LMP1 each use the germinal center kinase TNIK to activate TRAF6 (26), CD40 does not signal through TRADD. Taken together, these observations indicate that TES2 initiates canonical NF-κB and MAPK signaling by a unique mechanism, and a key future objective will be to identify LMP1-selective chemical antagonists.
TRAF6 catalyzes the assembly of lysine 63-linked polyubiquitin (K63 pUb) chains, which recruit and activate TAK1, a key MAPK and canonical NF-κB pathway kinase (22, 27, 28). We recently found that LCL LMP1 signalosomes are highly K63-modified (29) and may serve to recruit TAK1 to LMP1 complexes. K63 pUb chains also recruit the linear ubiquitin assembly complex (LUBAC), which catalyzes methionine (or M1)-linked pUb chains. LMP1 complexes are also highly decorated by M1 pUb chains (29), and LUBAC is critical for TES2 NF-κB activation (22, 30). LMP1 also triggers M1 chain attachment to the IκB kinase (IKK) component NEMO (IKKγ), a key regulatory protein of the kinase IKKβ (30). The NEMO UBAN domain binds to M1 pUb chains and is necessary for LMP1 canonical NF-κB activation (31). RNA interference (RNAi) or CRISPR disruption of LUBAC subunit HOIP impairs LCL growth and triggers LCL apoptosis (29, 30). Ubiquitin chains may therefore juxtapose TAK1 with its target IKK-β to drive TES2-medaited NF-κB activation, which culminates in degradation of the IκBα inhibitor and nuclear translocation of RelA- or cRel-containing dimeric NF-κB transcription factors (Fig. 1).
TES1 independently activates canonical NF-κB (32). We recently reported that TES1 induces K63 pUb chain attachment to TRAF2, which together with TRAF1, recruits and activates LUBAC (29) (Fig. 1). Interestingly, the human T-cell leukemia virus TAX oncoprotein catalyzes mixed K63-M1 pUb chains that instead juxtapose TAK1 and IKK complexes (33, 34). Further studies are required to determine whether mixed K63-M1 pUb chains also signal downstream of LMP1.
LMP1 NONCANONICAL NF-κB PATHWAY
The noncanonical NF-κB pathway has important roles in B-cell development, activation, and survival. LMP1 TES1 strongly induces noncanonical NF-κB signaling (32, 35–37), perhaps underlying the observation that TES1 is required for EBV-mediated B-cell transformation (38). While the activation of additional pathways by TES1 may contribute to this phenotype (39), the key noncanonical pathway NF-κB subunit p52 is necessary for LCL growth and survival, underscoring LCL dependence on EBV-induced noncanonical pathway activity (40).
The mechanism by which LMP1 activates noncanonical NF-κB remains incompletely understood. In resting cells, noncanonical NF-κB activity is suppressed by TRAF3, which serves as an adaptor protein that binds to the cIAP1/2-TRAF2 Ub ligase complex and also to the key noncanonical pathway kinase NIK. In the presence of TRAF3, cIAP1/2-TRAF2 targets NIK for proteasomal degradation, preventing activation of its downstream target, IKKα. CD40 activates noncanonical activity by causing cIAP1/2-TRAF2 to instead target TRAF3 for degradation, which allows NIK to accumulate (41). NIK then phosphorylates the kinase IKKα, which induces proteasomal processing of the NF-κB subunit p100 precursor into the mature p52 form. p52-containing NF-κB dimers then translocate to the nucleus to modulate target gene expression (Fig. 1).
LMP1 appears to use a mechanism distinct from CD40 to activate noncanonical pathway activity, since TRAF3 binds tightly to LMP1 and is robustly expressed in LCLs. While LMP1 and CD40 use a similar “PXQXT” motif to recruit TRAF3, additional hydrogen bond interactions enable LMP1 to more tightly associate with TRAF3 (42). LMP1 may therefore induce noncanonical pathway signaling by sequestering TRAF3 to disrupt its association with cIAP1/2-TRAF2 and/or NIK. In support of this, TRAF3 overexpression blocks LMP1-mediated noncanonical NF-κB activation (43), and dominant negative TRAF3 expression inhibits LMP1-driven fibroblast transformation (39). LMP1 also binds to TRAF2 (44), yet molecular TRAF2 roles downstream of TES1 remain incompletely understood. Ultimately, LMP1 TES1 activates NIK and IKKα, which are each critical for TES1-induced noncanonical NF-κB activity (35, 45).
LYMPHOBLASTOID B-CELL NF-κB GENOMIC BINDING LANDSCAPE
Microarray profiling studies suggest that LMP1 targets thousands of host genes in epithelial and B lymphocytes. To systematically investigate how LMP1-activated NF-κB subunits target host genes, we performed chromatin immunoprecipitation of the LCL NF-κB transcription factor subunits RelA, RelB, cRel, p50, and p52 followed by deep sequencing (ChIP-seq) (46). An intriguing LCL NF-κB genomic binding landscape was identified, with 11 NF-κB distinct subunit binding patterns (SBPs) at enhancers and 10 at promoters. Subunits activated by both the LMP1 canonical and noncanonical pathways each contributed to most SBPs. This complexity may stem from the fact that concurrent LMP1 canonical and noncanonical pathway activities cause nuclear translocation of up to 13 distinct NF-κB dimeric transcription factors. Interestingly, exclusive occupancy by the prototypic NF-κB heterodimers RelA:p50, cRel:p50, and RelB:p52 are rare. Distinct gene ontology terms were enriched at each of these clusters, suggesting nonredundant LCL roles (46). These results provide insights into why LMP1 TES1 and TES2 domains may each be necessary to fully activate target genes during B-cell transformation, yet raise questions about the mechanisms that establish LCL enhancer and promoter SBPs, about when these patterns are established in the course of EBV-mediated B-cell transformation, and the extent to which individual NF-κB subunits are necessary for target gene regulation at these sites.
Nearly one-third of LCL genome NF-κB-occupied sites do not have an identifiable κB DNA motif, suggesting that NF-κB may be recruited to these sites via crosstalk with other EBV-activated pathways (46). For instance, the IRF4 DNA motif is highly enriched at promoter sites occupied only by LMP1/noncanonical pathway-activated p52. EBNA3C and LMP1 upregulate IRF4 expression (17, 47), which may then tether p52 to these sites. Similarly, an E-box motif is enriched at LCL promoters bound predominantly by LMP1/canonical pathway-activated cRel, suggesting that an E-box-bound host transcription factor such as c-Myc may tether cRel homodimers to these sites. Crosstalk may underlie key NF-κB subunit-specific roles downstream of LMP1, which await further characterization.
Crosstalk between NF-κB and other nuclear transcription factors likely shapes many LMP1 effects on host gene expression. Unexpectedly, ChIP-seq analysis revealed that the forkhead box transcription factor FoxM1 co-occupies nearly half of all LCL NF-κB sites (46). The κB DNA motif, rather than a forkhead box motif, is highly enriched at these sites, suggesting that DNA-bound NF-κB may recruit FoxM1. FoxM1 knockdown significantly impairs the expression of LCL NF-κB target genes and triggers rapid LCL apoptosis, suggesting that FoxM1 is a key cofactor at these sites. Further studies are required to identify how EBV upregulates and activates FoxM1, how FoxM1 is recruited to NF-κB-bound LCL sites, and in particular, how it coactivates the expression of key targets such as Tak1 and cIAP2 (46).
VIRAL SUPER-ENHANCERS TARGET KEY HOST DEPENDENCY FACTORS
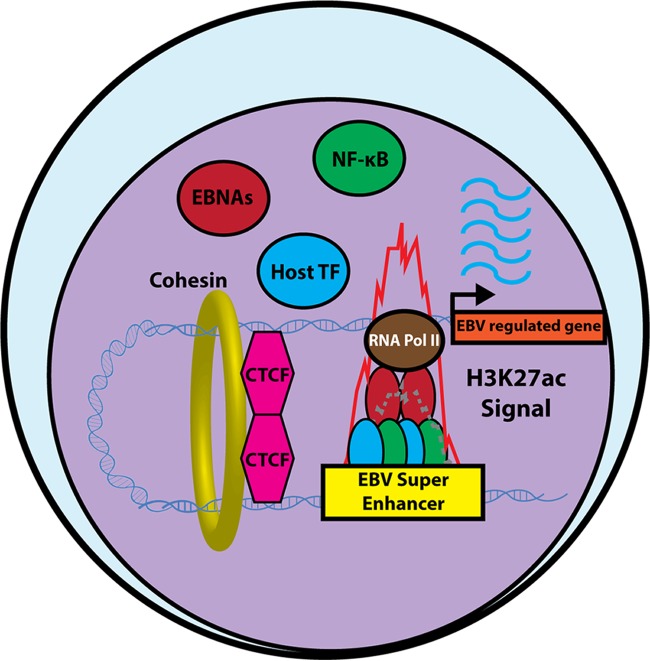
Super-enhancers (SE) are transcriptional elements that strongly upregulate genes important for cell identity and an oncogenic state (48). LCL ChIP-seq analysis identified the first viral super-enhancers, comprising five LMP1-activated NF-κB subunits and four EBNAs (49) (Fig. 2). Host genes targeted by the 187 EBV SE are expressed more highly than other LCL genes and include BCL2, c-Myc and the oncogenic micro-RNA miR-155. Interestingly, c-Myc is targeted by independent EBV SE, which loop to the c-Myc promoter from 525 and 428 kb upstream (50–52). CRISPR/Cas9-mediated knockout of either SE strongly impairs c-Myc expression and induces LCL death, underscoring EBV SE roles in lymphoblastoid B-cell growth and survival. EBV SE are exquisitely sensitive to NF-κB or BET bromodomain antagonists, such as the small molecule JQ1, which arrest LCL growth and induces apoptosis (49).
FIG 2.
LMP1-activated NF-κB transcription factors and EBNAs form EBV super-enhancers. In B cells with the EBV latency III expression program, LMP1-activated NF-κB, as well as EBNA2, EBNALP, EBNA3A, and EBNA3C form 187 EBV super-enhancers, together with other host transcription factors. Histone 3 lysine 27 acetylation (H3K27ac, shown in red) and polymerase (Pol) II ChIP-seq signals are markedly elevated at these sites. CTCF and cohesin-mediated DNA looping enables a subset of EBV super-enhancers to target host promoters by long-range DNA interactions. EBV super-enhancer targets are more highly expressed than LCL genes targeted by typical enhancers and drive the expression of key EBV-induced host dependency factors.
We used genome-wide CRISPR/Cas9 loss-of-function screens to systematically identify key EBV-induced host dependency factors. The top screen hits were enriched for EBV SE targets and included cFLIP, BATF, IRF4, and IRF2, which were found to be critical for evasion of EBV-induced tumor suppressor responses (40). Perhaps as an oncogene-induced stress response, the EBV latency III program stimulates lymphoblastoid B-cell TNF-α production, which necessitates LMP1-induced cFLIP to block extrinsic apoptosis and necroptosis pathways. LMP1-induced cFLIP may also protect cells in vivo from TNF-α produced by cell-mediated immune responses to EBV infection.
EBV oncogenic stress is a potent stimulus for the transcriptional upregulation of the intrinsic apoptosis executioner protein BIM. To circumvent BIM induction, LMP1 and EBNA proteins upregulate the transcription factors BATF and IRF4, which together with EBNA3 repressor complexes, block BIM induction. CRISPR knockout of IRF4 or BATF strongly induces BIM and triggers rapid LCL apoptosis. BATF/Jun and IRF4 cooperatively bind to a composite BIM promoter AP-1-interferon site, which they co-occupy with EBNA3 proteins. Taken together with the finding that the BATF/IRF4 motif is enriched at LCL EBNA3A and EBNA3C binding sites (53, 54), we hypothesize that BATF/IRF4 tethers EBNA3 proteins to DNA to enable the assembly of repressor complexes at key LCL genomic sites, including BIM.
EBV IRF4 dependency triggers another tumor suppressor response, since a key IRF4 B-cell target gene is Blimp1, a master regulator of plasma cell differentiation that blocks c-Myc. To overcome this barrier, LMP1, EBNA3, and EBV miRNAs each downmodulate Blimp1 (40, 55, 56). LMP1 upregulates IRF2 and its corepressor IRF2BP2 via EBV SE, which may form repressor complexes at a Blimp1 promoter interferon response element to counteract IRF4 effects on Blimp1 (40). EBNA3A and EBNA3C also block LCL Blimp1 upregulation, and withdrawal of EBNA3A and EBNA3C expression triggered plasma cell differentiation (56). Further studies are required to determine whether LMP1-activated IRF4, BATF4, and perhaps IRF2 tether EBNA3 corepressor complexes at the Blimp1 promoter. Underscoring the EBV need to counteract Blimp1, EBV-miR-BHRF1-2 further reduces Blimp1 expression (57).
LMP1 PROMOTES AEROBIC GLYCOLYSIS
LMP1 acts as a metabolism master regulator in lymphoblastoid and nasopharyngeal carcinoma (NPC) cells by promoting aerobic glycolysis, the so-called Warburg effect that supplies transformed cells with ATP and anabolic building blocks (58). In a pioneering study, LMP1-mediated NF-κB and AKT/PI3K pathways were found to enhance the transcription and plasma membrane translocation of glucose transporter 1 (GLUT1) (58). LMP1 signaling also increases the GLUT1 half-life to further upregulate GLUT1 (59) and also enhances glycolytic flux by upregulating the first glycolysis pathway enzyme, hexokinase 2 (60). NF-κB inhibition represses glucose uptake and induces autophagy and caspase-independent cell death (58).
LMP1 plays a key role in establishing the Warburg effect in newly infected primary human B cells, where glycolytic flux correlates with LMP1 levels during growth transformation (61). LMP1 is sufficient to induce aerobic glycolysis in NPC cells (60), and GLUT1 depletion suppresses nasopharyngeal epithelial cell aerobic glycolysis, proliferation, and colony formation (62). Further studies are required to determine whether additional EBV latency proteins contribute to this phenotype. Interestingly, LMP1-mediated aerobic glycolysis drives the expansion of myeloid-derived suppressor cells in the NPC tumor microenvironment (59), which may facilitate immune escape. LMP1 is also secreted from NPC and B cells in exosome vesicles and further shapes the tumor microenvironment (63–65).
MOUSE MODELS OF LMP1 LYMPHOMAGENESIS
Mouse models have provided key insights into LMP1 oncogenicity in vivo. In classic studies, LMP1 was found to transform rodent fibroblasts, to drive epithelial xenograft tumor formation (7, 66), and to accelerate B-cell lymphomagenesis (9). When expressed alone or together with LMP2A from an early stage of B-cell development, immune surveillance prevents lymphomagenesis (8, 67). Notably, the loss of T-cell surveillance results in LMP1-driven fatal lymphoproliferative disease within 60 days (8).
Most EBV-driven lymphomas arise from germinal center (GC) B cells, where LMP1 and LMP2A are coexpressed together with EBNA1 in the EBV latency II expression program. EBV latency II is observed in Hodgkin lymphoma (HL) and also in NPC. GC B-cell LMP1 and LMP2A coexpression causes rapidly fatal lymphoproliferative disease in T-cell deficient mice (68), further highlighting the key role of immune surveillance in countering LMP1 oncogenicity. NK cells also have key host defense roles necessary for host defense against EBV, and combined T/NK-cell depletion results in massive LMP1/LMP2A-driven GC B-cell outgrowth, plasmablast differentiation, and death within 12 days (69).
HL is thought to arise from crippled GC B cells, rescued from apoptosis by oncogenic mutations or by EBV infection, perhaps explaining why nearly 40% of HLs are EBV infected (70). In support of a pathogenic LMP1 role in HL, NF-κB activation mutations are observed at significantly higher frequencies in EBV-negative HL tumors then in EBV-infected samples (70). GC B-cell LMP1 and LMP2A coexpression induces transcriptional changes that overlap gene signatures found in classical Hodgkin Reed-Sternberg (HRS) cells. These include the expression of the therapeutic target CD30 (69) and the non-B-cell lineage markers perforin and granzyme, which are uniquely coexpressed in HRS cells.
LMP1 knockout markedly impairs EBV lymphomagenesis in a cord-blood humanized mouse model. In the absence of LMP1, low frequency development of EBV+ lymphomas was dependent on CD40 signaling induced by T cells, which likely provides similar growth and survival signals as LMP1, albeit at lower levels (71). Simultaneous LMP1 and LMP2A deletion further reduced and delayed the onset of tumors but did not completely eliminate lymphoma development (72).
It will be interesting to identify key CD40-induced host dependency factors in these models, the extent to which they overlap identified EBV-induced LCL dependency factors, and whether CD40-mediated NF-κB activation can substitute for that of LMP1 in driving assembly of NF-κB- and EBNA-containing SE at key sites, such as cFLIP, IRF4, and IRF2. A limitation of currently available humanized mouse models is that germinal center development is impaired, and consequently, cells with EBV latency II expression are rarely observed. As humanized models continue to develop, it will be of significant interest to study LMP1 roles in latency II GC B cells.
IMMUNOTHERAPEUTIC APPROACHES TARGETING LMP1
LMP1 lacks enzymatic function and has not yet proven to be a druggable target. However, the expression of LMP1 by epithelial and B-cell malignancies is increasingly being targeted in adoptive T-cell immunotherapies. LMP1-specific cytotoxic T lymphocytes (CTLs) can be found in most EBV+ individuals at a low frequency and can be expanded in vitro for adoptive immunotherapy. Adoptive CTL approaches were first used to treat lymphoproliferative diseases in patients receiving transplants of hematopoietic stem cells, which express the full complement of EBV latency III antigens (73, 74).
Adoptive immunotherapy approaches are also in development to overcome the less immunogenic EBV latency II antigens. LMP1- and LMP2A-specific cytotoxic T cells have induced remission in patients with high-risk or relapsed EBV+ Hodgkin and non-Hodgkin lymphomas (74, 75). Adoptive CTL approaches targeting latency II antigens are in development for the treatment of NPC (76). An adenoviral vector-based vaccine has shown promise for targeting LMP2A, EBNA1, and, when expressed, LMP1 in nasopharyngeal carcinoma (77–79). LMP1 drives the expression of the T-cell inhibitory receptor PD-L1 (80–82), and PD-1-PD-L1 immune checkpoint blockade has improved EBV+ lymphoma and NPC treatment responses (19, 83–85). It will be of interest to determine whether checkpoint-blockade, perhaps together with strategies to induce EBV lytic gene expression, synergizes with adoptive immunotherapy approaches.
FUTURE DIRECTIONS
Despite significant advances in the understanding of LMP1-mediated oncogenicity, many key questions remain. Within the cytosol, a more complete understanding of how LMP1 TES1 and TES2 initiate NF-κB signaling is an important objective, as this appears to significantly differentiate LMP1 from CD40. It is likely that additional pathway components remain to be identified, and focused genetic and proteomic approaches promise to identify LMP1 signalosome components selectively important for TES1-mediated NIK stabilization and for TES2-mediated TRAF6 activation.
CRISPR/Cas9 engineering should enable focused studies of NF-κB pathway-specific roles, including at early time points after primary B-cell infection by EBV. For instance, recent studies indicate that LMP1 is expressed with delayed kinetics after primary B-cell infection (86). The knockout of key pathway components promises to reveal why LMP1-induced canonical and noncanonical NF-κB are each critical for LCL growth and survival, when they first become critical dependency factors, and how the crosstalk between LMP1-activated NF-κB, MAPK, PI3 kinase, and interferon regulatory factor pathways sculpts target gene regulation. Similarly, LMP1 and LMP2A are typically coexpressed and colocalize in cell membranes, yet mechanisms by which they synergistically regulate a wide range of GC B-cell target genes are yet to be identified (69).
The major principles of NF-κB nuclear regulation downstream of LMP1 await further studies. CRISPR/Cas9 analysis suggests that multiple NF-κB subunits are critical for lymphoblastoid B-cell survival, yet little is presently known about their unique dependency factor roles. For instance, defining the mechanisms by which up to 13 distinct LMP1-activated NF-κB transcription factor dimers are targeted to distinct LCL enhancer and promoter sites will be an important objective.
EBV SE are highly sensitive to perturbation and are potentially druggable therapeutic targets. It will therefore be of significant interest to determine how LMP1-activated NF-κB and EBNA establish EBV SE, why they form at particular LCL sites, and whether EBNAs and NF-κB interact within viral SE and to define other host transcription factors necessary for their formation. Similarly, how EBV SE target LCL promoters by long-range DNA interactions and, specifically, whether NF-κB promotes DNA looping at these sites are open questions. Do LMP1 and LMP2A trigger the formation of super-enhancers in HL and NPC, where they are expressed in the absence of EBNA2, EBNA3A, EBNA3C, or EBNALP? If so, what are their genomic targets? What are the key EBV-induced host dependency factors in these tumors?
EBV latency III expression causes transformed B-cell addiction to host cell factors. It may ultimately be possible to use small molecule approaches to exploit these LMP1-induced synthetic lethal dependencies, including LMP1-mediated induction of cFLIP, IRF2, and IRF4. Indeed, transcription factors have increasingly become potentially druggable targets, for instance, as the substrates for small molecule-induced ubiquitin proteasome pathway degradation.
ACKNOWLEDGMENTS
We apologize to colleagues whose work we could not cite due to space limitations.
This work, including the efforts of Benjamin E. Gewurz, was funded by a Burroughs Wellcome Foundation Career Award in Medical Sciences and by the Brigham & Women's Hospital. S.J. is funded by a Howard Hughes Predoctoral Fellowship. L.W. is supported by a National Science Scholarship (Ph.D.) from Singapore’s Agency for Science, Technology and Research (A*STAR).
REFERENCES
- 1.Epstein A. 2015. Why and how Epstein-Barr Virus was discovered 50 years ago. Curr Top Microbiol Immunol 390:3–15. doi: 10.1007/978-3-319-22822-8_1. [DOI] [PubMed] [Google Scholar]
- 2.Gewurz BE, Mar JC, Padi M, Zhao B, Shinners NP, Takasaki K, Bedoya E, Zou JY, Cahir-McFarland E, Quackenbush J, Kieff E. 2011. Canonical NF-kappaB activation is essential for Epstein-Barr virus latent membrane protein 1 TES2/CTAR2 gene regulation. J Virol 85:6764–6773. doi: 10.1128/JVI.00422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pope JH, Horne MK, Scott W. 1968. Transformation of foetal human leukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int J Cancer 3:857–866. doi: 10.1002/ijc.2910030619. [DOI] [PubMed] [Google Scholar]
- 4.Pattengale PK, Smith RW, Gerber P. 1973. Selective transformation of B lymphocytes by E.B. virus. Lancet ii:93–94. doi: 10.1016/S0140-6736(73)93286-8. [DOI] [PubMed] [Google Scholar]
- 5.Longnecker R, Kieff E, Cohen JI. 2013. Epstein-Barr virus, p 1898–1959. In Knipe DM, Howley P.M. (ed), Fields virology, 6th ed, vol 2 Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Feederle R, Haar J, Bernhardt K, Linnstaedt SD, Bannert H, Lips H, Cullen BR, Delecluse HJ. 2011. The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes. J Virol 85:9801–9810. doi: 10.1128/JVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, Homig-Holzel C, Wang Z, Derudder E, Li S, Chakraborty T, Cotter SE, Koyama S, Currie T, Freeman GJ, Kutok JL, Rodig SJ, Dranoff G, Rajewsky K. 2012. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell 148:739–751. doi: 10.1016/j.cell.2011.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulwichit W, Edwards RH, Davenport EM, Baskar JF, Godfrey V, Raab-Traub N. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc Natl Acad Sci U S A 95:11963–11968. doi: 10.1073/pnas.95.20.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi M, Izumi KM, Kieff E. 2001. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc Natl Acad Sci U S A 98:4675–4680. doi: 10.1073/pnas.081075298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meckes DG Jr, Menaker NF, Raab-Traub N. 2013. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol 87:1301–1311. doi: 10.1128/JVI.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentz GL, Whitehurst CB, Pagano JS. 2011. Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J Virol 85:10144–10153. doi: 10.1128/JVI.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentz GL, Moss CR II, Whitehurst CB, Moody CA, Pagano JS. 2015. LMP1-induced sumoylation influences the maintenance of Epstein-Barr virus latency through KAP1. J Virol 89:7465–7477. doi: 10.1128/JVI.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumi KM, Cahir-McFarland ED, Riley EA, Rizzo D, Chen Y, Kieff E. 1999. The residues between the two transformation effector sites of Epstein-Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation. J Virol 73:9908–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieser A, Sterz KR. 2015. The latent membrane protein 1 (LMP1). Curr Top Microbiol Immunol 391:119–149. doi: 10.1007/978-3-319-22834-1_4. [DOI] [PubMed] [Google Scholar]
- 17.Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. 2004. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol 78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller SA, Hernandez-Hopkins D, Vider J, Ponomarev V, Hyjek E, Schattner EJ, Cesarman E. 2006. NF-kappaB is essential for the progression of KSHV- and EBV-infected lymphomas in vivo. Blood 107:3295–3302. doi: 10.1182/blood-2005-07-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman A, Patel SP, Kurzrock R. 2017. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol 14:203–220. doi: 10.1038/nrclinonc.2016.168. [DOI] [PubMed] [Google Scholar]
- 20.Schneider F, Neugebauer J, Griese J, Liefold N, Kutz H, Briseno C, Kieser A. 2008. The viral oncoprotein LMP1 exploits TRADD for signaling by masking its apoptotic activity. PLoS Biol 6:e8. doi: 10.1371/journal.pbio.0060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi KM, Cahir-McFarland ED, Ting AT, Riley EA, Seed B, Kieff ED. 1999. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-kappaB activation. Mol Cell Biol 19:5759–5767. doi: 10.1128/MCB.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gewurz BE, Towfic F, Mar JC, Shinners NP, Takasaki K, Zhao B, Cahir-McFarland ED, Quackenbush J, Xavier RJ, Kieff E. 2012. Genome-wide siRNA screen for mediators of NF-kappaB activation. Proc Natl Acad Sci U S A 109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue J, Nakano H, Mak TW, Yeh WC, Li X, Akira S, Suzuki N, Suzuki S, Mosialos G, Kieff E. 2003. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci U S A 100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultheiss U, Puschner S, Kremmer E, Mak TW, Engelmann H, Hammerschmidt W, Kieser A. 2001. TRAF6 is a critical mediator of signal transduction by the viral oncogene latent membrane protein 1. EMBO J 20:5678–5691. doi: 10.1093/emboj/20.20.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arcipowski KM, Stunz LL, Bishop GA. 2014. TRAF6 is a critical regulator of LMP1 functions in vivo. Int Immunol 26:149–158. doi: 10.1093/intimm/dxt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shkoda A, Town JA, Griese J, Romio M, Sarioglu H, Knofel T, Giehler F, Kieser A. 2012. The germinal center kinase TNIK is required for canonical NF-kappaB and JNK signaling in B cells by the EBV oncoprotein LMP1 and the CD40 receptor. PLoS Biol 10:e1001376. doi: 10.1371/journal.pbio.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Nakano H, Wu Z. 2006. The C-terminal activating region 2 of the Epstein-Barr virus-encoded latent membrane protein 1 activates NF-kappaB through TRAF6 and TAK1. J Biol Chem 281:2162–2169. doi: 10.1074/jbc.M505903200. [DOI] [PubMed] [Google Scholar]
- 28.Arcipowski KM, Bishop GA. 2012. Roles of the kinase TAK1 in TRAF6-dependent signaling by CD40 and its oncogenic viral mimic, LMP1. PLoS One 7:e42478. doi: 10.1371/journal.pone.0042478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenfeld H, Takasaki K, Walsh MJ, Ersing I, Bernhardt K, Ma Y, Fu B, Ashbaugh CW, Cabo J, Mollo SB, Zhou H, Li S, Gewurz BE. 2015. TRAF1 coordinates polyubiquitin signaling to enhance Epstein-Barr virus LMP1-mediated growth and survival pathway activation. PLoS Pathog 11:e1004890. doi: 10.1371/journal.ppat.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Wang Y, Zhao J, Ren J, Hall KH, Moorman JP, Yao ZQ, Ning S. 2017. The linear ubiquitin assembly complex modulates latent membrane protein 1 activation of NF-kappaB and Interferon regulatory factor 7. J Virol 91:e01138-16. doi: 10.1128/JVI.01138-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehm D, Gewurz BE, Kieff E, Cahir-McFarland E. 2010. Epstein-Barr latent membrane protein 1 transformation site 2 activates NF-kappaB in the absence of NF-kappaB essential modifier residues 133–224 or 373–419. Proc Natl Acad Sci U S A 107:18103–18108. doi: 10.1073/pnas.1011752107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kung CP, Raab-Traub N. 2010. Epstein-Barr virus latent membrane protein 1 modulates distinctive NF-kappaB pathways through C terminus-activating region 1 to regulate epidermal growth factor receptor expression. J Virol 84:6605–6614. doi: 10.1128/JVI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata Y, Tokunaga F, Goto E, Komatsu G, Gohda J, Saeki Y, Tanaka K, Takahashi H, Sawasaki T, Inoue S, Oshiumi H, Seya T, Nakano H, Tanaka Y, Iwai K, Inoue JI. 2017. HTLV-1 tax induces formation of the active macromolecular IKK complex by Generating Lys63- and Met1-linked hybrid polyubiquitin chains. PLoS Pathog 13:e1006162. doi: 10.1371/journal.ppat.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Long W, Peng C, Hu L, Zhang Q, Wu A, Zhang X, Duan X, Wong CC, Tanaka Y, Xia Z. 2016. HTLV-1 tax functions as a ubiquitin E3 ligase for direct IKK activation via synthesis of mixed-linkage polyubiquitin chains. PLoS Pathog 12:e1005584. doi: 10.1371/journal.ppat.1005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc Natl Acad Sci U S A 101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eliopoulos AG, Caamano JH, Flavell J, Reynolds GM, Murray PG, Poyet JL, Young LS. 2003. Epstein-Barr virus-encoded latent infection membrane protein 1 regulates the processing of p100 NF-kappaB2 to p52 via an IKKgamma/NEMO-independent signalling pathway. Oncogene 22:7557–7569. doi: 10.1038/sj.onc.1207120. [DOI] [PubMed] [Google Scholar]
- 37.Saito N, Courtois G, Chiba A, Yamamoto N, Nitta T, Hironaka N, Rowe M, Yamamoto N, Yamaoka S. 2003. Two carboxyl-terminal activation regions of Epstein-Barr virus latent membrane protein 1 activate NF-kappaB through distinct signaling pathways in fibroblast cell lines. J Biol Chem 278:46565–46575. doi: 10.1074/jbc.M302549200. [DOI] [PubMed] [Google Scholar]
- 38.Izumi KM, Kaye KM, Kieff ED. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci U S A 94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mainou BA, Everly DN Jr, Raab-Traub N. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J Virol 81:9680–9692. doi: 10.1128/JVI.01001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, Jiang S, Jiang C, Zhao B, Root DE, Doench JG, Gewurz BE. 2017. CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors. Cell Host Microbe 21:580.e7–591.e7. doi: 10.1016/j.chom.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M. 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol 9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Xie P, Welsh K, Li C, Ni CZ, Zhu X, Reed JC, Satterthwait AC, Bishop GA, Ely KR. 2005. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J Biol Chem 280:33620–33626. doi: 10.1074/jbc.M502511200. [DOI] [PubMed] [Google Scholar]
- 43.Song YJ, Kang MS. 2010. Roles of TRAF2 and TRAF3 in Epstein-Barr virus latent membrane protein 1-induced alternative NF-kappaB activation. Virus Genes 41:174–180. doi: 10.1007/s11262-010-0505-4. [DOI] [PubMed] [Google Scholar]
- 44.Mosialos G, Birkenbach M, Yalamanchili R, vanArsdale T, Ware C, Kieff E. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 45.Sylla BS, Hung SC, Davidson DM, Hatzivassiliou E, Malinin NL, Wallach D, Gilmore TD, Kieff E, Mosialos G. 1998. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc Natl Acad Sci U S A 95:10106–10111. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, Zhou H, Mollo SB, Shi TT, Takasaki K, Jiang S, Cahir-McFarland E, Kellis M, Bulyk ML, Kieff E, Gewurz BE. 2014. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell Rep 8:1595–1606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee S, Lu J, Cai Q, Saha A, Jha HC, Dzeng RK, Robertson ES. 2013. The EBV latent antigen 3C inhibits apoptosis through targeted regulation of interferon regulatory factors 4 and 8. PLoS Pathog 9:e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H, Schmidt SC, Jiang S, Willox B, Bernhardt K, Liang J, Johannsen EC, Kharchenko P, Gewurz BE, Kieff E, Zhao B. 2015. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 17:205–216. doi: 10.1016/j.chom.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, Mar JC, Morton CC, Freedman ML, Blacklow SC, Aster JC, Bernstein BE, Kieff E. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A 108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood CD, Veenstra H, Khasnis S, Gunnell A, Webb HM, Shannon-Lowe C, Andrews S, Osborne CS, West MJ. 2016. MYC activation and BCL2L11 silencing by a tumour virus through the large-scale reconfiguration of enhancer-promoter hubs. eLife 5:e18270. doi: 10.7554/eLife.18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang S, Zhou H, Liang J, Gerdt C, Wang C, Ke L, Narita Y, Colson T, Gewurz BG, Li G, Kieff E, Zhao B. 3D genome landscape of Epstein-Barr virus oncoproteins and virus activated NF-kB in lymphoblastoid cells. Cell Host Microbe, in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt SC, Jiang S, Zhou H, Willox B, Holthaus AM, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. 2015. Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes. Proc Natl Acad Sci U S A 112:554–559. doi: 10.1073/pnas.1422580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang S, Willox B, Zhou H, Holthaus AM, Wang A, Shi TT, Maruo S, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. 2014. Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci U S A 111:421–426. doi: 10.1073/pnas.1321704111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vrzalikova K, Vockerodt M, Leonard S, Bell A, Wei W, Schrader A, Wright KL, Kube D, Rowe M, Woodman CB, Murray PG. 2011. Down-regulation of BLIMP1alpha by the EBV oncogene, LMP-1, disrupts the plasma cell differentiation program and prevents viral replication in B cells: implications for the pathogenesis of EBV-associated B-cell lymphomas. Blood 117:5907–5917. doi: 10.1182/blood-2010-09-307710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Styles CT, Bazot Q, Parker GA, White RE, Paschos K, Allday MJ. 2017. EBV epigenetically suppresses the B cell-to-plasma cell differentiation pathway while establishing long-term latency. PLoS Biol 15:e2001992. doi: 10.1371/journal.pbio.2001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma J, Nie K, Redmond D, Liu Y, Elemento O, Knowles DM, Tam W. 2016. EBV-miR-BHRF1-2 targets PRDM1/blimp1: potential role in EBV lymphomagenesis. Leukemia 30:594–604. doi: 10.1038/leu.2015.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sommermann TG, O'Neill K, Plas DR, Cahir-McFarland E. 2011. IKKbeta and NF-kappaB transcription govern lymphoma cell survival through AKT-induced plasma membrane trafficking of GLUT1. Cancer Res 71:7291–7300. doi: 10.1158/0008-5472.CAN-11-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai TT, Ye SB, Liu YN, He J, Chen QY, Mai HQ, Zhang CX, Cui J, Zhang XS, Busson P, Zeng YX, Li J. 2017. LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathog 13:e1006503. doi: 10.1371/journal.ppat.1006503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao L, Hu ZY, Dong X, Tan Z, Li W, Tang M, Chen L, Yang L, Tao Y, Jiang Y, Li J, Yi B, Li B, Fan S, You S, Deng X, Hu F, Feng L, Bode AM, Dong Z, Sun LQ, Cao Y. 2014. Targeting Epstein-Barr virus oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal carcinoma to radiation therapy. Oncogene 33:4568–4578. doi: 10.1038/onc.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McFadden K, Hafez AY, Kishton R, Messinger JE, Nikitin PA, Rathmell JC, Luftig MA. 2016. Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization. Proc Natl Acad Sci U S A 113:E782–E790. doi: 10.1073/pnas.1517141113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Jia L, Lin W, Yip YL, Lo KW, Lau VM, Zhu D, Tsang CM, Zhou Y, Deng W, Lung HL, Lung ML, Cheung LM, Tsao SW. 2017. Epstein-Barr virus-encoded latent membrane protein 1 upregulates glucose transporter 1 transcription via the mTORC1/NF-kappaB signaling pathways. J Virol 91:e02168-16. doi: 10.1128/JVI.02168-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG Jr. 2017. CD63 regulates Epstein-Barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-kappaB signaling. J Virol 91:e02251-16. doi: 10.1128/JVI.02251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, Middeldorp JM, Pegtel DM. 2011. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-kappaB activation. EMBO J 30:2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meckes DG Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baichwal VR, Sugden B. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461–467. [PubMed] [Google Scholar]
- 67.Vrazo AC, Chauchard M, Raab-Traub N, Longnecker R. 2012. Epstein-Barr virus LMP2A reduces hyperactivation induced by LMP1 to restore normal B cell phenotype in transgenic mice. PLoS Pathog 8:e1002662. doi: 10.1371/journal.ppat.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wirtz T, Weber T, Kracker S, Sommermann T, Rajewsky K, Yasuda T. 2016. Mouse model for acute Epstein-Barr virus infection. Proc Natl Acad Sci U S A 113:13821–13826. doi: 10.1073/pnas.1616574113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minamitani T, Ma Y, Zhou H, Kida H, Tsai CY, Obana M, Okuzaki D, Fujio Y, Kumanogoh A, Zhao B, Kikutani H, Kieff E, Gewurz BE, Yasui T. 2017. Mouse model of Epstein-Barr virus LMP1- and LMP2A-driven germinal center B-cell lymphoproliferative disease. Proc Natl Acad Sci U S A 114:4751–4756. doi: 10.1073/pnas.1701836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weniger MA, Kuppers R. 2016. NF-kappaB deregulation in Hodgkin lymphoma. Semin Cancer Biol 39:32–39. doi: 10.1016/j.semcancer.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 71.Ma SD, Xu X, Plowshay J, Ranheim EA, Burlingham WJ, Jensen JL, Asimakopoulos F, Tang W, Gulley ML, Cesarman E, Gumperz JE, Kenney SC. 2015. LMP1-deficient Epstein-Barr virus mutant requires T cells for lymphomagenesis. J Clin Invest 125:304–315. doi: 10.1172/JCI76357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma SD, Tsai MH, Romero-Masters JC, Ranheim EA, Huebner SM, Bristol JA, Delecluse HJ, Kenney SC. 2017. Latent membrane protein 1 (LMP1) and LMP2A collaborate to promote Epstein-Barr virus-induced B cell lymphomas in a cord blood-humanized mouse model but are not essential. J Virol 91:e01928-16. doi: 10.1128/JVI.01928-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doubrovina E, Oflaz-Sozmen B, Prockop SE, Kernan NA, Abramson S, Teruya-Feldstein J, Hedvat C, Chou JF, Heller G, Barker JN, Boulad F, Castro-Malaspina H, George D, Jakubowski A, Koehne G, Papadopoulos EB, Scaradavou A, Small TN, Khalaf R, Young JW, O'Reilly RJ. 2012. Adoptive immunotherapy with unselected or EBV-specific T cells for biopsy-proven EBV+ lymphomas after allogeneic hematopoietic cell transplantation. Blood 119:2644–2656. doi: 10.1182/blood-2011-08-371971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heslop HE, Slobod KS, Pule MA, Hale GA, Rousseau A, Smith CA, Bollard CM, Liu H, Wu MF, Rochester RJ, Amrolia PJ, Hurwitz JL, Brenner MK, Rooney CM. 2010. Long-term outcome of EBV-specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood 115:925–935. doi: 10.1182/blood-2009-08-239186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bollard CM, Gottschalk S, Torrano V, Diouf O, Ku S, Hazrat Y, Carrum G, Ramos C, Fayad L, Shpall EJ, Pro B, Liu H, Wu MF, Lee D, Sheehan AM, Zu Y, Gee AP, Brenner MK, Heslop HE, Rooney CM. 2014. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J Clin Oncol 32:798–808. doi: 10.1200/JCO.2013.51.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Chen QY, He J, Li ZL, Tang XF, Chen SP, Xie CM, Li YQ, Huang LX, Ye SB, Ke M, Tang LQ, Liu H, Zhang L, Guo SS, Xia JC, Zhang XS, Zheng LM, Guo X, Qian CN, Mai HQ, Zeng YX. 2015. Phase I trial of adoptively transferred tumor-infiltrating lymphocyte immunotherapy following concurrent chemoradiotherapy in patients with locoregionally advanced nasopharyngeal carcinoma. Oncoimmunology 4:e976507. doi: 10.4161/23723556.2014.976507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith C, Tsang J, Beagley L, Chua D, Lee V, Li V, Moss DJ, Coman W, Chan KH, Nicholls J, Kwong D, Khanna R. 2012. Effective treatment of metastatic forms of Epstein-Barr virus-associated nasopharyngeal carcinoma with a novel adenovirus-based adoptive immunotherapy. Cancer Res 72:1116–1125. doi: 10.1158/0008-5472.CAN-11-3399. [DOI] [PubMed] [Google Scholar]
- 78.Ngo MC, Ando J, Leen AM, Ennamuri S, Lapteva N, Vera JF, Min-Venditti A, Mims MP, Heslop HE, Bollard CM, Gottschalk S, Rooney CM. 2014. Complementation of antigen-presenting cells to generate T lymphocytes with broad target specificity. J Immunother 37:193–203. doi: 10.1097/CJI.0000000000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin X, Gudgeon NH, Hui EP, Jia H, Qun X, Taylor GS, Barnardo MC, Lin CK, Rickinson AB, Chan AT. 2008. CD4 and CD8 T cell responses to tumour-associated Epstein-Barr virus antigens in nasopharyngeal carcinoma patients. Cancer Immunol Immunother 57:963–975. doi: 10.1007/s00262-007-0427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O'Donnell E, Neuberg D, Shipp MA. 2012. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res 18:1611–1618. doi: 10.1158/1078-0432.CCR-11-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang W, Zhang J, Hong S, Zhan J, Chen N, Qin T, Tang Y, Zhang Y, Kang S, Zhou T, Wu X, Liang W, Hu Z, Ma Y, Zhao Y, Tian Y, Yang Y, Xue C, Yan Y, Hou X, Huang P, Huang Y, Zhao H, Zhang L. 2014. EBV-driven LMP1 and IFN-gamma up-regulate PD-L1 in nasopharyngeal carcinoma:implications for oncotargeted therapy. Oncotarget 5:12189–12202. doi: 10.18632/oncotarget.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bi XW, Wang H, Zhang WW, Wang JH, Liu WJ, Xia ZJ, Huang HQ, Jiang WQ, Zhang YJ, Wang L. 2016. PD-L1 is upregulated by EBV-driven LMP1 through NF-kappaB pathway and correlates with poor prognosis in natural killer/T-cell lymphoma. J HematolOncol 9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma SD, Xu X, Jones R, Delecluse HJ, Zumwalde NA, Sharma A, Gumperz JE, Kenney SC. 2016. PD-1/CTLA-4 blockade inhibits Epstein-Barr virus-induced lymphoma growth in a cord blood humanized-mouse model. PLoS Pathog 12:e1005642. doi: 10.1371/journal.ppat.1005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P. 2015. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med 372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chua ML, Wee JT, Hui EP, Chan AT. 2016. Nasopharyngeal carcinoma. Lancet 387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 86.Price AM, Tourigny JP, Forte E, Salinas RE, Dave SS, Luftig MA. 2012. Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-kappaB activation. J Virol 86:11096–11106. doi: 10.1128/JVI.01069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Izumi KM, Kieff ED. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc Natl Acad Sci U S A 94:12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dirmeier U, Neuhierl B, Kilger E, Reisbach G, Sandberg ML, Hammerschmidt W. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res 63:2982–2989. [PubMed] [Google Scholar]