Abstract
The marine bacterium Vibrio vulnificus causes food-borne diseases, which may lead to life-threatening septicemia in some individuals. Therefore, identifying virulence factors in V. vulnificus is of high priority. We performed a transcriptome analysis on V. vulnificus after infection of human intestinal HT29-methotrexate cells and found induction of plpA, encoding a putative phospholipase, VvPlpA. Bioinformatics, biochemical, and genetic analyses demonstrated that VvPlpA is a phospholipase A2 secreted in a type II secretion system-dependent manner. Compared with the wild type, the plpA mutant exhibited reduced mortality, systemic infection, and inflammation in mice as well as low cytotoxicity toward the human epithelial INT-407 cells. Moreover, plpA mutation attenuated the release of actin and cytosolic cyclophilin A from INT-407 cells, indicating that VvPlpA is a virulence factor essential for causing lysis and necrotic death of the epithelial cells. plpA transcription was growth phase–dependent, reaching maximum levels during the early stationary phase. Also, transcription factor HlyU and cAMP receptor protein (CRP) mediate additive activation and host-dependent induction of plpA. Molecular biological analyses revealed that plpA expression is controlled via the promoter, PplpA, and that HlyU and CRP directly bind to PplpA upstream sequences. Taken together, this study demonstrated that VvPlpA is a type II secretion system-dependent secretory phospholipase A2 regulated by HlyU and CRP and is essential for the pathogenicity of V. vulnificus.
Keywords: gene regulation, microbial pathogenesis, Phospholipase A, transcription factor, virulence factor, CRP, HlyU, PlpA, Vibrio vulnificus
Introduction
The pathogenic marine bacterium Vibrio vulnificus is a causative agent of food-borne diseases such as gastroenteritis in healthy people and life-threatening septicemia in individuals with underlying predisposed conditions (for recent reviews, see Refs. 1 and 2). V. vulnificus infections are notable for their invasiveness, severe tissue destruction, and rapidly fulminating course of disease, leading to high mortality rates (>50%) and immediate death (within 1–2 days after the first symptoms appear). These pathological features of the disease indicate that the pathogenicity of V. vulnificus is a multifactorial and complex phenomenon that involves numerous virulence factors (3). The characterization of somatic and secreted products of V. vulnificus has yielded a large list of putative virulence factors, including a carbohydrate capsule, iron-sequestering systems, a cytolysin/hemolysin, an elastolytic metalloprotease, a multifunctional-autoprocessing repeats-in-toxin (MARTX)3 toxin, a lipopolysaccharide, a lipase, pili, and flagella to account for the destructive nature of V. vulnificus infections (1, 3). Of these putative virulence factors, however, only a few, such as cytolysin/hemolysin and MARTX toxin, have been confirmed as virulence factors that contribute to the lysis and necrotic death of the host cells by using the molecular version of Koch's postulates (4–6). Therefore, extensive screening and characterization of more virulence factors are still required for understanding the molecular pathogenesis of the multifaceted host–pathogen interaction of V. vulnificus.
Phospholipases, lipolytic enzymes hydrolyzing one or more ester linkages in phospholipids, are found in diverse bacterial pathogens, implying that these enzymes play essential roles in the pathogenesis of bacteria (7). Phospholipases are classified into four major groups (A–D) based on their site of action on the phospholipid (7). Among them, phospholipase A (PLA) is able to hydrolyze a fatty acid from the glycerol backbone and is subdivided into PLA1 and PLA2, each hydrolyzing the fatty acid from the sn-1 and -2 position of the glycerol moiety, respectively (7). Accordingly, PLAs are considered to be bacterial virulence factors disrupting the phospholipid membrane, thus leading to the lysis of human epithelial cells (7). In addition, the products generated by epithelial cell lysis, such as lysophosphatidylcholine, may further act as secondary messengers that induce apoptotic death of human endothelial cells (8). Nevertheless, very little is known about the regulatory mechanisms used by the bacteria to modulate the expression of PLAs (9).
To the best of our knowledge, no definitive analysis of the role of the PLAs in the pathogenesis of V. vulnificus has been reported until now. Furthermore, no genes encoding the PLAs have been identified from V. vulnificus, and thus the molecular mechanisms by which the bacterium modulates the expression of the genes have not yet been characterized. In the present study, we conducted a transcriptome analysis and identified the plpA gene, which is preferentially expressed in V. vulnificus cells exposed to human intestinal HT29-methotrexate (MTX) cells (10, 11). The characteristics of V. vulnificus PlpA (VvPlpA), the product of plpA, were verified experimentally. Construction of the plpA mutant and evaluation of its phenotypes provided strong evidence that VvPlpA contributes to the pathogenesis of V. vulnificus. To elucidate the regulation of the plpA expression, we compared the plpA transcript levels in the wild type and in the mutants lacking various transcription factors. Moreover, the transcriptional unit and promoter of plpA were determined, and the roles of HlyU, a transcription factor regulating virulence genes in Vibrio species (12–14), and cAMP receptor protein (CRP) (15) were analyzed at the molecular level.
Results
Identification and sequence analysis of VvPlpA
The transcriptomes of the V. vulnificus MO6-24/O harvested after either the infection of human intestinal HT29-MTX cells or grown with basal medium Eagle (Gibco-BRL) alone were compared as described under “Experimental procedures.” Transcriptome analysis revealed 650 differentially expressed genes in the host cell-exposed V. vulnificus compared with the unexposed one; 319 genes were up-regulated, and 331 were down-regulated (supplemental Tables S1 and S2). Among the up-regulated, virulence-related genes, a gene VVMO6_03257 encoding a putative phospholipase showed the highest-fold change of expression level (20.5-fold, p = 0.0001). In contrast, the expressions of several other putative phospholipase genes (VVMO6_00112, 00550, 00661, 01752, 03024, 03759, and 04072) were not significantly induced (data not shown), suggesting that the VVMO6_03257 gene product plays an important role in host–microbe interaction, at least under our experimental conditions. Real-time quantitative PCR (qPCR) confirmed that the expression of VVMO6_03257 is increased about 17.8-fold (p = 0.0005) after exposure of V. vulnificus to the host cells (data not shown).
The amino acid sequence deduced from the VVMO6_03257 nucleotide sequence predicted a protein with an N-terminal signal peptide for the type II secretion system (T2SS) (Fig. 1A) (16). The mature protein is composed of 398 amino acids with a theoretical mass of 45.22 kDa and pI of 5.38. When aligned using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo)4 (66), the deduced amino acid sequence of the mature protein was 69% identical to that of the Vibrio anguillarum phospholipase (VaPlpA) (17), leading us to name the protein VvPlpA. Amino acid sequence analysis further revealed that VvPlpA possesses five conserved blocks for the SGNH hydrolases as well as a GDSL motif found in typical lipases (Fig. 1A) (18). Also, the predicted profile of the hydrophobicity was significantly similar to that of VaPlpA, indicating that VvPlpA is a soluble protein like VaPlpA (data not shown) (17). Meanwhile, we found that the genetic environment of plpA in V. vulnificus is distinct from that in V. anguillarum. Unlike the V. anguillarum plpA located upstream of and sharing the promoter region with hemolysin gene, vah1 (19), V. vulnificus plpA has its own promoter region, and presents at a different location from hemolysin gene, vvhA, or MARTX toxin gene, rtxA1 (data not shown).
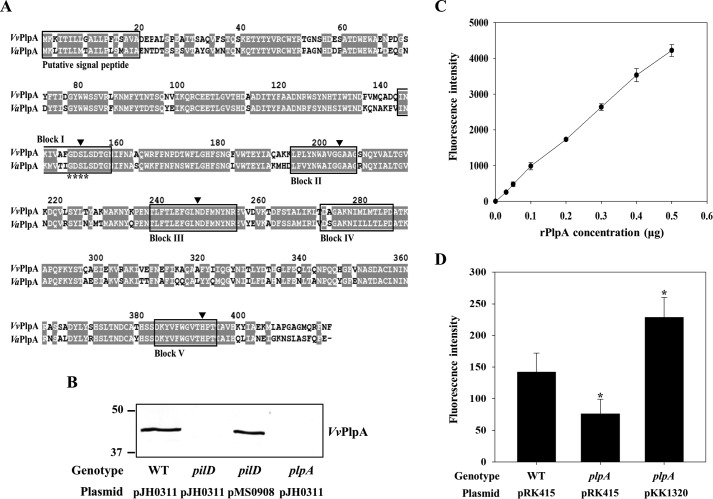
Figure 1.
Analysis of amino acid sequence, secretion, and enzymatic activity of VvPlpA. A, the amino acid sequences retrieved from the NCBI protein database (http://www.ncbi.nlm.nih.gov) (accession numbers ADV88279 for VvPlpA and AAY26144 for VaPlpA) were aligned using the Clustal Omega and Genedoc program. Identical residues (gray boxes) and missing sequence (dash) are indicated. The putative signal peptides and consensus blocks for SGNH hydrolases are boxed by a black line. The GDSL motif is indicated by asterisks. The four residues conserved in the SGNH hydrolases are indicated by black-filled inverse triangles. B, VvPlpA in the supernatants of the V. vulnificus strains grown to an A600 of 0.9 was detected by Western blot analysis. Molecular size markers (Bio-Rad) are shown in kDa. C and D, PLA2 activity was examined by measuring fluorescence intensity emitted from the fluorogenic phosphatidylcholine substrate (red/green BODIPY PC-A2) after incubation with various amounts of the rPlpA (C) or supernatants of the V. vulnificus strains (D) for 0.5 h. Error bars, S.D. Statistical significance was determined by Student's t test. *, p < 0.05 relative to the wild type. WT pJH0311 and WT pRK415, wild type; pilD pJH0311, pilD mutant; plpA pJH0311 and plpA pRK415, plpA mutant; pilD pMS0908 and plpA pKK1320, complemented strains.
The secretion of VvPlpA to the cell exterior through T2SS as predicted above was experimentally verified (Fig. 1B). To accomplish this, an isogenic mutant in which the pilD gene encoding the PilD component of T2SS was specifically deleted (20) was used, and the VvPlpA secretion from the mutant was compared with that from the parental wild type. As shown in Fig. 1B, VvPlpA was not detected from the culture supernatant of the pilD mutant. This lack of VvPlpA secretion was restored by the introduction of pMS0908 (20), carrying a pilD gene, to the pilD mutant. These results confirmed that VvPlpA is indeed an extracellular protein, and its secretion is T2SS-dependent.
The lipase activity of VvPlpA, predicted from amino acid sequence analysis, was also experimentally verified. Fluorescence was emitted from the red/green BODIPY PC-A2 (Invitrogen), a fluorescently labeled phosphatidylcholine (PC), in the presence of 0.02–0.5 μg of recombinant VvPlpA protein (rPlpA). The fluorescence emission intensity increased as greater amounts of rPlpA were added (Fig. 1C), suggesting that rPlpA contains the PLA2 activity cleaving specifically the sn-2 bond of the red/green BODIPY PC-A2 substrate (Invitrogen). Based on the standard curve developed using the honey bee venom PLA2 (21), the specific activity of rPlpA was calculated to be ∼247 units/mg (data not shown) (1 unit = the amount of protein that cleaves 1 μmol of red/green BODIPY PC-A2 per min).
We further examined whether VvPlpA indeed confers the PLA2 activity to V. vulnificus. For this, an isogenic mutant KK131 lacking a functional plpA gene was constructed (Table 1). When culture supernatant of the wild type, plpA mutant, or complemented strain was added to the reaction as a source for the enzyme, significantly lower fluorescence was emitted from the sample containing that of the plpA mutant compared with the other two samples (Fig. 1D). Therefore, the combined results indicated that VvPlpA is a T2SS-dependent secretory enzyme conferring the PLA2 activity to V. vulnificus.
Table 1.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| MO6-24/O | Wild type; clinical isolate; virulent | Ref. 65 |
| KK131 | MO6-24/O with ΔplpA | This study |
| ZW141 | MO6-24/O with ΔhlyU | This study |
| DI0201 | MO6-24/O with Δcrp | Ref. 49 |
| KK151 | MO6-24/O with ΔhlyUΔcrp | This study |
| MS023 | MO6-24/O with ΔpilD | Ref. 20 |
| E. coli | ||
| S17-1λpir | λ-pir lysogen; thi pro hsdR hsdM+ recA RP4–2 Tc:: Mu-Km::Tn7;Tpr Smr; host for π-requiring plasmids; conjugal donor | Ref. 55 |
| BL21 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Laboratory collection |
| Plasmids | ||
| pDM4 | R6K γ ori sacB; suicide vector; oriT of RP4; Cmr | Ref. 54 |
| pKK1301 | pDM4 with ΔplpA; Cmr | This study |
| pZW1401 | pDM4 with ΔhlyU; Cmr | This study |
| pBS0907 | pDM4 with Δcrp; Cmr | Ref. 56 |
| pRK415 | Broad-host-range vector; IncP ori oriT RK2; Tcr | Ref. 57 |
| pKK1320 | pRK415 with plpA; Tcr | This study |
| pJH0311 | Broad-host-range vector; Apr, Cmr | Ref. 58 |
| pZW1510 | pJH0311 with hlyU; Apr, Cmr | This study |
| pKK1502 | pJH0311 with crp; Apr, Cmr | This study |
| pMS0908 | pJH0311 with pilD; Apr, Cmr | Ref. 20 |
| pMBP parallel 1 | MBP tag fusion expression vector; Apr | Ref. 59 |
| pKK1503 | pMBP parallel 1 with plpA-his6; Apr | This study |
| pET28a(+) | His6 tag fusion expression vector; Kmr | Novagen |
| pYU1317 | pET28a (+) with hlyU; Kmr | This study |
| pKK1504 | pET28a (+) with truncated plpA; Kmr | This study |
| pHK0201 | pRSET A with crp; Apr | Ref. 49 |
| pGEM-T Easy | PCR product cloning vector; Apr | Promega |
| pKK1505 | pGEM-T Easy with 461-bp fragment of plpA upstream region; Apr | This study |
| pBBR-lux | Broad-host-range vector with luxCDABE operon; Cmr | Ref. 64 |
| pKK1514 | pBBR-lux with 378-bp fragment of plpA upstream region; Cmr | This study |
| pKK1515 | pBBR-lux with 308-bp fragment of plpA upstream region; Cmr | This study |
| pKK1516 | pBBR-lux with 228-bp fragment of plpA upstream region; Cmr | This study |
| pKK1517 | pBBR-lux with 129-bp fragment of plpA upstream region; Cmr | This study |
a Tpr, trimethoprim-resistant; Smr, streptomycin-resistant; Cmr, chloramphenicol-resistant; Tcr, tetracycline-resistance; Apr, ampicillin-resistant; Kmr, kanamycin-resistant.
VvPlpA is essential for virulence
Next we confirmed the induction of plpA expression under conditions closer to those in vivo rather than those under in vitro-cultured host cells. When V. vulnificus was inoculated into the blood of uninfected mice, the expression of plpA was dramatically elevated (Fig. 2A). This result suggested that VvPlpA may be highly expressed in vivo and take part in the pathogenesis of V. vulnificus. To test this hypothesis, the virulence of wild type and plpA mutant KK131 was compared in mice. As shown in Fig. 2B, mice intragastrically infected with the wild type started to die at 5 h postinfection. In contrast, the survival time of mice infected with KK131 was consistently and significantly prolonged (p = 0.0063, log-rank test). At 20 h postinfection, the percentages of surviving mice were 28 and 57% for the wild type– and KK131–infected group, respectively (Fig. 2B).
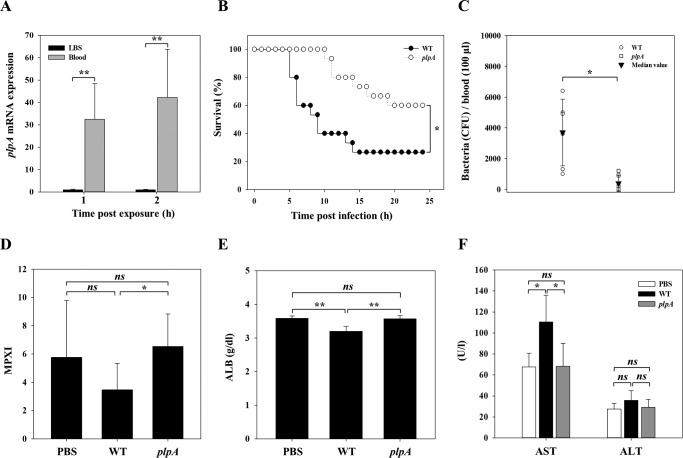
Figure 2.
VvPlpA is essential for pathogenesis of V. vulnificus. A, the wild type grown to an A600 of 0.5 was exposed to either LBS or blood of uninfected mice (n = 3) for 1 or 2 h and then used to isolate total RNAs. The plpA mRNA was determined by real-time qPCR analyses, and the plpA mRNA level in the bacteria exposed to LBS was set as 1. B, 7-week-old female ICR mice (n = 15 per group), injected intraperitoneally with iron-dextran, were intragastrically infected with the wild type and plpA mutant at doses of 108 cfu. C–F, the mice, without an iron-dextran administration, were intraperitoneally infected with the wild type (n = 11), plpA mutant (n = 11), or PBS (n = 5, control) at doses of 106 cfu and then sacrificed to obtain blood at 4 h postinfection. C, the number of either the wild type or plpA mutant in the blood of infected mice (n = 6 per group) was enumerated as cfu/100 μl of blood. Each open symbol represents an individual mouse. Black-filled inverted triangles indicate median values. D–F, the levels of MPXI (D), ALB (E), and AST/ALT (F) in the wild type– or plpA mutant–infected or PBS-treated mice (n = 5 per group) were determined by hematological analyses. Error bars, S.D. Statistical significance was determined by Student's t test for A and C–F, and by log-rank test for B. *, p < 0.05; **, p < 0.005; ns, not significant. WT, wild type; plpA, plpA mutant; PBS, control.
To further elucidate the role of VvPlpA during pathogenesis, we compared the number of V. vulnificus in the blood and various hematological indexes of either the wild type– or plpA mutant–infected mice. To deliver exactly the same number of V. vulnificus into the host without any colonization problems, we adopted an intraperitoneal infection model for these analyses. First, we confirmed the importance of VvPlpA in this infection model by comparing the survival rates of mice infected with the wild type or plpA mutant (data not shown). When the bacteria in the blood of the infected mice were enumerated, about 4 times more wild-type cells, compared with the plpA mutant cells, were recovered (Fig. 2C). Consistent with this, hematological analyses revealed that the levels of myeloperoxidase index (MPXI) and albumin (ALB), the biomarkers for septicemia (in inverse correlation; see Refs. 22 and 23), were significantly lower in the wild type–infected mice compared with those in the plpA mutant–infected or PBS-treated mice (Fig. 2, D and E). Also, mice infected with the wild type exhibited significantly higher levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) activities, the indexes for liver damage (24), compared with those infected with the plpA mutant or treated with PBS (Fig. 2F). Combined with the fact that plpA mutation does not attenuate the bacterial fitness in vitro (data not shown), these results clearly demonstrated the crucial role of VvPlpA in V. vulnificus systemic infection, and the accompanying inflammation resulted in host death.
VvPlpA is essential for the lysis and necrotic death of host cells
To investigate the role of VvPlpA in the cytotoxicity toward host cells, the activities of lactate dehydrogenase (LDH) released from INT-407 cells infected with the wild type and plpA mutant KK131 were determined. As shown in Fig. 3A, KK131 exhibited significantly decreased LDH-releasing activity compared with that of the wild type, when the V. vulnificus strains were used to infect the INT-407 cells at multiplicity of infections (MOIs) of 1, 5, and 20 for 2 h. Similarly, INT-407 cells infected with KK131 at an MOI of 10 and then incubated for 1, 2, and 3 h released lower levels of LDH compared with those infected with the wild type (Fig. 3B), indicating that VvPlpA plays an important role in V. vulnificus infecting and injuring host cells (Fig. 3, A and B). Complementation of the plpA mutation with a functional plpA gene (pKK1320) restored the LDH-releasing activity of KK131 to levels comparable with those of the wild type (Fig. 3, A and B). Therefore, the decreased cytotoxicity of KK131 resulted from the inactivation of plpA rather than the reduced expression of other genes downstream of plpA.
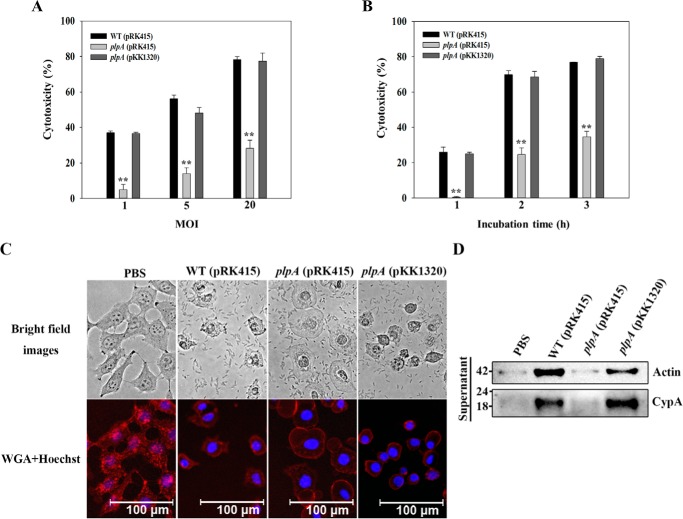
Figure 3.
VvPlpA is essential for necrotic cell death in tissue culture. A and B, INT-407 cells were infected with the V. vulnificus strains at various MOIs for 2 h (A) or at an MOI of 10 for various incubation times (B). The cytotoxicity was expressed using the total LDH activity of the cells completely lysed by 1% Triton X-100 as 100%. Error bars, S.D. Statistical significance was determined by Student's t test. **, p < 0.005 relative to groups infected with the wild type at each MOI or incubation time. C, INT-407 cells were infected with the V. vulnificus strains at an MOI of 10 for 1.5 h as indicated, stained using Texas Red®-X–conjugated WGA (for membrane, red) and Hoechst 33342 (for nucleus, blue), and then photographed using a fluorescence microscope. D, INT-407 cells were infected with the V. vulnificus strains at an MOI of 10 for 2 h, and actin and CypA released in the culture supernatants were analyzed by Western blot analysis. Molecular size markers (GenDEPOT, Barker, TX) are shown in kDa. WT (pRK415), wild type; plpA (pRK415), plpA mutant; plpA (pKK1320), complemented strain; PBS, control (uninfected).
To further understand the effects of VvPlpA on host cells, INT-407 cells were stained with Texas Red®-X–conjugated wheat germ agglutinin (WGA) (Thermo Fisher Scientific, San Jose, CA) and Hoechst® 33342 (Thermo Fisher Scientific), after which their morphological changes caused by infection with the V. vulnificus strains were examined. As shown in Fig. 3C, INT-407 cells exhibited typical morphology of surface-attached epithelial cells with regularly shaped cell margins and rounded nuclei in the absence of infection (PBS). Infection with the wild type injured INT-407 cells and resulted in severely shrunken cell morphology with irregular cell margins, presumably accompanied by the release of cellular materials (WT (pRK415)). Notably, however, infection with the plpA mutant KK131 resulted in cells with less severely shrunken and swollen morphology (plpA (pRK415)). The morphology of INT-407 cells infected with the complemented strain (plpA (pKK1320)) was similar to that infected with the wild type, suggesting that VvPlpA is responsible for the injury and lysis of INT-407 cells.
To examine this possibility further, the release of actin protein, acting as a biomarker of cell lysis, was determined by Western blot analysis (Fig. 3D). A significant amount of actin was detected from the supernatant of INT-407 cells infected with the wild type and complemented strains of V. vulnificus. In contrast, the level of actin from the supernatant of the host cells infected with the plpA mutant KK131 was almost undetectable and comparable with that of the cells treated with PBS (Fig. 3D), confirming that VvPlpA is responsible for host cell lysis. Furthermore, in addition to actin, cyclophilin A (CypA, a peptidyl-prolyl cis-trans isomerase), which is used as a biomarker of necrotic cell death (25), was also released from INT-407 cells infected with the wild type and complemented strains but not from the host cells infected with KK131 (Fig. 3D). These results indicated that VvPlpA mediates host cell lysis as well as necrotic death by releasing cytosolic components. Therefore, these results, combined with the previous results of decreased mouse mortality and LDH-releasing activity in the absence of VvPlpA, indicated that VvPlpA is a virulence factor essential for cell lysis and necrotic cell death, thereby contributing to the pathogenesis of V. vulnificus.
Transcription of plpA is growth phase–dependent and regulated by HlyU and CRP
To extend our understanding of plpA regulation, levels of the plpA transcript in the wild type grown under various conditions (oxidative stress, iron limitation, and different temperatures) were compared, but none of the tested conditions influenced plpA expression (data not shown). However, when the levels of plpA mRNA were analyzed at various growth phases, the plpA transcript level increased to maximum levels in the early stationary phase (A600 = 1.0) and then decreased in the late stationary phase (A600 = 2.5) (Fig. 4A), indicating growth phase–dependent expression of plpA.
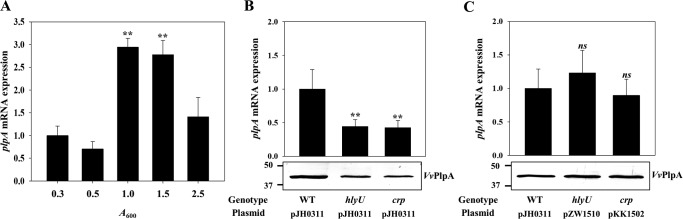
Figure 4.
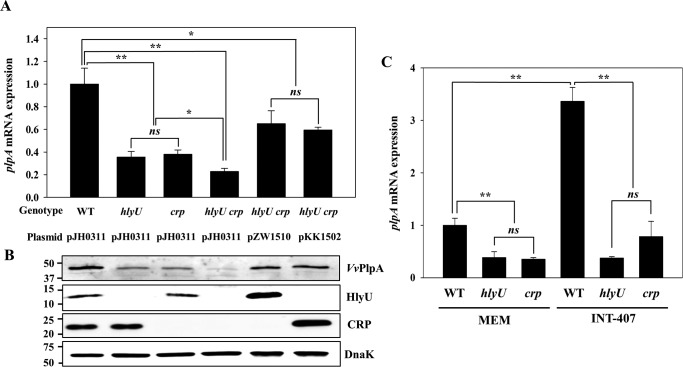
Expression of plpA in V. vulnificus under different growth phases (A) or with different genetic backgrounds (B and C). A, total RNAs from the V. vulnificus wild type under different growth phases were isolated, and the plpA mRNA levels were determined by real-time qPCR analyses. The plpA mRNA level in the cells grown to an A600 of 0.3 was set as 1. B and C, V. vulnificus strains were grown to an A600 of 0.9 and used to determine the plpA mRNA and VvPlpA levels. The plpA mRNA levels in the total RNAs isolated from the cultures were determined by real-time qPCR analyses, and the plpA mRNA level in the wild type was set as 1. The VvPlpA levels in the supernatants harvested form the cultures were determined by Western blot analyses. Molecular size markers (Bio-Rad) are shown in kDa. Error bars, S.D. Statistical significance was determined by Student's t test. **, p < 0.005 relative to the cells grown to an A600 of 0.3 (for A) or to the wild type (for B and C); ns, not significant. WT pJH0311, wild type; hlyU pJH0311, hlyU mutant; crp pJH0311, crp mutant; hlyU pZW1510 and crp pKK1502, complemented strains.
To identify transcription factor(s) associated with the plpA regulation, the levels of plpA mRNA were compared in the wild type and several mutants lacking a transcription factor known to regulate the virulence of V. vulnificus. The transcription factor-lacking mutants consisted of fur, hlyU, iscR, aphB, crp, leuO, and smcR mutants (13, 26–30). Expression of plpA did not differ in the wild type or mutants lacking Fur, IscR, AphB, LeuO, or a quorum-sensing master regulator SmcR (27) (data not shown). This result suggested that plpA might not be regulated by iron availability or by quorum sensing. In contrast, the hlyU and crp mutants showed almost 2.5-fold lower levels of plpA transcript compared with those in the wild type (Fig. 4B), indicating that both HlyU and CRP may act as positive regulators. The levels of VvPlpA determined by Western blot analyses also decreased in the culture supernatants of the mutants (Fig. 4B). The magnitudes of decrease in VvPlpA did not significantly differ from those observed in the plpA transcript, indicating that the regulation of plpA expression occurs mostly at the transcriptional level. The decreased levels of the plpA transcript and VvPlpA protein in the hlyU and crp mutants were restored to levels comparable with those in the wild type by introducing pZW1510 and pKK1502, each carrying the hlyU and crp gene, respectively (Fig. 4C). Taken together, these results suggested that both HlyU and CRP positively regulate plpA expression at the transcriptional level.
Additive activation of plpA by HlyU and CRP
To characterize the functions of HlyU and CRP in plpA expression, hlyU crp double mutant KK151 was constructed and used to determine plpA expression. Lack of both hlyU and crp in KK151 resulted in a significant reduction of plpA expression, as the transcript level was only one-quarter of that in the wild type (Fig. 5A). Furthermore, this residual plpA transcript level in KK151 was considerably lower than that in either the hlyU or crp single mutant (Fig. 5A), suggesting that both transcriptional regulators activate plpA additively. Indeed, the cellular level of HlyU in the crp mutant (or CRP in the hlyU mutant) was comparable with that in the wild type (Fig. 5B), confirming that neither CRP nor HlyU controls the expression of the other in a sequential regulatory cascade.
Figure 5.
HlyU and CRP mediate additive activation and host-dependent induction of plpA. A and B, V. vulnificus strains were grown to an A600 of 0.9 and used to determine the plpA mRNA levels and VvPlpA, HlyU, CRP, and DnaK protein levels. A, the plpA mRNA levels were determined by real-time qPCR analyses, and the plpA mRNA level in the wild type was set as 1. B, the secreted VvPlpA and cellular HlyU, CRP, and DnaK (as an internal control) levels were determined by Western blot analyses. Molecular size markers (Bio-Rad) are shown in kDa. C, V. vulnificus strains were exposed to MEM (control) or INT-407 cells at an MOI of 10 for 1 h and then used to isolate total RNAs. The plpA mRNA levels were determined by real-time qPCR, and the plpA mRNA level in the wild type exposed to MEM was set as 1. Error bars, S.D. Statistical significance was determined by Student's t test. *, p < 0.05; **, p < 0.005; ns, not significant. WT and WT pJH0311, wild type; hlyU and hlyU pJH0311, hlyU mutant; crp and crp pJH0311, crp mutant; hlyU crp pJH0311, hlyU crp double mutant; hlyU crp pZW1510 or hlyU crp pKK1502, hlyU crp double mutant overexpressing HlyU or CRP, respectively.
To further determine whether increasing the level of cellular HlyU could compensate for the lack of CRP in the activation of plpA, KK151 was complemented with pZW1510 expressing hlyU under the lac promoter. When hlyU was induced by isopropyl-β-d-thiogalactopyranoside, the HlyU level in KK151 (pZW1510) was significantly increased, exceeding the level in the wild type or crp single mutant (Fig. 5B). Nonetheless, the plpA transcript level in KK151 (pZW1510) was still lower than that in the wild type, corresponding to only about 60% of that in the wild type (Fig. 5A). This result suggested that if CRP is absent, overexpressed HlyU is insufficient in increasing plpA expression levels comparable with those in the wild type. Similarly, overproduced CRP (Fig. 5B) was unable to completely compensate for the lack of HlyU in the activation of plpA (Fig. 5A). Also, the changes in the plpA transcript were directly reflected on the secreted VvPlpA protein levels (Fig. 5B).
Next, we verified whether HlyU and CRP are involved in the induction of plpA during exposure to host cells by comparing the plpA transcript levels in the V. vulnificus strains exposed to either minimal essential medium (MEM, control) or INT-407 cells for 1 h. As shown in Fig. 5C, the plpA transcript level in the hlyU or crp mutant was significantly lower than that in the wild type, regardless of host cell exposure. Importantly, the host-dependent induction of plpA was evident in the wild type (3.36-fold induction; p = 0.0002), but not in either the hlyU or crp mutant (p = 0.67 or 0.054, respectively). Taken together, the results indicated that HlyU and CRP additively activate plpA expression, and thus both regulators are simultaneously required for the full activation of plpA. Furthermore, these regulators are essential for the induction of plpA in V. vulnificus during infection of host cells.
Determination and deletion analyses of the plpA promoter, PplpA
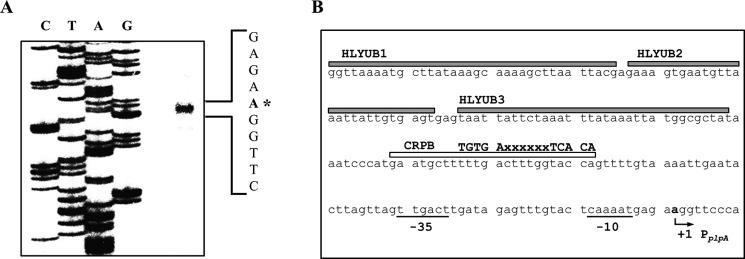
To map the promoter of plpA, the transcription start site of plpA was determined by primer extension analysis. A single reverse transcript was produced from the primer extension of the RNA isolated from the wild type grown to an A600 of 0.9 (Fig. 6A). The 5′-end of the plpA transcript was located 65 bp upstream of the translational initiation codon of plpA and was subsequently designated +1 (Fig. 6B). The putative promoter constituting this transcription start site was named PplpA to represent the plpA promoter, and the sequences for −10 and −35 regions of PplpA were assigned on the basis of similarity to the consensus sequences of the Escherichia coli σ70 promoter (Fig. 6B). Several attempts to identify other transcription start sites using different sets of primers hybridizing to the plpA mRNA were not successful (data not shown), indicating that a single promoter, PplpA, is used for plpA transcription.
Figure 6.
Transcription start site and sequences of the plpA-regulatory region. A, the transcription start site of plpA was determined by primer extension of the RNA isolated from the wild type grown to an A600 of 0.9. Lanes C, T, A, and G, nucleotide sequencing ladders of pKK1505. The asterisk indicates the transcription start site of plpA. B, the transcription start site of plpA is indicated by a bent arrow, and the positions of the putative −10 and −35 regions are underlined. The sequences for binding of HlyU (HLYUB1, HLYUB2, and HLYUB3; gray boxes) and CRP (CRPB; white box) were determined later in this study (Fig. 8, C and D). The consensus sequences for binding of CRP (32) are indicated above the V. vulnificus DNA sequence. x, any nucleotide.
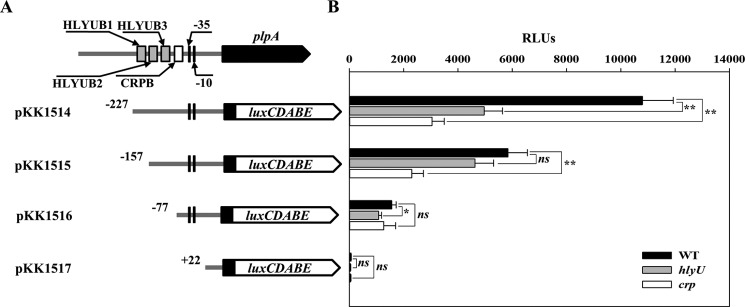
To delineate the cis-DNA sequences in PplpA required for HlyU- and CRP-mediated activation, pKK reporters carrying the PplpA regulatory region, which was deleted up to different 5′-ends and fused transcriptionally to luxCDABE, were constructed (Fig. 7A). The reporters were transferred into the V. vulnificus strains, and cellular luminescence was used to quantify the ability of each PplpA regulatory region to activate the promoter (Fig. 7B). The luminescence produced by pKK1514 carrying PplpA deleted up to −227 was ∼1.1 × 104 relative light units (RLUs) in the wild type but significantly reduced in the hlyU and crp mutants, supporting our previous observation that HlyU and CRP coactivate plpA expression. Compared with pKK1514, pKK1515 carrying PplpA deleted up to −157 produced significantly reduced RLUs in the wild type. Moreover, the RLUs of the wild type and the hlyU mutant containing pKK1515 did not significantly differ, indicating that the cis-DNA sequences required for HlyU to activate PplpA are deleted in pKK1515. Similarly, the comparable levels of RLUs produced by pKK1516 in the wild type and crp mutant indicated that deletion of the PplpA regulatory region up to −77 probably impairs CRP (in addition to HlyU) activation of PplpA. The undetectable luminescence in the wild type (and the hlyU or crp mutant) containing pKK1517 indicated that deletion of the PplpA regulatory region up to +22 results in a complete loss of PplpA activity. These results implied that cis-DNA sequences essential for HlyU and CRP to activate PplpA encompass the −157 and −77 regions, respectively.
Figure 7.
Deletion analysis of the PplpA regulatory region. A, construction of plpA-lux fusion pKK reporters. PCR fragments carrying the plpA-regulatory region with 5′-end deletions were subcloned into pBBR-lux (64) to create each pKK reporter. Solid lines, the upstream region of plpA; black blocks, the plpA coding region; white blocks, luxCDABE. The wild-type plpA-regulatory region is shown on top with the proposed −10 and −35 regions, and the binding sites for HlyU (HLYUB1, HLYUB2, and HLYUB3; gray boxes) and CRP (CRPB; white box) were determined later in this study (Fig. 8, C and D). B, cellular luminescence was determined from the wild type (black bars), hlyU mutant (gray bars), and crp mutant (white bars) containing each pKK reporter as indicated. Cultures grown to an A600 of 0.9 were used to measure the cellular luminescence. Error bars, S.D. Statistical significance was determined by Student's t test. *, p < 0.05; **, p < 0.005; ns, not significant. WT, wild type; hlyU, hlyU mutant; crp, crp mutant.
HlyU and CRP regulate by binding directly to PplpA
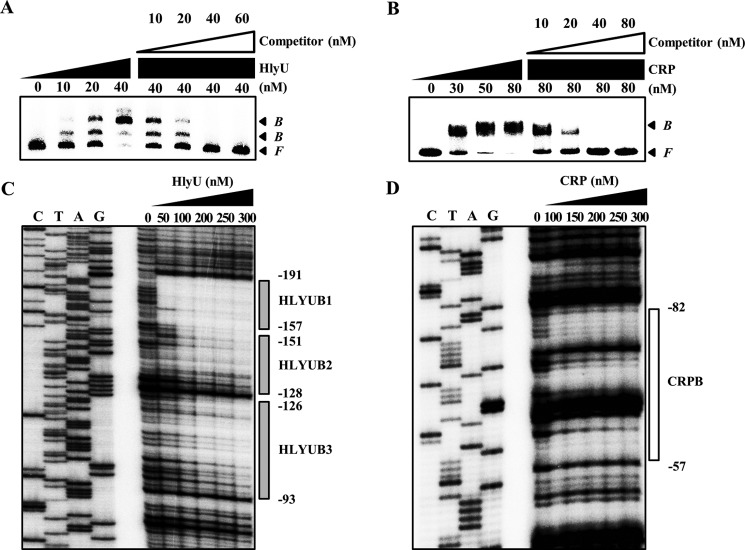
To examine whether HlyU and CRP bind directly to the promoter PplpA, EMSAs were performed as shown in Fig. 8 (A and B). For this purpose, the 461-bp labeled DNA probe encompassing the PplpA regulatory region (from −345 to +116) was incubated with increasing amounts of HlyU or CRP and then subjected to electrophoresis. EMSA revealed that the addition of HlyU to the DNA probe resulted in two retarded bands in a concentration-dependent manner, indicating that HlyU binds directly to at least two binding sites with different affinities (Fig. 8A). Furthermore, this binding of HlyU was specific because the EMSA was performed in the presence of a nonspecific competitor, poly(dI-dC) (0.1 μg; Sigma). In the second EMSA, the same but unlabeled 461-bp DNA probe was used as a self-competitor, which competed for the binding of HlyU in a dose-dependent manner (Fig. 8A), confirming that HlyU binds specifically as well as directly to PplpA. Similarly, EMSAs demonstrated that CRP also binds directly and specifically to the PplpA regulatory region (Fig. 8B). The combined results suggested that HlyU and CRP regulate plpA by directly binding to their specific regions within PplpA rather than by modulating the cellular levels of unidentified trans-acting factor(s), which in turn binds directly to PplpA.
Figure 8.
Specific bindings of HlyU and CRP to PplpA. A 461-bp DNA fragment of the plpA-regulatory region was radioactively labeled and then used as a probe DNA. A and B, the radiolabeled probe DNA (5 nm) was incubated with increasing amounts of HlyU (A) and CRP (B) as indicated. For competition analysis, the same but unlabeled 461-bp DNA fragment was used as a self-competitor DNA. Various amounts of the self-competitor DNA were added to a reaction mixture containing the 5 nm labeled DNA before the addition of 40 nm HlyU (A) and 80 nm CRP (B) as indicated. B, bound DNA; F, free DNA. C and D, the same radiolabeled probe DNA (25 nm) was reacted with increasing amounts of HlyU (C) and CRP (D) as indicated. The regions protected from DNase I cleavage by HlyU and CRP are indicated by gray boxes (HLYUB1, HLYUB2, and HLYUB3) and a white box (CRPB), respectively. Lanes C, T, A, and G represent the nucleotide sequencing ladders of pKK1505. Nucleotide numbers shown are relative to the transcription start site of plpA.
DNase I protection assays were performed using the same 461-bp DNA probe to identify the HlyU-binding sequences in the PplpA regulatory region (Fig. 8C). When HlyU was used up to 50 nm, the HlyU footprint extended from −191 to −157 (HLYUB1, centered at −174) (Fig. 8C). Upon increasing HlyU levels, two additional sequences extending from −151 to −128 (HLYUB2, centered at −139.5) and −126 to −93 (HLYUB3, centered at −109.5) were protected from DNase I digestion (Fig. 8C). Inspection of the HLYUBs revealed AT-rich sequences with an (imperfect) inverted repeat (Fig. 6B), indicating that HlyU binds to the sequences in the form of a dimer, as proposed previously (13, 31). Similarly, DNase I protection assays revealed the sequences of the region protected by CRP binding (Fig. 8D). The sequences of the protected region extended from −82 to −57 (CRPB, centered at −69.5) and scored 81% similarity to a consensus sequence for the binding of CRP (the TGTGAx6–8TCACA; see Ref. 32) (Fig. 6B). The positioning of HLYUBs and CRPB suggested that both HlyU and CRP may act as class I activators interacting with the C-terminal domain of RNA polymerase α subunits (33). Taken together, these results demonstrated that HlyU and CRP regulate plpA directly by binding to the specific sequences of PplpA.
Discussion
The epithelial surface of the gastrointestinal tracts is the major site of entry for V. vulnificus, an enteropathogen, to invade the host. Several methods, such as in vivo-induced antigen technology and in vivo expression technology, have been developed and adopted to discover V. vulnificus genes expressed preferentially in the host (34, 35). Although previous transcriptome analyses identified V. vulnificus genes induced specifically during infection, functional and regulatory features of the induced genes have been barely characterized (36, 37). In this study, plpA encoding a PLA2 of V. vulnificus, VvPlpA, was identified among the transcriptome profile expressed preferentially during infection of human intestine HT29-MTX cells (supplemental Tables S1 and S2) and then further characterized at molecular levels. As a part of the results, the ability of VvPlpA to cleave the sn-2 bond in the fluorescently labeled PC was demonstrated (Fig. 1C). Consistent with this, rPlpA showed hemolytic activity on human and horse erythrocyte containing about 29 and 38% PC in its membrane (38, 39), respectively, but not to PC-deficient sheep erythrocyte (38) (data not shown). These results suggested that PC is a preferential substrate for VvPlpA, as is the case for VaPlpA (17). Moreover, compared with the wild type, the plpA mutant, KK131, showed significantly attenuated virulence; it showed reduced systemic infection, liver damage, and mortality in mice (Fig. 2, B–F). These results led us to conclude that VvPlpA, which is secreted in a T2SS-dependent manner, is an essential virulence factor contributing to V. vulnificus pathogenesis along with other toxins, such as cytolysin/hemolysin and MARTX toxin (5, 40, 41). Indeed, the expression of neither the cytolysin/hemolysin gene, vvhA, nor the MARTX toxin gene, rtxA1, was affected by plpA mutation in V. vulnificus (data not shown). Furthermore, we found that the genes encoding VvPlpA homologues are highly conserved and expressed well in various clinical and environmental isolates of V. vulnificus (data not shown), further supporting its crucial role in the adaption of V. vulnificus to variety of environments, including the host interior. One such role of VvPlpA during infection is that this lipolytic enzyme disrupts membranes of intestine epithelial cells and thereby allows the pathogen to invade other tissues (or organs), such as blood vessels, where the pathogen most likely secretes additional toxins.
It has been reported that many pathogenic bacteria, including Salmonella spp. and Yersinia enterocolitica, induce necrotic death of the host epithelial barrier and immune cells to survive and spread easily in the host (42, 43). Consistent with this, V. vulnificus infection also manifests robust necrotic cell death and tissue destruction, which are caused by toxins such as MARTX, resulting in skin lesions (6, 44, 45). This study found that VvPlpA, in addition to the MARTX toxin, also contributes to the induction of necrotic death of INT-407 epithelial cells (Fig. 3D). Therefore, the combined results suggested that VvPlpA is essential for the necrotic death of human epithelial cells (Fig. 3).
Many pathogenic bacteria continually encounter environmental changes and cope with harsh conditions in the host. Therefore, the ability to constantly sense and respond to the host environment is important for the pathogens to establish successful infection (46, 47). Accordingly, pathogens have evolved sophisticated mechanisms to extensively regulate their gene expressions, which consequently plays a key role in the adaptation and pathogenesis of the bacteria to the host (46, 47). This study revealed that V. vulnificus regulates the expression of plpA in a growth phase–dependent manner and to the maximum level in the early stationary phase (Fig. 4A). Because this growth-phase dependent plpA expression was further confirmed by using a PplpA-luxCDABE transcriptional fusion reporter (data not shown), it seems that the regulation of plpA expression occurred at a transcriptional level rather than a post-transcriptional level. Consistent with this, HlyU and CRP were found to directly activate plpA transcription both in vitro and ex vivo (Figs. 5 and 8). Because the expression of HlyU is induced specifically during the infection (34), the positive control of VvPlpA by HlyU could enable V. vulnificus to express this critical virulence factor at the right place (i.e. inside the host). In addition, CRP, which is a central regulator of energy (catabolic) metabolism (15) may recognize host environments by sensing the starvation of specific nutrients imposed by the host cells and endogenous bacterial flora (47). Accordingly, CRP might contribute to the production of VvPlpA at the appropriate time when V. vulnificus encounters such nutrient starvation, probably leading to immune deficiency in the host. Taken together, V. vulnificus could express plpA in a spatially and temporally regulated fashion to obtain maximum effectiveness during pathogenesis.
Although a few examples of transcriptional coactivation of bacterial virulence genes by multiple activators have been reported (11, 48), coactivation by HlyU and CRP has not yet been reported. Each HlyU and CRP binds to the upstream sequence of PplpA and activates the promoter as a solitary regulator, albeit not to levels comparable with that of the wild type (Figs. 5 and 8). Overproduced HlyU (or CRP) failed to fully compensate for a lack of CRP (or HlyU), indicating that both HlyU and CRP are required for the expression of VvPlpA to the wild-type level (Fig. 5, A and B). Obviously, this coactivation of PplpA by both HlyU and CRP, which probably sense and incorporate different signals from the environment, as mentioned above, could allow the tight regulation and fine tuning of VvPlpA expression during pathogenesis. It is also noteworthy that HlyU and/or CRP are involved in the activation of the expression of other virulence factors, such as the MARTX toxin, cytolysin/hemolysin, VvpE (an elastolytic protease), GbpA (a mucin-binding protein), and HupA (a heme receptor protein), in addition to VvPlpA (11, 13, 48–50). It is likely that regulation by common regulatory proteins provides precisely coordinated expression of multiple virulence factors, thereby facilitating the cooperation of the virulence factors crucial for the overall success of the organism during pathogenesis.
In summary, a transcriptome analysis identified V. vulnificus plpA among the genes, which are specifically induced when the pathogen is exposed to human intestine HT29-MTX cells. VvPlpA is a T2SS-dependent secretory PLA2 and promotes lysis and necrotic death of host cells, thereby contributing to the pathogenesis of V. vulnificus, as determined in the mouse infection model. The expression of plpA, which reaches maximum levels at the early stationary growth phase, is activated by the global regulators HlyU and CRP at the transcriptional level. HlyU and CRP mediate the host-dependent induction of plpA and exert their effects by directly binding to the regulatory region of PplpA. The collaborative regulation of plpA by the global regulators could facilitate cooperation of VvPlpA with other virulence factors to obtain maximum effectiveness, thereby enhancing the overall success of V. vulnificus during infection.
Experimental procedures
Strains, plasmids, and culture conditions
The strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, the V. vulnificus strains were grown aerobically in LB medium supplemented with 2% (w/v) NaCl (LBS) at 30 °C, and their growth was monitored spectrophotometrically at 600 nm (A600).
Transcriptome analyses
V. vulnificus MO6-24/O (wild type) cells were used either to infect HT29-MTX intestinal epithelial cells at an MOI of 10 or to grow in the basal medium Eagle (Gibco-BRL) alone (as a negative control) for 2 h and were then harvested. Total RNAs were isolated from the bacteria using RNAprotect® bacteria reagent and the miRNeasy® minikit (Qiagen, Valencia, CA), fragmented, and used to synthesize double-stranded cDNA, as described previously (TruSeq® Stranded mRNA Sample Prep Kit, Illumina, San Diego, CA) (51). The cDNA library was amplified by PCR, sequenced, and then counted as the numbers of reads per kilobase of transcript per million mapped reads to quantify the expression levels of specific genes as described previously (52). These values, normalized using the quantile normalization procedure, were statistically analyzed by t tests to identify the genes expressed differentially (>4-fold change with p ≤ 0.05) when exposed to the HT-29 MTX cells.
Generation and complementation of the plpA, hlyU, and hlyU crp mutants
The plpA gene was inactivated in vitro by deletion of the ORF of plpA (353 of 1254 bp) using the PCR-mediated linker-scanning mutation method as described previously (53). Briefly, pairs of primers PLPA01-F and -R (for amplification of the 5′ amplicon) or PLPA02-F and -R (for amplification of the 3′ amplicon) were designed and used (Table 2). The resulting ΔplpA was amplified by PCR using the mixture of both amplicons as the template and PLPA01-F and PLPA02-R as primers. Similar experimental procedures were adopted for amplification of the ΔhlyU in vitro, except that primers HLYU01-F, HLYU01-R, HLYU02-F, and HLYU02-R (for 249-bp deleted hlyU) were used as indicated in Table 2. The resulting ΔplpA and ΔhlyU were ligated into SpeI-SphI–digested pDM4 (54) to generate pKK1301 and pZW1401, respectively (Table 1). E. coli S17-1 λpir, tra strain (55) containing pKK1301 or pZW1401 was used as a conjugal donor to V. vulnificus MO6-24/O to generate either the plpA mutant KK131 or the hlyU mutant ZW141, respectively (Table 1). Similarly, E. coli S17−1 λpir, tra containing pBS0907, which was constructed previously to carry a mutant allele of V. vulnificus crp on pDM4 (Table 1) (56), was used as a conjugal donor to ZW141 to generate the hlyU crp double mutant KK151 (Table 1). The conjugation and isolation of the transconjugants were conducted using the method described previously (56).
Table 2.
Oligonucleotides used in this study
| Name | Oligonucleotide sequence (5′ → 3′)a,b | Use |
|---|---|---|
| For mutant construction | ||
| PLPA01-F | ATGAAGAAGATAACTATTCTGTTGGGTGC | Deletion of plpA ORF |
| PLPA01-R | GTAGAATTCACAACGTTGCTTAATCACG | |
| PLPA02-F | GTGAATTCTACATCATTCAGGGTTACAACATC | Deletion of plpA ORF |
| PLPA02-R | TGAGCTGTGAGTGGCACAATCG | |
| HLYU01-F | CTTGTTTTGGTCGCGATGG | Deletion of hlyU ORF |
| HLYU01-R | CAGAATTCTTTATTGCGAAGAATAATGC | |
| HLYU02-F | AAAGAATTCTGCTCCATATCTTTTAAGTTC | Deletion of hlyU ORF |
| HLYU02-R | CGACACAAAGTCCCGGCC | |
| For mutant complementation | ||
| PLPA03-F | GGTACCAATAATCAAAACATAAGAAGATAG | Amplification of plpA ORF |
| PLPA03-R | GGTACCCTAAAAATTAAAGCG | |
| HLYU03-F | GAGCTCTCTCAAAGAAGAGTATTACGTG | Amplification of hlyU ORF |
| HLYU03-R | GGTACCTTACACCGATCCTAGACC | |
| CRP01-F | GAGCTCACACCCTTGTGGTGCC | Amplification of crp ORF |
| CRP01-R | GGTACCTTAACGAGTACCGTAAACAACG | |
| For protein overexpression | ||
| PLPA04-F | CATATGGAGCCAGCTCTCTCTCCTGAAGC | Amplification of plpA-his6 ORF |
| PLPA04-R | ACTAGTCTAGTGATGGTGATGGTGATGAAAATTAAAGCGTTG | |
| PLPA05-F | CCATGGAGCCAGCTCTCTCTCC | Amplification of truncated plpA ORF |
| PLPA05-R | GTCGACGTTGCCGGTATCAGAG | |
| HLYU04-F | GGATCCATGAACTTAAAAGATATGGAGC | Amplification of hlyU ORF |
| HLYU04-R | CTCGAGTTATTCTTCGCAATAAAGA | |
| For real-time qPCR | ||
| PLPA-qRT-F | TTGTTGGTATCGAACGGGCA | |
| PLPA-qRT-R | CGAGCTCCACCAATAACCGT | |
| 16S-qRT-F | CGGCAGCACAGAGAAACTTG | Quantification of the16S rRNA expression |
| 16S-qRT-R | CCGTAGGCATCATGCGGTAT | |
| For primer extension analysis, EMSA, and DNase I protection assay | ||
| PLPA06-F | GCGTAAATGCGCAATGAAAAG | Amplification of plpA upstream region |
| PLPA06-R | CTGCAGATGTGAAGGGCAACAGA | Amplification of plpA upstream region, extension of plpA transcript |
| For promoter deletion analysis | ||
| PLPAR001 | GAGCTCTAAAATGCCAAAAAAATCGC | Deletion of plpA regulatory region |
| PLPAR002 | GAGCTCGAGAAAGTGAATGTTAAATTATTG | |
| PLPAR003 | GAGCTCTTTTTGACTTTGGTACCAGTTTT | |
| PLPAR004 | GAGCTCCAACTGGTGATTAATAATCAAAAC | |
| PLPAR005 | ACTAGTGTGATCGCTTCAGGAGAGAGA | |
a The oligonucleotides were designed using the V. vulnificus MO6-24/O genomic sequence (GenBankTM accession number CP002469 and CP002470; www.ncbi.nlm.nih.gov).
b Regions of oligonucleotides not complementary to the corresponding genes are underlined.
To complement the plpA, hlyU, crp, and pilD (constructed previously (20)) mutations, each ORF of plpA, hlyU, crp, and pilD was amplified by PCR using a pair of specific primers, as listed in Table 2. The amplified plpA, hlyU, crp, and pilD ORFs were cloned into the broad-host-range vector pRK415 (for plpA) (57) and pJH0311 (for hlyU, crp, and pilD) (58) under the lac promoter to create pKK1320, pZW1510, pKK1502, and pMS0908, respectively (Table 1). The plasmids were transferred into the appropriate mutants by conjugation as described above. For complementation tests, when the cultures reached an A600 of 0.3, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.1 mm to induce the expression of the recombinant genes on the plasmids.
Protein purification and Western blot analysis
The ORF of plpA with a His6 tag was amplified by PCR using a pair of primers, PLPA04-F and -R (Table 2), digested with NcoI and SalI, and ligated into pMBP parallel 1 (59), resulting in pKK1503 (Table 1). The maltose-binding protein-PlpA-His6 protein was expressed in E. coli BL21 (DE3), the maltose-binding protein portion was removed, and the His6-tagged rPlpA was further purified using a HiLoad Superdex 200 gel filtration column according to the manufacturer's procedures (GE Healthcare). The purified rPlpA was used to perform a PLA2 activity assay. To raise a polyclonal antibody against PlpA, an N-terminal region of PlpA (amino acids 1–150) was subcloned into pET-28a(+) (Novagen, Madison, WI) using primers PLPA05-F and -R (Table 2), resulting in pKK1504 (Table 1). The ORF of hlyU was subcloned into pET-28a(+) using a pair of primers HLYU04-F and -R (Table 2), resulting in pYU1317 (Table 1). The plasmid pHK0201 containing an ORF for His6-tagged CRP was previously constructed (Table 1) (49). The His6-tagged truncated PlpA, HlyU, and CRP were expressed in E. coli BL21 (DE3) and purified by affinity chromatography (Qiagen).
The purified His6-tagged truncated PlpA, HlyU, and CRP proteins were used to raise rabbit polyclonal antibodies to PlpA, HlyU, and CRP of V. vulnificus, respectively (AB Frontier, Seoul, South Korea). A mouse antibody to E. coli DnaK was purchased (Enzo Lifescience, Farmingdale, NY) and used to detect V. vulnificus DnaK as a loading control (31). The V. vulnificus cultures grown to various A600 values were harvested and fractionated into cells and supernatants by centrifugation. The cells were lysed using cOmpleteTM Lysis-M EDTA-free buffer (Roche, Mannheim, Germany) for 10 min, and residual cell debris was removed by centrifugation to obtain clear lysates. The supernatants were filtered through a PuradiscTM 25-mm syringe filter (pore size 0.2 μm; GE Healthcare) and concentrated using Amicon Ultra-15 (cut-off 10 kDa; Millipore, Temecula, CA). HlyU, CRP, and DnaK in the clear cell lysates (equivalent to 10 μg of total protein) or VvPlpA in the supernatant concentrates (equivalent to 2 μg of total protein) were determined by Western blot analyses, as described previously (60). Similarly, culture medium of INT-407 (ATCC® CCL-6) human epithelial cells, infected with the V. vulnificus strains at an MOI of 10 for 2 h, were 5-fold concentrated and resolved by SDS-PAGE, and then the actin and CypA were detected by Western blot analyses as described previously (61).
Proteins of interest were detected with the following antibodies and reagent. Rabbit anti-VvPlpA (1:1,000), rabbit anti-V. vulnificus HlyU (1:1,000), rabbit anti-V. vulnificus CRP (1:5,000), mouse anti-E. coli DnaK (1:10,000; Enzo Lifescience), rabbit anti-CypA (1:1,000; Cell Signaling Technology, Beverly, MA), rabbit anti-actin (1:10,000; Cell Signaling Technology), HRP-conjugated goat anti-rabbit IgG (1:10,000; Sigma), and HRP-conjugated rabbit anti-mouse IgG (1:10,000, Sigma) antibodies and ECL SelectTM Western blotting detection reagent (GE Healthcare).
PLA2 activity assay
PLA2 activity was measured using the EnzChek® phospholipase A2 assay kit (Invitrogen) that consists of a fluorogenic PC substrate (red/green BODIPY PC-A2) specific to PLA2 (21). Briefly, the substrate/liposome mix was prepared by mixing 25 μl of 1 mm red/green BODIPY PC-A2 substrate in dimethyl sulfoxide, 25 μl of 10 mm 1,2-dioleoyl-sn-glycero-3-phosphocoline, and 25 μl of 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) in 5 ml of PLA2 assay buffer (Invitrogen). Various amounts of rPlpA were added to 50 μl of the substrate/liposome mix and incubated at room temperature for 0.5 h. When necessary, the culture supernatants of the V. vulnificus strains grown to an A600 of 0.9 were filtered and concentrated as described above and then added to the reaction mixture instead of rPlpA (equivalent to 2 μg of total protein). The cleavage of the sn-2 bond of the red/green BODIPY PC-A2 by the PLA2 results in an increase of fluorescence emission, which was measured by using a microplate reader with an excitation at 480 nm and emission at 515 nm (Tecan Infinite M200 reader, Männedorf, Switzerland). Data were normalized by subtracting the fluorescence emissions from appropriate negative control samples (mixed with rPlpA storage buffer or blank medium concentrate). The activity of the rPlpA was determined using the standard curve developed based on the fluorescence emission by the various units of honey bee venom PLA2.
RNA purification and transcript analysis
Total RNAs from the V. vulnificus strains grown to various A600 values were isolated using an RNeasy® minikit (Qiagen). For real-time qPCR, the concentrations of total RNAs from the strains were measured by using a Nano-Vue Plus spectrophotometer (GE Healthcare). cDNA was synthesized from 1 μg of the total RNA by using the iScriptTM cDNA synthesis kit (Bio-Rad), and real-time PCR amplification of the cDNA was performed by using the Chromo 4 real-time PCR detection system (Bio-Rad) with pairs of specific primers (Table 2), as described previously (62). Relative expression levels of the plpA mRNA were calculated by using the 16S rRNA expression level as the internal reference for normalization (50).
If required, the V. vulnificus strains were exposed to blood of uninfected mice (n = 3), MEM, or INT-407 cells for various incubation times. The mixture of V. vulnificus and the mouse blood (or V. vulnificus and the host cells) was centrifuged at 250 × g for 10 min to precipitate eukaryotic cells and debris in the mouse blood (or the host cells), and then the V. vulnificus cells were harvested from the supernatant and used to isolate total RNAs, as described previously (29).
For primer extension analysis, a 23-base primer PLPA06-R (Table 2) complementary to the coding region of plpA was end-labeled with [γ-32P]ATP and added to the RNA. The primer was then extended with SuperScript II RNase H− reverse transcriptase (Invitrogen). The cDNA products were purified and resolved on a sequencing gel alongside sequencing ladders generated from pKK1505 with the same primer. The plasmid pKK1505 was constructed by cloning the 461-bp plpA upstream region extending from −345 to +116, amplified by PCR using a pair of primers PLPA06-F and -R (Table 2), into pGEM-T Easy (Promega, Madison, WI). The primer extension product was visualized using a phosphor image analyzer (BAS1500, Fuji Photo Film Co., Ltd., Tokyo, Japan).
Mouse infection experiments
Three different experiments were conducted using female ICR mice (7-week-old specific-pathogen free; Seoul National University). For this purpose, the V. vulnificus strains grown to an A600 of 0.5 were harvested and suspended in PBS to 108 or 106 cfu/100 μl for intrasgastric or intraperitoneal infection, respectively. Mouse lethality of the V. vulnificus strains was compared as described previously (n = 15 per group) (11). The mice were intraperitoneally injected with 30 μg of iron-dextran for each gram of body weight, immediately after which they were administered with 100 μl of the inoculum intragastrically. Survival of the mice was recorded for 24 h.
To determine the number of V. vulnificus strains in the blood of infected mice and to conduct hematological analyses, the mice, without an iron-dextran pretreatment, were intraperitoneally infected with 100 μl of the inoculum (n = 11 for each wild type and plpA mutant and n = 5 for PBS control). The mice were sacrificed at 4 h postinfection to obtain blood samples. Equal amounts of diluted blood samples of wild type– and plpA mutant–infected mice (n = 6) were spread on LBS agar containing polymyxin B (100 units/ml) to specifically count the V. vulnificus cells. The recovered bacterial cells were enumerated as cfu/100 μl of blood. The remaining blood samples were used for hematological analyses, as described previously (63). Briefly, MPXI was analyzed by using a hematological autoanalyzer (AVIDA 2120, Bayer Diagnostics, Giessen, Germany). The serum levels for ALB, AST, and ALT were measured by using a biochemistry autoanalyzer (Hitachi 7180 autoanalyzer, High-Technologies Corp., Tokyo, Japan). All manipulations of mice were approved by the Animal Care and Use Committee at Seoul National University.
Cytotoxicity and cell morphology change
Two different assays were performed using INT-407 cells. To determine cytotoxicity, the monolayers of INT-407 cells were prepared and infected with the V. vulnificus strains in a 96-well tissue culture plate (Nunc, Roskilde, Denmark), and LDH activities in the supernatant were measured as described previously (29). Morphological changes were also examined by staining the INT-407 cells, which were seeded onto 6-well culture dishes (Nunc) and infected with the V. vulnificus strains at an MOI of 10 for 1.5 h. Briefly, INT-407 cell membranes and nuclei were stained with Texas Red®-X–conjugated WGA (final 5 μg/ml; Thermo Fisher Scientific) and with Hoechst® 33342 (final 5 μg/ml; Thermo Fisher Scientific), respectively, for 10 min and then photographed using a fluorescence microscope (FLoid® cell imaging station, Thermo Fisher Scientific).
Construction of a set of plpA-luxCDABE transcriptional fusions
The primer PLPAR005 (Table 2) carrying an SpeI restriction site was used in conjunction with one of the primers carrying an SacI restriction site, PLPAR001, PLPAR002, PLPAR003, or PLPAR004, to amplify the DNA of plpA extending up to −227, −157, −77, and +22 bp, respectively. The amplified DNA fragments were inserted into the SpeI-SacI–digested pBBR-lux carrying promoterless luxCDABE genes (64) to create four plpA-lux reporter constructs: pKK1514, pKK1515, pKK1516, and pKK1517. The constructs were then transferred into the V. vulnificus strains by conjugation. The cellular luminescences of the cultures grown to an A600 of 0.9 were measured using a microplate reader (Tecan Infinite M200 reader) and expressed in arbitrary RLUs as described previously (48).
EMSA and DNase I protection assay
The 461-bp plpA upstream region, extending from −345 to +116, was amplified by PCR using unlabeled PLPA06-F and [γ-32P]ATP-labeled PLPA06-R and as primers (Table 2). The binding of HlyU to the labeled DNA and electrophoretic analysis of the HlyU–DNA complexes have already been described (13, 56). The protein–DNA binding reactions with CRP were performed in the same manner as those with HlyU, except that the 1× CRP-binding buffer was used (49).
The same labeled 461-bp DNA was used for DNase I protection assays. The binding of HlyU or CRP to the labeled DNA and DNase I digestion of the protein–DNA complexes followed the procedures described previously (56). The digested DNA products were precipitated with ethanol and then resolved on a sequencing gel beside the sequencing ladders generated as described above. The gels were visualized using a phosphor image analyzer (BAS1500).
Data analyses
Averages and S.D. values were calculated from at least three independent experiments. Mouse mortality was evaluated using the Log Rank Test program (http://bioinf.wehi.edu.au/software/russell/logrank/).4 All other data were analyzed by Student's t tests using the SAS program (SAS Institute Inc.). Significance of differences between experimental groups was accepted at a p value of <0.05.
Transcriptome data accession number
All raw transcriptome data were deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/)4 (67) under accession number E-MTAB-5629.
Author contributions
K. K. J., M. H. K., B. S. K., and S. H. C. designed the research; K. K. J., Z.-W. L., B. K., and B. S. K. performed the research; K. K. J., M. H. K., B. S. K., Y. H. J., H. J. H., and S. H. C. analyzed the data; and K. K. J., B. S. K., and S. H. C. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank all members of the Choi laboratories for valuable discussions and technical support.
This work was supported by National Research Foundation Mid-career Researcher Program Grant 2015R1A2A1A13001654 from the Ministry of Science, ICT, and Future Planning and the R&D Convergence Center Support Program of the Ministry of Agriculture, Food, and Rural Affairs, Republic of Korea (to S. H. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MARTX
- multifunctional-autoprocessing repeat-in-toxin
- PLA
- phospholipase A
- MTX
- methotrexate
- CRP
- cAMP receptor protein
- VvPlpA
- V. vulnificus PlpA
- qPCR
- quantitative PCR
- T2SS
- type II secretion system
- VaPlpA
- V. anguillarum PlpA
- PC
- phosphatidylcholine
- rPlpA
- recombinant VvPlpA protein
- MPXI
- myeloperoxidase index
- ALB
- albumin
- AST
- aspartate aminotransferase
- ALT
- alanine aminotransferase
- LDH
- lactate dehydrogenase
- MOI
- multiplicity of infection
- WGA
- wheat germ agglutinin
- CypA
- cyclophilin A
- MEM
- minimal essential medium
- RLU
- relative light unit.
References
- 1. Oliver J. D. (2015) The biology of Vibrio vulnificus. Microbiol. Spectr. 10.1128/microbiolspec.VE-0001-2014 [DOI] [PubMed] [Google Scholar]
- 2. Phillips K. E., and Satchell K. J. (2017) Vibrio vulnificus: from oyster colonist to human pathogen. PLoS Pathog. 13, e1006053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones M. K., and Oliver J. D. (2009) Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77, 1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Falkow S. (1988) Molecular Koch's postulates applied to microbial pathogenicity. Rev. Infect. Dis. 2, S274–S276 [DOI] [PubMed] [Google Scholar]
- 5. Jeong H. G., and Satchell K. J. (2012) Additive function of Vibrio vulnificus MARTXVv and VvhA cytolysins promotes rapid growth and epithelial tissue necrosis during intestinal infection. PLoS Pathog. 8, e1002581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim Y. R., Lee S. E., Kang I. C., Nam K. I., Choy H. E., and Rhee J. H. (2013) A bacterial RTX toxin causes programmed necrotic cell death through calcium-mediated mitochondrial dysfunction. J. Infect. Dis. 207, 1406–1415 [DOI] [PubMed] [Google Scholar]
- 7. Schmiel D. H., and Miller V. L. (1999) Bacterial phospholipases and pathogenesis. Microbes Infect. 1, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 8. Takahashi M., Okazaki H., Ogata Y., Takeuchi K., Ikeda U., and Shimada K. (2002) Lysophosphatidylcholine induces apoptosis in human endothelial cells through a p38-mitogen-activated protein kinase-dependent mechanism. Atherosclerosis 161, 387–394 [DOI] [PubMed] [Google Scholar]
- 9. Walthers D., Li Y., Liu Y., Anand G., Yan J., and Kenney L. J. (2011) Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 286, 1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de los Reyes-Gavilán C. G., Suárez A., Fernández-García M., Margolles A., Gueimonde M., and Ruas-Madiedo P. (2011) Adhesion of bile-adapted Bifidobacterium strains to the HT29-MTX cell line is modified after sequential gastrointestinal challenge simulated in vitro using human gastric and duodenal juices. Res. Microbiol. 162, 514–519 [DOI] [PubMed] [Google Scholar]
- 11. Jang K. K., Gil S. Y., Lim J. G., and Choi S. H. (2016) Regulatory characteristics of Vibrio vulnificus gbpA gene encoding a mucin-binding protein essential for pathogenesis. J. Biol. Chem. 291, 5774–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williams S. G., Attridge S. R., and Manning P. A. (1993) The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol. Microbiol. 9, 751–760 [DOI] [PubMed] [Google Scholar]
- 13. Liu M., Naka H., and Crosa J. H. (2009) HlyU acts as an H-NS antirepressor in the regulation of the RTX toxin gene essential for the virulence of the human pathogens Vibrio vulnificus CMCP6. Mol. Microbiol. 72, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li L., Mou X., and Nelson D. R. (2011) HlyU is a positive regulator of hemolysin expression in Vibrio anguillarum. J. Bacteriol. 193, 4779–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Green J., Stapleton M. R., Smith L. J., Artymiuk P. J., Kahramanoglou C., Hunt D. M., and Buxton R. S. (2014) Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr. Opin. Microbiol. 18, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 17. Li L., Mou X., and Nelson D. R. (2013) Characterization of Plp, a phosphatidylcholine-specific phospholipase and hemolysin of Vibrio anguillarum. BMC Microbiol. 13, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akoh C. C., Lee G. C., Liaw Y. C., Huang T. H., and Shaw J. F. (2004) GDSL family of serine esterase/lipase. Prog. Lipid Res. 43, 534–552 [DOI] [PubMed] [Google Scholar]
- 19. Rock J. L., and Nelson D. R. (2006) Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 74, 2777–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim M. S., Kim J. A., Lim J. G., Kim B. S., Jeong K. C., Lee K. H., and Choi S. H. (2011) Identification and characterization of a novel serine protease, VvpS, that contains two functional domains and is essential for autolysis of Vibrio vulnificus. J. Bacteriol. 193, 3722–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rahman M. S., Ammerman N. C., Sears K. T., Ceraul S. M., and Azad A. F. (2010) Functional characterization of a phospholipase A2 homolog from Rickettsia typhi. J. Bacteriol. 192, 3294–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leibovici L., Samra Z., Konigsberger H., Drucker M., Ashkenazi S., and Pitlik S. D. (1995) Long-term survival following bacteremia and fungemia. JAMA 274, 807–812 [PubMed] [Google Scholar]
- 23. Yonezawa K., Horie O., Yoshioka A., Matsuki S., Tenjin T., Tsukamura Y., Yoneda M., Shibata K., Koike Y., Nomura T., Yokoyama M., Urahama N., and Ito M. (2010) Association between the neutrophil myeloperoxidase index and subsets of bacterial infections. Int. J. Lab. Hematol. 32, 598–605 [DOI] [PubMed] [Google Scholar]
- 24. Xie D. L., Zheng M. M., Zheng Y., Gao H., Zhang J., Zhang T., Guo J. C., Yang X. F., Zhong X. P., and Lou Y. L. (2017) Vibrio vulnificus induces mTOR activation and inflammatory responses in macrophages. PLoS One 12, e0181454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christofferson D. E., and Yuan J. (2010) Cyclophilin A release as a biomarker of necrotic cell death. Cell Death Differ. 17, 1942–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeong H. G., and Choi S. H. (2008) Evidence that AphB, essential for the virulence of Vibrio vulnificus, is a global regulator. J. Bacteriol. 190, 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim Y., Kim B. S., Park Y. J., Choi W. C., Hwang J., Kang B. S., Oh T. K., Choi S. H., and Kim M. H. (2010) Crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription. J. Biol. Chem. 285, 14020–14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H. J., Kim J. A., Lee M. A., Park S. J., and Lee K. H. (2013) Regulation of haemolysin (VvhA) production by ferric uptake regulator (Fur) in Vibrio vulnificus: repression of vvhA transcription by Fur and proteolysis of VvhA by Fur-repressive exoproteases. Mol. Microbiol. 88, 813–826 [DOI] [PubMed] [Google Scholar]
- 29. Lim J. G., and Choi S. H. (2014) IscR is a global regulator essential for pathogenesis of Vibrio vulnificus and induced by host cells. Infect. Immun. 82, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim J. A., Park J. H., Lee M. A., Lee H. J., Park S. J., Kim K. S., Choi S. H., and Lee K. H. (2015) Stationary-phase induction of vvpS expression by three transcription factors: repression by LeuO and activation by SmcR and CRP. Mol. Microbiol. 97, 330–346 [DOI] [PubMed] [Google Scholar]
- 31. Liu M., Rose M., and Crosa J. H. (2011) Homodimerization and binding of specific domains to the target DNA are essential requirements for HlyU to regulate expression of the virulence gene rtxA1, encoding the repeat-in-toxin protein in the human pathogen Vibrio vulnificus. J. Bacteriol. 193, 6895–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cameron A. D., and Redfield R. J. (2006) Non-canonical CRP sites control competence regulons in Escherichia coli and many other γ-proteobacteria. Nucleic Acids Res. 34, 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Browning D. F., and Busby S. J. (2016) Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 14, 638–650 [DOI] [PubMed] [Google Scholar]
- 34. Kim Y. R., Lee S. E., Kim C. M., Kim S. Y., Shin E. K., Shin D. H., Chung S. S., Choy H. E., Progulske-Fox A., Hillman J. D., Handfield M., and Rhee J. H. (2003) Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71, 5461–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee K. E., Bang J. S., Baek C. H., Park D. K., Hwang W., Choi S. H., and Kim K. S. (2007) IVET-based identification of virulence factors in Vibrio vulnificus MO6–24/O. J. Microbiol. Biotechnol. 17, 234–243 [PubMed] [Google Scholar]
- 36. Bisharat N., Bronstein M., Korner M., Schnitzer T., and Koton Y. (2013) Transcriptome profiling analysis of Vibrio vulnificus during human infection. Microbiology 159, 1878–1887 [DOI] [PubMed] [Google Scholar]
- 37. Williams T. C., Blackman E. R., Morrison S. S., Gibas C. J., and Oliver J. D. (2014) Transcriptome sequencing reveals the virulence and environmental genetic programs of Vibrio vulnificus exposed to host and estuarine conditions. PLoS One 9, e114376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nouri-Sorkhabi M. H., Agar N. S., Sullivan D. R., Gallagher C., and Kuchel P. W. (1996) Phospholipid composition of erythrocyte membranes and plasma of mammalian blood including Australian marsupials; quantitative 31P NMR analysis using detergent. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 113, 221–227 [DOI] [PubMed] [Google Scholar]
- 39. Virtanen J. A., Cheng K. H., and Somerharju P. (1998) Phospholipid composition of the mammalian red cell membrane can be rationalized by a superlattice model. Proc. Natl. Acad. Sci. U.S.A. 95, 4964–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee S. E., Ryu P. Y., Kim S. Y., Kim Y. R., Koh J. T., Kim O. J., Chung S. S., Choy H. E., and Rhee J. H. (2004) Production of Vibrio vulnificus hemolysin in vivo and its pathogenic significance. Biochem. Biophys. Res. Commun. 324, 86–91 [DOI] [PubMed] [Google Scholar]
- 41. Gavin H. E., Beubier N. T., and Satchell K. J. (2017) The effector domain region of Vibrio vulnificus MARTX toxin confers biphasic epithelial barrier disruption and is essential for systemic spread from the intestine. PLoS Pathog. 13, e1006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zong W. X., and Thompson C. B. (2006) Necrotic death as a cell fate. Genes Dev. 20, 1–15 [DOI] [PubMed] [Google Scholar]
- 43. Hering N. A., Richter J. F., Krug S. M., Günzel D., Fromm A., Bohn E., Rosenthal R., Bücker R., Fromm M., Troeger H., and Schulzke J. D. (2011) Yersinia enterocolitica induces epithelial barrier dysfunction through regional tight junction changes in colonic HT-29/B6 cell monolayers. Lab. Invest. 91, 310–324 [DOI] [PubMed] [Google Scholar]
- 44. Kim B. S., Gavin H. E., and Satchell K. J. (2015) Distinct roles of the repeat-containing regions and effector domains of the Vibrio vulnificus multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin. MBio 6, e00324–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horseman M. A., and Surani S. (2011) A comprehensive review of Vibrio vulnificus: an important cause of severe sepsis and skin and soft-tissue infection. Int. J. Infect. Dis. 15, e157–e166 [DOI] [PubMed] [Google Scholar]
- 46. Green J., Rolfe M. D., and Smith L. J. (2014) Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide. Virulence 5, 794–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fang F. C., Frawley E. R., Tapscott T., and Vázquez-Torres A. (2016) Bacterial stress responses during host infection. Cell Host Microbe 20, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong H. S., Lee M. H., Lee K. H., Park S. J., and Choi S. H. (2003) SmcR and cyclic AMP receptor protein coactivate Vibrio vulnificus vvpE encoding elastase through the RpoS-dependent promoter in a synergistic manner. J. Biol. Chem. 278, 45072–45081 [DOI] [PubMed] [Google Scholar]
- 49. Choi H. K., Park N. Y., Kim D. I., Chung H. J., Ryu S., and Choi S. H. (2002) Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J. Biol. Chem. 277, 47292–47299 [DOI] [PubMed] [Google Scholar]
- 50. Oh M. H., Lee S. M., Lee D. H., and Choi S. H. (2009) Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect. Immun. 77, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kirk J. S., Schaarschuch K., Dalimov Z., Lasorsa E., Ku S., Ramakrishnan S., Hu Q., Azabdaftari G., Wang J., Pili R., and Ellis L. (2015) Top2a identifies and provides epigenetic rationale for novel combination therapeutic strategies for aggressive prostate cancer. Oncotarget 6, 3136–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mortazavi A., Williams B. A., McCue K., Schaeffer L., and Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 53. Kim S., Bang Y. J., Kim D., Lim J. G., Oh M. H., and Choi S. H. (2014) Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93, 992–1009 [DOI] [PubMed] [Google Scholar]
- 54. Milton D. L., O'Toole R., Horstedt P., and Wolf-Watz H. (1996) Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178, 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Simon R., Priefer U., and Pühler A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1, 784–791 [Google Scholar]
- 56. Kim B. S., Hwang J., Kim M. H., and Choi S. H. (2011) Cooperative regulation of the Vibrio vulnificus nan gene cluster by NanR protein, cAMP receptor protein, and N-acetylmannosamine 6-phosphate. J. Biol. Chem. 286, 40889–40899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Keen N. T., Tamaki S., Kobayashi D., and Trollinger D. (1988) Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70, 191–197 [DOI] [PubMed] [Google Scholar]
- 58. Goo S. Y., Lee H. J., Kim W. H., Han K. L., Park D. K., Lee H. J., Kim S. M., Kim K. S., Lee K. H., and Park S. J. (2006) Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect. Immun. 74, 5586–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sheffield P., Garrard S., and Derewenda Z. (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 60. Lim J. G., Bang Y. J., and Choi S. H. (2014) Characterization of the Vibrio vulnificus 1-Cys peroxiredoxin Prx3 and regulation of its expression by the Fe-S cluster regulator IscR in response to oxidative stress and iron starvation. J. Biol. Chem. 289, 36263–36274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tian X., Zhao C., Zhu H., She W., Zhang J., Liu J., Li L., Zheng S., Wen Y. M., and Xie Y. (2010) Hepatitis B virus (HBV) surface antigen interacts with and promotes Cyclophilin A secretion: possible link to pathogenesis of HBV infection. J. Virol. 84, 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim S. M., Lee D. H., and Choi S. H. (2012) Evidence that the Vibrio vulnificus flagellar regulator FlhF is regulated by a quorum sensing master regulator SmcR. Microbiology 158, 2017–2025 [DOI] [PubMed] [Google Scholar]
- 63. Lee S. J., Jung Y. H., Ryu J. M., Jang K. K., Choi S. H., and Han H. J. (2016) VvpE mediates the intestinal colonization of Vibrio vulnificus by the disruption of tight junctions. Int. J. Med. Microbiol. 306, 10–19 [DOI] [PubMed] [Google Scholar]
- 64. Lenz D. H., Mok K. C., Lilley B. N., Kulkarni R. V., Wingreen N. S., and Bassler B. L. (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82 [DOI] [PubMed] [Google Scholar]
- 65. Wright A. C., Simpson L. M., Oliver J. D., and Morris J. G. (1990) Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58, 1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T., Megy K., Pilicheva E., Rustici G., Tikhonov A., Parkinson H., et al. (2015) ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 43, D1113–D1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.