Abstract
The bacterial condensin MukB and the cellular decatenating enzyme topoisomerase IV interact. This interaction stimulates intramolecular reactions catalyzed by topoisomerase IV, supercoiled DNA relaxation, and DNA knotting but not intermolecular reactions such as decatenation of linked DNAs. We have demonstrated previously that MukB condenses DNA by sequestering negative supercoils and stabilizing topologically isolated loops in the DNA. We show here that the MukB–topoisomerase IV interaction stabilizes MukB on DNA, increasing the extent of DNA condensation without increasing the amount of MukB bound to the DNA. This effect does not require the catalytic activity of topoisomerase IV. Cells carrying a mukB mutant allele that encodes a protein that does not interact with topoisomerase IV exhibit severe nucleoid decompaction leading to chromosome segregation defects. These findings suggest that the MukB-topoisomerase IV complex may provide a scaffold for DNA condensation.
Keywords: DNA, DNA structure, DNA topoisomerase, DNA topology, nucleic acid enzymology
Introduction
Structural maintenance of chromosome (SMC)2 proteins (1) and DNA topoisomerases play important roles in proper chromosome compaction (2, 3). In Escherichia coli, the SMC condensin protein is MukB that associates with the kleisin MukF, which binds the head domains of MukB, and another accessory protein MukE, which binds the kleisin (4). Strains deleted of mukB display decondensed nucleoids, chromosome segregation defects, and generate anucleate cells (5, 6).
Topoisomerase IV (Topo IV) is the cellular decatenase (7–9). It is a heterotetramer of a dimer of ParC (the DNA cleavage subunit) and ParE (the ATPase subunit) (10, 11). Mutations in either parC or parE are conditionally lethal, and cells carrying them display a classic par phenotype at the non-permissive temperature where a large mass of unsegregated, replicating DNA accumulates in the center of a filamenting cell (7).
The ParC subunit of Topo IV and MukB were shown to interact physically via amino acid residues in the MukB hinge region and the C-terminal β-propeller blade of the C-terminal domain of ParC (12, 13). Initial reports differed as to which activities of Topo IV were stimulated by the interaction with MukB. The Berger/Oakley laboratories reported a stimulation of DNA decatenation (13, 14). In contrast, we reported that the interaction stimulated only intramolecular reactions catalyzed by Topo IV, supercoiled DNA relaxation and knotting, but not intermolecular reactions of Topo IV such as DNA decatenation (12, 15). We therefore proposed that the MukB–Topo IV interaction played a direct role in chromosome condensation rather than in chromosome decatenation per se (15).
MukB can be found in foci in vivo that are associated with oriC (16, 17). Topo IV is also found associated with these foci (18). Depletion of MukB causes disruption of Topo IV foci, whereas the inverse is not the case, suggesting that MukB recruits Topo IV to the origin region. Live imaging of Topo IV intracellular dynamics showed that the amount of Topo IV associated with MukB foci increased during DNA replication, suggesting that Topo IV-catalyzed decatenation was ongoing in the foci (19).
In the accompanying article (45), we demonstrated that MukB alone was sufficient to condense DNA by sequestering negative supercoils and stabilizing topologically isolated loops via hinge–hinge interactions. The gel assays we developed to assess MukB DNA condensation allowed us to examine the effect of Topo IV directly. We find that the MukB-Topo IV complex leads to greater DNA compaction than by MukB alone, most likely because Topo IV stabilizes MukB on the DNA. This effect does not require the catalytic activity of Topo IV and can be mediated by ParC alone. Disruption of the MukB–Topo IV interaction in vivo leads to nucleoid decondensation and chromosome segregation defects. These findings raise the possibility that the MukB-Topo IV complex may provide a scaffold for DNA condensation in the cell.
Results
Topo IV and MukB are required to maximally compact DNA
In the accompanying article (45), we used agarose gel electrophoresis and a nicked plasmid DNA to identify two types of MukB-mediated DNA condensation: a fast-moving protein-DNA complex (FMcx) formed at low concentrations of MukB where the dominant electrophoretic determinant was MukB sequestration of negative supercoils, and a slow-moving protein-DNA complex (SMcx) formed at higher concentrations of MukB where the protein was stabilizing topologically isolated loops in the DNA (45). MukB alone was sufficient to observe this DNA compaction, neither MukEF nor ATP was required.
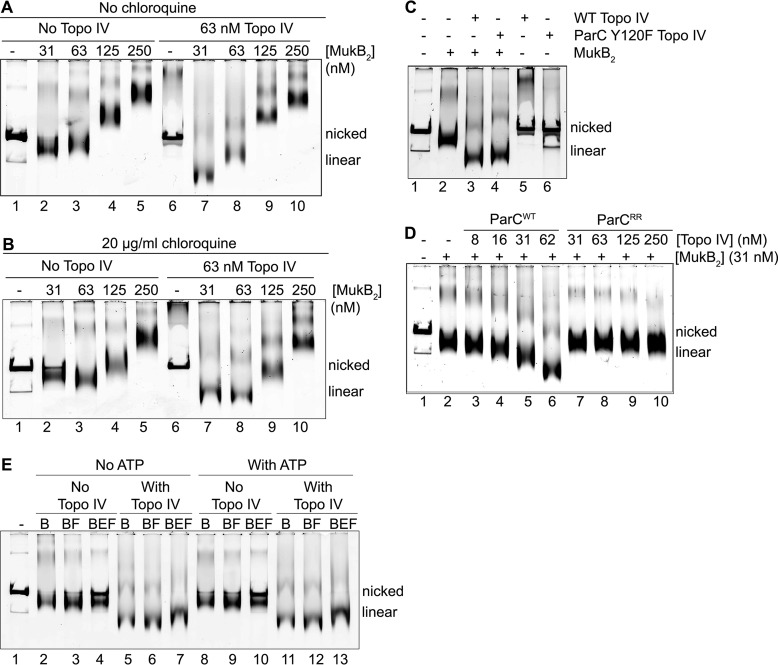
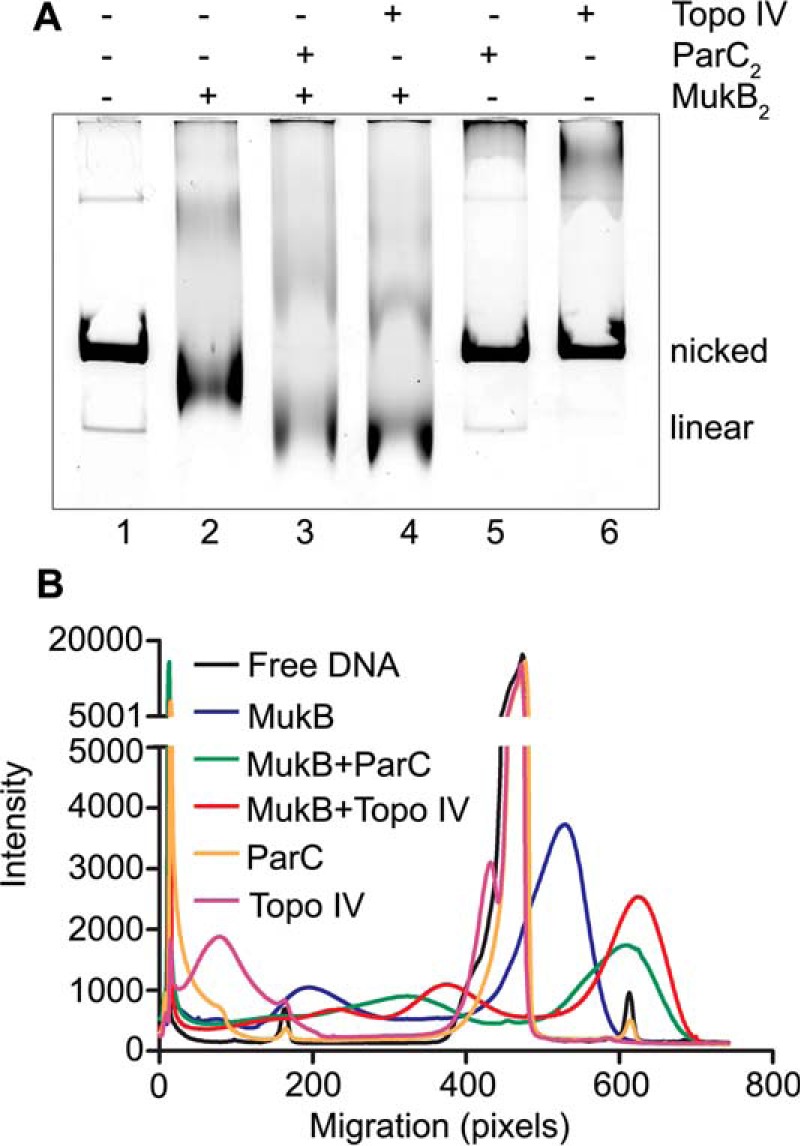
The addition of Topo IV caused an increase in the electrophoretic mobility of the FMcx formed at 31 nm MukB (compare lanes 2 and 7 in Fig. 1A and supplemental Fig. S1A). The mobility of the 31 nm MukB–Topo IV FMcx decreased relative to that of the nicked DNA in the presence of chloroquine (compare lanes 2 and 7 in Fig. 1, A and B, and supplemental Fig. S1A) indicating an increase in negative supercoiling compared with the respective MukB-DNA complex. The mobility of the FMcx formed at 63 nm MukB also increased in the presence of Topo IV (compare lanes 3 and 8 in Fig. 1A and supplemental Fig. S1A). The mobilities of the SMcxs formed at 125 and 250 nm did not change substantively in the presence of Topo IV (compare lanes 4 and 9 and 5 and 10 in Fig. 1A and supplemental Fig. S1A), likely because their electrophoretic mobilities placed these species at the exclusion limit of the gel. Topo IV alone had little effect on the mobility of the nicked DNA (Fig. 1C). However, the mobilities of the 63, 125, and 250 nm MukB-Topo IV-DNA complexes all increased relative to that of the nicked DNA in the presence of chloroquine, indicating an increase in topologically isolated loop formation compared with the respective MukB-DNA complexes (compare lanes 3 and 8, 4 and 9, and 5 and 10 in Fig. 1, A and B, and supplemental Fig. S1A).
Figure 1.
MukB and Topo IV cooperate to maximally condense DNA. The indicated concentrations of MukB and Topo IV were incubated with the nicked DNA substrate for 5 min at 37 °C, and the DNA complexes formed were analyzed by electrophoresis through 0.8% agarose gels either in the absence (A) or presence (B) of 20 μg/ml chloroquine in the gel and running buffer as described under “Experimental procedures.” Nicked, the pCG109 nicked plasmid DNA substrate; linear, linear DNA present in the substrate preparation. C, catalytic activity of Topo IV is not required for maximal DNA condensation. MukB (31 nm) and either 63 nm wild-type or ParCY120F Topo IV, as indicated, were incubated with nicked pCG109 DNA, and the reactions were processed and analyzed as in the legend to A. D, MukB–Topo IV interaction is required to achieve maximal condensation. MukB and either wild-type Topo IV or ParC R705E/R729A Topo IV, as indicated, were incubated with nicked pCG109 DNA, and the reactions were processed and analyzed as in the legend to A. E, maximal DNA condensation is independent of both MukEF and ATP. MukB2 (B), MukB2F2 (BF), or MukB2E4F2 (BEF) complexes (all at 31 nm) were incubated either in the presence or absence of 63 nm Topo IV and 2 mm ATP, as indicated, and the reactions were processed and analyzed as in the legend to A.
Thus, the presence of Topo IV improved the DNA compacting activities of MukB significantly. We refer to this effect of increased DNA condensation as “maximal compaction” to distinguish it from the extent of DNA condensation observed with MukB alone. In the accompanying article (45), we used scanning force microscopy to image the condensation of DNA by MukB. To determine whether we could glean any additional information about the maximal compaction of DNA by Topo IV and MukB, we also examined those protein-DNA complexes by scanning force microscopy (supplemental Fig. S2). However, we observed similar aggregates of mass with looped DNA as we did with MukB alone; thus, this mode of analysis was not pursued further.
Because there was no ATP present in the reaction mixtures, achieving maximal condensation did not require Topo IV catalytic activity. To confirm that this was the case, we used Topo IV reconstituted with ParC Y120F, carrying an inactivating mutation in the active-site tyrosine (Fig. 1C). The inactive Topo IV was nearly as efficient as the wild type in stimulating MukB DNA condensation, indicating that the ability of Topo IV to alter DNA topology was not required for this effect.
We next examined the effect of Topo IV reconstituted with ParC R705E/R729A on MukB DNA condensation (Fig. 1D). These amino acid substitutions disrupt the interaction between ParC and MukB (12). As clearly shown in Fig. 1D, the ParC–MukB interaction was required for maximal compaction of the DNA.
Because the tripartite condensin also contains MukEF and MukB is an ATPase, we assessed the effect of MukEF and ATP on maximal DNA compaction. The addition of MukF alone, which does not require MukE to bind to MukB and stimulates its ATPase activity (20), had a slight stabilizing effect on DNA condensation both in the presence and absence of ATP and in the presence and absence of Topo IV (compare lanes 2 and 3, 5 and 6, 8 and 9, and 11 and 12 in Fig. 1E). However, the addition of MukEF clearly destabilized MukB on the DNA in both the presence and absence of ATP, as observed by the appearance of free DNA in the lanes (compare lanes 2–4 and 8–10 in Fig. 1E). This effect is consistent with previous observations on the effect of MukEF on MukB DNA binding (21). In the presence of Topo IV, the inhibition of binding of MukB to DNA by MukEF was not observed (compare lanes 5–7 and 11–13 in Fig. 1E); however, maximal DNA compaction was decreased in the presence of MukEF (compare lanes 5 and 7 and lanes 11 and 13 in Fig. 1E). These observations suggest that the effect of MukEF on MukB may be modulated by Topo IV. We are actively investigating this possibility.
Thus, in a fashion that was independent of its catalytic activity, MukEF, and ATP, Topo IV increased the efficiency by which MukB could compact DNA. Because Topo IV itself was not capable of similar DNA compaction, these observations suggested that Topo IV was either stabilizing MukB on the DNA and/or rearranging how it bound DNA. We investigated these possibilities as described below.
Topo IV stabilizes MukB on DNA
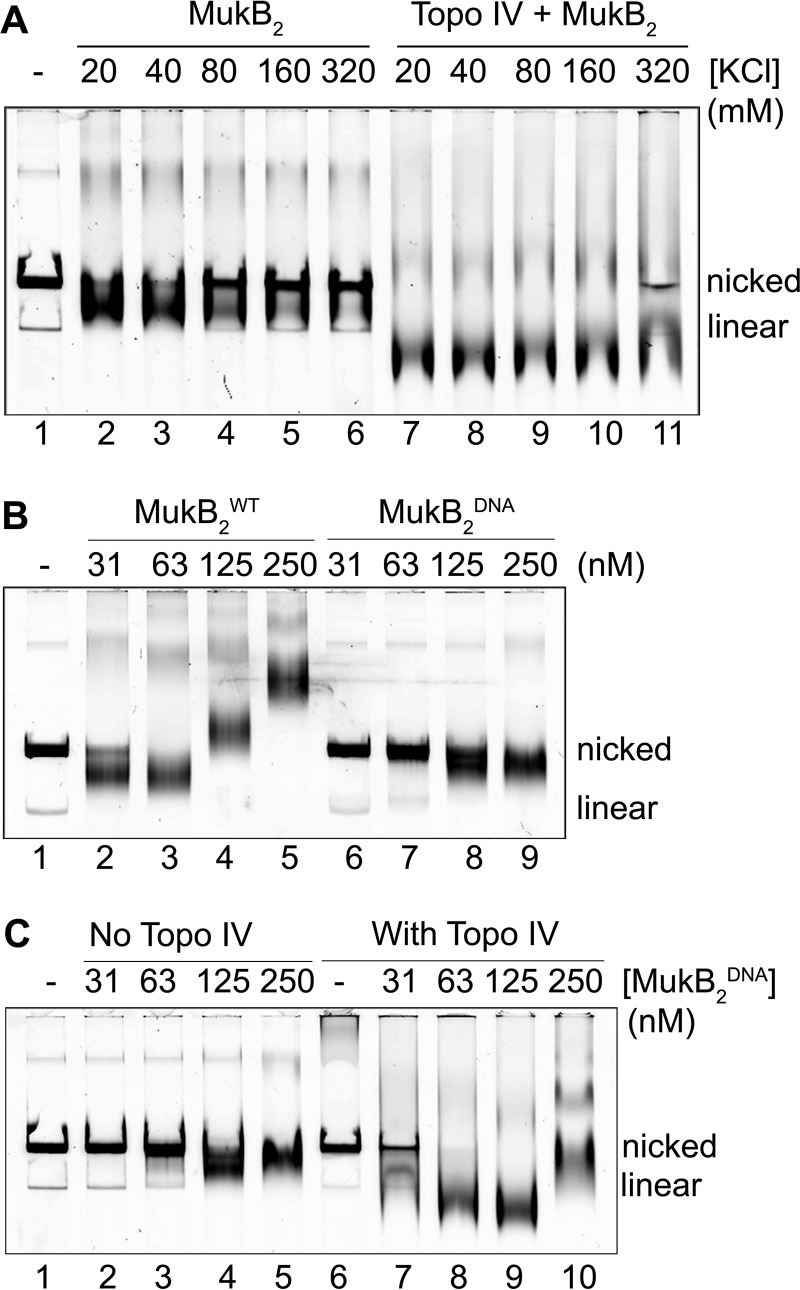
To assess whether Topo IV was stabilizing MukB on DNA, we tested the effect of salt on the stability of MukB- and MukB-Topo IV protein-DNA complexes (Fig. 2A). The FM-MukB-DNAcx formed at low MukB concentration was fairly salt-sensitive, with about one-half of the complex dissociated at 80 mm and nearly all of it dissociated at 160 mm (Fig. 2A, lanes 2–6). However, the Topo IV-MukB-DNA complex formed at the same concentration of MukB was fairly insensitive to salt, with the protein-DNA complex still clearly evident even at 320 mm (Fig. 2A, lanes 7–11). To investigate this issue further, we turned to a variant MukB protein, MukB R187E/R189E (MukBDNA) that is defective for binding DNA with a KD value roughly one-quarter that of the wild type (20). MukBDNA was similarly inefficient at DNA condensation, with some FMcx forming at 125 nm MukB compared with 31 nm for the wild type (compare lanes 2 and 8, Fig. 2B). The addition of Topo IV clearly stabilized the variant MukB on DNA, with FMcx present at 63 nm MukB in the presence of Topo IV, whereas in its absence almost no protein-DNA complex could be observed (compare lanes 3 and 8 in Fig. 2C).
Figure 2.
Topo IV stabilizes MukB on DNA. A, Topo IV-MukB-DNA complex is less salt-sensitive than the MukB-DNA complex. Standard reaction mixtures containing nicked DNA, MukB (31 nm), and Topo IV (63 nm) were incubated for 5 min at 37 °C. KCl was then added to the final concentrations indicated, and the incubation continued for 5 min. Protein-DNA complexes formed were then analyzed by gel electrophoresis as described in the legend to Fig. 1A. B, MukBDNA variant is inefficient at DNA condensation. The indicated concentrations of either wild-type or MukB R187E/R189E (MukBDNA) were incubated with nicked DNA for 5 min at 37 °C, and protein-DNA complexes formed were analyzed by electrophoresis as described in the legend to Fig. 1A. C, in the presence of Topo IV, the MukBDNA variant can condense DNA. Reaction mixtures containing nicked DNA, either in the presence (63 nm) or absence of Topo IV, and containing the indicated concentrations of the MukBDNA variant were incubated for 5 min at 37 °C, and the protein-DNA complexes formed were analyzed as described in the legend to Fig. 1A.
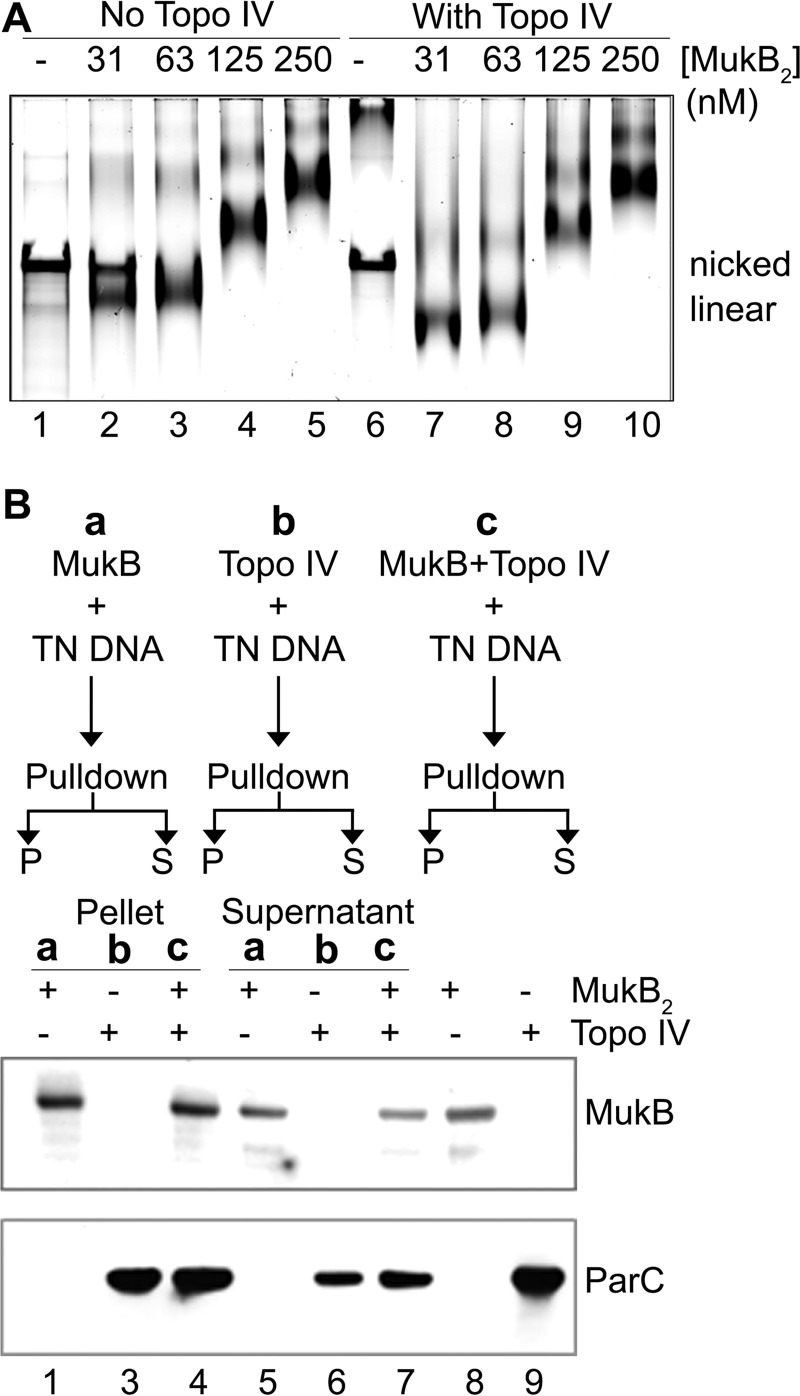
To address whether the presence of Topo IV on the DNA was recruiting additional MukB as opposed to stabilizing MukB that was already present, we used a slightly different DNA substrate where the nicked DNA had a 5′-biotinylated single-stranded tail contiguous with the 5′-side of the nick so that the DNA could be bound to streptavidin-coated magnetic beads (Fig. 3). This tailed, nicked DNA can be condensed by MukB alone to form the FMcx and SMcx (lanes 1–5, Fig. 3A), as well as maximally compacted DNA in the presence of Topo IV and MukB (lanes 6–10, Fig. 3A). Either MukB or MukB and Topo IV were bound to tailed, nicked DNA, bound to magnetic streptavidin beads, and pulled down on a magnet. Bound protein was then determined by SDS-PAGE followed by Western blotting (Fig. 3, A and C). The amount of MukB bound was essentially the same (n = 3) either in the absence (70 ± 11%) or presence (67 ± 12%) of Topo IV. Similarly, the amount of bound ParC was the same (n = 3) either in the absence (70 ± 3%) or presence (61 ± 5%) of MukB. Thus, we conclude that the presence of Topo IV on the DNA was not recruiting additional MukB to the DNA but instead was stabilizing or rearranging the MukB already bound to the DNA.
Figure 3.
Presence of Topo IV does not increase the amount of MukB on the DNA. A, tailed, nicked DNA is an effective substrate for maximal DNA condensation. Reaction mixtures containing the tailed, nicked DNA, the indicated concentrations of MukB, and either in the presence (63 nm) or absence of Topo IV were incubated for 5 min at 37 °C, and the protein-DNA complexes formed were analyzed by gel electrophoresis as described in the legend to Fig. 1A. B, schematic of the pulldown experiment. TN DNA is tailed, nicked DNA C, pulldown assay to determine the amount of MukB on the DNA. Reaction mixtures contained the tailed, nicked DNA substrate bound to streptavidin-coated magnetic beads, MukB (63 nm), and Topo IV (63 nm) as indicated. After a 5-min incubation at 37 °C, the beads were pulled down on a magnet, and the supernatants (S) were removed. The pellet (P) was resuspended in SDS-PAGE loading buffer, and the supernatants were mixed 1:1 with 2× SDS-PAGE loading buffer. After heating at 100 °C for 5 min, identical volume equivalents of the reaction mixtures were subjected to SDS-PAGE. The proteins were transferred to membranes and Western-blotted with either anti-MukB or anti-ParC antibodies as described under “Experimental procedures.”
ParC alone is sufficient to maximally compact DNA with MukB
A question at this juncture was whether intact Topo IV was required for the observed effects or whether the MukB-interacting subunit, ParC, alone was sufficient. As shown in Fig. 4, ParC in combination with MukB was sufficient to observe maximal compaction of the DNA; whereas at an equivalent concentration to Topo IV, ParC was slightly less efficient, suggesting that interaction of ParC with ParE may provide additional stabilization to the MukB-ParC protein-DNA complex.
Figure 4.
ParC is sufficient to form maximally compacted DNA with MukB. A, reaction mixtures containing nicked DNA, MukB (31 nm), and either Topo IV (63 nm) or ParC (63 nm), as indicated, were incubated for 5 min at 37 °C, and the protein-DNA complexes formed were analyzed as described in the legend to Fig. 1A. B, lane profiles of the gel shown in A.
Characterization of a MukB variant that does not interact with ParC
We have previously described a MukB variant, MukB D697K/D745K/E753K (MukBtriple), that does not interact with ParC in pulldown assays (15). MukBtriple also did not stimulate either Topo IV-catalyzed negatively supercoiled DNA relaxation or DNA knotting, unlike wild-type MukB (15). As a prelude to engineering an E. coli mutant strain defective in the interaction between MukB and ParC, we examined the ability of the MukBtriple variant to condense DNA (Fig. 5).
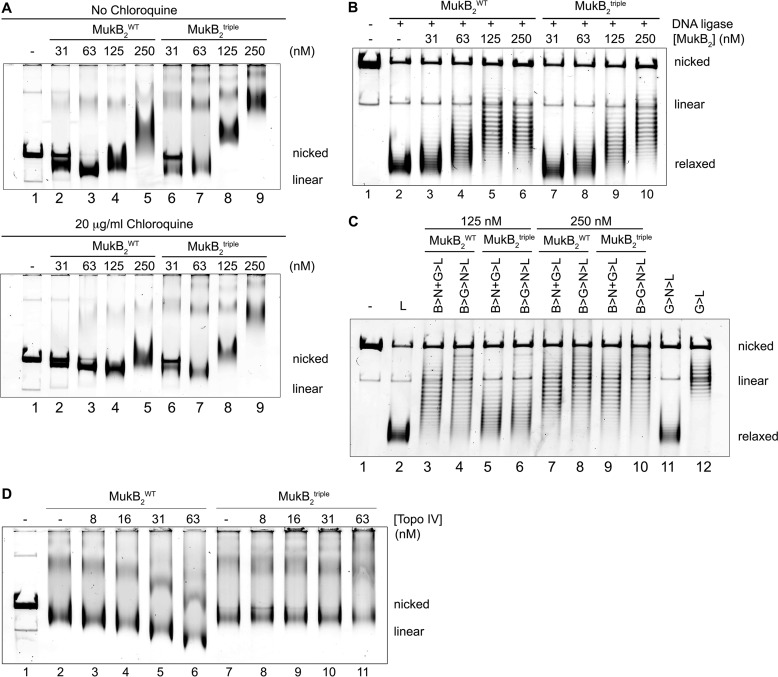
Figure 5.
MukB D697K/D745K/E753K variant condenses DNA but does not form maximally compacted DNA with Topo IV. A, DNA condensation by MukBtriple and wild-type proteins. The indicated concentrations of either MukBtriple or wild-type MukB were incubated with nicked DNA for 5 min at 37 °C, and the protein-DNA complexes formed were analyzed by gel electrophoresis either in the absence (upper panel) or presence (lower panel) of chloroquine as described in the legend to Fig. 1A. B, protection of negative supercoils by MukBtriple and wild-type proteins. The indicated concentrations of either MukBtriple or wild-type MukB were incubated with nicked DNA, E. coli DNA ligase, and NAD for 30 min at 37 °C. The samples were deproteinized, and the DNA was analyzed by electrophoresis through a 0.8% agarose gel in the presence of 10 μg/ml chloroquine in the gel and running buffer. C, loop stabilization by MukBtriple and wild-type proteins. MukB proteins were incubated with the nicked DNA substrate, 2 mm ATP, and 20 nm DNA gyrase for 30 min at 37 °C. Novobiocin was added to 10 μm, and the incubation was continued for 5 min. Bacteriophage T4 DNA ligase was then added, and the incubation was continued for 30 min. The products were deproteinized before analysis by electrophoresis through agarose gels containing 10 μg/ml chloroquine in both the gel and the running buffer. Relaxed, the nicked DNA sealed by DNA ligase to give a closed DNA ring that does not contain supercoils. The order of addition of components is outlined at the top of the gel. B, MukB; N, novobiocin; G, DNA gyrase; and L, DNA ligase. D, MukBtriple cannot form maximally condensed DNA with Topo IV. The indicated concentrations of Topo IV and either MukB or MukBtriple were incubated with the nicked DNA for 5 min at 37 °C, and the protein-DNA complexes formed were analyzed by gel electrophoresis as described in the legend to Fig. 1A.
The MukBtriple variant could condense DNA as the wild type, forming both the SMcx and FMcx (Fig. 5A and supplemental Fig. S3A). MukBtriple tended to aggregate at high concentration at 4 °C; thus the gels were run at room temperature, accounting for the lesser overall extent of DNA compaction observed compared with Fig. 1. The MukBtriple variant formed half as much of the FMcx than the wild-type protein (compare lanes 2–6 and 3–7 in Fig. 5A and see supplemental Fig. S4, A and B), whereas formation of the SMcx was comparable (compare lanes 4–8 and 5–9 in Fig. 5A and supplemental Fig. S3A), although there is a difference in the extent of loop formation (see below).
The formation of negative supercoils by MukB can be assayed by sealing the nick in the DNA substrate once MukB is bound and analyzing the products by agarose gel electrophoresis (45). MukBtriple was not as efficient as the wild type at low concentrations in protecting negative supercoils (compare lanes 3–7, 4–8, and 5–9 in Fig. 5B and see supplemental Fig. S5). This may account for the decrease in the amount of the fast-moving complex for the MukBtriple variant compared with the wild type in Fig. 5A. We have shown that supercoiling of DNA by MukB is affected by mutations in the hinge domain (45). The mutations in MukB that disrupt the interaction with ParC are in the hinge region and thus could also interfere with supercoiling. However, the FMcx formed at 31 nm MukBtriple was still dominated by negative supercoiling, as demonstrated by the decrease in mobility compared with the nicked DNA in the presence of chloroquine (compare lanes 6 in Fig. 5A and supplemental Fig. S3A), and the extent of protected supercoils formed at the highest concentration was similar (compare lanes 6 and 10 in Fig. 5B and see supplemental Fig. S5).
To assay the topological loops stabilized by MukB that dominate the SMcx, DNA gyrase and ATP are added to supercoil the loops; novobiocin is then added to inhibit DNA gyrase (22); the nick is then sealed by DNA ligase, and the products are analyzed by agarose gel electrophoresis (45). Supercoiling of the loops stabilized by MukB is obvious as an increase in negative supercoils observed when a reaction where novobiocin is added before DNA gyrase, preventing it from acting, is compared with one where gyrase is added first, allowed to act, and then inhibited by addition of novobiocin (compare even lanes with odd lanes in lanes 3–10, Fig. 5C and supplemental Fig. S6). Similar to the extent of protection of negative supercoils (Fig. 5B), less supercoiling of loops was observed with MukBtriple compared with the wild type at the lower concentration (125 nm, compare lanes 5 and 6 to lanes 3 and 4, Fig. 5C and supplemental Fig. S6); however, equivalent loop supercoiling was observed at the higher concentration of protein (250 nm, compare lanes 9 and 10 to lanes 7 and 8, Fig. 5C and supplemental Fig. S6).
Whereas MukBtriple could condense DNA by itself, it was profoundly defective in the maximal compaction observed when paired with Topo IV (compare lanes 7–11 to lanes 2–6 Fig. 5D and supplemental Fig. S7). No maximally compacted form of the DNA could be observed (for example, compare lanes 5 and 6 to lanes 10 and 11 in Fig. 5D and supplemental Fig. S7).
The data presented in this section indicates that MukBtriple is slightly defective in DNA condensation at low concentration compared with the wild-type protein. We note that the concentration of MukB in the cell ranges from about 200 molecules of dimer per cell in minimal medium (23) to 500 dimers in rich medium (21) or roughly 200-500 nm. (The level of MukBtriple expression was consistently slightly greater (14%) than the wild type in vivo (supplemental Fig. S8).) Thus, the slight condensation defect observed is not likely to manifest in vivo. We conclude that the primary defect of the MukBtriple variant is loss of interaction with Topo IV.
Loss of the MukB–Topo IV interaction in vivo results in decondensed nucleoids and chromosome segregation defects
We replaced the wild-type mukB allele in BW30270 with the mutant mukBD697KD745KE753K allele. The entire genome of the resulting strain, PN143, was sequenced to confirm that there were no additional mutations.
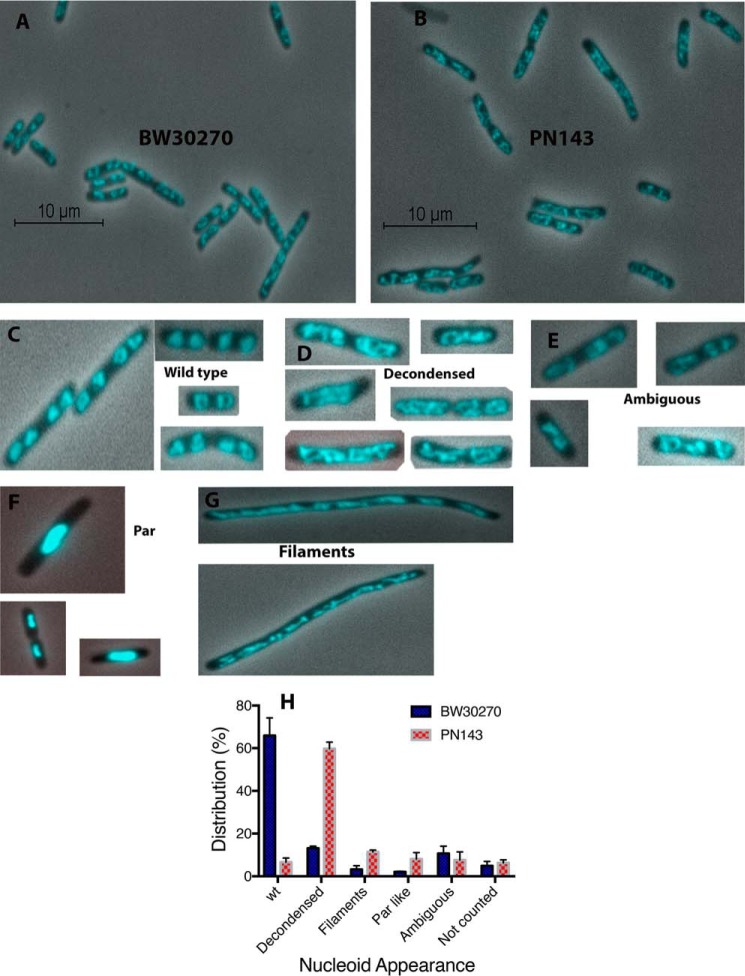
The cell and nucleoid morphology of the two strains grown to early log phase in LB medium were compared (Fig. 6). These conditions of rapid growth, where chromosomes re-initiate before replication is completed, would be expected to exacerbate any phenotype of the mutated allele. Representative fields of early log-phase cells that were briefly stained with DAPI, quickly spread on polylysine-coated slides, and observed immediately in the microscope are shown Fig. 6, A and B (N.B., the cells were not fixed). In the majority of wild-type cells, nucleoids appeared mostly spherical or U-shaped and were present in even-numbered pairs (wild type, Fig. 6C). By contrast, a small fraction of cells had nucleoids that had elongated, irregular shapes, occasionally with crossings of the DNA fiber obvious (decondensed, Fig. 6D, and see below). Some filamented cells were also evident (“filaments,” Fig. 6G), as well as a small fraction of cells that appeared par-like (“par-like,” Fig. 6F; dense nucleoids in the center of the cell), and cells where nucleoid appearance could not be accurately characterized (“ambiguous,” Fig. 6E) (Fig. 6H). However, the distribution of nucleoid morphology in PN143 was quite different (Fig. 6H). The fraction of cells with decondensed nucleoids increased by 500% compared with cells with wild-type nucleoids. Corresponding increases of roughly 400% for PN143 were observed in cells showing par-like nucleoids and filamentation (Fig. 6H).
Figure 6.
MukB–Topo IV interaction is required for proper chromosome compaction in vivo. A and B, fields of wild-type (BW30270, A) and mukBD697KD745KE753K mutant (PN143, B) cells grown in LB medium to early log phase, stained with DAPI (cells were not fixed), spread on polylysine-coated slides, and imaged immediately. C--G, examples of nucleoid and cell types. C, cells with wild-type nucleoids. D, cells with decondensed nucleoids. E, cells where the state of the nucleoids was ambiguous. F, cells with par-like nucleoids. G, filamented cells. H, distribution of cells with different nucleoid types. Three laboratory members independently categorized cells for different nucleoid types. The same images were viewed by each laboratory member. About 500 cells were categorized from each of three independent experiments. Shown is the mean of the means. Not counted, cells that were either obscured because other cells lie across them or that were not completely in the frame of the image.
To determine whether the more elongated nucleoids that we had labeled “decondensed” were, in fact, less compact than the wild-type nucleoids, we isolated spermidine nucleoids from the two strains and compared their sedimentation through sucrose gradients. Nucleoids from the mukBD697KD745KE753K strain sedimented significantly more slowly than nucleoids from the wild-type strain (Fig. 7). Thus, disrupting the MukB–Topo IV interaction led to overall nucleoid decompaction.
Figure 7.
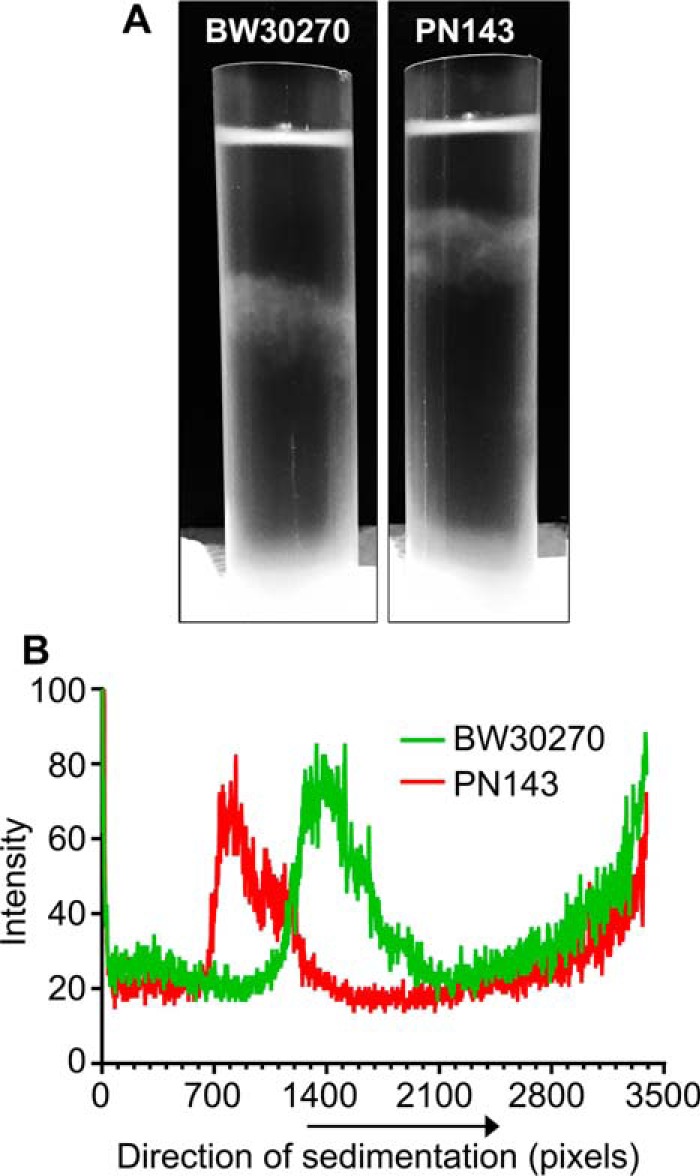
Nucleoids in the mukBD697KD745KE753K mutant strain are decondensed. Wild-type (BW30270) and mukBtriple (PN143) E. coli cells were grown to mid-log phase in MOPS medium. Cells were harvested, and spermidine nucleoids were prepared and sedimented through sucrose gradients as described under “Experimental procedures.” This figure is identical to Fig. 8 of the accompanying article (45) except that the gradient for PN141 has been removed from this image. A, photograph of typical sucrose gradients showing the positions of the sedimented nucleoids (white arrows). B, densitometric trace of A260 absorbing material in each gradient.
Whereas the fraction of filamented cells and cells containing par-like nucleoids increased in PN143, most of the cells underwent cell division and remained typically sized, suggesting that disruption of chromosome segregation was not the primary defect of the mukBtriple allele. To examine the chromosome segregation process directly, we constructed two strains where MukB was tagged with GFP and the nucleoid-associated protein HU was tagged with mCherry (PN124, BW30270 mukB-gfp hupA-mCherry, and PN139, BW30270 mukBD697KD745EE753K-gfp hupA-mCherry) and imaged both MukB localization and nucleoid division live in cells growing in MOPS medium (see supplemental Movies S1 and S2) for each strain. The mean of the wild-type nucleoid division cycle was 32.6 ± 0.7 min, and 90% of the nucleoids had divided by 41.4 ± 3.1 min; whereas the mean for the mukBtriple cells had increased to 43.5 ± 2.0 min, and it took 65.9 ± 11.1 min before 90% of the nucleoids had divided (Fig. 8). Thus, chromosome segregation in the mukBtriple cells was delayed for the majority of the nucleoids and significantly delayed for a smaller fraction. We conclude that the interaction between Topo IV and MukB is required for proper nucleoid organization. Disrupting this organization leads to delayed segregation of the daughter chromosomes.
Figure 8.
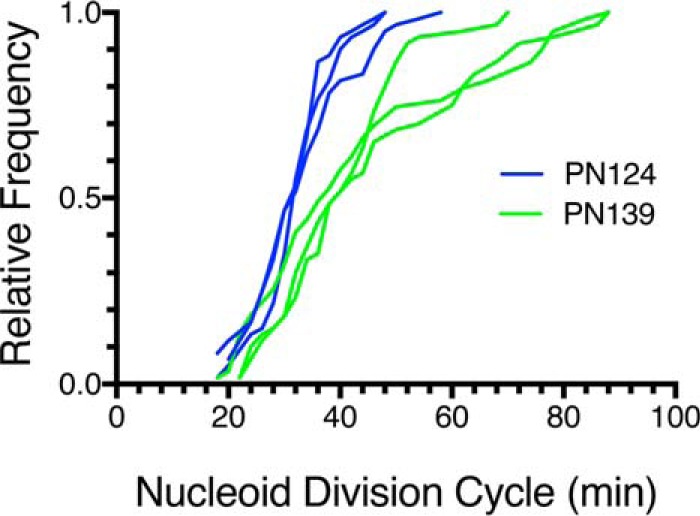
Nucleoid division cycle is elongated in mukBD697KD745KE753K mutant cells compared with wild type. Wild-type (PN124) and mukBtriple mutant (PN139) cells were grown to early log phase in MOPS medium, spread on a grooved agarose pad, and imaged every 2 min for MukB-GFP and HU-mcherry. Three laboratory members independently counted the nucleoid division cycles for 60 nucleoids from three independent experiments (20 from each supplemental movie). Each laboratory member selected nucleoids to follow at random. Shown are the cumulative distributions for PN124 (blue) and PN139 (green). See supplemental movie S1 (PN124) and supplemental movie S2 (PN139) for examples.
MukB is found in foci at the quarters of the cell that are associated with the oriC region of the chromosome (17). Formation of these foci require MukE and MukF (24), and they also contain Topo IV (18). Occupancy of the foci by both MukB and Topo IV molecules has been reported to be dynamic, with any one focus containing on average 8–10 dimers of MukBEF (MukB2(MukE2MukF)2) (23) and 15 Topo IV tetramers (19). Imaging of MukB foci in the mukBtriple mutant strain suggested that MukB had become more dispersed compared with the wild type, but quantification of this possible effect was unsatisfactory and is thus not included here.
Discussion
We have demonstrated that the interaction of MukB with the ParC subunit of Topo IV is required for maximal compaction of the DNA, amplifying DNA condensation by MukB that occurs by virtue of its ability to sequester negative supercoils and stabilize topologically isolated loops of DNA (45). This maximal condensation of DNA does not require Topo IV catalytic activity and, in fact, proceeds with the ParC subunit alone. Mutations in MukB that do not disrupt its ability to condense DNA but do disrupt its ability to interact with ParC and maximally compact DNA lead to nucleoid decompaction and chromosome segregation defects in vivo.
Topo IV and MukB cooperate to compact DNA
We showed (15) that the MukB–ParC interaction resulted in the stimulation of activities of Topo IV that involved only one DNA molecule (and thus referred to them as intramolecular), knotting of DNA, and relaxation of negative, but not positive, supercoils. Intermolecular reactions (i.e. involving more than one DNA molecule) catalyzed by Topo IV such as catenation and decatenation were not stimulated. However, Li et al. (13) argued that MukB did stimulate decatenation of kinetoplast DNA networks. Although we observed this stimulation as well, it was quite modest. By contrast, we showed that MukB did not stimulate Topo IV-catalyzed decatenation of catenated sister DNA molecules that arise during DNA replication and are linked multiple times. Because these multiply-linked DNA dimers are the actual substrate for sister chromosome unlinking, we concluded that the MukB–Topo IV interaction was not stimulating chromosome decatenation by Topo IV directly. Instead, given that MukB did stimulate the intramolecular activities of Topo IV, we proposed that the MukB–Topo IV interaction was required for proper condensation of the chromosome. Thus, disrupting this interaction would lead to chromosome decompaction that would itself lead to chromosome segregation defects. Our model has been borne out by the results reported here.
The maximal DNA compaction results from stabilization of MukB on the DNA via its interaction with Topo IV. Whereas MukE and MukF may provide some additional stabilization to the Topo IV-MukB-DNA complex, they are not required for the gain in DNA compaction provided by Topo IV and actually inhibit binding of MukB to DNA in the absence of Topo IV. MukB ATP hydrolysis has been shown to cause a re-arrangement of how MukF is bound to the MukB head domains and may regulate head domain contact in the dimeric protein (25); however, maximal DNA compaction in the presence of Topo IV was unaffected by ATP. Topo IV DNA cleavage and religation activity are also not required for maximal DNA compaction, indicating that it is not necessary for the Topo IV to modulate DNA topology to stimulate MukB-mediated DNA compaction. Thus, MukB and Topo IV/ParC could serve as the core of the DNA compacting complex, whereas the action of MukEF and the ATPase of MukB may modify its activity or placement on the DNA. Indeed, using FRAP, Nolivos et al. (26) concluded that MukB ATPase activity was required for recycling of MukB dimers to and from the DNA.
A scaffold for DNA compaction?
The requirement for association of MukB and Topo IV to achieve maximal DNA compaction was supported by our analysis of the effect of the mukBtriple allele in vivo. Disrupting the MukB–Topo IV interaction caused prominent restructuring of the nucleoids in vivo, with the DNA becoming elongated, disorganized, and tangled in appearance. We argue that it is this disorganization that leads to the observed delay in chromosome segregation, not a defect per se in Topo IV DNA-decatenating activity. In other studies where the MukB–Topo IV interaction was disrupted either by overexpressing the C-terminal domain of ParC (19) or replacing wild-type ParC with a variant defective in interaction with MukB (18), similar chromosome segregation defects were also noted based on sister chromatid cohesion time for specific tagged regions of the DNA. However, in those studies neither the state of the DNA nor the actual nucleoid division cycle was examined directly. We also note that in our hands, this variant, ParC R705E/R729A, forms a Topo IV with ParE that is defective in DNA-binding activity and the ability to complement parC temperature-sensitive strains (data not shown).
The association of a condensin and a type II topoisomerase has considerable precedent. These enzymes have been found to be major components of the nuclear matrix (27, 28). There is no evidence in bacteria of such a structure, but it has been shown that 15–20% of the MukB in the cell can be found in foci that associate with the oriC region of the chromosome. These foci require MukEF for formation, and half of them also contain Topo IV (18, 19, 23). Interestingly, in these studies, whereas Topo IV was not required for formation of MukB foci, in its absence (when it was degraded) MukB foci became delocalized along the long axis of the cell. But when wild-type ParC was substituted with the MukB non-interacting variant, Topo IV-MukB association in vivo was not disrupted (18), suggesting that co-residence of Topo IV and MukB in a focus might not be dependent on their interaction.
Cooperation of Topo IV and MukB in vivo to compact DNA suggests, but does not require, that association of the two proteins would be detectable by cell biological techniques. In fact, MukB (26) and Topo IV localizations (29) by ChIP do not overlap extensively (and there is also no obvious concentration of MukB observed in the oriC region (26)). Furthermore, very few (19 sites) Topo IV-binding sites were actually observed, whereas hundreds of norfloxacin-induced, Topo IV-mediated DNA cleavage sites were observed, although only 10–20 of these may be active in any one cell at one time (29). In these cases epitope-tagged versions of the proteins were substituted for wild type, so although it was the case that these tagged proteins did not appear to affect the growth rate of the cells, one must consider the possibility that the epitopes were hidden in Topo IV-MukB complexes on the DNA. Moreover, it is possible that the Topo IV–MukB interaction is transient, required initially to organize the condensin on DNA but not to maintain it.
Given that ParC alone could also achieve maximal DNA compaction with MukB as Topo IV, we also have to consider whether ParC might play a dual role in the cell: partners with ParE to form an active decatenating enzyme and with MukB to organize and compact DNA. Vos et al. (14) were able to dissect these two activities genetically, and ParC is generally thought to be in excess of ParE in vivo. Our own estimates indicated about twice as much ParC as ParE (30). Similarly, some comprehensive proteome-wide protein abundance surveys also indicate a similar excess of ParC over ParE, although these results vary widely (see the Protein Abundance Database, PaxDb, pax-db.org)3 (46). In contrast, counting of molecules of ParC and ParE by photobleaching of tagged versions in vivo indicated that the two subunits were roughly equal in abundance (19). Clearly further clarification is in order.
MukB, nucleoid organization, and chromosome segregation
Residence of Topo IV in MukB foci appears to increase its catalytic activity, leading to the suggestion that these foci are the location for decatenation of sister chromosomes and loading of MukB to the DNA (19). Our finding that disrupting the MukB–Topo IV interaction increases the time required for the nucleoid cycle of replication, chromosome segregation, and partition supports the previous conclusions about the effect of disrupting the MukB–Topo IV interaction on chromosome segregation.
Binding of MukB to the DNA is one of a number of forces that act to compact the chromosome. Binding of nucleoid-associated proteins clearly plays a role, as does DNA topology modulated through the action of topoisomerases (2, 3) and transcription and translation (31). Recent studies have indicated that the nucleoid-associated protein H-NS may be responsible for condensing DNA along adjacent segments of the chromosome (32), whereas changes in the multimerization state of HU brought about by the differential expression of the two alleles can allow dynamic changes in nucleoid architecture (33).
Confinement of the nucleoid has come to the forefront as a major organizing force. Here, it has been considered whether the nucleoidal fiber could be treated as a randomly oriented polymer with entropy driving newly formed sisters to occupy space as the cellular periphery grows (34). Alternatively, it has been argued that the sister nucleoidal domains were coherent and non-interacting, with separation coming about because of mechanical pushing forces possibly generated by release of internal tethers (35). Variations of the latter argument invoke intranucleoidal interactions organizing the chromosomal fiber into stacks of plectonemic loops (36) and as a compact self-adhering object with a definite, although dynamic, shape (37).
The profound change in nucleoid appearance observed in PN143 compared with the wild-type strain is supportive of the arguments that intranucleoidal interactions are important for overall nucleoid organization. Interestingly, in some instances what may be sister chromatids could be observed in parallel segments along the longitudinal length of the cell. Such a conformation would seem to be inconsistent with the model of Fisher et al. (35). Nevertheless, even with the significant disruption of nucleoid organization apparent when the MukB–Topo IV interaction is ablated, chromosome segregation, while delayed, did occur in the vast majority of cells. This observation could be taken in support of the entropic model. Further analysis of the dynamics of nucleoids in the PN143 strain should therefore prove informative.
Experimental procedures
DNAs, proteins, and cell growth and imaging were all as described in the accompanying article (45). MukB D697K/D745K/E753K was prepared as for the wild type (12) from E. coli strain BL21(DE3)pET11d-mukBD697KD745KE753K. Topo IV was reconstituted from purified ParE and ParC subunits as described (38). ParC R705E/R729A and MukBDNA were prepared as described (12). Concentrations of Topo IV and ParC used in all assays are indicated in the figure legends. All biochemical assays and in vivo studies shown were repeated at least three times.
Gel assays
Gel assays are described in detail in the accompanying article (45). A brief outline follows. MukB- and MukB-Topo IV (or ParC)-DNA complexes were assayed by incubating the components indicated in the figure legends with a singly nicked plasmid DNA (11 kbp) at 37 °C. Reaction mixtures were then applied directly to 0.8% agarose gels that were electrophoresed at 23 V for 18 h at 4 °C using 50 mm Tris-HCl (pH 7.8 at 23 °C), 40 mm NaOAc, 1 mm EDTA, and 1.5 mm Mg(OAc)2 as the electrophoresis buffer. Chloroquine di-phosphate (20 μg/ml) was added to the gel and the electrophoresis buffer as indicated in the figures. Induction of DNA supercoiling was measured by adding NAD and E. coli DNA ligase to the DNA-binding reactions to seal the nick in the DNA substrate and continuing the incubation at 37 °C. Reactions were deproteinized by proteinase K digestion and phenol-CHCl3 extraction before analyzing the products by agarose gel electrophoresis as above. To assess formation of topologically isolated loops in the DNA after binding of MukB or MukB and Topo IV to the nicked DNA, DNA gyrase was added in the presence of ATP, and the incubation was continued. Novobiocin was then added to inhibit DNA gyrase, and bacteriophage T4 DNA ligase was then added to seal the nick in the DNA. The DNA products were then analyzed after deproteinization by gel electrophoresis as above.
MukB–Topo IV pulldown and Western blotting
Reaction mixtures (20 μl) containing MukB2 and Topo IV (62.5 nm each), 50 ng of 5′-biotin tailed, nicked DNA (M13mp18) immobilized on M-280 streptavidin beads, 50 mm HEPES-KOH (pH 7.5), 20 mm KCl, 10 mm DTT, 0.5 mm Mg(OAc)2, 7.5% glycerol, and 0.05% Nonidet P-40 were incubated for 5 min at 37 °C. DNA beads were separated from the supernatant and resuspended in the same volume of reaction buffer. An equal volume of 2× SDS-PAGE sample buffer was added to both supernatant and pellet. Samples were heated for 10 min at 95 °C and separated on NuPAGE Novex 4–12% BisTris protein gels (Invitrogen) using 1× MES buffer. Proteins were then electrophoretically transferred to a PVDF membrane using Tris glycine buffer (25 mm Tris, 192 mm glycine and 20% v/v methanol) at 4 °C and 30 V for 15 h. Affinity-purified anti-MukB and anti-ParC antibodies, goat anti-rabbit IGG-HRP-conjugated secondary antibodies, and an Amersham Biosciences ECL kit were used to visualize the proteins as recorded by exposure to X-ray film. Films were quantified using Multigauge software, version 2.3 (Fuji).
Determination of MukB levels in vivo
BW30270 and PN143 were grown in MOPS EZ rich defined medium with 2% glucose to an A600 = 0.5. Cells (100 ml) were harvested and resuspended in 0.5 ml of 50 mm Tris-HCl (pH 8.0 at 4 °C), 10% sucrose. Cells were diluted to 2.0 ml in a final resuspension containing 150 mm NaCl, 20 mm EDTA, 10 mm DTT, 0.1 mm PMSF, 0.05% Triton X-100, and 0.2 mg/ml lysozyme, incubated at 37 °C for 10 min, and then chilled. Cleared lysates were recovered after centrifugation at 100,00 × g for 60 min at 4 °C. The indicated amounts of extract were subjected to SDS-PAGE using 4–12% NUPAGE BisTris gels (Invitrogen) for 2 h at 120 V. Protein was transferred to a NitroBind Cast nitrocellulose membrane, 0.45 μm (GVS North America) at 30 V for 18 h at 4 °C using 25 mm Tris base, 192 mm glycine, and 20% methanol as the transfer buffer. MukB was visualized using affinity-purified, anti-MukB antibody as above.
Isolation of spermidine nucleoids
BW30270 and PN143 were grown at 37 °C in MOPS EZ rich defined medium with 2% glucose to an A600 = 0.5. Cells (25 ml) were pelleted by centrifugation, lysed, and sedimented through 10–40% sucrose gradients as described by Murphy and Zimmerman (39) with the exception that the gradients were centrifuged for 20 min.
E. coli strains
BW30270 is an rph+ derivative of MG1655 (E. coli Genetic Stock Center). RY158 is BW30270ΔmukB::Kan. BW30270 was transduced to KanR using a P1 phage stock grown on AZ5372 (F−, trpC9941, ΔmukB::Kan) (National BioResource Project; NIG, Japan). C600mukB-ecgfps:frt-kan-frt (40) carrying a MukB-GFP fusion where GFPuv (41) was modified for optimum codon expression in E. coli and the gene was straightened by reducing AT runs as described (42). JC13509, hupA::mcherry:frtKan,Δ(attB)::sulAp-gfp (a gift from Steven Sandler, University of Massachusetts, Amherst). PN124 (BW30270mukB-ecgfpshupAmCherry:kan), as described in the accompanying article (45). PN143 (BW30270 mukBD697KD745KE753K). The ΔmukB::Kan allele in RY158 was replaced with the mukBD697KD745KE753K allele using the method of Link et al. (43); RY158 was transformed with the unstable plasmid pRC7-mukB, which is rapidly lost in the absence of selection for the ampicillin marker, and pKO3mukB2301, carrying a fragment of the mukB gene (nucleotides 499–2800) with the D697K/D745K/E753K mutations. This plasmid has a temperature-sensitive replicon and carries a sacB gene so that it is evicted when grown on sucrose. In the presence of 100 μm isopropyl 1-thio-β-d-galactopyranoside to induce expression of MukB from pRC7-mukB, RY158 carrying both plasmids was incubated at 43 °C to force integration of pKO3mukB2301 followed by resolution of the cointegrate by growth on sucrose at 30 °C. Cells were then grown in the absence of ampicillin, and AmpS and KanS cells were checked for restoration of full-length mukB by PCR. Clones carrying the mutated gene were identified by sequencing the PCR products. PN143, unlike RY158, could grow at 37 °C. The genome of the final isolate was completely sequenced. PN139 (BW30270mukBD697KD745KE753K-ecgfpshupA-mCherry:Kan) carries the mukBtriple allele tagged with our modified version of gfp (gfps, see above) and one subunit of the nucleoid associated protein HU tagged with mCherry. The mukBD697KD745KE753K allele in PN139 was fused to ecgfps using the method of Datsenko and Wanner (44) as described by Lee and Marians (40). The Kan marker was then deleted using the FLP recombinase as described, and the strain was transduced to KanR using a P1 phage stock grown on JC13509.
Author contributions
S. B. discovered the effect of Topo IV on MukB DNA condensation. R. K. performed all the experiments in Figs. 1–5 and 7. P. N. performed all the experiments in Figs. 6 and 8. C. M. L. and K. J. M. assisted in the quantification of the data in Figs. 6 and 8. R. K., S. B., P. N., and K. J. M. conceived and designed the experiments. All authors analyzed the data. K. J. M. wrote the manuscript.
Supplementary Material
Acknowledgment
The Memorial Sloan Kettering Cancer Center was recipient of National Institutes of Health Cancer Center Core Support Grant P30CA008748 from NCI.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34558 (to K. J. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S8 and supplemental Movies S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- SMC
- structural maintenance of chromosome protein
- Topo IV
- topoisomerase IV
- FMcx
- fast-moving complex
- SMcx
- slow-moving complex
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Uhlmann F. (2016) SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 [DOI] [PubMed] [Google Scholar]
- 2. Thanbichler M., and Shapiro L. (2006) Chromosome organization and segregation in bacteria. J. Struct. Biol. 156, 292–303 [DOI] [PubMed] [Google Scholar]
- 3. Luijsterburg M. S., White M. F., van Driel R., and Dame R. T. (2008) The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43, 393–418 [DOI] [PubMed] [Google Scholar]
- 4. Yamazoe M., Onogi T., Sunako Y., Niki H., Yamanaka K., Ichimura T., and Hiraga S. (1999) Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18, 5873–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., and Jaffé A. (1989) Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 171, 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niki H., Jaffé A., Imamura R., Ogura T., and Hiraga S. (1991) The new gene mukB codes for a 177-kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., and Suzuki H. (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404 [DOI] [PubMed] [Google Scholar]
- 8. Adams D. E., Shekhtman E. M., Zechiedrich E. L., Schmid M. B., and Cozzarelli N. R. (1992) The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell 71, 277–288 [DOI] [PubMed] [Google Scholar]
- 9. Peng H., and Marians K. J. (1993) Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 90, 8571–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peng H., and Marians K. J. (1993) Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268, 24481–24490 [PubMed] [Google Scholar]
- 11. Kato J., Suzuki H., and Ikeda H. (1992) Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 267, 25676–25684 [PubMed] [Google Scholar]
- 12. Hayama R., and Marians K. J. (2010) Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 18826–18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y., Stewart N. K., Berger A. J., Vos S., Schoeffler A. J., Berger J. M., Chait B. T., and Oakley M. G. (2010) Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci. U.S.A. 107, 18832–18837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vos S. M., Stewart N. K., Oakley M. G., and Berger J. M. (2013) Structural basis for the MukB–topoisomerase IV interaction and its functional implications in vivo. EMBO J. 32, 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hayama R., Bahng S., Karasu M. E., and Marians K. J. (2013) The MukB–ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV. J. Biol. Chem. 288, 7653–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohsumi K., Yamazoe M., and Hiraga S. (2001) Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40, 835–845 [DOI] [PubMed] [Google Scholar]
- 17. Danilova O., Reyes-Lamothe R., Pinskaya M., Sherratt D., and Possoz C. (2007) MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 65, 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicolas E., Upton A. L., Uphoff S., Henry O., Badrinarayanan A., and Sherratt D. (2014) The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. mBio 5, e01001–01013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zawadzki P., Stracy M., Ginda K., Zawadzka K., Lesterlin C., Kapanidis A. N., and Sherratt D. J. (2015) The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex, MukBEF. Cell Rep. 13, 2587–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bahng S., Hayama R., and Marians K. J. (2016) MukB-mediated catenation of DNA is ATP and MukEF independent. J. Biol. Chem. 291, 23999–24008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petrushenko Z. M., Lai C. H., and Rybenkov V. V. (2006) Antagonistic interactions of kleisins and DNA with bacterial condensin MukB. J. Biol. Chem. 281, 34208–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gellert M., O'Dea M. H., Itoh T., and Tomizawa J. (1976) Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. U.S.A. 73, 4474–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Badrinarayanan A., Reyes-Lamothe R., Uphoff S., Leake M. C., and Sherratt D. J. (2012) In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. She W., Wang Q., Mordukhova E. A., and Rybenkov V. V. (2007) MukEF Is required for stable association of MukB with the chromosome. J. Bacteriol. 189, 7062–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woo J. S., Lim J. H., Shin H. C., Suh M. K., Ku B., Lee K. H., Joo K., Robinson H., Lee J., Park S. Y., Ha N. C., and Oh B. H. (2009) Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136, 85–96 [DOI] [PubMed] [Google Scholar]
- 26. Nolivos S., Upton A. L., Badrinarayanan A., Müller J., Zawadzka K., Wiktor J., Gill A., Arciszewska L., Nicolas E., and Sherratt D. (2016) MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat. Commun. 7, 10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Earnshaw W. C., Halligan B., Cooke C. A., Heck M. M., and Liu L. F. (1985) Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100, 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., and Laemmli U. K. (1986) Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 188, 613–629 [DOI] [PubMed] [Google Scholar]
- 29. El Sayyed H., Le Chat L., Lebailly E., Vickridge E., Pages C., Cornet F., Cosentino Lagomarsino M., and Espéli O. (2016) Mapping topoisomerase IV binding and activity sites on the E. coli genome. PLoS Genet. 12, e1006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng H. (1995) Molecular Cloning, Purification, and Characterization of Escherichia coli topoisomerase IV. Ph.D. thesis, Cornell University, Ithaca, NY [Google Scholar]
- 31. Bakshi S., Choi H., and Weisshaar J. C. (2015) The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol. 6, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Helgesen E., Fossum-Raunehaug S., and Skarstad K. (2016) Lack of the H-NS protein results in extended and aberrantly positioned DNA during chromosome replication and segregation in Escherichia coli. J. Bacteriol. 198, 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hammel M., Amlanjyoti D., Reyes F. E., Chen J. H., Parpana R., Tang H. Y., Larabell C. A., Tainer J. A., and Adhya S. (2016) HU multimerization shift controls nucleoid compaction. Sci. Adv. 2, e1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jun S., and Mulder B. (2006) Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 103, 12388–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fisher J. K., Bourniquel A., Witz G., Weiner B., Prentiss M., and Kleckner N. (2013) Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153, 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiggins P. A., Cheveralls K. C., Martin J. S., Lintner R., and Kondev J. (2010) Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc. Natl. Acad. Sci. U.S.A. 107, 4991–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hadizadeh Yazdi N., Guet C. C., Johnson R. C., and Marko J. F. (2012) Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol. Microbiol. 86, 1318–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng H., and Marians K. J. (1999) Overexpression and purification of bacterial topoisomerase IV. Methods Mol. Biol. 94, 163–169 [DOI] [PubMed] [Google Scholar]
- 39. Murphy L. D., and Zimmerman S. B. (1997) Isolation and characterization of spermidine nucleoids from Escherichia coli. J. Struct. Biol. 119, 321–335 [DOI] [PubMed] [Google Scholar]
- 40. Lee C., and Marians K. J. (2013) Characterization of the nucleoid-associated protein YejK. J. Biol. Chem. 288, 31503–31516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crameri A., Whitehorn E. A., Tate E., and Stemmer W. P. (1996) Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14, 315–319 [DOI] [PubMed] [Google Scholar]
- 42. Corcoran C. P., Cameron A. D., and Dorman C. J. (2010) H-NS silences gfp, the green fluorescent protein gene: gfpTCD is a genetically Remastered gfp gene with reduced susceptibility to H-NS-mediated transcription silencing and with enhanced translation. J. Bacteriol. 192, 4790–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Link A. J., Phillips D., and Church G. M. (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar R., Grosbart M., Nurse P., Bahng S., Wyman C. L., and Marians K. J. (2017) The bacterial condensin MukB compacts DNA by sequestering supercoils and stabilizing topologically isolated loops. J. Biol. Chem. 292, 16904–16920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang M., Weiss M., Simonovic M., Haertinger G., Schrimpf S. P., Hengartner M. O., and von Mering C. (2012) PaxDb, a database of protein abundance averages across all three domains of life. Mol. Cell. Proteomics 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.