Abstract
Background
The association of HLA mismatching with kidney allograft survival has been well established. We examined whether amino acid (AA) mismatches (MMs) at the antigen recognition site of HLA molecules represent independent and incremental risk factors for kidney graft failure (GF) beyond those MMs assessed at the antigenic (2-digit) specificity.
Methods
Data on 240 024 kidney transplants performed between 1987 and 2009 were obtained from the Scientific Registry of Transplant Recipients. We imputed HLA-A, -B, and -DRB1 alleles and corresponding AA polymorphisms from antigenic specificity through the application of statistical and population genetics inferences. GF risk was evaluated using Cox proportional-hazards regression models adjusted for covariates including patient and donor risk factors and HLA antigen MMs.
Results
We show that estimated AA MMs at particular positions in the peptide-binding pockets of HLA-DRB1 molecule account for a significant incremental risk that was independent of the well-known association of HLA antigen MMs with graft survival. A statistically significant linear relationship between the estimated number of AA MMs and risk of GF was observed for HLA-DRB1 in deceased donor and living donor transplants. This relationship was strongest during the first 12 months after transplantation (hazard ratio, 1.30 per 15 DRB1 AA MM; P < 0.0001).
Conclusions
This study shows that independent of the well-known association of HLA antigen (2-digit specificity) MMs with kidney graft survival, estimated AA MMs at peptide-binding sites of the HLA-DRB1 molecule account for an important incremental risk of GF.
In a population of 240 024 kidney transplant recipients using the data of the Scientific Registry of Transplant recipients, the authors demonstrate that, independently of HLA antigen mismatches, estimated amino-acid mismatches at peptide-binding sites of the HLA-DRB1 molecule, accounts for an increased graft failure risk. Supplemental digital content is available in the text.
Genes encoding classical HLA molecules are extremely polymorphic, such that as of October 2015 classical HLA loci include over 13 800 allelic variants.1 HLA allele names are defined as 4 digit numbers that represent a unique protein sequence; the first 2 digits describe each allele’s serologic specificity and the last 2 digits identify successive protein variants within that specificity. In 2010, the nomenclature was updated by defining each domain as a colon-delimited field.1 Most polymorphisms are associated with the peptide-binding residues of the HLA class I and class II molecules.2 Allelic variations among these HLA molecules are the source of differential peptide binding, thymic selection, and alloreactivity.2-6 Until now, renal transplant HLA matching has focused on antigenic determination. These serologic specificities depend largely upon polymorphic solvent exposed amino acid (AA) residues of the antigen recognition site but may also be influenced by unexposed peptide binding sites impacting T cell allorecognition4-6 and alloantibody reactivity.7,8
HLA matching between donors and recipients improves the outcome of kidney transplantation. The best outcomes are observed when the donor organ has no mismatches (MMs) with the recipient at any of the 6 loci of the HLA-A, −B, and -DRB1 antigens.9,10 The relative risk of graft failure (GF) is weakly related to the number of HLA-A or -B antigen MMs, but more strongly associated with increases in the number of antigen MMs at HLA-DRB1.11-15
This conventional approach to HLA matching does not take into account how closely the mismatched HLA specificities are at AA polymorphic sites; with any given HLA antigen (2-digit specificity) MM, there is significant variability in the degree of AA MMs at the antigen recognition site that could dramatically impact allorecognition.5,16-20 Increasing attention is being focused on the importance of HLA epitope matching strategy to improve kidney transplantation outcomes.21-29 A few of these studies suggested that the degree of HLA disparity between a given donor and recipient pair is better expressed at the level of AA disparity than by the conventional antigen matching approach.21,22,25-27,29 However, the validity of these studies has been questioned, and it was suggested instead that the apparent benefit of epitope matching is dependent on underlying HLA serologic matching.26,30 We postulated that HLA MMs at the level of antigenic specificities do not fully describe the risk of kidney GF that may be related to biologically important AA substitutions in HLA molecules between donors and recipients.
In this study, we applied a novel approach using multiple imputations to infer HLA-A, -B, and -DRB1 alleles and corresponding AA polymorphisms from antigenic specificity through the application of computational and statistical methods. We examined the association of HLA-A, -B, and -DRB1 MMs at the AA level with kidney allo GF after adjusting for HLA antigen MMs. We discuss our findings in the context of structural and functional correlates of AA polymorphism at the antigen recognition site of HLA-DRB1 molecules, and the potential application of this approach to the complexity of HLA polymorphism in relation to kidney transplantation.
MATERIALS AND METHODS
Imputation of HLA Alleles and HLA AA MMs
We used a multistep process to impute from 2-digit HLA specificity, HLA-A, -B and -DRB1 alleles (4-digit specificity) and the corresponding AA polymorphisms localized in the antigen recognition site as described in the Supplemental Digital Content, pages 5-6, SDC, http://links.lww.com/TP/B402. During this process, we used a previously validated computational method for estimating HLA alleles based on HLA haplotype frequencies data set for broad US racial groups (African Americans, Caucasians, Hispanic, and Asians).31 Using this method, we imputed HLA-A, -B, and -DRB1 alleles (4-digit specificity) corresponding to each HLA antigen of recipient and donors obtained from the Scientific Registry of Transplant Recipients (SRTR). We then produced the corresponding assignment of AA polymorphic sites of these HLA alleles by converting each allele to its constituent AA variants using the allele alignment data from the International ImMunoGeneTics Project /HLA database (https://www.ebi.ac.uk/ipd/imgt/hla/; Release 2.28.0, January 2010). Details about this imputation method and the likelihood of AA MMs are described in the Methods, SDC, http://links.lww.com/TP/B402.
Patient Population and Source of Clinical Data
The SRTR compiles data on all solid organ donors, wait-listed candidates, and transplant recipients in the United States as submitted by transplant programs to the Organ Procurement and Transplantation Network. The Health Resources and Services Administration, U.S. Department of Health and Human Services, provides oversight to the activities of the Organ Procurement and Transplantation Network and SRTR contractors. SRTR data have been described elsewhere.32
Using SRTR Standard Analysis Files, we evaluated patients who received a kidney-alone transplant between January 1, 1987, and December 31, 2009. Graft function was assessed through June 30, 2012. The study sample numbered 240 024, which included 156 049 deceased donor transplants; 23 858 living unrelated donor transplants; and 60 117 living related transplants. The racial distribution in this population and the exclusion criteria are provided in the SDC (http://links.lww.com/TP/B402) on page 8-9.
Graft and patient status were reported on each recipient at hospitalization discharge, at 6 months posttransplant, at 1 year posttransplant, and annually thereafter. The completeness of follow-up both with and without extra ascertainment for GF and death was previously published.33,34
Statistical Analysis
All statistical analyses were performed using SAS version 9.3. We determined GF rates using Cox proportional-hazards regression. Models were fitted to investigate predictors of early GF (eg, during the first year after transplant), conditional GF predicated on 1-year graft survival, and overall GF. All of the primary analyses considered any instance of GF, including recipient death, as an outcome.
All analyses were adjusted for known confounding patient and donor variables including HLA antigen MMs (SDC, http://links.lww.com/TP/B402 on page 9-11). Model building started with factors shown to be predictive of GF in the 2010 Program Specific Reports (PSR) published by the SRTR. The development and use of these models has been reported previously.35 The same process that was used for PSR model refinements was adapted for the purposes of this study. PSR models were modified to allow for follow-up longer than 3 years.35 Because we were testing a large number of variables and interactions on an extremely large data set, we used Bonferroni-style correction on the many HLA AA MM indicators to avoid the multiple-comparisons problem. Additional details about these analyses are provided in the Methods, SDC, http://links.lww.com/TP/B402.
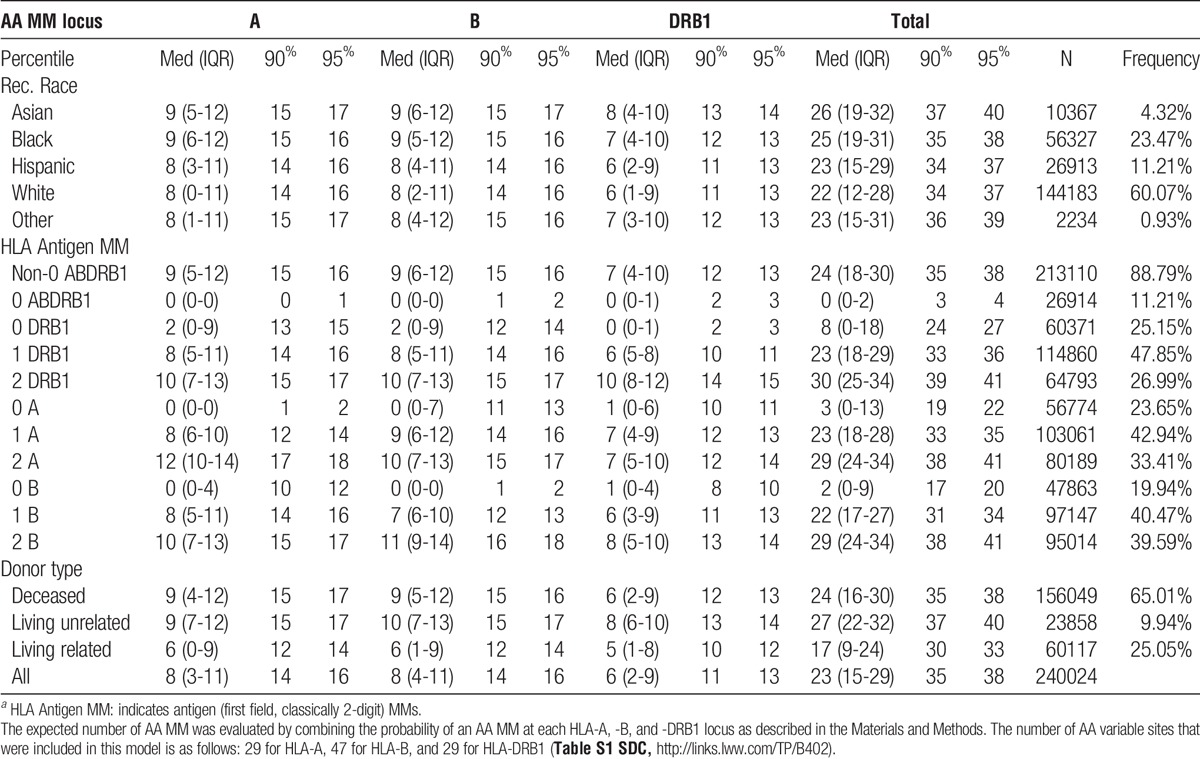
Separate statistical analyses involving the imputed AA MMs at antigen recognition sites were carried out to test the sensitivity and reproducibility of the analyses. The first analysis used a MM variable for each transplant which represented the expected number of AA MM. This variable was constructed by combining the probability of an AA MM at each HLA-A, -B, and -DRB1 locus using the formula: X = 0 * P(0MM) + 1 * P(1MM) + 2 * P(2MM). A total of 125 sites had a mean composite X ≥ 0.01 within at least 1 racial group. The mean frequency of expected HLA-A, -B, and -DRB1 AA mismatched sites by race is shown in Table 1. The GF model, using the AA MM variables adjusted for HLA antigen MM variables (0 ABDRB1 MM; 0 A MM, 1 A MM; 0 B MM, 1 B MM; 0 DRB1 MM, and 1 DRB1 MM; with 2 MM being the reference range for each HLA locus) and comorbidity factors was run one at a time for each of the 125 AA variable sites of the HLA-A, -B, and -DRB1 molecules. A Cox proportional hazards regression model of time to GF was then fitted to estimate the cumulative risk of GF associated with each additional statistically significant AA MM site identified above; models were analyzed separately for deceased donors and for living donors and by donor/recipient race, adjusted for all donor/recipient characteristics including the HLA antigen MM variables (SDC, http://links.lww.com/TP/B402 on page 9-11).
TABLE 1.
Estimated AA MM count distribution by locus, race, HLA antigen MMa, and donor type
In the second approach, we used summarized data over 10 separate analyses of imputed AA MMs at the antigen recognition sites based on the estimated probabilities of AA MM. Because the estimated probabilities of the dominant alleles are very high, we performed Cox regression analysis 10 times based on the randomly imputed HLA alleles and combined the results using the SAS MIANALYZE procedure.36,37 This approach provides an overall estimate with a combined confidence interval (CI) of the association of AA MM with GF based on the within-imputation standard errors and the between-imputation variability. Because the variances of the estimated risk across the 10 imputations were very small, we therefore chose to report our results based on 10 imputations. The established use of up to 10 imputations suffices under most realistic circumstances; it is unnecessary to use hundreds of imputations.38-40 To refine analyses using the sum of the MMs, a double MM among each 2 highly correlated MM sites was not counted as 2 separate MMs because we cannot distinguish their association with GF. The HLA-DRB1 variable sites included in this model are listed in Figure 2 and Table 3.
FIGURE 2.
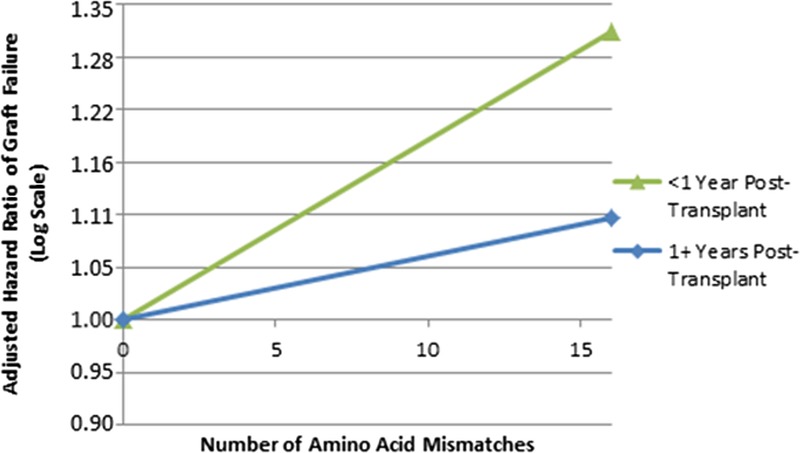
Association of HLA-DRB1 AA MMs with GF in separate posttransplantation intervals. This figure shows adjusted, log-scale HRs of GF by expected number of HLA-DRB1 AA MMs; models are based on 10 imputations as described in the Materials and Methods; figure shows results of models of all races combined. Zero HLA-A, -B, -DRB1 antigen mismatched transplants were excluded from these analysis. Estimated HRs were calculated based on 0 MMs as the reference using the modeled linear slopes in the log hazard, in a single model adjusted for patient risk factors, donor risk factors, and HLA-A, -B, and -DRB1 antigen MMs. The linear terms for HLA-DRB1 were statistically significant (P < 0.0001) for both models of GF in the first year after transplant as well as 1+ years posttransplant (all transplants combined). The AA MM variables evaluated in this analysis were selected by arbitrarily including only 1 site for each 2 highly correlated AA mismatched sites (r2 > 0.70) at each HLA locus. The following 23 variable sites were used for HLA-DRB1: positions 9, 10, 13, 14, 16, 26, 28, 31, 32, 33, 37, 38, 40, 47, 57, 58, 67, 70, 71, 74, 77, 85, and 86.
TABLE 3.
Adjusted HR for HLA-DRB1 AA MM site variables identified using the stepwise model based on 10 separate HLA imputations
Stepwise Model Using Individual AA-Mismatched Sites
A stepwise Cox model using imputed individual AA mismatched at the antigen recognition site was run separately for deceased donor and living donor transplants. A forward selection was applied; the demographics, comorbidity conditions, and HLA antigen MM variables were forced, and individual sites were added and subtracted according to a threshold P value of 0.05. A stepwise approach was separately performed in the model-building process in the 2 statistical analyses described above. In the data set generated from 10 imputations, the adjusted hazard ratio (HR) using the stepwise model was calculated by combining the estimates of AA variables that were consistently selected by the procedure in all 10 of the imputations using the SAS MIANLAYZE procedure (SAS software, version 9.3). Model results were adjusted for different covariates in each imputation including recipient factors, donor factors, HLA antigen MMs, and the other significant AA MM variables for each HLA-A, -B, DRB1 locus.
RESULTS
Distribution of HLA-A, -B, and -DRB1 AA MM Counts by Recipient Race, HLA Antigen (Classically 2-Digit Specificity) MMs and Donor Type
The distribution of the estimated HLA-A, -B, and -DRB1 AA MM count at the antigen recognition sites by recipient race, HLA antigen MMs, and donor type are shown in Table 1. The expected number of MM sites for each HLA locus was very similar among different recipient races. The bulk of AA MMs was a consequence of antigen MMs; additional MMs due to allele-level (first 2 fields, classically 4-digit specificity) disparity were limited to 1 to 3 sites. This observation is not unanticipated because some recipients with zero HLA-A, -B, -DRB1 antigen MMs were likely to be matched at the allele (4-digit specificity) level, and that for recipients who had MMs only at the allele (4-digit specificity) level, the expected number of AA MMs is known to be very small.42,43 Although overall the estimated number of AA MM is higher in 2 antigen MM compared with 1 antigen MM, there is a significant overlap in the estimated number of AA MM between these 2 categories of antigen MMs (Table 1, IQR, 90 and 95 percentiles). Notably, with any given HLA antigen MM, there is significant variability in the degree of AA MMs at peptide-binding sites including pockets in the binding groove (SDC, http://links.lww.com/TP/B402 on page 12-13 and Table S2). Thus, AA MMs at biologically important sites provide a more accurate assessment of the structural disparity in the HLA molecules than counting mismatched antigens (0, 1, or 2 MMs); this is consistent with a previous study using the HLA-DRB1 eplet matching algorithm.29
The Association of HLA-A, -B, and -DRB1 AA MMs With GF
Graft survival based on a Cox proportional-hazards model using mean numbers of expected AA MMs was run one at a time for each HLA-A, -B, -DRB1 locus and AA polymorphic site (without any other locus/position variables in the model) adjusted for comorbidity factors (Table S3, SDC, http://links.lww.com/TP/B402). Afterward, a Cox proportional-hazards regression model of time to GF was fitted to estimate the cumulative risk of the statistically significant AA MM sites for each HLA locus identified in the analysis above, adjusted for the other HLA-A, -B, and -DRB1 AA variables per locus/position. A statistically significant linear relationship was observed between the estimated number of HLA-DRB1 AA MMs and GF for both deceased and living related donor kidney transplants (Figure 2). The initial slope of HLA-DRB1 (P < 0.0001) indicates increasing risk through the entire range of AA MM with no significant change in the curve at 5 MMs. For living donor transplants, the initial slope of HLA-DRB1 (P = 0.06) suggests possible increasing risk from 0 to 5 MM with no significant change at 5 MMs (Figure 2). For living unrelated donor transplants, none of the initial slopes or spline factors reached significance (data not shown). Estimated HLA-DRB1 amino MMs were generally consistently associated with increased risk of GF in several groupings defined by AA loci, donor type (living and deceased donors), donor race, recipient race, and recipient panel-reactive antibody level (<10% and >10%). There were however, exceptions when dealing with smaller categories, for example, transplants limited to deceased Asian donors with Asian recipients (see page 13 and 14 and Table S3, SDC, http://links.lww.com/TP/B402).
In a second analysis, Cox models of each of 10 separate data sets with imputed AA variables were used to evaluate the adjusted HR of GF by sum of AA MMs in HLA-A, -B, and -DRB1 loci, using a continuous sum of AA MMs and splines at 5 and 10 MMs, for HLA-A, B and DRB1. Each imputed data set was analyzed separately in a Cox model and the data generated from the 10 imputation sets was combined as described in the Materials and Methods. As in the previous model, in deceased donor transplants, a statistically significant (P = 0.04) linear relationship between the imputed HLA-DRB1 AA MMs and increased risk of GF was found. For living related donors, the initial DR slope was borderline significant (P = 0.07), and no other slopes or splines were significant (Figures S1A and S1B, SDC, http://links.lww.com/TP/B402). The results obtained from the 2 Cox models of deceased donor transplants were similar and showed an increased risk of GF associated with increasing numbers of estimated DRB1 AA MMs (HR, 1.15 per 10 DRB1 MMs in the first model, and 1.10 per 10 DRB1 MMs in this second model). Additionally, a Cox model adjusted for each donor type showed that the interaction between HLA AA MMs versus deceased donor and living donor transplants was highly significant in all 10 imputations; estimates of the HRs (and 95% CIs) adjusted for all confounding factors included in the model are provided on pages 14-15 and Table S4, SDC, http://links.lww.com/TP/B402.
The Association of HLA AA MMs with GF by Posttransplantation Interval
The relationship between the imputed HLA-DRB1 AA MMs and the adjusted HR of GF was stronger in the first posttransplant year than it was after the first year (Figure 1, and Table S5, SDC, http://links.lww.com/TP/B402). In the first year posttransplant interval, the linear terms for HLA-DRB1 (P < 0.0001) were statistically significant (HR, 1.30 per 15 DRB1 AA MMs; P < 0.0001). After the first posttransplant year, the linear terms for HLA-DRB1 (P < 0.0001) were statistically significant; but the HR was weaker than in the first year posttransplant (HR, 1.10 per 15 DRB1 AA MMs; P < 0.0001).
FIGURE 1.
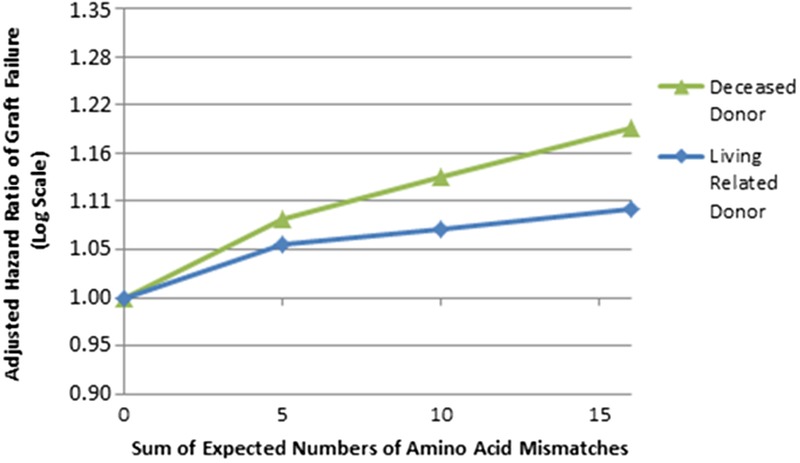
Adjusted HR of GF by sum of expected number of AA MMs at HLA-DRB1. This figure shows the adjusted, log-scale HR of GF by expected number of HLA-DRB1 AA MMs; figure shows results of models of all races combined. Zero HLA-A, -B, -DRB1 antigen mismatched transplants were excluded from these analyses. Estimated HRs for all loci were calculated based on 0 MMs as the reference using the initial slopes plus spline factors to model the degree of change in slope at 5, 10, and 15 in a single model adjusted for patient risk factors, donor risk factors, and HLA-A, -B, and -DRB1 antigen MMs. Splines at 5, 10, and 15 AA MMs were chosen primarily due to the distribution of the number of AA MMs. The initial slope of HLA-DRB1 AA MMs had a P < 0.0001 and 0.06 for deceased donor and living related donor transplants, respectively.
The association between the imputed DRB1 AA MM and increased risk of GF by calendar year of transplant, restricted to when molecular typing was introduced, is reasonably consistent (ie, positive and of similar magnitude) within each year of transplant suggesting that differences in demographics or HLA typing methods over time had no significant effect (Figure S2, SDC, http://links.lww.com/TP/B402).
Joint Risk of GF Associated With HLA AA MMs at Multiple Sites
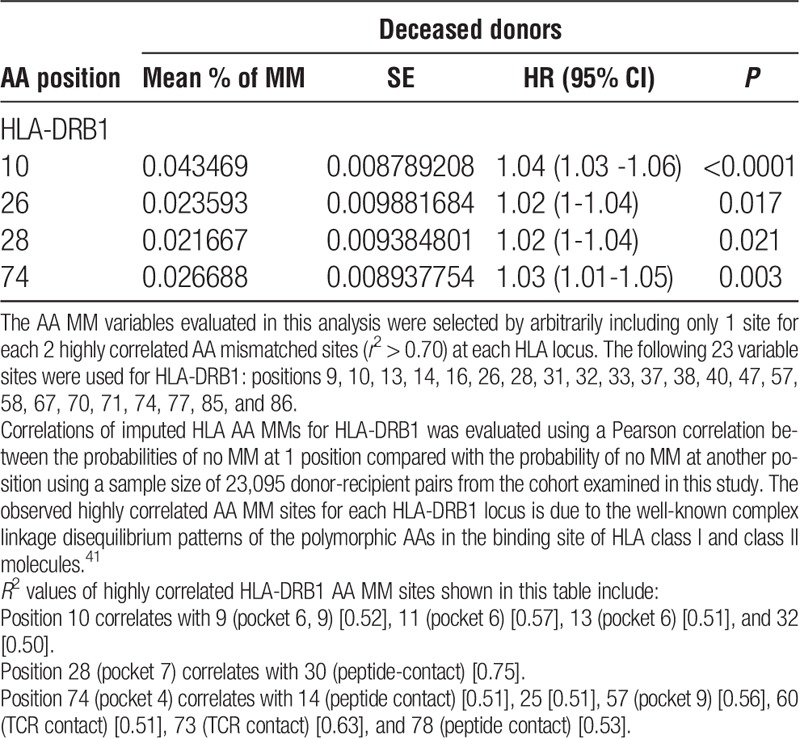
A stepwise approach was separately performed in the model-building process in the 2 statistical analyses used in this study as described in the Materials and Methods. In the first approach, the expected AA MM variables that were found to be statistically significant (see pages 15-16 and Table S3, SDC, http://links.lww.com/TP/B402) were entered in the model. A Cox proportional-hazards regression model of time to GF was then fitted to estimate the cumulative risk associated with the statistically significant AA MM sites identified for each HLA locus. This analysis showed an increased risk of GF associated with MMs at AA positions that make up the pockets in the peptide-binding groove of HLA-DRB1 molecules (Table 2). These findings were confirmed using a stepwise model based on 10 separate analyses of the imputed HLA alleles (see page 17 and Table S6, SDC, http://links.lww.com/TP/B402). The combined estimates of the HLA AA MM sites that were consistently selected in all 10 of the imputations using the stepwise model are summarized in Table 3. In deceased donor transplants, HLA-DRB1 sites include (in numerical order): position 28 (pocket 7) (HR, 1.02; 95% CI, 1-1.04; P = 0.021) and 74 (pocket 4) (HR, 1.03; 95% CI, 1.01-1.05; P = 0.003). In addition, position 10 has also a significant association with GF (HR, 1.04; 95% CI, 1.03-1.06; P < 0.0001); it is noteworthy that MMs at these positions are highly correlated with other MMs at AA sites that are located in pockets 4, 6, and 9 in the peptide-binding groove of HLA-DRB1 molecule (Table 3 and Figure 3). In living donor transplants, position 13 (pockets 4 and 6) was significantly associated with increased risk of GF (HR, 1.06; 95% CI, 1.02-1.1; P = 0.002).
TABLE 2.
Adjusted HR for expected HLA-DRB1 AA MM site variables using the stepwise modela
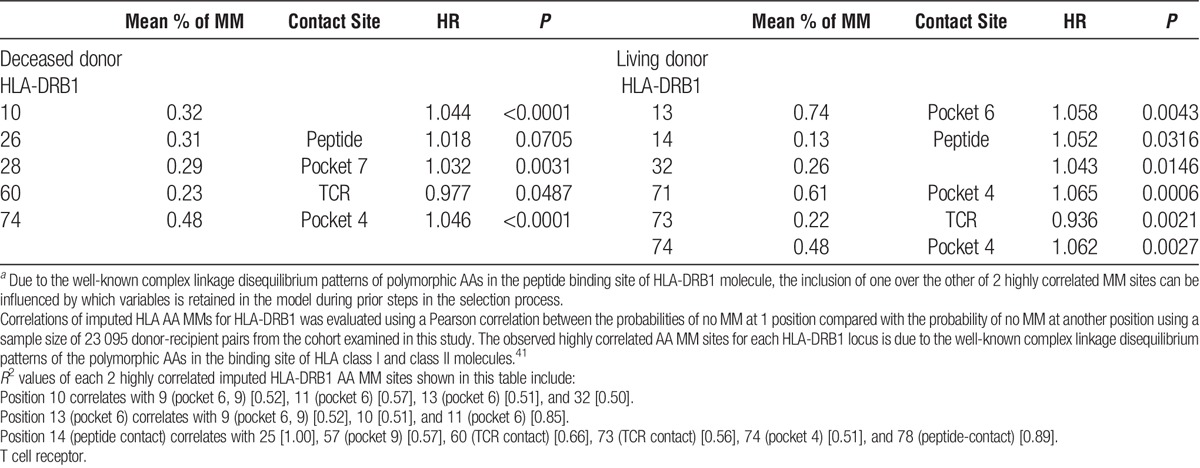
FIGURE 3.
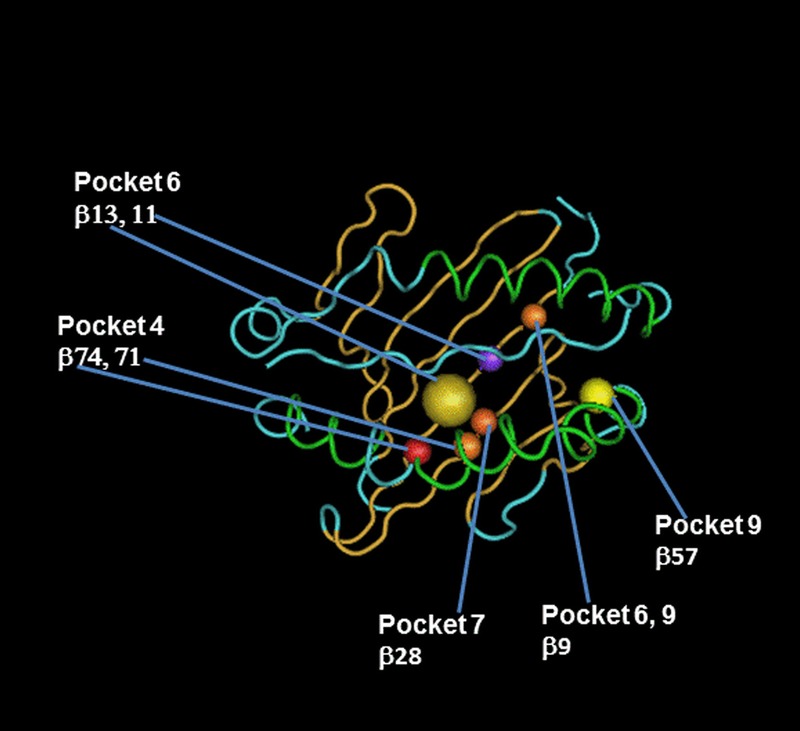
HLA-DRB1 AA substitutions in peptide-binding pockets account for the highest risk of GF attributable to HLA AA MMs. Figure illustrates the localization of HLA-DRB1 AA MMs found to be significantly associated with GF using a stepwise model as described in the Materials and Methods. AA sequences of HLA-DRB1 α and β chain are mapped onto a worm representation. AA mismatched sites of the HLA-DRB1 beta chain that account for the highest risk of GF attributable to HLA AA MM are labeled and numbered, with positions 71 and 74 occupying the P4 pocket, positions 11 and 13 occupying the P6 pocket, positions 28 occupying the P7 pocket, and position 57, occupying the P9 pocket. Arrows show these AA positions and pockets. A molecular model of the HLA-DRB1 α1β1 domain was derived by homology modeling from the 3D coordinates of HLA-DRB1*04 (Pdb_id 1:4MDJ_B and 4MDJ_A) with the bound peptide derived from Vimentin (Pdb_id 1:4MDJ_C).44 A worm representation model of AA sequences was produced using a Cn3D macromolecular structure viewer to view 3-dimensional structures from the NCBI's Entrez Structure database. (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). The functional and structural correlates of AA polymorphisms and interacting residue positions were based on a previously published report.2 Correlations plot of imputed HLA AA MMs for HLA-DRB1 was evaluated using a Pearson correlation between the probabilities of no MM at 1 position compared with the probability of no MM at another position using a sample size of 23,095 donor-recipient pairs from the cohort examined in this study. The observed highly correlated AA MM sites for each HLA- DRB1 locus is due to the well-known complex linkage disequilibrium patterns of the polymorphic AAs in the binding site of HLA class II molecule. R2 values of highly correlated HLA-DRB1 AA MM sites forming pockets 4, 6, 7, and 9 are listed below. Position 9 (pockets 6, 9) correlates with 10 [0.52], 11 (pocket 6) [0.62], and 13 (pocket 6) [0.52]. Position 13 (pocket 6) correlates with 9 (pockets 6, 9) [0.52], 10 [0.51], and 11 (pocket 6) [0.85]. Position 28 (pocket 7) correlates with 30 (peptide-contact) [0.75]. Position 57 (pocket 9) correlates with 14 (peptide contact) [0.57], 25 [0.57], 60 (TCR contact) [0.86], 74 (pocket 4) [0.56], 78 (peptide contact) [0.64]. Position 74 (pocket 4) correlates with 14 (peptide contact) [0.51], 25 [0.51], 57 (pocket 9) [0.56], 60 (TCR contact) [0.51], 73 (TCR contact) [0.63], and 78 (peptide contact) [0.53].
DISCUSSION
We showed consistently statistically significant linear associations between the number of imputed HLA-DRB1 AA MMs and the risk of GF in kidney transplant recipients based on a very large registry data set. Models were adjusted for key donor and recipient covariates that influence outcome including antigen mismatching (HLA-A, -B, and -DRB1 2-digit specificity). Thus, this incremental risk was independent of the well-known association of HLA-DRB1 antigen MM with graft survival.45 The linear relationship between the number of imputed AA MMs and GF suggests a synergy between mismatched AAs associated with allograft survival. Although overall, this incremental risk appears to be modest, it is actually much stronger when the number of imputed AA MM exceeds 10 during the first year after transplantation (Figure 1; HR = 1.30 per 15 DRB1 AA MM; P < 0.0001); this incremental risk is not insignificant compared to 1.15 and 1.26 relative risk of GF with 1 or 2 DRB1 MMs, respectively.45 These findings are consistent with previous studies showing that the association of HLA-DRB1 antigen matching with graft survival is strongest during the first 6 to 12 months period after transplantation.13,46 Overall, the imputed HLA-DRB1 AA MMs had a stronger association with GF than that of HLA-A and HLA-B on AA MMs (see page 14-18 and Table S6 and S7, SDC, http://links.lww.com/TP/B402); the influence of AA MMs of these 3 HLA loci on graft survival may be additive as previously reported for HLA antigen matching.46
Importantly, estimated AA MMs at particular positions in the HLA-DRB1 peptide-binding groove including the pockets account for a significant incremental risk of GF that was independent of the well-known association of HLA antigen MMs. Within any given HLA-DRB1 antigen MM, there is variability in the degree of nonself AA at peptide-binding sites that can influence alloreactivity leading to GF; this structural variability cannot be assessed by the simple count of DRB1 antigen MM (1 or 2 MM) alone. However, the distinction between the associations of specific AAs with GF is not simple, due to the well-known complex linkage disequilibrium patterns of polymorphic AAs in the binding site of HLA class I and class II molecules.41,47 In addition, this linkage disequilibrium makes it difficult to evaluate the interactions between individual AA MMs that were significantly associated with GF.
Our findings demonstrate important clinical ramifications of prior structural and functional studies highlighting the importance of AA substitutions at peptide-binding sites of HLA molecules in altering T cell allorecognition5,17,19 and alloantibody specificity.7,8 Both direct T cell allorecognition as well as indirect T cell allorecognition could play an important role in GF.48 A major source of alloreactivity is thought to be T cell receptor cross-reactivity between distinct HLA allelic variants from the same HLA class I (or class II) gene.2,4–6 Furthermore, HLA AA substitutions can significantly impact the indirect pathway of T cell allorecognition wherein the allogeneic graft HLA molecule itself is degraded into peptides and presented on host HLA molecules; the antigenic peptides can arise from cleft and noncleft regions of the HLA molecules secondary to AA polymorphisms in these regions.
Our finding is reminiscent of several reports describing the importance of HLA-DRB1 AA substitution affecting peptide-binding pockets in conferring susceptibility or resistance to several immune-mediated diseases by restricting the nature of peptides that can be accommodated in the HLA pockets of the antigen binding groove,41,44,47,49,50 further underlying the clinical importance of HLA AA polymorphisms at peptide-binding pockets.
Only a few studies to date have evaluated the association of HLA class II AA MMs with the risk of kidney allograft survival using the epitope/eplet matching algorithm; these studies were single-center studies and were based on a small sample size.21,25 A key strength of our study is the use of a very large registry of the United Network for Organ Sharing data set. The very large sample size serves to mitigate possible errors to a large extent and provides a more comprehensive coverage of HLA AA polymorphism associated with a racially/ethnically diverse transplant population. In addition, these previous studies picked the most likely allele corresponding to an HLA 2-digit specificity, ignoring other HLA alleles which are present with a lower frequency in a particular population; thus, introducing a high degree of uncertainty as compared to the multiple imputations approach described in our study. Limitation in defining HLA epitopes/eplets without allele level data is upheld by Duquesnoy et al.26,51,52
Other studies used the epitope/eplet algorithm to assess the magnitude of humoral sensitization. In these studies, the algorithm, based on AA polymorphisms in antibody accessible sites of the HLA molecules, was used for the definition of acceptable MMs and HLA compatibility for highly sensitized patients.26
The multiple imputation of HLA alleles we applied in this study is a well-accepted tool to use when high-resolution HLA typing is not feasible due to the high cost and lack of access to a sufficient number of archived samples, as in our study.37 This imputation is at the core of the National Marrow Donor Program Haplostats algorithm that has been widely used during the past several years to convert donor and recipient low-resolution HLA haplotypes to the most likely high-resolution alleles when HLA high resolution typing is not available53; a cross validation of an imputation-based HLA allele matching algorithm from 7 different allogeneic hematopoietic stem cell donor registries was recently reported, supporting the reproducibility of this imputation algorithm.54 Although this cross validation was done in the context of stem cell transplantation, imputation of HLA alleles using the Haplostats algorithm has been previously used in the setting of solid organ transplantation.21,25 These findings lend additional strength to the interpretation of our results and the ensuing consequences regarding kidney transplantation.
A potential limitation remains in the current study related to the uncertainty of the estimate of HLA AA MMs due to the imputation process. This was relatively small for most HLA-A, B, and DRB1 AA sites (see page 7 and 8 and Figures S3 and S4, SDC, http://links.lww.com/TP/B402). This was not unexpected because the bulk of AA MMs depends mainly on the mismatched HLA antigens, which were not imputed. Because some degree of uncertainty exists at a few imputed AA MM sites (mainly positions 57, 71, 74, and 86 of DRB1), the measurements of HR associated with these sites are likely to have a higher standard error. However, these measurement errors would generally result only in a power reduction, underestimating the degree of the association of AA MM with GF but not result in an increased type 1 error (detecting an association that is not present).
The computational approach we used may not efficiently capture the magnitude of the association of HLA MMs with GF because we examined HLA AA MMs at single AA variable sites and did not consider combinations of AAs that might be better suited to explain the biological differences among various HLA molecules.41,47 Furthermore, in this study, we did not interrogate polymorphic AA sites outside the HLA binding groove; some of these sites could also have a functional role. We also did not evaluate structural differences secondary to biochemical differences from AA substitutions for each variable site of HLA class I and class II molecules that could impact immunogenicity; immunogenicity associated with AA MMs could not be evaluated in this study. Additionally, given the strong linkage disequilibrium between HLA-DRB1, DQB1, and DQA1 alleles, DQA1 and/or DQB1 MMs could partially explain the observed association of DRB1 AA MM with GF. However, the constraint due to this linkage disequilibrium is likely to be minimized by the adjustment we have made for HLA-DRB1 antigen (2-digit specificity) MM.
In summary, estimated AA MMs at particular positions in the HLA-DRB1 peptide-binding groove including the pockets account for the main incremental risk of GF attributable to the HLA AA MMs beyond those MMs assessed at the antigenic specificity. This study highlights the importance of considering AA MMs at solvent unexposed/peptide-binding sites of HLA-DRB1 molecule in the assessment of the risk of GF. Although our findings are not definitive and merit further investigation, we believe it is important to apply the newest scientific concepts and the most precise technologies to define HLA compatibility barriers so we can advance this field and improve transplantation outcomes. The introduction of high-resolution typing and next-generation sequencing are moving the transplantation field towards practical clinical applications for AA sequence based matching. Our data provide a reasonable justification for future studies to confirm our findings and to assess the incremental risk of GF associated with DQA1 and DQB1 AA variation at individual positions in these molecules.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the excellent scientific input provided by Drs. Harold I. Feldman and Marshall M. Joffe, and the excellent administrative support and editing provided by Insuk Choe, JD.
Footnotes
The data reported here have been supplied by the Minneapolis Medical Research Foundation (MMRF) as the contractor for the SRTR. The interpretation and reporting of these data are the responsibility of the author(s) and in no way should be seen as an official policy of or interpretation by the SRTR or the U.S. Government.
The authors would be willing to communicate and/or collaborate with other researchers interested in this topic.
The study was submitted for IRB review and was approved.
This study was presented in part at the meeting of the American Transplant Congress, May 8-22, 2013 (Seattle, WA).
This work is dedicated to the memory of Professor Jean Dausset.
The authors declare no funding or conflicts of interest.
M.K. participated in research design, data analysis and writing of the article. K.P.M. participated in the performance of the research, data analysis, and writing of the article. M.M. participated in research design, performance of the research, and data analysis. M.A.F. participated in research design and data analysis. H.L. participated in data analysis. V.T. participated in the performance of the research. A.B.L. participated in research design, data analysis and writing of the article. R.M.M. participated in research design, data analysis, and writing of the article.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Institute ANR. Nomenclature for Factors of the HLA System. 2015; http://hla.alleles.org/announcement.html. Accessed October 26, 2015.
- 2.Reche P, Reinherz E. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol. 2003;331:623–641. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum ME, Mendoza JL, Sethi DK, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colf LA, Bankovich AJ, Hanick NA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. [DOI] [PubMed] [Google Scholar]
- 5.Lombardi G, Barber L, Sidhu S, et al. The specificity of alloreactive T cells is determined by MHC polymorphisms which contact the T cell receptor and which influence peptide binding. Int Immunol. 1991;3:769–775. [DOI] [PubMed] [Google Scholar]
- 6.Macdonald WA, Chen Z, Gras S, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Vina M, Maiers M, Gutierrez M, et al. 13th IHWS Serology and HLA Phenotypes Joint Report. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Congress; May 2002, 2005; Seattle, WA.
- 8.Mulder A, Eijsink C, Kester MG, et al. Impact of peptides on the recognition of HLA class I molecules by human HLA antibodies. J Immunol. 2005;175:5950–5957. [DOI] [PubMed] [Google Scholar]
- 9.Held PJ, Kahan BD, Hunsicker LG, et al. The impact of HLA mismatches on the survival of first cadaveric kidney transplants. N Engl J Med. 1994;331:765–770. [DOI] [PubMed] [Google Scholar]
- 10.Takemoto S, Terasaki P, Cecka J, et al. Survival of nationally shared, HLA-matched kidney transplants from cadaveric donors. The UNOS Scientific Renal Transplant Registry. N Engl J Med. 1992;327:834–839. [DOI] [PubMed] [Google Scholar]
- 11.Moen T, Albrechtsen D, Flatmark A, et al. Importance of HLA-DR matching in cadaveric renal transplantation: a prospective one-center study of 170 transplants. N Engl J Med. 1980;303:850–854. [DOI] [PubMed] [Google Scholar]
- 12.Opelz G, Mytilineos J, Scherer S, et al. Analysis of HLA-DR matching in DNA-typed cadaver kidney transplants. Transplantation. 1993;55:782–785. [PubMed] [Google Scholar]
- 13.Thorogood J, Persijn GG, Schreuder GM, et al. The effect of HLA matching on kidney graft survival in separate posttransplantation intervals. Transplantation. 1990;50:146–150. [DOI] [PubMed] [Google Scholar]
- 14.Zantvoort F, D'Amaro J, Persijn G, et al. The impact of HLA-A matching on long-term survival of renal allografts. Transplantation. 1996;61:841–844. [DOI] [PubMed] [Google Scholar]
- 15.Reisaeter AV, Leivestad T, Vartdal F, et al. A strong impact of matching for a limited number of HLA-DR antigens on graft survival and rejection episodes: a single-center study of first cadaveric kidneys to nonsensitized recipients. Transplantation. 1998;66:523–528. [DOI] [PubMed] [Google Scholar]
- 16.Araujo H, Dole K, Lazaro A, et al. Multiple epitopes of HLA-DRB1*0411 are recognized by T-cell clones originated from individuals carrying other DR4 subtypes. Hum Immunol. 1998;59:561–570. [DOI] [PubMed] [Google Scholar]
- 17.Archbold J, Macdonald W, Miles J, et al. Alloreactivity between disparate cognate and allogeneic pMHC-I complexes is the result of highly focused, peptide-dependent structural mimicry. J Biol Chem. 2006;281:34324–34332. [DOI] [PubMed] [Google Scholar]
- 18.Baxter-Lowe L, Eckels D, Ash R, et al. The predictive value of HLA-DR oligotyping for MLC responses. Transplantation. 1992;53:1352–1357. [DOI] [PubMed] [Google Scholar]
- 19.Coppin H, Carmichael P, Lombardi G, et al. Position 71 in the alpha helix of the DR beta domain is predicted to influence peptide binding and plays a central role in allorecognition. Eur J Immunol. 1993;23:343–349. [DOI] [PubMed] [Google Scholar]
- 20.Santos-Aguado J, Crimmins M, Mentzer S, et al. Alloreactivity studied with mutants of HLA-A2. Proc Natl Acad Sci U S A. 1989;86:8936–8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiebe C, Nevins T, Robiner W, et al. The synergistic effect of class II HLA epitope-mismatch and nonadherence on acute rejection and graft survival. Am J Transplant. 2015;15:2197–2202. [DOI] [PubMed] [Google Scholar]
- 22.Kosmoliaptsis V, Sharples LD, Chaudhry A, et al. HLA class I amino acid sequence-based matching after interlocus subtraction and long-term outcome after deceased donor kidney transplantation. Hum Immunol. 2010;71:851–856. [DOI] [PubMed] [Google Scholar]
- 23.Duquesnoy RJ, Howe J, Takemoto S. HLAmatchmaker: a molecularly based algorithm for histocompatibility determination. IV. An alternative strategy to increase the number of compatible donors for highly sensitized patients. Transplantation. 2003;75:889–897. [DOI] [PubMed] [Google Scholar]
- 24.Kosmoliaptsis V, Chaudhry AN, Sharples LD, et al. Predicting HLA class I alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation. 2009;88:791–798. [DOI] [PubMed] [Google Scholar]
- 25.Wiebe C, Pochinco D, Blydt-Hansen T, et al. Class II HLA epitope matching—a strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant. 2013;13:3114–3122. [DOI] [PubMed] [Google Scholar]
- 26.Duquesnoy RJ, Claas FH. Is the application of HLAMatchmaker relevant in kidney transplantation? Transplantation. 2005;79:250–251. [DOI] [PubMed] [Google Scholar]
- 27.Duquesnoy RJ, Takemoto S, de Lange P, et al. HLAmatchmaker: a molecularly based algorithm for histocompatibility determination. III. Effect of matching at the HLA-A, B amino acid triplet level on kidney transplant survival. Transplantation. 2003;75:884–889. [DOI] [PubMed] [Google Scholar]
- 28.Takemoto S, Terasaki PI, Gjertson DW, et al. Equitable allocation of HLA-compatible kidneys for local pools and minorities. N Engl J Med. 1994;331:760–764. [DOI] [PubMed] [Google Scholar]
- 29.Duquesnoy RJ, Askar M. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. V. Eplet matching for HLA-DR, HLA-DQ, and HLA-DP. Hum Immunol. 2007;68:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laux G, Mytilineos J, Opelz G. Critical evaluation of the amino acid triplet-epitope matching concept in cadaver kidney transplantation. Transplantation. 2004;77:902–907. [DOI] [PubMed] [Google Scholar]
- 31.Madbouly A, Gragert L, Freeman J, et al. Validation of statistical imputation of allele-level multilocus phased genotypes from ambiguous HLA assignments. Tissue Antigens. 2014;84:285–292. [DOI] [PubMed] [Google Scholar]
- 32.Leppke S, Leighton T, Zaun D, et al. Scientific Registry of Transplant Recipients: collecting, analyzing, and reporting data on transplantation in the United States. Transplant Rev (Orlando). 2013;27:50–56. [DOI] [PubMed] [Google Scholar]
- 33.HHS/HRSA/HSB/DOT, UNOS, URREA. 2004 Annual Report of the U.S. Organ Procurement and Transplantation Network and the Scientific Registry of Transplant Recipients: Transplant Data 1994–2003. 2004.
- 34.Dickinson DM, Dykstra DM, Levine GN, et al. Transplant data: sources, collection and research considerations, 2004. Am J Transplant. 2005;5(4 Pt 2):850–861. [DOI] [PubMed] [Google Scholar]
- 35.Dickinson DM, Arrington CJ, Fant G, et al. SRTR program-specific reports on outcomes: a guide for the new reader. Am J Transplant. 2008;8(4 Pt 2):1012–1026. [DOI] [PubMed] [Google Scholar]
- 36.SAS Institute I. SAS/STAT 13.1 User's Guide: The MIANALYZE Procedure. Cary, NC: SAS Institute, Inc.; 2013. [Google Scholar]
- 37.Kamoun M, Holmes J, Israni A, et al. HLA-A amino acid polymorphism and delayed kidney allograft function. Proc Natl Acad Sci U S A. 2008;105:18883–18888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubin D. Multiple Imputatioin for Nonresponse in Surveys. New York: John Wiley & Sons; 1987. [Google Scholar]
- 39.von Hippel PT. How Many Imputations are Needed? A Comment on Hershberger and Fisher (2003). In: Structural Equation Modeling. Vol 12(2): Lawrence Erlbaum Associates, Inc; 2005:334–335. [Google Scholar]
- 40.Hayati Rezvan P, Lee KJ, Simpson JA. The rise of multiple imputation: a review of the reporting and implementation of the method in medical research. BMC Med Res Methodol. 2015;15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson G, Marthandan N, Hollenbach JA, et al. Sequence feature variant type (SFVT) analysis of the HLA genetic association in juvenile idiopathic arthritis. Pac Symp Biocomput. 2010:359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamoun M, Israni AK, Joffe MM, et al. Assessment of differences in HLA-A, -B, and -DRB1 allele mismatches among African-American and non-African-American recipients of deceased kidney transplants. Transplant Proc. 2007;39:55–63. [DOI] [PubMed] [Google Scholar]
- 43.Baxter-Lowe LA, Maiers M, Spellman SR, et al. HLA-A disparities illustrate challenges for ranking the impact of HLA mismatches on bone marrow transplant outcomes in the United States. Biol Blood Marrow Transplant. 2009;15:971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scally S, Petersen J, Law S, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med. 2013;210:2569–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts J, Wolfe R, Bragg-Gresham J, et al. Effect of changing the priority for HLA matching on the rates and outcomes of kidney transplantation in minority groups. N Engl J Med. 2004;350:545–551. [DOI] [PubMed] [Google Scholar]
- 46.Opelz G, Wujciak T, Döhler B, et al. HLA compatibility and organ transplant survival. Collaborative Transplant Study. Rev Immunogenet. 1999;1:334–342. [PubMed] [Google Scholar]
- 47.Karp DR, Marthandan N, Marsh SG, et al. Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis. Hum Mol Genet. 2010;19:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Game DS, Lechler RI. Pathways of allorecognition: implications for transplantation tolerance. Transpl Immunol. 2002;10:101–108. [DOI] [PubMed] [Google Scholar]
- 49.Illing P, Vivian J, Dudek N, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. [DOI] [PubMed] [Google Scholar]
- 50.Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duquesnoy RJ. The eplet load concept in clinical transplantation. Pediatr Transplant. 2016;20:884–885. [DOI] [PubMed] [Google Scholar]
- 52.Duquesnoy RJ, Kamoun M, Baxter-Lowe LA, et al. Should HLA mismatch acceptability for sensitized transplant candidates be determined at the high-resolution rather than the antigen level? Am J Transplant. 2015;15:923–930. [DOI] [PubMed] [Google Scholar]
- 53.Dehn J, Setterholm M, Buck K, et al. HapLogic: a predictive human leukocyte antigen-matching algorithm to enhance rapid identification of the optimal unrelated hematopoietic stem cell sources for transplantation. Biol Blood Marrow Transplant. 2016;22:2038–2046. [DOI] [PubMed] [Google Scholar]
- 54.Bochtler W, Gragert L, Patel Z, et al. A comparative reference study for the validation of HLA-matching algorithms in the search for allogeneic hematopoietic stem cell donors and cord blood units. HLA. 2016;87:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


