Significance
Teosinte, the wild ancestor of maize, is a highly branched and low-yielding plant. Branch suppression in maize was achieved through selection for overexpression of the teosinte branched1 (tb1) transcription factor that acts as a repressor of axillary branching. Here, we show a molecular mechanism for how TB1 transformed teosinte into a viable crop plant. The tassels replace upper ears1 (tru1) mutant causes maize to revert back to its highly branched ancestral state, much like tb1. We demonstrate that tru1 is a direct target of TB1 and is overexpressed in modern maize. The genetic mechanism underlying the tb1 and tru1 pathway reveals a blueprint for domesticating new grass species.
Keywords: maize, teosinte, evolution, branch, tiller
Abstract
Axillary branch suppression is a favorable trait bred into many domesticated crop plants including maize compared with its highly branched wild ancestor teosinte. Branch suppression in maize was achieved through selection of a gain of function allele of the teosinte branched1 (tb1) transcription factor that acts as a repressor of axillary bud growth. Previous work indicated that other loci may function epistatically with tb1 and may be responsible for some of its phenotypic effects. Here, we show that tb1 mediates axillary branch suppression through direct activation of the tassels replace upper ears1 (tru1) gene that encodes an ankyrin repeat domain protein containing a BTB/POZ motif necessary for protein–protein interactions. The expression of TRU1 and TB1 overlap in axillary buds, and TB1 binds to two locations in the tru1 gene as shown by chromatin immunoprecipitation and gel shifts. In addition, nucleotide diversity surveys indicate that tru1, like tb1, was a target of selection. In modern maize, TRU1 is highly expressed in the leaf trace vasculature of axillary internodes, while in teosinte, this expression is highly reduced or absent. This increase in TRU1 expression levels in modern maize is supported by comparisons of relative protein levels with teosinte as well as by quantitative measurements of mRNA levels. Hence, a major innovation in creating ideal maize plant architecture originated from ectopic overexpression of tru1 in axillary branches, a critical step in mediating the effects of domestication by tb1.
Axillary meristems are produced at the base of each new leaf and can have different fates depending on their positions within the plant. In maize, those found aboveground have a reproductive fate, while those found at the base of the plant can become long branches that are vegetative clones of the main shoot and are called tillers. These extra branches often compete for valuable resources such as light (1) and can have a negative impact on plant growth if produced in excess. Indeed, suppression of axillary branching through an increase in apical dominance is a commonly bred trait among many domesticated crop plants (2), including maize (Zea mays ssp. mays), which was domesticated from teosinte (Z. mays ssp. parviglumis), its wild ancestor. While there are multiple genetic pathways necessary for suppression of axillary bud production in plants (3–5), in maize, this process has been found to operate through a specialized pathway mediated by the teosinte branched1 (tb1) locus first identified by Charles Burnham over 50 y ago (6). tb1 loss of function mutants overproduce tillers and have long aerial branches tipped by male tassels that replace the normally female ears (7), indicating that it functions as a repressor of both axillary bud growth and inflorescence sexual fate (8). Previous quantitative trait locus studies mapping the genetic basis for differences in plant morphology between maize and teosinte showed that selection at the tb1 locus was responsible for the increase in apical dominance found in modern maize (7). tb1 is one of only a few loci with large effect that are responsible for several key innovations that distinguish maize from teosinte (5). Cloning of the tb1 gene revealed that it encodes a class II TCP transcription factor (9) known to bind DNA (10). In situ expression of tb1 in domesticated maize was found in axillary buds, axillary branches, husk leaves, aborting florets, and floral organs of the ear (8). This expression pattern in tissues and organs that fail to grow indicates that tb1 acts as a general negative regulator of organ growth, consistent with the role of TCPs in diverse species (11).
A comparison of the tb1 locus in teosinte and domesticated maize revealed that the maize allele contains a HOPSCOTCH retrotransposon insertion ∼58 kb upstream of the promoter (12). Expression analysis revealed that this insertion in the maize tb1 allele increased its expression in both the ear primordium and shank that comprises the internodes below the ear (9). Thus, domesticated maize contains a natural gain of function allele of tb1, and the reduction of both basal and aerial axillary branches is due to overexpression of tb1. Natural variation in tb1 also correlates with the diversity of plant architectures found in different populations of teosinte (13) as well as other grasses. For example, selection on the pearl millet ortholog of tb1 is also associated with domestication (14).
The derepression of axillary branch growth in tb1 mutants is reminiscent of mutations that alter metabolism of hormones (15) such as auxins and strigolactones. For example, the rice tb1 ortholog, fine culm1, suppresses axillary bud growth downstream of strigolactone signals (16). In Arabidopis, the TCP gene most closely related to tb1 is BRANCHED1 (BRC1), which appears to have a similar function as tb1 in repressing axillary bud growth (17). Expression of BRC1 is strongly down-regulated by mutations in the MORE AXILLARY GROWTH 1–4 genes that function in strigolactone metabolism, leading to speculation that tb1 activity may also be dependent on strigolactone levels (17). BRC1 homologs appear to integrate several external signals such as those caused by increased planting density, as well as internal signals including hormones, to mediate axillary bud suppression. For example, in potato, the ratio of red to far red light and auxin levels appear to cause alternative splicing of the BRC1 homolog, leading to production of dominant negative forms of the protein and subsequent changes in branching and plant morphology (18). Finally, in other grasses such as sorghum, the tb1 ortholog is sensitive to far red light signals that induce the shade avoidance response known to suppress axillary bud growth (19). Thus, several factors may influence tb1 gene activity including natural variation, transposons, hormones, and light, but none have elucidated a precise molecular mechanism for how tb1 itself is able to effect axillary branch suppression in maize.
To understand the molecular pathway in which tb1 functions, a search was initiated for mutants with similar phenotypes. In maize, the tassels replace upper ears1 (tru1) mutation causes phenotypes similar to the aerial portion of the tb1 mutant in that long axillary branches tipped by tassels replace the ear primordia. The tru1 gene was cloned by chromosome walking and found to encode an ankyrin repeat domain gene containing a BTB/POZ motif involved in protein–protein interactions. Orthologs of tru1 in monocots and dicots are necessary for the specification of basal leaf identities (20), indicating that tru1 may play a role in tissue patterning and the acquisition of cell fates. Immunolocalization using an antibody raised to TRU1 showed that the protein localizes to axillary buds in a pattern overlapping with TB1, consistent with it being a direct target. This was confirmed by both epistasis and chromatin immunoprecipitation (ChIP) using a TB1 antibody. Both TRU1 protein and transcripts are overexpressed in ear shanks of domesticated maize compared with teosinte, and immunolocalization showed ectopic expression in the shank vasculature. Taken together, these data indicate that ectopic overexpression of tru1 was critical for mediating the effects of tb1 overexpression during maize domestication.
Results
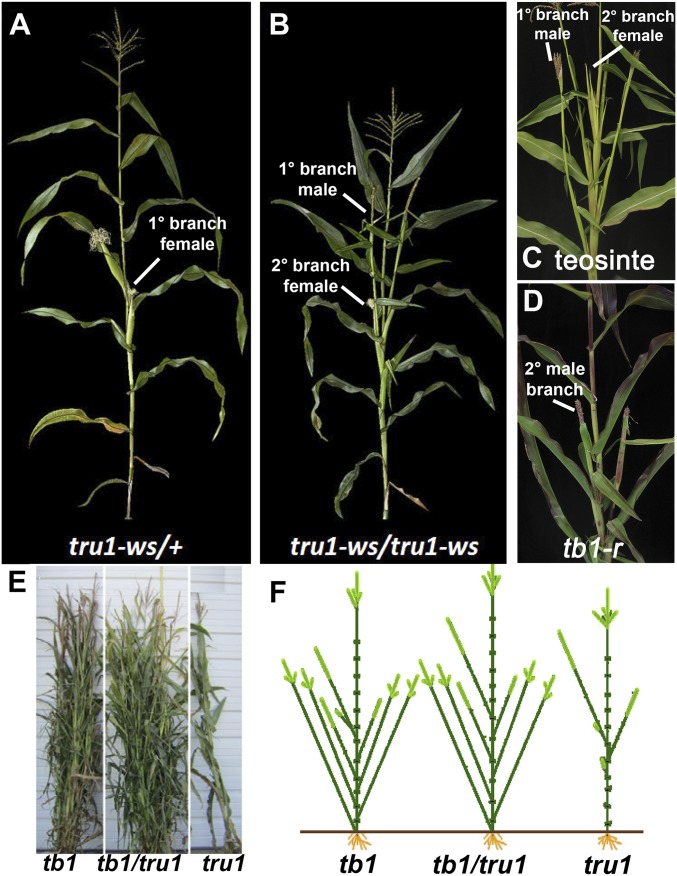
The recessive tru1-ws allele was first isolated and described by Sheridan (21) in 1988. In tru1 mutants, at the nodes that would normally produce ears, the short female axillary branches are derepressed to become long branches tipped by staminate tassel-like inflorescences (Fig. 1 A and B). The internodes of the branches that bear these inflorescences (which comprise the shank) are significantly longer than those in wild type, indicating the tru1 gene may have a role in suppression of internode growth. The tru1 aerial growth habit is reminiscent of teosinte, which displays a similar phenotype (Fig. 1C), as does the tb1-reference mutant (6, 7) (Fig. 1D). tb1 and tru1 differ, however, in that many extra tillers are produced in tb1 mutants (6, 7), while tru1-ws mutants produce almost none. Comparisons between the lengths of several different lateral organs revealed that the shanks of tru1 mutants were significantly elongated nearly fourfold compared with wild type (Fig. S1A). Even when introgressed into an inbred background, the elongated lateral branches showed a range of inflorescence sexes, ranging from fully female to a mixture of male and female, or fully male (Fig. S1B). Moreover, the uppermost derepressed axillary branch was the longest while branches at lower nodes were progressively shorter, until a short branch tipped by an ear usually formed at a lower node (Fig. 1F). Since in maize lateral branch length appears to correlate with sex identity (22), it is possible that this change from female to male could be a byproduct of distance of the axillary branch terminus away from the main stalk, a theory that explains the sex difference in teosinte branches compared with maize (23). This idea is supported by measurements of the shank lengths of tru1 branches that display different sexual identities, and it was confirmed that the longer shanks were in fact more male (Fig. S1C).
Fig. 1.
The tru1 phenotype and double mutants with tb1. (A) tru1-ws heterozygote with short primary axillary branch consisting of a female ear. (B) tru1-ws homozygote with derepressed long primary male axillary branches bearing secondary ears. (C) Z. mays parviglumis (teosinte) showing similar aerial growth habit as tru1-ws. Secondary female ear branches are in the axils of the leaves on the male primary branch. (D) tb1-ref mutant showing similar aerial growth habit as tru1-ws. (E) Double mutant phenotype of tb1-ref; tru1-ws (Center) compared with tb1-ref (Left) and tru1-ws (Right). Double mutant resembles tb1-ref in terms of axillary branching habit. (F) Schematic of axillary branching phenotypes in tru1-ws and tb1-ref double and single mutants. The overall branching pattern of the double mutant resembles that of the tb1 single mutant.
Fig. S1.
Phenotypic analysis of tru1. (A–C) A backcross population (BC1) was generated by crossing the original tru1-ws stock to the inbred A619 followed by backcrossing to the tru1-ws stock. Over 1,200 plants were grown to maturity in the summer maize nursery at Ames, Iowa. Phenotypes were recorded ∼110–120 d after sowing (DAS). (A) Measurement of shank, inflorescence, prophyll, husk leaf, sheath, leaf blade, and internode lengths of heterozygous and homozygous tru1 mutants. Highly significant differences were seen in the shank and inflorescence lengths. For each genotype, 25 representative, sibling plants were selected and all traits were measured on the same plants with the exception of shank length, which was measured on the whole population (C); inflorescence, located in the uppermost ear position, measured from last husk leaf insertion to tip; prophyll, associated with the same inflorescence; husk, associated with the same inflorescence, the outermost intact husk leaf; sheath and leaf blade, for the leaf subtending the same inflorescence; internode, immediately beneath the same inflorescence. (B) Range of sex determination of primary lateral branches, ranging from fully female (Left), mixed (Center), to fully male (Right), imaged 120 DAS. (C) Correlation between shank length and sex identity. The longer the shank, the more frequent the male sex identity. All inflorescences in the ear position still intact at 120 DAS were scored and measured; ear, n = 621; mixed, n = 28; male, n = 424. t test: *P < 0.05; **P < 0.005; ***P < 0.0005, ****P < 0.0001. (D) Measurements of shank lengths of tb1 and tru1 mutants compared with normal siblings (normal = 9.5 ± 6.3 cm, n = 28; tb1-ref = 39.2 ± 11.2 cm, n = 15; P < 0.0001; tru1-ws = 37.4 ± 21.6 cm, n = 12; P < 0.0001) and tb1-ref; tru1-ws double mutant (tb1-ref; tru1-ws = 40.3 ± 11.6 cm, n = 7). P value is not significantly different from that of tb1-ref single mutant (P = 0.84) or tru1-ws single mutant (P = 0.76), indicating that tru1 and tb1 function in the same pathway to inhibit shank elongation.
Since the tru1 phenotype resembles that of the upper shoot section of tb1 mutants, double mutants were made to establish epistasis (Fig. 1E). The double mutants resembled tb1 with respect to tillering and aerial lateral branch number and morphology (Fig. 1F). In addition, the shank length was the same in the double mutant compared with the two single mutants (Fig. S1D), consistent with an epistatic relationship. This finding is consistent with tru1 acting downstream of tb1 to suppress axillary branch elongation. Taken together, these results indicate that tru1 and tb1 function in the same pathway to repress internode elongation in aerial axillary branches, directly or indirectly resulting in a range of sex determination phenotypes.
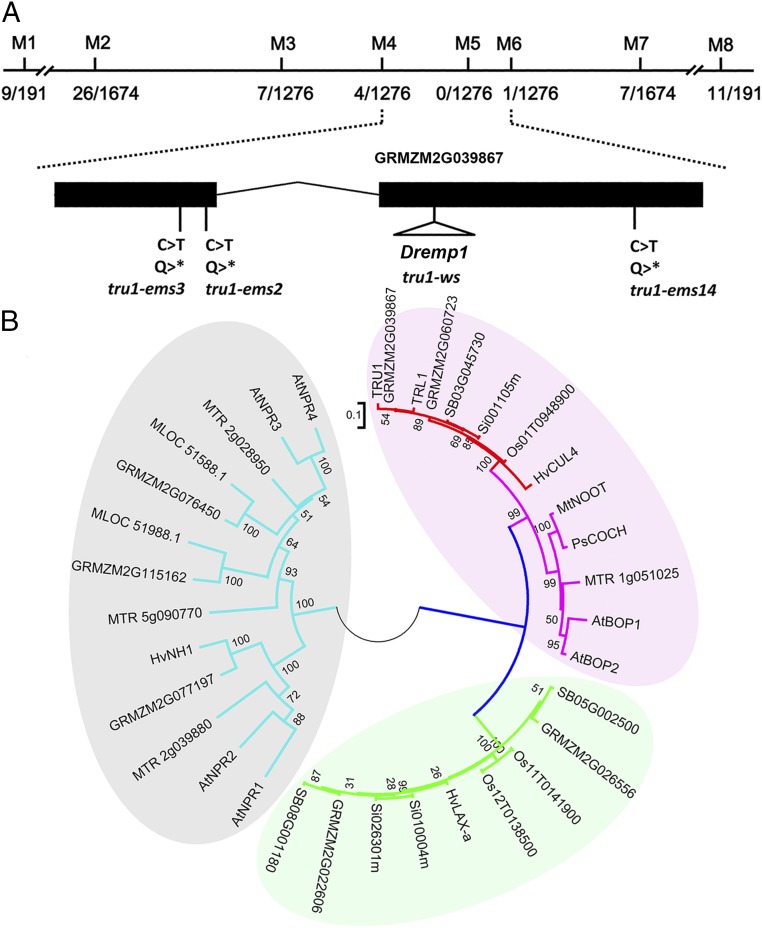
To understand the molecular function of tru1, the gene was cloned by chromosome walking. The tru1 gene was previously mapped to the long arm of chromosome three (21), where by bulked segregant mapping we identified flanking markers (M1 and M8) for fine mapping (Fig. 2A). A total of 5,399 plants from several segregating families were rescreened with additional PCR markers, narrowing the interval to a 1.6-Mb region between markers GDB201 (M4) and GDB190 (M6). Within this interval, a gene model (GRMZM2G039867, Maize Genetics and Genomics Database, Set 5b+) displaying homology to the Arabidopsis BLADE-ON-PETIOLE (BOP1) gene was identified as a candidate. Sequencing the tru1-ws allele of this candidate revealed a Dremp1 retrotransposon in the second exon (Fig. 2A). To confirm the identity of the tru1 gene, a targeted mutagenesis was performed using ethyl methane sulphonate (EMS)-treated wild-type pollen on tru1-ws homozygotes and heterozygotes. Three additional tru1 mutant alleles were recovered that contained C to T nonsense mutations resulting in premature stop codons (Fig. 2A). These results confirm that tru1 encodes a BOP1 homolog.
Fig. 2.
Cloning the tru1 gene. (A) Map-based cloning of tru1. The name of markers used for screening the recombinants are simplified as M1 to M8. Numbers of recombinants are indicated by fractions. Black boxes represent exons, and positions of transposon and ems-induced mutations are below. (B) Phylogenetic tree of BTB/POZ domain-containing homologs from selected dicots and monocots. Genes shaded in pink indicate the BOP clade, while red branches represent the monocot BOPs, including barley CUL4, maize tru1 (GRMZM2G039867), and the tru1 duplicated gene tru1-like1 (trl1) (GRMZM2G060723). Bar indicates the substitution rate per site.
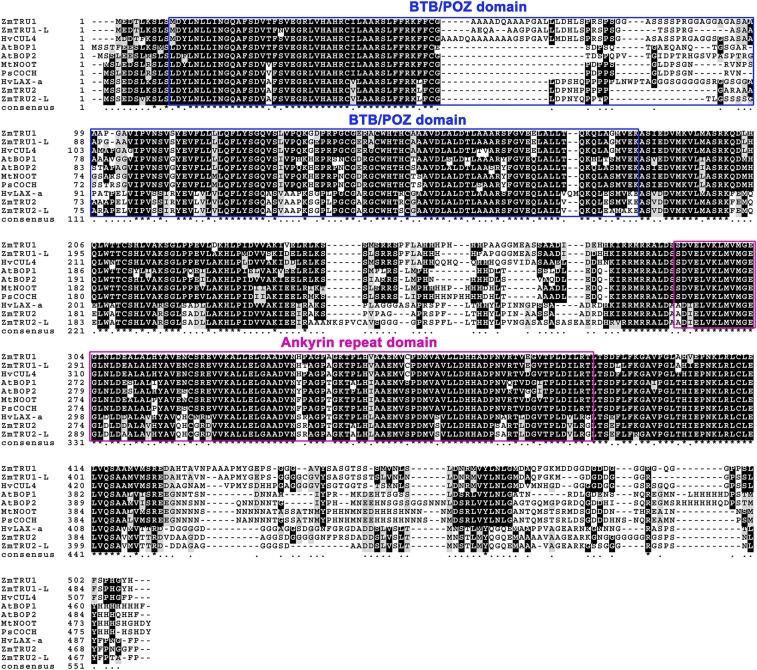
Mutants of the BOP1 gene in Arabidopsis display a shift of distal leaf identities to proximal compartments, resulting in the formation of ectopic leaf flaps growing along the petiole (24). BOP1 contains a BTB/POZ motif as well as an ankyrin repeat domain (24) (Fig. S2), both of which may be necessary for protein–protein interactions. This has been supported by two hybrid analyses of the BOP1 duplicate gene BOP2 that was shown to interact with the TGA transcription factor PERIANTHIA (25). Similar to Arabidopsis, a duplicate of tru1 is present in maize (Fig. 2B), which may function redundantly. Homologs of tru1 occupy their own clade (Fig. 2B), and several members have been functionally characterized and shown to be important for plant development and tissue patterning. Interestingly, two BOP1 orthologs in the dicots pea and Medicago, called COCHLEATA and NOOT, respectively (Fig. 2 and Fig. S2), also cause a shift in distal leaf identities to proximal locations, manifesting as leaves with altered stipule development in the mutants (26, 27). Recently, a barley knockout mutant of the tru1 ortholog uniculme4 (CUL4) was found to display a shift of distal leaf identities to proximal locations (28), relocating auricle tissue to the base of the sheath instead of at the sheath blade boundary. Moreover, CUL4 mutants lack ligules, a result which indicates that stipules and ligules are in fact homologous structures (20). Interestingly, CUL4 mutants also exhibit a dramatic decrease in axillary branching and tillering (28), which is almost the exact opposite of both the tb1 and tru1 phenotypes in maize. Taken together, it appears that members of the BOP1 clade primarily function to specify proximal leaf zone identities, and that in their absence upper leaf zone identities are shifted proximally. This results in alterations in ligule and stipule development within the blade, but also defective axillary branch growth.
Fig. S2.
Amino acid alignment of tru1 and homologs. Amino acid alignments of full-length BOP1 homologs from monocots and dicots.
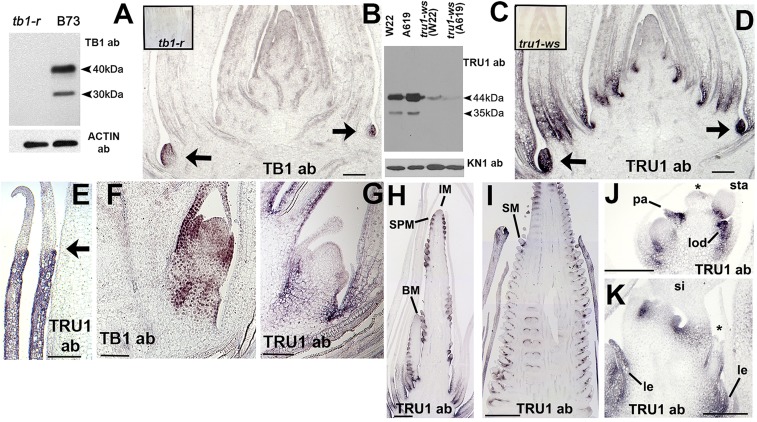
To understand where tru1 is expressed, antibodies were raised to full-length TB1 and TRU1 proteins. The TB1 serum was purified against the 3′ end of TB1 outside the TCP domain while the TRU1 serum was purified against full-length protein. Both antibodies were tested on nuclei protein preps by Western blotting, and by immunolocalization. The TB1 antibody recognized two bands on Western blots, both of which were absent in the tb1-ref mutant background (Fig. 3A). Since the upper band is the correct size for endogenous TB1, the lower band could possibly be an alternatively spliced product. The specificity of the antibody was confirmed by immunolocalization (Fig. 3B), where staining was observed in the axillary buds of 3-wk-old shoot apices, but was absent from tb1-r shoots (Fig. 3B, Inset). The TRU1 antibody also recognized two bands on Western blots, again with a larger band corresponding to the correct size protein (Fig. 3C) and a lower band that may correspond to an alternatively spliced product. The lower band was not detected in tru1-ws mutants while the upper band’s signal was greatly reduced. Similar to TB1, TRU1 protein was found in axillary meristems as seen in immunolocalization of adjacent sections of shoot apices (compare Fig. 3B to Fig. 3D) and was absent from tru1-ws sections (Fig. 3D, Inset). In addition, TRU1 was also found in leaf bases (Fig. 3D), extending up to and including the ligule (Fig. 3E), but not into the blade. This is similar to the expression of the barley ortholog, cul4 (28), and indicates that tru1 may also play a role in specification of basal leaf compartments. In sections of older axillary buds, both TB1 (Fig. 3F) and TRU1 (Fig. 3G) were expressed in the nodes below the axillary meristem and its surrounding leaves. TRU1 was not found in the younger leaves or the axillary meristem itself (Fig. 3G), similar to its pattern in the shoot apex. The expression overlap between TB1 and TRU1 in the axillary buds, combined with the genetic epistasis between the two, indicates a possible regulatory interaction between the two genes.
Fig. 3.
Expression of TRU1 in relation to TB1. (A) Western blot containing whole protein extracts from 3-wk-old axillary buds of wild-type B73 and tb1-ref probed with an anti-TB1 antibody. Full-length TB1 protein is ∼40 kDa. Blot was stripped and reprobed with anti-ACTIN antibody as a loading control. (B) Immunolocalization of 3-wk-old shoot apex with anti-TB1 showing expression in young axillary branch meristems. Control probing on tb1-ref mutant is in Inset. (C) Western blot containing whole protein extracts from 3-wk-old isolated axillary buds of wild-type W22 and A619 as well as tru1-ws introgressed into both backgrounds probed with anti-TRU1 antibody. Full-length TRU1 protein is 44 kDa. Blot was stripped and reprobed with anti-KNOTTED as a loading control. (D) Immunolocalization of 3-wk-old shoot apex with anti-TRU1 showing expression in young axillary branch meristems and leaf bases. Section is adjacent to that shown in B. Control probing on tru1-ws mutant is in Inset. Arrows indicate young axillary buds. (E) Immunolocalization of immature plastochron 8 leaf (Right) with anti-TRU1. Expression is in sheath and ligule (arrow) but not in blade. (F) Immunolocalization on developing axillary buds with anti-TB1. (G–K) Immunolocalization with anti-TRU1. (G) Immunolocalization on developing axillary buds. (H) Tassel primordia showing expression in initiating spikelet pair meristem (SPM), but not in inflorescence meristem (IM) and branch primordia (BM). (I) Ear primordium showing expression at bases of young spikelet meristems (SM). (J) Maturing tassel floret showing expression in palea (pa), base of stamens (sta), and lodicules (lod), but not in degenerating gynoecium (asterisk). (K) Maturing ear floret showing expression in lemma (le), and the base of carpels that comprise the silk (si), but not in the degenerating stamen (asterisk). (Scale bars: B–G, ∼200 μm; H, J, and K, ∼500 μm; and I, 1,000 μm.)
After the floral transition, TRU1 protein was expressed in the bases of several leafy lateral floral organs in both the tassel (Fig. 3 H and J) and the ear (Fig. 3 I and K). Interestingly, TRU1 is not expressed in several meristems, including the branch, spikelet and floral meristems, despite the fact that the products of many of these meristems are affected in the mutant. After the acquisition of sex identity in the male (Fig. 3J) and female florets (Fig. 3K), TRU1 expression was found in the bases of several different types of leafy floral organs, including the palea, lemma, lodicules, and glumes, but also in bases of the carpels of the gynoecium as well as the stamens. Thus, expression of TRU1 appears to mark basal compartments of a variety of lateral organs during both the vegetative and floral phase, but is absent from several meristems. Interestingly, TRU1 was not expressed in the degenerating carpels of the tassel (Fig. 3J) or the degenerating stamens of the ear (Fig. 3K), despite the fact that a sex reversal phenotype is seen in the mutant, consistent with the sex reversal phenotypes being a secondary, indirect consequence of mutation.
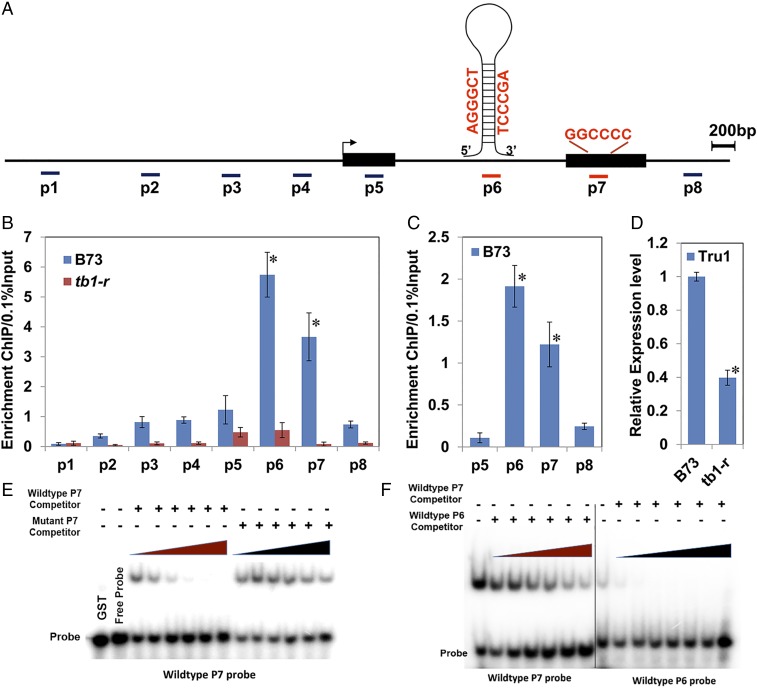
Since the expression of TRU1 in axillary buds overlapped with that of TB1, and the two genes shared an epistatic relationship, we used the TB1 antibody to determine if the tru1 gene is a direct target of TB1. The TB1 antibody was used to pull down chromatin fragments from approximately 3-wk-old tiller buds as well as young ear primordia. Eight regions across the tru1 gene were amplified from the purified chromatin (Fig. 4A) and compared with chromatin isolated from the tb1-r mutant background. Two strong peaks of enrichment were identified in the first intron (P6) as well as the last exon (P7) in wild-type tiller buds (Fig. 4B) as well as ears (Fig. 4C), demonstrating that tru1 is a direct target of TB1 in vivo. Expression of tru1 in a tb1-r mutant background was significantly reduced (Fig. 4D), consistent with tru1 being activated by TB1.
Fig. 4.
Tru1 is a direct target of and positively regulated by TB1. (A) Diagram of the tru1 genic region. Black boxes indicate exons, and lines between boxes represent the intron. Locations of the amplicons (P1 to P8) used for ChIP-qPCR are marked below. Two potential TB1-binding motifs are highlighted in red: a strong one (GGCCCC) in the P7 amplicon and a weaker one in P6 as part of an 80-bp inverted repeat. (B and C) ChIP-qPCR on anti-TB1 precipitated chromatin isolated from wild-type and tb1-r tiller buds (B) and wild-type ear primordia (C) showing that TB1 binds to tru1 in vivo. Enrichment was calculated by percentage of the immunoprecipitated DNA relative to 0.1% of the input. (D) qRT-PCR showing that tru1 mRNA is significantly down-regulated in the tb1-r mutant. Values in B–D are means ± SD of three biological replicates. * indicates P < 0.001 by Student's t test. (E) Gel shifts using TB1 protein against labeled P7 fragments. Competition was performed with either steadily increasing concentrations of unlabeled wild-type P7 oligos (red triangle) or mutated P7 oligo (black triangle). (F) Gel shifts using TB1 protein against labeled P7 fragments and competed with steadily increasing concentrations of cold P6 probe (red triangle), or labeled P6 probe competed with steadily increasing concentrations of cold P7 probe (black triangle).
Class II TCP proteins are known to heterodimerize and homodimerize and bind the consensus sequence GTGGNCCC (29, 30). A similar binding site containing GGCCCC was found in the P7 exon peak and tested for binding to TB1 using gel shifts. A 27-bp labeled oligo including the potential binding site was mixed with recombinant TB1 and found to bind (Fig. 4E). This same mix was competed against increasing concentrations of either unlabeled wild-type competitor or unlabeled mutant competitor. The oligo with the mutated binding site could not compete, while the wild-type oligo did, indicating that binding of TB1 to the sequence was specific (Fig. 4E). Although no clear consensus-like binding site was present in the intron P6 site, a weakly similar sequence containing TCCCGA was found as part of an 80-bp stem loop inverted repeat structure. While TB1 is able to bind to the labeled P6 sequence (Fig. 4F), it does so with reduced affinity since the P7 site competes very strongly with it and vice versa. It is possible that TB1 may require a different partner to bind the P6 site in vivo with stronger affinity, which is not surprising in light of the propensity of TCP proteins to heterodimerize.
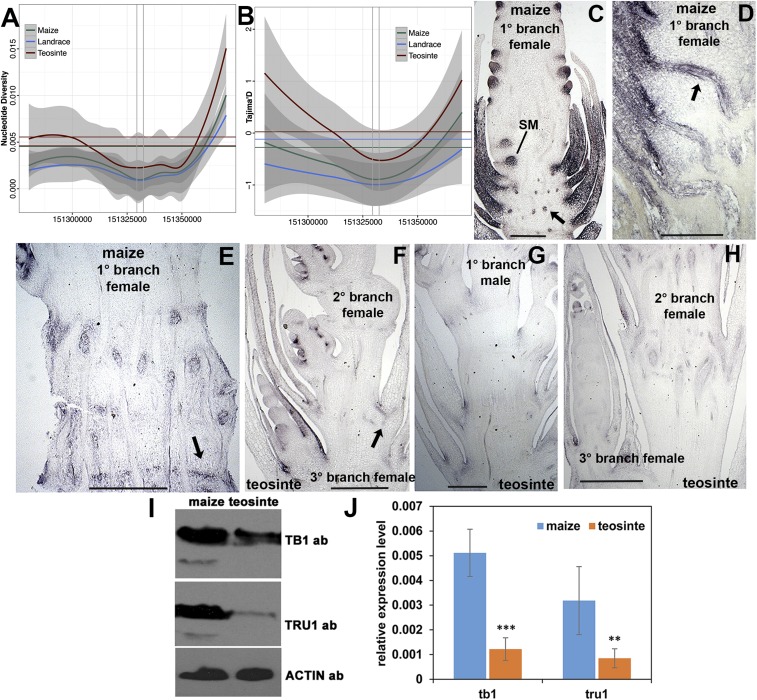
Since tru1 appears to be a target of TB1, which was under strong selection during maize domestication (31), we examined the interval around the tru1 locus for evidence of directional selection in modern maize, early maize landraces, and wild teosinte (32). Nucleotide diversity decreases dramatically within the tru1 transcription unit for maize, maize landraces, and teosinte (Fig. 5A) by an average of 67%, 73%, and 56%, respectively, compared with genome-wide averages. This result is strongly suggestive of a history of positive selection. Therefore, we conducted tests for departure from a model for neutral evolution. First, we computed Tajima’s D statistic (33). Tajima’s D values are significantly negative from the transcription start to beyond the end point of the tru1 gene, especially in maize landraces and modern maize inbreds (Fig. 5B), consistent with the tru1 gene being under stronger positive selection in maize compared with teosinte. As a stringent test of selection, we conducted maximum likelihood Hudson–Kreitman–Aguadé (MLHKA) tests of neutrality (34, 35). These tests rejected the neutral model for improved maize (P = 0.03) as well as the maize landraces (P = 0.03), but not for teosinte (P = 0.06), further supporting the notion that the tru1 alleles were under positive selection during maize domestication.
Fig. 5.
Analysis of tru1 in maize and Z. mays parviglumis. (A) Nucleotide diversity and Tajima’s D test (B) for the region surrounding the tru1 locus of improved maize, maize landraces, and teosinte. Base-pair positions are relative to AGPv4 of the maize reference genome. Vertical gray lines indicate the transcript start and end points of tru1. Gray bands denote 95% confidence intervals. Horizontal lines indicate genome-wide averages. (C–H) TRU1 immunolocalization. (C) Immature primary axillary branch of maize corresponding to the female ear primordium showing expression at base of SM, husk leaves, and transverse sections of leaf traces (arrow). (D) Close-up of shank of immature maize ear primordium showing expression in longitudinal sections of vascular traces (arrow) connecting to husk leaves. (E) Older maize ear shank showing continued expression in leaf traces and internode cells (arrow). (F) Immature female secondary and tertiary axillary branches of teosinte at floret and spikelet pair stages, respectively. Reduced expression is seen in leaf trace (arrow) as well as leaves. (G) Immature male primary axillary branch of teosinte with reduced expression in shank. (H) Closeup of older secondary and tertiary female branches of teosinte shank showing reduced expression in husk leaves and differentiated vasculature. (I) Western blot of whole protein extracts from flowering maize and teosinte ear shank tissue probed with TB1 and TRU1 antibodies. Anti-actin antibody was used as a loading control. (J) qRT-PCR of the same tissue in I. Actin was used as an internal control. **P < 0.01, ***P < 0.001 by Student’s t test. (Scale bars: C and D, ∼500 μm; E–H, ∼1,000 μm.)
Mechanistic consequences of this positive selection were examined using the TRU1 antibody. Given that tb1 became differentially expressed during domestication (9), we examined whether its target, tru1, is also differentially expressed in maize versus teosinte. No substantial difference in TRU1 expression was seen between teosinte and maize by both Western blots and immunolocalization on vegetative shoot apices. In tissues of the axillary branches, however, a clear difference was observed in the shanks of teosinte versus domesticated maize. In young ear primordia of maize, strong TRU1 expression was observed at the base of the spikelet meristems, and in husk leaves, as well as ear shank vasculature (Fig. 5C). This vasculature corresponds to the leaf traces that ultimately connect to husk leaves (Fig. 5D). In older elongated ear shanks, TRU1 expression is maintained in the leaf traces, but is also found in the compressed, unelongated internodes (Fig. 5E). In contrast, in the internodes and leaf traces below the young female inflorescence of teosinte, TRU1 protein was highly reduced or absent (Fig. 5F). Moreover, we also found that TRU1 protein was absent in these same regions within teosinte’s terminal male inflorescence (Fig. 5G) as well as its older female inflorescence (Fig. 5H). To confirm this overexpression of TRU1 and TB1 in domesticated maize shanks compared with teosinte, we performed Western blots (Fig. 5I) as well as by qRT-PCR analysis (Fig. 5J).
Discussion
Ideal plant architecture for row crops consists of few or no axillary branches to reduce overcrowding and lodging. To achieve this in maize, ancient farmers selected for gain of function mutations in tb1 (12), a repressor of axillary branch growth that is expressed at higher expression levels in the shank compared with teosinte (9). How elevated levels of tb1 are able to repress internode elongation and alter axillary branch fate is unknown. Here, we show that a BTB/POZ domain ankyrin repeat protein is a direct target of TB1 and functions to repress internode elongation in axillary branches as well as determine axillary branch fate. We show that TRU1 protein and RNA levels are higher in ear shank tissue of domesticated maize (Fig. 5 I and J), where the protein is ectopically localized to vascular leaf traces (Fig. 5C). Thus, ectopic overexpression of TRU1 inhibits axillary branch growth and alters meristem fates, a process that was critical during the domestication of maize from teosinte. Interestingly, tru1 affects both axillary branch growth and inflorescence sex determination in a progressive pattern along the stem axis, and yet it is not expressed in the axillary meristem itself at later stages (Fig. 3G) or in the male inflorescence meristem (Fig. 3H). These observations are consistent with tru1 functioning to control the activity of diffusible substance(s) that affect meristem activity at a distance.
Plant hormones are potential candidate substances that may be affected by the ectopic overexpression of TRU1 in ear shanks. Several hormones, including auxins (36, 37) and strigolactones (SL) (38), have been shown to repress axillary bud outgrowth in a manner consistent with the domesticated maize branching phenotype. SLs, however, have been shown to function independently of tb1 (39), so its target gene tru1 may function independently as well. Jasmonic acid (JA) could be an additional hormone affected by TRU1 since knockouts of the JA biosynthetic genes OPR7/OPR8 in maize phenocopy tru1 ear shanks (40). It is possible that in domesticated maize, the ectopic expression of TRU1 in vascular leaf traces may move additional JA that minimizes internode elongation through the known effects of JA on suppression of mitosis (41). Moreover, another consequence of reduced JA activity in maize inflorescences is a lack of carpel abortion within the tassel, leading to the formation of female instead of male flowers (42). This sex conversion is reminiscent of tru1 mutants where the primary axillary branches produce male inflorescences instead of female ones (Fig. 1B), along with secondary branches that produce female inflorescences similar to teosinte (Fig. 1C). However, double mutants between tru1 and the JA biosynthetic mutant tasselseed1 (42) appear additive, indicating that the two genes may operate in separate pathways. In addition, expression of tru1 is not specific to the male or female floral organs that undergo the abortion process that occurs during sex determination (Fig. 3 J and K). Thus, JAs by themselves may not explain every aspect of the tru1 phenotype, making it conceivable that TRU1 may act on several plant growth regulators.
Since a major function of tru1 is to repress internode elongation, it is possible that the elongated internodes of the tru1 mutant remove the axillary inflorescence from the regional control of several diffusible sex determination and branching factors acting in a combinatorial fashion. A similar theory has been invoked previously to explain how the short and compact female axillary inflorescence of domesticated maize was derived from the elongated male axillary inflorescence of teosinte (23). This theory speculates that the increase in apical dominance during domestication of teosinte secondarily brought the long, normally male, axillary branches closer to the main shoot, which is under local control of a feminizing hormone, resulting in a dramatic sex conversion (43). In support of this, there is a clear correlation between ear shank length and sex identity in maize (22). In addition, we observed a similar correlation in tru1 mutants where inflorescences on shorter shanks were usually female, but male on longer ones (Fig. S1 B and C). It is also possible that sex determination is dependent on several other additional hormones working in a position-dependent manner as seen in other species (44). Thus, we speculate that the elongated internodes of tru1 mutants place the inflorescence in an altered context of plant growth regulators of sex identity and axillary branching, resulting in alterations in both processes.
Members of the BTB/POZ clade of ankyrin repeat proteins have a wide range of functions (25) including establishment of floral and inflorescence meristem determinacy (45–47), but those most similar to tru1 appear to have a clear function in specifying leaf cell identities. The founding members of this clade, BLADE ON PETIOLE 1 and 2, display ectopic distal blade tissue growing down the petiole and lack of stipules (24, 48). Mutations in the dicot BOP1 orthologs COCHLEATA (coch) and NOOT in pea and Medicago, respectively, also affect stipule development (26). In coch mutants, the stipule is either absent or transformed into leaflets that are normally found in the distal portion of the leaf. Based on these phenotypes, it appears that members of this clade play roles in establishing boundaries in the proximal compartment of the leaf, including the stipule, where they function to repress distal leaf identities. Although BOP-like genes do not function as transcription factors, they have been shown to interact with them in Arabidopsis (25) as well as tomato. For example, the nuclear transport of tomato BTB/POZ genes is mediated by their interaction with transcription factors such as TERMINATING FLOWER (46), indicating that tru1 may be part of a transcriptional complex. Thus, the complex developmental changes conditioned by bop1 mutants in different species may be due to misexpression of multiple downstream target genes.
The phenotype of the only other known BOP mutant in grasses is perplexing compared with tru1. The barley cul4 mutant lacks ligules and has a proximal shift in leaf identities to distal locations, similar to bop and coch mutants (28). In addition, cul4 mutants display a reduction in axillary branching, the exact opposite of both the tb1 and tru1 phenotypes. This contradiction may be based in the fact that in modern domesticated maize, the tb1 gene is a natural gain of function mutation, caused by a transposon insertion in the promoter (12), while in barley, there is no evidence that the tb1 ortholog is similarly overexpressed. This dominant mutation in maize causes increased tb1 expression in the ear shank (9) (Fig. 5 I and J) and subsequent repression of axillary bud outgrowth. Thus, when the tb1 target gene tru1 is mutated, it is possible that this natural gain of function phenotype is suppressed and reverts maize back to its more ancestral, highly branched state. While tru1 mutants do not affect axillary meristem initiation and ligule development in a similar manner as cul4 in barley, we speculate that this may be a consequence of functional redundancy of the tru1 duplicate gene (Fig. 2B).
Previous work identified five major loci necessary for maize domestication, as well as other minor loci with lesser effects (5, 49). For example, a minor locus was identified that functions with tb1 and controls many of the same traits, including the presence of staminate inflorescences and increased lateral branch length (50). Interestingly, this second locus maps to a region on chromosome 3 where tru1 maps (50). Hence, it is very likely that the tru1 gene is responsible for this aspect of tb1 function, an idea that was proposed nearly 22 y ago (7). In addition, a second locus called grassy tillers1 (gt1) was found to be a domestication gene that affects the number and arrangement of ears on the derepressed axillary branches (51). gt1 was also found to be genetically downstream of tb1 (52), and is likely another TB1 target. In addition, recent work in Arabidopsis showed that gt1 homologs are directly targeted by BRC1, and regulate ABA levels and signaling (53). Both gt1 and tru1 independently control a subset of phenotypes seen in tb1, where gt1 represses tiller number at ground level, while tru1 represses axillary bud growth aboveground, without affecting tillering (Fig. 1B). Together, the combined activity of tru1 and gt1 as targets of tb1 may explain most of the axillary branch differences between domesticated maize and teosinte. This indicates that domestication genes such as tb1 have multifaceted functions that may differ depending on the position within the plant. A thorough analysis of all the targets of TB1 will likely uncover even more complex functions and help us understand why ancient farmers chose this gene as a domestication target.
Materials and Methods
Plant Materials.
The tru1-ws and tb1-ref mutants were obtained from the Maize Genetics Cooperation Stock Center. tru1-ws was backcrossed into W22 and A619 four times, while tb1-r was backcrossed into B73 four times. The primers used for genotyping are listed in Table S1. Z. mays parviglumis teosinte lines Til01 were obtained from John F. Doebley, University of Wisconsin-Madison, Madison, WI.
Table S1.
Primers used in this study
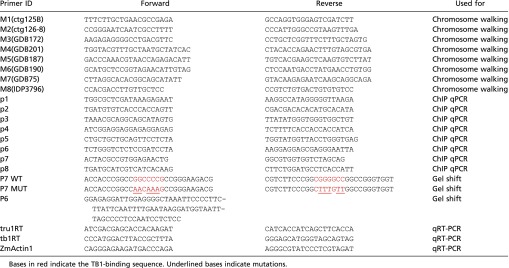 |
Bases in red indicate the TB1-binding sequence. Underlined bases indicate mutations.
Three new alleles, tru1-ems2, ems3, and ems14, were identified by a noncomplementation screen of M1 families using EMS mutagenesis of the A619 maize inbred. In brief, tru1-ws mutant plants were crossed to the A619 inbred line, and then the F1 plants were backcrossed to tru1-ws mutant plants. The backcross population was crossed as female by EMS-treated A619 pollen to obtain M1. From 28,500 M1 plants, we selected 34 plants that resembled a strong tru1 mutant phenotype and outcrossed them to B73 or Mo17 inbreds. The resulting F1 plants were genotyped by the tru1-linked marker M8, to distinguish new tru1 mutant alleles (A619 background) from self-contaminants of the original tru1-ws allele (undefined background). Allelism was finally confirmed by performing six pairwise complementation tests among the three EMS alleles and tru1-ws.
Map-Based Cloning of tru1.
The original tru1-ws plants were crossed to the A619 inbred and then backcrossed to tru1-ws to generate F1BC1 mapping populations. tru1 was mapped to bin 3.05 in the genetic map between marker M1 (ctg125B) and M8 (IDP7873) using 191 F1 BC1 homozygous mutants, and further narrowed into a 1.6-Mb region after chromosome walking using 2,258, 1,674, and 1,276 individual mutants in consecutive growing seasons. For the selected candidate gene, we corrected a large rearrangement in the genome assembly (AGPv3) and then amplified the gene region by PCR (Table S1) and sequenced the DNA.
Phylogenic Analysis.
Protein sequences for BOP orthologs were obtained by BLAST searching in EnsemblPlants (plants.ensembl.org) and by BLASTp in National Center for Biotechnology Information. Multiple sequence alignments were performed using CLUSTALW, and following phylogenic analyses were conducted through MEGA5 using the maximum likelihood method (54).
qPCR.
Total RNA was isolated from tiller buds harvested as described for ChIP. For qRT-PCR analysis, cDNA was synthesized from DNase I-treated total RNA as described previously (55). SD was calculated among three biological replicates for each sample. ZmActin1 was used as the internal reference to normalize the expression data. The primers used for qPCR are listed in Table S1.
ChIP.
Plant tissue including 3-wk-old tiller buds as well as young ear primordia smaller than 5 mm were dissected from field grown B73 or tb1-r. About 1 g of tissue per biological replicate was then fixed in 1% formaldehyde. Nuclei extraction and chromatin immunoprecipitation using α-TB1 antibody were performed as described previously (56). Normal goat anti-Guinea pig IgG (sc-2711; Santa Cruz Biotechnology) was used as a negative control. Three biological replicates were applied in ChIP-qPCR using primers listed in Table S1.
Antibody Generation and Immunolocalization.
Full-length TB1 and TRU1 coding sequence were cloned into the pET21d expression vectors (Novagen) for antigen production. The purified recombinant proteins were injected into guinea pigs, and the serum affinity purified using the 3′ end of TB1 and full-length TRU1 protein, respectively, as described by Chuck et al. (57) using standard protocols approved by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC 000868). Immunolocalization was performed as previously described (57), except for epitope retrieval by boiling paraffin sections slides submerged in a sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 8 min.
EMSA.
The 5′ end of TB1, including the TCP domain from amino acids (112–180) was cloned into the pDEST15 and expressed and purified from Escherichia coli using glutathione Sepharose beads. Approximately 100 ng of recombinant protein was added to 1 pmol of double-stranded oligonucleotide probes labeled with gamma ATP using polynucleotide kinase and run on nondenaturing PAGE gels using previously described methods (58). For competition experiments, 2, 10, 25, 50, 100, and 200 pmol, respectively, of unlabeled competitor oligo was added to the binding reactions.
Population Genetics.
We obtained population genetics statistics for maize (n = 35 improved maize lines, n = 23 traditional landraces), teosinte (n = 16), and the outgroup Tripsacum (n = 1) from ref. 32 for a region 50 kb upstream and 50 kb downstream of tru1. Nucleotide diversity and Tajima’s D were calculated using a 10-kb nonoverlapping window. Curved lines were smoothed using the Loess function in the R package. The HKA test was performed using a MLHKA approach (35) with 100,000 simulations on the same region. Significance was estimated by the likelihood test, where twice the difference in log likelihood between the neutral model and selection model was considered as the χ2 distribution. The number of degrees of freedom was equal to the difference in the number of parameters.
Acknowledgments
We thank China Lunde, Marisa Rosa, and Sarah Hake for critical reading of this manuscript. This work was supported by National Science Foundation Grants PGRP IOS-1339332 (to G.C.) and IOS-1238202 (to E.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714960114/-/DCSupplemental.
References
- 1.Holalu SV, Finlayson SA. The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J Exp Bot. 2017;68:943–952. doi: 10.1093/jxb/erw479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross-Ibarra J, Morrell PL, Gaut BS. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA. 2007;104:8641–8648. doi: 10.1073/pnas.0700643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ongaro V, Leyser O. Hormonal control of shoot branching. J Exp Bot. 2008;59:67–74. doi: 10.1093/jxb/erm134. [DOI] [PubMed] [Google Scholar]
- 4.Durbak A, Yao H, McSteen P. Hormone signaling in plant development. Curr Opin Plant Biol. 2012;15:92–96. doi: 10.1016/j.pbi.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 6.Burnham CR, Yagyu P. Linkage relations of teosinte branched. Maize Genet Coop News Lett. 1961;35:87. [Google Scholar]
- 7.Doebley J, Stec A, Gustus C. Teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 10.Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 11.Nicolas M, Cubas P. TCP factors: New kids on the signaling block. Curr Opin Plant Biol. 2016;33:33–41. doi: 10.1016/j.pbi.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 13.Studer AJ, Doebley JF. Evidence for a natural allelic series at the maize domestication locus teosinte branched1. Genetics. 2012;191:951–958. doi: 10.1534/genetics.112.138479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remigereau M-S, et al. Cereal domestication and evolution of branching: Evidence for soft selection in the Tb1 orthologue of pearl millet (Pennisetum glaucum [L.] R. Br.) PLoS One. 2011;6:e22404. doi: 10.1371/journal.pone.0022404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McSteen P. Hormonal regulation of branching in grasses. Plant Physiol. 2009;149:46–55. doi: 10.1104/pp.108.129056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minakuchi K, et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010;51:1127–1135. doi: 10.1093/pcp/pcq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilar-Martínez JA, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas M, Rodríguez-Buey ML, Franco-Zorrilla JM, Cubas P. A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr Biol. 2015;25:1799–1809. doi: 10.1016/j.cub.2015.05.053. [DOI] [PubMed] [Google Scholar]
- 19.Kebrom TH, Brutnell TP, Finlayson SA. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 2010;33:48–58. doi: 10.1111/j.1365-3040.2009.02050.x. [DOI] [PubMed] [Google Scholar]
- 20.Chuck G, Dong Z. BTB/POZ ankyrin repeat genes identify leaf homologies in monocots and eudicots. Trends Dev Biol. 2016;9:71–76. [Google Scholar]
- 21.Sheridan WF. Maize developmental genetics: Genes of morphogenesis. Annu Rev Genet. 1988;22:353–385. doi: 10.1146/annurev.ge.22.120188.002033. [DOI] [PubMed] [Google Scholar]
- 22.Irish EE. Regulation of sex determination in maize. BioEssays. 1996;18:363–369. [Google Scholar]
- 23.Iltis HH. Homeotic sexual translocations and the origin of maize (Zea mays, Poaceae): A new look at an old problem. Econ Bot. 2000;54:7–42. [Google Scholar]
- 24.Ha CM, et al. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development. 2003;130:161–172. doi: 10.1242/dev.00196. [DOI] [PubMed] [Google Scholar]
- 25.Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell. 2005;17:1434–1448. doi: 10.1105/tpc.104.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couzigou J-M, et al. NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE genes. Plant Cell. 2012;24:4498–4510. doi: 10.1105/tpc.112.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaxley JL, Jablonski W, Reid JB. Leaf and flower development in pea (Pisum sativum L.): Mutants cochleata andunifoliata. Ann Bot. 2001;88:225–234. [Google Scholar]
- 28.Tavakol E, et al. The barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol. 2015;168:164–174. doi: 10.1104/pp.114.252882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosugi S, Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002;30:337–348. doi: 10.1046/j.1365-313x.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 30.Franco-Zorrilla JM, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hufford MB, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright SI, Charlesworth B. The HKA test revisited: A maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 38.Shinohara N, Taylor C, Leyser O. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 2013;11:e1001474. doi: 10.1371/journal.pbio.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan JC, et al. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 2012;160:1303–1317. doi: 10.1104/pp.112.204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan Y, et al. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell. 2012;24:1420–1436. doi: 10.1105/tpc.111.094151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acosta IF, et al. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- 43.Iltis HH. From teosinte to maize: The catastrophic sexual transmutation. Science. 1983;222:886–894. doi: 10.1126/science.222.4626.886. [DOI] [PubMed] [Google Scholar]
- 44.Chuck G. Molecular mechanisms of sex determination in monoecious and dioecious plants. Adv Bot Res. 2010;54:53–83. [Google Scholar]
- 45.Khan M, et al. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis. Plant Physiol. 2015;169:2166–2186. doi: 10.1104/pp.15.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu C, Park SJ, Van Eck J, Lippman ZB. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016;30:2048–2061. doi: 10.1101/gad.288415.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jost M, et al. A homolog of blade-on-petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol. 2016;171:1113–1127. doi: 10.1104/pp.16.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ha CM, Jun JH, Nam HG, Fletcher JC. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1361–1370. doi: 10.1093/pcp/pch201. [DOI] [PubMed] [Google Scholar]
- 49.Doebley J. Mapping the genes that made maize. Trends Genet. 1992;8:302–307. doi: 10.1016/0168-9525(92)90261-2. [DOI] [PubMed] [Google Scholar]
- 50.Briggs WH, McMullen MD, Gaut BS, Doebley J. Linkage mapping of domestication loci in a large maize teosinte backcross resource. Genetics. 2007;177:1915–1928. doi: 10.1534/genetics.107.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wills DM, et al. From many, one: Genetic control of prolificacy during maize domestication. PLoS Genet. 2013;9:e1003604. doi: 10.1371/journal.pgen.1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whipple CJ, et al. Grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA. 2011;108:E506–E512. doi: 10.1073/pnas.1102819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.González-Grandío E, et al. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc Natl Acad Sci USA. 2017;114:E245–E254. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong Z, et al. Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol. 2013;163:1306–1322. doi: 10.1104/pp.113.227314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuda K, Kurata N, Ohyanagi H, Hake S. Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell. 2014;26:3488–3500. doi: 10.1105/tpc.114.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chuck G, Whipple C, Jackson D, Hake S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–1250. doi: 10.1242/dev.048348. [DOI] [PubMed] [Google Scholar]
- 58.Smith HM, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA. 2002;99:9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]