Advances of analytical methods and emerging microfluidic tools have made it possible to investigate biological processes in living organisms in detail and reach sensitivities sufficient for single-cell analysis. The term “single-cell analysis” typically refers to the elucidation of cell-to-cell differences in large cell populations, such as size, morphology, growth rate, or molecular content like the composition of lipids, proteins, metabolites, DNA/RNA, etc. Many different techniques have been developed to address the effects of cell heterogeneities.1–4
As far as we know today, heterogeneities appear in all cell populations (bacteria, yeast, and mammalian cells) and even within cell lineages, where all cells are derived from the very same mother cell. Besides the fundamental research questions (such as, why are cells different and how does the difference affect cell physiology and fate?), single-cell analysis has practical applications in many research fields.5 As will be covered in this Review, the examples include cancer biology, stem cells and regenerative medicine, microbiology and pathogenesis, neuroscience, immunology, and many more.
The biggest challenges of single-cell analysis arise from the small size of cells, the tiny absolute number of target molecules, the large number of different molecules present in a wide range of concentrations and, last but not least, the complexity imposed by many related intra- or intercellular dynamic processes. To follow these dynamic processes at the single cell level, due to the response to environmental changes or drugs, cell differentiation, or metabolic changes, methods with a high time resolution and high throughput are required in addition to high sensitivity and specificity. Quantification with highly precise and accurate read-out is essential to ensure that the revealed heterogeneities indeed originate from the cell population and are not methodical artifacts.
To date, various chemical and physical techniques are applied in the field of single-cell analysis. They typically address selected aspects of the single cells and may be complementary to each other. In the following, we focus on new developments in the fields of fluorescence microscopy, electrochemical analysis, mass spectrometry, and qPCR based technologies in the last two years. As microfluidic methods are employed in numerous analytical studies of single cells with either methodology, we introduce microfluidic devices for cell capture, cell isolation, and fluid handling in separate sections.
Microfluidic Tools for Single Cell Capture and Isolation
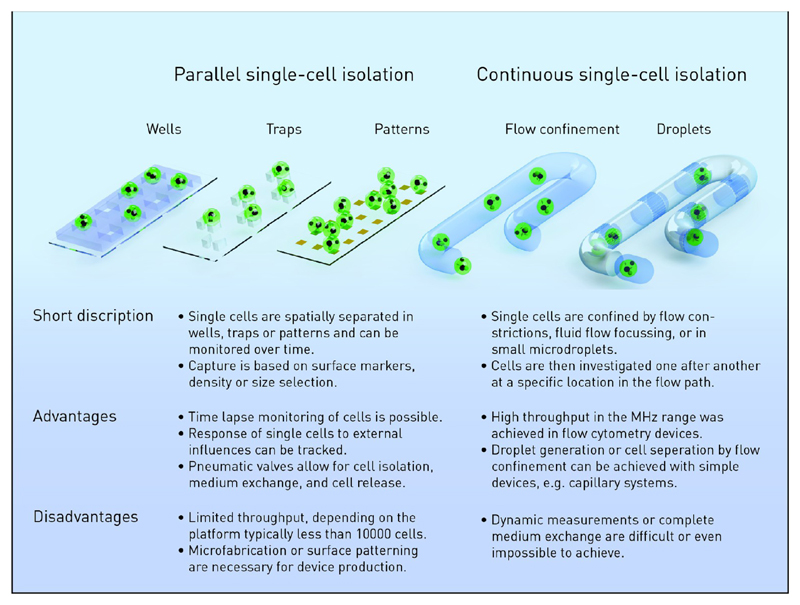
In many research questions that can be solved by single-cell analysis, a significant number of cells has to be analyzed. This can be done either in parallel or sequentially by employing methods for single cell and fluid handling (A brief comparison between parallel and continuous methods can be found in Figure 1). Microsystems technology is most valuable as it allows for building small devices for cell manipulation and isolation that can be combined with many analytical methods6–8 as will be evident in this Review. In the following, we discuss the various recent microfluidic developments to capture, position, isolate, and lyse single cells.
Figure 1. Comparison of parallel and continuous methods for single-cell positioning and analysis.
Wells, Traps, and Patterns: Parallel Processing of Single Cells
Parallel immobilization of cells is well suited to investigate the response of single cells to environmental parameters or drug treatment. A parallel setup enables the use of advanced closed microfluidic systems and valves to separate single cells in small volumes and chambers and actively exchange the media.
One possibility to realize the spatial arrangement of single cells with high occupancy rates is the use of microwells.9,10 Microwells allow for passive capture by sedimentation of cells and take advantage of the fact that cells have a higher density than the surrounding medium. The capture efficiency is adjusted to the organism of interest by varying the well’s geometry, size, depth, and material properties.11 Since sedimentation occurs on a relatively large time scale, new approaches focus on microwell techniques that are not only based on self-seeding effects.
Swennenhuis et al. presented a platform to isolate single cells by flushing them through a 6400 microwell chip acting as a microsieve.12 This fast and efficient cell individualization was coupled to the optical investigation of the cells by fluorescence microscopy. They were able to release the cells from the microwell chip for downstream analysis by punching out the well of interest. In another concept, Sun et al. used photopolymerization to capture and release cells that were trapped in wells.13
Wang et al. substituted the sedimentation based capture by a selective method by using magnetic labeling of cells to pull them toward microwells located at the top of an open microfluidic channel.14 This configuration benefits from the highly selective labeling possibilities of magnetic beads and allows simultaneous cell selection and isolation. In a first application, they isolated circulating tumor cells (CTCs) from whole blood samples of lung adenocarcinoma patients and retrieved information on genomic, proteomic, and metabolic levels.
Alternatively, a number of further cell trapping methods have been introduced, where cells are selected by size or by their mechanical, electrical, magnetic, acoustic, or optical properties.15,16 A frequently used method is mechanical entrapment, where cells are immobilized in flow constrictions. The method can be adapted to various cell sizes and types and has been employed very recently for sperm cells17 as well as for tracking host–microbe interactions.18 Traps with high efficiencies close to 100% are used for the analysis of small numbers of target cells19 and for monitoring of multigenerational cell lineages from primary, activated murine T-cells and lymphocytic leukemia cells.20 Chen et al. applied mechanical single cell traps to capture single breast cancer cells.21 They investigated dynamic sphere formation in vitro and used this information to identify cancer stem-like cells that play a significant role in cancer metastasis. Monitoring lymphocyte interactions on the single cell level was conducted by Dura et al.22 They employed a microfluidic platform to bring one T-cell and one lymphocyte in direct contact and monitored the heterogeneities in the activation of T-cells. As this was done for hundreds of single cell pairs, they could cluster the response in different subcategories to get a better understanding of immune responses that begin with the interaction of lymphocytes and antigen-presenting cells. Polarized cell growth is typical for many different organisms, and knowledge of the underlying principles of hyphen growth is of great interest in biotechnology and bioenergy production. It is technologically challenging to direct the cellular branching and control the environmental conditions in a conventional assay. Geng et al. tackled these difficulties and introduced a microfluidic platform for dynamic observations of polarized cell growth over extended periods.23 With the model organism filamentous fungus, they could visualize and analyze the distribution of nuclei in the hyphens and quantify gene expression levels with genetic markers.
While, usually, the number of cell traps is below 1000, large trap arrays for many thousands of single cells are also possible.24,25 A recently presented device for the capture of tumor cell clusters from blood samples uses triangular-shaped pillars that act as low shear stress inducing traps for cell clusters but permits smaller single cells and other blood components to pass. With this system, Sarioglu et al. were able to reliably detect tumor cell clusters from whole blood and to analyze them by immunocytochemical staining.26 Another technique incorporates design features to induce stable microvortices, which can be used to trap cells. As shown by Che et al., larger cells are caught in vortices arising at reservoirs placed along the channel.27 This system benefits from an easy setup and the fact that high flow rates can be applied.
Furthermore, dielectrophoresis (DEP) can be employed in open or closed systems. In this technique, polarizable objects such as cells can be aligned by a nonuniform electric field.28 Cells suspended in a culture medium that exhibits a relatively high electrical conductivity undergo negative DEP and are stably captured. With transparent ITO electrodes, optical transparency measurement techniques can be achieved.29 DEP traps have been used to measure the biomechanical properties of red blood cells30 and 3D embryonic bodies.31 In addition to DEP, optoelectronic tweezers can be utilized for cell isolation at optical intensities far below those of standard optical tweezers.32–34
Finally, alignment of cells in arrays can be achieved with chemical surface patterns. Cell-adhesive spots with diameters in the micrometer dimension, surrounded by cell-repellent surfaces, can be produced by microcontact printing,35–37 inkjet printing,38 or photopatterning.39 To be caught, cells have to get in contact with the adherent spots. However, the homogeneous distribution of cells over the entire patterned area is difficult in microfluidic systems with laminar flow. Fuchs and colleagues improved the capture efficiency on micropatterned antibody spots by facilitating chaotic mixing of the solution.40 Nonetheless, most micropatterning techniques suffer from the fact that they do not allow for the release of cells after the initial capture. Some newer examples have shown that there are ways to overcome this limitation and allow realization of micropatterns with the capability to capture and release the target cell. One example is glucose and pH-responsive capture sites as presented by Liu et al.41
An interesting application of microcontact printing was presented by Saliba et al.38 They used it to pattern magnetic ink onto a glass substrate and assembled a microfluidic chamber atop of this magnetic array. This setup allowed them to use a simple permanent magnet to self-assemble magnetic bead chains guided by the printed micropattern.
In addition to the aforementioned techniques, scaffolds integrated into the channel that are coated with a chemical linker can be used. This can be aptamer coated micropillars to capture cancer cells from blood samples42 or more complex 3D matrixes. Cheng et al. used such porous 3D matrixes to trap free-floating single cells from a biological sample making use of the increased contact between the cells and the capture sites.43 They coated the PDMS scaffold with anti-EpCAM antibodies and employed their method for the capture of CTCs from whole blood samples.
Cytometric Methods: Continuous Processing of Single Cells
In the above-mentioned approaches, up to a few thousand cells are captured for further analysis and many microfluidic devices are for single-use only. In approaches based on flow cytometry, suspended cells are continuously delivered through a small capillary and analyzed by multicolor fluorescence analysis as well as the detection of scattered signals to reveal information on cell size and expression of cell surface markers.44,45 In commercial instruments for fluorescence-activated cell sorting (FACS), the cells of interest are suspended in one single aqueous phase and not separated into individual compartments. However, many questions of single-cell analysis require the isolation of cells within a confined volume, e.g., when secretion of metabolites or proteins is under investigation or when cell lysis is needed for analysis and fixation cannot be done. In these cases, so-called droplet microfluidics is of particular interest.46,47 Highly monodisperse aqueous microdroplets separated by an oil or gas phase are formed in microfluidic channels either with crossed channels or with T-junctions. The results are discrete drops or plugs like water in oil droplets (W/O) that can be stabilized by surfactants.48,49 The size and shape of the droplets can be tuned by a set of parameters including, channel dimensions, flow rates, and viscosities, and the droplets are typically in the femto- to nanoliter range. Once stabilized, these systems produce droplets continuously at kHz frequencies, although recently it was shown that ultrasmall droplets can be generated at rates up to 1.3 MHz.50,51
The cell encapsulation in individual droplets is not homogeneous but follows a Poisson distribution; i.e., many droplets remain empty, while others encapsulate multiple cells. Additional techniques such as cell ordering prior to encapsulation can be implemented to improve the distribution.52 After droplet generation, droplet splitting, merging, and sorting or protocols for cell lysis, binding assays, or live/dead staining can be performed. However, protocols that require a complete exchange of medium or washing steps, e.g., immunoassays, cannot be conducted.47,53,54 In some cases, it is possible to adapt the assay, e.g., by the use of beads. An example is given by Mazutis et al. for the detection of antibodies produced in hybridoma cells.55 The cells are encapsulated in 50 pL droplets together with 6 μm microbeads that were labeled with anti-mouse capture antibodies, and fluorescently labeled detection antibodies were also incorporated into the droplets. The secreted molecules accumulate at the beads together with the detection antibodies. The signal on the bead surface then corresponds to the amount of secreted IgG, and a throughput of up to 200 cells per second can be achieved. In a more advanced approach for optimizing the production of vitamin B2 in Bacillus subtilis by directed evolution, Meyer et al. coencapsulated the cells in gel droplets with genetically modified E. coli sensor cells.56 Secreted vitamin from Bacillus subtilis initiated a cascade reaction in E. coli leading to the expression of GFP. Fluorescent analysis of the individual gel beads enabled them to select cells that produced vitamin B2 most efficiently.
One particular advantage of droplet microfluidics over conventional flow cytometry is the possibility to analyze the single cells at several time points. This is achievable because the droplets can be stored off-chip for incubation and reinserted into the cytometer afterward. However, this usually goes with a loss of the droplet order, and different strategies for tracing and labeling of the cells or droplets are required.57,58 Alternatively, a capture site for droplets can be integrated into the microchip as done on the DropSpot system, developed by Schmitz and co-workers,59 or the droplets are individually seeded on an agar plate as shown by Dong et al.60 Nevertheless, these solutions come with the disadvantage that the throughput of the total system is limited.
Optical Analysis
Microscopic techniques are extensively applied in single-cell analysis to visualize the cells or to monitor cell growth and morphological changes dynamically. Fluorescence methods reveal spatial distribution of molecules, cellular constituents, or temporal dynamics of biological processes. In recent years, sophisticated techniques based on fluorescence spectroscopy have been developed that enable extremely fast analysis and tracking of individual molecules. Two- or three-photon excitation or light-sheet microscopy is available to image thick specimens,61 and super-resolution microscopy permits one to take images with resolutions down to 10 nm.62
Besides these improvements of the instruments, the choice of the fluorescent label is critical. In the following, we highlight recent advances in fluorescent techniques including the labeling strategies, fluorescent molecular sensors, and the use of fluorescently tagged antibodies for specific labeling of targeted analytes. At the end of the chapter, we include the recent advances in label-free techniques as well.
Live-Cell Imaging
Live-cell imaging refers to long-term analysis of cells to study proliferation and metabolic processes that take change in the time course of minutes up to days. Live-cell imaging can be performed with or without a fluorescent label and is compatible with super-resolution microscopy.67 Several new reviews have recently discussed the use of external and genetic labeling techniques for live-cell imaging in detail.68,69 Live-cell monitoring of single bacteria and bacterial communities with time-lapse studies over 30 to 40 generations was achieved by Moffitt et al.70 They captured bacteria cells on agarose pads that were patterned in tracks on the submicrometer scale and constrained the cell growth in tracks. These tracks are flushed at both ends with buffer solution to wash away excess cells and provide the culture with fresh nutrients. Their system was used to cultivate multiple stains within the same microfluidic device. As the agarose confinements were porous and as small molecule exchange was preserved, this allowed them to study the intercellular communication. In another study, Hoffman et al. used live-cell imaging of GFP transfected bacteria to differentiate between reversible and irreversible adhesion.71
Multidrug resistant pathogens and antibiotic resistances are a great concern of today’s global public health. To address this issue, Hsieh et al. presented a microfluidic platform for testing of antibiotic susceptibility.72 They used a vacuum sealed chip to initiate “self-filling” of picoliter chambers with the sample solution and used oil to compartmentalize the individual chambers. Performing fluorescence based bacterial growth assays, they were able to test the antibiotic susceptibility of different bacterial strains against several antibiotics in 3 h.
With normal cells that do not express fluorescent proteins, quantitative dynamic long-term imaging of cell vitality with fluorescent labels is hindered by the fact that the intercalating dyes used to probe the perfusion through cell membranes to indicate cell death are often toxic and cannot be applied continuously. In many applications, where fluorescent stains are employed to measure the influences of drugs or other substances, this limits the vitality testing to single time-points. Krämer et al. have addressed this problem by establishing a protocol for noninvasive propidium iodide (PI) and counterstain perfusion for single cell vitality assessment.73 They altered the conventional PI staining concentrations, tested the PI-induced effect on the cell vitality, and were able to establish a nontoxic method for continuous cell vitality monitoring. They applied the method for testing of antibiotic resistances, and they were also able to differentiate between apoptotic and necrotic cell deaths.
A rather new concept to culture and monitor single stem cells and spheroid formation are hanging-drop networks. Birchler et al. combined their system with FACS to sort cells and subsequently seed single stem cells into hanging droplets.74 Afterwards, they were able to monitor the spheroid formation and growth for 125 h.75 In contrast to spheroids that stay at their predefined location during the measurement, long-term time-lapse microscopy of mobile cells over several generations requires automated cell tracking algorithms as presented by Hilsenbeck et al.76
Specialized Fluorescence Techniques
Advanced optical imaging techniques are available to analyze certain target molecules and image single cells with submicrometer resolution. We selected techniques that can be of interest as follows.
Förster Resonance Energy Transfer
FRET describes the transfer of energy from an excited donor fluorophore to an acceptor molecule and is extensively used for studies of biochemical reactions and cell biology.77 The advantage of FRET based probes is that they can be supplied in excess without the need for washing steps as fluorescence is only emitted upon binding to its target. An important intracellular target that activates intracellular enzymes is calcium. Miyamoto et al. developed a genetically encoded sensor to image cytosolic calcium signals. The probe uses FRET signals that record the activity of caspase-3 in single DT40 lymphocytes.78 They observed differences in the cytosolic calcium levels of apoptotic and surviving lymphocytes upon stimulation of B-cell receptors. Surviving cells reacted with a higher spike followed by more elevated levels of Ca2+ concentration than apoptotic cells.
FRET sensors with different excitation and emission wavelengths can be designed to allow for parallel analysis of several parameters. Ng et al. used four FRET sensors for analysis of several breast cancer cell lines on a microdroplet based platform.79 They were able to analyze live-cell and in situ cell lysis assay formats with respect to different metalloproteinases that are of importance in cancer progression on the single cell level. The study of metalloproteinases was also the aim of a study by Son et al., who reported a microfluidic system to measure the secreted levels of one of these enzymes (MMP9).80 They integrated photodegradable hydrogel capture sites for single cell capture in individual microwells. The secreted target was detected by using FRET probes that were cleaved in the presence of MMP9. The fluorescence signal was used to quantify the secretion rate, and the cells of interest could even be retrieved from the chip after photodegradation of the cell trap.
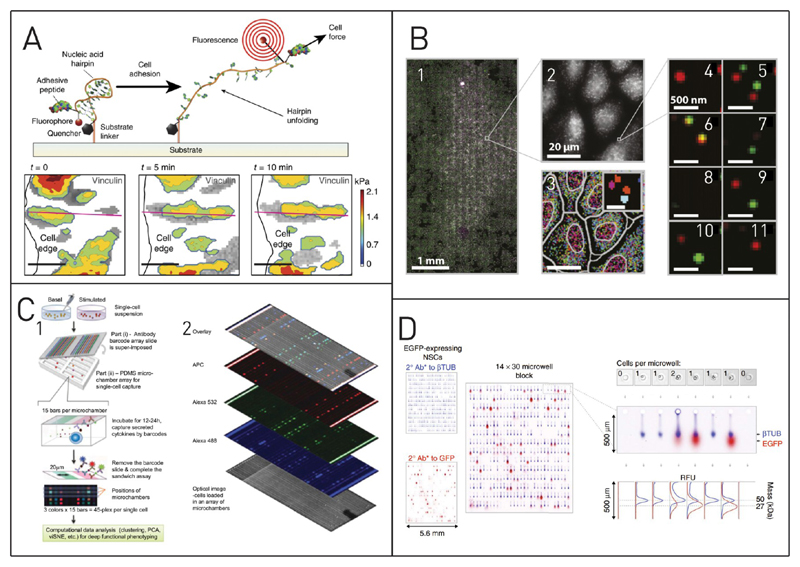
Several research groups reported FRET measurements to determine the forces that occur during the attachment of cells to a given surface.81 Blakely et al. have used a DNA based molecular probe to detect cellular traction forces of MEF cells.63 The DNA-FRET probe was elongated in the case of mechanical tension, and this leads to an increase in fluorescence with increasing force (see Figure 2A). Using these probes, they could see that cellular traction forces are not homogeneous throughout the cell but localized at their distal edges and differences between individual cells were found.
Figure 2. Specialized fluorescence techniques for single-cell analysis.
(A) Determination of traction forces that a cell is exerting on a substrate. Here, a technique based on Förster resonance energy transfer (FRET) is used for the determination of cell traction forces exerted on the surface. Besides localization of the force variation within a single cell, the differences between cells can be identified. Adapted with permission from Blakely, B. L.; Dumelin, C. E.; Trappmann, B.; McGregor, L. M.; Choi, C. K.; Anthony, P. C.; Duesterberg, V. K.; Baker, B. M.; Block, S. M.; Liu, D. R.; Chen, C. S. Nat. Methods 2014, 11, 1229–1232 (ref 63). Copyright 2014 Nature Publishing Group. (B) Analysis of the single cell transciptome. Multiplexed error-robust fluorescence in situ hybridization (MERFISH) of roughly 15 00 cells allows for sequential analysis of 130 RNA targets. A small portion of these measurements is depicted in the images numbered 4–11. Adapted with permission from Moffitt, J. R.; Hao, J.; Wang, G.; Chen, K. H.; Babcock, H. P.; Zhuang, X. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 11046–11051 (ref 64). Copyright 2016 National Academy of Sciences. (C) Multiplexed analysis of biomolecules. The combination of single-cell capture in microchambers, the use of antibody barcode arrays and three-color fluorescence microscopy facilitated the parallel detection of up to 45 parameters on the single-cell level. Adapted with permission from Lu, Y.; Xue, Q.; Eisele, M. R.; Sulistijo, E. S.; Brower, K.; Han, L.; Amir, E. D.; Pe’er, D.; Miller-Jensen, K.; Fan, R. Proc. Natl. Acad. Sci. U.S. A. 2015, 607–615 (ref 65). Copyright 2015 National Academy of Sciences. (D) Single-cell Western blotting for analysis of selected proteins of single cells. Adapted with permission from Hughes, A. J.; Spelke, D. P.; Xu, Z.; Kang, C.-C.; Schaffer, D. V; Herr, A. E. Nat. Methods 2014, 11, 749–755 (ref 66). Copyright 2014 Nature Publishing Group.
Fluorescence in Situ Hybridization
FISH is used to probe and localize specific sequences in DNA or RNA molecules in single cells or tissue samples. Even though the method is limited in throughput, it is particularly useful to identify spatial and temporal patterns or heterogeneities in gene expression within individual cells or complex tissues. Reduction of costs for analytes and sample was achieved by Perez-Toralla et al.82 They established a protocol that can entirely be performed in the liquid phase. This protocol permits the capture and chemical fixation of cells, followed by quantitative characterization with FISH. The target was the ERBB2 gene which is used as a biomarker for the monitoring of HER2+ breast cancer progression. They conclude that their system provides the necessary robustness for fully automated use in clinical settings with a 10-fold reduction of sample and analyte consumption and furthermore decreases the detection time by a factor of 2.
Moffitt et al. invented a new method for multiplexed error-robust fluorescence in situ hybridization (MERFISH) with a 100-fold higher throughput than traditional methods.64 They incorporated chemical cleavage sites to remove previously bound probes for subsequent measurement of multiple targets. This enabled them to quantify 130 different RNAs in several tenths of thousands of cells in 24 h (see Figure 2B).
Fluorescent Super-Resolution Microscopy
Super-resolution microscopy (SRM) has lately enabled optical imaging with resolutions down to ten nanometers. Several SRM techniques have been reported and produced spectacular two- and three-dimensional images of subcellular components, from individual biomolecules to entire organelles.62,83 SRM can be useful in single cell studies to localize target molecules in the cell. In particular, the recent approaches for multiplexed analysis in SRM are of interest for single cell studies.
Stimulated emission depletion microscopy (STED) reduces the illuminated volume with a depletion laser in order to achieve high resolution, and it permits acquisition of images with very high temporal resolution of up to 5 ms.84 The method suffers from high light exposure of the sample but has multiplexing capabilities as shown by Jungmann et al.85 They employed a sequential labeling and image acquisition protocol (Exchange-PAINT) and were able to achieve resolutions below 10 nm. Belousov and co-workers presented a live-cell STED microscopy method for applications with dynamic biosensors in single cells.86 Using the fluorescent H2O2 sensor HyPer2 for SRM imaging, they were able to image filaments and locally quantify H2O2 production in living cells as well as differences in the cell population. A strength of STED is that it can be combined with many advanced imaging techniques like fluorescence correlation spectroscopy87 or fluorescent lifetime imaging (FLIM), as presented by Niehörster et al.88 With spectrally resolved FLIM on a STED device, they were able to visualize up to nine targets simultaneously in mouse cells.
In contrast to STED, stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM) utilize photoswitchable probes to determine the centroid position of each fluorescent label. Dudok et al. applied STORM imaging to determine the exact location and amount of signaling molecules and pathways in single neurons.89 They were able to monitor nanoscale organization of cannabinoid signaling and could reveal the extent and time course of molecular changes induced by different doses of chronic THC treatment.
Recent Advances in Fluorescent Labels
The choice of fluorescent tags and the optimization of labeling protocols are critical for the quality of the analytical results. In particular, the measurement of intracellular molecules in living cells that are not genetically modified requires labels or sensor molecules that can permeate the cell membrane and are ideally not harmful to the cell. For dyes that are not taken up efficiently into intact cells, Henning et al. suggested the use of a glass nanopipette with an inner diameter of only 100 nm which is much smaller than microinjection needles.90 With the help of this pipet, they were able to deliver functionalized fluorescent probes directly into individual cells before analysis. Although it was labor intensive and had a low throughput, they successfully validated their method with a number of different dyes.
Chemosensors for Intracellular Measurements
Many research projects aim at the sensitive detection of toxic chemicals such as heavy metal ions to understand their effect on human health or the environment. With the intention to quantify Zn2+ ions in living cells, Wang et al. have synthesized a chemosensor with a high cell permeability and low toxicity that allowed for the detection of Zn2+ in single cells down to a detection limit of less than 100 nM.91 Moreover, a sensor for the sequential fluorescent detection of Cu2+ and S2− based on a fluorescein derivative was described by Meng et al.92 In aqueous solution, Cu2+ binding leads to quenching of the chemosensor, whereas the presence of S2− results in an increase in the fluorescence intensity. The biocompatibility of the dye for copper and sulfide monitoring was verified with MDA-MB-231 breast carcinoma cells. Another molecular probe for Cu2+ and pH was reported by Han et al.93 They developed a single fluorescent probe, capable of measuring pH, Cu2+, and pH/Cu2+ with different ratiometric fluorescent signals. The probe is not sensitive to other analytes like reactive oxygen species, copper containing proteins, nucleic acids, and other metal ions and presents good cell permeability and biocompatibility as tested with HeLa cells. In addition, toxic metal ions like Hg2+ and Cd2+ can be detected intracellularly using a probe that was reported by Maity et al.94 Apart from the detection of metal ions, a fluorescent probe for CO2 in single living cells was presented by Chen et al.95 The dye emits fluorescence upon aggregation caused by CO2 and can be used to monitor intracellular CO2 levels dynamically. The dye has shown high sensitivity and biocompatibility and was tested for quantitative detection of CO2 generated in single living MCF-7 and MEF cells.
Quantum Dots and Carbon Dots
Quantum dots (QDs) have become an alternative to dye based systems as they provide high photostability and their luminescence spectra are tunable by the particle size. Initially, QDs have been simple colloidal nanoparticles made from semiconductors like CdSe, CdS, CdTe, ZnS, and PbSe. Today, the luminescent semiconductor QDs are encapsulated within a polymeric shell, combined with various surface coatings, and widely commercially available. Being biocompatible and photostable and having a high quantum yield makes them suitable for applications in single-cell analysis.96,97 Recently, Ren et al. used multicolor QDs to study the functions of epidermal growth factors. Thereby, they also investigated the impact of QD labeling on the cell vitality.98 They found that the Quantification was independent of the color/size of QDs they used and that QD labeling did not impair cell vitality 24 h after labeling. This proves that QDs are suitable for a broad range of applications in single and multiplexed studies of single cells.
Blinking is a physical phenomenon that is frequently seen with QDs, and many applications are adversely affected by this effect. Although the underlying mechanism is still not fully understood, a new kind of QDs that avoids this effect has recently been introduced and there is still a lot of progress in this field.99 These particles are called carbon quantum dots or simply carbon dots (c-dots). c-dots are small carbon nanoparticles coated with organic molecules or biomolecules and exhibit superior aqueous solubility and chemical stability compared to classical QDs. Several reviews explain the current status of this technique in detail,100,101 and Jiang et al. showed that these c-dots can be prepared in various colors and successfully used to label MCF-7 cells for quantitative optical analysis.102
Immunoassays and Related Methods with Optical Read-Out
Fluorescent probes can directly bind to the target molecules via chemical interaction, but this often lacks specificity. Alternatively, fluorescently tagged antibody labels can be used to bind to secreted biomolecules or targets from a lysed cell. When ultrasensitive detection is needed, amplification strategies can be employed to enhance the signal.103 Enzymatic labels acting as biocatalyzers are ideally suited for such applications as they can be attached to the antibody and amplify the signal by catalyzing biochemical reactions with fluorescent products.104,105
Enzyme-Linked Immunosorbent Assays (ELISAs)
ELISAs are among the most sensitive techniques to precisely detect molecules in a complex matrix. Together with thoroughly conducted calibration measurements, they enable Quantification of the target molecules in subnanomolar concentrations. Eyer et al. have used ELISA in individual subnanoliter microchambers to analyze intracellular levels of GADPH in single HEK293 cells.106 They successfully demonstrated detection levels of less than one attomole per chamber and were able to determine the response of lutropin on murine leydig tumor cells. Their system was further miniaturized by Stratz et al. to capture and analyze single bacteria.107 This enabled quantification of β-galactosidase levels down to a few hundred enzymes in single E. coli cells dependent on the cell culture media used. Li et al. recently used a nicking enzyme and substituted the antibody with an aptamer in an assay to detect membrane proteins from single living cells.108 Herein, selective binding of the aptamer results in changes in its conformation and ultimately initiates enzymatic catalysis. As the conversion to the fluorescent product occurs only upon binding, this eliminates the need for washing steps as compared to other enzymatic techniques or fluorescent probes. This allowed them to use their system in microdroplets and analyze single cells sequentially with cytometric methods.
A related immunoassay technique, referred to as immunospot (ELISPOT, enzyme linked immunospot assay), was applied in several studies. Thereby, single cells are seeded in a microwell format and secretion of molecules such as cytokines can be investigated. As the wells are coated with specific antibodies, the secreted cytokines are directly bound to the well plate surface at the location of their release. Subsequent enzymatic labeling results in conversion of a precipitating substrate to a colored product, and this corresponds to the cytokine expression of an individual cell.109–111 Saletti et al. recently published a protocol for a modified ELISPOT that is capable of detecting virtually any vaccine antigen after magnetic enrichment of circulating plasmablasts from blood samples.112
To further increase the sensitivity of ELISA, Giri et al. have reported a new approach, where the product of an enzymatically catalyzed reaction is concentrated by applying an electric field.103 Their microfluidic platform was able to increase the detection limit of a TNF-α ELISA assay by a factor of 60 and could potentially be used with many other assays with charged products as well.
Several research groups worked toward multiplexing of immunoassays by combining spatial and spectral separation for multiparametric analysis of single cells in their microfluidic immunosensing platforms. Multiplexing with only one single fluorophore by spatial separation for the analysis of up to ten proteins in parallel was shown by Ramirez et al.113 Lu et al. have described a single cell platform for secretion analysis of 5044 cells in parallel.65 They succeeded in measuring up to 42 different proteins in parallel on the single cell level with a combination of spatial and spectral separation of individual assays (Figure 2C). They entrapped single cells in microwells and covered the wells with a barcode array slide. Thereby, they connect every single cell chamber with 15 different locally separated spots for protein analysis. To enhance the multiplexing capabilities even further, three different assays were executed on each spot. Labeling with three different fluorophores enabled them to then measure 42 protein targets and three control spots simultaneously at the single cell level.
Spatial separation can be done not only by using the surface of a microfluidic chip to separate different spots for multiparametric measurement but also by using affinity beads together with single cell isolation methods. Junkin et al. used four independent beads brought in proximity to the cell to measure the secreted cytokine levels from single macrophages.114 Instead of measuring different parameters, they measured the same parameter multiple times and correlated the change in the fluorescent signal on the microbead surfaces with the dynamic single cell secretory activity.
A separation technique that uses immunolabeling for detection is Western blotting. Hughes et al. have transferred this macroscopic technique to the single cell level with a microfabricated polyacrylamide gel (Figure 2D).66 They seeded single cells into 6720 microwells and then performed capillary electrophoresis upon cell lysis. After electrophoretic separation, the proteins were photo-cross-linked and fixed at their current location. After that, they were stained for analysis. Detection limits of <30 000 molecules were achieved, and separation of up to 11 different proteins from one cell is possible. Lately, a method to modulate the pore size of the gel matrix used for electrophoretic separation was developed by Duncombe et al.115 This allowed them to analyze proteins with largely varying sizes from 25 to 289 kDa on the same chip.
Label-Free Optical Methods
Raman Spectroscopy
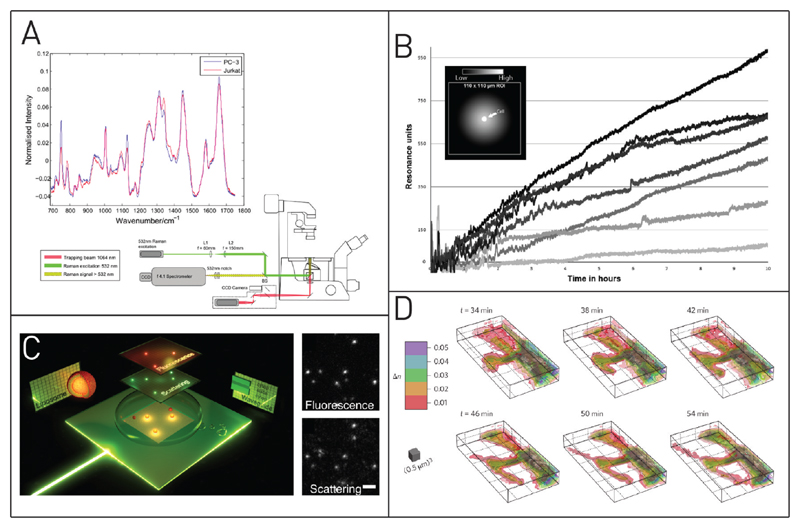
A number of optical microcopy techniques are available to probe single cells specifically, without the need for fluorescent labels or staining. Raman spectroscopy is one of these techniques that was recently used to perform analysis on the single-cell level.116,117 Casabella et al. reported an automated microfluidic platform for single-cell Raman spectroscopy (Figure 3A).118 The first step was the realization of an alternating flow in a simple microfluidic channel. Mounted on an inverted microscope, cells could be trapped by an optical tweezer during the intervals of low flow and were successfully removed once the fluid flow was increased. During each capture period, a Raman spectrum was measured for the individual cell. With this setup, they could enhance the throughput and reduce the manual work of the single-cell Raman measurements and detect differences in the Raman signals of PC3 and Jurkat cells. Kang et al. used Raman spectroscopy in combination with fluorescence microscopy to probe drug delivery dynamics in single cells.119 They functionalized gold nanoparticles with doxorubicin via pH-sensitive hydrazine linkers and monitored the pH-responsive doxorubicin release upon encapsulation in the acidic environment of lysosomes. Additionally, Raman spectroscopy has been employed for single cell sorting.120
Figure 3. Label-free optical analysis methods for single cells.
(A) Raman spectroscopy on the single-cell level discriminates live epithelial prostate cells and lymphocytes. Adapted with permission from Casabella, S.; Scully, P.; Goddard, N.; Gardner, P. Analyst 2016, 141, 689–696 (ref 118). Published by The Royal Society of Chemistry. (B) Single-cell secretion of anti-EpCAM antibodies quantified by surface plasmon resonance. The slopes of the curves represent the differences in the production rate of the individual cells. Adapted with permission from Stojanović I.; Van Der Velden, T. J. G.; Mulder, H. W.; Schasfoort, R. B. M.; Terstappen, L. W. M. M. Anal. Biochem. 2015, 485, 112–118 (ref 121). Copyright 2015 Elsevier. (C) Evanescent light scattering microscope for detection of fluorescent and label-free particles. Adapted from Agnarsson, B.; Lundgren, A.; Gunnarsson, A.; Rabe, M.; Kunze, A.; Mapar, M.; Simonsson, L.; Bally, M.; Zhdanov, V. P.; Höök, F. ACS Nano 2015, 9, 11849–11862 (ref 124). Copyright 2015 American Chemical Society. (D) Time-lapsed 3D live-cell tomography showing the refractive index change during filopodia formation of a neuronal spine. Adapted with permission from Cotte, Y.; Toy, F.; Jourdain, P.; Pavillon, N.; Boss, D.; Magistretti, P.; Marquet, P.; Depeursinge, C. Nat. Photonics 2013, 7, 113–117 (ref 125). Copyright 2013 Nature Publishing Group.
Surface Plasmon Resonance
To elucidate the binding/dissociation constant of two molecules, surface plasmon resonance (SPR) imaging is frequently applied. However, it may also find applications for single-cell analysis as shown by Stojanović et al.121 They used SPR to detect and quantify the secreted antibodies of individual hybridoma cells. The cells produced monoclonal antibodies against the epithelial cell adhesion molecule (EpCAM) that was preimmobilized on the SPR sensor surface. The antibody production of single cells was successfully measured, and excretion levels between 0.02 and 1.19 pg h−1 were determined (see Figure 3B).
Interferometric Scattering Microscopy
Interferometric scattering microscopy (iSCAT) is another nonfluorescent optical microscopy technique. Thereby, light is scattered by an object leading to a change in the detected light intensity based on interference with a reference light field (Figure 3C). iSCAT is mainly used in biochemical applications to visualize nanodomains on cell or lipid membranes or to track the movement of molecular motors.122,123
Agnarsson et al. have presented a new form of light scattering microscopy based on evanescent fields to measure the binding kinetics of single cells to a given surface.124 With their method, they were able to visualize the attachment process of single platelets and detect differences in the binding between cells and silica surfaces and could divide the binding process into several independent steps, from the first contact to complete resting.
Live-Cell Tomography
A new label-less super-resolution microscopy technique called live-cell tomography was recently presented by Cotte et al. from the EPFL Lausanne.125 They developed a microscopic method that uses phase contrast in unlabeled single cells for live-cell 3D imaging with resolutions below 100 nm. The capabilities of their system were tested with long-time neuronal observations for synaptic remodeling in 3D as well as for monitoring of individual bacteria cells (Figure 3D).
Electrochemical Analysis and Related Methods
For a couple of decades, electrochemical methods with microelectrodes have been employed for electrical measurements and stimulation of neurons and neurotransmitter secretion, but the technological advances in recent years enabled the use for other applications in the field of single cells.126,127 Electrochemistry is ideally suited for miniaturization as electrodes for the signal acquisition, and analysis can be integrated on a miniaturized platform. Furthermore, it broadens the choice of materials as there is no need for optical transparency. Microelectrodes can easily be fabricated on various substrates like polymers, silicon, or glass. They are sensitive toward a wide range of electrochemically active molecules and are particularly useful for the investigation of neuronal cells and networks by measuring neurotransmitter release. In this context, it is currently the only method to measure neuronal communication in situ quantitatively. The high sensitivity of the technique allows one to investigate influences of external parameters like drugs on exocytosis. One focus of current research is the fabrication of ultrasmall electrodes to enable measurement of neurotransmitter release of single vesicles from living cells. Liu et al. have successfully fabricated Au nanoelectrodes with only 6 nm in diameter and revealed the dopamine release of rat pheochromocytoma cells with high spatial resolution.128 Carbon fiber microelectrodes were used for transmitter release monitoring from single vesicles of individual cells as well.129 The disadvantage of this method is that the electrodes have to be punched into the cell. This procedure is difficult, requires a lot of experience, and is very low in throughput. An improved geometry was introduced by Robinson et al.130 They fabricated vertical nanowire-electrode arrays on silicon substrates with a silicon dioxide isolation, and they could track the response of multiple interconnected neurons on the grid (Figure 4A). Seeding of cells on these arrays enabled them to get a better insight into neuronal data storage and information processing. They used patch clamping to prove that the nanoelectrodes had no influence on the cell behavior and finally employed rat cortical neurons and mapped multiple synaptic connections in parallel. With amperometric measurements of neurotransmitter release, Li et al. conducted time-resolved quantitative measurements of catecholamine transmitters in PC12 cells.131 They found that 2 μM cisplatin treatment enhances the frequency of exocytotic events but reduces their extent. On the contrary, 100 μM cisplatin lowered the frequency of exocytotic catecholamine release and increased the duration. Furthermore, the cisplatin treatment affected the fusion of the pore that is formed in the lipid membrane during exocytosis.
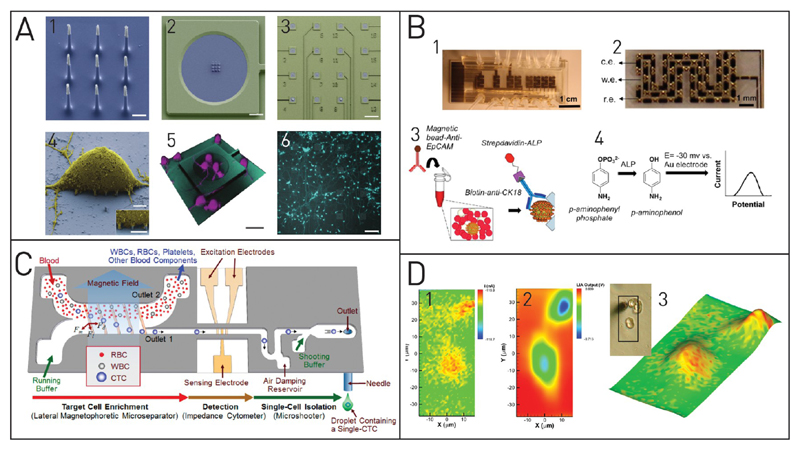
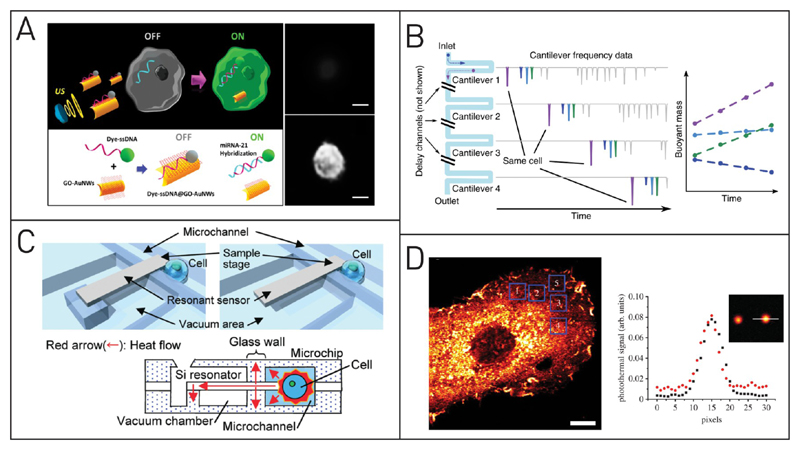
Figure 4. Electrochemical single-cell analysis techniques.
(A) Nanowires (1: SEM image, scale bar 1 μm) can penetrate single cells for electrical measurements. Sixteen individual measurement arrays are placed on one chip (2 and 3, scale bars 10 and 120 μm). Images 4–6: SEM image of rat cortical cell on the vertical electrode array, confocal reconstruction, and top view on calcein AM stained cells. Adapted with permission from Robinson, J. T.; Jorgolli, M.; Shalek, A. K.; Yoon, M.-H.; Gertner, R. S.; Park, H. Nat. Nanotechnol. 2012, 7, 180–184 (ref 130). Copyright 2012 Nature Publishing Group. (B) A microfluidic chip with eight independent sensors (1) comprising of X-shaped posts (2) and on-chip electrodes are used to capture cancer cells from a given sample. Dielectrophoretic cell capture is followed by cell labeling (3) and electrochemical detection (4). Adapted from Safaei, T. S.; Mohamadi, R. M.; Sargent, E. H.; Kelley, S. O. ACS Appl. Mater. Interfaces 2015, 7, 14165–14169 (ref 136). Copyright 2015 Americal Chemical Society. (C) Impedance spectroscopy is used in this microfluidic platform to detect single CTCs after magnetic separation. If a cell is detected, external processing evokes an actuation of the microshooter to print this cell onto a microtiter plate for further analysis. Adapted from Kim, J.; Cho, H.; Han, S.-I.; Han, K.-H. Anal. Chem. 2016, 88, 4857–4863 (ref 141). Copyright 2016 American Chemical Society. (D) Scanning electrochemical microscopy images of PC12 cells. To generate the images, a microelectrode is scanned over the sample and the amperometric current and the impedance signals are measured. Analysis of the topography (1) and oxygen consumption (2) of the cell can be achieved at the same time (3). Adapted from Koch, J. A.; Baur, M. B.; Woodall, E. L.; Baur, J. E. Anal. Chem. 2012, 84, 9537–9543 (ref 142). Copyright 2012 American Chemical Society.
Optical lithography is rarely used for the realization of these nanoelectrodes, merely because the use of light in standard lithographic processes cannot provide resolution of submicrometer structures with high aspect ratios. Wigström et al. nonetheless employed this technique and were able to realize a flexible thin film microelectrode array (MEA) probe with 16 platinum band electrodes to record single cell exocytosis release of bovine chromaffin cells.132 The exocytosis events were simultaneously detected by several electrodes. Thus, the two-dimensional localization of neurotransmitter release was possible.
Another common parameter for analysis with electrochemical methods on the single cell level is oxidative stress. Ions such as free radicals and peroxides that are released upon oxidative stress can be detected and investigated on a platinum electrode. Although several different molecular targets can be determined with the same setup, the method is limited with respect to the selectivity. Detection of reactive oxygen species (ROS) in single cells was recently presented by Jeffrey E. Dick.133 He used a macroscopic setup and detected the amperometric signal upon collision of single cells with a measurement electrode under the presence of surfactants. He detected a thousand-fold difference between the electrochemical responses of acute lymphoblastic lymphoma T-cells and healthy thymocytes. Major concerns here are the effect on cell behavior induced by the surfactant and the large drift of the measurement electrode due to adsorption of debris and surfactant. Sadeghian et al. monitored the superoxide release from skeletal muscle cells.134 Their electrochemical biosensor used thick film nanoporous gold to increase the sensitivity 14-fold in comparison to non-nanoporous electrodes, and they found a 1.90 nA nM−1 cm−2 limit of detection. They applied the system for the time-resolved ROS secretion measurement of mouse myoblast C2C12 cells upon drug stimulation, and they validated their system with fluorescent methods. In a study from Piskounova and co-workers, they revealed that oxidative stress inhibits distant metastasis in a study conducted on human melanoma cells.135
Enzymatic Assays with Electrochemical Read-out
Although electrochemical analysis can be very sensitive, an amplification step using enzymatic labels can be introduced to improve the detection limit further and get faster and more reliable signals. A good example of such a system is the platform presented by Safaei et al.136 They used a microfluidic chip to capture CTCs with magnetic labeling methods (Figure 4B). After having successfully immobilized single tumor cells, they used amperometric detection in a three-electrode setup. Therefore, they enzymatically labeled the cells with alkaline phosphatase. This enzyme catalyzes the reaction of p-aminophenyl phosphate to an electrochemically active reagent p-aminophenol that was subsequently measured. The number of captured CTCs was optically verified for cross-validation of their technique, and they proved that the platform was reliably detecting single CTCs. An aptamer biosensor for cell surface N-glycan evaluation with an integrated enzymatic amplification strategy was presented by Chen et al.137 As a proof-of-concept for their system, the detection of human acute lymphoblastic leukemia cells with respect to the presence of surface N-glycan was established. The biosensor was used for dynamic evaluation of cell surface N-glycan expression with a detection limit of 10 cells mL−1 in their open system. Decreasing the sample volume with a microfluidic platform might further enable this system to measure N-glycan on the single cell level.
Impedance Spectroscopy
In the previously mentioned applications, amperometric detection or measurement of the resistance was used to reveal information on the investigated sample. At a closer look, the resistance of a system is not a fixed value but is dependent on parameters like the frequency and phase of the input signal. Electrochemical impedance spectroscopy (EIS) describes the measure of this complex resistance and is nowadays frequently applied for the analysis of type, size, and dielectric properties in single cells without the need for labeling.138 There are two different ways to employ EIS on the single cell level, analysis of immobilized single or only a few cells and cytometric flow-through analysis. The former is suitable for real time investigations and focuses on a limited number of cells to be dynamically investigated. As an example, Zhu et al. have built a microfluidic platform in order to trap and monitor up to ten individual yeast cells.139 A standard Pt electrode was employed for stimulation of the cells, and ten individual recording electrodes for simultaneous measurements were included as well. They used the EIS signal to monitor the budding of yeast cells at a temporal resolution of 1 min and could differentiate between bud growth and cell motion. With fission yeast, they used an EIS system to monitor the cell cycle with 250 nm resolution for the cell length at a 5 min temporal resolution.140
Guo and Zhu used DEP to trap and electroporate individual cells in a microfluidic chip.143 Additional incorporation of quadrupole electrode units around each DEP cell trap allowed them to maneuver individual cells to predefined traps. Successively, they conducted a real-time investigation of electroporation with impedance monitoring and were able to perform transfection of HeLa cells with GFP-encoding vector DNA.
Highly conductive metal nanoparticles are used in many industrial applications, and the toxicity of nanoparticles got into the public focus. In order to improve the toxicity test for nanoparticles that is traditionally done on well-plate format, Shah et al. developed an electrochemical on-chip protocol.144 They implemented electrodes for electrochemical analysis and DEP single cell capture. After exposure to CuO or TiO2 nanoparticles, they monitored the vitality of single PC12 cells for 6 h with EIS. The second large application field for EIS is flow-through systems. Similar to other cytometric devices, single cells are flushed through a microchannel, thereby individually passing multiple microelectrodes. The electrolyte in which the cells are suspended (medium or PBS) allows electrical current to pass from one electrode to the other. When a cell passes by, it induces a change of the electrical signal and this can reveal the dielectric properties of the individual cells. In comparison with the time-resolved measurements achieved with local entrapment, the flow through EIS systems are capable of much higher throughput. Haandbæk et al. presented a resonance-enhanced EIS spectrometer for single bacteria cell detection with a speed of up to 500 MHz.145 Although such high throughput is rarely achieved, many systems are capable of detecting single cells fast and reliably. Unfortunately, simultaneous single-cell analysis at these speeds is still not possible. Therefore, the possibility to cross-validate the system with optical means is incorporated in some systems. Frankowski et al. have utilized a new gold electroplating technique to realize electrodes with 22 μm high posts.146 These electrode posts function similarly to the conventional planar electrodes used in many EIS devices, but they can be moved out of the optical axes. Simultaneous optical and electrochemical impedance monitoring was conducted to analyze leukocytes and discriminate between different subpopulations (lymphocytes, monocytes, and two subpopulations of granulocytes). In contrast to standard immunohistochemical staining, which is selective to certain markers, one has to consider here inhomogeneities in size or shape of the cells which limits quantitative read-out. An interesting application of EIS in a flow-through system was presented by Kim et al.141 They labeled CTCs in whole blood samples with commercially available magnetic nanobeads (Figure 4C). On their microfluidic platform, magnetic separation of the magnetically labeled cells from whole blood was done. The CTCs were directly counted as they passed the EIS on their way toward the outlet. Real-time analysis of the EIS signal was then used to trigger printing of the single CTC cells from the microfluidic platform through a microneedle. Furthermore, the EIS signal served for automated control of a xyz-stage that moves a microwell plate for dispensing a single cell per microwell. Although throughput and capture efficiency of the CTCs have to be improved, this miniaturized system nicely shows the strength of integrated single cell sorting and analysis and even allows for downstream single-cell analysis or culturing on a well plate format.
Alberti et al. presented a microfluidic platform for impedance measurements on single cells using a setup that resembles patch clamping.147 They fabricated microchannels that incorporated a microhole to probe the electrical impedance of a single cell that was trapped above the hole. The capacitance of HeLa cells was measured under physiological conditions to be 38 pF. Unfortunately, the system influences the behavior of the cells. Cells that were attached to the microhole did not survive in a 24 h time course while nonattached cells remained vital.
Scanning Electrochemical Microscopy
When an electrode is not integrated on the chip but is freely movable, it can theoretically be employed for 2D scanning of electrochemical properties of the sample underneath. In scanning electrochemical microscopy (SECM), this is used to map topology and chemical compounds on surfaces or biological matter.
Koch et al. used SECM with fast-scan cyclic voltammetry (CV) to simultaneously measure impedance and amperometric current of PC12 cells.142 The amperometric signal was converted to a topographic image as the current density is dependent on the distance between the tip and the cells (Figure 4D). CV was used to reveal information about the spatial distribution of the oxygen consumption with a minimal resolution of 500 nm. With a comparable setup, Zhang et al. measured cisplatin-induced permeation on single human T24 cells and measured the topography simultaneously.148 With these methods, it is possible to understand the response of individual cells to exogenous stimuli on both a morphological and a secretomic level. Here, the change in the impedance upon variations in membrane permeation is quite high.149 Quantification of individual species with SCEM is still difficult. SECM was nonetheless successfully applied for the time-resolved measurement of redox mediators across a cell monolayer. Berger et al. investigated rat epithelial cells that were grown to confluence on permeable membrane filters.150 This facilitated the study of redox mediator transportation through the cellular barrier. They switched the measurement potential to detect different mediators with the same electrode instead of using a setup with multiple electrodes. Para- and trans-cellular transport mechanisms could then be distinguished on the basis of the time it took between the mediator’s insertion below the membrane and its arrival at the electrode.
Mass Spectrometric Analysis
Mass spectrometry (MS) generates high-dimensional data by ionization of a chemical or biological sample and subsequent determination of the mass to charge ratios of the ionized molecules. MS instruments are applied in single-cell analysis for many years as their ability to differentiate hundreds of biomolecules from a single specimen without the need for labeling renders MS extremely attractive. In single-cell analysis, MS still suffers from the fact that unlabeled detection is possible only for highly abundant cellular targets and quantification needs internal references. This is the reason why many studies focus on highly abundant metabolites or investigate large cellular species.151
Single cell MS techniques can either profile or image the sample. The profiling techniques have a limited spatial resolution, and only one spectrum is generated from an individual cell. Although this does not allow for investigation of subcellular structures, individual profiling of adjacent cells or spots reveals important information on the heterogeneities in a given set of cells.152 Other MS techniques have high spatial resolutions that allow label-free imaging. Recently, the spatial resolution of many MS methods has improved and even three-dimensional image acquisition with MS has become possible. A special approach for mass spectrometry based single-cell analysis is mass cytometry. It employs rare earth metal tags as labels to improve the systems sensitivity.153
Electrospray Ionization MS
Electrospray ionization MS (ESI-MS) allows one to investigate small sample volumes. However, the lysate of a single cell must be supplied to the instrument. In a recent approach, Fujii et al. used micro-capillaries to extract sample from plant cells by punching the cell with a microneedle and analysis with ESI-MS.154 Zhang et al. investigated single plant epidermal cells by the withdrawal of the intracellular solution and found 23 metabolites and lipids in single A. thaliana epidermal cells.155 They were able to discriminate different cell types and compare their metabolic differences. Gong et al. used capillary microsampling to extract and analyze the cellular stress of healthy and wounded plant leave cells.156 They found clear differences in the levels of abscisic acid and compared their results with other MS methods. They concluded that capillary microsampling can be successfully used to monitor single living cells. Nonetheless, microneedles may hamper normal cellular functions, as it is very likely that the cells react to these needles as well as to other wounds (Figure 5A).
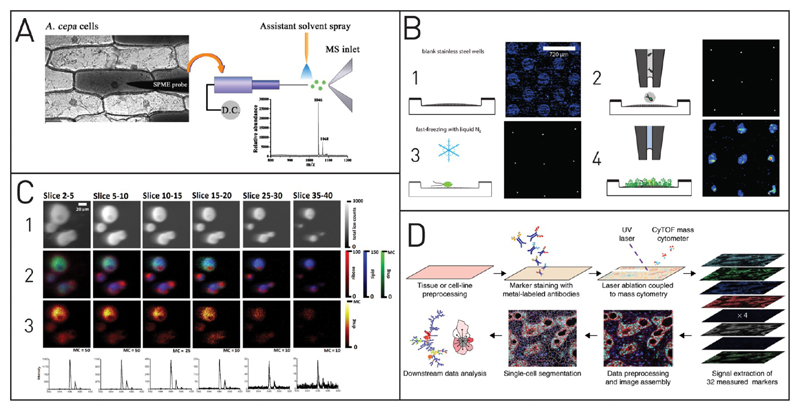
Figure 5. Mass spectrometry for single-cell analysis.
(A) Cytosol analysis by ESI-MS. A tiny microcapillary withdraws part of the cytosol and transfers it to the MS, where it is ionized and analyzed. Reprinted from Gong, X.; Zhao, Y.; Cai, S.; Fu, S.; Yang, C.; Zhang, S.; Zhang, X. Anal. Chem. 2014, 86, 3809–3816 (ref 156). Copyright 2014 American Chemical Society. (B) MALDI-MS platform for investigations of single cells that were spotted into microwells. Adapted with permission from Krismer, J.; Sobek, J.; Steinoff, R. F.; Fagerer, S. R.; Pabst, M.; Zenobi, R. Appl. Environ. Microbiol. 2015, 81, 5546–5551 (ref 159). Copyright 2015 American Society for Microbiology. (C) Label-free 3D-TOF-SIMS measurement of amiodarone-doped macrophages at different sputter depths. Many different molecules can be visualized by selecting the corresponding m/z ratio (1–3). The different slice numbers represent the sputtered z-stacks for the 3D imaging. Adapted from Passarelli, M. K.; Newman, C. F.; Marshall, P. S.; West, A.; Gilmore, I. S.; Bunch, J.; Alexander, M. R.; Dollery, C. T. Anal. Chem. 2015, 87, 6696–6702 (ref 160). Copyright 2015 American Chemical Society. (D) Mass cytometry achieves high sensitivities by employing rare earth metal tags. The isotopically pure tags allow simultaneous detection of more than 40 different targets. Besides cytometers, imaging systems based on this approach have been developed as well. Reprinted with permission from Giesen, C.; Wang, H. A. O.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schuffler, P. J.; Grolimund, D.; Buhmann, J. M.; Brandt, S.; Varga, Z.; Wild, P. J.; Günther, D.; Bodenmiller, B. Nat. Methods 2014, 11, 417–422 (ref 161). Copyright 2014 Nature Publishing Group.
Fujita et al. studied single cell secretomes after the cells were isolated on a microwell format, and an oil layer was used to prevent the liquid from evaporation.157 After separation, they independently measured the cells secretome by retrieval of small volumes from the surrounding medium with the help of a microcapillary. They detected 154 different metabolites in the individual extracellular fluid of undifferentiated and differentiated PC12 cells. Gasilova et al. have introduced another technique for measurement of secreted proteins from individual cells.158 Their approach uses droplet microfluidics to generate a continuous stream of discrete water plugs in an oil phase. At one particular point of the microchannel, a small opening (a “spyhole”) to the surrounding environment is realized. The droplets are passing underneath, and surface tension prevents them from leaking out of the system. When an electrode is located under the spyhole, application of high voltages results in the electrostatic release of spray from the passing aqueous droplets. The electrospray is then transferred to the MS device and analyzed while the remaining droplets can be further processed. This technique facilitates the combination of droplet based cell isolation and manipulation techniques with ESI-MS and could potentially also be used in single-cell analysis.
Laser Desorption/Ionization MS
MALDI-MS is a soft ionization technique that utilizes the power of a laser to desorb and ionize sample molecules which are embedded in matrix crystals. Here, the majority of molecules is not fragmented but maintain their original size and weight. In MALDI, the matrix assists desorption, ablation, and ionization of the sample by a pulsed laser.
Several groups have employed MALDI-MS in single-cell analysis over the last few years and improved the spatial resolution and the interference of the matrix material with the measurement decreased. Krismer et al. used MALDI to screen different strains of Chlamydomonas reinhardtii. They used a stainless steel plate with embedded microwells to spatially separate individually spotted cells. After fast-freezing and matrix deposition, mass spectra of single cells were obtained. On the basis of the MS results, the native strain with two different types of chlorophyll could be differentiated from a mutant strain that was lacking one of these chlorophyll subtypes (see Figure 5B).159 Instead of localizing single cells in a well format, Ong et al. combined MALDI with optical imaging. At first, they imaged the sample and identified single cells. In a second step, they performed MALDI experiments at these locations.162 With a lateral resolution of ten micrometers, this technique enabled the acquisition of single cell spectra and the determination of the peptide content of different cells. Moreover, they detected cellular subtypes and identified rare cells with non-normal peptide patterns. A similar system was developed by Jansson et al. to investigate the heterogeneity of peptide composition in rat islets of Langerhans.163 Another strength of MALDI is the possibility to combine it with droplet microfluidic techniques as it has been recently reported by Küster et al.164 but not yet applied for single-cell analysis.
The application of the matrix in MALDI provides a tool to enable soft ionization of large molecules. Still, the investigation of living cells or dynamic processes is almost impossible. Lee et al. have tackled this problem with a new method referred to as laser desorption/ionization droplet delivery mass spectrometry (LDIDD-MS).165 Here, a pulsed UV laser is focused on a surface covered with target cells to trigger desorption and ionization of molecules at ambient conditions. Simultaneously, a spray of liquid droplets is focused onto the illuminated spot to capture the ionized analytes and transport them to the MS instrument. The high sensitivity and temporal resolution of the system allowed them to perform multiple MS measurements in a sequence and perform time-resolved MS on single cell level. Another approach to conduct dynamic single-cell analysis with MS methods was presented by Lee at al.166 They placed single neurons in a capillary and investigated the neuronal secretome before and after stimulation with KCl.
Apart from dynamic measurements, laser desorption/ionization without the use of any matrix has been the focus of research for several years. Recently, so-called desorption/ionization from nanoporous silicon (DIOS) and silicon nanopost arrays (NAPAs) were presented with ultrahigh sensitivities, but applications on the single cell level are rare. In 2012, Walker et al. were able to detect down to 800 zeptomoles of verapamil in artificial samples and 24 biochemicals in single yeast cells.167 For example, Stopka et al. have reported a NAPA-LDI-MS setup for analysis of mouse kidney slices and single human hepatocarcinoma cells, but their analysis suffered from a low lateral resolution of 40 μm.168
Secondary Ion Mass Spectroscopy (SIMS)
SIMS is a technique to analyze the composition of solid surfaces and thin films. In SIMS, the surface is sputtered with an ion beam and the release of secondary ions is then investigated by MS. Its main advantages are the low detection limit and the high spatial resolution, but the sample has to be placed in an ultrahigh vacuum for analysis, which inhibits the investigation of living organisms. Bobrowska et al. have published a detailed description of the necessary steps for pretreatment of biological samples, with emphasis on single cells.169 This includes fixation, washing, and dehydration before insertion in the SIMS device.
In addition, time-of-flight SIMS (TOF-SIMS) has frequently been applied for analysis of single cells. Robinson et al. recently used it to compare different breast cancer cell lines and found distinct differences like an 18-carbon chain fatty acid that was only present in the BT-474 cell line.170 Vanbellingen et al. have proven that delayed extraction can be an efficient solution to extend the mass resolution at a high spatial resolution of 400 nm.171 This improvement could turn out to be very useful when lipids on tissue sections or rare samples are investigated. Additionally, numerous groups have reported studies conducted with 3D TOF-SIMS. Thereby, SIMS is used not only for the generation of ions but also for sputtering the sample. This enables the user to take several MS images in different heights and create a z-scan of the sample. Passarelli et al. employed this technique for visualization of pharmaceutical compounds and metabolites in single cells.160 They treated macrophages with therapeutic dosages of amiodarone and quantified its amount and the distribution of the cells (Figure 5C). From their spatial analysis, they followed that amiodarone was preferentially located in the cell membrane and subsurface regions and it was completely absent in the nucleus. Graham et al. used 3D-TOF-SIMS to localize polymer nanoparticles in individual HeLa cells.172 They compared the TOF-SIMS results with optical images of cells incubated with similar polymer nanoparticles that were fluorescently labeled. They found nanoparticle clusters and correlated this with endosomal encapsulation. The current resolution limit of 3D-TOF-SIMS is 250 nm, and the technique is able to detect multiple unlabeled targets in the cell simultaneously.173 Angelo et al. used SIMS with isotope labels to increase the sensitivity of the method even further.174 They referred to their method as multiplexed ion bead imaging (MIBI) and successfully imaged up to 100 different targets on fixed tissue slices with a comparably low resolution of five micrometers.
Liquid- or Gas-Chromatography Coupled MS
Combinations of MS with liquid or gas chromatography enable improved separation and identification of compounds in single cells. Onjiko et al. used LC-MS for molecular identification in single cells from the South African clawed frog.175 They reported that they were able to identify 40 different metabolites that were related to the central metabolic network. They could also track the differences in activities between different cell types in the unperturbed embryos. ElAzzouny et al. reported the effects of 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) on the synthesis of glycerolipids, ceramides, and nucleotides in INS-1 cells.176 They measured the variations in 66 metabolites and their dependency on AICAR.
Inductively Coupled Plasma MS
The use of inductively coupled plasma mass spectrometry (ICPMS) for analysis of unlabeled single cells is a recently growing research field. Nonetheless, the applications are still far from routine.153 ICPMS can be used either with or without labeling cells prior to analysis. Nonlabeled phytoplankton Scrippsiella trochoidea was investigated and imaged by Van Malderen et al. using laser ablation ICPMS (LA-ICPMS).177 In a case study, they investigated the Cu uptake of the single cells after exposure to Cu concentrations between 0 and 650 μg L−1. For cross-validation, subcellular LA-ICPMS imaging was compared to synchrotron radiation confocal X-ray fluorescence of single cells. Recently, Verboket et al. presented single cell profiling for elemental analysis of single red blood cells.178 Using a microfluidic adapter, they were able to inject cell-laden droplets into an ICPMS instrument and detected the iron content of the individual cells. In addition, Wang and co-workers reported a system to investigate single cell mineral element contents with time-resolved ICPMS.179
Mass Cytometry
The detection of unlabeled target molecules is difficult, and there is currently no MS method available for quantitative single-cell analysis with LODs down to a few hundred targets. These low detection limits are necessary for proteomics or microRNA studies. The group of Gary Nolan and DVS Sciences (acquired by Fluidigm in 2014) developed a mass cytometry system that uses isotopically pure metal nanoparticles as labels.180 Similar to staining with fluorescent molecules and FACS, the stained individual cells are ionized and sensitively detected with cytometry and time-of-flight ICPMS. The quantified amount of each rare earth isotope can then be related to the corresponding target molecule, and sensitivities down to a LOD of 350 molecules/cell at a throughput of up to 2000 cells per second are achieved.181
Zunder et al. used flow cytometry recently to analyze three fibroblast reprogramming systems.182 They measured protein expression, cell-cycle status, and cellular signaling at the single cell level and thereby generated comprehensive references for dynamic changes during cellular reprogramming. Bodenmiller and Bendall and their co-workers have presented methods to use a mass cytometer for imaging of tissue slices by combining it with laser ablation or sputtering (see Figure 5D).161,183,184 Using seven mass tags as labels for a 96-well plate, mass cytometry can be used to label all cells from a particular well on a microtiter plate. Being identified by this label, all cells from the well plate can be analyzed with respect to more than 40 different parameters in one run and the results can later be assigned to the different wells. This mass-tag cellular barcoding (MCB) enables detection of multiple samples without the need for instrument cleaning in between different measurements and thereby minimizes the risk of inhomogeneous staining or instrumental drift during the analysis. To expand the multiplexing capabilities of mass cytometry beyond the currently available isotopic tags, Catena et al. have reported the use of OsO4 and RuO4 as fatty acid tags.185
PCR Based Methods and Single Cell Sequencing
Single cell genomics is a mature technology. Nowadays, several commercial devices are available with the capability of measuring at the single-cell level. In the last two years, genomic analysis has continuously evolved and particularly droplet microfluidic methods have further enabled high throughput sequencing applications at the single-cell level.
PCR Based Analysis
Polymerase from the thermophilic bacterium Thermus aquaticus (Taq) is probably the most frequently used enzyme due to its capability to amplify DNA. In quantitative reverse transcription PCR (RT-qPCR), PCR is employed to amplify the transcribed target RNA strands and the Quantification is done by monitoring the amplification steps with optical methods such as imaging or photon counting via chemiluminescence or fluorescent labeling. Duan et al. have presented a platform that is capable of detecting miRNA with detection limits of down to 10 fM at 37 °C and 1 aM at 4 °C.186 This corresponds to nine strands of miRNA in a 15 μL sample and would translate to single molecule sensitivity in microfluidic chambers or microdroplets with submicroliter volumes. It is quite interesting that they tested their method also with crude samples from MCF-7 and PC3 cells, where all intracellular molecules and debris could influence the analysis. However, there was no notable loss in sensitivity even in the crude samples or real patient tissues. Gracz et al. established a platform for clonogenic stem cell culturing.187 The platform allows the release of single cells or centroids for downstream RNA analysis to relate morphological or functional differences to changes in the gene expression profile. They used the Biomark system from the company Fluidigm for transcriptomic downstream analysis. Fluidigm has recently also commercialized a system for automated mRNA sequencing, DNA sequencing, epigenetics, and detection of miRNA expression levels in up to 800 cells in parallel.188 Chen et al. have employed this device to reveal new insight into hierarchical heterogeneity and complexity of single breast cancer and normal mammary cells or tumor stromal interactions in cocultures.189 Burns et al. used it to investigate sensory organs from the neonatal inner ear with RNA sequencing of single cells.190 Quantitative PCR and RT-qPCR can be extended to measure proteins and other biomolecules with the help of DNA labels. This can be exploited in measurements of targets with very low copy numbers as it allows for signal amplification. This method has become a versatile tool now in infectious disease diagnostics and immunosensing.191,192 Additionally, labeling with DNA enables the parallel Quantification of proteins, RNA, and DNA samples on a single platform. Darmanis et al. used this technique to study the responses of glioblastoma cells to putative therapeutic BMP4.193 They labeled the protein targets for this purpose with specific DNA sequences and used the Fluidigm array together with the Biomark HD system for qPCR.
All PCR methods with Taq-polymerase require cyclic heating and cooling for replication. To avoid this and allow easy integration into portable systems, isothermal amplification techniques can be chosen.194 Kunze et al. used isothermal nucleic acid amplification together with chemiluminescence detection.195 They achieved comparable sensitivities as reported for PCR based systems and could simultaneously amplify three different DNA targets in one measurement chamber. Likewise, Xu et al. used chemiluminescence to detect microRNA on an isothermal amplification platform.196 To further increase sensitivity, they implemented a cascade reaction to achieve exponential amplification of the signal resulting in very high sensitivities.
Single Cell Sequencing with Droplet microfluidics
Microdroplets are well suited for high throughput applications as they can be produced sequentially with kHz production speeds (See Cytometric Methods: Continuous Processing of Single Cells). Recently, droplet microfluidics was applied for single-cell sequencing. Therein, a part or the whole genome of single cells is decoded in a sequencing device. To be able to relate the measured sequences back to the individual cells, different barcoding techniques on droplet microfluidic platforms were employed.
Rotem et al. merged lysed single cells and barcoded droplets in a microfluidic chip. In the droplets, DNA barcodes were ligated to the chromatin fragments of the original cells.197 After sequencing, the DNA barcode could be used to assign the measured sequences to the individual cells. A similar principle was set up by Macosko et al.198 and Klein et al.199 to analyze the RNA of individual cells. They followed the same roadmap as presented before but additionally applied a reverse transcription step. Using this system, it is possible to analyze the transcriptomes of many thousand cells in parallel, create gene expression maps, and identify cell subpopulations from heterogeneous cell mixtures.
Other Techniques
While the above-mentioned analytical methods represent a major fraction of single-cell analysis techniques, several other methods are promising approaches to interrogate information on cell properties.
Nanomotors for miRNA Detection
Esteban-Fernández de Ávila et al. have presented nanomotors for real-time microRNA measurements.200 They used a dye that was labeled to single-stranded DNA and attached to a graphene oxide coated gold nanowire (Figure 6A). Externally applied ultrasound was used to propel the nanomotors and increase the probability of penetrating intact cancer cells. Once the robot was internalized and the ssDNA probe bound to its target, the quenching effect of the graphene oxide was lost and the labeled RNA target turned fluorescent. The system was tested with MCF-7 and HeLa cells by investigating the expression levels of microRNA-21.
Figure 6. Special single-cell analysis platforms.
(A) The detection of intracellular microRNA is initiated by application of ultrasound. It accelerates the nanomotor-tags, and they pass the cell membrane. Upon contact with the target microRNA sequence, the fluorescent label is released from the quencher and fluorescence arises. Fluorescent signal of a MCF-7 cell before (right top) and after (right bottom) ultrasound exposure. Scale bars, 10 μm. Adapted from Esteban-Fernández de Ávila, B.; Martín, A.; Soto, F.; Lopez-Ramirez, M. A.; Campuzano, S.; Vásquez-Machado, G. M.; Gao, W.; Zhang, L.; Wang, J. ACS Nano 2015, 9, 6756−6764 (ref 200). Copyright 2015 American Chemical Society. (B) Cantilever beam resonance is affected by changes in its mass. Serial mass measurements of single cells flowing in a hollow cantilever were then employed to measure mass changes of single cells over time and detect the cells growth rates. Adapted with permission from Cermak, N.; Olcum, S.; Delgado, F. F.; Wasserman, S. C.; Payer, K. R.; Murakami, M. A.; Knudsen, S. M.; Kimmerling, R. J.; Stevens, M. M.; Kikuchi, Y.; Sandikci, A.; Ogawa, M.; Agache, V.; Baleras, F.; Weinstock, D. M.; Manalis, S. R. Nat. Biotechnol. 2016, 34, 1052–1059 (ref 201). Copyright 2016 Nature Publishing Group. (C) Single cells were thermally analyzed in an ultrasensitive and thermally well isolated microfluidic setup. The resonant frequency of a cantilever beam depending on the temperature and effects of the heat production on the single cell level can be investigated. Adapted with permission from Inomata, N.; Toda, M.; Ono, T. Lab Chip 2016, 16, 3597–3603 (ref 202). Copyright 2016 Royal Society of Chemistry. (D) A new type of microscope that visualizes magnetism has been reported. The quantum diamond microscope was used to investigate magnetically labeled cells. Scale bars, 100 μm. Reprinted with permission from Glenn, D. R.; Lee, K.; Park, H.; Weissleder, R.; Yacoby, A.; Lukin, M. D.; Lee, H.; Walsworth, R. L.; Connolly, C. B. Nat. Methods 2015, 12 (8), 736–738 (ref 203). Copyright 2015 Nature Publishing Group.
Atomic Force Microscopy for Single Cells
Cantilever beams with a sharp tip are employed to raster surfaces and measure the topography with atomic resolution. As the mechanical properties of cancer cells differ from normal cell behavior, much effort has been put into the development of atomic force microscopy (AFM) based systems to probe cell mechanics on the single cell level.204 One possibility of using these systems is for the Quantification of microRNA as presented by Koo et al.205 They measured the binding forces of a functionalized AFM tip when scanning over a cell and could detect the presence of a complementary microRNA sequence. An approach presented by Guillaume-Gentil et al.206 is very attractive for the analysis of cytosolic molecules derived from live cells. They used hollow AFM cantilevers as nanopipettes to extract subpicoliter volumes from the cytoplasm or nucleoplasm of individual cells. This approach enabled them to measure cellular heterogeneities in living cells. The throughput of these methods is currently limited, and only one cell can be measured at a time.
Mass sensing of single cells was recently done by probing the resonance frequency of cantilever beams. The resonance frequency is affected by changes in its mass as well as by geometrical changes due to thermal expansion of the cantilever beam. Keeping the temperature constant, Cermak et al. supplied various cell types in suspension through a microfluidic channel with at least ten resonant mass sensors distributed along its length.201 As each cell was successively measured ten times, their setup allowed for Quantification of the mass related growth rate of single cells (see Figure 6B). The parallel configuration could measure more than one cell per minute and reached resolutions in growth rates of up to 0.2 pg h−1.
Thermal Measurements on Single Cell Level
The electrical resistance of a metal conductor is temperature dependent. To use this effect for precise quantitative calorimetric measurements of single cells, thermal insulation is crucial. Inomata et al. have lately produced highly sensitive miniaturized thermometers by encapsulating silicon cantilever based resonance temperature sensors in vacuum to minimize heat loss.202 Direct contact of the cell to the cantilever beam established a heat transfer from the cell to the isolated cantilever (see Figure 6C). Thereby, they could sensitively probe the temperature rise of single brown fat cells with a thermal resolution of 79 μK or 1.90 nW. Interestingly, they found spiked thermal response patterns in nonactivated cells and continuous heat production in cells stimulated by norepinephrine.
Photothermal Imaging
Detection based on an increase of temperature upon light irradiation can be done with photothermal microscopy. This enables detection of nanometer sized objects like gold nanoparticles in single cells due to their higher absorption coefficient, and it additionally benefits from stable signals as bleaching effects do not occur.207 Nieves et al. implemented raster image correlation spectroscopy for longterm photothermal imaging.208 They used the technique to measure the diffusion of fibroblast growth factor 2 (FGF2) labeled with gold nanoparticles when FGF2 was bound to heparan sulfate in live fibroblast cells.
Magnetic Single-Cell Analysis Using Hall Sensors
Magnetic Hall sensors are used to measure the magnetic field strength. Issadore et al. have recently employed such integrated Hall sensors for detection of magnetically labeled circulating tumor cells in a cytometric fashion.209 They achieved a high throughput of up to 107 cells per minute, which is comparable to the speed of state of the art FACS machines. They detected CTCs in samples from 20 ovarian cancer patients by using the epithelial cell adhesion molecule (EpCAM) surface marker. Their system benefits from the confined size and low costs of the analysis, but the validation and robustness of this instrument in the clinical routine have yet to be shown.
Magnetic Imaging with a Quantum Diamond Microscope
Glenn et al. have presented a quantum diamond microscope for imaging of magnetic properties.203 They employed their microscope for the detection of HER2 positive SKBR3 cells that were labeled with magnetic nanoparticles (see Figure 6D). The quantum diamond microscope consists of a dense layer of fluorescent quantum sensors, based on nitrogen-vacancy centers close to the surface of a diamond chip. When a sample of magnetically labeled cells is placed on the diamond chip, the electronic spins of the nitrogen vacancies are probed with a 532 nm laser and microwaves. The emitted fluorescence is used as a measure for the presence of magnetic nanoparticles, and imaging is conducted with conventional CCD or CMOS cameras.
Conclusions
Nowadays, sensitive analytical methods are available to identify heterogeneities in cell populations. The most important methods are based on optical microscopy and fluorescence based techniques and have reached incredibly high sensitivities and spatial resolutions far below the wavelength of the optical light. For high-throughput characterization of cell populations, methods based on cytometry are mature and used routinely.
However, many other analytical methods such as electrochemistry and mass spectrometry contributed to the emerging field of single-cell analysis in recent years and extended our knowledge of cellular processes. These techniques have the advantage of mostly having no need for labeling which potentially interferes with the biological processes that are under investigation. In addition, microfluidic methods provide valuable tools for cell positioning and treatment.
Nevertheless, some difficulties have remained. Techniques that allow label-free identification of cellular compounds like MS lack the sensitivity to detect or quantify low-abundant targets or cannot be applied for dynamic studies of living cells. Furthermore, live-cell imaging techniques and sophisticated fluorescence microscopy have limited multiplexing capabilities or are not (yet) developed for high throughput. In addition, the limited precision of many analytical methods and the lack of references such as artificial model cells with a defined composition question the reliability of quantitative results.
Due to the complexity of cellular processes, it is very promising to combine two or more methods that can provide complementary information. Therefore, new methods for sampling are valuable such as micro- or nanocapillaries154 and hollow AFM-tips206 that facilitate the withdrawal of small portions of the cytosol from living cells. This enables the use of destructive techniques like MS in dynamic studies and dynamic live-cell imaging. In addition, novel methods to capture selected cells directly from tissue samples, e.g., the microfluidic probe,210 open the way to study heterogeneities within (healthy) organs or tumors. We believe that further innovations of analytical instrumentation and computational methods for data interpretation will soon lead to new insights into single-cell heterogeneity and potentially find applications in personalized medicine.
Acknowledgments
We thank C. C. Lin, A. Kling, Y. Schmid, D. Hümmer, F. Kurth, S. Knobelspies, and R. Steinhoff (ETH Zurich, Switzerland) for critical reading of this manuscript and helpful suggestions. We also greatly acknowledge R. Tan (UC Berkeley, USA) for proofreading. P.S.D. acknowledges financial support by SystemsX, the ETH foundation, and the European Research Council (ERC Advanced Grant No. 681587).
Biographies
Biographies
Lucas Armbrecht studied microsystems technology at the University of Freiburg (Breisgau, Germany). Since June 2015, he has been a Ph.D. student in the Bioanalytics Group at ETH Zurich (Switzerland). In his doctoral studies, he is developing microfluidic methods for multiparametric single-cell analysis.
Petra Dittrich is an Associate Professor in the Department of Biosystems Science and Engineering at ETH Zurich (Switzerland). She studied Chemistry at Bielefeld University and the University of Salamanca (Spain). After completing her doctoral studies at the Max Planck Institute for Biophysical Chemistry in Göttingen (Germany) and her postdoctoral work at the Institute for Analytical Sciences in Dortmund, she was Assistant Professor of Bioanalytics at the Department of Chemistry and Applied Biosciences at ETH Zurich from 2008 to 2014. Her research focuses on the miniaturization of bioanalytical methods, especially for cell and membrane analysis and for the creation of artificial cells.
Footnotes
ORCID
Petra S. Dittrich: 0000-0001-5359-8403
Author Contributions
The manuscript was written through contributions of both authors. Both authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
Special Issue: Fundamental and Applied Reviews in Analytical Chemistry 2017
References
- (1).Konry T, Sarkar S, Sabhachandani P, Cohen N. Annu Rev Biomed Eng. 2016;18:259–284. doi: 10.1146/annurev-bioeng-090215-112735. [DOI] [PubMed] [Google Scholar]
- (2).Khamenehfar A, Gandhi MK, Chen Y, Hogge DE, Li PCH. Anal Chem. 2016;88:5680–5688. doi: 10.1021/acs.analchem.5b04446. [DOI] [PubMed] [Google Scholar]
- (3).Borland LM, Kottegoda S, Phillips KS, Allbritton NL. Annu Rev Anal Chem. 2008;1:191–227. doi: 10.1146/annurev.anchem.1.031207.113100. [DOI] [PubMed] [Google Scholar]
- (4).Saadatpour A, Lai S, Guo G, Yuan G-C. Trends Genet. 2015;31:576–586. doi: 10.1016/j.tig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Weaver WM, Tseng P, Kunze A, Masaeli M, Chung AJ, Dudani JS, Kittur H, Kulkarni RP, Di Carlo D. Curr Opin Biotechnol. 2014;25:114–123. doi: 10.1016/j.copbio.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P. Int J Mol Sci. 2015;16:16897–16919. doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chen Y, Li P, Huang P-H, Xie Y, Mai JD, Wang L, Nguyen N-T, Huang TJ. Lab Chip. 2014;14:626–645. doi: 10.1039/c3lc90136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hümmer D, Kurth F, Naredi-Rainer N, Dittrich PS. Lab Chip. 2016;16:447–458. doi: 10.1039/c5lc01314c. [DOI] [PubMed] [Google Scholar]
- (9).Yamamura S, Kishi H, Tokimitsu Y, Kondo S, Honda R, Rao SR, Omori M, Tamiya E, Muraguchi A. Anal Chem. 2005;77:8050–8056. doi: 10.1021/ac0515632. [DOI] [PubMed] [Google Scholar]
- (10).Lin C-H, Hsiao Y-H, Chang H-C, Yeh C-F, He C-K, Salm EM, Chen C, Chiu I-M, Hsu C-H. Lab Chip. 2015;15:2928–2938. doi: 10.1039/c5lc00541h. [DOI] [PubMed] [Google Scholar]
- (11).Wang Y, Shah P, Phillips C, Sims CE, Allbritton NL. Anal Bioanal Chem. 2012;402:1065–1072. doi: 10.1007/s00216-011-5535-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Swennenhuis JF, Tibbe AGJ, Stevens M, Katika MR, van Dalum J, Duy Tong H, van Rijn CJM, Terstappen LWMM. Lab Chip. 2015;15:3039–3046. doi: 10.1039/c5lc00304k. [DOI] [PubMed] [Google Scholar]
- (13).Sun T, Kovac J, Voldman J. Anal Chem. 2014;86:977–981. doi: 10.1021/ac403777g. [DOI] [PubMed] [Google Scholar]
- (14).Wang Z, Wu W, Wang Z, Tang Y, Deng Y, Xu L, Tian J, Shi Q. Analyst. 2016;141:3621–3625. doi: 10.1039/c5an02554k. [DOI] [PubMed] [Google Scholar]
- (15).Dusny C, Grünberger A, Probst C, Wiechert W, Kohlheyer D, Schmid A. Lab Chip. 2015;15:1822–1834. doi: 10.1039/c4lc01270d. [DOI] [PubMed] [Google Scholar]
- (16).Wyatt Shields C, IV, Reyes CD, López GP. Lab Chip. 2015;15:1230–1249. doi: 10.1039/c4lc01246a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).de Wagenaar B, Berendsen JTW, Bomer JG, Olthuis W, van den Berg A, Segerink LI. Lab Chip. 2015;15:1294–1301. doi: 10.1039/c4lc01425a. [DOI] [PubMed] [Google Scholar]
- (18).Delincé MJ-H, Bureau J-B, López-Jiménez AT, Cosson P, Soldati T, McKinney J. Lab Chip. 2016;16:3276–3285. doi: 10.1039/c6lc00649c. [DOI] [PubMed] [Google Scholar]
- (19).Kim H, Lee S, Lee J, Kim J. Lab Chip. 2015;15:4128–4132. doi: 10.1039/c5lc00904a. [DOI] [PubMed] [Google Scholar]
- (20).Kimmerling RJ, Szeto GL, Li JW, Genshaft AS, Kazer SW, Payer KR, de Riba Borrajo J, Blainey PC, Irvine DJ, Shalek AK, Manalis SR. Nat Commun. 2016;7:1–7. doi: 10.1038/ncomms10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chen Y-C, Ingram PN, Fouladdel S, McDermott SP, Azizi E, Wicha MS, Yoon E. Sci Rep. 2016;6:27301. doi: 10.1038/srep27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dura B, Dougan SK, Barisa M, Hoehl MM, Lo CT, Ploegh HL, Voldman J. Nat Commun. 2015;6:5940. doi: 10.1038/ncomms6940. [DOI] [PubMed] [Google Scholar]
- (23).Geng T, Bredeweg EL, Szymanski CJ, Liu B, Baker SE, Orr G, Evans JE, Kelly RT. Sci Rep. 2015;5:16111. doi: 10.1038/srep16111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chen H, Sun J, Wolvetang E, Cooper-White J. Lab Chip. 2015;15:1072–1083. doi: 10.1039/c4lc01176g. [DOI] [PubMed] [Google Scholar]
- (25).Wang Y, Tang X, Feng X, Liu C, Chen P, Chen D, Liu BF. Anal Bioanal Chem. 2015;407:1139–1148. doi: 10.1007/s00216-014-8325-3. [DOI] [PubMed] [Google Scholar]
- (26).Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, et al. Nat Methods. 2015;12:685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Che J, Yu V, Dhar M, Renier C, Matsumoto M, Heirich K, Garon EB, Goldman J, Rao J, Sledge GW, Pegram MD, et al. Oncotarget. 2016;7:12748–12760. doi: 10.18632/oncotarget.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Huang C, Liu C, Loo J, Stakenborg T, Lagae L. Appl Phys Lett. 2014;104:013703. [Google Scholar]
- (29).Kim SH, Antfolk M, Kobayashi M, Kaneda S, Laurell T, Fujii T. Lab Chip. 2015;15:4356–4363. doi: 10.1039/c5lc01065a. [DOI] [PubMed] [Google Scholar]
- (30).Du E, Dao M, Suresh S. Extrem Mech Lett. 2014;1:35–41. doi: 10.1016/j.eml.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ahadian S, Yamada S, Ramón-Azcón J, Ino K, Shiku H, Khademhosseini A, Matsue T. Lab Chip. 2014;14:3690–3694. doi: 10.1039/c4lc00479e. [DOI] [PubMed] [Google Scholar]
- (32).Yang Y, Mao Y, Shin K-S, Chui CO, Chiou P-Y. Sci Rep. 2016;6:22630. doi: 10.1038/srep22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang D, Zhao X, Gu Z. Opt Commun. 2016 doi: 10.1016/j.optcom.2016.03.047. [DOI] [Google Scholar]
- (34).Huang K-W, Wu Y-C, Lee J-A, Chiou P-Y. Lab Chip. 2013;13:3721–3727. doi: 10.1039/c3lc50607j. [DOI] [PubMed] [Google Scholar]
- (35).Yang T, Gao D, Jin F, Jiang Y, Liu H. Rapid Commun Mass Spectrom. 2016;30:73–79. doi: 10.1002/rcm.7628. [DOI] [PubMed] [Google Scholar]
- (36).Pan C-J, Qin H, Nie Y-D, Ding H-Y. Colloids Surf B. 2013;104:18–26. doi: 10.1016/j.colsurfb.2012.11.045. [DOI] [PubMed] [Google Scholar]
- (37).Knight GT, Klann T, McNulty JD, Ashton RS. J Visualized Exp. 2014;92:e52186. doi: 10.3791/52186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Saliba a-E, Saias L, Psychari E, Minc N, Simon D, Bidard F-C, Mathiot C, Pierga J-Y, Fraisier V, Salamero J, Saada V, et al. Proc Natl Acad Sci U S A. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Custódio CA, San Miguel-Arranz V, Gropeanu RA, Gropeanu M, Wirkner M, Reis RL, Mano JF, del Campo A. Langmuir. 2014;30:10066–10071. doi: 10.1021/la502688h. [DOI] [PubMed] [Google Scholar]
- (40).Brinkmann F, Hirtz M, Haller A, Gorges TM, Vellekoop MJ, Riethdorf S, Müller V, Pantel K, Fuchs H. Sci Rep. 2015;5:15342. doi: 10.1038/srep15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Liu H, Li Y, Sun K, Fan J, Zhang P, Meng J, Wang S, Jiang L. J Am Chem Soc. 2013;135:7603–7609. doi: 10.1021/ja401000m. [DOI] [PubMed] [Google Scholar]
- (42).Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH. Anal Chem. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Cheng S-B, Xie M, Xu J-Q, Wang J, Lv S-W, Guo S, Shu Y, Wang M, Dong W-G, Huang W-H. Anal Chem. 2016;88:6773–6780. doi: 10.1021/acs.analchem.6b01130. [DOI] [PubMed] [Google Scholar]
- (44).Kodzius R, Gojobori T. Gene. 2016;576:701–707. doi: 10.1016/j.gene.2015.10.031. [DOI] [PubMed] [Google Scholar]
- (45).Hosic S, Murthy SK, Koppes AN. Anal Chem. 2016;88:354–380. doi: 10.1021/acs.analchem.5b04077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Joensson HN, Andersson Svahn H. Angew Chem, Int Ed. 2012;51:12176–12192. doi: 10.1002/anie.201200460. [DOI] [PubMed] [Google Scholar]
- (47).Wong D, Ren CL. Lab Chip. 2016;16:3317–3329. doi: 10.1039/c6lc00626d. [DOI] [PubMed] [Google Scholar]
- (48).Anna SL. Annu Rev Fluid Mech. 2016;48:285–309. [Google Scholar]
- (49).Baret J-C. Lab Chip. 2012;12:422–433. doi: 10.1039/c1lc20582j. [DOI] [PubMed] [Google Scholar]
- (50).Shim J, Ranasinghe RT, Smith CA, Ibrahim SM, Hollfelder F, Huck WTS, Klenerman D, Abell C. ACS Nano. 2013;7:5955–5964. doi: 10.1021/nn401661d. [DOI] [PubMed] [Google Scholar]
- (51).Zinchenko A, Devenish SRA, Kintses B, Colin P-Y, Fischlechner M, Hollfelder F. Anal Chem. 2014;86:2526–2533. doi: 10.1021/ac403585p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Collins DJ, Neild A, DeMello A, Liu A-Q, Ai Y. Lab Chip. 2015;15:3439–3459. doi: 10.1039/c5lc00614g. [DOI] [PubMed] [Google Scholar]
- (53).Sciambi A, Abate AR. Lab Chip. 2015;15:47–51. doi: 10.1039/c4lc01194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lagus TP, Edd JF. RSC Adv. 2013;3:20512. [Google Scholar]
- (55).Mazutis L, Gilbert J, Ung WL, Weitz Da, Griffiths AD, Heyman Ja. Nat Protoc. 2013;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Meyer A, Pellaux R, Potot S, Becker K, Hohmann H-P, Panke S, Held M. Nat Chem. 2015;7:673–678. doi: 10.1038/nchem.2301. [DOI] [PubMed] [Google Scholar]
- (57).Werner M, Palankar R, Arm L, Hovius R, Vogel H. Small. 2015;11:2607–2613. doi: 10.1002/smll.201402650. [DOI] [PubMed] [Google Scholar]
- (58).Popova AA, Demir K, Hartanto TG, Schmitt E, Levkin PA. RSC Adv. 2016;6:38263–38276. [Google Scholar]
- (59).Schmitz CHJ, Rowat AC, Köster S, Weitz DA. Lab Chip. 2009;9:44–49. doi: 10.1039/b809670h. [DOI] [PubMed] [Google Scholar]
- (60).Dong L, Chen D-W, Liu S-J, Du W. Sci Rep. 2016;6:24192. doi: 10.1038/srep24192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Pampaloni F, Chang BJ, Stelzer EHK. Cell Tissue Res. 2015;360:129–141. doi: 10.1007/s00441-015-2144-5. [DOI] [PubMed] [Google Scholar]
- (62).Sydor AM, Czymmek KJ, Puchner EM, Mennella V. Trends Cell Biol. 2015;25:730–748. doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- (63).Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, Chen CS. Nat Methods. 2014;11:1229–1232. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. Proc Natl Acad Sci U S A. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, Amir ED, Pe’er D, Miller-Jensen K, Fan R. Proc Natl Acad Sci U S A. 2015;112:E607–E615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Hughes AJ, Spelke DP, Xu Z, Kang C-C, Schaffer DV, Herr AE. Nat Methods. 2014;11:749–755. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Lee K, Kim K, Jung J, Heo J, Cho S, Lee S, Chang G, Jo Y, Park H, Park Y. Sensors. 2013;13:4170–4191. doi: 10.3390/s130404170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Nienhaus K, Ulrich Nienhaus G. Chem Soc Rev. 2014;43:1088–1106. doi: 10.1039/c3cs60171d. [DOI] [PubMed] [Google Scholar]
- (69).van de Linde S, Heilemann M, Sauer M. Annu Rev Phys Chem. 2012;63:519–540. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- (70).Moffitt JR, Lee JB, Cluzel P. Lab Chip. 2012;12:1487. doi: 10.1039/c2lc00009a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Hoffman MD, Zucker LI, Brown PJB, Kysela DT, Brun YV, Jacobson SC. Anal Chem. 2015;87:12032–12039. doi: 10.1021/acs.analchem.5b02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hsieh K, Zec HC, Chen L, Kaushik A, Wang T. Rapid, Accurate, and General Single-Cell Antibiotic Susceptibility Test in Digital Bacteria Picoarray. The 20th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS 2016; Dublin, Ireland. October 9–13; 2016. pp. 174–175. [Google Scholar]
- (73).Krämer CEM, Wiechert W, Kohlheyer D. Sci Rep. 2016;6:32104. doi: 10.1038/srep32104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Birchler A, Berger M, Jäggin V, Lopes T, Etzrodt M, Misun PM, Pena-Francesch M, Schroeder T, Hierlemann A, Frey O. Anal Chem. 2016;88:1222–1229. doi: 10.1021/acs.analchem.5b03513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Schmid YRF, Bürgel SC, Misun PM, Hierlemann A, Frey O. ACS Sensors. 2016;1:1028–1035. doi: 10.1021/acssensors.6b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Hilsenbeck O, Schwarzfischer M, Skylaki S, Schauberger B, Hoppe PS, Loeffler D, Kokkaliaris KD, Hastreiter S, Skylaki E, Filipczyk A, Strasser M, et al. Nat Biotechnol. 2016;34:703–706. doi: 10.1038/nbt.3626. [DOI] [PubMed] [Google Scholar]
- (77).Hohng S, Lee S, Lee J, Jo MH. Chem Soc Rev. 2014;43:1007–1013. doi: 10.1039/c3cs60184f. [DOI] [PubMed] [Google Scholar]
- (78).Miyamoto A, Miyauchi H, Kogure T, Miyawaki A, Michikawa T, Mikoshiba K. Biochem Biophys Res Commun. 2015;460(1):82–87. doi: 10.1016/j.bbrc.2015.02.045. [DOI] [PubMed] [Google Scholar]
- (79).Ng EX, Miller MA, Jing T, Chen C-H. Biosens Bioelectron. 2016;81:408–414. doi: 10.1016/j.bios.2016.03.002. [DOI] [PubMed] [Google Scholar]
- (80).Son KJ, Shin D-S, Kwa T, You J, Gao Y, Revzin A. Lab Chip. 2015;15:637–641. doi: 10.1039/c4lc00884g. [DOI] [PubMed] [Google Scholar]
- (81).Cost AL, Ringer P, Chrostek-Grashoff A, Grashoff C. Cell Mol Bioeng. 2015;8:96–105. doi: 10.1007/s12195-014-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Perez-Toralla K, Mottet G, Guneri ET, Champ J, Bidard F-C, Pierga J-Y, Klijanienko J, Draskovic I, Malaquin L, Viovy J-L, Descroix S. Lab Chip. 2015;15:811–822. doi: 10.1039/c4lc01059k. [DOI] [PubMed] [Google Scholar]
- (83).Godin AG, Lounis B, Cognet L. Biophys J. 2014;107:1777–1784. doi: 10.1016/j.bpj.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Schneider J, Zahn J, Maglione M, Sigrist SJ, Marquard J, Chojnacki J, Kräusslich H-G, Sahl SJ, Engelhardt J, Hell SW. Nat Methods. 2015;12:827–830. doi: 10.1038/nmeth.3481. [DOI] [PubMed] [Google Scholar]
- (85).Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Mishina NM, Mishin AS, Belyaev Y, Bogdanova EA, Lukyanov S, Schultz C, Belousov VV. Nano Lett. 2015;15:2928–2932. doi: 10.1021/nl504710z. [DOI] [PubMed] [Google Scholar]
- (87).Andrade DM, Clausen MP, Keller J, Mueller V, Wu C, Bear JE, Hell SW, Lagerholm BC, Eggeling C. Sci Rep. 2015;5:11454. doi: 10.1038/srep11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Niehörster T, Löschberger A, Gregor I, Krämer B, Rahn H-J, Patting M, Koberling F, Enderlein J, Sauer M. Nat Methods. 2016;13:257–262. doi: 10.1038/nmeth.3740. [DOI] [PubMed] [Google Scholar]
- (89).Dudok B, Barna L, Ledri M, Szabó SI, Szabadits E, Pintér B, Woodhams SG, Henstridge CM, Balla GY, Nyilas R, Varga C, et al. Nat Neurosci. 2014;18:75–86. doi: 10.1038/nn.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Hennig S, van de Linde S, Lummer M, Simonis M, Huser T, Sauer M. Nano Lett. 2015;15:1374–1381. doi: 10.1021/nl504660t. [DOI] [PubMed] [Google Scholar]
- (91).Wang P, Wu J, Zhou P, Liu W, Tang Y. J Mater Chem B. 2015;3:3617–3624. doi: 10.1039/c5tb00142k. [DOI] [PubMed] [Google Scholar]
- (92).Meng Q, Zhang R, Jia H, Gao X, Wang C, Shi Y, Everest-Dass AV, Zhang Z. Talanta. 2015;143:294–301. doi: 10.1016/j.talanta.2015.04.072. [DOI] [PubMed] [Google Scholar]
- (93).Han Y, Ding C, Zhou J, Tian Y. Anal Chem. 2015;87:5333–5339. doi: 10.1021/acs.analchem.5b00628. [DOI] [PubMed] [Google Scholar]
- (94).Maity SB, Banerjee S, Sunwoo K, Kim JS, Bharadwaj PK. Inorg Chem. 2015;54:3929–3936. doi: 10.1021/acs.inorgchem.5b00106. [DOI] [PubMed] [Google Scholar]
- (95).Chen D, Wang H, Dong L, Liu P, Zhang Y, Shi J, Feng X, Zhi J, Tong B, Dong Y. Biomaterials. 2016;103:67–74. doi: 10.1016/j.biomaterials.2016.06.055. [DOI] [PubMed] [Google Scholar]
- (96).Zrazhevskiy P, Gao X. Nat Commun. 2013;4:1619. doi: 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Kairdolf BA, Smith AM, Stokes TH, Wang MD, Young AN, Nie S. Annu Rev Anal Chem. 2013;6:143–162. doi: 10.1146/annurev-anchem-060908-155136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Ren D, Xia Y, Wang B, You Z. Anal Chem. 2016;88:4318–4327. doi: 10.1021/acs.analchem.5b04471. [DOI] [PubMed] [Google Scholar]
- (99).Efros AL, Nesbitt DJ. Nat Nanotechnol. 2016;11:661–671. doi: 10.1038/nnano.2016.140. [DOI] [PubMed] [Google Scholar]
- (100).Zhao A, Chen Z, Zhao C, Gao N, Ren J, Qu X. Carbon. 2015;85:309–327. [Google Scholar]
- (101).Luo PG, Yang F, Yang S-T, Sonkar SK, Yang L, Broglie JJ, Liu Y, Sun Y-P. RSC Adv. 2014;4:10791. [Google Scholar]
- (102).Jiang K, Sun S, Zhang L, Lu Y, Wu A, Cai C, Lin H. Angew Chem, Int Ed. 2015;54:5360–5363. doi: 10.1002/anie.201501193. [DOI] [PubMed] [Google Scholar]
- (103).Giri B, Pandey B, Neupane B, Ligler FS. TrAC, Trends Anal Chem. 2016;79:326–334. [Google Scholar]
- (104).van de Linde S, Heilemann M, Sauer M. Annu Rev Phys Chem. 2012;63:519–540. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- (105).Thomas JA. Chem Soc Rev. 2015;44(14):4494–4500. doi: 10.1039/c5cs00070j. [DOI] [PubMed] [Google Scholar]
- (106).Eyer K, Stratz S, Kuhn P, Küster SK, Dittrich PS. Anal Chem. 2013;85:3280–3287. doi: 10.1021/ac303628j. [DOI] [PubMed] [Google Scholar]
- (107).Stratz S, Eyer K, Kurth F, Dittrich PS. Anal Chem. 2014;86:12375–12381. doi: 10.1021/ac503766d. [DOI] [PubMed] [Google Scholar]
- (108).Li L, Wang Q, Feng J, Tong L, Tang B. Anal Chem. 2014;86:5101–5107. doi: 10.1021/ac500881p. [DOI] [PubMed] [Google Scholar]
- (109).Liao M, Yang Q, Zhang J, Zhang M, Deng Q, Liu H, Graner MW, Kornfeld H, Zhou B, Chen X. Clin Vaccine Immunol. 2014;21:347–353. doi: 10.1128/CVI.00680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Nesher L, Shah DP, Ariza-Heredia EJ, Azzi JM, Siddiqui HK, Ghantoji SS, Marsh LY, Michailidis L, Makedonas G, Rezvani K, Shpall EJ, et al. J Infect Dis. 2016;213:1701–1707. doi: 10.1093/infdis/jiw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Maggioli MF, Palmer MV, Vordermeier HM, Whelan AO, Fosse JM, Nonnecke BJ, Waters WRJ. Visualized Exp. 2015;101:e52833. doi: 10.3791/52833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Saletti G, Çuburu N, Yang JS, Dey A, Czerkinsky C. Nat Protoc. 2013;8:1073–1087. doi: 10.1038/nprot.2013.058. [DOI] [PubMed] [Google Scholar]
- (113).Ramirez L, Herschkowitz JI, Wang J. Sci Rep. 2016;6:32505. doi: 10.1038/srep32505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Junkin M, Kaestli AJ, Cheng Z, Jordi C, Albayrak C, Hoffmann A, Tay S. Cell Rep. 2016;15:411–422. doi: 10.1016/j.celrep.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Duncombe TA, Kang CC, Maity S, Ward TM, Pegram MD, Murthy N, Herr AE. Adv Mater. 2016;28:327–334. doi: 10.1002/adma.201503939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Wang Y, Huang WE, Cui L, Wagner M. Curr Opin Biotechnol. 2016;41:34–42. doi: 10.1016/j.copbio.2016.04.018. [DOI] [PubMed] [Google Scholar]
- (117).Kann B, Offerhaus HL, Windbergs M, Otto C. Adv Drug Delivery Rev. 2015;89:71–90. doi: 10.1016/j.addr.2015.02.006. [DOI] [PubMed] [Google Scholar]
- (118).Casabella S, Scully P, Goddard N, Gardner P. Analyst. 2016;141:689–696. doi: 10.1039/c5an01851j. [DOI] [PubMed] [Google Scholar]
- (119).Kang B, Afifi MM, Austin LA, El-sayed MA. ACS Nano. 2013;7:7420–7427. doi: 10.1021/nn403351z. [DOI] [PubMed] [Google Scholar]
- (120).Song Y, Yin H, Huang WE. Curr Opin Chem Biol. 2016;33:1–8. doi: 10.1016/j.cbpa.2016.04.002. [DOI] [PubMed] [Google Scholar]
- (121).Stojanović I, Van Der Velden TJG, Mulder HW, Schasfoort RBM, Terstappen LWMM. Anal Biochem. 2015;485:112–118. doi: 10.1016/j.ab.2015.06.018. [DOI] [PubMed] [Google Scholar]
- (122).de Wit G, Danial JSH, Kukura P, Wallace MI. Proc Natl Acad Sci U S A. 2015;112:12299–12303. doi: 10.1073/pnas.1508483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Ortega Arroyo J, Andrecka J, Spillane KM, Billington N, Takagi Y, Sellers JR, Kukura P. Nano Lett. 2014;14:2065–2070. doi: 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Agnarsson B, Lundgren A, Gunnarsson A, Rabe M, Kunze A, Mapar M, Simonsson L, Bally M, Zhdanov VP, Höök F. ACS Nano. 2015;9:11849–11862. doi: 10.1021/acsnano.5b04168. [DOI] [PubMed] [Google Scholar]
- (125).Cotte Y, Toy F, Jourdain P, Pavillon N, Boss D, Magistretti P, Marquet P, Depeursinge C. Nat Photonics. 2013;7:113–117. [Google Scholar]
- (126).Bucher ES, Wightman RM. Annu Rev Anal Chem. 2015;8:239–261. doi: 10.1146/annurev-anchem-071114-040426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Vasdekis AE, Stephanopoulos G. Metab Eng. 2015;27:115–135. doi: 10.1016/j.ymben.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Liu Y, Li M, Zhang F, Zhu A, Shi G. Anal Chem. 2015;87(11):5531–5538. doi: 10.1021/ac5042999. [DOI] [PubMed] [Google Scholar]
- (129).Anderson SE, Bau HH. Nanotechnology. 2015;26:185101. doi: 10.1088/0957-4484/26/18/185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Robinson JT, Jorgolli M, Shalek AK, Yoon M-H, Gertner RS, Park H. Nat Nanotechnol. 2012;7:180–184. doi: 10.1038/nnano.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Li X, Majdi S, Dunevall J, Fathali H, Ewing AG. Angew Chem Int Ed. 2015;54:11978–11982. doi: 10.1002/anie.201504839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Wigström J, Dunevall J, Najafinobar N, Lovrić J, Wang J, Ewing AG, Cans A-S. Anal Chem. 2016;88:2080–2087. doi: 10.1021/acs.analchem.5b03316. [DOI] [PubMed] [Google Scholar]
- (133).Dick JE. Chem Commun. 2016;52:10906–10909. doi: 10.1039/c6cc04515d. [DOI] [PubMed] [Google Scholar]
- (134).Sadeghian RB, Ostrovidov S, Han J, Salehi S, Bahraminejad B, Bae H, Chen M, Khademhosseini A. ACS Sensors. 2016;1:921–928. [Google Scholar]
- (135).Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Safaei TS, Mohamadi RM, Sargent EH, Kelley SO. ACS Appl Mater Interfaces. 2015;7:14165–14169. doi: 10.1021/acsami.5b02404. [DOI] [PubMed] [Google Scholar]
- (137).Chen X, Wang Y, Zhang Y, Chen Z, Liu Y, Li Z, Li J. Anal Chem. 2014;86:4278–4286. doi: 10.1021/ac404070m. [DOI] [PubMed] [Google Scholar]
- (138).Chen J, Xue C, Zhao Y, Chen D, Wu M-H, Wang J. Int J Mol Sci. 2015;16:9804–9830. doi: 10.3390/ijms16059804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Zhu Z, Frey O, Franke F, Haandbæk N, Hierlemann A. Anal Bioanal Chem. 2014;406:7015–7025. doi: 10.1007/s00216-014-7955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Zhu Z, Frey O, Haandbæk N, Franke F, Rudolf F, Hierlemann A. Sci Rep. 2015;5:17180. doi: 10.1038/srep17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Kim J, Cho H, Han S-I, Han K-H. Anal Chem. 2016;88:4857–4863. doi: 10.1021/acs.analchem.6b00570. [DOI] [PubMed] [Google Scholar]
- (142).Koch JA, Baur MB, Woodall EL, Baur JE. Anal Chem. 2012;84:9537–9543. doi: 10.1021/ac302402p. [DOI] [PubMed] [Google Scholar]
- (143).Guo X, Zhu R. Sci Rep. 2016;6:31392. doi: 10.1038/srep31392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Shah P, Zhu X, Zhang X, He J, Li C. ACS Appl Mater Interfaces. 2016;8:5804–5812. doi: 10.1021/acsami.5b11409. [DOI] [PubMed] [Google Scholar]
- (145).Haandbæk N, With O, Bürgel SC, Heer F, Hierlemann A. Lab Chip. 2014;14:3313. doi: 10.1039/c4lc00576g. [DOI] [PubMed] [Google Scholar]
- (146).Frankowski M, Simon P, Bock N, El-Hasni A, Schnakenberg U, Neukammer J. Eng Life Sci. 2015;15:286–296. [Google Scholar]
- (147).Alberti M, Snakenborg D, Lopacinska JM, Dufva M, Kutter JP. Microfluid Nanofluid. 2014;17:263–274. [Google Scholar]
- (148).Zhang M-N, Ding Z, Long Y-T. Analyst. 2015;140:6054–6060. doi: 10.1039/c5an01148e. [DOI] [PubMed] [Google Scholar]
- (149).Filice FP, Li MSM, Henderson JD, Ding Z. Anal Chim Acta. 2016;908:85–94. doi: 10.1016/j.aca.2015.12.027. [DOI] [PubMed] [Google Scholar]
- (150).Bergner S, Wegener J, Matysik F-M. Anal Methods. 2012;4:623–629. [Google Scholar]
- (151).Zenobi R. Science. 2013;342:1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- (152).Heeren RMA. Int J Mass Spectrom. 2015;377:672–680. [Google Scholar]
- (153).Mueller L, Traub H, Jakubowski N, Drescher D, Baranov VI, Kneipp J. Anal Bioanal Chem. 2014;406:6963–6977. doi: 10.1007/s00216-014-8143-7. [DOI] [PubMed] [Google Scholar]
- (154).Fujii T, Matsuda S, Tejedor ML, Esaki T, Sakane I, Mizuno H, Tsuyama N, Masujima T. Nat Protoc. 2015;10:1445–1456. doi: 10.1038/nprot.2015.084. [DOI] [PubMed] [Google Scholar]
- (155).Zhang L, Foreman DP, Grant PA, Shrestha B, Moody SA, Villiers F, Kwak JM, Vertes A. Analyst. 2014;139:5079–5085. doi: 10.1039/c4an01018c. [DOI] [PubMed] [Google Scholar]
- (156).Gong X, Zhao Y, Cai S, Fu S, Yang C, Zhang S, Zhang X. Anal Chem. 2014;86:3809–3816. doi: 10.1021/ac500882e. [DOI] [PubMed] [Google Scholar]
- (157).Fujita H, Esaki T, Masujima T, Hotta A, Kim SH, Noji H, Watanabe TM. RSC Adv. 2015;5:16968–16971. [Google Scholar]
- (158).Gasilova N, Yu Q, Qiao L, Girault HH. Angew Chem, Int Ed. 2014;53:4408–4412. doi: 10.1002/anie.201310795. [DOI] [PubMed] [Google Scholar]
- (159).Krismer J, Sobek J, Steinhoff RF, Fagerer SR, Pabst M, Zenobi R. Appl Environ Microbiol. 2015;81:5546–5551. doi: 10.1128/AEM.01201-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Passarelli MK, Newman CF, Marshall PS, West A, Gilmore IS, Bunch J, Alexander MR, Dollery CT. Anal Chem. 2015;87:6696–6702. doi: 10.1021/acs.analchem.5b00842. [DOI] [PubMed] [Google Scholar]
- (161).Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, et al. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- (162).Ong T-H, Kissick DJ, Jansson ET, Comi T, Romanova EV, Rubakhin SS, Sweedler JV. Anal Chem. 2015;87:7036–7042. doi: 10.1021/acs.analchem.5b01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Jansson ET, Comi TJ, Rubakhin SS, Sweedler JV. ACS Chem Biol. 2016;11:2588–2595. doi: 10.1021/acschembio.6b00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Küster SK, Pabst M, Zenobi R, Dittrich PS. Angew Chem, Int Ed. 2015;54:1671–1675. doi: 10.1002/anie.201409440. [DOI] [PubMed] [Google Scholar]
- (165).Lee JK, Jansson ET, Nam HG, Zare RN. Anal Chem. 2016;88:5453–5461. doi: 10.1021/acs.analchem.6b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Lee CY, Fan Y, Rubakhin SS, Yoon S, Sweedler JV. Sci Rep. 2016;6:26940. doi: 10.1038/srep26940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Walker BN, Stolee JA, Vertes A. Anal Chem. 2012;84:7756–7762. doi: 10.1021/ac301238k. [DOI] [PubMed] [Google Scholar]
- (168).Stopka SA, Rong C, Korte AR, Yadavilli S, Nazarian J, Razunguzwa TT, Morris NJ, Vertes A. Angew Chem. 2016;128:4558–4562. doi: 10.1002/anie.201511691. [DOI] [PubMed] [Google Scholar]
- (169).Bobrowska J, Pabijan J, Wiltowska-Zuber J, Jany BR, Krok F, Awsiuk K, Rysz J, Budkowski A, Lekka M. Anal Biochem. 2016;511:52–60. doi: 10.1016/j.ab.2016.06.011. [DOI] [PubMed] [Google Scholar]
- (170).Robinson MA, Graham DJ, Morrish F, Hockenbery D, Gamble LJ. Biointerphases. 2016;11:02A303. doi: 10.1116/1.4929633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Vanbellingen QP, Elie N, Eller MJ, Della-Negra S, Touboul D, Brunelle A. Rapid Commun Mass Spectrom. 2015;29:1187–1195. doi: 10.1002/rcm.7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Graham DJ, Wilson JT, Lai JJ, Stayton PS, Castner DG. Biointerphases. 2016;11:02A304. doi: 10.1116/1.4934795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Vanbellingen QP, Castellanos A, Rodriguez-Silva M, Paudel I, Chambers JW, Fernandez-Lima FA. J Am Soc Mass Spectrom. 2016;27:2033–2040. doi: 10.1007/s13361-016-1485-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, et al. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Onjiko RM, Moody SA, Nemes P. Proc Natl Acad Sci U S A. 2015;112:6545–6550. doi: 10.1073/pnas.1423682112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).ElAzzouny MA, Evans CR, Burant CF, Kennedy RT. PLoS One. 2015;10:e0129029. doi: 10.1371/journal.pone.0129029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Van Malderen SJM, Vergucht E, De Rijcke M, Janssen C, Vincze L, Vanhaecke F. Anal Chem. 2016;88:5783–5789. doi: 10.1021/acs.analchem.6b00334. [DOI] [PubMed] [Google Scholar]
- (178).Verboket PE, Borovinskaya O, Meyer N, Günther D, Dittrich PS. J Visualized Exp. 2015;97:e52525. doi: 10.3791/52525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Wang H, Wang B, Wang M, Zheng L, Chen H, Chai Z, Zhao Y, Feng W. Analyst. 2015;140:523–531. doi: 10.1039/c4an01610f. [DOI] [PubMed] [Google Scholar]
- (180).Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Fluidigm. Helios. [accessed Oct 2, 2016]; https://www.fluidigm.com/products/helios.
- (182).Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (184).Bendall SC, Simonds EF, Qiu P, Amir E-aD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, et al. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Catena R, Özcan A, Zivanovic N, Bodenmiller B. Cytometry, Part A. 2016;89:491–497. doi: 10.1002/cyto.a.22848. [DOI] [PubMed] [Google Scholar]
- (186).Duan R, Zuo X, Wang S, Quan X, Chen D, Chen Z, Jiang L, Fan C, Xia F. J Am Chem Soc. 2013;135:4604–4607. doi: 10.1021/ja311313b. [DOI] [PubMed] [Google Scholar]
- (187).Gracz AD, Williamson IA, Roche KC, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza R, Gaynor LT, et al. Nat Cell Biol. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Fluidigm. C1. [accessed Oct 2, 2016]; https://www.fluidigm.com/products/c1-system.
- (189).Chen Y-C, Zhang Z, Fouladdel S, Deol Y, Ingram PN, McDermott SP, Azizi E, Wicha MS, Yoon E. Lab Chip. 2016;16:2935–2945. doi: 10.1039/c6lc00062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (190).Burns JC, Kelly MC, Hoa M, Morell RJ, Kelley MW. Nat Commun. 2015;6:8557. doi: 10.1038/ncomms9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Park S, Zhang Y, Lin S, Wang T-H, Yang S. Biotechnol Adv. 2011;29:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Adler M, Wacker R, Niemeyer CM. Analyst. 2008;133:702. doi: 10.1039/b718587c. [DOI] [PubMed] [Google Scholar]
- (193).Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, Westermark B, et al. Cell Rep. 2016;14:380–389. doi: 10.1016/j.celrep.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Li J, Macdonald J. Biosens Bioelectron. 2015;64:196–211. doi: 10.1016/j.bios.2014.08.069. [DOI] [PubMed] [Google Scholar]
- (195).Kunze A, Dilcher M, Abd El Wahed A, Hufert F, Niessner R, Seidel M. Anal Chem. 2016;88:898–905. doi: 10.1021/acs.analchem.5b03540. [DOI] [PubMed] [Google Scholar]
- (196).Xu Y, Li D, Cheng W, Hu R, Sang Y, Yin Y, Ding S, Ju H. Anal Chim Acta. 2016;936:229–235. doi: 10.1016/j.aca.2016.07.007. [DOI] [PubMed] [Google Scholar]
- (197).Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. Nat Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, et al. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Esteban-Fernández de Ávila B, Martín A, Soto F, Lopez-Ramirez MA, Campuzano S, Vásquez-Machado GM, Gao W, Zhang L, Wang J. ACS Nano. 2015;9:6756–6764. doi: 10.1021/acsnano.5b02807. [DOI] [PubMed] [Google Scholar]
- (201).Cermak N, Olcum S, Delgado FF, Wasserman SC, Payer KR, Murakami MA, Knudsen SM, Kimmerling RJ, Stevens MM, Kikuchi Y, Sandikci A, et al. Nat Biotechnol. 2016;34:1052–1059. doi: 10.1038/nbt.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Inomata N, Toda M, Ono T. Lab Chip. 2016;16:3597–3603. doi: 10.1039/c6lc00949b. [DOI] [PubMed] [Google Scholar]
- (203).Glenn DR, Lee K, Park H, Weissleder R, Yacoby A, Lukin MD, Lee H, Walsworth RL, Connolly CB. Nat Methods. 2015;12:736–738. doi: 10.1038/nmeth.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Sun Y, Kim D-H, Simmons CA. Integrative Mechanobiology - Micro- and Nano- Techniques in Cell Mechanobiology. 1st ed. Cambridge University Press; Cambridge, UK: 2015. [Google Scholar]
- (205).Koo H, Park I, Lee Y, Kim HJ, Jung JH, Lee JH, Kim Y, Kim J-H, Park JW. J Am Chem Soc. 2016;138:11664–11671. doi: 10.1021/jacs.6b05048. [DOI] [PubMed] [Google Scholar]
- (206).Guillaume-Gentil O, Grindberg RVV, Kooger R, Dorwling-Carter L, Martinez V, Ossola D, Pilhofer M, Zambelli T, Vorholt JAA. Cell. 2016;166:506–517. doi: 10.1016/j.cell.2016.06.025. [DOI] [PubMed] [Google Scholar]
- (207).Vermeulen P, Cognet L, Lounis B. J Microsc. 2014;254:115–121. doi: 10.1111/jmi.12130. [DOI] [PubMed] [Google Scholar]
- (208).Nieves DJ, Li Y, Fernig DG, Lévy R. R Soc Open Sci. 2015;2:140454. doi: 10.1098/rsos.140454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Issadore D, Chung J, Shao H, Liong M, Ghazani aa, Castro CM, Weissleder R, Lee H. Sci Transl Med. 2012;4:141ra92. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Kashyap A, Autebert J, Delamarche E, Kaigala GV. Sci Rep. 2016;6:29579. doi: 10.1038/srep29579. [DOI] [PMC free article] [PubMed] [Google Scholar]