Abstract
The methylation of cytosine at CG sites in the mammalian genome is dynamically reprogrammed during gametogenesis and preimplantation development. It was previously shown that oocyte-derived DNMT1 (a maintenance methyltransferase) is essential for maintaining and propagating CG methylation at imprinting control regions in preimplantation embryos. In mammalian somatic cells, hemimethylated-CG-binding protein UHRF1 plays a critical role in maintaining CG methylation by recruiting DNMT1 to hemimethylated CG sites. However, the role of UHRF1 in oogenesis and preimplantation development is unknown. In the present study, we show that UHRF1 is mainly, but not exclusively, localized in the cytoplasm of oocytes and preimplantation embryos. However, smaller amounts of UHRF1 existed in the nucleus, consistent with the expected role in DNA methylation. We then generated oocyte-specific Uhrf1 knockout (KO) mice and found that, although oogenesis was itself unaffected, a large proportion of the embryos derived from the KO oocytes died before reaching the blastocyst stage (a maternal effect). Whole genome bisulfite sequencing revealed that blastocysts derived from KO oocytes have a greatly reduced level of CG methylation, suggesting that maternal UHRF1 is essential for maintaining CG methylation, particularly at the imprinting control regions, in preimplantation embryos. Surprisingly, UHRF1 was also found to contribute to de novo CG and non-CG methylation during oocyte growth: in Uhrf1 KO oocytes, transcriptionally-inactive regions gained less methylation, while actively transcribed regions, including the imprinting control regions, were unaffected or only slightly affected. We also found that de novo methylation was defective during the late stage of oocyte growth. To the best of our knowledge, this is the first study to demonstrate the role of UHRF1 in de novo DNA methylation in vivo. Our study reveals multiple functions of UHRF1 during the global epigenetic reprogramming of oocytes and early embryos.
Author summary
The methylation of cytosine at CG sites in the mammalian genome is an epigenetic modification that is important for cell differentiation and embryonic development. During oocyte growth, the actively transcribed regions gain both CG and non-CG methylation. However, after fertilization, such methylation is globally erased, except for certain gene control regions and a subset of retrotransposons that retain CG methylation. We examined the role of UHRF1, a protein essential for the maintenance of CG methylation in somatic cells, in oocytes and preimplantation embryos by generating oocyte-specific Uhrf1 gene knockout mice. We found that oocyte-derived maternal UHRF1 protein was important for nuclear localization of DNMT1 (a maintenance DNA methyltransferase) and for CG maintenance methylation, particularly at the imprinting control regions, in preimplantation embryos. Unexpectedly, we found that the gain in CG and non-CG methylation in oocytes was also affected by Uhrf1 knockout in certain genomic regions. To the best of our knowledge, this is the first study to demonstrate a role of UHRF1 in de novo DNA methylation in vivo. Our study reveals multiple functions of UHRF1 during the global epigenetic reprogramming of oocytes and preimplantation embryos.
Introduction
DNA methylation is a key epigenetic modification that is involved in various cellular processes, including cell differentiation, transposon silencing, genomic imprinting, and carcinogenesis [1,2]. This covalent epigenetic modification normally occurs at the C5 position of the cytosine (C) ring. In most mammalian cell types, the product of this modification, 5-methylcytosine (5mC), almost exclusively occurs in a CG context (CG methylation). However, non-CG methylation exists in certain cells such as oocytes [3–5], brain cells [6], and embryonic stem cells (ESCs) [7,8].
CG methylation is dynamically reprogrammed during mammalian gametogenesis and preimplantation development. In mouse germ cell development, CG methylation is first erased in primordial germ cells, partly due to the low expression of UHRF1 (Ubiquitin-like with PHD and RING finger domains 1) [9], a factor that is essential for maintenance methylation at CG sites [10,11] (see below). The hydroxylation of 5mC also plays a role in removing CG methylation through active or passive demethylation, particularly at the imprinting control regions (ICRs) [12,13], which dictate the parental-origin-specific expression of the imprinted genes in somatic cells [14]. Subsequently, in the female germline, an oocyte-specific methylation pattern is established in GOs by a de novo DNA methyltransferase (DNMT) complex composed of DNMT3A and DNMT3L [4,5,15–17]. De novo methylation continues until the fully grown oocyte (FGO) stage, with the genome of the oocyte accumulating both CG methylation and non-CG methylation [3–5]. It is known that the highly methylated regions in oocytes correspond to the actively transcribed regions [4,18,19], which are marked by histone H3 lysine 36 trimethylation (H3K36me3) [20]. Since DNMT3A recognizes H3K36me3 through its PWWP domain [21], the enzyme appears to methylate regions that are premarked with this modification. De novo methylation in oocytes also requires histone replacement, since the disruption of the H3.3 chaperone HIRA results in a global reduction in CG methylation [22].
After fertilization, with the exception of certain regions such as the ICRs and a subset of retrotransposons, the gamete-specific methylation patterns are globally erased [4,23]. Notably, the paternal genome undergoes 5mC hydroxylation, followed by active or passive demethylation [24–27]. Several factors, including ZFP57, TRIM28, DPPA3, and maintenance-type DNA methyltransferase DNMT1 are required for maintaining CG methylation at the ICRs [28–33] against the wave of hydroxylation-dependent and hydroxylation-independent demethylation. In a study using oocyte-specific Dnmt1 knockout (KO) mice, we previously reported that, although DNMT1 is mainly localized in the cytoplasm, a small amount of oocyte-derived maternal DNMT1 in the cell nucleus engages in CG maintenance methylation at the ICRs in preimplantation embryos [31].
Mouse UHRF1 is a multi-domain protein that is essential for the CG maintenance methylation in proliferating somatic cells [10,11]. The conventional KO of Uhrf1 results in an embryonic lethal phenotype, which is accompanied by global genomic hypomethylation [10]. UHRF1 contains five major protein domains: an ubiquitin-like (UBL) domain, a tandem Tudor domain (TTD), a plant homeodomain (PHD) finger, a SET and RING associated (SRA) domain, and a really interesting new gene (RING) finger [34–36]. UHRF1 recognizes hemimethylated CG sites through its SRA domain [37–39], and ubiquitinates H3K18 and H3K23 via the E3 ubiquitin ligase activity of its RING finger [40,41]. Subsequently, DNMT1 recognizes H3K18 and/or other ubiquitinated lysine residues and methylates the unmodified C of hemimethylated CG sites to copy the existing methylation patterns. In addition, the TTD and PHD finger of this protein recognize H3K9me2/me3 [35,36,42,43] and unmodified H3R2 [44,45], respectively, suggesting their roles in the proper localization of this protein.
Unlike DNMT1, however, the role of UHRF1 during oogenesis and preimplantation development has not been explored. We therefore generated oocyte-specific Uhrf1 conditional KO mice and examined the impact on DNA methylation in oocytes and preimplantation embryos. Our study revealed an expected role of maternal UHRF1 in preimplantation embryos and an unexpected new function in GOs.
Results
The subcellular localization of UHRF1 in oocytes and preimplantation embryos
To reveal the role of UHRF1 proteins in mouse oocytes and preimplantation embryos, we first examined the expression and subcellular localization of UHRF1 by immunostaining. Like DNMT1 [31], UHRF1 was mainly localized in the cytoplasm of GOs and preimplantation embryos (Fig 1A and 1B). However, a weaker signal was detected in the nucleus. To examine the timing of the zygotic activation of the Uhrf1 and Dnmt1 genes, we reprocessed published polyA(+) mRNA-seq data from F1 hybrid embryos [46], where expressed alleles could be distinguished based on the presence of single nucleotide polymorphisms (SNPs). While the levels of maternal Uhrf1 and Dnmt1 mRNAs had decreased by the 16-cell stage, the total levels of both mRNAs increased after this stage (S1 Fig). The Uhrf1 and Dnmt1 mRNAs, which were derived from the paternal allele, were first detected at the mid 2-cell stage (S1 Fig), pinpointing the timing of the zygotic activation of these genes.
Fig 1. Expression and subcellular localization of UHRF1 in oocytes and preimplantation embryos.
(A,B) Immunostaining of C57BL/6J developing oocytes (A), zygote to blastocyst stage embryos (B), Uhrf1 KO oocytes (C), and Uhrf1 mat-KO embryos (D) performed with an anti-UHRF1 antibody (Th-10a [green]). Each experiment was done with at least two batches of oocytes/embryos, and the total number of oocytes/embryos examined at each stage exceeded 20. The cell nucleus was counterstained with DAPI (blue). NGO, <20 μm; GO, 20–50 μm; FGO, >70 μm. Scale bar, 20 μm.
We generated oocyte-specific Uhrf1 KO mice carrying a zona pellucida glycoprotein 3 (Zp3)-Cre transgene [Uhrf12lox/2lox, Zp3-Cre] (S2A and S2B Fig). The Zp3-Cre transgene is expressed exclusively in GOs [47]. No significant defects were observed in the morphology, size, or number of Uhrf1 KO [Uhrf11lox/1lox] FGOs (S2C Fig). As expected, virtually no Uhrf1 mRNA (by a quantitative reverse transcription-polymerase chain reaction; qRT-PCR) or UHRF1 protein (by Western blotting) was detected in the KO FGOs (S2D and S2E Fig). The predicted truncated protein (amino acid 1–133) was not expressed, as its coding exons (2–3) were not detected at the mRNA level (S2D Fig), probably due to nonsense-mediated mRNA decay [48]. The loss of UHRF1 protein was confirmed by immunostaining as early as the non-growing oocyte (NGO) stage (Fig 1C and S2F Fig). The expression of Dnmt1 was not affected by the Uhrf1 KO (S2E Fig). When Uhrf1 KO oocytes were fertilized with wild-type sperm in vitro (in vitro fertilization, IVF), only 20% of the embryos (Uhrf1 maternal-KO or mat-KO [Uhrf11lox/+]) developed to the expanded blastocyst stage (S3A Fig), suggesting that maternal UHRF1 plays a critical role in preimplantation development. This phenotype was much more severe than that of Dnmt1 mat-KO [Dnmt11lox/+] embryos [31,32] (S3A Fig). Virtually no UHRF1 protein was detected by immunostaining even in the surviving embryos (Fig 1D).
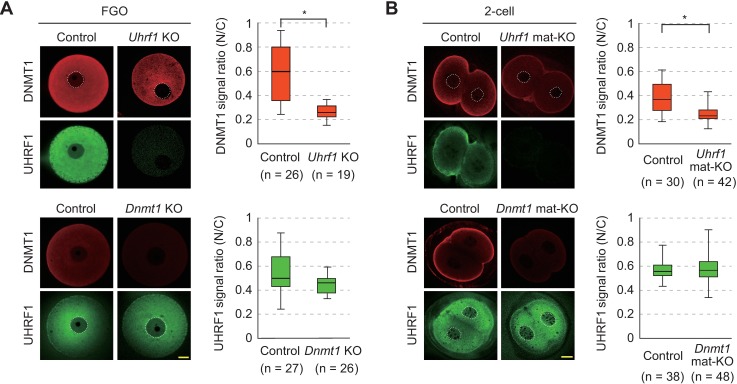
Interestingly, while the absence of DNMT1 did not affect the subcellular localization of UHRF1, the absence of UHRF1 clearly decreased the level of DNMT1 in the nucleus of FGOs (Fig 2A). The absence of maternal UHRF1 also decreased the level of nuclear DNMT1 in 2-cell embryos (Fig 2B), suggesting that the nuclear localization of DNMT1 is dependent on UHRF1.
Fig 2. Dependence of nuclear localization of DNMT1 on UHRF1.
(A,B) Immunostaining of control [Uhrf12lox/2lox or Dnmt12lox/2lox], Uhrf1 KO, and Dnmt1 KO FGOs (A) and control [Uhrf12lox/+ or Dnmt12lox/+], Uhrf1 mat-KO, and Dnmt1 mat-KO 2-cell embryos (B) was performed with anti-UHRF1 antibodies (green) or anti-DNMT1 antibodies (red). The nuclear/cytoplasmic signal ratios (N/C) were calculated using the mean signal intensities obtained by linear scanning across the cells. Each experiment was done with at least two batches of oocytes/embryos, and the total number (n) of oocytes/embryos is indicated. Each box indicates the 25–75 percentile and the bar in each box indicates the median. Asterisk, P <0.01 (Mann-Whitney U test).
The role of maternal UHRF1 in the CG maintenance methylation in preimplantation embryos
In preimplantation embryos, both the paternal and maternal genomes are globally demethylated, while the ICRs and certain retrotransposons (such as intracisternal A particle [IAP] elements) retain CG methylation [4,23,31,49,50]. To examine whether the maternal UHRF1 proteins play a role in CG maintenance methylation in these sequences, whole genome bisulfite sequencing (WGBS) was performed with control [Uhrf12lox/+], Uhrf1 mat-KO, and Dnmt1 mat-KO blastocysts generated by IVF. Only well-developed Uhrf1 mat-KO blastocysts were pooled (S3A Fig, indicated as expanded) and subjected to the analysis. Since KO females were of the C57BL/6J background (Mus musculus domesticus) and the wild-type sperm was from JF1 males (Mus musculus molossinus), the parental alleles were distinguishable where SNPs were available. We used the post-bisulfite adaptor tagging (PBAT) method, which was amplification-free and applicable to a limited amount of DNA, to construct the WGBS libraries [51].
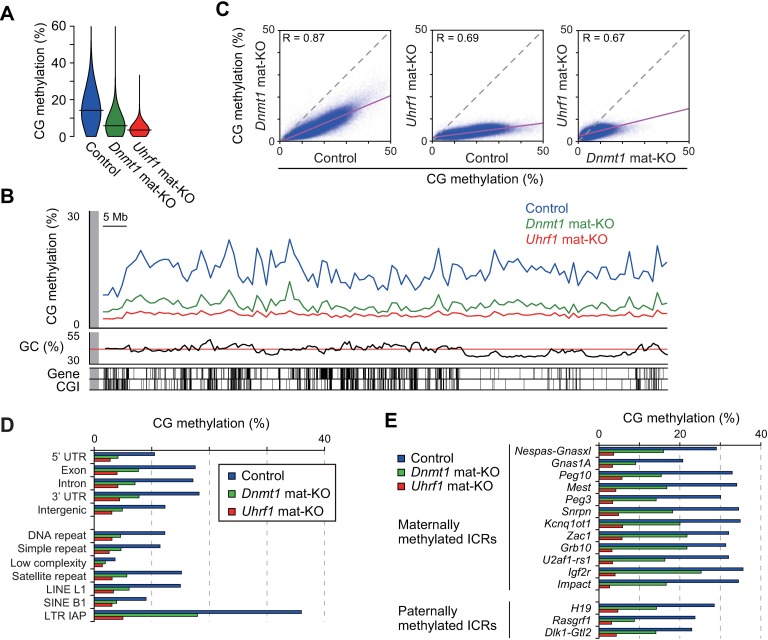
Our WGBS with replicate samples basically confirmed good reproducibility, but the Uhrf1 mat-KO blastocysts showed a lower correlation, likely due to slight differences in developmental stage and extremely low levels of CG methylation, which are sensitive to small variations (S1 and S2 Tables and S4 Fig). The global CG methylation levels of the control, Uhrf1 mat-KO, and Dnmt1 mat-KO blastocysts were 14.3%, 3.5%, and 5.7%, respectively (Fig 3A and S2 Table, combined replicate data). Thus, the absence of maternal UHRF1 reduced the level of global CG methylation to one-fourth of the control level. The fact that the global methylation decreased by more than 50% suggests that not only the maternal genome but also the paternal genome, which was derived from the wild-type sperm, was affected after fertilization. Indeed, this was confirmed by the allele-specific methylation analysis using SNPs (see later). The loss of CG methylation was milder in Dnmt1 mat-KO blastocysts. The non-CG methylation level was basically unaffected by the absence of either maternal UHRF1 or maternal DNMT1 (S2 Table).
Fig 3. The role of maternal UHRF1 in CG maintenance methylation in preimplantation embryos.
(A) Violin plots showing the distribution of the 10-kb genomic regions with different CG methylation levels in control [Uhrf12lox/+], Uhrf1 mat-KO, and Dnmt1 mat-KO blastocysts. The horizontal bars indicate the mean CG methylation levels. (B) The genomic distribution of CG methylation across chromosome 14 in control and mat-KO blastocysts. The CG methylation levels of 1-Mb windows are shown. GC contents, RefSeq genes, and CGIs are shown at the bottom. (C) The correlations between the CG methylation levels of the 10-kb genomic windows in the control and mat-KO blastocysts. (D) The CG methylation levels of respective portions of the genic regions, intergenic regions, and various repetitive sequences in control and mat-KO blastocysts. (E) The CG methylation levels of the ICRs in control and mat-KO blastocysts.
The analysis of CG methylation in 1-megabase (Mb) or 10-kilobase (kb) windows revealed that the absence of maternal UHRF1 or DNMT1 caused a basically global and proportional decrease in CG methylation across the genome (Fig 3B and 3C and S3B Fig). The genic regions, intergenic regions, repetitive elements (including IAPs), and CpG islands (CGIs) were all affected (Fig 3D and S3C Fig). The expected allele-specific CG methylation was confirmed in 12 (ten maternally methylated and two paternally methylated) of the 15 ICRs examined in the control blastocysts (S3D Fig); SNPs were not available for the rest of the ICRs. As we reported previously [31], the level of CG methylation was decreased to half that of the control blastocysts at the ICRs in Dnmt1 mat-KO blastocysts. In contrast, the CG maintenance methylation at the ICRs was more severely affected in Uhrf1 mat-KO blastocysts, to less than one-fourth the level observed in the control blastocysts (Fig 3E and S3D Fig). The loss of methylation from both parental genomes in mat-KO blastocysts indicates a role for maternal Uhrf1 in the maintenance of CG methylation.
The role of UHRF1 in de novo CG methylation during oocyte growth
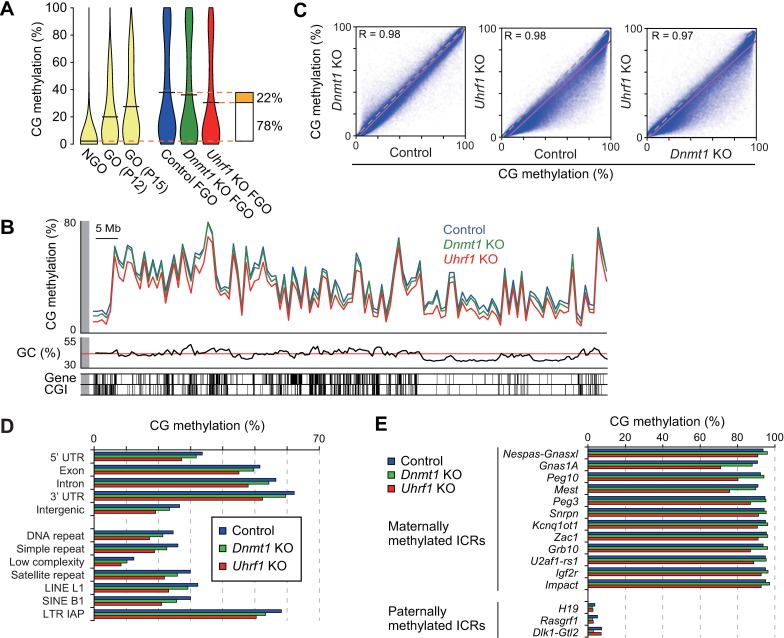
We next examined whether this protein has any effect on CG methylation in FGOs. We therefore performed WGBS with control [Uhrf12lox/2lox] and Uhrf1 KO FGOs (S1 and S2 Tables and S4 Fig). We found that the global CG methylation level was 30.8% in Uhrf1 KO FGOs, which was 7.9% lower in comparison to control FGOs (38.7%) (Fig 4A and S2 Table). This was surprising as no DNA replication occurs during oocyte growth (meiotic prophase I), during which only de novo methylation is observed. Since the CG methylation level in NGOs was reported to be 2.3% [5], these results suggested that UHRF1 was required for approximately one-fourth of the total increase in CG methylation (Fig 4A). We previously reported that the slight decrease in CG methylation in Dnmt1 KO FGOs (36.0% global CG methylation level compared to 38.7% in the control FGOs) (Fig 4A and S2 Table) could be explained by increased hemimethylation [5]. Although the proportion of hemimethylated sites among the highly methylated CG sites was 6.1% greater in Uhrf1 KO FGOs (12.9%) than it was in Dnmt1 KO FGOs (6.8%) (S5A Fig) (see Materials and Methods), this difference was not sufficient to explain the decrease in CG methylation that was observed in the Uhrf1 KO FGOs.
Fig 4. The role of UHRF1 in de novo CG methylation during oocyte growth.
(A) Violin plots showing the distribution of the 10-kb genomic regions with different CG methylation levels in NGOs [5], GOs (P12 and P15) [53], and control [Uhrf12lox/2lox], Dnmt1 KO, and Uhrf1 KO FGOs. The horizontal bars indicate the mean CG methylation levels. (B) The genomic distribution of CG methylation across chromosome 14 in control and KO FGOs. The CG methylation levels of the 1-Mb windows are shown. GC contents, RefSeq genes, and CGIs are shown at the bottom. (C) The correlations between the CG methylation levels of the 10-kb genomic windows in control and KO FGOs. (D) The CG methylation levels of respective portions of the genic regions, intergenic regions, and various repetitive sequences in control and respective KO FGOs. (E) The CG methylation levels of the ICRs in control and KO FGOs.
We previously reported that, in FGOs, nearly 65% of all 5mCs occurred at non-CG sites [5]. This was confirmed in our control FGOs (S5B Fig and S2 Table) and, surprisingly, non-CG methylation was decreased to 85% of the level of control FGOs in Uhrf1 KO FGOs (S5B–S5D Fig and S2 Table). This corresponded to a 0.6% reduction in non-CG methylation on a per site basis (from 3.2% to 2.6%) (S2 Table). Dnmt1 KO FGOs showed an increased non-CG methylation level, possibly due to the compensatory upregulation of Dnmt3a [5]. With regard to the sequence context, the CHG and CHH sites (where H represents the base A, C, or T) were equally affected (S5B Fig). These results strongly suggest that UHRF1 plays a role in de novo CG and non-CG methylation, not only through the recognition of hemimethylated CG sites, but also through an unknown mechanism.
To examine whether any regions of the genome are preferred for the de novo CG methylation involving UHRF1, we determined the CG methylation levels in 1-Mb windows and found that the levels in Uhrf1 KO FGOs decreased across the genome in comparison to control FGOs (Fig 4B and S6A Fig). However, subsequent analyses in 10-kb windows revealed that the loss of CG methylation was most remarkable in genomic regions that showed moderate levels of CG methylation (Fig 4C). In addition, there was a positive correlation between the losses in CG and non-CG methylation in the 10-kb genomic windows (S6B Fig). Various repetitive elements, including IAPs, were moderately affected (Fig 4D). Many CGIs were known to be methylated in FGOs [49] and, again, moderately methylated CGIs were the most affected in Uhrf1 KO FGOs (S6C Fig). In contrast, the maternally methylated ICRs, which represent the most highly methylated regions in the FGO genome, were only slightly affected by the absence of UHRF1, except the Gnas1A, Peg10, and Mest ICRs, which showed significant reduction in CG methylation (p = 6.49 x 10−121, 1.85 x 10−41, and 2.68 x 10−81, respectively; kai-squared test) (Fig 4E).
UHRF1 facilitates de novo methylation in inactive regions during late oocyte growth
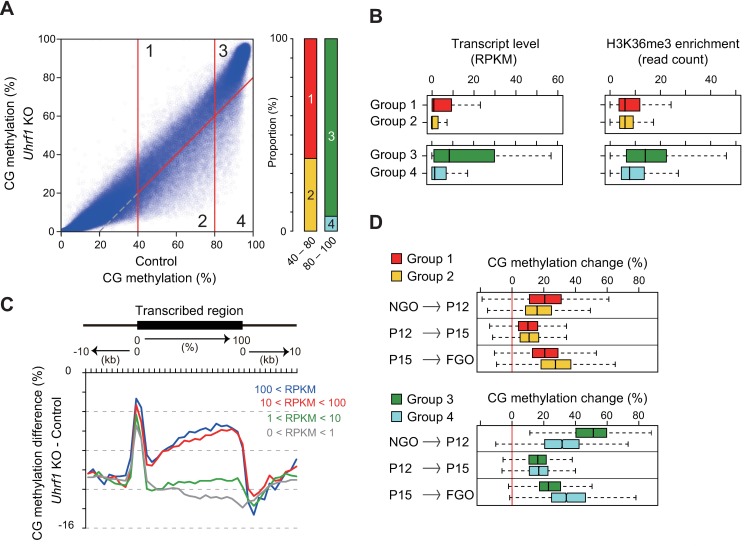
As a first step to understand how UHRF1 is involved in de novo CG and non-CG methylation in oocytes, we attempted to explore the features of the regions affected in Uhrf1 KO FGOs in greater detail. We first identified the genomic regions (10-kb windows) with a CG methylation level of ≥40% in control FGOs and divided them into four groups: moderately (40–80%) methylated regions that were unaffected (decrease <20%) by Uhrf1 KO (Group 1); moderately methylated regions that were affected (decrease ≥20%) by KO (Group 2); highly (≥80%) methylated regions that were unaffected by KO (Group 3); highly methylated regions that were affected by KO (Group 4) (Fig 5A and S3 Table). Of the moderately methylated regions (Groups 1 and 2), 35% of the cases were affected (Group 2) but, in contrast, only 9% of the highly methylated regions (Groups 3 and 4) were affected (Group 4) (Fig 5A), confirming that the moderately methylated regions were preferentially affected.
Fig 5. UHRF1 facilitates de novo methylation in inactive regions during late oocyte growth.
(A) The classification of the 10-kb genomic regions that were moderately (40–80%) or highly (≥80%) methylated in the control FGOs, based on the extent of methylation change (≥20% or not) in Uhrf1 KO FGOs (Groups 1–4). The right panel shows the proportion of moderately and highly methylated 10-kb regions belonging to each group. (B) The distribution of the transcript levels (RPKM) and H3K36me3 enrichment levels [20] (corrected read count) of the 10-kb regions belonging to each group. The bar in each box indicates the median value. (C) Differences in CG methylation across the transcribed regions between control and Uhrf1 KO FGOs. The transcribed regions were classified into four categories based on the expression levels (RPKM) in control FGOs and the results for the respective groups are shown separately. (D) The changes in CG methylation occurring in the three different stages of oocyte growth were determined for the 10-kb regions of the respective groups.
Transcription-coupled de novo CG methylation is predominant in oocytes [4,18,19]. To examine the relationship between transcription and UHRF1-dependent de novo CG methylation, we performed RNA-seq with replicate samples from control [Uhrf12lox/2lox] and Uhrf1 KO FGOs (S4 Table). Overall, the transcription profiles of these FGOs were very similar (R = 0.969, combined replicate data, S6D Fig), and we confirmed that the highly methylated regions produced more transcripts (Fig 5B). Also, using previously published H3K36me3 chromatin immunoprecipitation-sequencing data from GOs (≥30–65 μm) [20], we found that the highly methylated regions were preferentially marked with this transcription-coupled histone mark [52] (Fig 5B). We then examined the CG methylation changes in Uhrf1 KO FGOs in the transcribed regions and found that untranscribed or only lowly transcribed regions were the most severely affected (Fig 5C).
Lastly, we used the published WGBS data from NGOs collected at the newborn stage [5] and GOs collected on postnatal days 12 (P12) and P15 [53] to examine the timing of de novo CG methylation in affected and unaffected regions. In comparison to the regions that were unaffected by Uhrf1 KO (Groups 1 and 3), the affected regions gained less methylation in the early phase of oocyte growth (between newborn NGO and P12 GO) and underwent greater methylation in the later phase (between P15 GO to FGO) (Fig 5D). Interestingly, the three maternally methylated ICRs more affected by Uhrf1 KO (the Gnas1A, Peg10, and Mest ICRs) compared to others (Fig 4E) also gained more methylation in the later phase (S6E Fig). These results indicate that UHRF1 accelerates de novo CG methylation in untranscribed or lowly transcribed regions during the late stage of oocyte growth.
The relative contribution to de novo CG methylation and maintenance CG methylation
Since UHRF1 was found to be involved in de novo CG (and non-CG) methylation in GOs, we attempted to determine what proportion of the loss of CG methylation in Uhrf1 mat-KO blastocysts (Fig 3) was explained by the de novo activity and what proportion was explained by the maintenance activity. The de novo activity only affects the maternal genome, while the maintenance activity affects both parental genomes: we therefore determined the CG methylation level of each parental genome in control and Uhrf1 mat-KO blastocysts using SNPs. In the control blastocysts, the CG methylation level of the maternal genome was 14.3%, which was 2.3% lower than that in the paternal genome (16.6%) (S7A Fig). (The observed levels were lower than those of the inner cell mass cells (20% and 21% for the maternal and paternal genome, respectively) [50], perhaps because trophectoderm cells display lower methylation.) In Uhrf1 mat-KO blastocysts, the CG methylation levels of the maternal and paternal genomes were decreased to 3.1% and 4.5%, respectively (S7A Fig). (Since SNP-associated CG sites were more frequently found in the genic regions, where the CG methylation level was high, the average CG methylation level detected in the parental genomes (3.8%) was slightly higher than that in the whole genome (3.5%).)
The CG methylation level in control and Uhrf1 KO FGOs was 38.7% and 30.8%, respectively (S2 Table). These levels correspond to those of the maternal genome at the time of fertilization. Since the methylation level of the maternal genome was 14.3% in control blastocysts (see above), this genome undergoes a ×0.37 (14.3%/38.7%) methylation change, mainly due to replication-coupled dilution during preimplantation development (S7B Fig). (Hydroxylation-dependent demethylation selectively occurs on the paternal genome [24–27].) If there was no maternal effect caused by oocyte-specific Uhrf1 KO and if we apply the demethylation rate determined above to the maternal genome derived from Uhrf1 KO FGOs, then the expected methylation level would be 11.4% (= 30.8% ×0.37) (S7B Fig). Thus, the de novo activity would account for 2.9% (= 14.3%–11.4%) of the total methylation loss of the maternal genome (11.2% [= 14.3%–3.1%]) in Uhrf1 mat-KO blastocysts). The rest of the loss (8.3% [= 11.2%–2.9%]) would be attributable to the maintenance activity in preimplantation embryos (S7B Fig). Although the precise values may be subject to variations between the samples (S4 Fig), these results show that the loss of CG methylation in the Uhrf1 mat-KO blastocysts can largely be explained by the loss of the maintenance activity in early embryos.
Discussion
In the present study, we found that the UHRF1 produced in oocytes has an important role in shaping the DNA methylation landscape in oocytes and preimplantation embryos. We first showed that, like DNMT1 [31], UHRF1 is mainly, but not exclusively, localized in the cytoplasm during oocyte growth and preimplantation development. Why should epigenetic regulators, which in most cases exert their function in the nucleus, exist in the cytoplasm during this period? For DNMT1, a few studies have addressed the mechanism of cytoplasmic localization in early embryos [54]; however its function in the cytoplasm is still unknown. Furthermore, CG maintenance methylation and other events that are known to involve UHRF1 (for example, DNA repair [55–57]) occur in the nucleus and thus the biological significance of cytoplasmic UHRF1 remains to be clarified. As expected, however, a small amount of UHRF1 did exist in the nucleus of oocytes and early embryos, as did DNMT1 [31]. Interestingly, the nuclear localization of DNMT1 was dependent on UHRF1, as oocyte-specific Uhrf1 KO resulted in a greatly reduced DNMT1 signal in the nucleus. A previous study in ESCs showed that DNMT1 recruitment to specific target loci, but not nuclear localization, is dependent on UHRF1 [10,11]. Thus, the nuclear entry and/or retention of DNMT1 is regulated differently in different cell types.
Uhrf1 mat-KO embryos showed partial preimplantation lethality, a phenotype that is much more severe than that of Dnmt1 mat-KO embryos. Our WGBS showed that the global CG methylation level is more greatly reduced in Uhrf1 mat-KO blastocysts (×0.25 in comparison to control blastocysts) than it is in Dnmt1 mat-KO blastocysts (×0.36 in comparison to control). The loss of CG methylation at the ICRs and IAP elements in the Dnmt1 mat-KO embryos was approximately ×0.50 (see also [31]); however, the loss in Uhrf1 mat-KO embryos was greater (×0.25 or less). These results strongly suggest that maternal UHRF1 is essential for imprint maintenance and IAP repression in preimplantation embryos. It is known that the repressed alleles of the ICRs and IAP elements are marked with H3K9me3 in ESCs and preimplantation embryos [58,59]. These results suggest that UHRF1 may be recruited to specific targets through H3K9me3 recognition via its TTD [35,36,42,43]. However, mice expressing a mutated UHRF1 protein with a defective TTD exhibit normal viability/fertility with an only ~10% reduction in CG methylation [60]. Thus, the mechanism through which UHRF1 is recruited to the targets (particularly the ICRs and IAP elements) in the preimplantation embryos remains an open question.
An unexpected discovery of the present study was the involvement of UHRF1 in de novo CG and non-CG methylation in GOs. We previously reported that DNMT1 is important for the completion of de novo CG methylation in oocytes: DNMT1 appears to act on the hemimethylated CG sites left behind during the de novo methylation process and make them fully methylated [5]. UHRF1 may contribute to this process by efficiently recognizing hemimethylated CG sites since the proportion of hemimethylated CG sites among the highly methylated sites was increased in Uhrf1 KO FGOs. This increase in hemimethylated CG sites was greater than that observed in Dnmt1 KO FGOs. Unexpectedly, but consistent with the above-mentioned findings, we found that not only CG methylation, but also non-CG methylation, was affected in Uhrf1 KO FGOs. Thus we speculate that UHRF1 may be directly involved in de novo CG and non-CG methylation, which are both mediated by the DNMT3A-DNMT3L complex in GOs [5].
Since UHRF1 can interact with DNMT3A and DNMT3B in ESCs and silence exogenous promoters [61], it may interact with these enzymes in oocytes as well and facilitate de novo methylation. Interestingly, however, UHRF1 only played a limited role in the establishment of CG methylation at the maternally methylated ICRs: rather, the protein was important for de novo CG and non-CG methylation in the moderately methylated genomic regions. These regions were mostly untranscribed, lacked H3K36me3 marking, and tended to be de novo methylated in a late stage of oocyte growth. Interestingly, among the maternally methylated ICRs, those that were less resistant to UHRF1 depletion normally gained CG methylation in the late stage of oocyte growth. At present, the mechanism through which UHRF1 is recruited to these regions and through which it engages in de novo CG and non-CG methylation remains to be elucidated.
In conclusion, our study showed the importance of maternal UHRF1 in CG maintenance methylation during preimplantation development and in de novo CG and non-CG methylation in transcriptionally inactive regions during oocyte growth. To the best of our knowledge, this is the first study to show the involvement of UHRF1 in de novo methylation in vivo. We believe that our findings give further insight into the epigenetic reprogramming in early development and provide a basis for further improvements in reproductive technologies and regenerative medicine.
Materials and methods
Ethics statement
Mouse husbandry and all of the mouse experiments were carried out under the ethical guidelines of Kyushu University. The mice were euthanized by carbon dioxide asphyxiation. The protocols were approved by the Institutional Animal Care and Use Committee of Kyushu University.
KO mice
We generated ESCs carrying a loxP site in intron 3 and a neomycin selection cassette flanked by loxP sites in intron 5 of Uhrf1 [Uhrf13lox/+]. Uhrf13lox/+ mice were obtained by chimera formation and germline transmission. Because the Uhrf13lox/3lox males were infertile, we crossed females of this genotype with EIIA-Cre males [62] to remove the neomycin cassette and generated Uhrf12lox/+ mice. Uhrf12lox/2lox mice were crossed with mice carrying the Zp3-Cre transgene [47] to generate oocyte-specific Uhrf1 KO mice [Uhrf12lox/2lox, Zp3-Cre]. The oocyte-specific Dnmt1 KO mice [Dnmt12lox/2lox, Zp3-Cre] have been described previously [31,63]. The mice were genotyped by PCR under standard conditions using the primers listed in S5 Table. All of the KO mice were basically of the C57BL/6J background (Mus musculus domesticus).
Oocyte collection, IVF, and embryo culture
NGOs and GOs were obtained from the ovaries on P10. FGOs were obtained from the ovaries at 8–12 weeks. Preimplantation embryos were obtained by IVF. Females aged ≥8 weeks were injected with 7.5 U of pregnant mare serum gonadotropin; 48 h later, they were injected with 7.5 U of human chorionic gonadotropin to induce super ovulation. Cumulus-oocyte complexes were collected from the oviduct and fertilized with C57BL/6J sperm for immunostaining and Western blotting or with JF1 (Mus musculus molossinus) sperm for WGBS. Cumulus cells were carefully removed by washing with phosphate-buffered saline (PBS), and fertilized embryos were cultured in KSOM medium (EmbryoMax KSOM Medium (1X) w/ 1/2 Amino Acids, Merck Millipore) at 37°C with 5% CO2.
Western blotting
Twenty FGOs were lysed in a sample buffer (62.5 mM Tris-HCl (pH 6.8), 0.5× PBS, 2% SDS, 10% glycerol, and 5% 2-mercaptoethanol) and sonicated. Proteins were denatured by heating at 95°C for 5 min, separated by electrophoresis on a 10% SDS polyacrylamide gel, and transferred onto a nitrocellulose membrane (Amersham). The blots were blocked with 5% skimmed milk, incubated with anti-UHRF1 rabbit polyclonal antibody (M-132, Santa Cruz), anti-DNMT1 rabbit polyclonal antibody (a kind gift from Shoji Tajima), or anti-β-actin mouse monoclonal antibody (AC-15, Santa Cruz) (1:10,000 dilution each). After several washes, the blots were incubated with HRP-conjugated anti-rabbit or mouse IgG antibody (1:10,000 dilution), and detected using Chemi-Lumi One Ultra reagent (11644–40, Nacalai Tesque) and an LAS-3000 lumino-image analyzer (Fujifilm).
Immunostaining
Embryos and oocytes were fixed with 4% paraformaldehyde in PBS at room temperature for 30 min and washed with PBS three times. After incubation with a pretreatment buffer (1% bovine serum albumin and 2% Triton-X100 in PBS) at room temperature for 15 min, the embryos and oocytes were incubated at 4°C overnight with anti-UHRF1 rat monoclonal (Th-10a, MBL) antibody (1:500 dilution) or anti-DNMT1 rabbit polyclonal antibody (1:1000 dilution). After washing three times, the embryos and oocytes were incubated with fluorescence-labeled secondary antibody (1:1000 dilution) at room temperature for 30 min. The secondary antibody was CF488 donkey anti-rabbit IgG (H+L) antibody (20015, Biotium Inc), CF488A donkey anti-rat IgG (H+L) antibody (20027, Biotium Inc), or CF594 donkey anti-rabbit IgG (H+L) antibody (20152, Biotium Inc). After washing three times, oocytes and embryos were mounted in VECTASHIELD medium with DAPI (Vector Laboratory) and observed using an LSM510 or LSM700 confocal laser scanning microscope (Carl Zeiss) with a 63× objective lens and a 10× ocular lens. The signal intensities were measured using the ZEN 2012 (blue edition) software program (Carl Zeiss). The nuclear/cytoplasmic signal ratios were calculated using the mean nuclear and cytoplasmic signal intensities.
The qRT-PCR
Total RNA was extracted from 100 FGOs using AllPrep DNA/RNA Mini kit (QIAGEN) and reverse transcribed using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). A qRT-PCR was carried out using a KAPA SYBR Fast qPCR kit (Kapa Biosystems) in Thermal Cycler Dice Real Time System Single (TaKaRa) in accordance with the manufacturer's protocol. The primers that were used are listed in S5 Table. The relative gene expression was quantified using the comparative cycle threshold method, with the Gapdh expression used for normalization.
WGBS and the data analysis
WGBS libraries were prepared using the PBAT method as described in our previous reports [5,51,64]. Five hundred to 1,000 oocytes or 30–40 blastocysts were spiked with 0.1 ng of lambda phage DNA (Promega) and subjected to bisulfite conversion. The concentrations of the PBAT libraries were measured by a qPCR using a KAPA Illumina Library Quantification kit (Kapa Biosystems). Cluster generation and sequencing were performed in a single-read mode using a TruSeq SR Cluster kit v3-cBot-HS (Illumina) and TruSeq SBS kit v3-HS (Illumina) according to manufacturer’s protocols. The libraries were sequenced on a HiSeq 2500 or HiSeq 1500 equipped with HCS v2.0.5 and RTA v1.17.20 (suitable for WGBS) [65] to generate 101-nucleotide single-end reads. We trimmed the raw sequence reads to 96 bases by removing the adapter sequences from the 5’ end and one base from the 3’ end. The resulting reads were aligned to the reference mouse genome (mm10) using Bismark v0.14.2 [66]. The seed length was 28, the maximum number of mismatches permitted in the seed was 1, and the “—pbat” option was selected. Only uniquely aligned reads were analyzed. We estimated the bisulfite conversion rate using reads that were uniquely aligned to the lambda phage genome (accession no. J02459). The sequences and information of the chromosomes, RefSeq genes, CGIs, and repetitive elements were downloaded from the UCSC genome browser [67]. Throughout this paper, the global CG methylation levels refer to the weighted levels, which take the sequencing depth into account [68].
Estimating the frequency of hemimethylated CG sites
The proportion of hemimethylated sites among the highly methylated CG sites was estimated as previously described [5]. In brief, we selected CG sites with a methylation level of ≥70% on either strand that showed a read depth of ≥10. The CG sites with a significant difference in methylation (Fisher’s exact test, p <0.05) were defined hemimethylated.
RNA-seq and the data analysis
Total RNA was extracted from 30 oocytes. Whole transcript amplification and library construction were performed as described previously [69]. The libraries were sequenced on a HiSeq 2000 to generate 50-nucleotide single-end reads. We trimmed the raw sequence reads by removing poly-A tails and low quality bases from the 3’ end. The resulting sequences were aligned to the reference mouse genome (mm10) using the TopHat software program (v2.0.14) [70]. We used a gtf file of published oocyte transcripts [19] for mapping. The mapped reads were counted using featureCounts [71] to calculate the reads per kb per million mapped reads (RPKM) values.
Accession numbers
The sequence data sets supporting the results of this article are available in the DDBJ Sequence Read Archive under accession number DRA005849.
Supporting information
The expression of Uhrf1 and Dnmt1 mRNAs from the maternal and paternal alleles was analyzed separately using published data from (CAST/EiJ × C57BL/6J) F1 hybrid embryos (GSE45719) [46]. An enlarged view of the Uhrf1 and Dnmt1 expression from the zygote to the late 4-cell stage is shown on the right. The expression levels are shown by the RPKM values. Error bar, standard error.
(EPS)
(A) A schematic representation of the genomic organization of the wild-type (WT), 2lox, and 1lox Uhrf1 alleles. The filled boxes indicate the protein-coding regions. (B) The predicted domain architecture of the WT and mutated UHRF1 proteins. The immunogen used to generate the anti-UHRF1 antibody M-132 (used for Western blotting) is shown. (C) The morphology of control [Uhrf12lox/2lox] and Uhrf1 KO FGOs. Scale bar, 100 μm. (D) The expression of Uhrf1 and the Dnmt family mRNAs in control and Uhrf1 KO FGOs was measured by qRT-PCR. Two portions of Uhrf1 mRNA were amplified for quantification. The expression of Gapdh was used for normalization. (E) The detection of UHRF1 and DNMT1o proteins in control and Uhrf1 KO FGOs by Western blotting using M-132 and anti-DNMT1 antibodies. β-actin was used as a loading control. (F) The immunostaining signal intensities obtained with an anti-UHRF1 antibody (Th-10a) in control and Uhrf1 KO developing oocytes.
(EPS)
(A) The morphology of control [Uhrf12lox/+], Dnmt1 mat-KO, and Uhrf1 mat-KO embryos at 96 h after IVF. Among the Uhrf1 mat-KO embryos, those that developed to the expanded blastocyst stage are shown on the left. Only well-developed blastocysts were used for WGBS. Scale bar, 100 μm. (B) The genomic distribution of CG methylation across all chromosomes in the control and mat-KO blastocysts. The CG methylation levels of the 1-Mb windows are shown. (C) The correlations between the CG methylation levels of the CGIs in control and mat-KO blastocysts. (D) Allele-specific CG methylation at the ICRs in control and Uhrf1 mat-KO blastocysts.
(EPS)
The correlations of the CG methylation levels of the biological replicates were examined in 10-kb genomic windows.
(EPS)
(A) The proportions of hemimethylated sites among highly methylated CG sites in control [Uhrf12lox/2lox], Dnmt1 KO, and Uhrf1 KO FGOs. (B) The proportion of 5mCs in all Cs in each sequence context. H represents the base A, C, or T. (C) Violin plots showing the distribution of the 10-kb genomic regions with different non-CG methylation levels in control [Uhrf12lox/+], Dnmt1 KO, and Uhrf1 KO FGOs. The horizontal bars indicate the mean non-CG methylation levels. (D) The correlation between the non-CG methylation levels of the 10-kb genomic windows in the control and KO FGOs.
(EPS)
(A) The genomic distribution of CG methylation across all chromosomes in control [Uhrf12lox/2lox] and Uhrf1 KO FGOs. The CG methylation levels of the 1-Mb windows are shown. (B) Box plots showing the correlation between the impact of Uhrf1 KO on CG and non-CG methylation in the 10-kb windows. (C) The correlation between the CG methylation levels of CGIs in control and Uhrf1 KO FGOs. (D) The correlation between the (RPKM+1) values in control and Uhrf1 KO FGOs. (E) The CG methylation levels of the ICRs in NGOs, P15 GOs, and FGOs.
(EPS)
(A) The levels of CG methylation in the respective parental genomes in control [Uhrf12lox/+], Dnmt1 mat-KO and Uhrf1 mat-KO blastocysts. (B) The contribution of UHRF1 produced in oocytes to de novo CG methylation in GOs and maintenance methylation in preimplantation embryos.
(EPS)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Isao Suetake and Shoji Tajima (Osaka University) for providing the anti-DNMT1 antibody, Kyohei Arita, Shin-ichi Tomizawa (Yokohama City University), Takashi Sado (Kinki University), Kenji Ichiyanagi (Nagoya University), Wan Kin Au Yeung (Kyushu University), and Kenjiro Shirane (Kyushu University and University of British Columbia) for their useful advice, and Miho Miyake, Tomomi Akinaga, and Junko Oishi (Kyushu University) for providing technical assistance.
Data Availability
The sequence data sets supporting the results of this article are available in the DDBJ Sequence Read Archive under accession number DRA005849.
Funding Statement
This work was supported by KAKENHI from JSPS (24613005 to MU, 15K06803 to MU, and 25112010 to HS), AMED-CREST from AMED to HK and HS, Research Grant from Naito Foundation to MU, and Program & Project Fund from Kyushu University to MU. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14: 204–220. doi: 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- 2.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153: 38–55. doi: 10.1016/j.cell.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138: 811–820. doi: 10.1242/dev.061416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8: e1002440 doi: 10.1371/journal.pgen.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, Ito T, et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013;9: e1003439 doi: 10.1371/journal.pgen.1003439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148: 816–831. doi: 10.1016/j.cell.2011.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97: 5237–5242. doi: 10.1073/PNAS.97.10.5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462: 315–322. doi: 10.1038/nature08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 2012;32: 340–353. doi: 10.1038/emboj.2012.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo T a, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450: 908–912. doi: 10.1038/nature06397 [DOI] [PubMed] [Google Scholar]
- 11.Bostick M, Kim JK, Estève P- O, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317: 1760–1764. doi: 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504: 460–464. doi: 10.1038/nature12805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339: 448–452. doi: 10.1126/science.1229277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14: 188–195. doi: 10.1016/j.gde.2004.01.005 [DOI] [PubMed] [Google Scholar]
- 15.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129: 1983–1993. [DOI] [PubMed] [Google Scholar]
- 16.Kaneda M, Hirasawa R, Chiba H, Okano M, Li E, Sasaki H. Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells. 2010; 169–179. doi: 10.1111/j.1365-2443.2009.01374.x [DOI] [PubMed] [Google Scholar]
- 17.Bourc’his D. Dnmt3L and the Establishment of Maternal Genomic Imprints. Science. 2001;294: 2536–2539. doi: 10.1126/science.1065848 [DOI] [PubMed] [Google Scholar]
- 18.Chotalia M, Smallwood S a, Ruf N, Dawson C, Lucifero D, Frontera M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23: 105–117. doi: 10.1101/gad.495809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veselovska L, Smallwood S a., Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas, et al. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015;16: 209 doi: 10.1186/s13059-015-0769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, et al. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 2015;29: 2449–2462. doi: 10.1101/gad.271353.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285: 26114–26120. doi: 10.1074/jbc.M109.089433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nashun B, Hill PWS, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, et al. Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol Cell. 2015;60: 611–625. doi: 10.1016/j.molcel.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484: 339–344. doi: 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu T-P, Guo F, Yang H, Wu H-P, Xu G-LG-FG, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477: 606–610. doi: 10.1038/nature10443 [DOI] [PubMed] [Google Scholar]
- 25.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2: 241 doi: 10.1038/ncomms1240 [DOI] [PubMed] [Google Scholar]
- 26.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334: 194 doi: 10.1126/science.1212483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PWS, D’Souza Z, et al. supplement De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15: 547–557. doi: 10.1016/j.devcel.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335: 1499–1502. doi: 10.1126/science.1216154 [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9: 64–71. doi: 10.1038/ncb1519 [DOI] [PubMed] [Google Scholar]
- 31.Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22: 1607–1616. doi: 10.1101/gad.1667008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104: 829–838. doi: 10.1016/S0092-8674(01)00280-X [DOI] [PubMed] [Google Scholar]
- 33.Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8: 9 doi: 10.1186/1471-213X-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23: 7601–7610. doi: 10.1038/sj.onc.1208053 [DOI] [PubMed] [Google Scholar]
- 35.Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010;38: 1796–1804. doi: 10.1093/nar/gkp1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nady N, Lemak A, Walker JR, Avvakumov G V, Kareta MS, Achour M, et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286: 24300–24311. doi: 10.1074/jbc.M111.234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455: 818–821. doi: 10.1038/nature07249 [DOI] [PubMed] [Google Scholar]
- 38.Avvakumov G V, Walker JR, Xue S, Li Y, Duan S, Bronner C, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455: 822–825. doi: 10.1038/nature07273 [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455: 826–829. doi: 10.1038/nature07280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502: 249–253. doi: 10.1038/nature12488 [DOI] [PubMed] [Google Scholar]
- 41.Qin W, Wolf P, Liu N, Link S, Smets M, Mastra F La, et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015;25: 911–929. doi: 10.1038/cr.2015.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28: 705–717. doi: 10.1128/MCB.01598-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A. 2012;109: 12950–12955. doi: 10.1073/pnas.1203701109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Shen J, Yang Z, Chen P, Zhao B, Hu W, et al. Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger. Cell Res. 2011;21: 1379–1382. doi: 10.1038/cr.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43: 275–284. doi: 10.1016/j.molcel.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343: 193–196. doi: 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]
- 47.de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, et al. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26: 110–112. doi: 10.1002/(SICI)1526-968X(200002)26:2<110::AID-GENE2>3.0.CO;2–8 [PubMed] [Google Scholar]
- 48.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5: 89–99. doi: 10.1038/nrm1310 [DOI] [PubMed] [Google Scholar]
- 49.Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43: 811–814. doi: 10.1038/ng.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157: 979–991. doi: 10.1016/j.cell.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miura F, Enomoto Y, Dairiki R, Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40: e136 doi: 10.1093/nar/gks454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13: 115–126. doi: 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537: 548–552. doi: 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147: 25–32. doi: 10.1083/jcb.147.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang C-C, Zhan B, Yoshikawa Y, Haas W, Gygi SP, Cohn MA. UHRF1 is a sensor for DNA interstrand crosslinks and recruits FANCD2 to initiate the Fanconi anemia pathway. Cell Rep. 2015;10: 1947–1956. doi: 10.1016/j.celrep.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian Y, Paramasivam M, Ghosal G, Chen D, Shen X, Huang Y, et al. UHRF1 contributes to DNA damage repair as a lesion recognition factor and nuclease scaffold. Cell Rep. 2015;10: 1957–1966. doi: 10.1016/j.celrep.2015.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Liu H, Chen Y, Yang X, Wang P, Liu T, et al. A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat Commun. 2016;7: 10201 doi: 10.1038/ncomms10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44: 361–372. doi: 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, et al. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci U S A. 2015;112: 14641–14646. doi: 10.1073/pnas.1512775112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Q, Zhang J, Chen R, Wang L, Li B, Cheng H, et al. Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals. Nat Commun. 2016;7: 12464 doi: 10.1038/ncomms12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WEF, et al. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009;10: 1259–1264. doi: 10.1038/embor.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X, Li C, Garrett-Beal L, Larson D, Wynshaw-Boris A, Deng CX. Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis. 2001;30: 1–6. doi: 10.1002/gene.1025 [DOI] [PubMed] [Google Scholar]
- 63.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27: 31–39. doi: 10.1038/83730 [DOI] [PubMed] [Google Scholar]
- 64.Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, et al. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics. 2015;16: 624 doi: 10.1186/s12864-015-1833-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toh H, Shirane K, Miura F, Kubo N, Ichiyanagi K, Hayashi K, et al. Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing. BMC Genomics. 2017;18: 31 doi: 10.1186/s12864-016-3392-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27: 1571–1572. doi: 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44: D717–D725. doi: 10.1093/nar/gkv1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schultz MD, Schmitz RJ, Ecker JR. “Leveling” the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012;28: 583–585. doi: 10.1016/j.tig.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14: R31 doi: 10.1186/gb-2013-14-4-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14: R36 doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30: 923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression of Uhrf1 and Dnmt1 mRNAs from the maternal and paternal alleles was analyzed separately using published data from (CAST/EiJ × C57BL/6J) F1 hybrid embryos (GSE45719) [46]. An enlarged view of the Uhrf1 and Dnmt1 expression from the zygote to the late 4-cell stage is shown on the right. The expression levels are shown by the RPKM values. Error bar, standard error.
(EPS)
(A) A schematic representation of the genomic organization of the wild-type (WT), 2lox, and 1lox Uhrf1 alleles. The filled boxes indicate the protein-coding regions. (B) The predicted domain architecture of the WT and mutated UHRF1 proteins. The immunogen used to generate the anti-UHRF1 antibody M-132 (used for Western blotting) is shown. (C) The morphology of control [Uhrf12lox/2lox] and Uhrf1 KO FGOs. Scale bar, 100 μm. (D) The expression of Uhrf1 and the Dnmt family mRNAs in control and Uhrf1 KO FGOs was measured by qRT-PCR. Two portions of Uhrf1 mRNA were amplified for quantification. The expression of Gapdh was used for normalization. (E) The detection of UHRF1 and DNMT1o proteins in control and Uhrf1 KO FGOs by Western blotting using M-132 and anti-DNMT1 antibodies. β-actin was used as a loading control. (F) The immunostaining signal intensities obtained with an anti-UHRF1 antibody (Th-10a) in control and Uhrf1 KO developing oocytes.
(EPS)
(A) The morphology of control [Uhrf12lox/+], Dnmt1 mat-KO, and Uhrf1 mat-KO embryos at 96 h after IVF. Among the Uhrf1 mat-KO embryos, those that developed to the expanded blastocyst stage are shown on the left. Only well-developed blastocysts were used for WGBS. Scale bar, 100 μm. (B) The genomic distribution of CG methylation across all chromosomes in the control and mat-KO blastocysts. The CG methylation levels of the 1-Mb windows are shown. (C) The correlations between the CG methylation levels of the CGIs in control and mat-KO blastocysts. (D) Allele-specific CG methylation at the ICRs in control and Uhrf1 mat-KO blastocysts.
(EPS)
The correlations of the CG methylation levels of the biological replicates were examined in 10-kb genomic windows.
(EPS)
(A) The proportions of hemimethylated sites among highly methylated CG sites in control [Uhrf12lox/2lox], Dnmt1 KO, and Uhrf1 KO FGOs. (B) The proportion of 5mCs in all Cs in each sequence context. H represents the base A, C, or T. (C) Violin plots showing the distribution of the 10-kb genomic regions with different non-CG methylation levels in control [Uhrf12lox/+], Dnmt1 KO, and Uhrf1 KO FGOs. The horizontal bars indicate the mean non-CG methylation levels. (D) The correlation between the non-CG methylation levels of the 10-kb genomic windows in the control and KO FGOs.
(EPS)
(A) The genomic distribution of CG methylation across all chromosomes in control [Uhrf12lox/2lox] and Uhrf1 KO FGOs. The CG methylation levels of the 1-Mb windows are shown. (B) Box plots showing the correlation between the impact of Uhrf1 KO on CG and non-CG methylation in the 10-kb windows. (C) The correlation between the CG methylation levels of CGIs in control and Uhrf1 KO FGOs. (D) The correlation between the (RPKM+1) values in control and Uhrf1 KO FGOs. (E) The CG methylation levels of the ICRs in NGOs, P15 GOs, and FGOs.
(EPS)
(A) The levels of CG methylation in the respective parental genomes in control [Uhrf12lox/+], Dnmt1 mat-KO and Uhrf1 mat-KO blastocysts. (B) The contribution of UHRF1 produced in oocytes to de novo CG methylation in GOs and maintenance methylation in preimplantation embryos.
(EPS)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The sequence data sets supporting the results of this article are available in the DDBJ Sequence Read Archive under accession number DRA005849.