Abstract
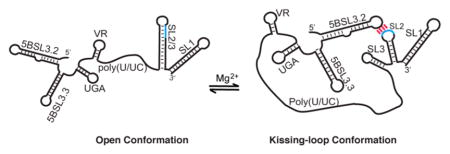
The 3′-end of the genomic RNA of the hepatitis C virus (HCV) embeds conserved elements that regulate viral RNA synthesis and protein translation by mechanisms that have yet to be elucidated. Previous studies with oligo-RNA fragments have led to multiple, mutually exclusive secondary structure predictions, indicating that HCV RNA structure may be context-dependent. Here we employed a nuclear magnetic resonance (NMR) approach that involves long-range adenosine interaction detection, coupled with site-specific 2H labeling, to probe the structure of the intact 3′-end of the HCV genome (385 nucleotides). Our data reveal that the 3′-end exists as an equilibrium mixture of two conformations: an open conformation in which the 98 nucleotides of the 3′-tail (3′X) form a two-stem–loop structure with the kissing-loop residues sequestered and a closed conformation in which the 3′X rearranges its structure and forms a long-range kissing-loop interaction with an upstream cis-acting element 5BSL3.2. The long-range kissing species is favored under high-Mg2+ conditions, and the intervening sequences do not affect the equilibrium as their secondary structures remain unchanged. The open and closed conformations are consistent with the reported function regulation of viral RNA synthesis and protein translation, respectively. Our NMR detection of these RNA conformations and the structural equilibrium in the 3′-end of the HCV genome support its roles in coordinating various steps of HCV replication.
Graphical Abstract
As a Hepacivirus from the Flaviviridae family, hepatitis C virus (HCV) is the major cause of chronic hepatitis, which leads to liver fibrosis, cirrhosis, liver failure, and hepatocellular carcinoma. The viral genome is a 9.6 kb single-stranded, positive-sense RNA, which serves as a template for both viral protein translation and minus-strand RNA synthesis. It contains a single open reading frame (ORF) flanked by highly conserved untranslated regions (UTRs) on both ends that regulate various viral functions (Figure 1A). The 5′-UTR contains a highly structured internal ribosomal entry site (IRES) that initiates cap-independent translation of the polyprotein encoded in the ORF. The IRES element recruits the 40S ribosomal subunit and eIF3, followed by incorporation of the eIF2–tRNAiMet ternary complex and the 60S subunit to form preinitiation complexes.1–7 The polyprotein is co- and post-translationally processed by host and viral proteases to produce structural proteins (core, E1 and E2), ion channel protein (p7), and nonstructural proteins (NS2, NS3A, NS4A, NS5A, and NS5B) (Figure 1A).8 Nonstructural protein NS5B is a RNA-dependent RNA polymerase that replicates viral minus-strand RNA from the positive-strand genomic RNA (gRNA), as well as the positive-strand RNA using the minus-strand RNA as a template.
Figure 1.
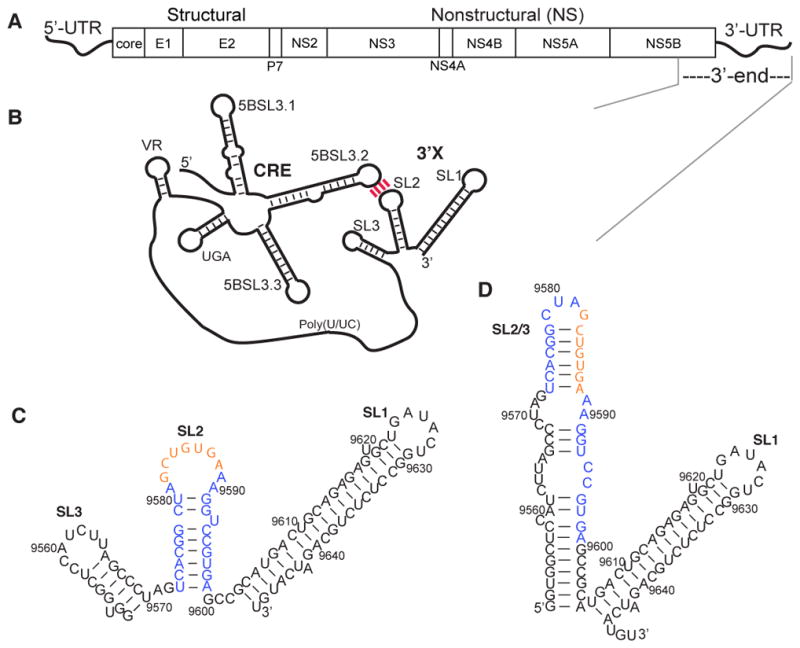
HCV genome and predicted secondary structures for the 3′X region and the 3′-end of the HCV gRNA. (A) The HCV genome contains 5′- and 3′-UTRs and a polyprotein-coding ORF. (B) The 3′X region is proposed to form a long-range interaction with the upstream CRE element via the complementary loop residues between 5BSL3.2 and SL2. (C) The SL2 kissing-loop residues (orange) are readily exposed in the proposed 3SL conformation of the 3′X region. (D) The extended SL2/3 stem–loop structure is shown in the 2SL conformation, which sequesters the SL2 kissing-loop residues.
The compact HCV gRNA contains many functional RNA structures that modulate various stages of the viral life cycle,9,10 and some of the RNA structures participate in dynamic RNA:RNA interaction networks that contribute to regulation of viral replication. Many of the reported RNA:RNA interactions consist of RNA elements at the 3′-end of gRNA. Cell culture-based assays have indicated a potential long-range RNA:RNA interaction between the highly conserved 3′-tail element on the very 3′-end of the HCV gRNA (3′X) and an upstream cis-acting regulatory element (CRE) in the NS5B coding region (5BSL3.2) (Figure 1B). The base pair complementarities between 5BSL3.2 and 3′X are essential for HCV replication.11–13 Although HCV protein translation is initiated by the IRES in the 5′-UTR, its efficiency is affected by RNA elements at the 3′-end of the gRNA. The CRE in the NS5B protein coding region negatively regulates translation efficiency, whereas the 3′-UTR seems to exert a positive impact.14–19 The translation regulation of the 3′-UTR might be achieved by forming a closed-loop topology with the 5′-UTR, allowing the 3′-UTR to deliver ribosome complexes to the 5′-UTR for efficient initiation of subsequent rounds of translation.20 Such end-to-end RNA interactions could occur in the absence of proteins and are likely to be mediated by the IIId loop of the IRES and the bulge of 5BSL3.2.21–23
Despite its essential roles in viral replication, no high-resolution structural information for the 3′-end of gRNA is currently available. It has been proposed to adopt multiple conformations for regulation of different functions, and the structural heterogeneity creates challenges in biophysical studies. The highly conserved 98-nucleotide 3′X region directs viral RNA replication initiation24–27 and plays crucial roles in regulating viral protein translation.14,28 Multiple structural models of 3′X have been proposed on the basis of data from site-directed mutagenesis, chemical probing, enzymatic probing, and free energy calculation.29–31 There is a general consensus among the secondary structure predictions of 3′X that the 46 nucleotides at the 3′-end fold into a stable stem–loop SL1, but different secondary structures have been proposed for the 5′-residues.25,30,32 The three-stem–loop (3SL) model (Figure 1C) is believed to be the dominant conformation because the kissing-loop residues in SL2 (nucleotides 9582–9588) are readily exposed and thus supports the long-range 5BSL3.2:3′X interaction.11–13 The interaction between the SL2 and 5BSL3.2 RNA fragments has been observed by in vitro RNA:RNA binding assays,23,33 but not by nuclear magnetic resonance (NMR) analysis.11 Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) and chemical probing studies showed that the nucleotide activities of 3′X residues were different in the presence of 5BSL3.2,19,34 supporting the proposed 5BSL3.2:3′X interaction. However, a recent NMR study of the 98-nucleotide 3′X RNA examined the base pair patterns based on imino proton assignment and concluded that it adopts a two-stem–loop (2SL) structure with an extended SL2/3 stem–loop structure.35 The 2SL structure is further confirmed by a follow-up small-angle X-ray crystallography (SAXS) study.36 The palindromic CUAG residues are exposed in the apical loop of SL2/3, which directs RNA dimerization, even though it is unclear whether such a dimeric genome exists in the HCV replication cycle (Figure 1D).37,38 Therefore, it is possible that the 3′-end of gRNA adopts multiple conformations and utilizes a structural equilibrium to fine-tune various functions.
In this study, we employed an adenosine-based NMR approach to probe RNA secondary structures at the 3′-end of the HCV gRNA. The method was validated by probing the reported 2SL structure of the 98-nucleotide 3′X35,36 and then applied to investigate RNA structures at the 3′-end. Our data demonstrate that 5BSL3.2 induces structural rearrangement of 3′X and forms intramolecular long-range interactions with 3′X at the 3′-end. We have also obtained direct NMR evidence demonstrating the structural heterogeneity of the 3′-end of the gRNA, which provides a structural basis for a Mg2+-dependent RNA structural equilibium that regulates multiple viral replication steps.
MATERIALS AND METHODS
Plasmid Constructs
The templates for synthesizing 3′X and 3′-end were amplified from the corresponding region of the monocistronic HCV genotype 1a (H77) replicon (Apath LLC)39 using forward primers 5′-GGC AGG AAT TCT AAT ACG ACT CAC TAT AGG TGG CTC CAT CTT AGC CCT AGT CAC GG-3′ and 5′-GGC AGG AAT TCT AAT ACG ACT CAC TAT AGA CAG CGG GGG AGA CAT TTA TCA CAG-3′, respectively, and a common reverse primer, 5′-GCC TCG GAT CC G TAT CCG CTA GCA CAT GAT CTG CAG AGA GGC CAG TAT CAG C-3′. The polymerase chain reaction products were then cloned into the pUC19 plasmid with a 3′-BciVI site for template linearization without introducing non-native 3′-residues during RNA transcription by T7 polymerase. The mutant RNAs were generated using the QuickChange XL Site-Directed Mutagenesis Kit (Agilent). All the plasmid sequences were confirmed by Sanger sequencing at the University of Missouri DNA core facility. The DNA templates for transcribing 5BSL3.2 and control RNAs were ordered directly from Integrated DNA Technologies (IDT).
In Vitro RNA Transcription
Template plasmid DNA constructs were amplified by DH5α cells and extracted with a Plasmid Mega Kit (Qiagen) according to the manufacturer’s instructions. The DNA plasmids were then linearized by restriction enzyme BciVI prior to in vitro RNA transcription. The ordered DNA constructs were annealed to Top17 (5′-TAA TAC GAC TCA CTA TA-3′) to serve as templates for in vitro T7 transcriptions. Trial transcription reactions using varying concentrations of MgCl2 and NTPs were performed to determine optimal conditions prior to large scale RNA transcription. The remaining components included transcription buffer [40 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol, 10 mM spermidine, and 0.01% (v/v) Triton X-100], 0.03% (v/v) DNA (1500 ng/μL), 10% (v/v) DMSO, and doubly distilled H2O in a total volume of 8–10 mL. The transcription reactions were performed at 37 °C for 3 h and quenched with 25 mM EDTA and 1 M urea, and the solutions were mixed with 10% (v/v) glycerol and run on denaturing acrylamide gels at 20 W overnight. RNA bands were visualized by ultraviolet (UV) shadowing, extracted from the gel through electro-elution (Elutrap, Whatman), and washed in Amicon ultracentrifugal filters.
Gel Shift Assays
RNA construct 3′X and its lr-AID mutants were prepared in low-ionic strength buffer [10 mM Tris-HCl (pH 7.5)]. The RNA samples were heated at 95 °C for 3 min, snap-cooled at 4 °C, mixed with 5BSL3.2 in a 1:1 ratio in physiological ionic strength buffer {PI buffer [10 mM Tris-HCl (pH 7.5), 140 mM KCl, 10 mM NaCl, and 1 mM MgCl2]}, and incubated at 37 °C for 1 h. The RNA samples were then mixed with a 10% (v/v) glycerol solution (50%) and loaded onto 8% native polyacrylamide gels containing 2 mM MgCl2. Gels were run at 200 V for 2 h in TBM buffer [45 mM Tris-HCl (pH 7.5), 45 mM boric acid, and 2 mM MgCl2] at 4°C. Gels were stained with stains-all and destained in doubly distilled H2O. Dimerization analysis of the 3′-end RNA was performed by incubating 1 μM 3′-end RNA in low-ionic strength buffer, PI buffer, and PI buffer containing 6 mM MgCl2 at 37 °C overnight to reach structural equilibrium. The RNAs were then loaded on a 2% native agarose gel in 1× TB buffer [45 mM Tris-HCl (pH 7.5) and 45 mM boric acid]. RNA bands were visualized by ethidium bromide staining, and gel images were recorded with a Kodak camera.
NMR Spectroscopy
Purified RNA samples were lyophilized overnight and dissolved in 2H2O under various buffer conditions as described in the Results. NMR data were collected at 308 K on a Bruker Avance III 800 MHz spectrometer equipped with a TCI cryoprobe (University of Missouri). Two-dimensional 1H–1H NOESY data were processed with NMRPipe and NMR Draw40 and analyzed with NMRViewJ (One Moon Scientific).
Monte Carlo Tightly Bound Ion (MCTBI) Model
The MCTBI model was used to predict the effects of ions on the changes in RNA conformation. In the MCTBI model, the ion correlation and fluctuation effects were taken into account. According to the ion correlation strength,41 the ions around a RNA are classified as tightly bound ions and diffusively bound ions for strongly and weakly correlated ions, respectively. For the tightly bound ions, we used the Monte Carlo insertion deletion algorithm,42 a newly developed Monte Carlo-based method, to sample the discrete tightly bound ion distributions. For the diffusively bound ions, we used the nonlinear Poisson–Boltzmann method to calculate the electrostatic free energy. The validity of the MCTBI model has been demonstrated through extensive comparisons of theory versus experiment.42
To calculate the ion binding effects for a given two-dimensional (2D) (base pair) structure, we first used the Vfold3D43 model to generate an ensemble of three-dimensional (3D) structures followed by molecular dynamics (MD) simulations. For each given 2D structure, 250, 450, and 1050 conformations were generated for 2SL, 3SL, and the 3′X:5BSL3.2 kissing complex, respectively. According to the root-mean-square deviations (RMSDs) between the conformations, we classified the conformational ensemble for each 2D structure into 10 clusters. For the top-ranked structure in each cluster, we applied the MCTBI model to evaluate the electrostatic free energy and the ion binding properties. The electrostatic free energy for a given 3D structure in a salt solution was given by the formula ΔG = ΔGion + ΔGRNA, where ΔGion and ΔGRNA denote the ion-induced electrostatic free energy and the Coulombic interaction energy between RNA backbone charges, respectively. The free energy of a given 2D structure was calculated as the Boltzmann average of the 10 corresponding structures. In the MCTBI calculation for this work, the bulk concentration of K+ was fixed at 150 mM and the bulk concentration of Cl− was set to satisfy the ionic neutrality condition: [Cl−] = [K+] + 2[Mg2+]. For the given RNA structures, the box size for the nonlinear Poisson–Boltzmann (NLPB) calculation was set as 8λD + 200 Å. If the RNA dimension is larger than 8λD, we set the box boundary with a minimum distance of 200 Å from the RNA surface. The ions were assumed to be fully hydrated. The hydrated ionic radii of Mg2+, K+, and Cl− are 4.5, 4.0, and 4.0 Å, respectively.42
RESULTS
The 3′X RNA Adopts the 2SL Conformation
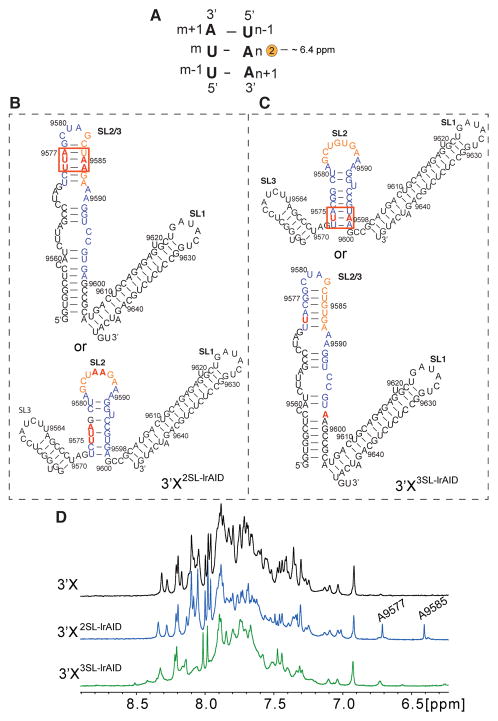
Although cell-based viral replication assays have suggested that 3′X adopts a 3SL structure with the SL2 kissing-loop residues exposed to form a long-range interaction with 5BSL3.2 (Figure 1B),11–13 the Gallego group reported both NMR and SAXS data showing that the 98-nucleotide 3′X forms a 2SL structure with the SL2 kissing-loop residues sequestered (Figure 1D).35,36 To probe the reported 3′X 3SL and 2SL structures by NMR, we employed the long-range adenosine interaction detection (lr-AID) strategy, which has been successfully applied in large RNA structural studies.44 When the triplet Um−1UmAm+1 base pairs with Un−1AnAn+1 in an A-form helix (Figure 2A), the C2-H signal of An appears at approximately 6.4 ppm, which is upfield of most aromatic protons and thus considered diagnostic for such an AU cassette.44,45 To examine the 2SL and 3SL models of 3′X, two mutants, 3′X2SL-lrAID and 3′X3SL-lrAID (panels B and C of Figure 2, respectively), were engineered with consecutive AU base pairs. 3′X2SL-lrAID was designed on the basis of the 2SL model in which A9585 in the consecutive AU base pairs (Figure 2B, top) is expected to give rise to a shifted adenosine C-H2 signal at approximately 6.4 ppm. If the 3′X adopts the 3SL conformation, then the mutation introduced cannot form the lr-AID cassette (Figure 2B, bottom) and no diagnostic 6.4 ppm peak would be detected. On the other hand, 3′X3SL-lrAID was designed so that A9598 would give rise to a diagnostic lr-AID signal only if the RNA adopts the 3SL conformation (Figure 2C, top). The mutations would not form a lr-AID cassette if the RNA folds into the 2SL structure and are expected to destabilize or even disrupt the 2SL folding (Figure 2C, bottom). One-dimensional (1D) proton spectra of 3′X, 3′X2SL-lrAID, and 3′X3SL-lrAID were recorded. No proton signals were detected near the 6.4 ppm region in the wild type 3′X spectrum (Figure 2D, black), demonstrating that the folding of the 98-nucleotide 3′X does not contain UAA:UUA base pairs. A proton signal at 6.4 ppm together with another proton signal at 6.7 ppm was observed in the 3′X2SL-lrAID spectrum (Figure 2D, blue), while most of the remaining signals remained similar to that of wild type 3′X. The slight spectral differences were caused by a change in the chemical environment of nucleotides near the mutation sites. Assignment of the 3′X2SL-lrAID spectrum confirmed that it adopted the same secondary structure as 3′X (Figure S1). The two new signals in the 1D spectrum of 3′X2SL-lrAID were assigned to the C2-H of A9585 and A9577 (Figure 1D and Figure S1). On the other hand, no 6.4 ppm signal was detected in the 3′X3SL-lrAID spectrum (Figure 2D, green), and the proton pattern was different from that of the 3′X, indicating that the mutated nucleotides in 3′X3SL-lrAID did not form the lr-AID cassette and disrupted the native folding of 3′X. These lr-AID probing data suggest that the 98-nucleotide 3′X by itself adopts a 2SL structure.
Figure 2.
The 98-nucleotide 3′X RNA adopts the 2SL conformation. (A) Proposed lr-AID probing strategy. The C2-H of An possesses an unusual chemical shift at approximately 6.4 ppm. (B) The nucleotide mutations in 3′X2SL-lrAID are expected to form the lr-AID base pairs if the RNA adopts the 2SL conformation (top), but not the 3SL conformation (bottom). (C) The nucleotide mutations in 3′X3SL-lrAID are expected to form the lr-AID base pairs if the RNA adopts the 3SL conformation (top), but not the 2SL conformation (bottom). (D) One-dimensional proton spectra of 3′X (black), 3′X2SL-lrAID (blue), and 3′X3SL-lrAID (green). The lr-AID diagnostic proton signal at 6.4 ppm was observed in the 3′X2SL-lrAID spectrum.
To validate the lr-AID results, we then prepared two RNA segments, SL1 (nucleotides 9603–9648, with a non-native 5′-G to enhance T7 transcription yield) and SL2/3 (nucleotides 9551–9604). Two-dimensional NOESY spectra were recorded for 3′X, SL1, and SL2/3. All the RNA samples afforded narrow 1H NMR signals without signs of exchanging peaks, demonstrating the structural homogeneity under our experimental conditions. As expected, similar spectral patterns were identified between the RNA segments and 3′X RNA. Figure 3A shows a representative region where the 3′X signals are found in either the SL1 spectra or the SL2/3 spectrum. The chemical shift of the A9640 C2-H is sensitive to sample conditions as it is close to unpaired A9642 and thus exhibited slight shifts in the SL1 spectrum. Assignment of the SL2/3 nuclear Overhauser effects (NOEs) has led us to conclude that it folded into an extended stem–loop structure, and the same NOE patterns were observed in the 3′X spectrum. For example, in the SL2/3 spectrum, the C2-H proton of A9560 gave a NOE to ribose proton H1′ of its subsequent residue, U9561, demonstrating base stacking between these two residues (Figure 3B, orange lines). An additional NOE was observed to correlate with the A9560 C2-H, which was assigned to G9598 H1′. Such a NOE pattern suggests that these two protons are within 5 Å and thus supports the 2SL model in which A9560 base pairs with U9587 and is adjacent to the C9558-G9598 base pair (Figure 1D). It would not be possible to detect such a NOE if the RNA adopts the 3SL structure in which A9560 and G9598 are far apart (Figure 1C). Similarly, the A9588 C2-H was observed to give a NOE to H1′ of C9574, supporting the 2SL model but not the 3SL model (Figure 3B, purple lines). Therefore, our lr-AID probing results, as well as aromatic proton assignment, demonstrate that the 98-nucleotide 3′X by itself adopts only the 2SL structure. These data validate the lr-AID strategy as an efficient tool for probing secondary structures in large RNA molecules.
Figure 3.
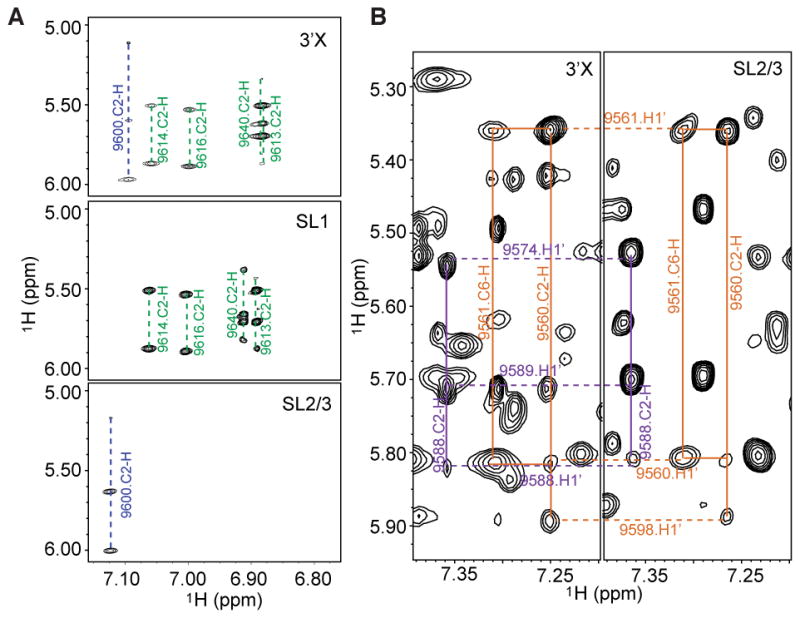
2D NOESY spectra of SL1 and SL2/3 segment RNAs exhibit NOE patterns similar to that of the spectrum of 3′X. (A) Portions of the NOESY spectra of 3′X (top), SL1 (middle), and SL2/3 (bottom). (B) Similar NOE patterns of SL2/3 were observed in the 3′X spectrum. A9560 C2-H NOE connectivities with nearby ribose protons from U9561 and G9598 are colored orange. A9588 C2-H NOE connectivities with nearby ribose protons from A9589 and C9574 are colored purple.
5BSL3.2 Induces Structural Rearrangement of 3′X To Form a Complex
The SL2 kissing-loop residues of 3′X are sequestered in the 2SL structure and not available to form base pairs with the complementary sequences in 5BSL3.2. Thus, we hypothesized that 3′X undergoes structural rearrangement to exposes the SL2 kissing-loop residues and form a complex with 5BSL3.2. Previous gel electrophoresis and surface plasmon resonance (SPR) studies have shown that in vitro-transcribed 5BSL3.2 and 3′X can form a RNA:RNA complex,23,33 and Mg2+ is required in the gel and running buffer for detection of the RNA complex band.38 We used gel electrophoresis to test if 3′X and its lr-AID mutants could form a RNA:RNA complex with 5BSL3.2. As shown in Figure 4, both 3′X3SL-lrAID and 3′X were able to form a complex with 5BSL3.2, whereas no interactions between 3′X2SL-lrAID and 5BSL3.2 were detected. Both 3′X3SL-lrAID and 3′X contained the native SL2 kissing-loop residues, which were complementary to the loop residues of 5BSL3.2. The complementarity between the kissing loops was disrupted by the lr-AID mutation in 3′X2SL-lrAID. As expected, no RNA:RNA interactions were observed between 5BSL3.2 and 3′X2SL-lrAID, demonstrating that complex formation requires sequence complementarity between the kissing-loop residues. Because the SL2/3 loop of the 2SL structure is palindromic, a weak dimer band of 3′X was observed (Figure 4, lane 6). Because the kissing-loop residues are sequestered in the 2SL structure, a small amount of free 3′X was detected in the 3′X +5BSL3.2 lane (Figure 4, lane 7), indicating that the 2SL and the kissing-loop-exposed structures are mutually exclusive. Interestingly, although 3′X3SL-lrAID did not adopt the same structure as wild-type 3′X, it formed a complex with 5BSL3.2 and the free 3′X3SL-lrAID RNA band was almost undetectable (Figure 4, lane 3). A possible explanation is that 3SL-lrAID was designed on the basis of the 3SL model. The mutations disrupted the two-stem–loop folding of 3′X in the absence of 5BSL3.2 and may stabilize the complex conformation as the 3SL model of 3′X exposes the kissing-loop residues to form base pairs with 5BSL3.2. Hence, our data support the hypothesis that 5BSL3.2 introduces structural rearrangement of 3′X by switching from a 2SL structure to a kissing-loop-exposed conformation to form a RNA:RNA complex.
Figure 4.
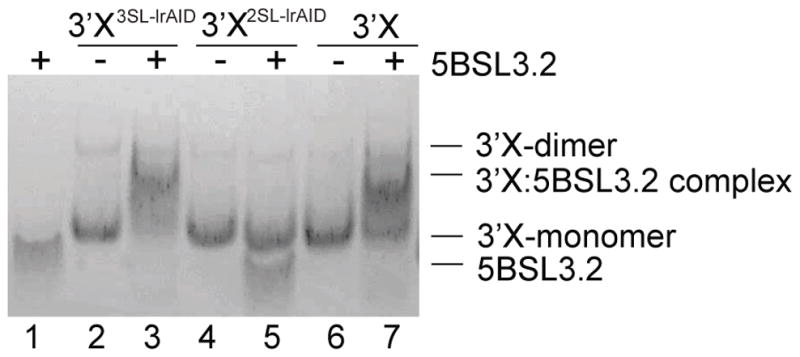
Kissing-loop complementarity is necessary for 3′X and 5BSL3.2 to form a RNA:RNA complex. The mixtures of 5BSL3.2 with 3′X and its lr-AID mutants were examined by native polyacrylamide gel electrophoresis. Both 3′X3SL-lrAID and 3′X formed a complex with 5BSL3.2, whereas no interactions between 3′X2SL-lrAID and 5BSL3.2 were detected.
Two Mutually Exclusive Conformations at the 3′-End of the HCV gRNA
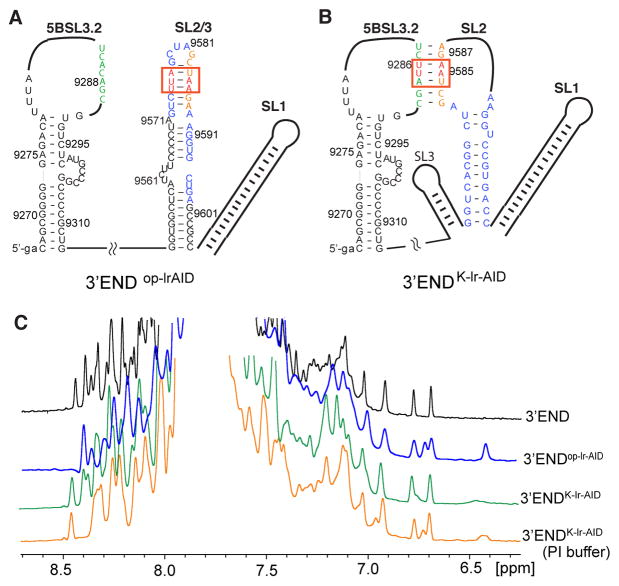
Our in vitro biophysical assays suggest that 3′X mainly adopts a 2SL structure by itself and rearranges its structure to form a complex with 5BSL3.2 mediated by the complementary kissing-loop residues. At the 3′-end of the gRNA, 5BSL3.2 and 3′X are approximately 200 nucleotides apart, connected by several small RNA hairpins, including 5BSL3.3, UGA, and VR, and a long poly(U/UC) linker (Figure 1B). Thus, the question of whether 3′X adopts the 2SL conformation or readily forms long-range kissing-loop interactions with 5BSL3.2 at the 3′-end of the gRNA remained interesting. To investigate the long-range RNA:RNA interaction, we prepared a RNA construct spanning 5BSL3.2 and 3′X to recapitulate the 3′-end of the gRNA (3′-end, 385 nucleotides). Non-native GA residues were added to the 5′-end to enhance the RNA transcription yield. Because no 6.4 ppm signals were detected in the wild type 3′-end spectrum (Figure 5C), the lr-AID strategy was employed to probe the secondary structures within the wild type 3′-end RNA (Figure 5A,B). We first engineered the 3′X2SL-lrAID mutations into the 3′-end to make 3′-endop-lrAID (op stands for open, no kissing-loop interactions between 5BSL3.2 and 3′X). As expected, a new proton signal at 6.4 ppm, as well as a signal at ~6.7 ppm, appeared (Figure 5A,C), suggesting that 3′X adopts the same 2SL conformation at the 3′-end of the gRNA in buffer with a low ionic strength [10 mM Tris-HCl (pH 7.5)]. To probe the interactions between 5BSL3.2 and 3′X kissing loops, 3′-endK-lrAID was designed with an lr-AID cassette engineered into the kissing-loop base pairs (Figure 5B). When the sample was prepared in low-ionic strength buffer, a very weak and broad peak was detected at approximately 6.4 ppm (Figure 5C), indicating that a small portion of the RNA formed the long-range kissing-loop interaction. We then inquired whether salts could promote and stabilize the kissing-loop interaction and added PI buffer (see Materials and Methods for buffer conditions) to the 3′-endK-lrAID sample. As expected, the intensity of the 6.4 ppm peak substantially increased (Figure 5C), indicating that salts stabilize the kissing-loop conformation of the 3′-endK-lrAID RNA. However, the intensity of the lr-AID peak was still relatively weaker than those of other proton signals, suggesting that probably not all of the RNA formed the kissing-loop conformation. Collectively, the lr-AID probing data indicate that both the open and closed (kissing-loop) conformations could occur at the 3′-end of the HCV gRNA. It is likely that the 3′X 2SL conformation is favored in low-ionic strength buffer, and PI buffer promotes the long-range kissing-loop interaction.
Figure 5.
Structural probing of the 3′-end RNA by the lr-AID NMR approach. (A) Proposed secondary structure of the 3′-end open conformation in which 3′X folds into the 2SL conformation. The lr-AID cassette was engineered into the top SL2/3 stem in 3′-endop-lrAID. (B) Proposed secondary structure of the kissing-loop conformation of the 3′-end. The lr-AID cassette was introduced into the kissing-loop base pairs of 3′-endK-lrAID to probe the long-range interaction. Lowercase letters denote non-native nucleotides added to the RNA constructs to promote in vitro RNA synthesis by T7 polymerase. (C) One-dimensional NOESY spectra of 3′-end (black), 3′-endop-lrAID (blue), 3′-endK-lrAID in low-ionic strength buffer (green), and 3′-endK-lrAID in PI buffer (orange).
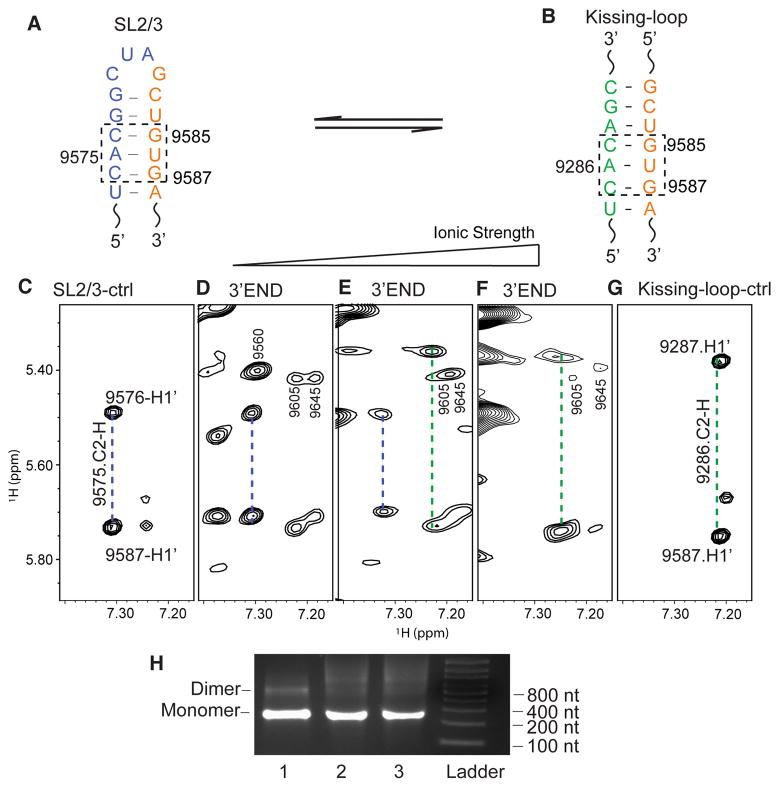
The lr-AID strategy enables a quick NMR detection of RNA conformations at the 3′-end of the HCV gRNA by simple 1D proton experiments. However, it does not provide structural equilibrium information as both lr-AID RNA samples contained mutations that strongly favored one of the conformations. To directly detect the structural equilibrium, we collected 1H–1H NOESY spectra for the 3′-end RNA under various buffer conditions. To decipher secondary structures of large RNA molecules, we heavily rely on the adenosine signals because their C2-H protons give NOEs to the H1′ protons of cross-strand nucleotides in A-form helices (see Figure 2A for illustration) and thus provide important base pairing information. The adenosine C2-H fingerprint spectra of the 3′-end were obtained by selectively 2H labeling the C8-H of adenosines and guanosines. This labeling strategy partially overcomes the signal overlap problem and enhances the quality of the spectra by making the purine C8 proton signals NMR invisible.46 Residues G9582–A9588 in SL2 either base pair with residues U9573–C9579 in the 2SL conformation of 3′X or form a long-range base pair with residues U9284–C9290 in 5BSL3.2 (Figure 6A,B). In both cases, there is a common CAC:GUG base pairing in which the C2-H of adenosine is expected to give NOEs to H1′ of its subsequent cytidine (C9576 in SL2/3 or C9287 in the kissing loop) and the cross-strand G9587 (Figure 6A,B). Because of the base pairing differences between SL2/3 and the kissing-loop interaction (G9577:U9584 vs A9288:U9584), the H1′ chemical shift of C9576 is expected to be slightly different from that of C9287.45 This is confirmed by peak assignment of control oligonucleotides (Figure 6C,G and Figures S2 and S3). For the same reason, a slightly upfield-shifted C2-H of A9575 was observed as compared with the chemical shift of the A9286 C2-H. These two distinct NOE patterns (Figure 6C,G) provide important references for examination of the RNA conformations at the 3′-end of the HCV gRNA. At low ionic strengths, only the SL2/3 A9575 C2-H correlated NOE pattern was observed (Figure 6D), demonstrating that the open conformation was dominant. When the NMR data were collected in PI buffer, both NOE features were detected (Figure 6E), demonstrating the coexistence of the open and closed conformations. Slight chemical shift differences are due to the control RNAs being much simpler than the full 3′-end RNA. This is consistent with the relatively weak lr-AID signal of 3′-endK-lrAID observed in Figure 5C because the ions in PI buffer were not sufficient to promote complete formation of the closed conformation in the 3′-end RNA. Because Mg2+ has been reported to promote and stabilize the kissing-loop interaction between 5BSL3.2 and 3′X segment RNAs,23 we continued to titrate Mg2+ into the 3′-end RNA sample to examine whether the kissing-loop interaction can be further promoted by Mg2+. At 6 mM Mg2+, the SL2/3 NOE pattern became undetectable and the kissing-loop NOE signals were dominant (Figure 6F). This result suggests that the long-range kissing-loop conformation is favored by high Mg2+ concentrations. Additional spectral changes were observed to support the RNA switch. For example, the A9560 C2-H signal detected at ~7.3 ppm in low-ionic strength buffer (Figure 6D) matches the signals in the spectrum of isolated 3′X adopting the 2SL conformation. The signal shifted as the buffer ionic strength increased, consistent with the RNA switch model. It is worth mentioning that the 3′-end RNA remained monomeric under these buffer conditions (Figure 6H), demonstrating that the observed long-range kissing-loop interaction was intramolecular.
Figure 6.
NMR detection of the 3′-end RNA structural equilibrium. (A) In the 3′-end open conformation, G9585UG9587 base pairs with C9574AC9576 in SL2/3 (dashed box). (B) In the 3′-end kissing-loop conformation, G9585UG9587 base pairs with C9285AC9287 in 5BSL3.2 (dashed box). (C) Portion of the SL2/3 control oligonucleotide NOESY spectrum showing that the C2-H of A9575 gives NOEs to H1′ of C9576 and G9587 (blue dashed line). (D–F) NOESY spectra of the 3′-end RNA in 10 mM Tris-HCl (D), PI buffer (E), and PI buffer containing 6 mM Mg2+ (F). (G) NOESY spectrum of the kissing-loop control oligonucleotides showing that the C2-H of A9286 gives NOEs to H1′ of C9287 and G9587 (green dashed line). (H) The 3′-end RNA remained monomeric under various buffer conditions: left lane, low-ionic strength buffer; middle lane, PI buffer; right lane, PI buffer containing 6 mM MgCl2.
In summary, the lr-AID probing data show that the 3′-end could adopt both open and closed conformations. By referencing the spectra recorded for SL2/3 and kissing-loop control RNA samples, we were able to recognize NMR signals from each conformation in the 3′-end RNA. In addition to the characteristic NOEs shown in Figure 6, other NOE signals representing the open and closed conformations were also observed (data not shown). Identification of these signals allows us to trace the change in RNA structural equilibrium under different buffer conditions. The NMR results show that salts, especially Mg2+, promote and stabilize the long-range kissing-loop interaction at the 3′-end of the HCV gRNA, even in the absence of chaperone proteins.
The Intervening Sequences Do Not Contribute to the Structural Equilibrium
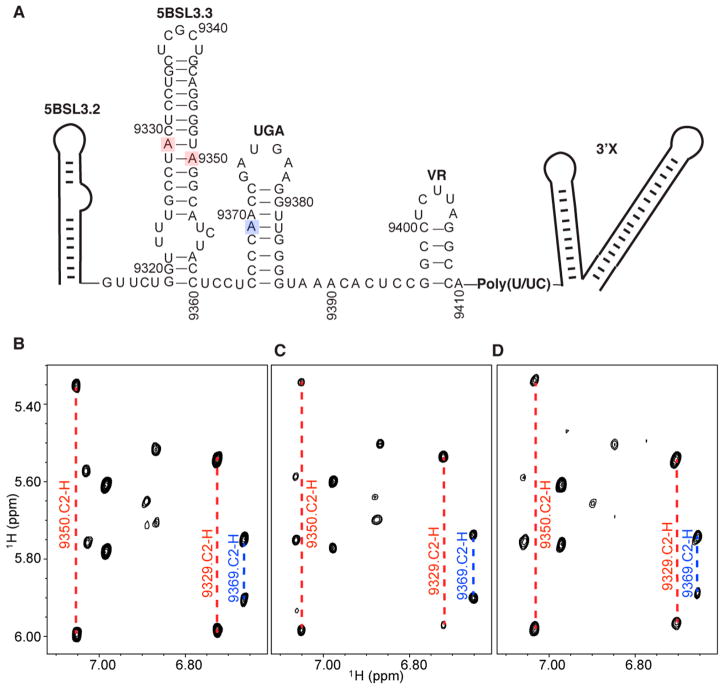
The non-poly(U/UC) sequences connecting 5BSL3.2 and 3′X are predicted to fold into 5BSL3.3, UGA, and VR stem–loop structures (Figure 1B), and the formation of these structures was confirmed by the characteristic adenosine C2-H signals in the NMR spectra of the 3′-end RNA (Figure 7). These signals remained unchanged when the structural equilibrium shifted from the open to closed conformation as the Mg2+ concentration increased, demonstrating that these residues do not participate in regulation of the Mg2+-dependent structural equilibrium at the 3′-end of the HCV genome. Our results are consistent with previous reports that synonymous mutations in 5BSL3.3 that maintained the protein sequence but disrupted the predicted RNA secondary structure maintained a replication efficiency similar to that of the wild type virus.11,13
Figure 7.
NMR signals of intervening sequences at the 3′-end RNA remained unchanged at different Mg2+ concentrations. (A) Predicted secondary structures of 5BSL3.3, UGA, and VR that connect 5BSL3.2 and 3′X. A9329 and A9350 in 5BSL3.3 (boxed in red) and A9369 in UGA (boxed in blue) give rise to characteristic adenosine C2-H signals in NOESY spectra. (B–D) Portions of the NOESY spectra of the 3′-end in 10 mM Tris-HCl (B), low-ionic strength PI buffer (C), and PI buffer containing 6 mM MgCl2 (D). C2-H signals of A9329 and A9350 in 5BSL3.3 (red dashed lines) and A9369 in UGA (blue dashed lines) are marked.
Mg2+ Stabilizes the Kissing-Loop Conformation
To investigate why the 5BSL3.2:3′X interaction is dependent on Mg2+, we calculated the Mg2+ concentration dependence of the free energies for the structures. The calculations were based on the MCTBI method42 combined with the Vfold3D RNA folding model.43 The Vfold3D model followed by MD simulation gives a total of 250, 450, and 1050 conformations for 2SL, 3SL, and the 5BSL3.2:3′X kissing complex, respectively. The MCTBI model predicts the Mg2+ concentration dependence of the electrostatic free energy and the ion binding properties for each given 3D structure. Here, an electrostatic free energy of a given 2D structure, such as ΔG2SL and ΔG3SL, is the Boltzmann-weighted average for the respective 3D conformational ensemble.
The 5BSL3.2 structure, which contains two large loops, the U9280–G9291 hairpin loop and the C9298–G9305 bulge loop (Figure 5A,B), can be flexible. The Vfold3D model generates an ensemble of 3D conformations for 5BSL3.2. According to the RMSD between the conformations, we classified the conformational ensemble into 10 ranked clusters, where conformations in each cluster are similar to each other. In each cluster, we chose the top-ranked structure. The procedure returned with 10 candidate structures for the calculations of the ion effects. The MCTBI model predicted ion-induced free energy ΔGion of each structure (Figure 8A). The free energy result shows that without Mg2+ ions, structure 9 (marked with a circle) has the lowest ΔGion, with a competing structure 3, which has the next lowest ΔGion. As the Mg2+ concentration increases, structure 10 (marked with a circle) may become the most stable structure (see panels B and C of Figure 8 for structures 9 and 10, respectively). The main difference between the two structures is the relative positions of the loops (labeled with red and yellow) to the main body (helix). For structure 10, which tends to be stabilized at 2 mM Mg2+, the bending of the loops leads to the formation of the pockets, which can capture more binding Mg2+ ions (the green spheres). The result demonstrates that the folding of 5BSL3.2 is sensitive to Mg2+. Meanwhile, similar to 5BSL3.2, we also generated 10 ranked clusters for the 2SL structure and then selected the top-ranked one in each cluster. We noticed that in the absence of 5BSL3.2, the 2SL structure of 3′X is less sensitive to the binding of Mg2+ ions because the ΔGion profiles are very similar at different Mg2+ concentrations for each predicted 3D structure (Figure S4).
Figure 8.
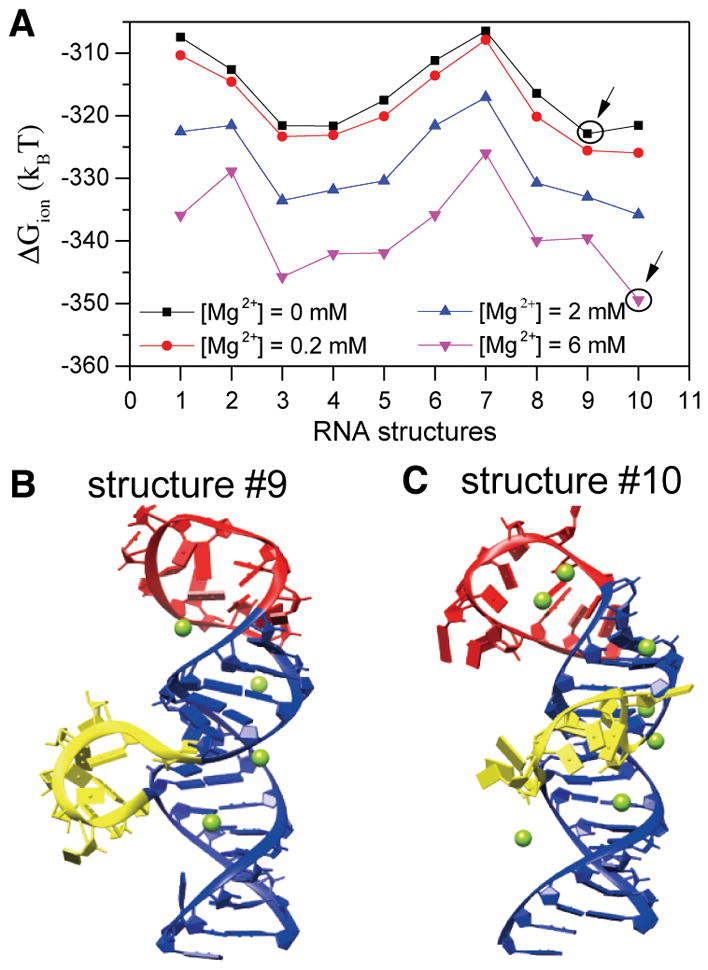
Structure of 5BSL3.2 is sensitive to Mg2+ concentration. (A) Ion electrostatic free energies for the different 3D structures of 5BSL3.2 at different Mg2+ concentrations. The structures that have the lowest free energy are labeled with circles at 0 and 6 mM Mg2+. (B and C) Structures 9 and 10, respectively, with the predicted bound Mg2+ ions at 2 mM Mg2+. Loop residues U9280–G9291 are colored red, bulge residues A9298–G9304 yellow, and residues in the main helix blue.
Furthermore, to investigate the effect of ion binding on the conformational switch between 2SL and 3SL for 3′X with and without 5BSL3.2, we computed electrostatic free energy difference ΔΔG between the two structures (see Figure 9A). In the case of 3′X without 5BSL3.2, adding Mg2+ ions can slightly promote the change in the structure of 3′X from 2SL to 3SL. However, such a minor ion binding effect is not sufficient to induce the structural switch. The structural switch is likely caused by other interactions such as H-bond and base stacking, such as the interactions associated with the kissing-loop interaction with the 5BSL3.2.47 In the case of 3′X with 5BSL3.2, at low Mg2+ concentrations, the negative charges on the RNA backbones cause strong Coulombic repulsion between 3′X and 5BSL3.2, preventing the formation of the 3′X:5BSL3.2 kissing complex. With the increase in Mg2+ concentration, more Mg2+ ions are trapped at the kissing interface, where significant RNA negative charge buildup occurs (Figure 9B,C). Here the 3D structure shown in panels B and C of Figure 9 is the top-ranked structure out of all the predicted structures for the 3′X:5BSL3.2 complex. These bound Mg2+ ions would significantly weaken 3′X:5BSL3.2 Coulombic repulsion, resulting in a drastic decrease in ΔΔG. Compared with that of 3′X (without 5BSL3.2), the ion binding effect is much more pronounced for the 3′X:5BSL3.2 kissing-loop interaction, causing an ion-induced stabilization of the kissing complex.
Figure 9.
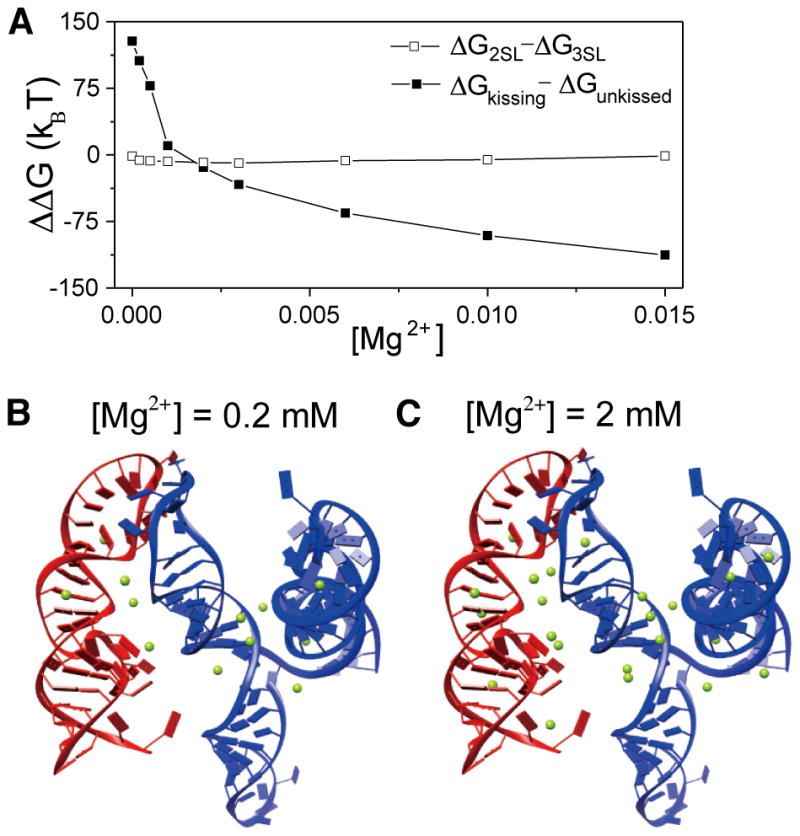
Mg2+ stabilizes the kissing-loop interaction. (A) The free energy differences between the 2SL and 3SL structures for 3′X (□) are not sensitive to Mg2+ concentration, indicating that Mg2+ is not sufficient to promote the structural switch from 2SL to 3SL in 3′X. The free energy difference between the 5BSL3.2:3′X kissing complex and the unkissed structure (■) decreases significantly as the Mg2+ concentration increases, indicating the kissing-loop interaction is favored at high Mg2+ concentrations. (B and C) Structures of the 5BSL3.2:3′X kissing complex with the predicted bound Mg2+ ions at 0.2 and 2 mM Mg2+, respectively. 5BSL3.2 and 3′X are colored red and blue, respectively. Mg2+ ions are shown as green spheres.
DISCUSSION
Here we present direct structural evidence showing that the 3′-end of the HCV genome exists in an equilibrium of open and closed conformations, which has been previously proposed to modulate viral protein translation and RNA synthesis.11–13,48 By employing an adenosine-based structural probing NMR approach, we show that the 98-nucleotide 3′X by itself mainly adopts the 2SL conformation. At the 3′-end of gRNA, 3′X rearranges its structure and exposes the SL2 kissing-loop residues to form a long-range interaction with 5BSL3.2, in the absence of proteins. To avoid structural bias caused by lr-AID mutations, we also recorded NMR spectra of the 3′-end RNA under different buffer conditions and were able to detect a structural equilibrium between the open and closed conformations. The structural equilibrium is sensitive to buffer ionic strength, especially to Mg2+. As the most abundant multivalent cation in cells, Mg2+ stabilizes the RNA tertiary structure by nonspecific long-range electrostatic interactions with the highly negatively charged phosphate backbone of RNA.49 Therefore, the long-range intramolecular kissing-loop interaction is favored at high Mg2+ concentrations. Meanwhile, the charges of SL2 and SL3 in the 3SL model are rather clustered compared to those of the extended SL2/3 stem–loop structure and thus require cations, such as Mg2+, to neutralize the condensed charge and stabilize the RNA structure. In addition, 5BSL3.2 is likely to contain a specific Mg2+ binding pocket. Computational structure calculations show that folding of 5BSL3.2 is sensitive to Mg2+ and favors the kissing-loop residue-exposed conformation in the presence of high Mg2+ concentrations (Figure 8). Interestingly, a recent atomic force microscopy study of IRES reported that Mg2+ induces a structural switch from an “open” and elongated structure at 0–2 mM Mg2+ to a closed comma-shaped conformation at 4–6 mM Mg2+.50 The observed Mg2+-sensitive IRES structural equilibria could be related to the reported optimal activity in translation competent extracts containing 1–2.5 mM Mg2+,51 and initiation factor-independent translation at 5 mM Mg2+.52 While the intracellular Mg2+ concentration is estimated to range from 1 to 10 mM, liver cells contain Mg2+ concentrations relatively higher than those of other tissue cells.53,54 Thus, the observed Mg2+-dependent structural equilibrium is likely an adaptive property of the 3′-end of gRNA, and the equilibrium is optimized to the intracellular Mg2+ concentration in liver cells for efficient viral replication.
The lr-AID method was first employed to detect a long-range interaction in the dimeric 5′-UTR of the HIV-1 genome.44 It uses the unusual adenosine C2-H chemical shift of a “UUA-UAA” base pair. Our data demonstrate that this efficient NMR approach can be widely applied to other large RNA systems. The NMR signals of SL2 residues U9573–C9579 are expected to be very similar when they are in the 2SL or 3SL conformation, as the base pairings are almost identical. Thus, the lr-AID approach provides a very efficient way to confirm the 2SL folding of the 98-nucleotide 3′X (Figure 2D), and the same 2SL conformation remains at the 3′-end when RNA was folded in low-ionic strength buffer (Figure 5C). Formation of the kissing-loop interaction appears to be salt-dependent, and the lr-AID strategy has enabled us to monitor the formation of such long-range RNA:RNA interactions by quick 1D scans under various buffer conditions (Figure 5C). Previous studies have indicated through gel electrophoresis that the monomeric band of a 3′X segment (termed X55, spanning SL2–SL3) can be resolved into two bands, supposedly corresponding to two different conformations. The weak and main monomeric bands were proposed to correspond to the 3SL and 2SL structures, respectively.23 However, it is also possible that this weaker band represents RNA adopting the same secondary structure but exhibiting a different tertiary conformation.35 Our effort to probe the 3SL conformation in the 98-nucleotide 3′X RNA alone was unsuccessful, and our follow-up aromatic proton-based 2D NMR experiments with 3′X (Figure 3) confirmed the 2SL conformation. If both conformations exist, similar to the case of open and closed conformations in the 3′-end RNA, lr-AID probing should work for both conformations (Figure 5C). However, we could not exclude the possibility that the 3SL conformation exists only at concentrations much lower than the concentrations used for NMR studies.
Many positive-strand RNA viruses use long-range RNA:RNA interaction networks in their genomes to regulate gene expression and genome replication. For example, long-range interactions between the 5′-end and 3′-end to circularize gRNA are required for replication of positive-strand plant RNA viruses,55–61 as well as animal RNA viruses.62–68 Interactions between the UTR and coding region have also been reported to modulate viral replication. For example, barley yellow dwarf virus employs a long-range interaction between the frameshift element and the 3′-UTR to regulate viral protein translation.69 In this study, our NMR data confirmed the existence of the 5BSL3.2:3′X long-range kissing-loop interaction at the 3′-end of HCV gRNA, consistent with the previous reports that the conserved sequence complementarity between 5BSL3.2 and 3′X is essential for viral replication.11–13 We also report that the equilibrium of the open and closed conformations at the 3′-end of the gRNA is sensitive to buffer ionic strength, which might explain some unsuccessful attempts to detect such interactions under in vitro conditions.11,34 The structural equilibrium may also be tuned by other long-range RNA:RNA interactions. SHAPE analysis of the HCV gRNA structure suggests a complex dynamic interaction among IRES, 5BSL3.2, and 3′X.19,21,22,70 The eight-nucleotide bulge loop of 5BSL3.2 can form base pairs with an upstream element in the coding region on SL9110, as well as the IIId loop of IRES on the 5′-UTR under in vitro conditions.34,70 Thus, the interplay among these complementary sequences is expected to modulate the dynamics of the RNA:RNA network, which fine-tunes the functional regulation in viral replication. Our assay focused on only the RNA structures within the 3′-end of the HCV gRNA and did not include either IRES or SL9110. We believe the 5BSL3.2 bulge-loop residues are not involved in the kissing-loop conformation as characteristic NMR signals of the bulge loop remained unchanged when the RNA experienced a switch from the open to the closed conformation upon titration with salt (Figure S5). Consistent with previous reports,33,34 we expect that the 5BSL3.2 bulge involves interactions that, if they exist, should not affect the kissing-loop interaction at the 3′-end of gRNA. Future efforts will be made to investigate the preference of IRES IIId and SL9110 for different 3′-end structures and to explore the biological significance.
In the open conformation of the 3′-end, our NMR lr-AID probing demonstrates that the 3′X segment adopts the 2SL conformation, consistent with a recent NMR study of the 98-nucleotide 3′X based on imino proton assignments and SAXS studies.35,36 The 2SL conformation exposes the palindromic sequence 5′-CUAG-3′ in SL2, which promotes gRNA dimerization.23,37,38 However, no dimer stage of the HCV gRNA has been observed, and its biological relevance remains unknown. Although we were not able to distinguish the NMR signals between the monomeric and dimeric 3′X 2SL conformations, as most of the palindromic residues are expected to have the same chemical environment, our electrophoresis analysis showed that majority of the 3′X RNA in PI buffer was monomeric (Figure 4). Similarly, despite the fact that the open:closed conformation equilibrium varied under different buffer conditions (Figure 6D–F), the 3′-end RNA mainly migrated as a monomer band (Figure 6H). Thus, in the absence of proteins, our biophysical data demonstrate the structural equilibrium between the open- and kissing-loop conformations at the 3′-end of the HCV gRNA but do not support the model of RNA dimerization.
Because both viral protein translation and RNA synthesis occur on the gRNA, the dynamic RNA:RNA interaction networks within gRNA are believed to play an essential role in spatial and temporal regulation of these replication steps. Translation of reporter RNA with an authentic HCV IRES in the 5′-UTR was negatively regulated by introducing CRE at the 3′-end, whereas adding the 3′-UTR downstream of CRE rescued the inhibitory effect of CRE.19 A recent study using antisense locked nucleic acids that target the functional domain of CRE and its proposed interacting RNA elements showed that blocking the 5BSL3.2:3′X kissing-loop interaction suppressed translation without affecting RNA synthesis in transfected cells.48 The 3′-UTR specifically interacts with the 40S ribosomal subunit and supports multiround translation. Such interaction can be disrupted by the IRES, suggesting that the 3′-UTR could retain translation machinery and facilitate initiation of subsequent rounds of translation through end-to-end RNA:RNA interactions.20 In agreement with these cell-based assays, our NMR studies of the 3′-end of the HCV gRNA provide direct evidence that the 5BSL3.2:3′X kissing-loop base pairings are formed under in vitro conditions. Our structural model shows that SL1 remains relatively unchanged, whereas residues in SL2 and SL3 undergo substantial rearrangement from the open to the closed conformation. As the kissing-loop residues of SL2 contribute significantly to the formation of the long-range base pairing with 5BSL3.2, SL3 is expected to regulate the structural equilibrium, as well. Murakami et al. reported that deletion of SL3 enhanced translation efficiency, even in the presence of host polypyrimidine tract binding protein,72 which has been shown to downregulate HCV translation by binding to 3′X residues, including SL3.28,31 Although we cannot exclude the possibility of other unidentified host factors being involved, our structural model would predict a favorable structural equilibrium toward the closed conformation at the 3′-end of the gRNA in the absence of SL3, which thus promotes translation. On the other hand, mass spectrometry analysis of CRE binding proteins has identified EWSR1 as a host protein that promotes HCV RNA synthesis by preferentially binding to 5BSL3.2 in the absence of 3′X.71 Therefore, the open conformation at the 3′-end of the HCV gRNA with disabled long-range kissing-loop interaction is likely to be required for RNA synthesis.
Our studies have provided direct NMR evidence demonstrating a RNA switch at the 3′-end of the HCV gRNA, which supports many cell-based functional analyses. It is possible that the structural equilibrium is also modulated by other viral RNA elements and protein factors, and each RNA conformation recruits viral and host factors at different viral replication stages. Continuous investigations of the dynamic RNA:RNA interaction networks are needed to unravel their essential roles in the viral life cycle.
Supplementary Material
Acknowledgments
Funding
This research was supported by University of Missouri startup funds and MU Research Board funds (to X.H.) and National Institutes of Health Grants R01-GM117059 and R01-GM063732 (to S.-J.C.).
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.7b00573.
NMR spectra of 3′X and 3′X2SL-lrAID (Figure S1), NMR spectrum and assignment of the SL2/SL3 control (Figure S2), NMR spectrum and assignment of the kissing interaction control (Figure S3), ion-induced free energy for each predicted 3D structure of the 3′X 2SL conformation (Figure S4), and NMR proton signals of 5BSL3.2 bulge residues (Figure S5) (PDF)
References
- 1.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 7.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 8.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 9.Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, Martin DP, Weeks KM. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci U S A. 2015;112:3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol Cell. 2016;62:111–120. doi: 10.1016/j.molcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You S, Rice CM. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J Virol. 2008;82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T, Tahara SM, Lai MM. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- 16.Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bung C, Bochkaeva Z, Terenin I, Zinovkin R, Shatsky IN, Niepmann M. Influence of the hepatitis C virus 3′-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett. 2010;584:837–842. doi: 10.1016/j.febslet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Romero-Lopez C, Berzal-Herranz A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell Mol Life Sci. 2012;69:103–113. doi: 10.1007/s00018-011-0729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai Y, Zhou K, Doudna JA. Hepatitis C virus 3′UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 2013;41:7861–7874. doi: 10.1093/nar/gkt543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero-Lopez C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero-Lopez C, Barroso-Deljesus A, Garcia-Sacristan A, Briones C, Berzal-Herranz A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3′-end of the viral genome. Nucleic Acids Res. 2012;40:11697–11713. doi: 10.1093/nar/gks927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shetty S, Stefanovic S, Mihailescu MR. Hepatitis C virus RNA: molecular switches mediated by long-range RNA-RNA interactions? Nucleic Acids Res. 2013;41:2526–2540. doi: 10.1093/nar/gks1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi M, Lemon SM. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol. 2003;77:3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci U S A. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blight KJ, Rice CM. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T, Lai MM. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutkiewicz M, Ciesiolka J. Structural characterization of the highly conserved 98-base sequence at the 3′ end of HCV RNA genome and the complementary sequence located at the 5′ end of the replicative viral strand. Nucleic Acids Res. 2005;33:693–703. doi: 10.1093/nar/gki218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palau W, Masante C, Ventura M, Di Primo C. Direct evidence for RNA-RNA interactions at the 3′ end of the Hepatitis C virus genome using surface plasmon resonance. RNA. 2013;19:982–991. doi: 10.1261/rna.037606.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuplin A, Struthers M, Simmonds P, Evans DJ. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012;40:6908–6921. doi: 10.1093/nar/gks370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantero-Camacho A, Gallego J. The conserved 3′X terminal domain of hepatitis C virus genomic RNA forms a two-stem structure that promotes viral RNA dimerization. Nucleic Acids Res. 2015;43:8529–8539. doi: 10.1093/nar/gkv786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantero-Camacho A, Fan L, Wang YX, Gallego J. Three-dimensional structure of the 3′X-tail of hepatitis C virus RNA in monomeric and dimeric states. RNA. 2017;23:1465–1476. doi: 10.1261/rna.060632.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin F, Fosse P, Darlix JL. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34:2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shetty S, Kim S, Shimakami T, Lemon SM, Mihailescu MR. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913–925. doi: 10.1261/rna.1960410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Tan ZJ, Chen SJ. Electrostatic correlations and fluctuations for ion binding to a finite length polyelectrolyte. J Chem Phys. 2005;122:044903. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun LZ, Chen SJ. Monte Carlo Tightly Bound Ion Model: Predicting Ion-Binding Properties of RNA with Ion Correlations and Fluctuations. J Chem Theory Comput. 2016;12:3370–3381. doi: 10.1021/acs.jctc.6b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X, Zhao P, Chen SJ. Vfold: a web server for RNA structure and folding thermodynamics prediction. PLoS One. 2014;9:e107504. doi: 10.1371/journal.pone.0107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barton S, Heng X, Johnson BA, Summers MF. Database proton NMR chemical shifts for RNA signal assignment and validation. J Biomol NMR. 2013;55:33–46. doi: 10.1007/s10858-012-9683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boeras I, Song Z, Moran A, Franklin J, Brown WC, Johnson M, Boris-Lawrie K, Heng X. DHX9/RHA Binding to the PBS-Segment of the Genomic RNA during HIV-1 Assembly Bolsters Virion Infectivity. J Mol Biol. 2016;428:2418–2429. doi: 10.1016/j.jmb.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun LZ, Zhang D, Chen SJ. Theory and Modeling of RNA Structure and Interactions with Metal Ions and Small Molecules. Annu Rev Biophys. 2017;46:227–246. doi: 10.1146/annurev-biophys-070816-033920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuplin A, Struthers M, Cook J, Bentley K, Evans DJ. Inhibition of HCV translation by disrupting the structure and interactions of the viral CRE and 3′ X-tail. Nucleic Acids Res. 2015;43:2914–2926. doi: 10.1093/nar/gkv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misra VK, Draper DE. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Sacristan A, Moreno M, Ariza-Mateos A, Lopez-Camacho E, Jaudenes RM, Vazquez L, Gomez J, Martin-Gago JA, Briones C. A magnesium-induced RNA conformational switch at the internal ribosome entry site of hepatitis C virus genome visualized by atomic force microscopy. Nucleic Acids Res. 2015;43:565–580. doi: 10.1093/nar/gku1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shenvi CL, Dong KC, Friedman EM, Hanson JA, Cate JH. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations–implications for the study of ribosome dynamics. RNA. 2005;11:1898–1908. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swaminathan R. Magnesium metabolism and its disorders. Clin Biochem Rev (Ultimo, Aust) 2003;24:47–66. [PMC free article] [PubMed] [Google Scholar]
- 54.Romani AM. Cellular magnesium homeostasis. Arch Biochem Biophys. 2011;512:1–23. doi: 10.1016/j.abb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 56.Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Treder K, Pettit Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholson BL, White KA. Context-influenced cap-independent translation of Tombusvirus mRNAs in vitro. Virology. 2008;380:203–212. doi: 10.1016/j.virol.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Nicholson BL, Zaslaver O, Mayberry LK, Browning KS, White KA. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J Virol. 2013;87:1872–1883. doi: 10.1128/JVI.02711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabian MR, White KA. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Na H, Fabian MR, White KA. Conformational organization of the 3′ untranslated region in the tomato bushy stunt virus genome. RNA. 2006;12:2199–2210. doi: 10.1261/rna.238606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez de Quinto S, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Serrano P, Pulido MR, Saiz M, Martinez-Salas E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J Gen Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- 64.Huang SW, Chan MY, Hsu WL, Huang CC, Tsai CH. The 3′-terminal hexamer sequence of classical swine fever virus RNA plays a role in negatively regulating the IRES-mediated translation. PLoS One. 2012;7:e33764. doi: 10.1371/journal.pone.0033764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alvarez DE, Filomatori CV, Gamarnik AV. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375:223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friebe P, Harris E. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J Virol. 2010;84:6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008;373:1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Barry JK, Miller WA. A -1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci U S A. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romero-Lopez C, Barroso-Deljesus A, Garcia-Sacristan A, Briones C, Berzal-Herranz A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′X-tail region. Nucleic Acids Res. 2014;42:567–582. doi: 10.1093/nar/gkt841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oakland TE, Haselton KJ, Randall G. EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J Virol. 2013;87:6625–6634. doi: 10.1128/JVI.01006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murakami K, Abe M, Kageyama T, Kamoshita N, Nomoto A. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3' untranslated region of RNA. Arch Virol. 2001;146:729–741. doi: 10.1007/s007050170142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.