Abstract
Translesion synthesis (TLS) is the mechanism in which DNA polymerases (TLS polymerases) bypass unrepaired template damage with high error rates. DNA polymerase η and ζ (Polη and Polζ) are major TLS polymerases that are conserved from yeast to humans. In this study, we quantified frequencies of basesubstitutions by yeast Polη and Polζ on undamaged and abasic templates in vitro. For accurate quantification, we used a next generation sequencing (NGS)-based method where DNA products were directly analyzed by parallel sequencing. On undamaged templates, Polη and Polζ showed distinct basesubstitution profiles, and the substitution frequencies were differently influenced by the template sequence. The base-substitution frequencies were influenced mainly by the adjacent bases both upstream and downstream of the substitution sites. Thus we present the base-substitution signatures of these polymerases in a three-base format. On templates containing abasic sites, Polη created deletions at the lesion in more than 50% of the TLS products, but the formation of the deletions was suppressed by the presence of Polζ. Polζ and Polη cooperatively facilitated the TLS reaction over an abasic site in vitro, suggesting that these two polymerases can cooperate in efficient and high fidelity TLS.
1. Introduction
DNA damages are introduced by variety of environmental and metabolic agents and impede DNA replication, thereby becoming lethal threats to the cell. The DNA lesions can be restored by various DNA repair pathways including base excision repair, nucleotide excision repair, mismatch repair, and homologous recombination (1). If not repaired, DNA lesions can be tolerated by a mechanism known as translesion synthesis (TLS), in which a special group of DNA polymerases (TLS polymerases) bypass the lesion without repairing it (2). Since damaged bases often cannot base-pair with incoming nucleotides, DNA damage tolerance is considered as a major mechanism of damage-induced mutagenesis.
An abasic (AP) site is one of the most common DNA lesions in normally growing cells. They are created by spontaneous hydrolysis of glycosyl bonds or by DNA glycosylases that remove damaged bases (3,4). Approximately 104 steady state AP sites were observed in a mammalian cell (5). Whereas the majority of AP sites are repaired, occasionally a DNA replication fork encounters an unrepaired AP site, leading to mutations (6,7).
DNA polymerases ζ (Polζ) and η (Polη) are key TLS polymerases that are conserved from yeast to humans (2,8,9). Polζ of yeast Saccharomyces cerevisiae is a complex of four-subunits (Rev3, Rev7, Pol31, and Pol32), and is a member of the B-family DNA polymerases (10–13). But unlike other family members, Polζ is a low fidelity polymerase that does not have 3′ to 5′ exonuclease activity (14–16). A purified Rev3-Rev7 complex has TLS activity on damaged templates, containing a cis-syn thymine dimer (14), thymine glycol (17), and AP site (16). Pol31 and Pol32, which had been known as subunits of Polδ, were recently identified as subunits of Polζ (18,19). Human Polζ has also been purified as a conserved four-subunit complex (20).
The REV3 gene of yeast, encoding the catalytic subunit of Polζ, is required for mutagenesis induced by UV (21–24), gamma-ray (25), psoralen-mediated interstrand crosslinking (26,27), several alkylating anti-cancer agents (28), and for approximately half of all spontaneous mutations (29). In mammals, REV3−/− knockout is embryonic lethal (30–32), and the conditional inactivation of mouse REV3 function shows severe UV-sensitivity, reduced spontaneous mutation rate, elevated sensitivity to cisplatin, and reduced immunoglobulin hypermutation (9,33–35).
Polη is a member of Y-family TLS polymerases (2,36). Yeast cells lacking Polη function (rad30) have increased sensitivity to UV (37). Loss of Polη function in humans causes xeroderma pigmentosum (38,39). Consistently, Polη has TLS activity over a cis-syn thymine dimer, which is a major damage caused by UV-irradiation (38,39). Polη can also bypass other DNA lesions including 8-oxoguanine (40), O6-methylguanine (41), and the cisplatin adduct (42). Polη is a low-fidelity polymerase even on undamaged template, with base-substitution frequencies between 10−1 and 10−4 (2,43–47). However, Polη mediates error-free TLS over cis-syn thymine dimer, by inserting two AMPs opposite to the damage (38,48–51).
Although Polζ and Polη can individually carry out TLS reactions in vitro, increasing evidence supports that TLS is mediated by more than one TLS polymerase (43,46,52–54). It has also been proposed that multiple polymerases are involved in TLS over AP sites (55,56).
Since the majority of mutagenesis involves error-prone DNA polymerases, it is fundamental to identify the profiles of mutations that are introduced by these polymerases. Detailed mutation signatures of individual TLS polymerases may be beneficial to understand the potential etiology of mutation signatures in human cancers (57), (58). In this report, taking an advantage of next generation DNA sequencing (NGS) technologies, we analyzed profiles of base-substitutions created by Polζ and Polη of S. cerevisiae.
2. Materials and methods
2.1. DNA
Synthetic DNA oligonucleotides (oligos) with AP sites were purchased from Integrated DNA Technology. Other oligos used in this study were purchased from Integrated DNA technology, Sigma, and Fisher. Sequences and uses of the oligos are summarized in Supplemental Table S1 and Fig. S1.
2.2. Purification of Proteins
Polη of S. cerevisiae was purified as described previously (59). The four-subunit complex of Polζ (Rev3/Rev7/Pol31/Pol32) was overexpressed in yeast BJ5465 harboring pESCTrp-POL31, pESCLeu-His6-POL32, and pESCUra-REV3/FLAG-REV7 by the same protocol used previously for the two-subunit Polζ purification (59). Cells were suspended in lysis buffer (50 mM Tris-HCl (pH7.5), 200 mM NaCl, 5% glycerol (vol/vol), and 10 mM imidazole) containing 1 mM PMSF and lysed using glass beads (0.5 mm) in a vortex mixer (30 sec of mixing followed by 30 sec of chilling on ice, repeated 10 times). The cell lysate was cleared by centrifugation at 20,000 rpm for 30 min at 4°C in a JA-25.50 rotor (Beckman). The supernatant was loaded onto an 8 ml Ni-Chelating Sepharose column (GE Healthcare) preequilibrated with cell lysis buffer. The column was washed with 100 ml of lysis buffer and with 100 ml of wash buffer (50 mM Tris-HCl (pH7.5), 100 mM NaCl, 5% glycerol (vol/vol), and 50 mM imidazole). Polζ was eluted with elution buffer (50 mM Tris-HCl (pH7.5), 200 mM NaCl, 5% glycerol (vol/vol), and 200 mM imidazole). The eluate was mixed with 0.5 ml of EZview Red ANTI-FLAG M2 Affinity Gel (Sigma) that was preequilibrated in TGE200 buffer (30 mM Tris-HCl (pH7.5), 200 mM NaCl, 5% glycerol (vol/vol), 1 mM EDTA) and incubated at 4°C for 2.5 hours with gentle shaking. The affinity beads were recovered in an Econo-column (BioRad) and washed with 7 ml TGE200 buffer 5 times. Polζ was eluted from the resin with 5 ml of TGE200 buffer containing 150 μg/ml FLAG peptide. The eluate was divided into small aliquots, and stored at −80°C. The four subunits of Polζ were identified using SDS-PAGE (Supplemental Fig. S1A), and the concentration of Polζ was determined by the band intensity of the catalytic subunit (Rev3) compared with the BSA standard (Pierce).
The replicative DNA polymerases Polδ and Polε of S. cerevisiae were used in control reactions. Polδ was purified as described previously (59). To purify the Polε catalytic subunit, the POL2 gene was amplified from yeast S288C genomic DNA, fused with N-terminal FLAG tag, and cloned on pESC-Trp (between Apal-Xhol) to produce pESC-Trp-FLAG-POL2. Polε (POL2 product) was expressed in freshly transformed BJ5465 cells harboring pESC-Trp-FLAG-POL2 by the same protocol that was used for Polζ preparation. Thawed cells were resuspended in TGE200 buffer (30 mM Tris-HCl (pH7.5), 200 mM NaCl, 5% glycerol (vol/vol), 1 mM EDTA) containing 1 mM PMSF. Cells were then lysed, cleared, and subjected to EZview Red ANTI-FLAG M2 Affinity Gel purification as described above. The eluate was aliquoted and stored at −80°C. The concentration of Pol2 was determined based on the band intensity on SDS-PAGE gel as described above.
2.3. DNA polymerase assay by denaturing polyacrylamide gel electrophoresis
The synthetic DNA primer TSO590 (34-mer) was labeled with 32P using T4-DNA kinase (New England Biolabs) and γ-32P-ATP (Perkin Elmer). The labeled primer (10 nM) was annealed with 11 nM of 99-mer template with an AP site (TSO526 or TSO589) or without AP site (TSO525), and incubated with Polδ (10 nM), Polε (30 nM), Polη (20 nM), or Polζ (20 nM) at 37°C in a 10 μl reaction containing 25 mM Tris-acetate (pH7.5), 4 mM MgCl2, 100 μg/ml BSA, 5 mM DTT, 1 mM ATP, 100 μM each of four dNTPs, and NaCl [73 mM (with Polδ), 87 mM (with Polε), 55 mM (with Polη), or 65 mM (with Polζ)]. At the times indicated in the Fig. 1 Caption, a small volume (typically 2.5 μl) of the reaction was withdrawn and mixed with 1.5× volume of stop buffer (20 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol in formamide) and heated at 95°C for 5 min. Labeled DNA products were separated by electrophoresis through 10% polyacrylamide gel (25 × 14.5 cm) in TBE buffer containing 7 M urea, and visualized with BioRad Molecular Imager Personal FX and quantified with Quantity-One software. When indicated in the figures, Polη and Polζ were mixed on ice for 3–5 min before adding to the reactions.
Fig. 1. Polη and Polζ can bypass the AP site in vitro.
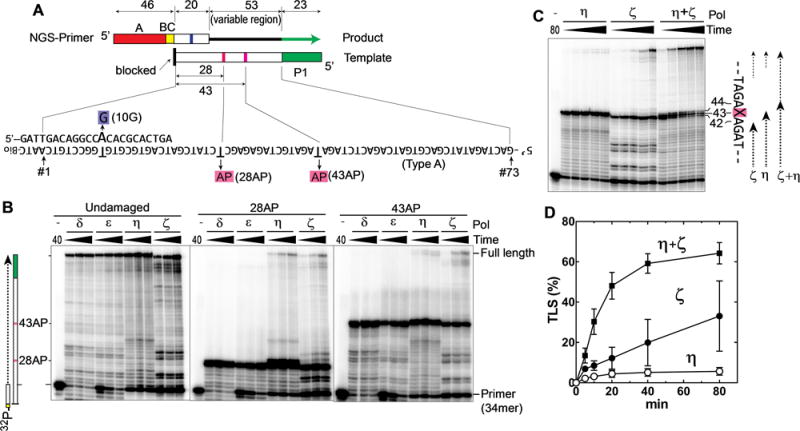
(A) Schematic drawing of Synthetic DNA substrates for TLS reactions. The NGS-primer contained an A-adaptor (red box) and a 6-nt barcode (yellow box, BC) for Ion Torrent PGM. Templates contained a P1-adaptor at the 5′-end (green box) and the complementary sequence to the primer (20-nt-long region) at the 3′ end. The product, which has both adaptors, is recognized by the NGS system that reads the sequences of the 73-nt region (position #1–73), in which the first 20-nt region (position #1–20) are part of the primer, and the remaining 53-nt region (position #21–73; variable region) is extended by yeast DNA polymerases. In addition to the template without lesion (type-A), we used templates with the same sequences but containing an AP site at the position #28 (28AP) or position #43 (43AP). All templates were modified with biotin at the 3′-OH group to prevent extension. The NGS primer had a one-base mismatch at position #10 (10G) to distinguish the primer-extension products from any template-extension product. (B) Primer extension reactions were carried out using the indicated polymerase and 32P-labeled primer (34mer) hybridized to the template with no damage, 28AP, or 43AP. After incubating for 2.5, 5, or 10 min (Polδ and ε), or 10, 20, or 40 min (Polη and ζ), a small portion was withdrawn and analyzed by denaturing polyacrylamide gel. (C) The same TLS reactions as in panel B were carried out on the template containing 43AP by Polη, Polζ, and a mixture of Polη and Polζ, and the reaction was stopped at 5, 10, 20, 40, and 80 min. (D) The TLS efficiency (% of the fully extended products in all DNA molecules that reached the AP site) was calculated from C and repeated experiments (error bar is SD (n=3)).
2.4. NGS sample preparation and analysis
The primer for each reaction contained an Ion-A adaptor and a specific barcode (see Fig. 1A, Supplemental Fig. S1B, and Table S1). In addition, the primers have a mismatched base “G” at the center of the primer-template hybridization region (Fig. 1A, “10G”). DNA synthesis reactions were carried out using indicated polymerases and templates under the same conditions as for gel electrophoresis assay above, except that the reaction volume was 50 μl. After 15 min (for Polδ and Polε reactions) or 60 min (for Polη and Polζ reactions), the reaction was stopped by adding 5 μl of 0.5 mM EDTA and treated with phenol/chloroform, and the DNA was precipitated with ethanol and resuspended in 10 μl of H2O. Since each reaction should produce DNA products with unique barcode, samples were pooled and analyzed by lon Torrent PGM system (Thermo Fisher Scientific). Same reactions were repeated two to three times.
All output read sequences were automatically sorted by barcodes. Output sequence files in FASTAQ format were analyzed by tools in Galaxy (https://usegalaxy.org/)(60) local server set-up as follows. First, quality format was converted to Sanger and Illumina 1.8+ quality score type using the FASTAQ Groomer tool. Then, using Filter FASTAQ tool, we eliminated all read sequences that had any base call with quality score lower than p=0.05, and that were longer by 7-nt or shorter by 13-nt than the expected error-free product. The numbers of read sequences after this screening (N) are shown in Supplemental Table S2 and S3. The sequence format was converted to FASTA using the fastaq_to_fasta tool. The FASTA files were analyzed using Lastz sequence alignment tool (Galaxy tool version 1.1.1), where base-substitutions were reported in the “Polymorphisms” format. Output of the Lastz analysis was processed by Microsoft Excel, selecting read sequences that contained the “10G”. This process eliminated the sequences that were created by extension of the template, not primer. Read sequences after this point are considered “qualified reads”. The numbers of the qualified reads (n) are shown in Supplemental Table S2 and S3. To avoid sampling biases, the error rate (%) in the qualified reads was calculated first at individual base as [the numbers of error at each position/the number of the qualified reads at the position] ×100% in each reaction. Then the % values of base-substitutions at individual bases were averaged in repeated reactions (Fig. 2 and Supplemental Fig. S2). Thus all base-substitution frequencies shown in Figs. 2 to 6 are the % of substitutions per synthesis of the particular base.
Fig. 2. Profiles of base-substitutions on NGS-primer-extension products.
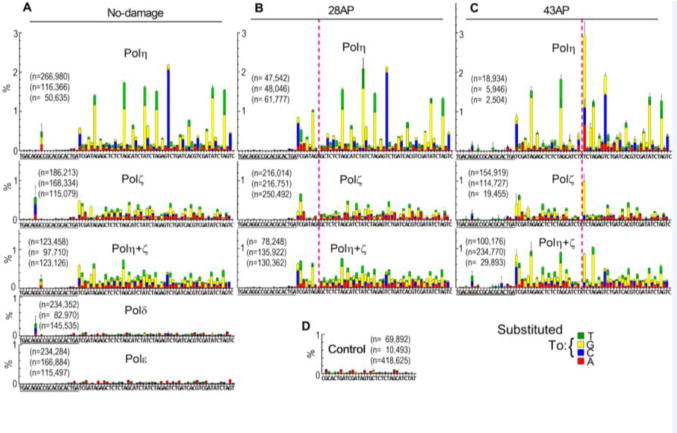
(A–C) In vitro primer extension reactions were carried out with the substrates shown in Fig. 1A, containing no lesion (A), 28AP (B), or 43AP (C), and indicated DNA polymerases, and the products were analyzed by the NGS system. Base-substitutions in the output sequences were mapped on the reference sequence after barcode sorting and quality control. The reference sequence (sequence of growing strand) is given under each graph, in which underlined region belongs to the primer. Pink dashed lines indicate the positions of AP sites. Base-substitution data at the AP sites are omitted in this figure (shown in Fig. 5). Vertical axis (%) indicates the frequency of base-substitution per synthesis at indicated position of template. Reactions were repeated three times and mean data with SD of the triplicate experiments are shown. The number of qualified output sequences that was obtained from each reaction is given as “n”. See text and Materials and Methods for more details. (D) An oligo containing two adaptors (TSO561, Supplemental Fig. S1C and Table S1) was directly subjected to NGS analysis to quantitate background of the analysis.
Fig. 6. Base-substitution signatures of Polη and Polζ on undamaged template in a three-base format.
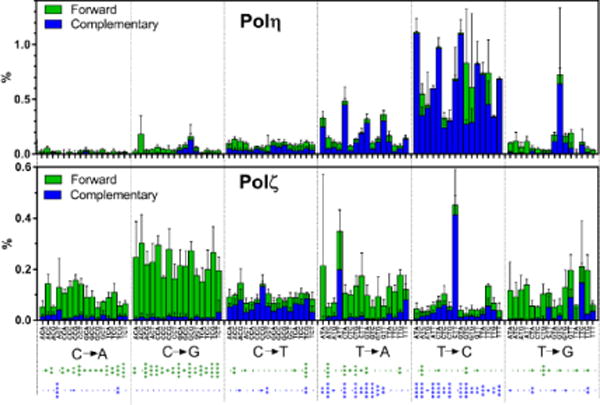
Substitution frequencies of Polη and Polζ at the center of three-nucleotide sequences were calculated from the data presented in Fig. 2A and Supplemental Fig. S2. Frequencies of complementary changes are organized in the same columns in different colors. For example, “5′ATC→5′AGC” substitution and “5′GAT→5′GCT” substitution are complementary to each other, thus their frequencies are categorized in the “T → G” group and stacked in the single column (ATC) as green (forward) and blue (complementary) bars, respectively. Asterisks below the graphs indicate significance of difference in the two polymerases making forward (green asterisks) and complementary (blue asterisks) base-substitutions (*, **, ***, and **** indicate p<0.05, p<0.01, p<0.001, and p<0.0001, respectively).
For Figs. 3, 4, and 6, we carried out reactions with six undamaged templates with different sequences (type A to F, Supplemental Fig. S1B), each of which were repeated two to three times. Base-substitution frequencies were calculated at individual bases in the 50-nt region of each template (position #22–71, Supplemental Fig. S1B). Thus substitution frequencies of each base in total 300-nt region were obtained. As a background, the base-substitution frequencies created by Polδ was subtracted from ones created by TLS polymerases at each bases in the 300-nt region. These subtracted % values were used to make average of entire region (Fig. 3A), analysis of sequence context (Fig. 3B, C and Fig. 4) and the three-base signatures (Fig. 6).
Fig. 3. Analysis of base-substitution by Polη and Polζ on undamaged templates.
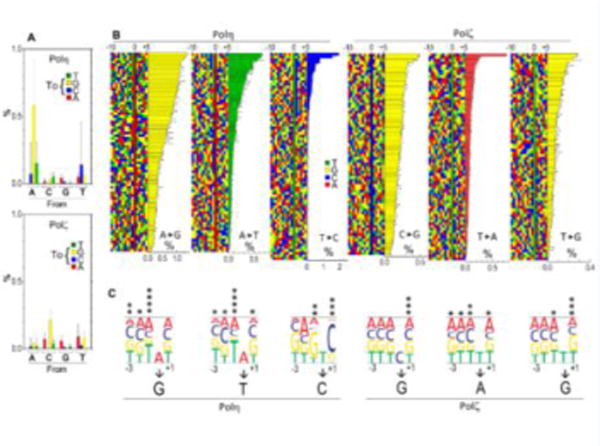
Base-substitution frequencies in total of 300-nt of undamaged template DNA, (position #22–71 of type A to F templates, constituted by 73 “A”, 74 “C”, 77 “G”, and 76 “T”) were analyzed. (A) Average base-substitution frequencies by Polη and Polζ. Error bars are SD (n are from 73 to 77). (B) The three most predominant base-substitution types by Polη and Polζ were selected from (A), and individual sites of the substitutions were sorted by frequencies. Colored squares indicate DNA sequence of the growing strand from 10-nt upstream (−10) to 5-nt downstream (+5) of the substitution sites. All substitution sites were sorted from highest (top) to lowest (bottom) frequency, which is indicated by the bar graphs. Positions of substitutions are indicated by “0”. (C) Effects of the bases from 3-nt upstream to 1-nt downstream of the substitution on the substitution frequencies. Height of each letters represents the relative substitution frequencies to the average value when the specific bases exist at given positions. Significance of difference between most and least prevalent bases at each position is indicated by asterisks (*, **, ***, and **** indicate p<0.05, p<0.01, p<0.001, and p<0.0001, respectively).
Fig. 4. Correlation of base-substitution frequencies created by Polη and Polζ on undamaged templates.
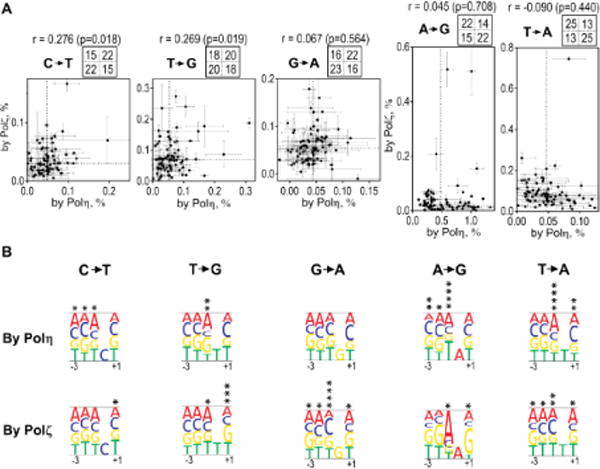
(A) Frequencies of indicated base-substitutions by Polη (x-axis) versus by Polζ (y-axis) at individual substitution sites are plotted. Error bars are SD (n=2–3). Dashed lines in the plotting area indicate medians, and numbers in four sections above each graph indicate the numbers of data points in corresponding sections in the plotting area. The value “r” is the Pearson correlation coefficient (with two-sided p values).
(B) Effects of the bases from 3-nt upstream to 1-nt downstream of the substitution on the substitution frequencies by each polymerase are expressed as heights of the letters, as in Fig. 3C.
2.5. Statistical analysis
When indicated, statistical significance was evaluated by two-tailed T-test and results were expressed by the number of asterisks (*, **, ***, and **** indicate p<0.05, p<0.01, p<0.001, and p<0.0001, respectively).
3. Results
3.1. Experimental design
To analyze base-substitution profiles that were created in vitro by two key TLS polymerases Polη and Polζ, we developed an NGS-based system consisting of synthetic DNA substrates (Fig. 1A, Primer and Template). Two NGS adaptors (A and P1 for Ion Torrent PGM) were separately located on the primer and template, thus only the fully extended products can be detected by the NGS system. Synthetic templates containing an AP site at 8-nt (28AP) or 23-nt (43AP) ahead of the 3′ end of the primer was used to mimic DNA damage. The 3′-OH group of the template DNA was modified with biotin to prevent it from undesired extension. In addition, to allow us to distinguish between the NGS-Primer-extension product and any Template-extension product produced, the NGS primers had a mismatched “G” at the center of the primertemplate hybridization region (Fig. 1A, “10G”). During NGS data analysis, we selected only the sequences that contained “10G” and calculated the base-substitution frequency in the DNA extension products containing the “10G”.
3.2. Polη and Polζ cooperatively bypass AP site
To confirm that our preparation of Polη and Polζ were enzymatically active and had TLS activity, we first carried out primer-extension of a 32P-labeled primer that was hybridized with a template with or without an AP site (Fig. 1B). As controls, we also analyzed Polδ and Polε that are high-fidelity replicative DNA polymerases. On undamaged template, all the polymerases extended the primer to the end of the template. When the template contained an AP site (28AP or 43AP), extension of the primer by Polδ and Polε were almost completely blocked at the AP site. In contrast, consistent with earlier studies (14,16,38,39), Polη and Polζ produced full-length products on the templates with AP site, demonstrating their TLS activity.
When both Polη and Polζ were present, the reaction produced greater amount of full-length product than the additive amount produced by each polymerase (Fig. 1C and D), indicating that these two polymerases cooperate for efficient TLS reaction. At the AP site, Polη predominantly produced a DNA molecule one-nucleotide longer than that was produced by Polζ, by incorporating a nucleotide opposite to the AP site (Fig. 1C, position 43). These results suggest that Polη incorporates one dNMP opposite to the AP site but unable to extend the unpaired nucleotide, and Polζ extends the 3′-OH of the unpaired nucleotide that is created by Polη.
3.3. Polη and Polζ have distinct base-substitution signatures on the undamaged template
Next we analyzed the nucleotide misincorporations in the fully extended products that were created by each polymerase. Non-labeled primers (NGS primers) were extended on templates (Fig. 1A) by the polymerases, and the products were directly subjected to NGS analysis (see Experimental Procedures for details). Fig. 2 shows the frequency of single base-substitutions on the template without lesion (A), with 28AP (B), and with 43AP (C), that are mapped on the 73-nt reference sequence. For simplicity, dAMP, dCMP, dGMP, and dTMP are abbreviated to “A”, “C”, “G”, and “T”, respectively. Misincorporation is expressed as resulting base-substitution. For example, misincorporation of “A” at “G” template results in substitution of “C” into “A”, which is expressed as “C→A” substitution. On the template without lesion, Polη and Polζ had significantly higher frequencies of base-substitution than Polδ or Polε. More importantly, Polη and Polζ showed distinct patterns of base-substitution, indicating that these TLS polymerases could be distinguished by their substitution patterns. Although the products of Polδ and Polε had detectable substitutions, their frequencies were similar to the control (Fig. 2D) where a synthetic oligonucleotide containing two adaptors was directly sequenced. This indicates that most of the observed base-substitutions in the Polδ- and Polε-mediated reactions were not created by these replicative DNA polymerases. Instead, we believe these changes were due to the errors in the synthetic DNA substrates or in NGS. Therefore, we considered these basesubstitution frequencies as background of the analysis, and subtracted them from the data obtained with TLS polymerases.
To obtain information about the influence of DNA sequence context on the base-substitution identity and frequency, we repeated the same analysis with five more templates with different sequences (type B to F; Supplemental Figs. S1B and S2). All templates had the P1-adaptor and the same primer-hybridization region but had different sequences in the intervening 53-nt region (position #21–73 in Fig. 1A and Supplemental Fig. S1B; variable region). The base-substitution frequencies at each base from position #22 to #71 (50-nt) of all six templates (total 300-nt) were determined. The average frequencies of the same base-substitution types (Fig. 3A and Supplemental Table S4) indicate that Polη introduced “A→G” substitution in the highest frequency (0.58%), and “A→T” (0.15%) and “T→C” (0.14%) had the second and third highest-frequencies, respectively. Polζ introduced “C→G” substitution most frequently (0.21%), and “T→A” (0.091%) and “T→G” (0.080%) had the second and third highest-frequencies, respectively. These results indicate that Polη and Polζ have different preferred base-substitutions types. Interestingly, the majority (66%) of base-substitutions by Polη occurred at the template base “T”, at which Polζ was most accurate (12%; Supplemental Table S4). Nearly half (47%) of the base-substitutions by Polζ occurred at the template base of “G”, at which Polη showed high fidelity (8.2%; Supplemental Table S4). In addition, the three most frequent substitutions made by Polζ were all substitutions of pyrimidines by purines, suggesting that Polζ more readily misincorporates a purine nucleotide at a template purine. These results indicate that Polη and Polζ have distinct base-substitution signatures, probably due to distinct mechanisms of nucleotide misincorporation.
Although Fig. 3A shows a trend in preferred base-substitution types of each polymerase, the error bars of the graphs show that there are considerable variations of frequencies in the same base-substitution types. This suggests that base-substitution frequencies are influenced by surrounding DNA sequences. To analyze the effects of the flanking DNA sequences on base-substitution frequency, we aligned the individual base-substitution sites in the order of substitution frequencies and compared the sequences (Fig. 3B). For each polymerase, we chose the three substitution types with the highest average frequencies (A→G, A→T, and T→C for Polη, and C→G, T→A, and T→G for Polζ) for the analysis. The alignments of Polη-mediated base-substitution sites show that some of the highly error-prone sites shared the same bases adjacent to the bases being substituted. For example, seventeen of the twenty most frequent sites for “A→G” substitutions by Polη had “T” at one nucleotide upstream of the base to be substituted (left of the asterisk on Fig. 3B; position −1). Similarly, “A→T” substitution by Polη also had “T” at position −1. In contrast, for “T→C” substitution by Polη, nineteen of the twenty most frequent sites had a “C” in the position +1 (downstream of the base to be substituted). To investigate further the contributions of surrounding nucleotides, we calculated the effect of each nucleotide at positions from −3 to +1 on substitution frequency (Fig. 3C). Both polymerases had influenced by sequence context. Although less extensively than Polη, Polζ was also influenced by flanking sequences. Most significantly, “T→G” substitution preferred “G” at +1 positions, and “T→A” substitution preferred “C” at −1 positions. In general, fidelity of both Polη and Polζ were most influenced by the nucleotides at positions −1 and +1, indicating that the fidelity of these polymerases was affected not only by the nucleotide providing the 3′-OH group, but also by the template base ahead of the growing DNA strand.
3.4. Polη and Polζ have distinct sequence-preferences when creating the same base-substitutions
Although the three most frequent base-substitutions created by Polη and Polζ were distinct, other basesubstitutions were produced above the background frequencies by both polymerases. This allowed us to compare the sequence-preferences of the two polymerases when they introduce the same basesubstitutions. Fig. 4 shows the correlations of frequencies of the Polη- and Polζ-mediated substitutions for the same substitution types. The correlation factor r for some substitutions (“C→T” and “T→G”) showed weak but significant positive correlations (r = 0.28 (p=0.018) and 0.27 (p=0.019), respectively), indicating that fidelity of Polη and Polζ were similarly affected to some extent by the same sequence context. Other substitutions (“G→A”, “A→G”, and “T→A”) showed almost no correlation (r = 0.067, 0.045, and −0.09, respectively), indicating that Polη and Polζ did not share the sequence-preferences for these substitutions. We also analyzed the effects of surrounding sequences on the substitution frequencies (Fig. 4B). With some base-substitutions, we found clear differences in preferred sequences. The most notable is that, for “A→G” substitution, Polη was most error-prone when the −1 position was “T”, whereas Polζ was least error-prone in this sequence. Polζ frequently introduced “A→G” substitution in the middle of “AAG” sequence. For both “T→A” and “G→A” substitutions, “C” at the −1 position was significantly preferred by Polζ but not by Polη. Even in “T→G” substitution, which showed positive correlation, Polζ showed strong preference of “G” at the +1 position, which was not preferred by Polη. These results indicate that, even when they are making same base-substitution, Polη and Polζ have distinct sequence-preferences. It is noteworthy that, based on the results shown in Fig. 3 and 4, base-substitution frequencies were most influenced by the sequence at +1 and −1 position. Thus we concluded that the base-substitution signatures could be best expressed in three nucleotide sequences (see Discussion).
3.5. TLS-associated base-substitutions at and around the AP site
Next we analyzed TLS products created on the AP-containing templates by NGS. As expected from their TLS activities (Fig. 1), both Polη and Polζ produced sufficient numbers of output in NGS analysis (Supplemental Table S2). Profiles of base-substitution frequencies on templates containing 28AP and 43AP are shown in Fig. 2B and C, respectively. Nucleotides inserted opposite to the 28AP and 43AP sites are shown in Fig. 5A. Remarkably, Polη-mediated TLS resulted in deletions in approximately 60% of products on both AP-containing templates. In contrast, Polζ products showed very few deletions at either AP site. The Polη-dependent deletions were clearly different at 28AP and 43AP. Whereas the vast majority (96%) of the deletions at 28AP were single-nucleotide deletions (open bar), 92% of deletions at 43AP were multi-nucleotide deletions (grey bar). Most of the deletions at 43AP were a four-nucleotide deletion (A43–46, Fig. 5B), and may have been the result of template-slippage between repeated “TCT” sequences 5′ and 3′ to the AP site. Many other multi-nucleotide deletions were also associated with tandemly repeated sequences (arrows in the Fig. 5B), indicating that template-slippage was a major mechanism of damage-induced deletions by Polη at 43AP.
Fig. 5. Polη and Polζ have distinct nucleotide incorporation and deletion spectra at AP site.
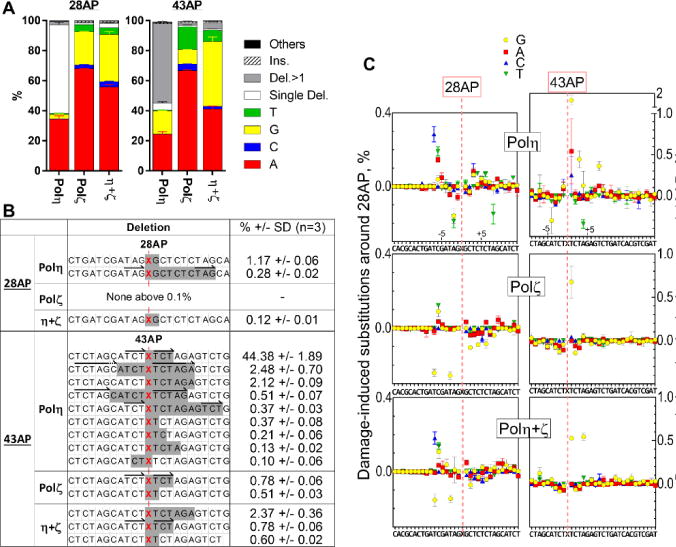
(A) Nucleotides opposite to the AP sites. TLS reaction was carried out on the templates containing 28AP or 43AP by Polη, Polζ, or a mixture of Polη and Polζ (η+ζ). The fully extended TLS products were analyzed by NGS, and the nucleotides inserted opposite to the AP sites (colored bars), single nucleotide and multi-nucleotide deletions to skip the lesion (open and grey bars, respectively), more than one nucleotide insertions over the lesion (striped bar) and others (black bar) were quantified. (B) Analysis of the multi-nucleotide deletions at AP sites. Shaded sequences indicate the sites of deletion at indicated percentages in the TLS products. The red “X” is the position opposite to the AP site. All the deletions produced at the frequencies above 0.1% in each TLS product were shown. (C) Effects of AP sites on base substitutions in the flanking regions. Base substitutions that were changed by the presence of AP sites were calculated by subtracting the substitution frequencies on the undamaged template from the frequencies on the AP-containing templates.
When inserting a nucleotide at the AP site, both polymerases predominantly used “A” and very little “C”. This phenomenon has been known as “A-rule” (61), which has been observed in E. coli and in some eukaryotic systems including a yeast strain that lacks Rev1 function (16,38,49,62). The frequency of the “A” insertion was different in two AP sites, indicating that surrounding sequences also affect the nucleotides inserted opposite the template AP sites. Interestingly, a preincubated mixture of Polη and Polζ incorporated distinct ratios of nucleotides (Fig. 5A, “η+ζ”). The two-polymerase reaction created few deletions, indicating that Polζ suppressed deletion formation by Polη. In addition, the two-polymerase reaction inserted “G” at significantly higher frequencies than either single-polymerase reaction (Fig. 5A, yellow bar). This was particularly evident at 43AP, where “G” was inserted at a higher frequency than “A”. This cannot be easily explained by a mechanism in which the polymerases work additively during the TLS. Potential mechanisms to explain this phenomenon are discussed below.
Fidelities of TLS polymerases synthesizing near but not exact opposite the AP site may be also influenced by the lesion. To highlight the effect of the AP sites on the base-substitution frequencies near the lesions, we subtracted the substitution frequencies on the undamaged template from ones on the AP-containing templates (Fig. 5C). As expected, the base-substitution frequencies downstream of 43AP were significantly increased compared to the control template. At one-nucleotide downstream of the 43AP site (+1 position), Polη misincorporated “G” (yellow circle) in significantly (p<0.005) higher frequencies than it did on undamaged template. Polζ also misincorporated “G” at the +1 position (p<0.005) but at a lower frequency than Polη. In addition, Polη-mediated substitution frequency continued to display elevated levels up to 5- to 8-nt downstream of 43AP. In contrast, Polη showed almost no AP-associated substitutions beyond the +1 position. Although fewer base-substitutions were observed downstream of 28AP than of 43AP, Polη also introduced substitutions slightly above background 3- to 6-nt downstream of the 28AP (e.g. misincorporation of “A”, “T”, and “T” at position +3, +4, and +6; p<0.01 and p<10−4, and p<0.05, respectively). Polζ showed little increase in base-substitution frequencies after 28AP. Unexpectedly, the AP sites also influenced the base-substitution frequencies upstream of the lesion. We observed reproducible (in triplicate) increases in base-substitution frequencies by Polη 6- to 7-nt upstream of 28AP (e.g. misincorporation of “C”, “A”, and “T” at position −7, −6, and −6; p<10−4, p<10−4, p<0.001, respectively), and similar trend in 43AP to a lesser extent. Furthermore, Polη-mediated base-substitution frequencies were decreased by the lesion (below 0% in the Fig. 5C) at 1- and 2-nt upstream of 28AP (misincorporation of “T”, “G”, and “A” at position −2, −2, and −1; p<0.001, p<0.05, p<0.05, respectively), suggesting that the lesion might increase the fidelity of Polη at these positions. Similar patterns (positive around 6-nt before and negative around 2-nt before the lesion) were also observed, though less pronounced, at the 43AP with Polη. The types and exact positions of the base-substitutions were different, indicating that these effects were not due to pre-existing errors on the synthetic DNA templates. Currently we do not have clear explanation for these observations.
4. Discussion
Since NGS technology allowed us to conveniently examine millions of independent DNA sequences, it has been used to quantify mutation frequencies in vivo and in vitro (63–65). In this paper, by analyzing in vitro DNA products directly with NGS system, we quantified base-substitution frequencies introduced by the error-prone DNA polymerases. This method did not use PCR amplification before loading the DNA product on the PGM beads, therefore output sequences should represent fully extended DNA products without bias. Thus, we believe that even low base-substitution frequencies that are above background reflect the fidelity of the TLS polymerases when the numbers of qualified reads (n) are large enough. This analysis not only provides images of base-substitution signatures of error-prone DNA polymerases, but can also be applied for further studies of other DNA polymerases, other forms of DNA damage, and other potential mutagenic biochemical processes. However, because the background misincorporation rate was approximately 0.02–0.05% per base, our method could not quantify the fidelity of replicative polymerases Polδ or Polε. In addition, we could not accurately quantify insertions/deletions, because of the high background of the signals on the undamaged templates (Supplemental Fig. S3). The observed background of insertion/deletion was likely caused by errors in the synthetic DNA templates and in the NGS system.
Our results reveal that the base-substitution frequencies by Polη and Polζ on undamaged DNA template were influenced by adjacent bases both upstream and downstream of the substitution sites (Fig. 3 and 4), though there were exceptions. Thus we expressed their base-substitution signatures in the format of substitution frequencies in the context of three-base sequences (Fig. 6). In addition, to simulate in vivo mutational analyses, we organized our data into the well-established format that has been used to classify mutation signatures in human cancers (57,66), in which all base-substitutions are reported as substitutions of pyrimidines. Although our in vitro analysis separately quantified “C→A” and “G→T” substitutions, for example, these complementary changes cannot normally be distinguished in vivo. Therefore, in Fig. 6, the frequencies of two complementary base-substitutions are added in a single column in separate colors. This approach may simulate in vivo mutation signatures by yeast Polη and Polζ in the absence of repair mechanisms. Since our in vitro signatures were created on undamaged template by yeast polymerases, it should not match any human cancer signature that should be created in the human genome by multiple mechanisms including errors in DNA repair, DNA replication, and error-prone DNA synthesis. However, we believe that this approach is useful for future research toward a mechanistic understanding of cancer-causing mutational processes.
Fig. 1 showed that Polη and Polζ facilitate TLS over AP sites to a higher degree than the combined efforts of each alone, supporting the two-polymerase mechanism (2,46,55,67,68) of TLS where Polη inserts a nucleotide opposite to the damage and Polζ extends the 3′OH of the unpaired nucleotide (15,16,69). Our results (Fig. 5A) indicate that Polζ can suppress Polη-mediated formation of deletions at AP site. Since deletion can destroy gene function by frame-shift in the cell, the reduction of deletion frequency demonstrated here might be an important component of the damage-tolerance mechanism. Polζ also suppressed the Polη-mediated base-substitutions downstream of the AP site (Fig. 5C), supporting the idea that Polζ can efficiently extend the 3′-OH of unpaired nucleotides at the damage site with a low error frequency. Together, these findings enforce the two-polymerase mechanism of TLS across AP sites.
Fig. 5A shows that the TLS product that was produced in the presence of both Polη and Polζ had a base-substitution signature distinct from that of either individual polymerase. More specifically, the two-polymerase TLS is more likely to have a “G” opposite to the AP site than either single-polymerase reactions. This may be due to protein-protein interaction of the two polymerases, or it may be caused by potential differences in extension efficiencies from different nucleotides (“G” or “A”) opposite to the lesion. As noted above, our system determined the sequences of fully extended products of TLS reactions, not intermediates that were stalled at the lesion. Therefore, if Polη incorporated more “G” than “A” at the AP site but could not extend the unpaired “G”, such intermediates would not be detected by the analysis. If Polζ can extend a mismatched “G” efficiently, then the final products of the two-polymerase reaction would contain more “G” than the Polη single-polymerase product. In fact this interpretation is consistent with independent steady-state kinetic studies showing that “G” is more efficiently incorporated than “A” opposite to AP site by Polη (70) and that unpaired “G” is most efficiently extended by Polζ (16).
Since we used a method that is not commonly used to analyze fidelity of DNA polymerases, it may be useful to compare our results with already published fidelity data. The fidelity of TLS polymerases on both un-damaged and damaged templates have been studied extensively (2,46,71). The average basesubstitution frequencies obtained in this study (Fig. 3A) are largely consistent with those reported in previous studies, in which error frequencies were measured by the gel fidelity assay in vitro or by sequencing individual plasmid isolates after genetic screening. For example, this study shows that Polη is prone to introduce “A→G” and “A→T” substitutions, consistent with previous reports (38,48,49). We also show that Polζ produces purine: purine mismatches at high frequency, consistent with previous reports (15,69,72). On templates containing AP sites, we show that Polη introduced deletions by traversing the lesion, or incorporated “A” or “G” opposite to the AP site, which is also roughly consistent with previous reports (55,67,70,73,74). These indicate that our system reliably quantified base-substitution frequencies.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Institutes of Health [R15GM116098]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Ronan Carol (Ohio University) for useful advice about NGS system, Bill Broach and Rachel Yoho (Ohio University Genomics Facility) for NGS operation. We also thank Kalen Robeson (Ohio University) for the bioinformatics Galaxy local server setup, Don Holzschu (Ohio University), Ronan Carol, and Noriko Kantake (Ohio University), and Frank Drews (Ohio University), for comments on the manuscript.
Appendix A. Accession number
NGS read data for this work have been deposited with the figshare (https://figshare.com) under “NGS-based analysis of base-substitution signatures created by yeast DNA polymerase eta and zeta on undamaged and abasic DNA templates in vitro”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
None declared.
References
- 1.Boiteux S, Jinks-Robertson S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013;193:1025–1064. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 6.Neto JB, Gentil A, Cabral RE, Sarasin A. Mutation spectrum of heat-induced abasic sites on a single-stranded shuttle vector replicated in mammalian cells. J Biol Chem. 1992;267:19718–19723. [PubMed] [Google Scholar]
- 7.Knobel PA, Marti TM. Translesion DNA synthesis in the context of cancer research. Cancer cell international. 2011;11:39. doi: 10.1186/1475-2867-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence CW, Maher VM. Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2001;356:41–46. doi: 10.1098/rstb.2000.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 10.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito J, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubscher U, Nasheuer HP, Syvaoja JE. Eukaryotic DNA polymerases, a growing family. Trends Biochem Sci. 2000;25:143–147. doi: 10.1016/s0968-0004(99)01523-6. [DOI] [PubMed] [Google Scholar]
- 14.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence CW, Gibbs PE, Murante RS, Wang XD, Li Z, McManus TP, McGregor WG, Nelson JR, Hinkle DC, Maher VM. Roles of DNA polymerase zeta and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb Symp Quant Biol. 2000;65:6169. doi: 10.1101/sqb.2000.65.61. [DOI] [PubMed] [Google Scholar]
- 17.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta (zeta) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc Natl Acad Sci U S A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YS, Gregory MT, Yang W. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemontt JF. Mutants of Yeast Defective in Mutation Induced by Ultraviolet Light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics. 1979;92:397–408. doi: 10.1093/genetics/92.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence CW, Christensen R. UV mutagenesis in radiation-sensitive strains of yeast. Genetics. 1976;82:207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKee RH, Lawrence CW. Genetic analysis of gamma-ray mutagenesis in yeast. II. Allele-specific control of mutagenesis. Genetics. 1979;93:375–381. doi: 10.1093/genetics/93.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques JA, Moustacchi E. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics. 1980;95:273–288. doi: 10.1093/genetics/95.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cassier-Chauvat C, Moustacchi E. Allelism between pso1-1 and rev3-1 mutants and between pso2-1 and snm1 mutants in Saccharomyces cerevisiae. Curr Genet. 1988;13:37–40. doi: 10.1007/BF00365754. [DOI] [PubMed] [Google Scholar]
- 28.Ruhland A, Brendel M. Mutagenesis by cytostatic alkylating agents in yeast strains of differing repair capacities. Genetics. 1979;92:83–97. doi: 10.1093/genetics/92.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96:819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- 31.Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 32.Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 33.Lange SS, Bedford E, Reh S, Wittschieben JP, Carbajal S, Kusewitt DF, DiGiovanni J, Wood RD. Dual role for mammalian DNA polymerase zeta in maintaining genome stability and proliferative responses. Proc Natl Acad Sci U S A. 2013;110:E687–696. doi: 10.1073/pnas.1217425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu F, Lin X, Okuda T, Howell SB. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res. 2004;64:8029–8035. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- 35.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 37.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 39.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 40.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 41.Haracska L, Prakash S, Prakash L. Replication past O(6)-methylguanine by yeast and human DNA polymerase eta. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry. 2000;39:45754580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 43.Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 45.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 46.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 48.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc Natl Acad Sci U S A. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 50.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. Embo J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu SL, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase eta for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesionbypass polymerases. Curr Opin Genet Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 54.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 55.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi JY, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV1. J Mol Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Holzschu DL, Sugiyama T. PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase delta, eta, and zeta activities. Proc Natl Acad Sci U S A. 2013;110:7672–7677. doi: 10.1073/pnas.1222241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, Galaxy T. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 62.Gibbs PE, Lawrence CW. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J Mol Biol. 1995;251:229–236. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 63.Chang SC, Fedeles BI, Wu J, Delaney JC, Li D, Zhao L, Christov PP, Yau E, Singh V, Jost M, et al. Next-generation sequencing reveals the biological significance of the N(2),3-ethenoguanine lesion in vivo. Nucleic Acids Res. 2015;43:5489–5500. doi: 10.1093/nar/gkv243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taggart DJ, Camerlengo TL, Harrison JK, Sherrer SM, Kshetry AK, Taylor JS, Huang K, Suo Z. A high-throughput and quantitative method to assess the mutagenic potential of translesion DNA synthesis. Nucleic Acids Res. 2013;41:e96. doi: 10.1093/nar/gkt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K, Ma X, Zhang X, Wu D, Sun C, Sun Y, Lu X, Wu CI, Guo C, Ruan J. Using ultra-sensitive next generation sequencing to dissect DNA damage-induced mutagenesis. Scientific reports. 2016;6:25310. doi: 10.1038/srep25310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 69.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase eta. J Biol Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 71.Rattray AJ, Strathern JN. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 72.Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 74.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


