Abstract
Human spermatogenesis includes three main stages, namely, the mitosis of spermatogonia, meiosis of spermatocytes, and spermiogenesis of spermatids, which are precisely regulated by epigenetic and genetic factors. Abnormality of epigenetic and genetic factors can result in aberrant spermatogenesis and eventual male infertility. However, epigenetic regulators in controlling each stage of normal and abnormal human spermatogenesis remain unknown. Here, we have revealed for the first time the distinct microRNA profiles in human spermatogonia, pachytene spermatocytes, and round spermatids between obstructive azoospermia (OA) patients and non-obstructive azoospermia (NOA) patients. Human spermatogonia, pachytene spermatocytes, and round spermatids from OA patients and NOA patients were isolated using STA-PUT velocity sedimentation and identified by numerous hallmarks for these cells. RNA deep sequencing showed that 396 microRNAs were differentially expressed in human spermatogonia between OA patients and NOA patients and 395 differentially expressed microRNAs were found in human pachytene spermatocytes between OA patients and NOA patients. Moreover, 378 microRNAs were differentially expressed in human round spermatids between OA patients and NOA patients. The differential expression of numerous microRNAs identified by RNA deep sequencing was verified by real-time PCR. Moreover, a number of novel targeting genes for microRNAs were predicted using various kinds of software and further verified by real-time PCR. This study thus sheds novel insights into epigenetic regulation of human normal spermatogenesis and the etiology of azoospermia, and it could offer new targets for molecular therapy to treat male infertility.
Keywords: microRNAs, human spermatogonia, pachytene spermatocytes, round spermatids, targeting genes
Introduction
Infertility affects 10%–15% of couples in the world, and half of them are attributed to male factors. Azoospermia comprises approximately 15% of male infertility,1, 2 and it can be classified into obstructive azoospermia (OA) and non-obstructive azoospermia (NOA). OA has been defined as the obstruction in any region of the sperm excurrent ductal system, with normal spermatogenesis in the seminiferous tubule. It may result from vasectomy, epididymis inflammation, or genetic disorders. Notably, spermatogenesis failure usually occurs in NOA patients, which can result from enorchia, microdeletion of Y chromosomes, chromosome abnormality, hypogonadotrophic hypogonadism, drug abuse, toxic exposure, and varicocele.3, 4, 5, 6, 7 Nevertheless, the etiology of NOA, especially the causes by epigenetic regulators, remains largely unknown.
Human spermatogenesis is a complex process that includes three main stages, i.e., the mitosis of spermatogonia, meiosis of spermatocytes, and spermiogenesis of spermatids. The Adark spermatogonia have been regarded as reserve stem cells in the seminiferous tubule. These cells can divide to retain the pool of stem cells, and they differentiate into the Apale spermatogonia that proliferate and produce type B spermatogonia and preleptotene spermatocytes.8, 9 After the induction of retinoic acid, preleptotene spermatocytes differentiate into primary spermatocytes.10, 11 Through two meiotic divisions, pachytene spermatocytes give rise to round spermatids. Finally, round spermatids transform to become elongated spermatids. Abnormality of genetic and epigenetic factors can lead to aberrant spermatogenesis and eventual male infertility. For instance, microdeletion of Y chromosome, chromosome abnormality, or CFTR gene mutation can result in male infertility.7 Epigenetic regulators include non-coding RNA, DNA methylation, and histone modifications. Small non-coding RNA is essential for spermatogenesis, and it consists of microRNA, endo-small interfering RNA (siRNA), Piwi-interacting RNA (piRNA), small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA). Notably, microRNAs have been demonstrated to be key regulators for cellular proliferation, differentiation, and apoptosis.12, 13, 14, 15 MicroRNAs act via inhibiting their binding targets through the base pairing of the seed sequence in mature microRNA and 3′ UTR of the targeting genes, which results in mRNA degradation of targeting genes or inhibition of their translation.16, 17 MicroRNA biogenesis consists of three steps: (1) primary microRNA transcripts (pri-microRNA) are cleaved into pre-microRNA (∼70 nt) by Drosha and DGCR8;18, 19 (2) pre-microRNA is exported from the nuclei to the cytoplasm by exportin 5; and (3) pre-microRNA in the plasma is cleaved into the mature microRNA by DICER.20 Specific knockout of these enzymes of mature microRNA biogenesis in germ cells can lead to severe disruption in spermatogenesis, implicating that microRNAs are required for spermatogenesis.21, 22 We have recently shown that microRNA-20 and microRNA-106a induce the self-renewal of mouse spermatogonial stem cells (SSCs) by targeting transcription factor STAT3 and cyclin D1.23 MicroRNA-21 has been found to play an important role in regulating the proliferation and maintenance of mouse SSCs by the control of transcription factor ETV5,24 whereas miRNA-221 and miRNA-222 maintain the undifferentiated status of mouse spermatogonia via inhibiting KIT expression.25 In addition, overexpression of microRNA let-7 may result in the decrease of germline stem cells in Drosophila.26 We have recently reported 599 differentially expressed microRNAs among normal human spermatogonia, pachytene spermatocytes, and round spermatids.27 However, epigenetic regulators, especially microRNAs, in controlling normal and abnormal human spermatogenesis remain unknown.
In the current study, we have uncovered for the first time distinct global microRNA profiles of human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients using RNA deep sequencing and identified their targets. Significantly, this study provides novel insights into mechanisms underlying azoospermia and offers new targets for treating male infertility.
Results
Morphological and Phenotypic Characteristics of Freshly Isolated Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids of OA Patients and NOA Patients
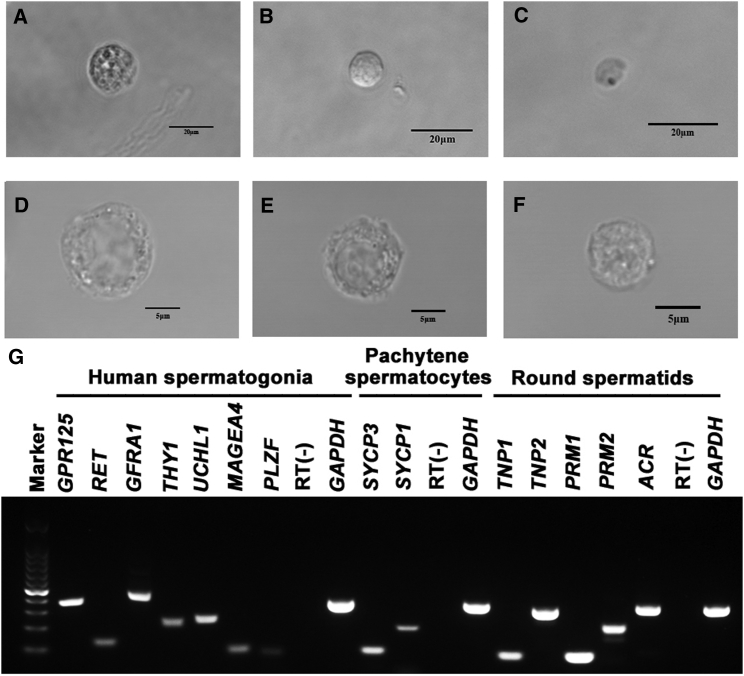
Human spermatogonia, pachytene spermatocytes, and round spermatids were isolated from OA patients and NOA patients using STA-PUT velocity sedimentation. In morphology, human spermatogonia (S2) (gradient fractions 10-15) were spherical and had round or oval nuclei (Figures 1B and 1E). The diameters of human spermatogonia were 9–12 μm under a phase-contrast microscope (Figure 1B) and a differential interference contrast (DIC) microscope (Figure 1E). Human pachytene spermatocytes (S1) (gradient fractions 3-8) had larger nuclei and condensed chromatin, with diameters of 14–16 μm (Figures 1A and 1D). Human round spermatids (S3) (gradient fractions 18-28) possessed round nuclei and diameters of 6–8 μm under the phase-contrast microscope (Figure 1C) and DIC microscope (Figure 1F).
Figure 1.
Morphological and Phenotypic Characterization of Freshly Isolated Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids
(A–C) Phase-contrast microscope revealed the morphology of the freshly isolated human pachytene spermatocytes (A), spermatogonia (B), and round spermatids (C) of OA patients. (D–F) DIC microscope showed the morphological characteristics of the freshly isolated human pachytene spermatocytes (D), spermatogonia (E), and round spermatids (F) of OA patients. Scale bars, 20 μm (A–C) and 5 μm (D–F). (G) RT-PCR revealed the transcripts of GPR125, RET, GFRA1, THY1, UCHL1, MAGEA4, and PLZF in the fleshly isolated spermatogonia, the expression of SYCP3 and SYCP1 in pachytene spermatocytes, and mRNA of TNP1, TNP2, PRM1, PRM2, and ACR in round spermatids. RNA without RT (RT-) but with PCR of GAPDH primers was utilized as negative controls, and GAPDH served as loading controls of total RNA.
The freshly isolated human spermatogonia, pachytene spermatocytes, and round spermatids were identified phenotypically. RT-PCR showed that transcripts of GPR125 (G protein-coupled receptor 125), RET (Ret proto-oncogene), GFRA1 (GDNF family receptor alpha 1), THY1, UCHL1 (ubiquitin C-terminal hydrolase L1), MAGEA4 (MAGE family member A4), and PLZF, markers for human spermatogonia or SSCs, were expressed in the freshly isolated human spermatogonia (Figure 1G). The mRNA of SYCP3 (synaptonemal complex protein 3) and SYCP1, hallmarks for spermatocytes, was detected in the freshly isolated human pachytene spermatocytes (Figure 1G), while the transcription of TNP1 (transition protein 1), TNP2, PRM1 (protamine 1), PRM2, and ACR (acrosin), markers for human spermatids, was present in the freshly isolated human round spermatids (Figure 1G). RNA without RT (RT-) but with PCR of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers served as negative controls (Figure 1G), and GAPDH was used as loading controls of total RNA (Figure 1G).
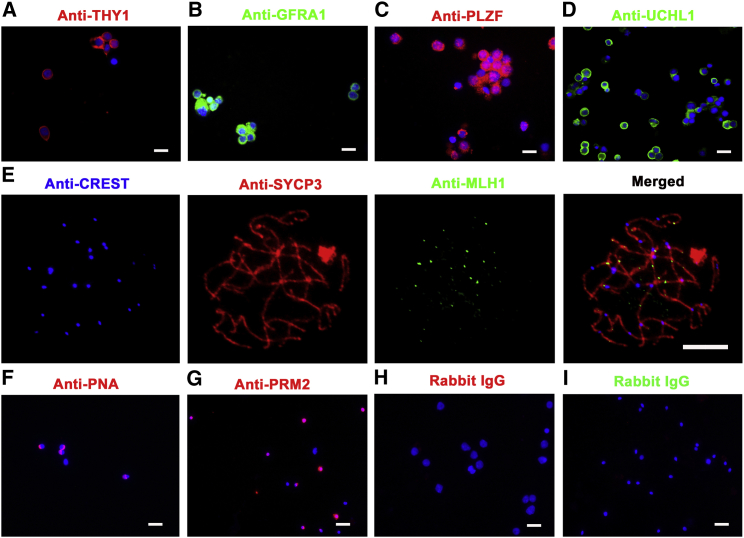
Immunocytochemistry further revealed that 90% of the freshly isolated human spermatogonia were positive for THY1 (Figure 2A), GFRA1 (Figure 2B), PLZF (Figure 2C), and UCHL1 (Figure 2D). Meiotic chromatin spread displayed that 92% of the freshly isolated human pachytene spermatocytes were co-expressing CREST, SYCP3, and MLH1 (Figure 2E). Immunocytochemistry demonstrated that 95% of the freshly isolated human round spermatids were positively stained for PNA (Figure 2F) and PRM2 (Figure 2G). Replacement of primary antibodies with isotype immunoglobulins G (IgGs) served as negative controls, and no immunostaining was observed in the cells (Figures 2H and 2I), thus verifying the specific immunostaining of the antibodies in these cells.
Figure 2.
Phenotypic Characterization of Freshly Isolated Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids
(A–D) Immunocytochemistry revealed the expression of THY1 (A), GFRA1 (B), PLZF (C), and UCHL1 (D) in the freshly isolated human spermatogonia. Scale bars, 10 μm. (E) Meiotic chromatin spread by triple immunostaining displayed the co-expression of CREST, SYCP3, and MLH1 in the freshly isolated human pachytene spermatocytes. Scale bar, 5 μm. (F–I) Immunocytochemistry demonstrated the expression of PNA (F), PRM2 (G), and rabbit IgG (I) in the freshly isolated human round spermatids and rabbit IgG (H) in the freshly isolated human spermatogonia. Scale bars, 10 μm.
RNA Deep Sequencing and Mapping of Reads in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids of OA Patients and NOA Patients to Human Reference Genome
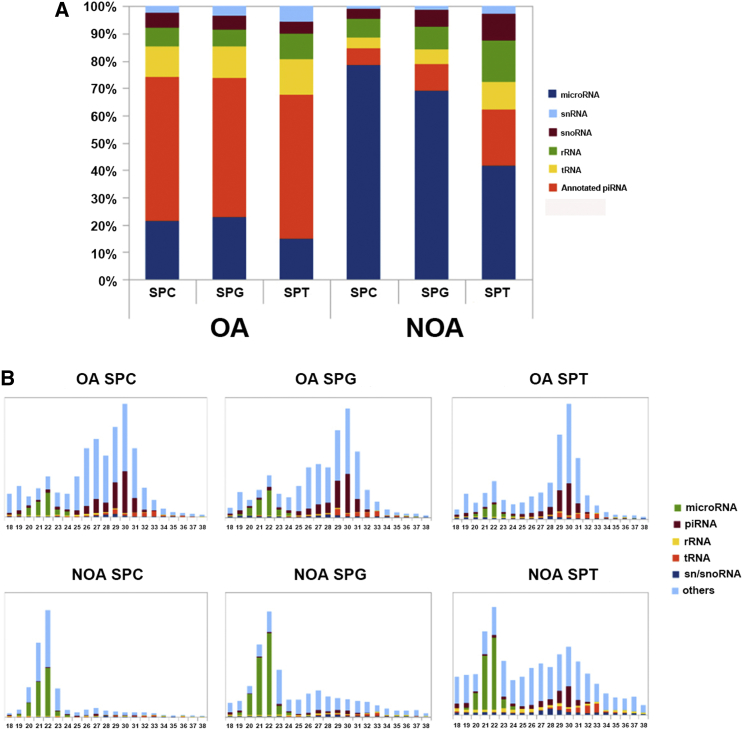
RNA deep sequencing was used to compare the small RNA profiles in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients. Over 6 million reads were detected in six kinds of cells, and the mapping ratio of each type of cells is illustrated in Table 1. Moreover, the reads were mapped to the annotated small non-coding RNAs as follows: microRNA, snRNA, snoRNA, tRNA, rRNA, and annotated piRNA. The expression levels of microRNAs were significantly higher in human spermatogonia, pachytene spermatocytes, and round spermatids of NOA patients compared to OA patients (Figure 3A). In contrast, the levels of piRNA were much lower in human spermatogonia, pachytene spermatocytes, and round spermatids of NOA patients than of OA patients (Figure 3A). As shown in Figure 3B, the reads were further mapped to the small non-coding RNAs in human spermatogonia, pachytene spermatocytes, and round spermatids of OA patients (upper panels) and NOA patients (lower panels) in terms of their nucleotide lengths.
Table 1.
Summary of the Reads in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids of OA Patients and NOA Patients
| Male Germ Cells | Total Reads | Quality Reads | Mapped Reads | Mapping Ratios (%) | |
|---|---|---|---|---|---|
| OA | pachytene spermatocytes | 10,036,603 | 9,883,597 | 4,614,721 | 50.62 |
| spermatogonia | 9,508,783 | 9,387,708 | 7,972,010 | 84.92 | |
| round spermatids | 7,789,625 | 7,694,720 | 6,595,277 | 85.71 | |
| NOA | pachytene spermatocytes | 10,299,748 | 10,157,255 | 8,696,852 | 85.62 |
| spermatogonia | 9,388,366 | 9,262,623 | 6,585,956 | 71.10 | |
| round spermatids | 7,818,228 | 7,698,708 | 3,869,551 | 50.26 |
Figure 3.
Composition and Length Distributions of Small Non-coding RNAs in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids of OA and NOA Patients
(A) Percentages of different kinds of small non-coding RNAs, including microRNA, snRNA, snoRNA, tRNA, rRNA, and annotated piRNA, mapped onto the human genome in human spermatogonia, pachytene spermatocytes, and round spermatids of OA and NOA patients. (B) Composition of small non-coding RNA categories according to their length distributions in human spermatogonia, pachytene spermatocytes, and round spermatids of OA and NOA patients. SPG, human spermatogonia; SPC, pachytene spermatocytes; SPT, round spermatids.
Distinct Global MicroRNA Profiles in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between NOA Patients and OA Patients with Normal Spermatogenesis
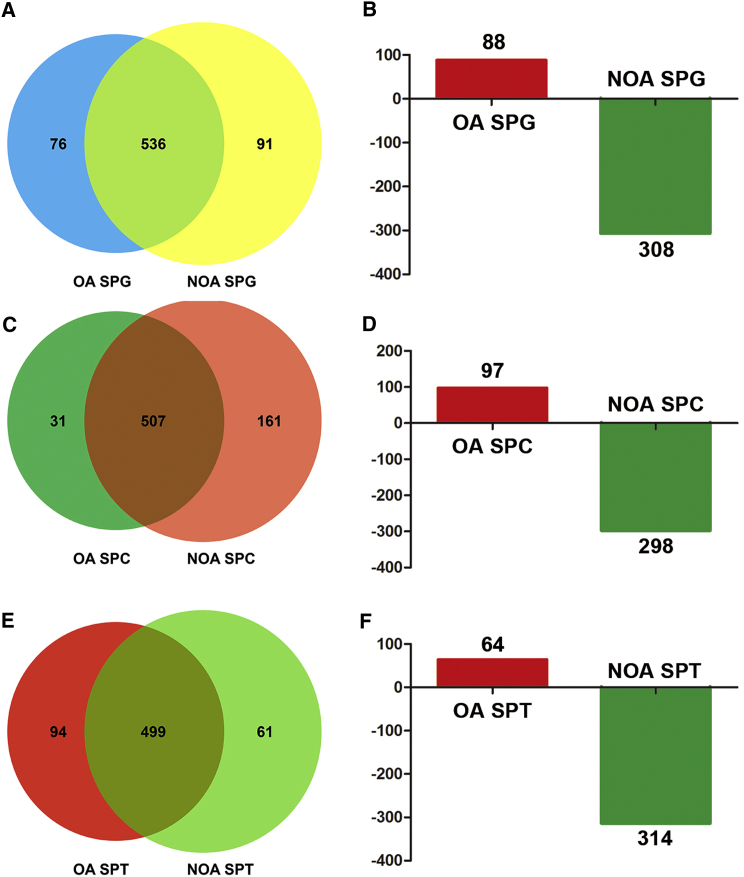
In total, 612 microRNAs were detected in human spermatogonia of OA patients, whereas there were 627 microRNAs in human spermatogonia of NOA patients. Notably, 76 microRNAs were expressed in human spermatogonia of OA patients but not in human spermatogonia of NOA patients (Figure 4A). Moreover, 91 microRNAs were expressed only in human spermatogonia of NOA patients (Figure 4A). We found that 88 microRNAs were upregulated in human spermatogonia of OA patients, whereas 308 microRNAs were downregulated in human spermatogonia of OA patients (Figure 4B). Meanwhile, 538 microRNAs were detected in pachytene spermatocytes of OA patients, and 668 microRNAs were present in human spermatocytes of NOA patients. There were 31 and 161 microRNAs that were found specifically in pachytene spermatocytes of OA and NOA patients, respectively (Figure 4C). In total, 395 microRNAs were differentially expressed in pachytene spermatocytes between OA patients and NOA patients (Figure 4D). In more detail, 97 microRNAs were upregulated in pachytene spermatocytes of OA patients compared to NOA patients (Figure 4D), whereas 298 microRNAs were downregulated in pachytene spermatocytes of OA patients compared with NOA patients (Figure 4D). Furthermore, we observed that 593 microRNAs were detected in round spermatids of OA patients and 560 microRNAs were found in round spermatids of NOA patients. There were 94 microRNAs and 61 microRNAs in round spermatids uniquely in OA patients and NOA patients (Figure 4E). Additionally, 64 microRNAs were upregulated in round spermatids of OA patients when compared to NOA patients, whereas 314 microRNAs were downregulated in round spermatids of OA patients when compared with NOA patients (Figure 4F). Hierarchical clustering analysis revealed distinct profiles of the top 200 differentially expressed microRNAs in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients (Figure 5).
Figure 4.
The Numbers and Differentially Expressed MicroRNAs in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between OA and NOA Patients
(A, C, and E) Pie charts showed the numbers of the expressed microRNAs in human spermatogonia (A), pachytene spermatocytes (C), and round spermatids (E) in OA patients and NOA patients. (B, D, and F) Histograms displayed the differentially expressed microRNAs in human spermatogonia (B), pachytene spermatocytes (D), and round spermatids (F) between OA patients and NOA patients.
Figure 5.
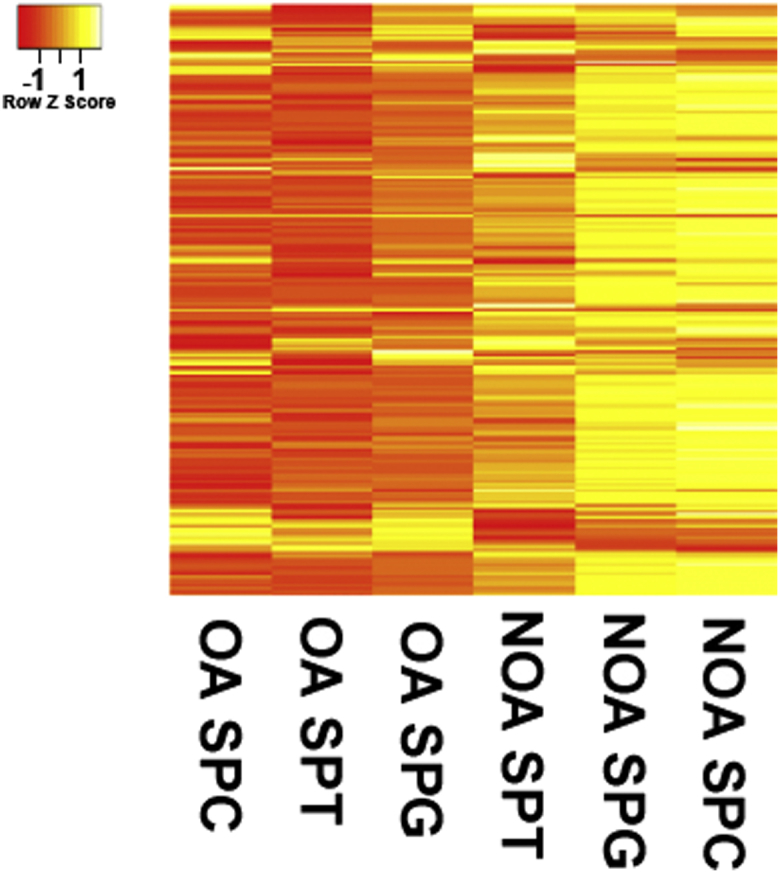
Distinct MicroRNA Expression Profiles among Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids in OA Patients and NOA Patients
Hierarchical clustering showed the top 200 differentially expressed microRNAs among human spermatogonia, pachytene spermatocytes, and round spermatids derived from OA patients and NOA patients.
Quantitative Real-Time PCR Verified the Expression of Differential MicroRNAs Identified by RNA Deep Sequencing in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between OA Patients and NOA Patients
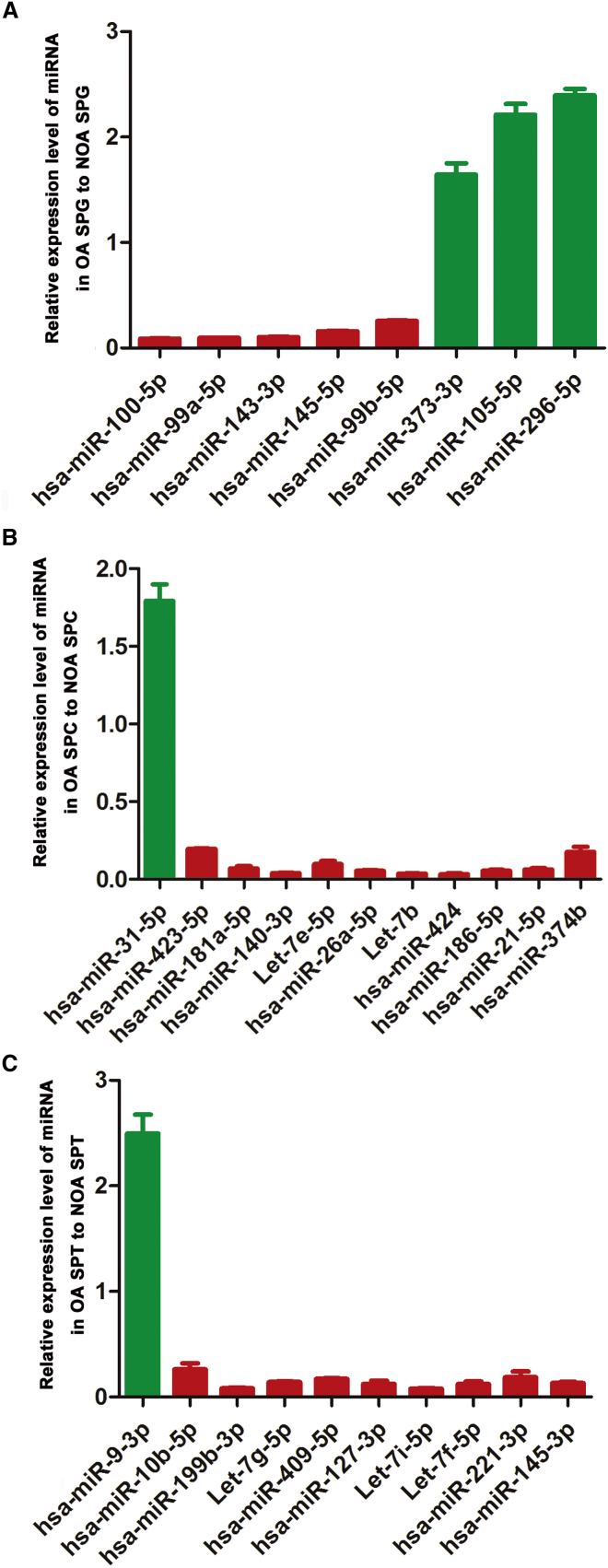
In order to verify the data of RNA deep sequencing, we further conducted real-time PCR for the expression of a number of the differential microRNAs in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients. To this end, 29 differentially expressed microRNAs identified by RNA deep sequencing were chosen randomly. Real-time PCR revealed that the expression levels of hsa-miR-100-5p, hsa-miR-99a-5p, hsa-miR-143-3p, hsa-miR-145-5p, and hsa-miR-99b-5p were upregulated in human spermatogonia of NOA patients compared to OA patients (Figure 6A), whereas the transcripts of hsa-miR-373-3p, hsa-miR-105-5p, and hsa-miR-296-5p were downregulated in human spermatogonia of NOA patients when compared with OA patients (Figure 6A). Moreover, the levels of hsa-miR-423-5p, hsa-miR-181a-5p, hsa-miR-140-3p, Let-7e-5p, hsa-miR-26a-5p, Let-7b, hsa-miR-424, hsa-miR-186-5p, hsa-miR-21-5p, and hsa-miR-374b were upregulated in pachytene spermatocytes of NOA patients compared to OA patients (Figure 6B).Conversely, the transcripts of hsa-miR-31-5p were downregulated in pachytene spermatocytes of NOA patients when compared with OA patients (Figure 6B). In addition, we found that the expression levels of hsa-miR-10b-5p, hsa-miR-199b-3p, Let-7g-5p, hsa-miR-409-5p, hsa-miR-127-3p, Let-7i-5p, Let-7f-5p, hsa-miR-221-3p, and hsa-miR-145-3p were upregulated, whereas hsa-miR-9-3p was downregulated in round spermatids of NOA patients when compared with OA patients (Figure 6C). Significantly, these results of the randomly chosen microRNAs by the real-time PCR assay were fully consistent with the data by RNA deep sequencing.
Figure 6.
Distinct Expression Patterns of MicroRNAs in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between OA Patients and NOA Patients
(A) Real-time PCR showed that hsa-miR-100-5p, hsa-miR-99a-5p, hsa-miR-143-3p, hsa-miR-145-5p, hsa-miR-99b-5p, hsa-miR-373-3p, hsa-miR-105-5p, and hsa-miR-296-5p were differentially expressed in human spermatogonia between OA patients and NOA patients. (B) Real-time PCR revealed the different expression profiles of hsa-miR-423-5p, hsa-miR-181a-5p, hsa-miR-140-3p, Let-7e-5p, hsa-miR-26a-5p, Let-7b, hsa-miR-424, hsa-miR-186-5p, hsa-miR-21-5p, hsa-miR-374b, and hsa-miR-31-5p in human pachytene spermatocytes between OA patients and NOA patients. (C) Real-time PCR demonstrated that hsa-miR-10b-5p, hsa-miR-199b-3p, Let-7g-5p, hsa-miR-409-5p, hsa-miR-127-3p, Let-7i-5p, Let-7f-5p, hsa-miR-221-3p, hsa-miR-145-3p, and hsa-miR-9-3p were differentially expressed in round spermatids between OA patients and NOA patients.
Identification of Novel Binding Targets of Differentially Expressed MicroRNAs among Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between OA Patients and NOA Patients
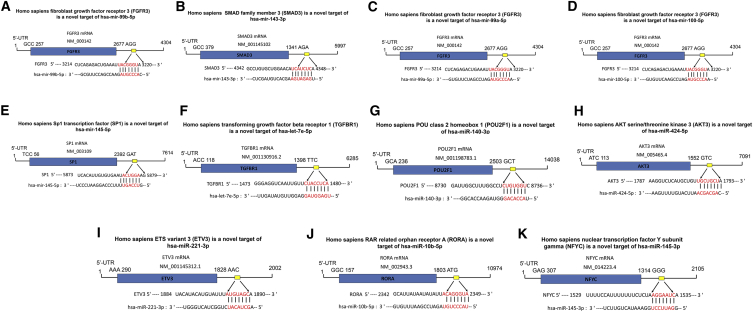
To explore the new mechanism of the differentially expressed microRNAs, we identified their binding targets. Novel binding targets were predicted using bioinformatics, including TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://mirdb.org/miRDB/index.html), and PicTar. Specifically, FGFR3 (fibroblast growth factor receptor 3), SMAD3 (SMAD family member 3), SP1 (Sp1 transcription factor), TGFBR1 (transforming growth factor beta receptor 1), POU2F1 (POU class 2 homeobox 1), AKT3 (V-Akt murine thymoma viral oncogene homolog 3), ETV3 (ETS variant 3), RORA (RAR related orphan receptor A), and NFYC (nuclear transcription factor Y subunit gamma) were predicted to be the targets of has-miR-99b-5p (Figure 7A), has-miR-143-3p (Figure 7B), has-miR-99a-5p (Figure 7C), has-miR-100-5p (Figure 7D), has-miR-145-5p (Figure 7E), has-Let-7e-5p (Figure 7F), has-miR-140-3p (Figure 7G), has-miR-424-5p (Figure 7H) has-miR-221-3p (Figure 7I), has-miR-10b-5p (Figure 7J), and has-miR-145-3p (Figure 7K), respectively.
Figure 7.
Predicted Targets and Binding Sites of the Differentially Expressed MicroRNAs in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between OA Patients and NOA Patients
(A–K) Bioinformatics illustrated that the binding sites of has-miR-99b-5p (A), has-miR-143-3p (B), has-miR-99a-5p (C), has-miR-100-5p (D), has-miR-145-5p (E), has-Let-7e-5p (F), has-miR-140-3p (G), has-miR-424-5p (H), has-miR-221-3p (I), has-miR-10b-5p (J), and has-miR-145-3p (K) to FGFR3, SMAD3, SP1, TGFBR1, POU2F1, AKT3, ETV3, RORA, and NFYC, respectively.
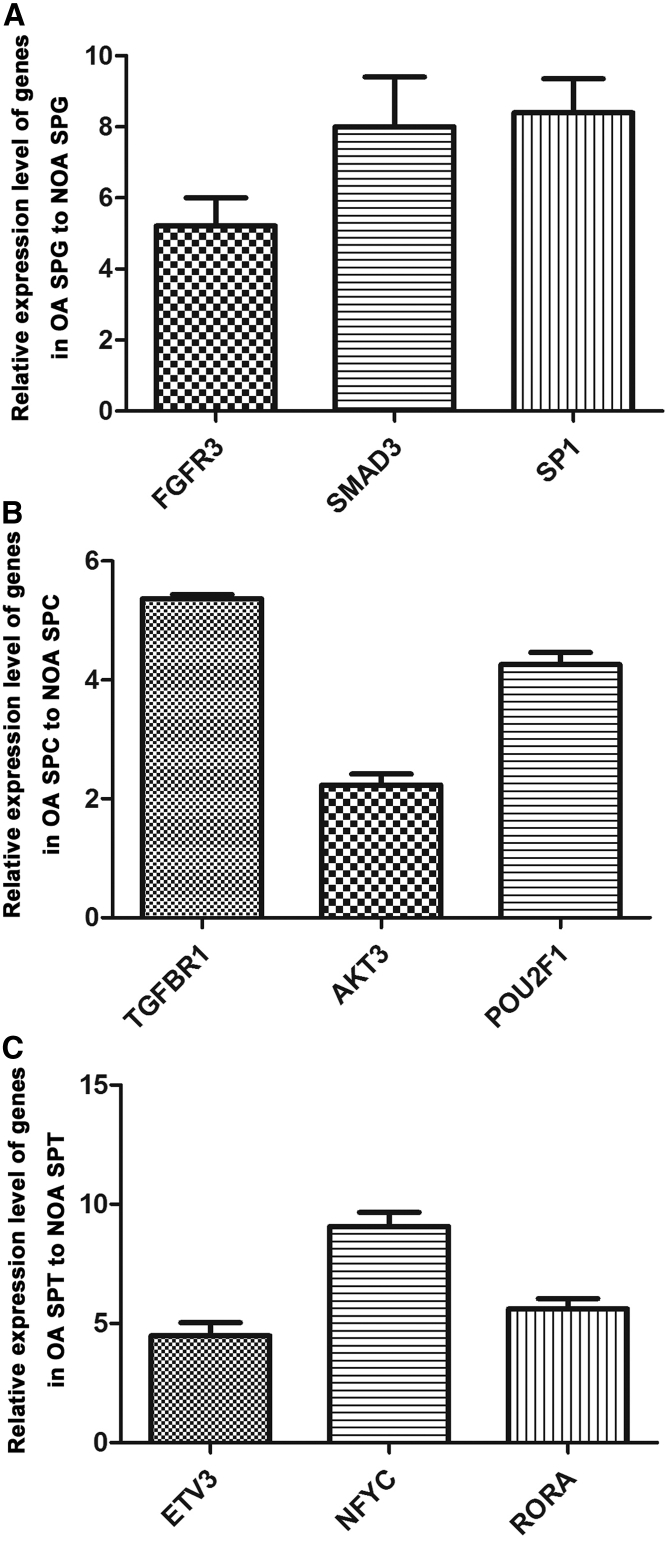
Furthermore, real-time PCR was conducted to verify the targeting genes predicted by microRNA software programs. We observed that the expression levels of FGFR3, SMAD3, and SP1 were higher in human spermatogonia of OA patients than of NOA patients (Figure 8A). The transcripts of TGFBR1, POU2F1, and AKT3 were much higher in pachytene spermatocytes in OA patients than in NOA patients (Figure 8B). Compared with round spermatids in NOA patients, the expression levels of ETV3, RORA, and NFYC genes were expressed at higher levels in round spermatids of OA patients (Figure 8C). Collectively, the transcripts of the genes mentioned above were conversely correlated with the levels of microRNAs in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients, confirming that these genes were binding targets of the microRNAs mentioned above.
Figure 8.
Distinct Expression Patterns of Targeting Genes of the Differentially Expressed MicroRNAs in Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids between OA Patients and NOA Patients
(A) Real-time PCR showed that the transcripts of FGFR3, SMAD3, and SP1 were expressed statistically at higher levels in human spermatogonia of OA patients compared to those of NOA patients. (B) Real-time PCR displayed higher expression levels of TGFBR1, POU2F1, and AKT3 in pachytene spermatocytes of OA patients compared with NOA patients. (C) Real-time PCR revealed that the expression levels of ETV3, RORA, and NFYC were higher in round spermatids of OA patients than of NOA patients.
Discussion
Although much progress has been made in the mechanisms of spermatogenesis in rodents, very little is known about epigenetic regulation in human normal and abnormal spermatogenesis. Thus, uncovering new epigenetic regulators that are involved in the mitosis of human spermatogonia, meiosis of spermatocytes, and spermiogenesis of spermatids is of particular significance. In this study, we isolated human spermatogonia, pachytene spermatocytes, and round spermatids using STA-PUT velocity sedimentation. To eliminate the difference in the inter-patient expression variation of microRNAs caused by individual patients, testicular tissues were combined from OA patients and NOA patients, respectively. Significantly, we have revealed, using RNA deep sequencing, a large scale of differentially expressed microRNA profiles in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients with normal spermatogenesis and NOA patients with aberrant spermatogenesis. Moreover, a number of new targeting genes have been predicted and verified by real-time PCR. These results could shed novel insights into epigenetic regulation underlying NOA and might provide new targets for gene therapy of male infertility.
Several approaches, including STA-PUT, fluorescence-activated cell sorting (FACS), magnetic-activated cell sorting (MACS), and elutriation, have been used to separate specific male germ cells in rodents.28, 29, 30, 31 In the current study, STA-PUT was chosen to isolate human spermatogonia, pachytene spermatocytes, and round spermatids, owing to high purity and cell viability for a subsequent study compared with other methods.29, 32 The identities of freshly isolated human spermatogonia, pachytene spermatocytes, and round spermatids were verified morphologically and phenotypically. Due to the scarce resource of human testicular tissue, the concentrations of RNA extracted from human male germ cells usually can’t meet the requirement for traditional RNA sequencing. Therefore, we utilized a highly sensitive profiling method by optimizing the conditions for the 5′ and 3′ adaptor ligation and PCR amplification steps,33 which allows only 10–100 ng total for RNA deep sequencing.
MicroRNAs play vital roles in mediating normal and aberrant spermatogenesis, and much progress has recently been made in this field in rodents. Specific deletion of DICER (key enzyme in biogenesis of mature microRNA) in male germ cells can result in an increase in spermatocyte apoptosis and the defect in chromatin organization and nuclear shaping of elongating spermatids.34, 35 Also, conditional knockout of Drosha in male germ cells leads to testis weight reduction and severe disruption in meiosis and spermiogenesis.36 Additionally, conditional deletion of DGCR8 in male germ cells can lead to disruption in the meiotic and haploid phases of spermatogenesis.37 These studies described above illustrate that microRNAs are indispensable for spermatogenesis in rodents. Nevertheless, it remains to be elucidated whether microRNAs are required for human normal spermatogenesis and if the abnormality of these microRNAs is associated with male infertility. In this study, we have revealed for the first time, using RNA deep sequencing, that 1,169 microRNAs were differentially expressed in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients with normal spermatogenesis and NOA patients with failure in the production of male germ cells. We hypothesized that the abnormal expression levels of microRNAs might be associated with aberrant human spermatogenesis, which eventually result in azoospermia and male infertility. Several lines of evidence suggest that abnormal expression of microRNAs can lead to spermatogenesis failure. For instance, overexpression of let-7 results in the inhibition of insulin growth factor II (IGF-II) messenger RNA binding protein (Imp). Subsequently, self-renewal factor Unpaired (upd) mRNA becomes unprotected and susceptible to degradation. Therefore, let-7 overexpression can induce the altered testicular stem niche in Drosophila.26 Also, overexpression of let-7 can reduce the numbers of male germ cells through targeting Lin28a, which can regulate the cyclic expansion of spermatogonial progenitors.38, 39 In addition, it has been reported that aberrant expression of miR-19b and let-7a in the seminal plasma can lead to spermatogenic failure.40 Here, we observed that the expression levels of microRNAs essential for rodent spermatogenesis were much lower in male germ cells in NOA patients compared to OA patients. As examples, we found that microRNA-34b/c and microRNA-449a were expressed at significantly lower levels in male germ cells in NOA patients than in OA patients. MicroRNA-34b/c and microRNA-449a have the similar seed sequence (microRNA-34b/c: AGGCAGU; microRNA-449a: GGCAGUG), and thus they assume the same effect on regulating spermatogenesis. In mice, individual knockout of microRNA-34b/c and microRNA-449a has no effect on spermatogenesis, whereas simultaneous inactivation of microRNA-34b/c and microRNA-449a can lead to male infertility.41
We next verified a number of the differentially expressed microRNAs between OA patients and NOA patients identified by RNA deep sequencing using real-time PCR. Among these microRNAs, miRNA-99 and miRNA-100 have been shown to induce cardiomyocyte dedifferentiation and heart regeneration in zebrafish by targeting Smarca5 and Fntb.42 miRNA-373 regulates the maintenance and differentiation of human embryonic stem cells,43 and it activates the Wnt/β-catenin pathway to control carcinogenesis.44 Also, miRNA-143 and miRNA-145 can modulate smooth muscle cell plasticity by regulating certain key transcription factors.45 Thus, these microRNAs may be involved in fate determinations, including the division and differentiation of human spermatogonia. Moreover, the expression levels of miRNA-140 and miRNA-424 in human pachytene spermatocytes as well as miRNA-221, miRNA-10b-5p, and miRNA-145-3p in round spermatids of OA patients were statistically lower than those of NOA patients, suggesting these microRNAs may be involved in the regulation of meiosis and spermiogenesis.
We further predicted a number of novel targeting genes of microRNAs using bioinformatics software and verified these targets through real-time PCR. We revealed that FGFR3, SMAD3, SP1, TGFBR1, AKT3, POU2F1, ETV3, NFYC, and RORA might be the binding genes of hsa-mir-99b-5p, hsa-mir-99a-5p, hsa-mir-100-5p, hsa-mir-143-3p, hsa-mir-145-5p, hsa-let-7e-5p, hsa-mir-424-5p, hsa-mir-140-3p, hsa-mir-221-3p, hsa-mir-145-3p, and hsa-mir-10b-5p, respectively. FGFR3 is a receptor for FGF2, which has been shown to be essential for the survival and proliferation of spermatogonial stem cells.23 The expression of FGFR3 has been shown to be restricted to type A spermatogonia in human testis, suggesting that the FGF2/FGFR3 signal pathway may be required for the self-renew and/or differentiation of human spermatogonia.46 SMAD3 belongs to the member of the SMAD super family, and it can be activated by transforming growth factor beta (TGF-β), which is vital for both the proliferation and differentiation of spermatogonia.47 In addition, SP1 has been reported to regulate Sohlh1 gene transcription, which is required for spermatogonial differentiation during early spermatogenesis.48, 49 It has been demonstrated that TGFBR1 is present in pachytene spermatocytes of rat and boar testis but absent in elongated spermatids, suggesting TGFBR1 may be involved in the control of meiosis.50 Moreover, AKT3 is a member in the PI3K-AKT pathway, and activation of this pathway is vital for inducing Stra8 expression elicited by retinoic acid for spermatogonial differentiation.51 Nevertheless, the functions of other microRNA targets, including POU2F1, ETV3, NFYC, and RORA, in regulating meiosis and spermiogenesis remain to be defined.
In summary, we have uncovered for the first time the distinct global microRNA profiles in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients. With regards to clinical applications, microRNA mimics can be utilized to enhance the functions of microRNAs that are expressed at lower levels in human male germ cells of NOA patients, with an aim to restore human normal spermatogenesis. Conversely, microRNA inhibitors may be employed to reduce the extra amounts of microRNAs that are expressed at higher levels in human male germ cells of NOA patients, which may achieve human normal spermatogenesis. We also predicted and verified a number of targeting genes for the differentially expressed microRNAs in spermatogenic cells between OA patients and NOA patients. Some of these genes, e.g., FGFR3, SMAD3, SP1, TGFBR1, and AKT3, have been shown to be essential for mammalian spermatogenesis, as discussed above. Gene corrections can be used to treat NOA patients with mutation of these genes. This study could thus offer new insights into mechanisms underlying the etiology of NOA patients and might offer new targets of molecular therapy for treating male reproductive disorders.
Materials and Methods
Procurement of Testicular Tissues from OA Patients and NOA Patients
The testicular tissues were obtained from 20 OA patients and 60 NOA patients (aged from 25 to 40 years) who underwent micro-testicular sperm extraction (micro-TESE) at Ren Ji Hospital, which is affiliated with the Shanghai Jiao Tong University School of Medicine. The OA patients had normal spermatogenesis, and these patients exhibited vasoligation and inflammation, but not congenital absence of the vas deferens or other congenital diseases. This study was approved by the Institutional Ethical Review Committee of Ren Ji Hospital (license number of ethics statement: 2012-01), Shanghai Jiao Tong University School of Medicine, and the informed consents for testicular biopsies were obtained from the donors for research only.
Isolation of Human Spermatogonia, Pachytene Spermatocytes, and Round Spermatids from OA Patients and NOA Patients by STA-PUT
Human spermatogonia, pachytene spermatocytes, and round spermatids were isolated by velocity sedimentation using the STA-PUT apparatus. Testicular tissues were combined from OA patients and NOA patients, respectively, in order to obtain enough male germ cells, and they were washed three times aseptically using DMEM/F12 (Gibco, Grand Island, NY, USA), with 1% penicillin and streptomycin (Gibco). The seminiferous tubules were isolated by enzyme I containing 2 mg/mL collagenase IV (Life Technology) and 1 μg/μL DNase I (Roche) in an oscillating water bath at 34°C, 100 rpm, for 15 min. Male germ cells and Sertoli cells were obtained from seminiferous tubules by enzyme II containing 4 mg/mL collagenase IV, 2.5 mg/mL hyaluronidase (Sigma), 2 mg/mL trypsin (Sigma), and 1 μg/μL DNase I for 10–12 min. Finally, male germ cells were obtained through the differential plating pursuant to the procedure as previously described.52 Around 106 male germ cells were obtained after the differential plating.
The STA-PUT method was used to separate human spermatogonia, pachytene spermatocytes, and round spermatids by linear BSA gradient and sedimentation in terms of cell size, mass, and gravity. After centrifugation at 1,000 rpm for 5 min, male germ cells were resuspended by 25 mL 0.5% BSA solution. Two cylinders were loaded with 300 mL 2% BSA solution and 300 mL 4% BSA solution, respectively. The cell chamber was loaded with cell suspension, and the stopcock was opened to allow the cells to flow slowly into the sedimentation chamber. The stirrer in the 2% BSA solution started to work, and artery clips were removed so that both 2% and 4% BSA solutions went into the sedimentation chamber. 30 min later, the stirrers were switched off, and artery clips were replaced and followed by around 3 hr sedimentation in the cell sedimentation chamber. In order to collect different cell types, the centrifuge tubes were used as the fraction collectors. The cells in each fraction were collected by centrifugation at 1,000 rpm for 5 min. An aliquot of each fraction was examined carefully under a phase-contrast microscope and a DIC microscope to assess cellular integrity and identify cell types. Fractions containing cells of similar size and morphology were pooled according to the procedure, as described previously.27
RNA Extraction and RT-PCR
Total RNA was extracted from the isolated human spermatogonia, pachytene spermatocytes, and round spermatids using the Trizol reagent (Invitrogen), and Nanodrop was used to measure the quality and concentrations of total RNA. RT was performed using the First Strand cDNA Synthesis Kit (Thermo Scientific, catalog no: K1622), and PCR of the cDNA was carried out according to the protocol, as described previously.53 The primers of the chosen genes, including GPR125, RET, GFRA1, THY1, UCHL1, MAGEA4, PLZF, SYCP3, SYCP1, TNP1, TNP2, PRM1, PRM2, ACR, and GAPDH, were designed and are listed in Table 2. The PCR reaction started at 94°C for 2 min and was performed as follows: denaturation at 94°C for 30 s, annealing at 55°C–60°C for 30 s, as listed in Table 2, and elongation at 72°C for 60 s. After 35 cycles, the samples were incubated for an additional 5 min at 72°C. PCR products were separated by electrophoresis on 1.5% agarose gel and visualized with gengreen nucleic acid staining (GENVIEW). Images were recorded by chemiluminescence (Chemi-Doc XRS, Bio-Rad). RNA without RT (RT-) but with PCR of GAPDH primers served as a negative control.
Table 2.
The Sequences of Gene Primers Used for RT-PCR and Real-Time PCR
| Genes | Primer Sequences (5′–3′) | Product Size (bp) | Temperature (°C) | |
|---|---|---|---|---|
| GPR125 | forward | CAGAACATTGGCAGGCATTAC | 367 | 62 |
| reverse | CATCACGTCACCTAGCTCTTT | |||
| RET | forward | CTCGTTCATCGGGACTTG | 126 | 60 |
| reverse | ACCCTGGCTCCTCTTCAC | |||
| GFRA1 | forward | GGACTCCTGCAAGACGAATTA | 543 | 62 |
| reverse | GAAGCACCGAGACCTTCTTT | |||
| THY1 | forward | CAGAAGGTGACCAGCCTAAC | 233 | 60 |
| reverse | TTGCTAGTGAAGGCGGATAAG | |||
| UCHL1 | forward | CCCGAGATGCTGAACAAAGT | 274 | 62 |
| reverse | TGGCCACTGCGTGAATAAG | |||
| MAGEA4 | forward | CCGAGAAGCACTCAGTAACAA | 107 | 60 |
| reverse | TGATGACTCTCTCCAGCATTTC | |||
| PLZF | forward | GATCCTCTTCCACCGCAATAG | 99 | 60 |
| reverse | TGCAGCGTGGCTGTATATG | |||
| SYCP1 | forward | AACTACTGTCTGCAGCTTGG | 124 | 60 |
| reverse | CATCTCTTCCAGCTCACTTGAT | |||
| SYCP3 | forward | TGCAGAAAGCTGAGGAACAA | 247 | 58 |
| reverse | TGCTGCTGAGTTTCCATCAT | |||
| TNP1 | forward | CGACCAGCCGCAAATTAAAG | 110 | 60 |
| reverse | CAGGTTGCCCTTACGGTATT | |||
| TNP2 | forward | AGACTCACAGCCTTCCTATCA | 359 | 62 |
| reverse | CTGGATCCTCTTGGCCATTT | |||
| PRM1 | forward | ATAGCACATCCACCAAACTCC | 120 | 58 |
| reverse | AGGCGGCATTGTTCCTTAG | |||
| PRM2 | forward | CATGGGCAAGAGCAAGGA | 105 | 60 |
| reverse | TCTGCGCCTATAGTGAGACT | |||
| ACR | forward | CATCGACCTGGACTTGTGTAA | 369 | 58 |
| reverse | CAAGGAAGGTGAGCAGAGATAG | |||
| GADPH | forward | AATCCCATCACCATCTTCC | 382 | 55 |
| reverse | CATCACGCCACAGTTTCC | |||
| TGFBR1 | forward | ACGGCGTTACAGTGTTTCTG | 167 | 60 |
| reverse | GCACATACAAACGGCCTATCTC | |||
| AKT3 | forward | TGTGGATTTACCTTATCCCCTCA | 79 | 60 |
| reverse | GTTTGGCTTTGGTCGTTCTGT | |||
| POU2F1 | forward | ATGAACAATCCGTCAGAAACCAG | 157 | 60 |
| reverse | GATGGAGATGTCCAAGGAAAGC | |||
| ETV3 | forward | GGTGGAGGGTATCAGTTTCCT | 346 | 60 |
| reverse | TGATGAATGGGTAGTTGGGCAT | |||
| NFYC | forward | GGAGGATTTGGTGGTACTAGCA | 109 | 60 |
| reverse | GCACTCGGAAGTCTTTCACTG | |||
| RORA | forward | ACTCCTGTCCTCGTCAGAAGA | 95 | 60 |
| reverse | CATCCCTACGGCAAGGCATTT | |||
| FGFR3 | forward | CCCAAATGGGAGCTGTCTCG | 109 | 60 |
| reverse | CCCGGTCCTTGTCAATGCC | |||
| SP1 | forward | TGGCAGCAGTACCAATGGC | 126 | 60 |
| reverse | CCAGGTAGTCCTGTCAGAACTT | |||
| SMAD3 | forward | TGGACGCAGGTTCTCCAAAC | 90 | 60 |
| reverse | CCGGCTCGCAGTAGGTAAC | |||
| ACTB | forward | CCTGGCACCCAGCACAAT | 144 | 60 |
| reverse | GGGCCGGACTCGTCATAC | |||
Immunocytochemistry
The freshly isolated human spermatogonia and round spermatids were fixed in 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.5% Triton X-100 for 30 min, blocked with 5% BSA for 1 hr, and incubated with primary antibodies. The primary antibodies included THY1 (Abcam, ab133350, 1:200), GFRA1 (Santa Cruz, sc-6156, 1:200), PLZF (Santa Cruz, sc-22839, 1:200), UCHL1 (Bio-Rad, MCA4750, 1:200), PNA (Life Technologies, L32458, 1:200), and Protamine 2 (PRM2) (Atlas Antibodies, HPA056386, 1:200). After incubation at 4°C overnight, cells were washed three times in PBS (Medicago, Uppsala, Sweden), followed by incubation with secondary antibodies for 1 hr at room temperature. Secondary antibodies were conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen). DAPI was used to label cell nuclei, and images were captured with a fluorescence microscope (Leica). Isotype IgGs replaced primary antibodies and served as negative controls.
Meiotic Chromatin Spread
Meiotic chromatin spread assays were performed to identify the purity of isolated human pachytene spermatocytes pursuant to the method described previously.27 Cells were lysed by a hypotonic solution and spread evenly over slides layered with 1% paraformaldehyde (PFA) and 0.15% Triton X-100. Slides were dried for 24 hr at room temperature in a humid chamber. Cells were treated with 0.04% photoflo for 5 min and blocked with 4% goat serum. Triple immunostaining was performed in these cells using primary antibodies, including SYCP3 (Abcam, ab15093, 1:100), CREST (Immunovision, HCT-0100, 1:100), and MLH1 (Abcam, ab14206, 1:50) at 37°C for 2 hr in a humid chamber. Goat anti-rabbit Alexa Fluor 594 (Invitrogen) and goat anti-human Alexa Fluor 488 secondary antibodies (Invitrogen) were applied at 1:200 dilution and incubated for 90 min at 37°C. Cells were washed three times with PBS, and images were captured with a fluorescence microscope (Leica).
RNA Deep Sequencing
Total RNA was extracted from human spermatogonia, pachytene spermatocytes, and round spermatids of OA patients and NOA patients by Trizol reagent (Ambion). The concentration and quantity of total RNA were measured by Nanodrop (Thermo), and the ratios of A260/A280 of total RNA were set as 1.9 to 2.0 to ensure great purity. The cDNA libraries of small RNAs were established using 100 ng of total RNA, and adaptor ligation, first-strand cDNA synthesis, PCR amplification, gel purification, and sequencing of small RNA libraries using an Illumina HiSeq 2000 were performed according to the procedure as described previously.33 Further analyses of small RNA expression and mapping of microRNAs, piRNA, endo-siRNA, and other small RNAs were conducted pursuant to the previous method.33
Quantitative Real-Time PCR
RNA was extracted from human spermatogonia, pachytene spermatocytes, and round spermatids of OA patients and NOA patients using Trizol reagent (Invitrogen). For microRNA real-time PCR, RT reaction was performed using the miScript II RT Kit (QIAGEN, catalog no: 218160). Each RT reaction was composed of 100 ng RNA, 4 μL miScript HiSpec Buffer, 2 μL Nucleics Mix, and 2 μL miScript Reverse Transcriptase Mix (QIAGEN) in a total volume of 20 μL. Reactions were performed in a Veriti 96-Well Thermal Cycler (Applied Biosystems) for 60 min at 37°C, followed by heat inactivation of RT for 5 min at 95°C. RT reaction mix was diluted 5 times in nuclease-free water and held at −20°C. Primer sequences of microRNAs used for real-time PCR are listed in Table 3.
Table 3.
The Sequences of miRNA-Specific Primers Used for Real-Time PCR
| Human MicroRNAs | Primer Sequences (5′–3′) | Temperature (°C) |
|---|---|---|
| hsa-miR-100-5p | AACCCGTAGATCCGAACTTGTG | 60 |
| hsa-miR-145-3p | GGATTCCTGGAAATACTGTTCT | 60 |
| hsa-miR-99a-5p | AACCCGTAGATCCGATCTTGTG | 60 |
| hsa-miR-143-3p | TGAGATGAAGCACTGTAGCTC | 60 |
| hsa-miR-145-5p | GTCCAGTTTTCCCAGGAATCCCT | 60 |
| hsa-miR-99b-5p | CACCCGTAGAACCGACCTTGCG | 60 |
| hsa-miR-373-3p | GAAGTGCTTCGATTTTGGGGTGT | 60 |
| hsa-miR-105-5p | TCAAATGCTCAGACTCCTGTGGT | 60 |
| hsa-miR-296-5p | AGGGCCCCCCCTCAATCCTGT | 60 |
| hsa-miR-423-5p | TGAGGGGCAGAGAGCGAGACTTT | 60 |
| hsa-miR-181a-5p | AACATTCAACGCTGTCGGTGAGT | 60 |
| hsa-miR-140-3p | TACCACAGGGTAGAACCACGG | 60 |
| Let-7e-5p | TGAGGTAGGAGGTTGTATAGTT | 60 |
| hsa-miR-26a-5p | TTCAAGTAATCCAGGATAGGCT | 60 |
| hsa-miR-424 | CAGCAGCAATTCATGTTTTGAA | 60 |
| hsa-miR-186-5p | CAAAGAATTCTCCTTTTGGGCT | 60 |
| hsa-miR-21-5p | CAAAGAATTCTCCTTTTGGGCT | 60 |
| hsa-miR-374 | TTATAATACAACCTGATAAGTG | 60 |
| Let-7b | TGAGGTAGTAGGTTGTGTGGTT | 60 |
| hsa-miR-31-5p | AGGCAAGATGCTGGCATAGCT | 60 |
| hsa-miR-10b-5p | TACCCTGTAGAACCGAATTTGTG | 60 |
| hsa-miR-199b-3p | ACAGTAGTCTGCACATTGGTTA | 60 |
| hsa-miR-409-5p | AGGTTACCCGAGCAACTTTGCAT | 60 |
| hsa-miR-127-3p | TCGGATCCGTCTGAGCTTGGCT | 60 |
| Let-7i-5p | TGAGGTAGTAGTTTGTGCTGTT | 60 |
| Let-7f-5p | TGAGGTAGTAGATTGTATAGTT | 60 |
| hsa-miR-221-3p | AGCTACATTGTCTGCTGGGTTTC | 60 |
| Let-7g-5p | TGAGGTAGTAGTTTGTACAGTT | 60 |
| hsa-miR-9-3p | ATAAAGCTAGATAACCGAAAGT | 60 |
| U6 | CAAGGATGACACGCAAATTCG | 60 |
Real-time PCR was performed in triplicates using the StepOnePlus Real-Time PCR System (Applied Biosystems) with 25 μL PCR reaction mixture containing 2 μL cDNA, 12.5 μL QuantiTect SYBR Green PCR Master Mix (QIAGEN), 2.5 μL universal primer (QIAGEN), 2.5 μL microRNA specific primers (Table 3), and 5.5 μL nuclease-free water. Reactions were incubated in a 96-well optical plate (Applied Biosystems) at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Individual samples were run in triplicates. At the end of the PCR cycles, a melting curve analysis was performed to validate the specific generation of the expected PCR products. The expression levels of microRNAs were normalized to U6 and calculated using the 2-ΔΔCt method.54 In addition, real-time PCR was conducted to evaluate the expression levels of the predicted target genes in freshly isolated human spermatogonia, pachytene spermatocytes, and round spermatids from OA patients and NOA patients. These genes included TGFBR1, AKT3, POU2F1, ETV3, NFYC, RORA, FGFR3, SP1, and SMAD3. The primers of these genes were designed and are listed in Table 2, and their expression levels were normalized to ACTB and calculated using the 2-ΔΔCt method.54
Statistical Analysis
The data were presented as mean ± SEM and analyzed by Student’s t test or one-way ANOVA with the appropriate post hoc tests (Dunnet’s test or Turkey’s multiple comparison) using Prism (version 5, GraphPad Software), and p < 0.05 was considered statistically significant.
Author Contributions
C.Y., Q.Y., M.N., H.F., F.Z., W.Z., H.W., L. Wen, and L. Wu performed the laboratory experiments and collected the data. Z.H. and C.Y. analyzed the data and wrote the manuscript. Z.H. and Z.L. conceived the project, designed the experiments, and supervised the entire study.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by grants from National Nature Science Foundation of China (31230048 and 31671550) and Chinese Ministry of Science and Technology (2016YFC1000600 and 2014CB943101), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152511), The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (2012.53), and Shanghai Hospital Development Center (SHDC12015122).
Contributor Information
Zheng Li, Email: lizhengboshi@163.com.
Zuping He, Email: zupinghe@sjtu.edu.cn.
References
- 1.De Kretser D.M., Baker H.W. Infertility in men: recent advances and continuing controversies. J. Clin. Endocrinol. Metab. 1999;84:3443–3450. doi: 10.1210/jcem.84.10.6101. [DOI] [PubMed] [Google Scholar]
- 2.Wosnitzer M., Goldstein M., Hardy M.P. Review of azoospermia. Spermatogenesis. 2014;4:e28218. doi: 10.4161/spmg.28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu Z., Xia Y., Guo X., Dai J., Li H., Hu H., Jiang Y., Lu F., Wu Y., Yang X. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat. Genet. 2011;44:183–186. doi: 10.1038/ng.1040. [DOI] [PubMed] [Google Scholar]
- 4.Docampo M.J., Hadziselimovic F. Molecular pathology of cryptorchidism-induced infertility. Sex Dev. 2015;9:269–278. doi: 10.1159/000442059. [DOI] [PubMed] [Google Scholar]
- 5.Ezeh U.I. Beyond the clinical classification of azoospermia: opinion. Hum. Reprod. 2000;15:2356–2359. doi: 10.1093/humrep/15.11.2356. [DOI] [PubMed] [Google Scholar]
- 6.Raman J.D., Schlegel P.N. Testicular sperm extraction with intracytoplasmic sperm injection is successful for the treatment of nonobstructive azoospermia associated with cryptorchidism. J. Urol. 2003;170:1287–1290. doi: 10.1097/01.ju.0000080707.75753.ec. [DOI] [PubMed] [Google Scholar]
- 7.O’Flynn O’Brien K.L., Varghese A.C., Agarwal A. The genetic causes of male factor infertility: a review. Fertil. Steril. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 8.Dym M., Kokkinaki M., He Z. Spermatogonial stem cells: mouse and human comparisons. Birth Defects Res. C Embryo Today. 2009;87:27–34. doi: 10.1002/bdrc.20141. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y., Hai Y., Gong Y., Li Z., He Z. Characterization, isolation, and culture of mouse and human spermatogonial stem cells. J. Cell. Physiol. 2014;229:407–413. doi: 10.1002/jcp.24471. [DOI] [PubMed] [Google Scholar]
- 10.Yang S., Ping P., Ma M., Li P., Tian R., Yang H., Liu Y., Gong Y., Zhang Z., Li Z. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Reports. 2014;3:663–675. doi: 10.1016/j.stemcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogarth C.A., Griswold M.D. The key role of vitamin A in spermatogenesis. J. Clin. Invest. 2010;120:956–962. doi: 10.1172/JCI41303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 13.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 14.Yi R., Poy M.N., Stoffel M., Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 16.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 17.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 19.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutvágner G., McLachlan J., Pasquinelli A.E., Bálint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi K., Chuva de Sousa Lopes S.M., Kaneda M., Tang F., Hajkova P., Lao K., O’Carroll D., Das P.P., Tarakhovsky A., Miska E.A. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maatouk D.M., Loveland K.L., McManus M.T., Moore K., Harfe B.D. Dicer1 is required for differentiation of the mouse male germline. Biol. Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- 23.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu Z., Goodyear S.M., Rao S., Wu X., Tobias J.W., Avarbock M.R., Brinster R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q.E., Racicot K.E., Kaucher A.V., Oatley M.J., Oatley J.M. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–290. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toledano H., D’Alterio C., Czech B., Levine E., Jones D.L. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Niu M., Yao C., Hai Y., Yuan Q., Liu Y., Guo Y., Li Z., He Z. Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Sci. Rep. 2015;5:8084. doi: 10.1038/srep08084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellvé A.R. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 29.Meistrich M.L., Bruce W.R., Clermont Y. Cellular composition of fractions of mouse testis cells following velocity sedimentation separation. Exp. Cell Res. 1973;79:213–227. [PubMed] [Google Scholar]
- 30.Getun I.V., Torres B., Bois P.R. Flow cytometry purification of mouse meiotic cells. J. Vis. Exp. 2011;50:2602. doi: 10.3791/2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastos H., Lassalle B., Chicheportiche A., Riou L., Testart J., Allemand I., Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- 32.Miller R.G., Phillips R.A. Separation of cells by velocity sedimentation. J. Cell. Physiol. 1969;73:191–201. doi: 10.1002/jcp.1040730305. [DOI] [PubMed] [Google Scholar]
- 33.Yang Q., Lin J., Liu M., Li R., Tian B., Zhang X., Xu B., Liu M., Zhang X., Li Y. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2016;2:e1501482. doi: 10.1126/sciadv.1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korhonen H.M., Meikar O., Yadav R.P., Papaioannou M.D., Romero Y., Da Ros M., Herrera P.L., Toppari J., Nef S., Kotaja N. Dicer is required for haploid male germ cell differentiation in mice. PLoS One. 2011;6:e24821. doi: 10.1371/journal.pone.0024821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero Y., Meikar O., Papaioannou M.D., Conne B., Grey C., Weier M., Pralong F., De Massy B., Kaessmann H., Vassalli J.D. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One. 2011;6:e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Q., Song R., Ortogero N., Zheng H., Evanoff R., Small C.L., Griswold M.D., Namekawa S.H., Royo H., Turner J.M. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J. Biol. Chem. 2012;287:25173–25190. doi: 10.1074/jbc.M112.362053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann C., Romero Y., Warnefors M., Bilican A., Borel C., Smith L.B., Kotaja N., Kaessmann H., Nef S. Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS One. 2014;9:e107023. doi: 10.1371/journal.pone.0107023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinoda G., De Soysa T.Y., Seligson M.T., Yabuuchi A., Fujiwara Y., Huang P.Y., Hagan J.P., Gregory R.I., Moss E.G., Daley G.Q. Lin28a regulates germ cell pool size and fertility. Stem Cells. 2013;31:1001–1009. doi: 10.1002/stem.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakraborty P., Buaas F.W., Sharma M., Snyder E., de Rooij D.G., Braun R.E. LIN28A marks the spermatogonial progenitor population and regulates its cyclic expansion. Stem Cells. 2014;32:860–873. doi: 10.1002/stem.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W., Hu Z., Qin Y., Dong J., Dai J., Lu C., Zhang W., Shen H., Xia Y., Wang X. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol. Hum. Reprod. 2012;18:489–497. doi: 10.1093/molehr/gas022. [DOI] [PubMed] [Google Scholar]
- 41.Wu J., Bao J., Kim M., Yuan S., Tang C., Zheng H., Mastick G.S., Xu C., Yan W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. USA. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguirre A., Montserrat N., Zacchigna S., Nivet E., Hishida T., Krause M.N., Kurian L., Ocampo A., Vazquez-Ferrer E., Rodriguez-Esteban C. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Zhou A.D., Diao L.T., Xu H., Xiao Z.D., Li J.H., Zhou H., Qu L.H. β-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/β-catenin-signaling pathway. Oncogene. 2012;31:2968–2978. doi: 10.1038/onc.2011.461. [DOI] [PubMed] [Google Scholar]
- 45.Neppl R.L., Wang D.Z. Smooth(ing) muscle differentiation by microRNAs. Cell Stem Cell. 2009;5:130–132. doi: 10.1016/j.stem.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Kopylow K., Staege H., Spiess A.N., Schulze W., Will H., Primig M., Kirchhoff C. Differential marker protein expression specifies rarefaction zone-containing human Adark spermatogonia. Reproduction. 2012;143:45–57. doi: 10.1530/REP-11-0290. [DOI] [PubMed] [Google Scholar]
- 47.Young J.C., Wakitani S., Loveland K.L. TGF-β superfamily signaling in testis formation and early male germline development. Semin. Cell Dev. Biol. 2015;45:94–103. doi: 10.1016/j.semcdb.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 48.Ballow D., Meistrich M.L., Matzuk M., Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev. Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 49.Toyoda S., Yoshimura T., Mizuta J., Miyazaki J. Auto-regulation of the Sohlh1 gene by the SOHLH2/SOHLH1/SP1 complex: implications for early spermatogenesis and oogenesis. PLoS One. 2014;9:e101681. doi: 10.1371/journal.pone.0101681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olaso R., Pairault C., Habert R. Expression of type I and II receptors for transforming growth factor beta in the adult rat testis. Histochem. Cell Biol. 1998;110:613–618. doi: 10.1007/s004180050324. [DOI] [PubMed] [Google Scholar]
- 51.Tassinari V., Campolo F., Cesarini V., Todaro F., Dolci S., Rossi P. Fgf9 inhibition of meiotic differentiation in spermatogonia is mediated by Erk-dependent activation of Nodal-Smad2/3 signaling and is antagonized by Kit Ligand. Cell Death Dis. 2015;6:e1688. doi: 10.1038/cddis.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y., Liu L., Sun M., Hai Y., Li Z., He Z. Expansion and long-term culture of human spermatogonial stem cells via the activation of SMAD3 and AKT pathways. Exp. Biol. Med. 2015;240:1112–1122. doi: 10.1177/1535370215590822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y., Hai Y., Yao C., Chen Z., Hou J., Li Z., He Z. Long-term culture and significant expansion of human Sertoli cells whilst maintaining stable global phenotype and AKT and SMAD1/5 activation. Cell Commun. Signal. 2015;13:20. doi: 10.1186/s12964-015-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao C., Sun M., Yuan Q., Niu M., Chen Z., Hou J., Wang H., Wen L., Liu Y., Li Z. MiRNA-133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget. 2016;7:2201–2219. doi: 10.18632/oncotarget.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]