Abstract
Isolates of Teratosphaeriaceae have frequently been found in the integument of attine ants, proving to be common and diverse in this microenvironment. The LSU phylogeny of the ant-isolated strains studied revealed that they cluster in two main lineages. The first was associated with the genus Xenopenidiella whereas the other represented two ant-isolated lineages sister to the taxa Penidiella aggregata and P. drakensbergensis, which are allocated to the new genus Penidiellomyces. The genus Penidiella is limited to the lineage containing P. columbiana, which is not congeneric with Penidiellomyces or Penidiellopsis, nor with Simplicidiella, a novel genus introduced here to accommodate a strain isolated from ants. For species level analysis, the final 26 aligned sequences of the ITS (498 characters), cmdA (389 characters), tef1 (342 characters) and tub2 (446 characters) gene regions lead to the introduction of six new species in Xenopenidiella, and one in respectively Penidiellopsis and Simplicidiella. The species described in this study were distinguished by the combination of morphological and phylogenetic data. Novelties on the integument of leaf-cutting ants from Brazil include: Penidiellopsis ramosus, Xenopenidiella clavata, X. formica, X. inflata, X. laevigata, X. nigrescens, X. tarda spp. nov., and Simplicidiella nigra gen. & sp. nov. Beta-tubulin is recommended as primary barcode for the distinction of species in Penidiellopsis, whereas ITS was sufficient to distinguish species of Xenopenidiella.
Keywords: Attini tribe, leaf-cutting ants, multi-gene analyses, systematics, Xenopenidiella
INTRODUCTION
Seen as important symbionts, fungi interact with a wide range of organisms, including insects (McLaughlin et al. 2009). According to Hawksworth (2012) it is estimated that at least 1.5–3 M species of fungi are living on Earth, of which only around 100 000 are presently known (Crous et al. 2015). Since the symbiotic relationships of attine ants with fungi are not fully understood and comprise a wide fungal diversity (Pagnocca et al. 2008, Rodrigues et al. 2011, Duarte et al. 2014), nests of ants are favourable environments to investigate for undescribed taxa.
The symbiosis between attine ants and their mutualistic fungi (Leucocoprineae and Pterulaceae), nourished as food source (Mueller & Rabeling 2008) exists for approximately 50 M years (Schultz & Brady 2008). In order to maintain the symbiont, attine ants utilise different agricultural systems (Schultz & Brady 2008). The leaf-cutter agriculture performed by genera Atta and Acromyrmex is the most common in Brazil and consists of cutting fresh plant material to be incorporated into the fungal garden and used as energy source for the growth of the mutualistic fungus Leucoagaricus gongylophorus (Weber 1972).
Recent studies of melanised fungi carried in the integument of leaf-cutting ants have revealed the existence of a wide diversity of undescribed fungal species (Duarte et al. 2014). Little & Currie (2007) were the first to show an association between a black yeast (phialophora-like), classified by the authors as the fifth symbiont, and Apterostigma ants (tribe Attini). Thus far, three new black fungal species isolated from leaf-cutting ants have been described: Phialophora attae, P. capiguarae (Attili-Angelis et al. 2014) and Ochroconis globalis (Samerpitak et al. 2015). In addition, Duarte et al. (2014) isolated several unknown species affiliated to Teratosphaeriaceae, which are the subject of the present study.
The Teratosphaeriaceae was originally separated from Mycosphaerellaceae (Crous et al. 2007) and circumscribed to include saprobic, extremophilic, human opportunistic and plant pathogenic fungi (Quaedvlieg et al. 2014). Although often linked to diseases in plants, especially Myrtaceae and Proteaceae hosts (Pérez et al. 2009, Carnegie et al. 2011, Crous & Groenewald 2011, Hunter et al. 2011), Teratosphaeriaceae species do not have reports of specific associations with ants. However, this family is also rich in genera and species associated with rocks and other extreme environments (Egidi et al. 2014), and its ecology remains insufficiently known.
In a recent revision to phylogenetically and morphologically classified lineages in Teratosphaeriaceae, Quaedvlieg et al. (2014) generated a multi-locus DNA sequence dataset and introduced 17 new genera within Teratosphaeriaceae, including Xenopenidiella, a saprobic fungus on leaf litter. Xenopenidiella was introduced based on a single species, X. rigidophora, which was represented by a single isolate (Quaedvlieg et al. 2014). The discovery of more species of Xenopenidiella will thus allow for a more robust generic circumscription of the genus and facilitate a better understanding of its ecology.
The present study aims to provide a taxonomic study of Teratosphaeriaceae species obtained from the integument of leaf-cutting ants from Brazil. Here, we describe a novel genus isolated from ants (Simplicidiella), a new species of Penidiellopsis (P. ramosus), and six new species of Xenopenidiella (X. clavata, X. formica, X. inflata, X. laevigata, X. nigrescens, X. tarda). A third genus, Penidiellomyces, is also introduced for two species of Penidiella s.lat.
MATERIALS AND METHODS
Isolation techniques
In order to isolate the fungi present in the integument of attine ants, three different methods were used as described below.
For the oil flotation technique (Satow et al. 2008) the bodies of sampled ants were immersed in 25 mL of saline solution (NaCl 0.85 %) with antibiotics (200 U penicillin, 200 μg/mL chloramphenicol, 200 μg/mL streptomycin and 500 μg/mL cycloheximide). Tubes were vortexed and after 30 min of incubation at room temperature, approximately 20 % sterile mineral oil was added, followed by vortexing for 5 min. After 20 min of settling, 100 μL aliquots were removed from the oil-solution interface and plated on Mycosel agar using a Drigalsky rod. Plates were incubated in the dark at 25 °C and monitored daily until the appearance of fungal colonies.
For the nitrogen-free cultivation, ants were immersed in distilled water and sonicated for 15 min. Then, 150 μL of the solution containing fungal cells in suspension were plated in a free-nitrogen medium described by Thanh (2006). Plates were incubated in the dark at 25 °C for 3 wk.
In the third method, a dextrose 50 % enrichment technique was performed by immersing the ants in tubes containing 50 % dextrose and 0.5 % yeast extract. After 10 d of incubation in the dark at 28 °C, tubes were shaken and 150 μL of the mixed solution was plated on malt-dextrose agar (1 % malt, 1 % dextrose, 0.01 % chloramphenicol, 2 % agar) and again incubated in the dark at 28 °C for 10 d (Thanh et al. 2013).
Fungal isolates
The 25 isolates analysed are listed in Table 1 and were all obtained from the integument of Atta capiguara and A. laevigata leaf-cutting ant gynes (primary reproductive female caste) and drones (male caste which role is to mate with a gyne) from Brazil.
Table 1.
Fungal isolates details and GenBank accession numbers of isolates included in this study.
| Species | Isolate number1 | Source | Isolation technique | GenBank Accession numbers2 |
||||
|---|---|---|---|---|---|---|---|---|
| tub2 | cmdA | tef1 | ITS | LSU | ||||
| Penidiellopsis radicularis | AP386 = CBS 131976 = CBMAI 1938 | A. capiguara gyne | Oil flotation | KU216267 | KU216292 | KU216339 | KT833148 | KU216314 |
| AP387 = CBS 131979 = CBMAI 1947 | A. capiguara gyne | Oil flotation | KU216268 | KU216293 | KU216340 | KT833149 | KU216315 | |
| AP389 | A. capiguara gyne | Oil flotation | KU216269 | KU216294 | KU216341 | KT833150 | KU216316 | |
| AP410 = CBS 131963 | A. capiguara gyne | Oil flotation | KU216272 | KU216297 | KU216344 | KT833153 | KU216319 | |
| AP418 | A. capiguara gyne | Oil flotation | KU216273 | KU216298 | KU216345 | KT833154 | KU216320 | |
| AP440 = CBS 132769 = CBMAI 1951 | A. capiguara gyne | Oil flotation | KU216276 | KU216301 | KU216348 | KT833157 | KU216323 | |
| DOC350 = CBMAI 1952 | A. capiguara drone | Nitrogen-free | KU216277 | – | KU216349 | KT833158 | KU216324 | |
| DOC363 = CBMAI 1953 | A. capiguara drone | Nitrogen-free | KU216278 | KU216302 | KU216350 | KT833159 | KU216325 | |
| Penidiellopsis ramosus | AP391 = CBMAI 1937ET | A. capiguara gyne | Oil flotation | KU216270 | KU216295 | KU216342 | KT833151 | KU216317 |
| AP392 = CBMAI 1948 | A. capiguara gyne | Oil flotation | KU216271 | KU216296 | KU216343 | KT833152 | KU216318 | |
| AP420 = CBMAI 1949 | A. capiguara gyne | Oil flotation | KU216274 | KU216299 | KU216346 | KT833155 | KU216321 | |
| AP421 = CBMAI 1950 | A. capiguara gyne | Oil flotation | KU216275 | KU216300 | KU216347 | KT833156 | KU216322 | |
| Simplicidiella nigra | AP 416 = CBMAI 1939ET | A. capiguara gyne | Oil flotation | KU216266 | KU216291 | KU216338 | KT833147 | KU216313 |
| Xenopenidiella clavata | DOC354 = CBMAI 1942ET | A. capiguara drone | Nitrogen-free | KU216287 | – | – | KT833168 | KU216334 |
| Xenopenidiella formica | 67N = CBMAI 1954 | A. laevigata drone | Oil flotation | KU216282 | KU216306 | KU216354 | KT833163 | KU216329 |
| AP380 = CBMAI 1946 | A. capiguara gyne | Oil flotation | KU216283 | KU216307 | KU216355 | KT833164 | KU216330 | |
| DOC323 = CBMAI 1941ET | A. capiguara drone | Nitrogen-free | KU216284 | KU216308 | KU216356 | KT833165 | KU216331 | |
| DOC349 | A. capiguara drone | Nitrogen-free | KU216285 | KU216309 | KU216357 | KT833166 | KU216332 | |
| DOC362 | A. capiguara drone | Nitrogen-free | KU216286 | KU216310 | KU216358 | KT833167 | KU216333 | |
| Xenopenidiella inflata | 87N = CBMAI 1945ET | A. laevigata drone | Oil flotation | KU216290 | KU216312 | KU216359 | KT833171 | KU216337 |
| Xenopenidiella laevigata | 89N = CBMAI 1944ET | A. laevigata gyne | Oil flotation | KU216289 | – | – | KT833170 | KU216336 |
| Xenopenidiella nigrescens | DOC356 = CBMAI 1943ET | A. capiguara drone | Nitrogen-free | KU216288 | KU216311 | – | KT833169 | KU216335 |
| Xenopenidiella tarda | DOC248 = CBMAI 1940ET | A. capiguara drone | Dextrose 50% | KU216279 | KU216303 | KU216351 | KT833160 | KU216326 |
| DOC317 | A. capiguara drone | Nitrogen-free | KU216280 | KU216304 | KU216352 | KT833161 | KU216327 | |
| DOC343 | A. capiguara drone | Nitrogen-free | KU216281 | KU216305 | KU216353 | KT833162 | KU216328 | |
1 CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; CBMAI: Brazilian Collection of Environmental and Industrial Microorganisms; all other codes are placed in the culture collection of the Microbial Resource Centre (UNESP, Brazil).
2 cmdA: partial calmodulin gene; ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial 28S nrRNA gene; tef1: partial translation elongation factor 1-alpha; tub2: partial beta-tubulin gene.
ET ex-type.
All isolates are maintained in the culture collection of the Microbial Resource Centre (CRM-UNESP, Brazil). Ex-type strains were deposited in the CBS-KNAW Fungal Biodiversity Centre (The Netherlands) and/or in the Brazilian Collection of Environmental and Industrial Microorganisms (CBMAI, Brazil). Sequences derived in this study were lodged at GenBank and taxonomic novelties with MycoBank (Crous et al. 2004).
DNA extraction
Isolates were transferred to fresh 2 % MA (Malt Agar) plates and incubated in the dark at 25 °C for 14–21 d. Approximately 1 cm2 of mycelium was transferred to a 2 mL Eppendorf tube containing 500 μL of lysis buffer (50 mmol/L Tris-HCl; 250 mmol/L NaCl; 50 mmol/L EDTA; 0.3 % w/v SDS; pH 8) and the equivalent to 100 μL of glass beads (SIGMA-ALDRICH catalogue G8893). After vortexing for 4 min, the microtubes were incubated for 1 h at 65 °C (two replicates). After centrifugation for 15 min at 13 000 rpm the aqueous phase with DNA was transferred into new microtubes. DNA extracts were stored at -20 °C prior to use.
Multi-locus PCR amplification and sequencing
Isolates were screened for five loci (ITS, LSU, cmdA, tef1 and tub2) using the primer sets listed in Table 2. Amplification reactions were performed in a total volume of 25 μL containing 1 μL of 50 mM MgCl2, 4 μL of 1.25 mM of dNTP Mix, 2.5 μL of 10× PCR buffer, 2 μL of 10 μM of each primer (100 μM stock concentration), 0.2 μL of 5 U/μL of Taq polymerase (Invitrogen), 5 μL of DNA template (diluted 1 : 100) and 8.3 μL PCR water. PCR conditions were set as follows: an initial denaturation temperature of 96 °C for 2 min, followed by 35 cycles of denaturation at 96 °C for 45 s, primer annealing at the temperature stated in Table 2, primer extension at 72 °C for 90 s and a final extension step at 72 °C for 2 min. PCR products were analysed on a 1 % agarose gel and a negative control (DNA free) was also included to check for possible contaminations. Amplicons were purified using a NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel), according to the manufacturer’s instructions. The PCR products were sequenced in both directions using the BigDye® Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems Life Technologies, Carlsbad, CA, USA) and an ABI Prism® 3130 Genetic Analyzer (Applied Biosystems). Consensus sequences were generated using the BioEdit Sequence Alignment Editor v. 7.0.5.3 (Hall 1999).
Table 2.
Primer combinations used for amplification and sequencing.
| Locus | Primer | Primer sequence 5′ to 3′ | Annealing temperature (°C) | Orientation | Reference |
|---|---|---|---|---|---|
| β-tubulin (tub2) | T1 | AACATGCGTGAGATTGTAAGT | 52 | Forward | O’Donnell & Cigelnik (1997) |
| β-Sandy-R | GCRCGNGGVACRTACTTGTT | Reverse | Stukenbrock et al. (2012) | ||
| Calmodulin (cmdA) | CAL-235F | TTCAAGGAGGCCTTCTCCCTCTT | 50 | Forward | Quaedvlieg et al. (2012) |
| CAL2Rd | TGRTCNGCCTCDCGGATCATCTC | Reverse | Groenewald et al. (2013) | ||
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | 52 | Forward | White et al. (1990) |
| ITS4 | TCCTCCGCTTATTGATATGC | Reverse | White et al. (1990) | ||
| LSU (28S nrDNA) | LSU1Fd | GRATCAGGTAGGRATACCCG | 52 | Forward | Crous et al. (2009a) |
| LR5 | TCCTGAGGGAAACTTCG | Reverse | Vilgalys & Hester (1990) | ||
| Translation elongation factor-1α (tef1) | EF1-728F | CATCGAGAAGTTCGAGAAGG | 52 | Forward | Carbone & Kohn (1999) |
| EF-2 | GGARGTACCAGTSATCATGTT | Reverse | O’Donnell et al. (1998) |
Alignment and phylogenetic reconstruction
A preliminary identification of the isolates to genus level was made by comparing the consensus LSU sequences against NCBIs GenBank nucleotide database using the megaBLAST algorithm. The most similar sequences were downloaded in FASTA format and subsequently combined with the Teratosphaeriaceae alignment of Quaedvlieg et al. (2014) (TreeBASE study S16145). A draft phylogeny was created from this alignment (data not shown), after which a reduced dataset was used to generate the final overview phylogeny presented here. The species level phylogeny was created from a concatenated alignment consisting of ITS, cmdA, tub2 and tef1 sequences. All loci were aligned individually using the MAFFT v. 7 online portal (http://mafft.cbrc.jp/alignment/server/index.html; Katoh & Standley 2013), after which they were manually checked and improved in MEGA v. 6.06 (Tamura et al. 2013). The phylogenetic reconstructions were conducted using the command line versions of MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003) and PAUP v. 4.0b10 (Swofford 2003), while MrModeltest v. 2.2 (Nylander 2004) was used to determine the best nucleotide substitution model settings for MrBayes. A parsimony analysis was performed on the overview LSU alignment and the combined 4-locus alignment; prior to concatenation of the four loci, individual gene trees derived from each locus were examined for topological conflict compared to the trees from the other loci. Gaps were treated as a new state, missing data were indicated as missing data in the analyses and statistics such as tree length, consistency index, retention index and rescaled consistency index (TL, CI, RI and RC, respectively) were calculated. For the Bayesian analysis of the overview LSU phylogeny, the heating parameter was set to 0.2 and the search was stopped when convergence was reached (stopval = 0.01). Trees were saved every 100 generations and the Markov Chain Monte Carlo (MCMC) analysis of 4 chains started in parallel from a random tree topology. The optimal substitution model under the Akaike’s Information Criterion was the GTR model with dirichlet (1,1,1,1) state frequency distribution and inverse gamma-shaped rate variation across sites. Sequences derived from this study were deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank) (Table 1), and the alignments and trees in TreeBASE (https://treebase.org/treebase-web/home.html). The resulting phylogenetic trees were imported into and printed with Geneious v. 7.1.8 (http://www.geneious.com, Kearse et al. 2012), and the layout of the tree for publication was done using Adobe Illustrator v. CS6.
Morphology
Colony characters and pigment production were observed on 2 % MA (Malt Agar), PDA (Potato Dextrose Agar, Acumedia®) and CMA (Corn Meal Agar, Himedia®) at 25 °C in the dark for 21 d. Microscopic observations were based on slide culture techniques using CMA due to the rapid induction of sporulation. Agar blocks of ~1 cm2 were placed on a sterile Petri dish and inoculated at the four edges. The block was subsequently covered with a sterile cover slip (~2 cm2). Plates were incubated at 25 °C in the dark for 14–21 d. Slides were made in distilled water. Micrographs were taken using a phase-contrast microscope (DM 750, Leica; software Leica Application Suite v. 3.5.0, Leica).
RESULTS
Phylogeny
The LSU alignment consisted of 75 sequences (including the outgroup sequence, Toxicocladosporium rubrigenum GenBank FJ790305) and 776 characters were included in the analysis. Of the 776 analysed characters, 174 were parsimony-informative, 85 variable characters were parsimony-uninformative and 517 characters were constant. A maximum of 1 000 equally most parsimonious trees were saved, one of which is shown in Fig. 1 (TL = 799 steps; CI = 0.458; RI = 0.777; RC = 0.356). The Bayesian analysis sampled 255 unique site patterns and lasted 775 000 generations, after which convergence was achieved. The posterior probability values mapped unto Fig. 1 were calculated from the 11 628 trees sampled from the 15 502 trees generated; the remainder of the trees were discarded as burn-in. A similar topology was observed between the Bayesian and parsimony analyses. The LSU phylogeny revealed that the strains from ants are present in two main clades, one of which is associated with the genus Xenopenidiella, whereas the other has as closest neighbour ‘Penidiella’ aggregata (GenBank JF499862), ‘Penidiella’ drakensbergensis (GenBank KC005792) and the recently described genus Penidiellopsis (GenBank LN834445). As the genus Penidiella is restricted to the lineage containing P. columbiana (represented here by GenBank EU019274, obtained from the ex-type culture CBS 486.80), which is not congeneric with the lineage containing P. aggregata, P. drakensbergensis and the ant strains, one novel genus, Simplicidiella, is described below to accommodate one of the strains isolated from ants and another new genus. Penidiellomyces, is proposed for the two ‘Penidiella’ species.
Fig. 1.
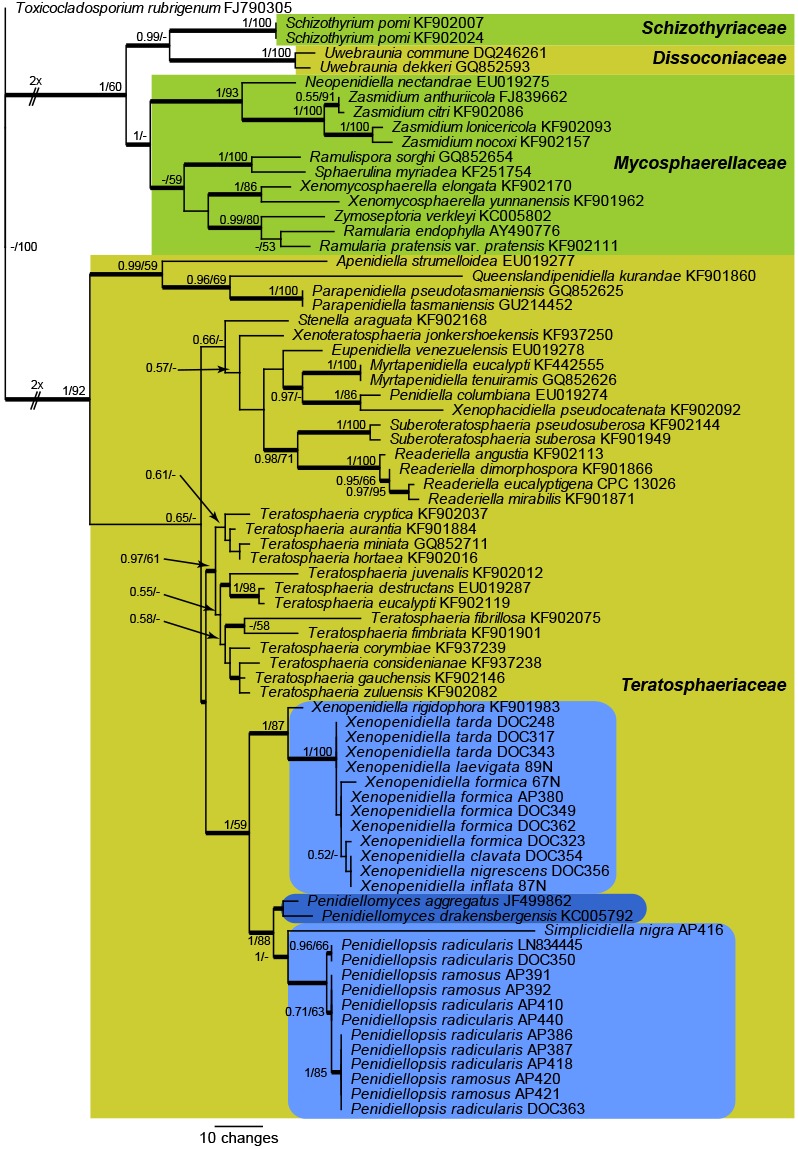
One of 1 000 equally most parsimonious trees obtained from a maximum parsimony analysis of the LSU sequence alignment. The scale bar shows 10 changes, and posterior probability values from a Bayesian analysis (PP) and parsimony bootstrap support (PBS) values from 1 000 replicates are shown at the nodes (PP/PBS). Thickened lines represent those branches also present in the strict consensus tree and families are indicated with coloured square blocks and the clades with ant isolates with light blue-coloured blocks with rounded corners. The darker blue block with rounded corners represents the novel genus introduced here to accommodate ‘P.’ aggregata and ‘P.’ drakensbergensis. The lengths of some branches were halved to facilitate easier layout. The tree was rooted to Toxicocladosporium rubrigenum (GenBank accession number FJ790305).
For the species level analysis, a concatenated sequence alignment from 26 strains (including the outgroup species Parapenidiella pseudotasmaniensis strain CBS 124991) was subjected to maximum parsimony analyses. The final aligned sequences of the ITS (498 characters), cmdA (389 characters), tef1 (342 characters) and tub2 (446 characters) gene regions had a total length of 1 675 characters (including alignment gaps) which were included in the analyses. The gaps in the alignment were treated as fifth base for the parsimony analyses. From the analysed characters, 902 were constant (ITS: 293; cmdA: 257; tef1: 152; tub2: 200), 255 were variable and parsimony-uninformative (ITS: 93; cmdA: 44; tef1: 65; tub2: 53) and 518 were parsimony-informative (ITS: 112; cmdA: 88; tef1: 125; tub2: 193). A total of 59 equally most parsimonious trees (TL = 1 343 steps; CI = 0.832; RI = 0.944; RC = 0.786) were saved from the parsimony analysis, one of which is shown in Fig. 2. Rerunning the analysis without the tef1 sequences (the least complete dataset) did not result in an overall topology that was much different from the topology depicted in Fig. 2; only some internal rearrangements were observed between the isolates representing each of the two multi-strain species in Penidiellopsis (data not shown). In addition, a comparison of the individual gene trees revealed the same generic and terminal species clades, with some rearrangements in the order of the species clades within the different genera (data not shown). The only exception was the genus Penidiellopsis, where the most support for the two species recognised in Fig. 2 was obtained from the tub2 sequences. The ITS sequence of strain DOC 350 was 100 % identical to Penidiellopsis radicularis (GenBank LN834441); unfortunately no other loci were available to include the ex-type culture of this species in the multi-gene phylogeny. Several unique lineages are present in the obtained phylogeny (Fig. 2), and these are described as novel species in the Taxonomy section below.
Fig. 2.
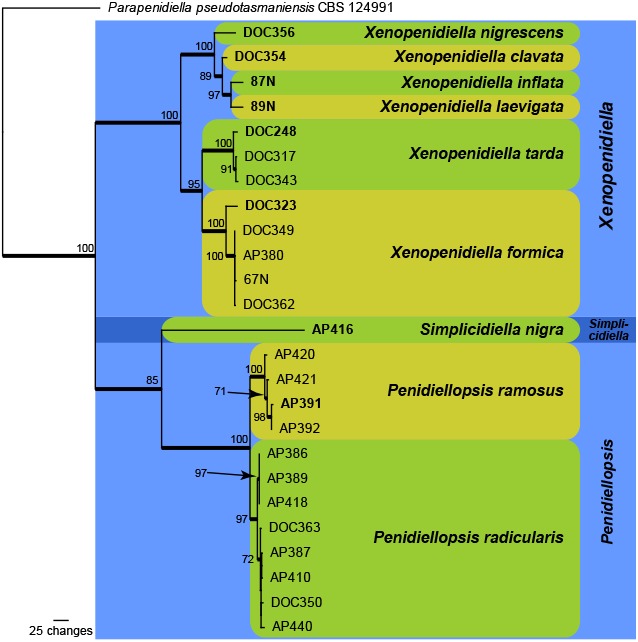
One of 59 equally most parsimonious trees obtained from a maximum parsimony analysis of the combined ITS, cmdA, tub2 and tef1 sequence alignment. The scale bar shows 100 changes, and bootstrap support values from 1 000 replicates are shown at the nodes. Thickened lines represent those branches also present in the strict consensus tree and species are indicated with coloured square blocks with rounded corners and the genera with coloured blocks. The tree was rooted to Parapenidiella pseudotasmaniensis strain CBS 124991 (GenBank accession numbers KF901522, KF902589, KF902855, KF903152, respectively).
Taxonomy
Penidiellomyces Crous, Attili-Angelis, A.P.M. Duarte, Pagnocca & J.Z. Groenew., gen. nov. — MycoBank MB817411
Etymology. Named after its morphological similarity to the genus Penidiella.
Type species. Penidiellomyces aggregatus (Crous) Crous & A.P.M. Duarte.
Hyphomycetous. Mycelium consisting of branched, septate, smooth, pale brown hyphae. Conidiophores solitary, arising from superficial mycelium, erect, brown, smooth, septate, straight to irregularly geniculate-sinuous. Conidiogenous cells terminal, subcylindrical, unbranched, medium brown, smooth, tapering to a flattened or rounded apical region, scars unthickened, aggregated, somewhat darkened, not refractive. Ramoconidia 0–1-septate, medium brown, smooth, ellipsoidal to obclavate, obovoid or subcylindrical, with 1–3-apical hila. Intermediate and terminal conidia subcylindrical to ellipsoid, 0–1-septate, brown, in chains of up to 6; hila truncate, unthickened, somewhat darkened.
Notes — Penidiellomyces resembles Penidiella in having solitary conidiophores with an apical apparatus that can appear penicillate, giving rise to branched chains of conidia. It differs, however, by having a terminal conidiogenous cell with a more well-developed apical rachis that gives rise to ramoconidia. The recent introduction of several penidiella-like genera in this complex (Quaedvlieg et al. 2014), however, will make it difficult to identify species of Penidiellomyces without the aid of molecular data. Ecologically, both known species share a foliicolous habitat on plants known from South Africa (Crous et al. 2007, 2012, Crous & Groenewald 2011), and are presumed to be saprobic.
Penidiellomyces aggregatus (Crous) Crous & A.P.M. Duarte, comb. nov. — MycoBank MB817412
Basionym. Penidiella aggregata Crous, Persoonia 26: 78. 2011.
Description and illustration — See Crous & Groenewald (2011).
Penidiellomyces drakensbergensis (Crous) Crous & Attili-Angelis, comb. nov. — MycoBank MB817413
Basionym. Penidiella drakensbergensis Crous, Persoonia 29: 161. 2012.
Description and illustration — See Crous et al. (2012).
Penidiellopsis Sandoval-Denis et al., Persoonia 36: 439. 2016
Type species. Penidiellopsis radicularis Sandoval-Denis et al.
Conidiophores differentiated, solitary, erect, straight to geniculate-sinuous, rarely branched, pale to medium brown, smooth- and thick-walled. Conidiogenous cells integrated, terminal or intercalary, pale to medium brown, smooth, mono- and polyblastic, giving rise to one or more sets of ramoconidia, scars truncate, slightly darkened, unthickened and not refractive. Ramoconidia 0–1-septate, obovoid, ellipsoid or slightly clavate, pale to medium brown, smooth- and thick-walled, apical part with denticle-like loci, basal scar flattened, slightly darkened, unthickened and not refractive. Conidia in branched acropetal chains, 0-septate, obovoid, ellipsoid or limoniform, pale to medium brown, smooth, thick-walled, with conidial scars truncate or protuberant, somewhat darkened, unthickened and not refractive. Sexual morph unknown.
Notes — Penidiellopsis was introduced as a new genus to accommodate an isolate from a human nail, collected in South Carolina, USA (Crous et al. 2016). Phylogenetically it is closely related to Penidiellomyces aggregatus and P. drakensbergensis. Conidiophores in Penidiella are penicillate with well-developed apical branches, characteristics that were absent in Penidiellopsis. Morphological data are only known from slow-growing colonies in artificial media.
Penidiellopsis radicularis Sandoval-Denis et al., Persoonia 36: 439. 2016 — Fig. 3
Fig. 3.
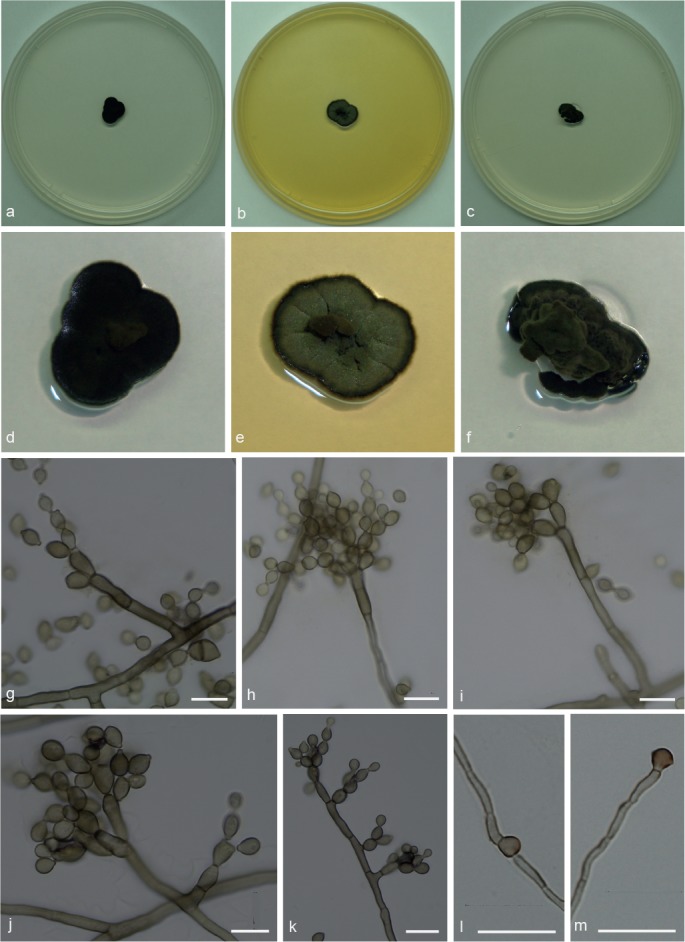
Penidiellopsis radicularis. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–k. conidiophores with conidia in chains; l–m. chlamydospores. — Scale bars = 10 μm.
Isolated from gynes and drones of Atta capiguara (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth-walled, pale brown, 1.5–3 μm wide hyphae. Conidiophores erect, solitary, from superficial mycelium, branched or not, 25–53 × 3.5–4 μm. Conidiogenous cells mostly elongate, aseptate, smooth-walled, 10.5–13 × 3.5–4 μm. Conidia catenate in branched chains, 0–1-septate, smooth, lemon-shaped with prominent hila, not thickened, slightly darkened, 4.5–6.5 × 3–4 μm. Chlamydospores terminal and intercalary. Hila present but neither thickened nor darkened.
Culture characteristics — Colonies were grown at 25 °C for 21 d. On CMA, colonies flat with velutinous central portion, dark olivaceous to black, lobed margin, reaching 10 mm diam. Colonies on MA greyish green with darker mycelium in the centre and margin, crenate surface, reaching 11.5 mm diam. On PDA, dark olivaceous folded colonies, with a raised, velutinous central portion, reaching 11 mm diam. Colonies sporulating on all media, with dark mycelium and olivaceous black in reverse on all three media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara gyne, Nov. 2009, F.C. Pagnocca, CBMAI 1938.
Notes — Similar to P. ramosus although colonies are smaller. Penidiellopsis radicularis (originally isolated and described from an infection of a human nail) was found on gynes and drones, while P. ramosus only occurs on gynes of Atta capiguara.
Penidiellopsis ramosus Attili-Angelis & A.P.M. Duarte, sp. nov. — MycoBank MB817886; Fig. 4
Fig. 4.
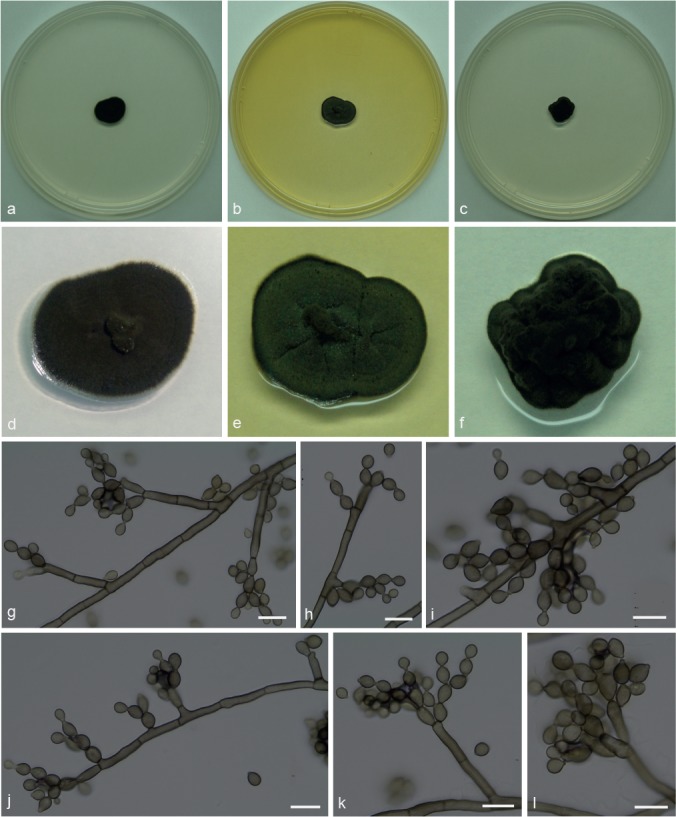
Penidiellopsis ramosus. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–l. conidiophores with conidia in chains. — Scale bars = 10 μm.
Etymology. Name refers to its apically branched conidiophores.
Isolate from gynes of Atta capiguara (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth-walled, pale olivaceous-brown, 2–3.5 μm wide hyphae. Conidiophores ascending to erect, apically branched, bearing conidia in short chains, irregularly geniculate-sinuous, 27–63.5 × 3–4 μm. Conidiogenous cells aseptate, smooth-walled, 7–17 × 3–4 μm; scars not thickened, slightly darkened. Conidia catenate in branched chains, aseptate, smooth, limoniform to fusiform, 4–7.5 × 3–4 μm. Chlamydospores not observed. Hila present but neither thickened nor darkened.
Culture characteristics — Colonies were grown at 25 °C for 21 d. On CMA, colonies flat with velutinous central portion, dark olivaceous to black, reaching 13 mm diam. Colonies on MA greyish green with darker mycelium in the centre and margin, sulcate surface, reaching 14 mm diam. On PDA, dark olivaceous folded colonies, raised with an entire edge, velutinous central portion, reaching 12 mm diam. Colonies sporulating on all media, with dark mycelium and olivaceous black in reverse on all three media.
Specimens examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara gyne, Nov. 2009, F.C. Pagnocca (holotype and culture ex-type CBMAI 1937, preserved as metabolically inactive).
Notes — Similar to P. radicularis, but chlamydospores were not observed. Although conidia are similar in shape and size, those of P. ramosus are aseptate.
Simplicidiella Crous, Attili-Angelis, A.P.M. Duarte, Pagnocca & J.Z. Groenew., gen. nov. — MycoBank MB817414
Etymology. Named after the presence of simple and poorly differentiated conidiophores (Simplici-, Latin = simple).
Type species. Simplicidiella nigra A.P.M. Duarte & Attili-Angelis.
Hyphomycetous. Mycelium consisting of septate, smooth-walled, pale brown hyphae. Conidiophores erect, solitary, poorly branched or unbranched. Conidiogenous cells elongate, aseptate, smooth-walled; scars not thickened, slightly darkened. Conidia catenate, in branched chains, 0–1-septate, smooth, ellipsoidal to broadly ellipsoidal with prominent hila. Sexual morph unknown.
Notes — The genus Simplicidiella is described to accommodate ant-isolated melanised fungi in Brazil, with simple and poorly differentiated reproductive structures. Its few phenotypic characteristics underly the need of molecular tools for identification. The genus is presumed here to be saprobic. Simplicidiella is distinct from Penidiella and other penidiella-like genera for not sharing the following characteristics: penicillate conidiophores with branches (as in Penidiella s.str. and Queenslandipenidiella); being described as foliicolous (as in Neopenidiella and Myrtapenidiella); associated with opportunistic human infections with dimorphic conidiophores (as reported for Eupenidiella); production of ramoconidia (as in Apenidiella), and mycelium strongly branched (as observed in Xenopenidiella).
Simplicidiella nigra A.P.M. Duarte & Attili-Angelis, sp. nov. — MycoBank MB817415; Fig. 5
Fig. 5.
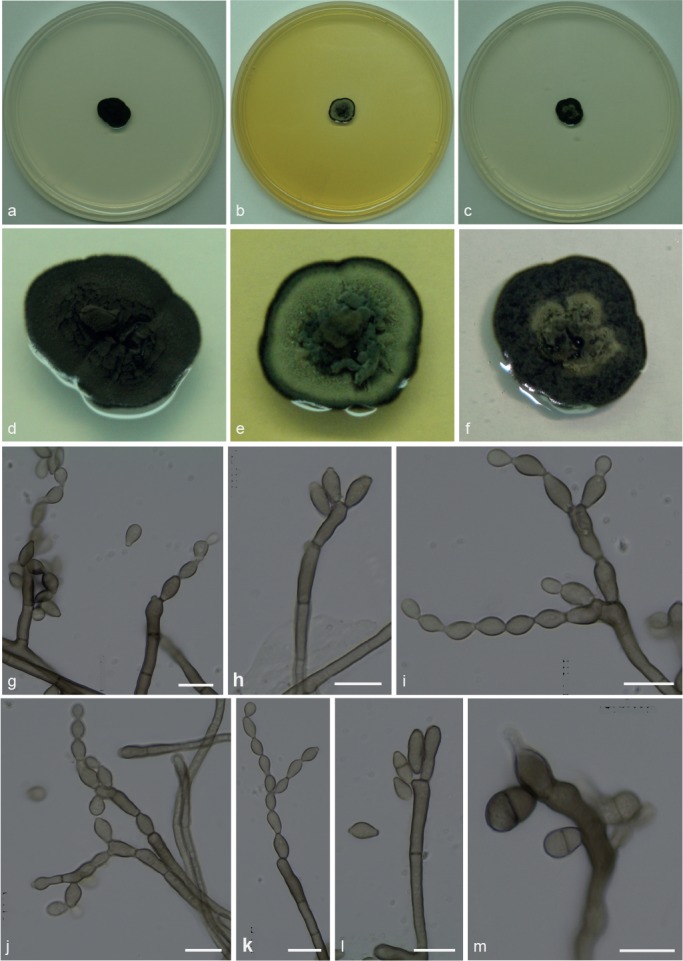
Simplicidiella nigra. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–k. conidiophores with conidia in chains; l–m. conidiogenous cells. — Scale bars = 10 μm.
Etymology. Name refers to very dark colonies on CMA.
Isolated from gyne of Atta capiguara (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth-walled, pale brown, 1.5–3 μm wide hyphae. Conidiophores erect, solitary, poorly branched or unbranched, 27–47 × 3–3.5 μm. Conidiogenous cells elongate, aseptate, smooth-walled, 8–18.5 × 3–4 μm; scars not thickened, slightly darkened. Conidia catenate in branched chains, 0–1-septate, smooth, ellipsoidal to broadly ellipsoidal with prominent hila, 5.5–9 × 3–4 μm. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. On CMA, colonies flat with velutinous central portion, colony margin dark olivaceous to black, reaching 14 mm diam. Colonies on MA flat with raised velutinous central portion, intermediate section with olivaceous green mycelium, and darker margins, reaching 11 mm diam. Exudate produced. On PDA, colonies with raised greyish central portion producing exudate, black mycelium and darker margins, reaching 11 mm diam. Sporulation and olivaceous black reverse were present in all media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara gyne, Nov. 2009, F.C. Pagnocca (holotype and culture ex-type CBMAI 1939, preserved as metabolically inactive).
Notes — Due to few phenotypic characteristics available, identification of S. nigra requires molecular data. This species is presumed to be saprobic and a potential hydrocarbon-degrader, based on the selective method used for isolation and due to the known presence of a complex mixture of hydrocarbons on the cuticle of insects (Howard & Blomquist 2005). The species is described based on a single isolate, although its phylogenetic placement was supported by all loci and analyses.
Xenopenidiella Quaedvlieg & Crous, Persoonia 33: 33. 2014
Type species. Xenopenidiella rigidophora (Crous et al.) Quaedvlieg & Crous.
Xenopenidiella clavata Attili-Angelis, A.P.M. Duarte & Pagnocca, sp. nov. — MycoBank MB817416; Fig. 6
Fig. 6.
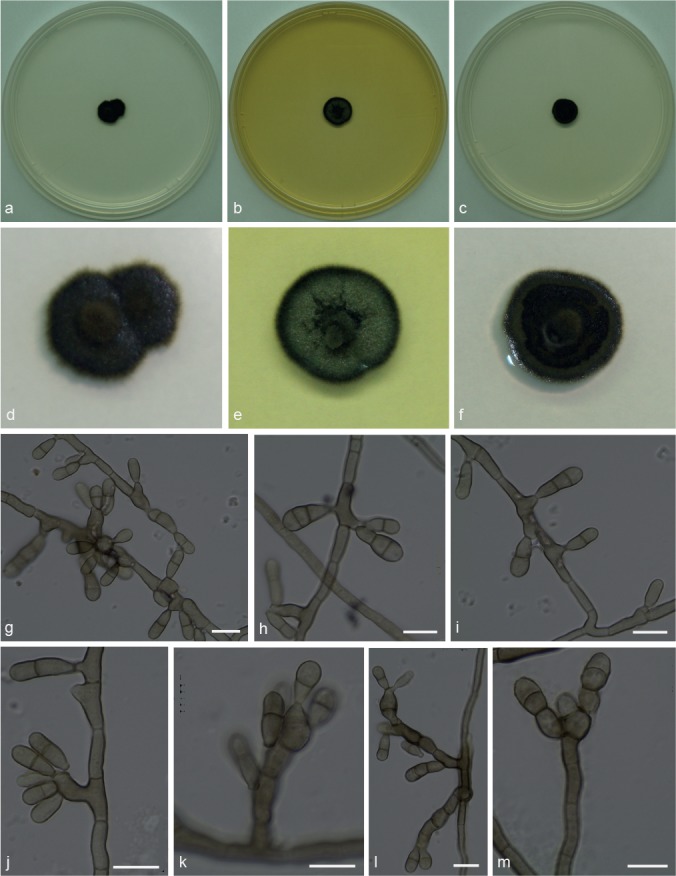
Xenopenidiella clavata. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–m. conidiophores with conidia. — Scale bars = 10 μm.
Etymology. Named after its clavate conidia.
Isolated from drone of Atta capiguara (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth-walled, pale brown, 3–4 μm wide hyphae. Conidiophores mostly reduced to conidiogenous cells arising directly from hyphae, rarely 1–5-septate, brown, smooth-walled, straight to geniculate-sinuous, 11–34 × 3.5–4.5 μm. Conidiogenous cells intercalary or terminal, brown, smooth-walled, 11–12 × 3.5–4 μm, with several aggregated terminal scars, not thickened nor darkened. Conidia 2–3-celled, smooth-walled to finely verrucose, clavate, apex obtuse, base truncate, constricted at both septa, 8–15.5 × 3–4 μm; hila not thickened, slightly darkened, no conidial scars observed. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. Colonies on CMA flat and dark olivaceous to brown, with a velutinous central portion, reaching 10 mm diam. Colonies on MA with a slightly elevated velutinous central portion, greenish mycelium and black margins, reaching 11 mm diam. On PDA, flat velutinous dark olivaceous to brown colonies, reaching 10 mm diam. Colonies with low to moderate sporulation and olivaceous black in reverse on all media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara drone, Nov. 2013, F.C. Pagnocca (holotype and culture ex-type CBMAI 1942, preserved as metabolically inactive).
Notes — Xenopenidiella clavata is ecologically distinct from X. rigidophora (type species) in its association to the leaf-cutting ants, which represents a new niche for the genus. Morphologically, conidiophores of X. clavata are not strongly branched, hyphae are not guttulate, and its mycelium is not strongly branched. Dimorphic conidiophores are present in both species, but in X. clavata conidiogenous cells are predominantly intercalary and conidia are only finely verrucose.
Xenopenidiella formica A.P.M. Duarte, Attili-Angelis & N.C. Baron, sp. nov. — MycoBank MB817417; Fig. 7
Fig. 7.
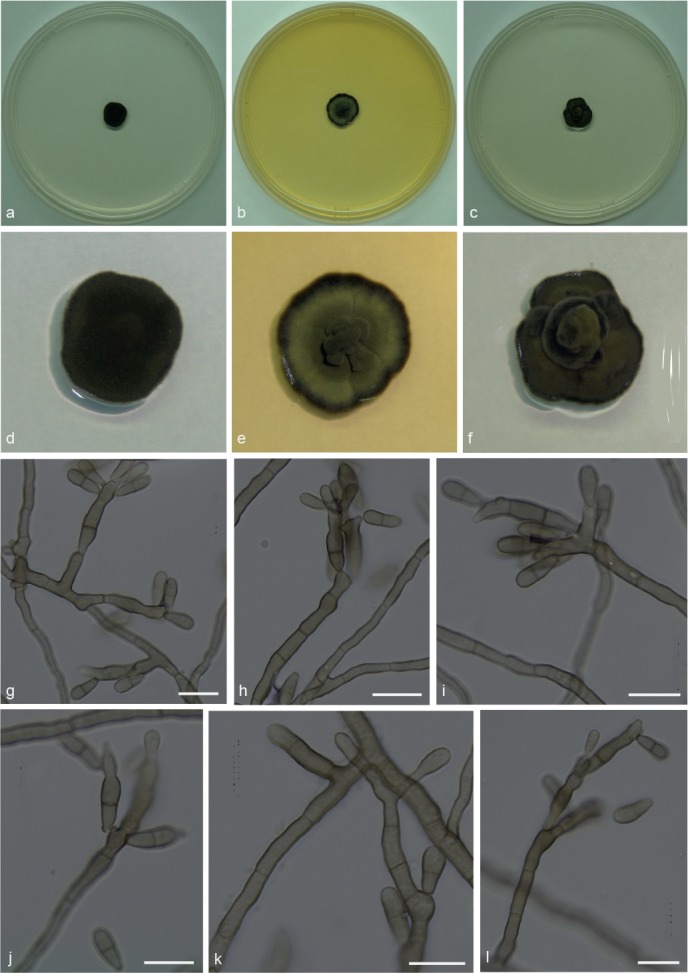
Xenopenidiella formica. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–l. conidiophores with conidia. — Scale bars = 10 μm.
Etymology. Named after the isolation source (ants).
Isolated from gynes and drones of Atta capiguara and A. laevigata (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth-walled, pale olivaceous, 2–3 μm wide hyphae, producing poorly differentiated conidiophores. Conidiophores erect, solitary, poorly branched or unbranched, smooth, brown, straight to geniculate-sinuous, 28.5–75 × 2–3.5 μm. Conidiogenous cells smooth-walled, bearing conidia from flat to semi-denticulate scars, 13–17 × 3.5–4 μm. Conidia 1-septate, smooth-walled, clavate, apex obtuse, base truncate, constricted at septum, 7–12 × 2.5–4 μm. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. Colonies on CMA were dark olivaceous and flat, velutinous, reaching 12 mm diam. On MA, flat colonies with velutinous central portion, greenish mycelium and darker margins, reaching 12.5 mm diam. On PDA, velutinous colonies with raised olivaceous green central portion, reaching 11 mm diam. Poor sporulation and olivaceous black in reverse on all media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara drone, Nov. 2013, F.C. Pagnocca (holotype and culture ex-type CBMAI 1941, preserved as metabolically inactive).
Notes — Phylogenetically, X. formica is closely related to X. tarda, but with a faster growth rate in culture, and smaller conidia. It is represented by five isolates from gynes and drones of A. capiguara and A. laevigata, while X. tarda was found only on A. capiguara. In contrast to X. rigidophora, X. formica does not exhibit strongly branched hyphae, and its conidia are not verrucose.
Xenopenidiella inflata A.P.M. Duarte, N.C. Baron, Pagnocca & Attili-Angelis, sp. nov. — MycoBank MB817418; Fig. 8
Fig. 8.
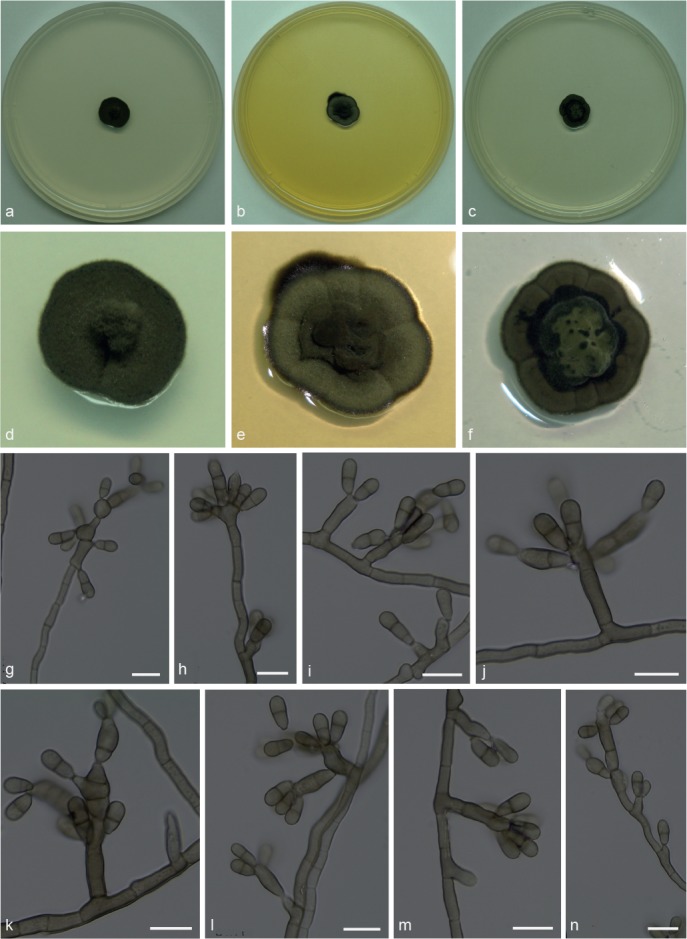
Xenopenidiella inflata. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–n. conidiophores with conidia. — Scale bars = 10 μm.
Etymology. Named after its swollen conidiogenous cells (Inflatus, Latin = swollen).
Isolated from drone of A. laevigata (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth, pale brown, 1.5–3 μm wide hyphae. Conidiophores reduced to conidiogenous cells, or subcylindrical, smooth, brown, up to 3-septate, straight to geniculous-flexuous, 16–52 × 4–5 μm. Conidiogenous cells swollen or elongated, arising from mycelium or terminal on conidiophores, sometimes rejuvenating and elongating to have lateral and terminal conidiogenous cells, smooth-walled, 9–14 × 3.5–4.5 μm; scars not thickened nor darkened. Conidia 1-septate, smooth-walled, clavate, constricted at septum, truncate hila, 7.5–10 × 3–4 μm. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. On CMA, colonies with black velutinous mycelium and pronounced central portion, reaching 12.5 mm diam. Colonies on MA greyish green, velutinous and sulcate, black margins, reaching 12 mm diam. On PDA colonies were greenish and raised at the centre, olivaceous green towards the sulcate and darker margin, reaching 12.5 mm diam. Poor to moderate sporulation and olivaceous black reverse on all media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta laevigata drone, Nov. 2011, F.C. Pagnocca (holotype and culture ex-type CBMAI 1945, preserved as metabolically inactive).
Notes — Xenopenidiella inflata is described based on a single isolate from an A. laevigata drone. Although phylogenetically closely related to X. laevigata, it differs based on its swollen conidiogenous cells and slower growth rate on cultural media. Xenopenidiella inflata differs from the X. rigidophora in the following morphological aspects: pale brown mycelium present (not pale olivaceous to medium brown); thinner hyphae (1.5–3 μm wide, not up to 6 μm diam); conidiogenous cells terminal and intercalary; conidia not verrucose (not appearing like small spines under light microscope); thicker hila.
Xenopenidiella laevigata N.C. Baron, A.P.M. Duarte, Pagnocca & Attili-Angelis, sp. nov. — MycoBank MB817419; Fig. 9
Fig. 9.
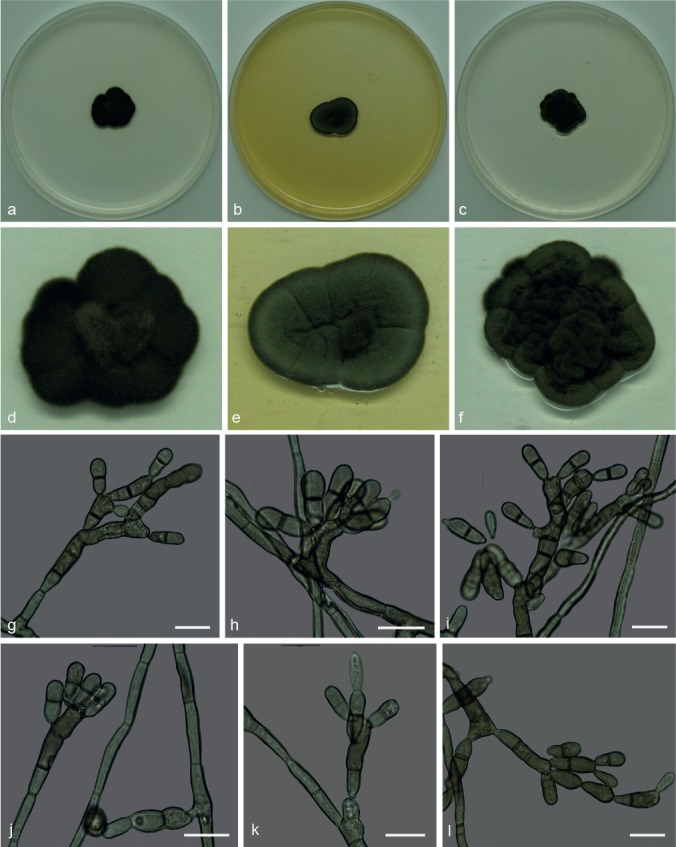
Xenopenidiella laevigata. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–l. conidiophores with conidia. — Scale bars = 10 μm.
Etymology. Name refers to the first isolation source: the ant species Atta laevigata.
Isolated from gyne of A. laevigata (Myrmicinae, Attini tribe). Mycelium consisting of septate, olivaceous-brown, smooth and thick-walled, 3–3.5 μm wide hyphae. Conidiophores brown, smooth to finely verrucose, thick-walled erect, straight to geniculous-sinuous, 1–10-septate, 10–64 × 2–4.5 μm. Conidiogenous cells in loose branches, smooth-walled to finely verrucose, terminal or lateral, 10.5–14 × 2.5–3.5 μm; scars unthickened and not darkened. Conidia 1-septate, smooth-walled to finely verrucose, clavate, constricted at somewhat darker septum, 9.5–17.5 × 3–5 μm; hila unthickened and not darkened. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. On CMA, colonies with black velutinous mycelium and aerial central portion, reaching 18 mm diam. Colonies on MA greyish green, velutinous and sulcate, black margins, reaching 19 mm diam. On PDA colonies were dark olivaceous, sulcate and folded, with raised central portion and velutinous mycelium, reaching 18 mm diam. Poor sporulation and olivaceous black reverse on all three media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta laevigata gyne, Nov. 2011, F.C. Pagnocca (holotype and culture ex-type CBMAI 1944, preserved as metabolically inactive).
Notes — In this study both species X. laevigata and X. inflata are known from single strains isolated from A. laevigata via the oil flotation method. The parsimony tree shows that they are closely related, but the former differs in the melanisation of the mycelium (colonies are darker and hyphae are thicker), growth rate and conidial size. A comparison with X. rigidophora shows differences such as less-branched and thinner hyphae, rare macronematous conidiophores that are loosely penicillate, and conidiogenous cells frequently intercalary.
Xenopenidiella nigrescens Attili-Angelis, A.P.M. Duarte & Pagnocca, sp. nov. — MycoBank MB817420; Fig. 10
Fig. 10.
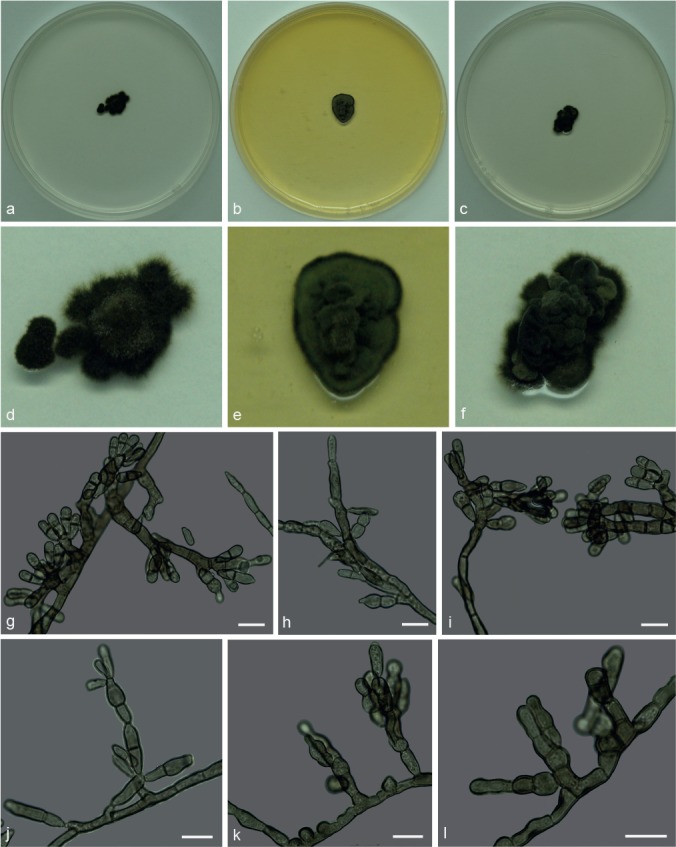
Xenopenidiella nigrescens. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–l. conidiophores with conidia. — Scale bars = 10 μm.
Etymology. Named after its dark brown mycelium.
Isolated from drone of Atta capiguara (Myrmicinae, Attini tribe). Mycelium consisting of septate, olivaceous-brown, smooth and thick-walled 3–4 μm wide hyphae. Conidiophores brown, smooth to finely verrucose, thick-walled, erect, straight to flexuous, 0–6-septate, 7–79 × 3.5–4.5 μm. Conidiogenous cells arising from mycelium, loosely branched, smooth-walled to verrucose, terminal and lateral, 13–18 × 3–3.5 μm. Conidia 1-septate, smooth-walled to finely verrucose, dark brown, clavate and constricted, 9–13 × 3.5–4 μm; hila unthickened and not darkened. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. Colonies on CMA were flat and dark olivaceous to brown, velutinous with raised central portion, irregular margins, reaching 14 mm diam. Colonies on MA with velutinous and slightly folded surface, raised central portion, olivaceous mycelium, reaching 13 mm diam. On PDA, dark olivaceous highly folded colonies, raised velutinous central portion, reaching 13 mm diam. Colonies presented low to moderate sporulation and olivaceous black reverse on all media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara drone, Nov. 2013, F.C. Pagnocca (holotype and culture ex-type CBMAI 1943, preserved as metabolically inactive).
Notes — Xenopenidiella nigrescens was found to consistently have the darkest colonies on all three media tested when compared to other Xenopenidiella species. A combination of previously mentioned characteristics is also found in this species: slow growth rate (as in X. tarda), clavate conidia (as in X. clavata), 1-septate, smooth-walled to finely verrucose conidia (resembling X. laevigata), conidiophores poorly differentiated (as in X. formica). In comparison to X. rigidophora, X. nigrescens differs in having darker mycelium, conidiogenous cells smooth to verrucose, and conidia rarely occurring in branched chains. Molecular data are required to confirm its identification.
Xenopenidiella tarda Pagnocca, A.P.M. Duarte & Attili-Angelis, sp. nov. — MycoBank MB817421; Fig. 11
Fig. 11.
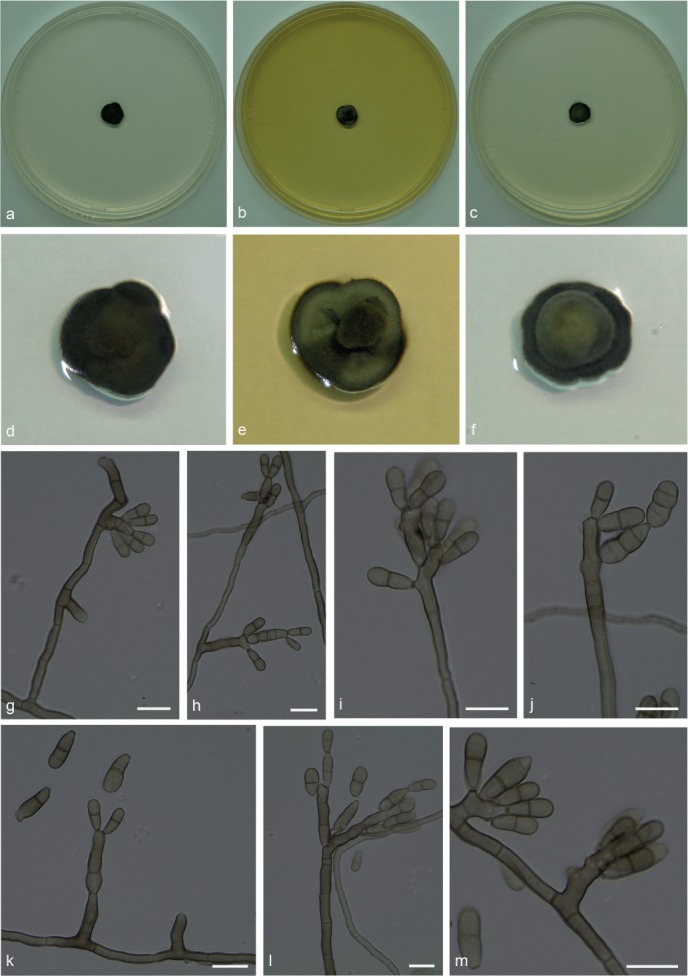
Xenopenidiella tarda. a, d. Colony on CMA; b, e. colony on MA; c, f. colony on PDA; g–m. conidiophores with conidia. — Scale bars = 10 μm.
Etymology. Named due to very slow growth rate (Tardus, Latin = slow).
Isolated from drones of Atta capiguara (Myrmicinae, Attini tribe). Mycelium consisting of septate, smooth-walled, pale brown, 2–3 μm wide hyphae. Conidiophores ascending to erect, solitary, poorly branched or unbranched, 0–4-septate, 5.5–51 × 3–4 μm. Conidiogenous cells elongate, aseptate, smooth-walled, 5.5–21.5 × 3–3.5 μm; scars unthickened and not darkened. Conidia 1-septate, smooth-walled to finely verrucose, clavate, constricted at septum, 8–11.5 × 3–5 μm; hila truncate, unthickened and not darkened. Chlamydospores not observed.
Culture characteristics — Colonies were grown at 25 °C for 21 d. On CMA, colonies dark olivaceous, velutinous, flat and small, reaching 9 mm diam. Colonies on MA flat with aerial velutinous central portion, olivaceous green mycelium and black margin, reaching 9 mm diam. On PDA colonies with raised greyish green central portion, and black margins, reaching 9 mm diam. Moderate sporulation and olivaceous black reverse present on all three media.
Specimen examined. BRAZIL, São Paulo (S22°50.6" W48°26.1", elev. 798 m), isolated from the integument of Atta capiguara drone, Nov. 2013, F.C. Pagnocca (holotype and culture ex-type CBMAI 1940, preserved as metabolically inactive).
Notes — Xenopenidiella tarda is represented by two isolates from drones of A. capiguara. It is morphologically similar to X. formica, although colonies have a slower growth rate in culture. This species can be distinguished from X. rigidophora because mycelium of X. tarda does not show to be strongly branched, not necessarily constricted at septa and hyphae are not so wide. Furthermore, conidiogenous cells are less wide and pale brown; conidia mostly smooth-walled or finely verrucose.
DISCUSSION
Leaf-cutting ants from the Attini tribe are social insects limited to the New World, widely distributed from Argentina to Southern USA, with a range of latitude from N10° to S25° (Mayhé-Nunes & Jaffé 1998). These ants have been investigated for the huge economic losses they can cause by damaging plant leaves, and for their unique ability for fungiculture, which has become a model system for coevolutionary studies (Chapela et al. 1994, Currie et al. 2003).
These insects were found to harbour a large and unknown diversity of microorganisms (Pagnocca et al. 2008, Rodrigues et al. 2008, 2011, Guedes et al. 2012, Duarte et al. 2014), and some novel species of filamentous fungi and yeasts were already described from the ants’ microenvironment (Middelhoven et al. 2003, Carreiro et al. 2004, Pagnocca et al. 2010, Augustin et al. 2013, Attili-Angelis et al. 2014, Melo et al. 2014, Masiulionis et al. 2015, Meirelles et al. 2015, Samerpitak et al. 2015, Montoya et al. 2016). However, the occurrence of species of Teratosphaeriaceae in the ants’ environment has never been documented.
The family Teratosphaeriaceae was established based on molecular phylogenetic analyses, which strongly supported its separation from Mycosphaerellaceae. Teratosphaeria and related genera were initially associated with leaf diseases of Eucalyptus (Myrtaceae) and Proteaceae, but their ecological habitat vary from saprobic, human opportunistic and plant pathogen to extremophilic species (Crous et al. 2007, Ruibal et al. 2009, Teodoro et al. 2012, Egidi et al. 2014, Quaedvlieg et al. 2014). Investigators in Brazil have isolated plant pathogenic fungi (Guedes et al. 2012) and opportunistic melanised representatives (Duarte et al. 2014) from workers of Atta laevigata and gynes of Atta spp., respectively, which were shown here to belong to the Teratosphaeriaceae.
One genus and eight new species of Teratosphaeriaceae were isolated when investigating the diversity of fungi occurring on the integument of pre-nuptial flight gynes and drones. The aim of these investigations was to further explore and better understand the primary microbial community that is dispersed into a young ant colony by these ants.
This is therefore the first study to provide a taxonomic treatment of Teratosphaeriaceae from ants, which has significantly expanded the number of Xenopenidiella species in addition to just the type species, X. rigidophora, thereby allowing for a more robust amended description of the genus. Previous reports on filamentous fungi from ant niches characterised similar isolates as found in the soil community (Pagnocca et al. 2008, Rodrigues et al. 2008), which is different from the results found in this study, where the fungi were isolated directly from the insects themselves. It can be hypothesized that the isolation methods used in the other studies may have contributed to this difference.
The lack of knowledge about ant nest microenvironments encouraged studies with a focus on this microenvironment, but even less is known about the ant integument from which the new species were obtained. One interesting ecological aspect of the Teratosphaeriaceae is that the family harbours isolates from extreme environments. However, the present scientific information is still insufficient to answer several questions about the studied substrate, for instance, if it should be treated as extreme or not. It is known that the pH of fungal gardens ranges from 4.35 to 5 (Powell & Stradling 1986), but nothing has been published about the pH of the integument itself. On the other hand, the presence of cuticular hydrocarbons and alkaloids on the integument was already reported (Roux et al. 2009).
The main lineages in the Teratosphaeriaceae remained obscured until the extensive revision published by Quaedvlieg et al. (2014). In this publication, a previous discussion point regarding the phylogenetic position of Piedraiaceae was once again raised as this fungal family appears to cluster within the Teratosphaeriaceae (Crous et al. 2009a). Other reports studying rock-inhabiting fungi (RIF) showed that these are phylogenetically highly diverse, and highlighted the importance of larger taxon samplings to define questions about generic boundaries (Ruibal et al. 2009, 2011). All these results demonstrate the importance of dedicated projects on fungal isolation from diverse niches.
Although this study still under-represents the total biodiversity on leaf-cutting ants, it does reveal yet another source of Teratosphaeriaceae species. It is known that the morphology of many genera of saprobic and plant pathogenic fungi have evolved several times independently (Crous et al. 2009b), which explains how these penidiella-like isolates could adapt to such an unlikely environment such as gynes and drones of attine ants.
The species phylogeny presented in Fig. 2 is based on four genomic loci (ITS, cmdA, tef1 and tub2 gene regions). For the species of Xenopenidiella, all loci for which data was available could distinguish the included species (data not shown, trees for individual loci were deposited in TreeBASE). However, the amplification success rate of tef1 and cmdA for this genus was not as high as for ITS and tub2. The ex-type culture of X. formica, CBMAI 1941, clusters slightly distant to the rest of the isolates of this species; this is due to the tub2 sequence which is different (93 % identical; 371/401 nucleotides) from the other isolates of the species. The phylogenetic placement of the single isolate of Simplicidiella was supported by all loci and analyses (Fig. 1, 2). Both species of Penidiellopsis are only reliably distinguishable by their tub2 sequences; all other loci either lack resolution to resolve the species or show an intermixing of isolates from the two species. It is quite possible that with a broader sampling these two species might turn out to be synonymous; however, on tub2 they are only 92 % identical (394/428 nucleotides) based on fixed nucleotide differences and therefore we maintain them as separate species for now. A broader sampling from wider geographic regions is being planned to determine whether this difference is significant or not. Since all four loci together did not provide 100 % amplification success rate, it is recommended that an ideal identification protocol includes both ITS and tub2 sequences.
Concluding remarks
Representatives of the Teratosphaeriaceae include species of agricultural importance and wide ecological plasticity with ability to colonise diverse substrates, including ants and extreme environments. Fungal richness among social insects is still underestimated despite the increasing number of new species found in this microenvironment. Over the last 10–15 yr it was concluded that knowledge on fungal diversity contributes greatly to safeguard global genetic resources, which will have a great impact on human lives in future (Hawksworth 2004, Lange et al. 2012). The present study contributes to this knowledge, which is a fundamental tool to plan strategies aimed at the protection of species and environments, especially in tropical and subtropical regions. A significant number of fungi are part of the complex interactions in leaf-cutting ant colonies and further studies will be required to determine their roles in this specific niche.
Acknowledgments
This study was supported by FAPESP (São Paulo Research Foundation; grant 2013/08540-4) and CNPq (National Council for Scientific and Technological Development; grants number: 560.682/2010-7 and 448941/2014-7). We also thank Dr. Nilson S. Nagamoto and Dr. Luiz Carlos Forti for ants identification and José Luiz Novelli, for permission to sample at Santana Farm, Botucatu, São Paulo State, Brazil.
REFERENCES
- Attili-Angelis D, Duarte APM, Pagnocca FC, et al. 2014. Novel Phialophora species from leaf-cutting ants (tribe Attini). Fungal Diversity 65: 65–75. [Google Scholar]
- Augustin JO, Groenewald JZ, Nascimento RJ, et al. 2013. Yet more “weeds” in the garden: Fungal novelties from nests of leaf-cutting ants. PLoS ONE 8: e82265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Carnegie AJ, Pegg GS, White D, et al. 2011. Species within Mycosphaerellaceae and Teratosphaeriaceae from eucalypts in eastern Australia. Australasian Plant Pathology 40: 366–384. [Google Scholar]
- Carreiro SC, Pagnocca FC, Bacci M, Jr, et al. 2004. Sympodiomyces attinorum sp.nov., a yeast species associated with nests of the leaf-cutting ant Atta sexdens. International Journal of Systematic and Evolutionary Microbiology 54: 1891–1894. [DOI] [PubMed] [Google Scholar]
- Chapela IH, Rehner SA, Schultz TR, et al. 1994. Evolutionary history of the symbiosis between fungus growing ants and their fungi. Science 266: 1691–1694. [DOI] [PubMed] [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Groenewald JZ. 2011. Why everlastings don’t last. Persoonia 26: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Summerell BA, et al. 2009a. Co-occurring species of Teratosphaeria on Eucalyptus. Persoonia 22: 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hawksworth DL, Wingfield MJ. 2015. Identifying and naming plant-pathogenic fungi: past, present, and future. Annual Review of Phytopathology 53: 247–267. [DOI] [PubMed] [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, et al. 2009b. Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Wingfield MJ, et al. 2012. Fungal Planet description sheets: 128–153. Persoonia 29: 146–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, et al. 2016. Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie CR, Bot ANM, Boomsma JJ. 2003. Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos 101: 91–102. [Google Scholar]
- Duarte APM, Attili-Angelis D, Baron NC, et al. 2014. Leaf-cutting ants: an unexpected microenvironment holding human opportunistic black fungi. Antonie van Leeuwenhoek 106: 465–473. [DOI] [PubMed] [Google Scholar]
- Egidi E, De Hoog GS, Isola D, et al. 2014. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothidemycetes based on multi-locus phylogenies. Fungal Diversity 65: 127–165. [Google Scholar]
- Groenewald JZ, Nakashima C, Nishikawa J, et al. 2013. Species concepts in Cercospora: spotting the weeds among the roses. Studies in Mycology 75: 115–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes FLA, Attili-Angelis D, Pagnocca FC. 2012. Selective isolation of dematiaceous fungi from the workers of Atta laevigata (Formicidae: Attini). Folia Microbiologica 57: 21–26. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hawksworth DL. 2004. Fungal diversity and its implications for genetic resource collections. Studies in Mycology 50: 9–18. [Google Scholar]
- Hawksworth DL. 2012. Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodiversity and Conservation 21: 2425–2433. [Google Scholar]
- Howard RW, Blomquist GJ. 2005. Ecological, behavioral and biochemical aspects of insect hydrocarbons. Annual Review of Entomology 50: 371–393. [DOI] [PubMed] [Google Scholar]
- Hunter GC, Crous PW, Carnegie AJ, et al. 2011. Mycosphaerella and Teratosphaeria diseases of Eucalyptus; easily confused and with serious consequences. Fungal Diversity 50: 145–166. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange L, Bech L, Busk PK, et al. 2012. The importance of fungi and of mycology for a global development of the bioeconomy. IMA Fungus 3: 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little AEF, Currie CR. 2007. Symbiotic complexity: discovery of a fifth symbiont in the attine-microbe symbiosis. Biology Letters 3: 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulionis VE, Cabello MN, Seifert KA, et al. 2015. Escovopsis trichodermoides sp. nov., isolated from a nest of the lower attine ant Mycocepurus goeldii. Antonie van Leeuwenhoek 107: 731–740. [DOI] [PubMed] [Google Scholar]
- Mayhé-Nunes AJ, Jaffé K. 1998. On the biogeography of Attini (Hymenoptera: Formicidae). Ecotropicos 11: 45–54. [Google Scholar]
- McLaughlin DJ, Hibbett DS, Lutzoni F, et al. 2009. The search for the fungal tree of life. Trends in Microbiology 17: 488–497. [DOI] [PubMed] [Google Scholar]
- Meirelles LA, Montoya QV, Solomon SE, et al. 2015. New light on the systematics of fungi associated with attine ant gardens and the description of Escovopsis kreiselii sp. nov. PLoS ONE 10: e0112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo WGP, Arcuri SL, Rodrigues A, et al. 2014. Starmerella aceti f.a., sp. nov., an ascomycetous yeast species isolated from fungus garden of the leafcutter ant Acromyrmex balzani. International Journal of Systematic and Evolutionary Microbiology 64: 1428–1433. [DOI] [PubMed] [Google Scholar]
- Middelhoven WJ, Fonseca A, Carreiro SC, et al. 2003. Cryptococcus haglerorum, sp. nov., an anamorphic basidiomycetous yeast isolated from nests of the leaf-cutting ant Atta sexdens. Antonie van Leeuwenhoek 83: 167–174. [DOI] [PubMed] [Google Scholar]
- Montoya QV, Meirelles LA, Chaverri P, et al. 2016. Unraveling Trichoderma species in the attine ant environment: description of three new taxa. Antonie van Leeuwenhoek 109: 633–651. [DOI] [PubMed] [Google Scholar]
- Mueller UG, Rabeling C. 2008. A breakthrough innovation in animal evolution. Proceedings of the National Academy of Sciences USA 105: 5287–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Sweden. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, et al. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnocca FC, Legaspe MFC, Rodrigues A, et al. 2010. Yeasts isolated from a fungus-growing ant nest, including the description of Trichosporon chiarellii sp.nov., an anamorphic basidiomycetous yeast. International Journal of Systematic and Evolutionary Microbiology 60: 1454–1459. [DOI] [PubMed] [Google Scholar]
- Pagnocca FC, Rodrigues A, Nagamoto NS, et al. 2008. Yeasts and filamentous fungi carried by the gynes of leaf-cutting ants. Antonie van Leeuwenhoek 94: 517–526. [DOI] [PubMed] [Google Scholar]
- Pérez CA, Wingfield MJ, Altier NA, et al. 2009. Mycosphaerellaceae and Teratosphaeriaceae associated with Eucalyptus leaf diseases and stem cankers in Uruguay. Forest Pathology 39: 349–360. [Google Scholar]
- Powell RJ, Stradling DJ. 1986. Factors influencing the growth of Attamyces bromatificus, a symbiont of attine ants. Transactions of the British Mycological Society 87: 205–213. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg W, Groenewald JZ, De Jesús Yáñez-Morales M, et al. 2012. DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia 29: 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A, Bacci M, Jr, Mueller UG, et al. 2008. Microfungal “weeds” in the leafcutter ant symbiosis. Microbial Ecology 56: 604–614. [DOI] [PubMed] [Google Scholar]
- Rodrigues A, Mueller UG, Ishak HD, et al. 2011. Ecology of microfungal communities in gardens of fungus-growing ants (Hymenoptera: Formicidae): a year-long survey of three species of attine ants in Central Texas. FEMS Microbiology Ecology 78: 244–255. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Roux O, Martin J-M, Ghomsi NT, et al. 2009. A non-lethal water-based removal-reapplication technique for behavioral analysis of cuticular compounds of ants. Journal of Chemical Ecology 35: 904–912. [DOI] [PubMed] [Google Scholar]
- Ruibal C, Gueidan C, Selbmann L, et al. 2009. Phylogeny of rock-inhabiting fungi related to Dothideomycetes. Studies in Mycology 64: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruibal C, Millanes AM, Hawksworth DL. 2011. Molecular phylogenetic studies on the lichenicolous Xanthoriicola physciae reveal Antarctic rock-inhabiting fungi and Piedraia species among closest relatives in the Teratosphaeriaceae. IMA Fungus 2: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samerpitak K, Duarte APM, Attili-Angelis D, et al. 2015. A new species of the oligotrophic genus Ochroconis (Sympoventuriaceae). Mycological Progress 14: 6. [Google Scholar]
- Satow MM, Attili-Angelis D, De Hoog GS, et al. 2008. Selective factors involved in oil flotation isolation of black yeast from the environment. Studies in Mycology 61: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TR, Brady SG. 2008. Major evolutionary transitions in ant agriculture. Proceedings of the National Academy of Sciences USA 105: 5435–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Quaedvlieg W, Javan-Nikhah M, et al. 2012. Zymoseptoria ardabilia and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola). Mycologia 104: 1397–1407. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro MG, Ferreira MA, Guimarães LMS, et al. 2012. Mycosphaerella and Teratosphaeria species associated with leaf diseases on Eucalyptus globulus in southern Brazil. Phytopathologia Mediterranea 51: 355–364. [Google Scholar]
- Thanh VN. 2006. Lipomyces orientalis sp. nov., a yeast species isolated from soil in Vietnam. International Journal of Systematic and Evolutionary Microbiology 56: 2009–2013. [DOI] [PubMed] [Google Scholar]
- Thanh VN, Hien DD, Thom TT. 2013. Moniliella byzovii sp. nov., a chlamydospore-forming black yeast isolated from flowers. International Journal of Systematic and Evolutionary Microbiology 63: 1192–1196. [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber NA. 1972. The fungus-culturing behavior of ants. American Zoologist 12: 577–587. [Google Scholar]
- White TJ, Bruns T, Lee J, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications. Academic Press, San Diego. [Google Scholar]


