Abstract
Rationale/Objective
Cardiomyopathy and arrhythmias are under significant genetic influence. Here, we studied a family with dilated cardiomyopathy and associated conduction system disease in whom prior clinical cardiac gene panel testing was unrevealing.
Methods/Results
Whole genome sequencing and induced pluripotent stem cells were used to examine a family with dilated cardiomyopathy and atrial and ventricular arrhythmias. We also characterized a mouse model with heterozygous and homozygous deletion of Mybphl.
Results
Whole genome sequencing identified a premature stop codon, R255X, in the MYBPHL gene encoding myosin-binding protein-H like (MyBP-HL), a novel member of the myosin binding protein family. MYBPHL was found to have high atrial expression with low ventricular expression. We determined that MyBP-HL protein was myofilament-associated in the atria, and truncated MyBP-HL protein failed to incorporate into the myofilament. Human cell modeling demonstrated reduced expression from the mutant MYBPHL allele. Echocardiography of Mybphl heterozygous and null mouse hearts exhibited a 36% reduction in fractional shortening and an increased diastolic ventricular chamber size. Atria weight normalized to total heart weight was significantly increased in Mybphl heterozygous and null mice. Using a reporter system, we detected robust expression of Mybphl in the atria as well as in discrete puncta throughout the right ventricular wall and septum. Telemetric ECG recordings in Mybphl mice revealed cardiac conduction system abnormalities with aberrant atrioventricular conduction and an increased rate of arrhythmia in heterozygous and null mice.
Conclusions
The findings of reduced ventricular function and conduction system defects in Mybphl mice support that MYBPHL truncations may increase risk for human arrhythmias and cardiomyopathy.
Keywords: Myofilament, myosin binding protein, cardiomyopathy, atria, conduction system, MYBPHL, mutation, mouse models, arrhythmias, genetics, contractile function
INTRODUCTION
Dilated cardiomyopathy (DCM) is often a genetic disease. DCM is also the more genetically diverse of the common cardiomyopathies which include hypertrophic (HCM), left ventricular non-compaction, and arrhythmogenic right ventricle (ARVC) 1. There are currently over 70 genes with mutations linked to DCM 2. These mutations are found in genes that contribute to diverse functions including sarcomere machinery, the nuclear membrane, calcium handling, metabolic processes, ion channels, and cytoskeletal networks. Phenotypically, DCM is characterized by ventricular dilation, and impaired cardiac function that can lead to heart failure.
Genetic testing for cardiomyopathies relies on gene panels, focusing on exons and the immediate intronic regions 3. The sensitivity of clinical gene panel testing is 40–50% in DCM, suggesting the involvement of additional genes or even combinations of genes. The most commonly mutated gene in DCM is TTN, which encodes the giant protein titin that confers elasticity and stability to the sarcomere. Heterozygous TTN truncations are found in approximately 20% of DCM4 and about 1% of the general population. Mutations in MYBPC3, which also encode a major component of the sarcomere, more commonly lead to HCM 5, 6. MYBPC3 encodes cardiac myosin binding protein-C (cMyBP-C), a protein that binds directly to myosin-containing thick filaments. The majority of gene mutations in DCM and HCM are autosomal dominant heterozygous variants.
Cardiac MyBP-C is a cardiac specific protein composed of repeating immunoglobulin (Ig) and fibronectin III (FnIII) domains. Many MYBPC3 mutations are heterozygous truncating genetic variants 6. Individuals with heterozygous MYBPC3 truncating mutations have been described with a range of severity of HCM and variable penetrance7, 8. Truncating MYBPC3 mutations may act through haploinsufficiency and improper actomyosin crossbridge regulation 9, although other mechanisms are may occur 10, 11. Truncating mutations remove the myosin binding, carboxyl terminus of cMyBP-C, resulting in improper myofilament incorporation and degradation of the truncated protein 8.
In this work, we describe the application of whole genome sequencing (WGS) to a family with DCM and associated cardiac conduction system disease. WGS was used to determine genetic variation because WGS applies more even sequencing coverage across exons and, importantly, samples more exons, including those that may not be present on whole exome sequencing arrays. We identified a premature stop codon (stop-gained) in MYBPHL. We found that myosin binding protein-H like (MyBP-HL) is related to other well-studied myosin binding proteins, and that it is a cardiac myofilament protein enriched in the atria and expressed in the ventricular conduction system. Because MYBPHL truncating variants are found at higher than expected population frequency for a rare disorder, we assessed both heterozygous and homozygous loss of Mybphl in the mouse. We found that disruption of Mybphl in the mouse leads to cardiomyopathy and associated arrhythmias, demonstrating the importance of MyBP-HL in normal cardiac function.
METHODS
Study Subjects
Peripheral blood was collected for WGS and confirmation Sanger sequencing. The study was approved by both the University of Chicago and Northwestern University Institutional Review Boards. Genetic counseling was provided.
WGS
Blood was collected from subjects and genomic DNA was extracted. Massively parallel sequencing was performed by Illumina (San Diego, CA) on a HiSeq2000 or X-Ten to generate >30X coverage WGS data. Paired end reads were mapped to hg19 and variants were called using MegaSeq 12, which employs the Burrows-Wheeler aligner (BWA), Genome Analysis Tool Kit (GATK), and Haplotype Caller for variant identification. SnpEff was used to classify variants according to predicted protein alteration into classes that include HIGH, MODERATE, MODIFIER and LOW effect variants 13. MODERATE (missense) variants were scored using Polyphen2 and GERP 14, 15. Left ventricular and atrial appendage expression data from the GTEx database was used to prioritize genes expressed in the heart 16. Variants were further annotated with data from ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), and the MalaCards database using cardiomyopathy specific keywords (http://www.malacards.org/). All variants were annotated with both global and continental ExAC frequencies 17, and sorted based on rare frequency in the global ExAC population (<0.002).
IACUC statement
All experiments were performed in accordance with protocols reviewed and approved by the IACUC at Northwestern University.
Mybphl targeted deletion in mice
A mouse line with a targeted deletion of Mybphl exons 2–6 was created by the NIH Knock-Out Mouse Project (www.komp.org) in association with the Welcome Sanger Trust and obtained from the European Mutant Mouse Archive. The targeted allele contained an LacZ reporter, loxP site, a neo cassette, a second FRT site and a second LoxP site between exons 1 and 2. A third loxP site was located between exon 6 and 7. This allele was bred to the Ella deleter mouse, causing recombination and resulting in the tm1b allele configuration, with exon 1 splicing into the lacZ reporter. These mice were archived on a C57/Bl6(N) background and re-derived from frozen sperm at Northwestern University on a C57/Bl6(J) line by the Transgenic and Targeted Mutagenesis Laboratory at Northwestern.
Echocardiography
Echocardiography was performed using a Vevo2100 echocardiography platform with a 550MHz solid-state probe (FujiFilm, Ontario, Canada). Mice were imaged on a heated table with surface ECG recordings. Isoflurane anesthesia was used at 1% during acquisition; if heart rate fell below 450 beats per minute the session was re-performed. Chamber dimensions were measured by short-axis M-mode echocardiography.
Mouse cardiac magnetic resonance imaging
MRI was performed on a 9.4T Bruker Biospec MRI system with a 30 cm bore, a 12 cm gradient insert, and an Autopac automated sample positioning system (Bruker Biospin Inc, Billerica, MA). Atria were imaged using a prospectively triggered flow compensated cine FLASH sequence (flcFLASH) with TR/TE/α = 10 ms / 2.4 ms / 15°, matrix size 192 x 192, field of view 3 cm x 3 cm, slice thickness 1 mm, and 12 frames per cardiac cycle. Analysis was performed on the coronal images using Segment version 2.0 R5450 (http://segment.heiberg.se) 18.
Antibodies, Immunofluorescence microscopy and Immunoblotting
Cardiomyocytes were isolated from 1.5-day-old neonatal mice using the Pierce Primary Cardiomyocyte Isolation Kit (Thermo 88281). Cells were transfected using Lipofectamine 3000 (Life Technologies L3000008) according to the manufactures specifications using endo-free pCMV6 plasmids encoding mouse Mybphl with and without the R255X variant with amino-terminal Myc-DDK tags. Cells were fixed in 4% PFA for 10 minutes, permeabilized in 0.25% Triton X-100 in PBS for 20 minutes and blocked using 20% FBS, 0.1% Triton X-100 in PBS for 30 minutes. Cells were incubated with Anti-Myc antibody (Millipore 05-724) and Phalloidin 488 (Molecular Probes A12379). Images were taken using a Zeiss Axio Observer epifluorescence microscope with an Apotome-2.
Atria and ventricle tissue was homogenized in urea buffer (50mM tris HCl, pH 7.5, 4M urea, 1M thiourea, 0.4% CHAPS, 20mM spermine, 20mM DTT) using silica beads and a homogenizer (BioSpec) for total protein. Myofilament fractions were isolated by homogenizing tissue in F60 buffer (60mM KCl, 30mM Imidazole, 2mM MgCl2) for total protein, then centrifuging at 12,000 rcf at 4°C to separate the soluble protein fraction in the supernatant. The myofilament fraction was washed by homogenizing three times in F60 with 1% Triton X-100 and 12,000 rcf centrifugations and finally solubilized in urea buffer 19. Antibodies used for immunoblotting were cMyBP-C E-7 (Santa Cruz sc137180, lot K3011), MyBP-HL N-16 (SC sc248025, lot G2412), Annexin 6 (AbCam ab31026).
Quantitative PCR
Gene expression was assessed by qPCR using SybrGreen chemistry measured by a CFX96 thermocycler (BioRad). Primers were designed against exon 8 and the 3′ UTR in exon 9 of Mybphl (forward 5′ TCACATCCACAAGGCAGATATT 3′, reverse 5′ TATAGCCAGGAGTGGAGGTATG 3′, efficiency 104.3%) and primers designed against Mybph (forward 5′ TCACATCCACAAGGCAGATATT3′, reverse 5′ TATAGCCAGGAGTGGAGGTATG 3′, efficiency 103.5%). Expression was normalized to Gapdh (forward 5′ TTGTGATGGGTGTGAACCACGA 3′, reverse 5′ AGCCCTTCCACAATGCCAAAGT3′, efficiency 102.7%). Expression was calculated using an efficiency-corrected ΔΔCq equation.
Detailed methods for mouse cardiac MRI, histology, continuous telemetry and iPSC generation and differentiation are found in the Supplemental Methods.
Statistical Methods
Statistical evaluation was performed using GraphPad Prism7. Significance was set a priori at p < 0.05. Two-way ANOVA was performed with a Bonferroni multiple comparison test for experiments for human RNA-seq data, mouse echocardiography, and mouse heart rates after isoproterenol. One-way ANOVA was performed with Tukey’s multiple comparison test for experiments using mouse RT-PCR, protein quantitation, and atrial weight to body weight. Assumed equality of variance for ANOVA calculations were tested by a Bartlett’s test. An unpaired t-test was performed to compare right atrial volumes by MRI. Statistical tests are indicated for each data set in the legend.
RESULTS
WGS identifies a premature stop in MYBPHL associated with conduction system abnormalities and DCM
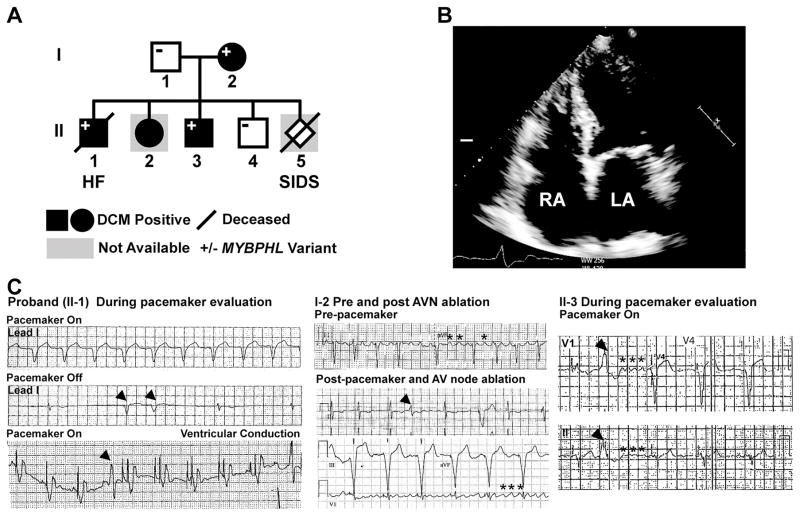
A family with a clinical history of conduction system abnormalities and DCM was evaluated (Figure 1). The proband (II-1) had congenital complete heart block with a pacemaker implanted at 12 years of age, followed by progressive dilated cardiomyopathy (DCM) and heart failure (HF). The proband died in his fourth decade from progressive HF and arrhythmia. Medical records from the year prior to death show severe left ventricular (LV) systolic dysfunction with an LV ejection fraction (LVEF) <25% and impaired conduction system function. Four-chamber echocardiography imaging of his mother (I-2) showed marked atrial dilation (Figure 1B), DCM and reduced systolic function. ECG recordings from the proband showed profound sinus and atrioventricular (AV) node dysfunction in the absence of pacing (Figure 1C, left panels). Premature ventricular contractions were also present. ECG’s from the proband’s mother (I-2) showed atrial flutter, PVCs and junctional beats; she required AV node ablation for management (Figure 1C, middle panels). Siblings II-2 and II-3 also had pacemaker implantation for complete heart block, both at 11 years of age, with DCM developing later (Table 1). ECG recordings from sibling II-3 also show atrial flutter and junctional ventricular arrhythmia (Figure 1C, right panels). The expanded pedigree of the mother (I-2) indicated other family members with DCM and arrhythmias (Figure S1). The clinical findings are summarized in Table 1.
Figure 1. Whole genome sequencing (WGS) identified a premature stop codon in MYBPHL with conduction system abnormalities and DCM.
A. WGS on the proband (II-1) and parents (I-1, I-2) identified the MYBPHL R255X variant which was found in multiple family members. B. Four-chamber echocardiogram of I-2 shows enlarged right and left atria (LA, RA). C. Abnormal heart rhythms in multiple family members. ECG recordings from the proband’s pacemaker test months prior to death showing paced rhythm (top, lead 1). In the middle tracing with the pacemaker off, underlying sinus and atrioventricular node dysfunction was evident, as were PVCs (arrow heads). The bottom tracing highlights a paced rhythm and revealed PVCs (arrow head). The center panels show ECG recordings from the affected parent (I-2) show atrial flutter (asterisks) and PVC in the middle tracing. The bottom tracing shows pacing after atrioventricular (AV) node ablation with persistent atrial flutter (asterisks). The right hand panels show ECG tracing from II-3 with a PVC (arrow head) and coarse atrial fibrillation (asterisks).
Table 1.
Clinical features of conduction system abnormalities and cardiomyopathy.
| FamilyMember | MYBPHLVariant | Cardiac Disease | Anatomical Findings | Events (age) |
|---|---|---|---|---|
| I-1 | No | None | None | |
| I-2 | Yes | DCM, AFSick sinus syndromeBundle branch block | Echocardiography:Dilated atria | Pacemaker (65)AV ablation (69) |
| II-1 | Yes | DCM, SCDComplete heart block (congenital) | On autopsy:Thickened, dilated atriaFatty, fibrotic SA & AV nodes Fibroelastosis in conducting fibers | Pacemaker (12)Died (32) |
| II-2 | Unknown | DCM (29)Complete heart block (congenital) | Pacemaker (11) | |
| II-3 | Yes | DCMComplete heart block (congenital) | Pacemaker (11) | |
| II-4 | No | None | None | |
| II-5 | Unknown | Unknown | SIDS |
AF, Atrial fibrillation; DCM, dilated cardiomyopathy; SCD, sudden cardiac death; SIDS, sudden infant death syndrome
Clinical genetic testing with a broad cardiomyopathy gene panel was unrevealing in this family. To identify the genetic etiology, WGS was performed on the proband (II-1) and parents (I-1, I-2). Variants were sorted by frequency (ExAc <0.002), and variant effects were determined using snpEff focusing on HIGH and MODERATE effect variants. HIGH effect variants disrupt protein length and include stop/start loss/gain, frameshifts, and splice site variants. MODERATE effect variants include missense and in-frame insertion/deletions. No mutations were seen in genes previously linked to DCM, so we evaluated the remainder of the genome for HIGH and MODERATE effect variants focusing on genes highly expressed in the heart. We identified a HIGH effect variant in MYBPHL (1:109838960 G>A) that substitutes a stop codon for the arginine at residue 255 (of 354) (Figure 2A). Sanger sequencing confirmed this presence of MYBPHL R255X in multiple affected family members (Figure 2). We identified MYBPHL R255X in one of 198 subjects with cardiomyopathy examined using WGS. Although the MYBPHL R255X variant is rare in the overall population, it is found at a higher than expected frequency based on population estimates and genetic heterogeneity of DCM (Table S1). Because of the observed population frequency in ExAc, we queried 900 genomes sequenced from the NUGene database, representing individuals who receive care at Northwestern Medicine. We identified MYBPHL R255X variant in one individual (1:1800 alleles). At age 30, this variant carrier had two echocardiograms with an LVIDd that exceeded normal values (3.0 and 2.8 cm/m2, with normal being 2.1–2.7 cm/m2). LVEF was normal on these studies, but the enlarged LV dimension is consistent with early phase DCM. Based on these findings, we investigated both cells and mice with MYBPHL truncations.
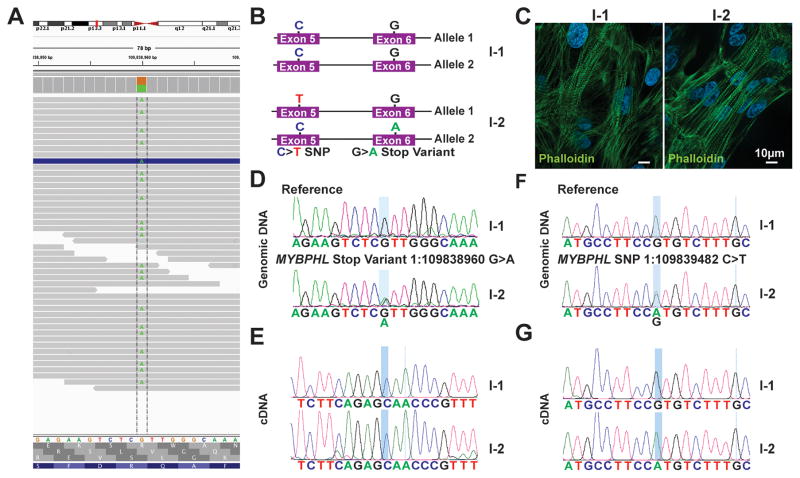
Figure 2. The mutant MYBPHL allele is suppressed by nonsense-mediated decay.
A. Genome viewer shows the alignment of reads at 45x converge over this variant with an even distribution of “A” or “G” reads, consistent with MYBPHL R255X being heterozygous. B. Schematic of both MYBPHL alleles from family members I-1 and I-2. I-2 has the G>A stop variant in exon 6, as well as a synonymous SNP on the opposite allele in exon 5. C. Representative phalloidin stained image of iPSC-derived cardiomyocytes generated from family members I-1 and I-2. D. Sanger sequencing of iPSC-cardiomyocyte genomic DNA confirms the presence of reference sequence in I-1 and the 1:109838960G>A variant in I-2. E. Direct sequencing of RT-PCR-generated cDNA from iPSC-cardiomyocytes from I-1 and I-2 shows expression of only the normal “G” allele (expressed as a “C”) in both I-1 and I-2. This is consistent with no detectable expression from the premature stop allele in I-2. F. Direct sequencing of genomic DNA from iPSC-cardiomyocytes from I-1 and I-2 showed the presence of a benign synonymous C>T SNP in exon 5 of I-2. This variant is found on the opposite allele from R255X. G. Direct sequencing of RT-PCR product generated from iPSC-cardiomyocyte cDNA showed that I-2 only expressed the allele with the C>T SNP, consistent with nonsense mediated decay of the mutant allele.
Since premature stop codons may be subject to nonsense-mediated decay, we assessed allele-specific expression of MYBPHL, taking advantage of polymorphisms that distinguish the mutant and normal alleles (Figure 2B). We generated cardiomyocytes from induced pluripotent stem cells (iPSCs) from family members I-1 (unaffected father) and I-2 (affected mother) (Figure 2C). Direct sequencing of MYBPHL cDNA from individual I-2 identified expression from only the normal G-containing allele (Figure 2D, E), and there was no detectable signal from the mutant MYBPHL allele (Figure 2E, lower panel). We also evaluated a second benign synonymous SNP (1:109839482 C>T) in individual I-2, where the “A” allele is found on the normal MYBPHL allele (Figure 2F). Direct sequencing of cDNA confirmed expression only from the normal allele (Figure 2G) with no detectable expression from the R255X allele. These findings demonstrate that MYBPHL R255X is subject to nonsense-mediated decay. Nonsense-mediated decay has been described as contributing to MYBPC3 truncations 20.
MYBPHL expression is enriched in human and mouse atria
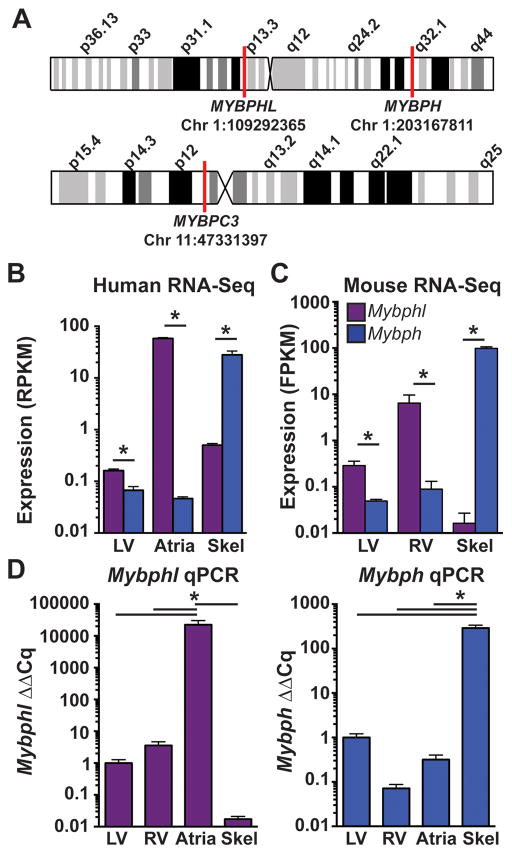
There are two related genes, MYBPHL and MYBPH, with both genes residing on human chromosome 1 (Figure 3), and on mouse chromosomes 3 and 1, respectively. Human RNA-Seq data demonstrated that MYBPHL is enriched in the heart while MYBPH is enriched in skeletal muscle (www.gtexportal.org) 16. Moreover, MYBPHL is much more highly expressed in left atria than in left ventricle (Figure 3B). These data are consistent with MYBPH as the primary H protein in skeletal muscle, and MYBPHL being the H protein in the heart, with higher expression in atria. RNA-Seq data from mouse tissues demonstrated a similar pattern, with significantly higher Mybphl expression in the right and left ventricles compared to Mybph and negligible expression in skeletal muscle. As in humans, mouse Mybph was highly expressed in skeletal muscle (Figure 3C). Expression levels of Mybphl and Mybph were confirmed by qPCR using mouse atria, left and right ventricle, and skeletal muscle. These data confirmed Mybphl expression in the ventricles, with over 10,000-fold enrichment in the atria. Mybph was detected in small amounts in the ventricles, but was increased over 100-fold in skeletal muscle (Figure 3D).
Figure 3. MYBPHL expression is enriched in human and mouse atria and is distinct from MYBPH.
A. Schematic of human chromosomes 1 and 11 show locations of the three genes MYBPHL, MYBPH, and MYBPC3. B. Human RNA-Seq data from the GTEx database of normalized MYBPHL and MYBPH reads in ventricle, atria, and skeletal muscle shows enriched MYBPHL expression in atrial tissue, and enriched MYBPH expression in skeletal muscle (MYBPHL n= 218 LV, 194 atria, 229 skel; MYBPH n= 218 LV, 194 atria, 232 skel). C. RNA-Seq data from mouse tissue shows enrichment of MYBPHL expression in ventricle and enrichment of MYBPH in skeletal muscle (n=6). D. qPCR analysis of wild-type mouse ventricle, atria, and skeletal muscle confirms the cardiac and atrial enrichment of MYBPHL and the enrichment of MYBPH in skeletal muscle (n=4). *= p<0.05 two-way ANOVA with a Bonferroni multiple comparison test (B,C) or one-way ANOVA with a Tukey’s post-hoc test (D).
MYBPHL interacts with the myofilament
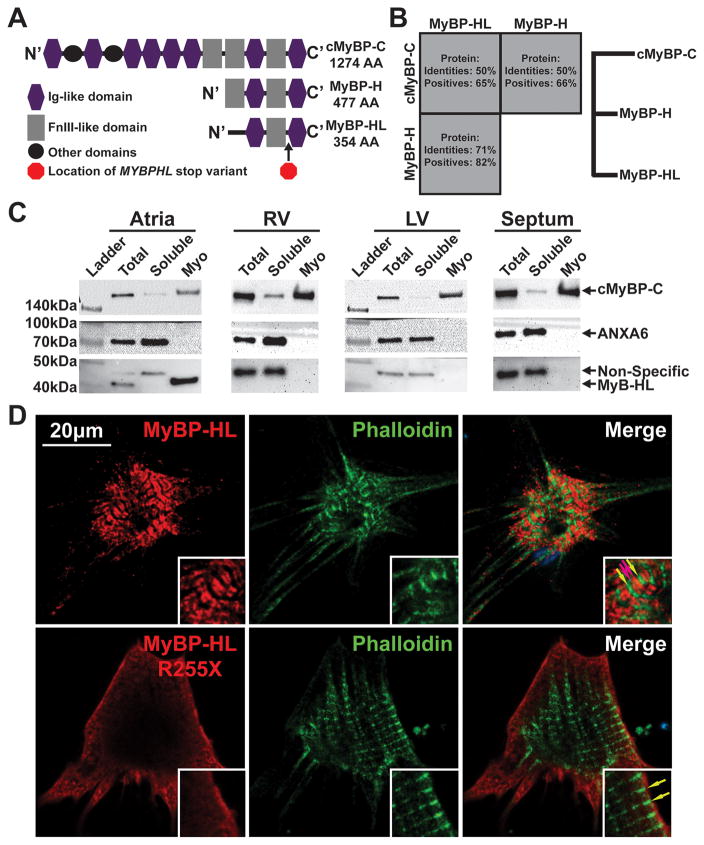
We compared MYBPHL with the better-studied myosin binding proteins. Protein alignment of cMyBP-C, MyBP-H, MyBP-HL shows homology between these three myosin binding proteins, with H and HL having the highest relatedness to each other (82% conserved amino acids) and HL having 65% similarity to cMyBP-C (Figure 4A, B). The three carboxy-terminal domains, seen as Ig-Fn-Ig domains in Figure 4A, are shared among these myosin binding proteins. In studies of cMyBP-C and H protein, these domains are necessary for proper sarcomere incorporation by direct binding to the myosin thick-filament 21–23. To determine whether MyBP-HL interacts with the myofilament, wildtype mouse atria, right and left ventricle, and ventricular septum were assessed for total, myofilament, and soluble protein fractions (Figure 4C). Cardiac MyBP-C was used as a myofilament positive control, as it was observed in the total and myofilament fractions from each chamber as expected 19. The membrane-associated protein annexin A6 was used as a positive control for the soluble fraction 24. MyBP-HL protein was observed in the total and myofilament fractions of atria reflecting its high expression in atrial tissue and its association with myofilaments.
Figure 4. MYBPHL is structurally similar to other myosin binding proteins and interacts with the myofilament.
A. Diagram of the Immunoglobulin (Ig)-like and fibronectin type 3 (FnIII)-like domains that comprise cMyBP-C, MyBP-H, and MyBP-HL; the location of the premature stop in MyBP-HL created by the identified variant is marked between the C’ Ig and FnIII domains. B. Protein sequence conservation and phylogeny between the myosin binding protein family members show that MyBP-H and MyBP-HL are more closely related to each other than to cMyBP-C. C. Immunoblot analysis of total tissue homogenate, soluble protein, and myofilament fractions from atria, right and left ventricle, and ventricular septum detect MyBP-HL in the atrial myofilament, with cMyBP-C and annexin-6 used as myofilament and soluble controls, respectively. D. Immunofluorescence microscopy of neonatal mouse cardiomyocytes transfected with N’-terminally Myc-tagged full-length and R255X MyBP-HL constructs showed myofilament patterned MyBP-HL localization that is not observed with the R255X variant (green and red arrows, insets).
In order to visualize the subcellular localization of MyBP-HL, a Myc-DDK epitope tag was added to the amino terminus, and full-length mouse Mybphl cDNA and truncated (R255X) constructs were introduced into neonatal mouse cardiomyocytes. Full length MyBP-HL was observed to localize to the myofilament, with distinct doublets flanked by phalloidin-stained actin M-lines (Figure 4D, red and green arrows). The truncated MyBP-HL protein showed diffuse staining at the periphery of cells without any discernable incorporation into the myofilament. This indicates that the premature stop codon disrupts normal localization, similar to how truncating MYBPC3 gene mutations disrupt cMyBP-C. These data indicate that MyBP-HL is natively myofilament-associated in the atria and when expressed in neonatal ventricular cardiomyocytes the protein is able to incorporate into the myofilament in a pattern similar to cMyBP-C C-zone doublets (Figure S2) 25, 26.
Mybphl is expressed in atria and in puncta throughout the ventricles
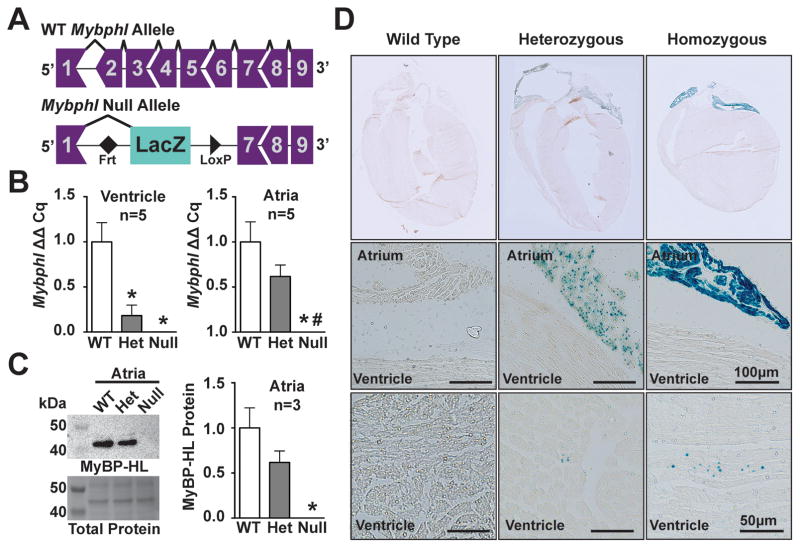
In order to study the physiological role of MyBP-HL, we obtained an Mybphl allele generated by the Knock-Out Mouse Project (www.komp.org) 27–30. This allele removes Mybphl exons 2 through 6, which includes the majority of the protein-coding region and inserts a LacZ reporter 3′ of exon 1 (Figure 5A). Mice were derived onto a C57Bl6/J background; heterozygous and homozygous mice were produced in normal Mendelian ratios, indicating no lethality on this background (Table S2). The Mybphl mRNA transcript was undetectable in homozygous animals, and decreased in heterozygous animals as assessed by qPCR with primers targeting exon 8 and the 3′UTR in exon 9 (Figure 5B). Furthermore, no compensatory changes were observed in Mybph expression by qPCR in the Mybphl heterozygous or null atria and ventricles (Figure S3). MyBP-HL protein was also undetectable in atrial homogenates from homozygous mice by immunoblotting (Figure 5C), confirming the deletion strategy generated a null allele.
Figure 5. Mybphl null mouse.
A. Schematic diagram of the WT Mybphl exons and the Knock-Out Mouse Project (KOMP) gene-trap deletion allele. B. qPCR shows reduction and elimination of Mybphl expression in Het and Null atria and ventricles, respectively (n=4). C. Immnoblot of atrial whole-tissue homogenates confirms reduction and elimination of MyBP-HL protein in Het and Null mice (n=3). D. LacZ staining confirms atrial enrichment of Mybphl expression, with small puncta of LacZ positive cells seen scattered throughout the ventricle. * = p<0.05 vs. WT, # = p<0.05 vs. Het (one-way ANOVA with a Tukey’s multiple comparison test).
The LacZ reporter was used to examine the expression of Mybphl in the mouse heart. Consistent with the RNA-seq data, Mybphl-LacZ was highly expressed in atria. We also identified LacZ positive puncta throughout the ventricle (Figure 5D). The nature of these ventricular puncta was studied using serial 10μm sections spanning an approximately 700μm thick anterior region of myocardium containing right ventricular free wall, septum, and left ventricular free wall (Figure S4). LacZ puncta were seen in Mybphl heterozygous hearts but not in controls. These LacZ puncta were mapped on a representative section, with one dot representing either a single LacZ positive focus, or a cluster of LacZ positive foci, examples of which are illustrated in Figure 5. LacZ puncta were seen accumulated in the right ventricular free wall and the ventricular septum, with fewer puncta on the left ventricular free wall.
Cardiac structural defects were not observed in Mybphl heterozygous and null mice. Assessment with Masson’s Trichrome staining for fibrosis did not reveal any notable differences between genotypes in the ventricle or atria (Figure S5). Sarcomere organization in atrial tissue was visualized by scanning electron microscopy and revealed normal looking sarcomeres in each genotype, with defined M- and Z-lines. (Figure S5, arrows). Taken together these data show that loss of Mybphl is compatible with life and support the atrial enrichment and low-level ventricular expression of Mybphl.
Loss of Mybphl results in ventricular dysfunction and atrial enlargement
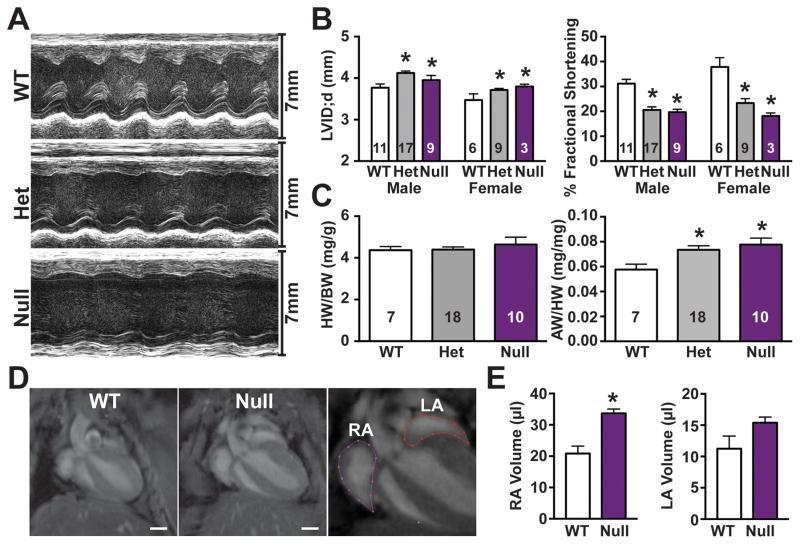
We evaluated the consequences of the loss of Mybphl on cardiac function in 12-week-old mice using echocardiography analysis. M-mode-derived data revealed significantly larger left ventricle internal diameter during peak diastolic filling in both Mybphl heterozygous and null mice (Male: WT = 3.77 ± 0.09mm, Het = 4.13 ± 0.06mm, Null= 3.95 ± 0.11mm; Female: WT = 3.47 ± 0.15mm, Het = 3.71 ± 0.04mm, Null = 3.80 ± 0.06mm) (Figure 6A, B), consistent with moderate dilation of the ventricle. Fractional shortening was reduced in both heterozygous and homozygous mice (Male: WT = 31.1 ± 1.7%, Het = 20.6 ± 1.2%, Null = 19.7 ± 1.18%; Female: WT = 37.8 ± 3.8%, Het = 23.4 ± 1.7%, Null = 18.7 ± 1.2%) (Figure 6A, B).
Figure 6. Loss of Mybphl causes ventricular dysfunction and atrial enlargement.
A. Representative short-axis M-mode echocardiography images. B. M-mode derived fractional shortening and diastolic left ventricular internal diameter shows systolic dysfunction and dilation in heterozygous and null groups, consistent between sexes. C. Heart weight to body weight and atria weight to total heart weight ratios show increased atrial mass in Mybphl heterozygous and null mice of mixed sex. D. MRI of WT and Mybphl null atria at peak systole (left, center) and representative segmentation analysis of atrial volume (right). E. Right atria volume is significantly increased in Mybphl null mice (n=3 female mice). *= p<0.05 vs. WT by two-way ANOVA with a Bonferroni multiple comparison test (B), one-way ANOVA with a Tukey’s multiple comparison test (C), or unpaired t-test (E).
Heart mass to body mass ratios were not significantly different among the groups (Figure 6C). However, the ratio of atrial mass normalized to total heart mass was significantly increased in both heterozygous and homozygous groups (WT = 0.058 ± 0.004, Het = 0.074 ± 0.003, Null = 0.078 ± 0.005) (Figure 6C). We used cardiac MRI on WT and Mybphl null mice to examine atrial morphology in vivo (Figure 6D). The atria were imaged at peak ventricular systole to capture peak atrial filling. Right atrium volume was significantly increased in Mybphl null animals compared to WT (WT= 20.9 ± 0.2μl, Mybphl null = 33.7 ± 1.4μl) (Figure 6E). Therefore, loss of Mybphl in the mouse impaired left ventricular function and caused atrial enlargement.
Mybphl deletion increases susceptibility to arrhythmias
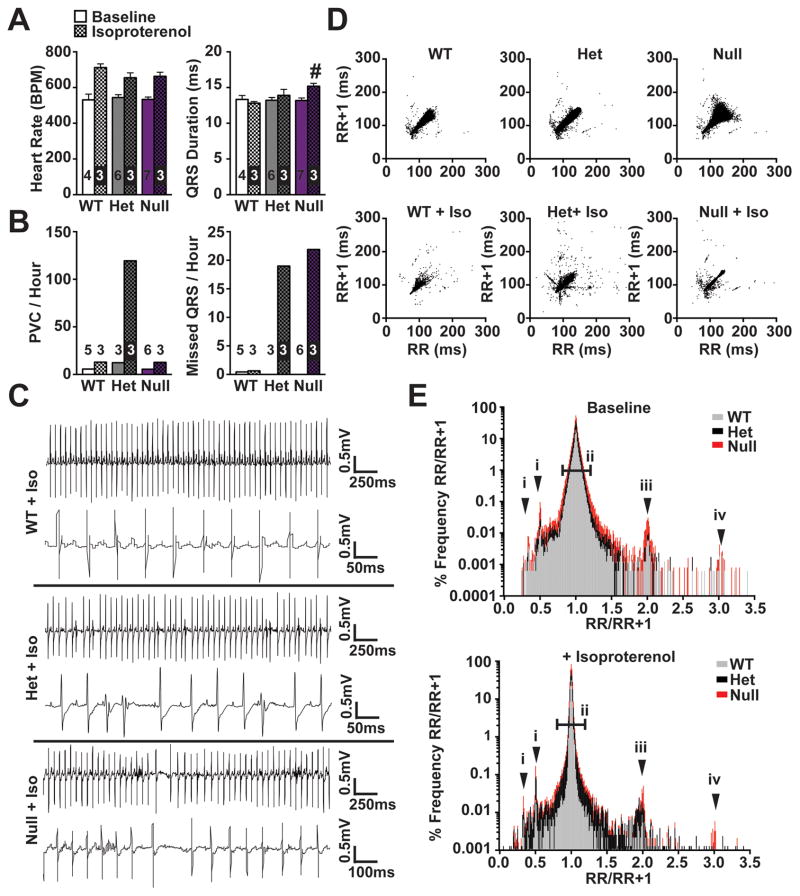
We performed conscious ambulatory ECG recordings to assess the arrhythmogenic risk in Mybphl heterozygous and homozygous mice. Average heart rate and QRS duration did not differ among the groups under normal conditions. However, after administration of isoproterenol, the Mybphl null mice showed a significant increase in QRS duration compared to WT mice treated with isoproterenol (Baseline: WT= 13.3 ± 0.6ms, Het= 13.2 ± 0.4ms, Null= 13.2 ± 0.4ms; isoproterenol: WT= 12.8 ± 0.2ms, Het= 13.9 ± 0.8ms, Null= 15.2 ± 0.4ms) (Figure 7A). Thus, ventricular conduction was significantly slowed in Mybphl null hearts following adrenergic stimulation, consistent with a role for the low level expression of MyBP-HL seen in a punctate pattern in the ventricle.
Figure 7. Increased frequency of arrhythmic events in Mybphl mutant mice.
A. Conscious ambulatory ECG analysis shows significantly prolonged QRS duration in Mybphl null mice following isoproterenol administration. B. Following isoproterenol, PVCs and nonconducted beats per hour were increased in heterozygous mice. Nonconducted beats were increased in both heterozygous and homozygous mice following isoproterenol. C. Representative ECG traces from each group following isoproterenol administration show an interval with a high rate of PVCs from heterozygous mice and a period with non-conducted beats and sinus arrest in the homozygous traces. D. Poincaré plots showing increased R-R variability in Mybphl heterozygous and null mice that is exacerbated with isoproterenol treatment. E. Histogram of the RR/RR+1 ratio illustrating peaks representing PVCs or early beats (i), single and double skipped beats in sinus rhythm (iii, iv), and overall RR variability illustrated by increased deviation around the 1.0 ratio point (ii) (n=3–5). # = p<0.05 vs. WT + Isoproterenol by two-way ANOVA with a Bonferroni multiple comparison test.
To assess if loss or reduction of MyBP-HL increased the likelihood of arrhythmia, ECG traces from these mice were analyzed blinded to genotype. Following isoproterenol injection the total number of PVCs normalized to time was higher in the heterozygous group, while both heterozygous and null mice showed an increased occurrence of nonconducted ventricular beats (Figure 7B, C, Figure S6). In addition to the frequently observed PVCs and missed QRS events, heterozygous and null mice also displayed instances of sinus arrest, gradual inversion of P-waves indicating possible ectopic atrial rhythms, and bigeminy resulting in increased P-P and R-R variability (Figure S6, arrows). None of these specific events were observed in WT recordings at baseline or following isoproterenol administration.
Under baseline conditions and following adrenergic stimulation, heterozygous and homozygous mice exhibited notable bigeminy and irregular rhythms. These irregularities were visualized by use of Poincaré plots, plotting the R-R interval of one beat with the R-R interval of the following beat as (x,y) pairs (Figure 7D) 31. Representative Poincaré plots from individual animals with and without isoproterenol treatment showed increased variability in the heterozygous and null animals (baseline ~60,000 beats per plot, isoproterenol ~30,000 beats per plot) (Figure 7D). Heart rate variability was also visualized by plotting the ratio of one R-R interval and the R-R interval of the following beat on a histogram (Figure 7E), with peaks at 2 and 3 corresponding with one or two non-conducted beats in sinus rhythm (arrows iii and iv), points below 1 corresponding to PVCs (arrow i), and the wider distribution of the heterozygous and null ratios illustrating increased beat-to-beat variability (bar ii). These data indicate that Mybphl regulates conduction in the atria and ventricular conduction systems.
DISCUSSION
A potential role for MYBPHL in DCM and cardiac arrhythmias
In this report, we used WGS to identify a gene associated with cardiac conduction defects and DCM in humans. Mice deleted for Mybphl provide corroborating evidence that MyBP-HL has important roles for cardiac function in the mammalian heart. In the human population, MYBPHL R255X has a population frequency of 0.1% in the ExAC database (Table S1), a database that was assembled including individuals with a number of clinical disorders including cardiac phenotypes. Notably, although rare, 0.1% is higher than expected for a rare disorder. DCM itself is reported as occurring in 1 in 250–500 people (0.2–0.4%) 32. Incomplete penetrance and variable expressivity are hallmarks of genetic cardiomyopathy, and this higher than expected frequency indicates that MYBPHL truncations may be insufficient on their own and instead act in concert with other stressors to express a cardiac phenotype. Additional studies documenting an enrichment of MYBPHL truncations in DCM populations are needed to develop statistical evidence for MYBPHL as a DCM gene.
TTN truncating variants (TTNtvs) are responsible for ~20% of inherited DCM, yet TTNtvs are found in ~1–3% of the general population 4, 33. While it appears that TTNtvs in cardiomyopathy enrich in the A band, the population prevalence of A-band TTNtvs is ~0.19%, considerably higher than the incidence of DCM and consistent with reduced penetrance 34. Despite this reduced penetrance, TTNtvs are a major genetic risk factor for developing DCM. Genetic and environmental effects are expected to influence the expression of pathogenic variants. Both cis-acting and trans-acting genetic modifiers likely contribute to disease manifestation. Thus, in humans, this truncating variant in MYBPHL may increase risk for developing arrhythmias and DCM.
The major myosin binding proteins share conserved myofilament-binding residues
Myosin binding proteins-C and -H were originally described from skeletal muscle extractions 35, 36. The structure of MyBP-C was described as a chain of immunoglobulin-like (Ig-like) and fibronectin type 3 (FnIII) domains that binds to myosin thick filaments and titin via four carboxyl-terminal FnIII-Ig-FnIII-Ig domains 37. These domains are recapitulated in MyBP-H and similarly bind myosin 23. Despite a similar domain structure, electron microscopy of skeletal muscle has shown skeletal MyBP-C and -H to localize differently in the sarcomere, with C-protein occupying 7–9 axial stripes within the A-band of cardiac and skeletal muscle, and H-protein occupying one stripe in the A-band 25.
In addition to cardiac MyBP-C, the fast and slow MyBP-C proteins exhibit the same basic conserved domain structure 38–41. Specifically, all three C-type proteins share the carboxy-terminal myosin binding residues, which bear resemblance to those found in MyBP-H and MyBP-HL. MyBP-HL lacks the extra FnIII domain at its amino-terminus. The highly enriched skeletal and cardiac muscle expression pattern of MyBP-H and MyBP-HL, suggest that these two genes should be viewed as skeletal and cardiac H protein, respectively. This classification is partially similar to the MyBP-C isoforms, except for the skeletal MyBP-C isoforms that have split into two proteins in mammals 41.
MyBP-H was described as being expressed in the cardiac conduction system, specifically in Purkinje cells of the conduction system 42. However, the methodologies may have been confounded by the existence of MyBP-HL. Given the high degree of similarity between these proteins, immunoreagents are likely to detect both proteins. A more recent study identified Mybph binding partners and subcellular localization in cardiac Purkinje cells, finding a protein similar in size to MyBP-HL on immunoblotting using a commercially available antibody that may detect both proteins (Abcam #ab55561) 43. Using a LacZ reporter in the Mybphl locus, we identified focal expression throughout the myocardium in a pattern consistent with Purkinje cell distribution. Moreover, the finding of both atrial and ventricular arrhythmias in Mybphl mutant mice provides a functional correlate. There are many cell types that comprise the conduction system, some with blurred boundaries with non-conduction system ventricular cardiomyocytes. The cardiac conduction system may express both Mybph and Mybphl; however, the low expression of Mybph in the heart suggests that this role may be more likely conferred by Mybphl. However, isoform specific reagents are needed to confirm these findings.
Enriched atrial expression of Mybphl
We detected high expression of Mybphl in atria in both human and mouse with little ventricular expression. Yet, deletion of Mybphl in mice leads to ventricular dysfunction, even in heterozygous mutant mice. The absence of ventricular hypertrophy and myofibrillar disarray in Mybphl null hearts stands in contrast to Mybpc3 null mouse models 21, 44, 45 and is consistent with MYBPHL’s low expression in the ventricle. The data presented here support a model where disruption of MYBPHL in the ventricle may disrupt the conduction system, which could lead to ventricular dysfunction. Alternatively, atrial dysfunction may predispose to ventricular dysfunction, which would be highly novel.
RNA-Seq evaluating gene expression in atrial cardiomyocytes and microdissected sinoatrial node cells showed Mybphl to be equally expressed in both cell types 46. Furthermore, RNA-Seq data from isolated cardiac Purkinje cells and working ventricular myocardium reported Mybphl expression as enriched in Purkinje cells (53.23 ± 26 FPKM in Purkinje cells vs 6.43 ± 3.37 FPKM in ventricular cells) 47. This data, along with the observation of ventricular arrhythmias in Mybphl null mice and ventricular Mybphl-driven LacZ puncta, support the hypothesis that Mybphl is expressed in specific and critical locations in the ventricular conduction system. The mechanism by which the loss of Mybphl elicits ventricular arrhythmias is unknown and could be due to structural and morphological changes leading to perturbations in conduction. Alternatively, MyBP-HL may be able to regulate intracellular signaling and non-muscle myosin, as reported for MyBP-H in non-cardiomyocytes 48.
DCM with conduction system defects
Several cardiomyopathy genes have been linked to a high risk for arrhythmias, notably LMNA, SCN5A, and more recently FLNC, supporting the utility of genetic assessment in evaluating accompanying risk in DCM. MYBPHL may also contribute to the development of arrhythmias in the setting of DCM. The presence of cardiac conduction system disease, especially when it occurs before the development of DCM, warrants a distinct clinical management strategy.
Supplementary Material
Clinical Perspective.
What is new?
We identified a low frequency premature stop codon in the MYBPHL gene.
MYBPHL encodes myosin binding protein H-like (MyBP-HL) and shares homology with other myosin binding proteins.
MYBPHL is expressed highly in atria and at low level in the ventricle, and is found in a punctate ventricular pattern suggestive of the conduction system.
MyBP-HL protein interacts with the cardiac myofilament.
We characterized mice with a heterozygous or homozygous deletion in Mybphl and found dilated cardiomyopathy and cardiac conduction system disease, recapitulating the clinical phenotype.
What are the clinical implications?
Heterozygous truncating mutations in the TTN and MYBPC3 genes cause inherited forms of cardiomyopathy with variable expressivity and penetrance.
TTN and MYBPC3 encode the myofilament proteins titin and myosin binding protein C, respectively.
We now identified a novel myofilament component, myosin binding protein H-like (MyBP-HL), and described mice with disruption in the Mybphl gene as having cardiomyopathy and arrhythmias.
MYBPHL’s unusual expression pattern, atria and punctate in the ventricle, suggests a novel mechanism for ventricular dysfunction.
Acknowledgments
ACKNOWLEDGEMENTS AND SOURCES OF FUNDING
Magnetic resonance imaging was performed at the Northwestern University Center for Advanced Molecular Imaging. We acknowledge the support from the Center for Advanced Microscopy and Mr. Lennell Reynolds, Jr. and Dr. Joshua Rappoport at Northwestern University. We acknowledge the technical support from the Transgenic and Targeted Mutagenesis Laboratory in the Center for Genetic Medicine at Northwestern University. We thank Maureen Smith and Jennifer Pacheco from NUGene.
SOURCES OF FUNDING
This work was supported by NIH HL128075 (EMM), NIH HG008673, NIH F32 HL131304 (DYB), and NIH T32 HD009007 (EYK). The Northwestern University Center for Advanced Molecular Imaging is supported by NIH National Cancer Institute P30 CA060553.
Nonstandard Abbreviations
- ARVC
arrhythmogenic right ventricular cardiomyopathy
- AV
atrioventricular
- DCM
dilated cardiomyopathy
- ECG
electrocardiogram
- ExAC
Exome aggregation consortium
- FKPM
fragments per kilobase of exon per million fragments mapped
- FNIII
fibronectin III domain
- FS
fractional shortening
- HCM
hypertrophic cardiomyopathy
- HF
heart failure
- Ig
immunoglobulin domain
- LV
left ventricle
- LVEF
left ventricle ejection fraction
- PVC
premature ventricular contraction
- RPKM
reads per kilobase of transcript per million mapped reads
- TTNtv
TTN truncating variant
- WGS
whole genome sequencing
- WT
wildtype
Footnotes
AUTHOR CONTRIBUTIONS
DYB designed experiments, conducted experiments, analyzed data, and drafted the manuscript; MRP conceived of experiments, conducted experiments, analyzed data, and edited the manuscript; EYK, EAW and LP conducted experiments and analyzed data, LDW, LDC and AHV analyzed data; MH assisted with experiments; EMM conceived of the study, designed experiments, analyzed data, and edited the manuscript.
AUTHOR CONFLICTS OF INTEREST/DISCLOSURES: None
References
- 1.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB American Heart A, Council on Clinical Cardiology HF, Transplantation C, Quality of C, Outcomes R, Functional G, Translational Biology Interdisciplinary Working G, Council on E Prevention. Contemporary Definitions and Classification of the Cardiomyopathies: An American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 2.McNally EM, Barefield DY, Puckelwartz MJ. The Genetic Landscape of Cardiomyopathy and Its Role in Heart Failure. Cell Metab. 2015;21:174–182. doi: 10.1016/j.cmet.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, Chung WK, Jones C, Psychogios A, Duffy E, Rehm HL, White E, Seidman JG, Seidman CE, Ho CY. Genetic Testing for Dilated Cardiomyopathy in Clinical Practice. J Card Fail. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of Titin Causing Dilated Cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE. Mutations in the Cardiac Myosin Binding Protein-C Gene on Chromosome 11 Cause Familial Hypertrophic Cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 6.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, Weissenbach J, Vosberg HP, Fiszman M, Komajda M, Schwartz K. Cardiac Myosin Binding Protein-C Gene Splice Acceptor Site Mutation Is Associated with Familial Hypertrophic Cardiomyopathy. Nat Genet. 1995;11:438–440. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 7.Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A Common Mybpc3 (Cardiac Myosin Binding Protein C) Variant Associated with Cardiomyopathies in South Asia. Nat Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J. Cardiac Myosin-Binding Protein C Mutations and Hypertrophic Cardiomyopathy: Haploinsufficiency, Deranged Phosphorylation, and Cardiomyocyte Dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 9.Barefield D, Kumar M, Gorham J, Seidman JG, Seidman CE, de Tombe PP, Sadayappan S. Haploinsufficiency of Mybpc3 Exacerbates the Development of Hypertrophic Cardiomyopathy in Heterozygous Mice. J Mol Cell Cardiol. 2015;79:234–243. doi: 10.1016/j.yjmcc.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahrudin U, Morisaki H, Morisaki T, Ninomiya H, Higaki K, Nanba E, Igawa O, Takashima S, Mizuta E, Miake J, Yamamoto Y, Shirayoshi Y, Kitakaze M, Carrier L, Hisatome I. Ubiquitin-Proteasome System Impairment Caused by a Missense Cardiac Myosin-Binding Protein C Mutation and Associated with Cardiac Dysfunction in Hypertrophic Cardiomyopathy. J Mol Biol. 2008;384:896–907. doi: 10.1016/j.jmb.2008.09.070. [DOI] [PubMed] [Google Scholar]
- 11.Helms AS, Davis FM, Coleman D, Bartolone SN, Glazier AA, Pagani F, Yob JM, Sadayappan S, Pedersen E, Lyons R, Westfall MV, Jones R, Russell MW, Day SM. Sarcomere Mutation-Specific Expression Patterns in Human Hypertrophic Cardiomyopathy. Circ Cardiovasc Genet. 2014;7:434–443. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puckelwartz MJ, Pesce LL, Nelakuditi V, Dellefave-Castillo L, Golbus JR, Day SM, Cappola TP, Dorn GW, 2nd, Foster IT, McNally EM. Supercomputing for the Parallelization of Whole Genome Analysis. Bioinformatics. 2014;30:1508–1513. doi: 10.1093/bioinformatics/btu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, Snpeff: Snps in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A Method and Server for Predicting Damaging Missense Mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. Distribution and Intensity of Constraint in Mammalian Genomic Sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium GT. The Genotype-Tissue Expression (Gtex) Project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG Exome Aggregation C. Analysis of Protein-Coding Genetic Variation in 60,706 Humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heiberg E, Sjogren J, Ugander M, Carlsson M, Engblom H, Arheden H. Design and Validation of Segment--Freely Available Software for Cardiovascular Image Analysis. BMC Med Imaging. 2010;10:1. doi: 10.1186/1471-2342-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac Myosin-Binding Protein-C Phosphorylation and Cardiac Function. Circ Res. 2005;97:1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Pointu H, Mougenot N, Guiard J, Reimer R, Hohenberg H, Schwartz K, Vernet M, Eschenhagen T, Carrier L. Nonsense-Mediated Mrna Decay and Ubiquitin-Proteasome System Regulate Cardiac Myosin-Binding Protein C Mutant Levels in Cardiomyopathic Mice. Circ Res. 2009;105:239–248. doi: 10.1161/circresaha.109.201251. [DOI] [PubMed] [Google Scholar]
- 21.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated Cardiomyopathy in Homozygous Myosin-Binding Protein-C Mutant Mice. J Clin Invest. 1999;104:1235–1244. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuster DW, Govindan S, Springer TI, Martin JL, Finley NL, Sadayappan S. A Hypertrophic Cardiomyopathy-Associated Mybpc3 Mutation Common in Populations of South Asian Descent Causes Contractile Dysfunction. J Biol Chem. 2015;290:5855–5867. doi: 10.1074/jbc.M114.607911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welikson RE, Fischman DA. The C-Terminal Igi Domains of Myosin-Binding Proteins C and H (Mybp-C and Mybp-H) Are Both Necessary and Sufficient for the Intracellular Crosslinking of Sarcomeric Myosin in Transfected Non-Muscle Cells. J Cell Sci. 2002;115:3517–3526. doi: 10.1242/jcs.115.17.3517. [DOI] [PubMed] [Google Scholar]
- 24.Demonbreun AR, Quattrocelli M, Barefield DY, Allen MV, Swanson KE, McNally EM. An Actin-Dependent Annexin Complex Mediates Plasma Membrane Repair in Muscle. J Cell Biol. 2016;213:705–718. doi: 10.1083/jcb.201512022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett P, Craig R, Starr R, Offer G. The Ultrastructural Location of C-Protein, X-Protein and H-Protein in Rabbit Muscle. J Muscle Res Cell Motil. 1986;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 26.Pepe FA, Drucker B. The Myosin Filament. Iii. C-Protein. J Mol Biol. 1975;99:609–617. doi: 10.1016/s0022-2836(75)80175-6. [DOI] [PubMed] [Google Scholar]
- 27.White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC, Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP Sanger Institute Mouse Genetics P. Genome-Wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A Conditional Knockout Resource for the Genome-Wide Study of Mouse Gene Function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Burger A, Bult CJ, Bushell W, Collins FS, Desaintes C, Doe B, Economides A, Eppig JT, Finnell RH, Fletcher C, Fray M, Frendewey D, Friedel RH, Grosveld FG, Hansen J, Herault Y, Hicks G, Horlein A, Houghton R, Hrabe de Angelis M, Huylebroeck D, Iyer V, de Jong PJ, Kadin JA, Kaloff C, Kennedy K, Koutsourakis M, Lloyd KC, Marschall S, Mason J, McKerlie C, McLeod MP, von Melchner H, Moore M, Mujica AO, Nagy A, Nefedov M, Nutter LM, Pavlovic G, Peterson JL, Pollock J, Ramirez-Solis R, Rancourt DE, Raspa M, Remacle JE, Ringwald M, Rosen B, Rosenthal N, Rossant J, Ruiz Noppinger P, Ryder E, Schick JZ, Schnutgen F, Schofield P, Seisenberger C, Selloum M, Simpson EM, Skarnes WC, Smedley D, Stanford WL, Stewart AF, Stone K, Swan K, Tadepally H, Teboul L, Tocchini-Valentini GP, Valenzuela D, West AP, Yamamura K, Yoshinaga Y, Wurst W. The Mammalian Gene Function Resource: The International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57bl/6n Embryonic Stem Cells for Mouse Genetic Resources. Nat Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huikuri HV, Seppanen T, Koistinen MJ, Airaksinen J, Ikaheimo MJ, Castellanos A, Myerburg RJ. Abnormalities in Beat-to-Beat Dynamics of Heart Rate before the Spontaneous Onset of Life-Threatening Ventricular Tachyarrhythmias in Patients with Prior Myocardial Infarction. Circulation. 1996;93:1836–1844. doi: 10.1161/01.cir.93.10.1836. [DOI] [PubMed] [Google Scholar]
- 32.Hershberger RE, Hedges DJ, Morales A. Dilated Cardiomyopathy: The Complexity of a Diverse Genetic Architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 33.Golbus JR, Puckelwartz MJ, Fahrenbach JP, Dellefave-Castillo LM, Wolfgeher D, McNally EM. Population-Based Variation in Cardiomyopathy Genes. Circ Cardiovasc Genet. 2012;5:391–399. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akinrinade O, Koskenvuo JW, Alastalo TP. Prevalence of Titin Truncating Variants in General Population. PLoS One. 2015;10:e0145284. doi: 10.1371/journal.pone.0145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Offer G, Moos C, Starr R. A New Protein of the Thick Filaments of Vertebrate Skeletal Myofibrils. Extractions, Purification and Characterization. J Mol Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 36.Starr R, Offer G. H-Protein and X-Protein. Two New Components of the Thick Filaments of Vertebrate Skeletal Muscle. J Mol Biol. 1983;170:675–698. doi: 10.1016/s0022-2836(83)80127-2. [DOI] [PubMed] [Google Scholar]
- 37.Einheber S, Fischman DA. Isolation and Characterization of a Cdna Clone Encoding Avian Skeletal Muscle C-Protein: An Intracellular Member of the Immunoglobulin Superfamily. Proc Natl Acad Sci U S A. 1990;87:2157–2161. doi: 10.1073/pnas.87.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Previs MJ, Michalek AJ, Warshaw DM. Molecular Modulation of Actomyosin Function by Cardiac Myosin-Binding Protein C. Pflugers Arch. 2014;466:439–444. doi: 10.1007/s00424-013-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barefield D, Sadayappan S. Phosphorylation and Function of Cardiac Myosin Binding Protein-C in Health and Disease. J Mol Cell Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ackermann MA, Patel PD, Valenti J, Takagi Y, Homsher E, Sellers JR, Kontrogianni-Konstantopoulos A. Loss of Actomyosin Regulation in Distal Arthrogryposis Myopathy Due to Mutant Myosin Binding Protein-C Slow. FASEB J. 2013;27:3217–3228. doi: 10.1096/fj.13-228882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaffer JF, Gillis TE. Evolution of the Regulatory Control of Vertebrate Striated Muscle: The Roles of Troponin I and Myosin Binding Protein-C. Physiol Genomics. 2010;42:406–419. doi: 10.1152/physiolgenomics.00055.2010. [DOI] [PubMed] [Google Scholar]
- 42.Alyonycheva T, Cohen-Gould L, Siewert C, Fischman DA, Mikawa T. Skeletal Muscle-Specific Myosin Binding Protein-H Is Expressed in Purkinje Fibers of the Cardiac Conduction System. Circ Res. 1997;80:665–672. doi: 10.1161/01.res.80.5.665. [DOI] [PubMed] [Google Scholar]
- 43.Mouton J, Loos B, Moolman-Smook JC, Kinnear CJ. Ascribing Novel Functions to the Sarcomeric Protein, Myosin Binding Protein H (Mybph) in Cardiac Sarcomere Contraction. Exp Cell Res. 2015;331:338–351. doi: 10.1016/j.yexcr.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Hypertrophic Cardiomyopathy in Cardiac Myosin Binding Protein-C Knockout Mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 45.Carrier L, Knoll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J, Jr, Schwartz K, Chien KR. Asymmetric Septal Hypertrophy in Heterozygous Cmybp-C Null Mice. Cardiovasc Res. 2004;63:293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Vedantham V, Galang G, Evangelista M, Deo RC, Srivastava D. Rna Sequencing of Mouse Sinoatrial Node Reveals an Upstream Regulatory Role for Islet-1 in Cardiac Pacemaker Cells. Circ Res. 2015;116:797–803. doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim EE, Shekhar A, Lu J, Lin X, Liu FY, Zhang J, Delmar M, Fishman GI. Pcp4 Regulates Purkinje Cell Excitability and Cardiac Rhythmicity. J Clin Invest. 2014;124:5027–5036. doi: 10.1172/JCI77495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosono Y, Usukura J, Yamaguchi T, Yanagisawa K, Suzuki M, Takahashi T. Mybph Inhibits Nm Iia Assembly Via Direct Interaction with Nmhc Iia and Reduces Cell Motility. Biochem Biophys Res Commun. 2012;428:173–178. doi: 10.1016/j.bbrc.2012.10.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.