Abstract
Protein array technology not only identifies a large number of proteins but also determines their expression levels. In the present study, antibody array analysis is used to decipher the proteins involved in hesperidin-induced cell death in HepG2 cells. Altered proteins in hesperidin treated cells were compared with that of untreated control cells by using a RayBio® Label-based (L series) human antibody array kit. The identified proteins were further confirmed using western blot analysis. STRING software based analysis was used to determine the protein-protein interactions. Many proteins related to signal transduction, cellular mechanisms, cell growth and proliferation regulatory proteins were identified. Among the proteins identified Hsp90, Smac/DIABLO, Prdx6 and FRK were significantly reduced in hesperidin treated cells. To the best of the authors' knowledge, the present study is the first to use antibody array for identifying proteins marker in hesperidin-induced cell death in HepG2 cells. The present study provides a novel insight into the anticancer mechanism of hesperidin.
Keywords: HepG2, hesperidin, antibody array, STRING
Introduction
Hesperidin is a flavone glycoside that is abundantly present in citrus fruits. Many have reported the anticancer activity of hesperidin on malignant cancer cells such as breast and prostate cancer (1) and colon cancer (2). It also inhibits tumor invasiveness of hepatocellular carcinoma (3,4). Recently, the authors reported that hesperidin induced paraptosis in human hepatoblastoma HepG2 cells (5). Paraptosis is a type of caspase independent programmed cell death that is characterized by extensive cytoplasmic vaculation with swelling of mitochondria or endoplasmic reticulum without cell membrane blebbing or nuclear fragmentation (6).
Proteomic profiling has become a promising technology and been widely used for clinical biomarker identification, pathogenesis investigation, new drug discovery, pharmacological research and toxicological examination (7). Antibody array enables parallel detection of multiple proteins with low sample volume. It is of two types: A label based assay and a sandwich assay. In a label-based assay, the targeted proteins are labeled with a tag and are then applied onto an antibody chip. The bound proteins can be visualized using a laser scanner. It is a competitive assay where the analytes in the test and reference solutions compete for binding with the antibodies leading to improvement in linearity of response and dynamic range compared with non-competitive assays, but the disadvantage of label-based assay is limited to sensitivity and specificity (8,9). However, there is no study of proteomic studies of hesperidin induced cell death in human hepatoblastoma HepG2 cells. The purpose of the study is to detect protein markers in HepG2 cells treated with or without hesperidin using RayBio® Label-based (L series) human antibody array.
The antibody array analysis identified many proteins that are signal transduction proteins, structural proteins, mitochondrial proteins and proteins related to cellular metabolism. The STRING database was used to identify the protein-protein interactions of the proteins identified by antibody array. As most of the biological functions are transmitted via proteins, identification of proteins in hesperidin induced cell death will provide a better understanding on the anticancer mechanism of hesperidin.
Materials and methods
Chemical and reagents
Dulbecco's modified Eagle's medium (DMEM) was purchased from HyClone; GE Healthcare Life Sciences (Chalfont, UK). Antibiotics (streptomycin/penicillin) and fetal bovine serum (FBS) were obtained from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Goat anti-rabbit IgG (no. ADI-SAB-300, 1:2,000) and horseradish peroxidase-conjugated goat anti-mouse IgG (no. ADI-SAB-100-J, 1:2,000) were both purchased from Enzo Life Sciences, Inc. (Farmingdale, NY, USA). The RayBio® Label-based (L series) human L-507 antibody array 1000 membrane kit was purchased from RayBiotech (Norcoss, GA, USA). Anti-Prdx6 (ab59543, 1:1,000) and Anti-Frk (ab64914, 1:1,000) were purchased from Abcam (Cambridge, UK). Anti-Smac (no. 15108S, 1:1,000) and Anti-Hsp90 (no. 4877S, 1:1,000) were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA).
Cell culture and treatment
HepG2 cells were obtained from Korean Cell Line Bank (Seoul, Korea) and were maintained in DMEM supplemented with 10% heat inactivated FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2 incubator. Cells were treated with vehicle alone (1% DMSO) or 1 mM hesperidin dissolved in 1% DMSO.
Antibody array
An antibody array was carried out according to the manufacturer's instructions. Overnight grown cells were treated/untreated with 1 mM hesperidin. Following a 24 h incubation, cells were trypsinized and dialyzed at 4°C. The dialyzed samples were then biotin-labeled using 1X labeling reagent provided in the assay kit. The membranes are then blocked in blocking buffer at room temperature with gentle shaking for 1 h. After aspirating the blocking buffer, 8 ml of the diluted sample is placed on each membrane and incubated at RT for 2 h with gentle shaking. The membranes are then washed with 1X washing buffer for 3 times. After washing, 1X horseradish peroxidase-conjugated streptavidin was added to each membrane and incubated at room temperature for 2 h with gentle shaking. After washing, the membranes were placed in the detection buffer for 2 min and then the signals were detected using a ChemiDoc imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). By comparing the signal intensities quantified by densitometry, relative expression levels of target proteins were made.
Protein-protein interaction
Protein-protein interactions were analyzed using Search Tool for the Retrieval of Interacting Genes (STRING database, version 10; http://string.embl.de).
Western blot analysis
Overnight grown HepG2 cells incubated with or without 1 mM hesperidin for 24 h at 37°C were lysed overnight with radioimmunoprecipitation assay lysis buffer containing protease inhibitor cocktail and EDTA. The extracts were then centrifuged at 14,500 × g for 30 min at 4°C to remove debris. The protein concentration in the supernatants was determined using a Bradford protein assay kit (Bio-Rad Laboratories, Inc.), according to the manufacturer's instructions. After being boiled with loading buffer, 50 µg protein samples were separated by 12% SDS-PAGE and then transferred onto polyvinylidene difluoride membrane, which was blocked with 5% non-fat milk for 1 h. Then, membranes were incubated with primary antibodies at 4°C overnight. After washing five times, the membranes were incubated with respective horseradish peroxidase-conjugated secondary antibodies at 37°C for 3 h. Western blot analyses were developed with enhanced chemiluminescence kit (GE Healthcare Life Sciences). Normalization was ensured by β-actin and each band was quantified using ImageJ software version 1.50i (National Institutes of Health, Bethesda, MD, USA).
Antibody array analysis
Independent replicate antibody array experiments were performed and the RayBio® Human L-507 array analysis tool software, provided with the array, was used to analyze the signal intensities of the identified proteins. The data were normalized after background subtraction with the positive control densities. The positive control of the first sample is considered as 1 and the signal intensities of other samples are calculated according to the formula: Normalized signal intensity of particular spot = (signal intensity of particular spot) × (positive signal intensity on reference array/positive signal intensity on sample array).
Statistical analysis
Data are expressed as the mean ± standard deviation of a minimum of three replicates independent experiments. A Student's t-test was performed using SPSS version 10.0 for Windows (SPSS Inc., Chicago, IL, USA). P<0.05 was determined to indicate a statistically significant difference.
Results
Protein identification by antibody array
Antibody array analysis of HepG2 cells treated with or without hesperidin was carried out in order to identify the proteins involved in hesperidin induced cell death. As described in our previous study, paraptotic cell death was induced in HepG2 cells by treating with 1 mM hesperidin for 24 h (5). Thus, for all the studies HepG2 cells treated with 1 mM hesperidin was compared with that of untreated control cells. Total cell extract of HepG2 cells treated with or without 1 mM hesperidin were subjected to human antibody array analysis [RayBio® Label-based (L series) RayBiotech]. RayBio® Label-based array contains duplicate spots of 1,000 proteins. The Fig. 1 demonstrates the protein spot and signal intensities of the proteins determined by antibody array. Antibody array identified many proteins that were signal transduction proteins, structural proteins, mitochondrial proteins and proteins related to cellular metabolism. The identified proteins are listed in Table I. Of the 19 identified proteins, the expression of seven proteins were upregulated while the remaining 12 proteins were downregulated in hesperidin-treated HepG2 cells. Among these proteins, the intensities of S100A4, S100A6, calbidin, ROS, Hsp90, FRK, Lyn, Csk and Prdx6 were significant.
Figure 1.
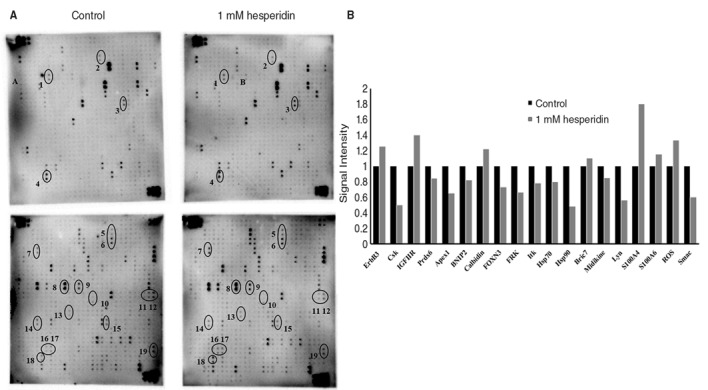
Antibody array analysis of HepG2 cells. (A) Altered protein spots detected during paraptosis. HepG2 cells were treated with or without 1 mM hesperidin and subjected to RayBio® Label-based array. Protein spots: 1, ErbB3; 2, Csk; 3, IGFIIR; 4, Prdx6; 5, calbindin; 6, Apex1; 7, BNIP2; 8, FOXN3; 9, FRK; 10, livin; 11, Itk; 12, Hsp70; 13, Hsp90; 14, Midkine; 15, S100A4; 16, S100A6; 17, ROS; and 18, Smac. (B) Signal intensities of the identified proteins. ERbB3, Receptor tyrosine-protein kinase erbB-3; IGFIIR, insulin-like growth factor type II receptor; Prdx6, peroxiredoxin 6; Apex1, DNA-(apurinic or apyrimidinic site) lyase; BNIP2, BCL2/adenovirus E1B 19 kDa protein-interacting protein 2; FOXN3, forkhead box protein N3; FRK, Fyn related tyrosine kinase; Itk, tyrosine-protein kinase; HSP, heat shock protein; ROS, proto-oncogene receptor tyrosine kinase; Smac, second mitochondria derived activator of caspases.
Table I.
Proteins identified by antibody array analysis. HepG2 cells were treated with/without 1 mM hesperidin and the extracted proteins were subjected to RayBio® Label-based antibody array.
| Uniprot ID | Symbol | Protein name | P-value | Up/down regulation | Biological function |
|---|---|---|---|---|---|
| P26447 | S100A4 | S100A4 | 0.002 | ↑↑ | Calcium binding protein, promotes cell migration |
| P05937 | – | Calbindin | 0.006 | ↑↑ | Calcium binding protein |
| P08922 | ROS | Proto-oncogene receptor tyrosine kinase | 0.01 | ↑↑ | Cell growth, proliferation regulator, activates PI3k/Akt pathway, MAPK cascade |
| P07900 | Hsp90 | Heat sock protein 90 | 0.03 | ↓↓ | Negative inhibitor of apoptosis, activates MAPK cascade |
| P06703 | S100A6 | S100A6 | 0.03 | ↑↑ | Calcium binding protein |
| P41240 | Csk | Tyrosine-protein kinase Csk | 0.04 | ↓↓ | Cell growth, differentiation, cell migration regulation |
| P42685 | FRK | Fyn related tyrosine kinase | 0.05 | ↓↓ | Suppress growth and function during G1 and S phase of cell cycle |
| P07948 | Lyn | Tyrosine-protein kinase Lyn | 0.05 | ↓↓ | Regulates activation of MAP kinase signaling cascade cascade, cell proliferation, survival, differentiation, migration, adhesion, degranulation, and cytokine release |
| P30041 | Prdx6 | Peroxiredoxin 6 | 0.05 | ↓↓ | Cellular protection, cell survival |
| O00409 | FOXN3 | Forkhead box protein N3 | 0.08 | ↓↓ | Transcriptional repressor, cell cycle arrest at G1 and G2 phase |
| Q9GK35 | IGF-IIR | Insulin-like growth factor type II receptor | 0.08 | ↑↑ | Suppresses tumor growth |
| P21860 | ErbB3 | Receptor tyrosine-protein kinase erbB-3 | 0.1 | ↑↑ | Activates MAPK and PI3/Akt pathway |
| Q08881 | Itk | Tyrosine-protein kinase | 0.1 | ↓↓ | Itk phosphorylation leads to Ca2+ release from ER to the cytoplasm |
| Q96CA5 | BRIC7 | Livin | 0.3 | ↑↑ | Inhibits apoptosis protein family member |
| P27695 | Apex1 | DNA-(apurinic or apyrimidinic site) lyase | 0.5 | ↓↓ | Activator of transcriptional repression |
| P0DMV8 | Hsp70 | Heat sock protein 70 | 0.5 | ↓↓ | Inhibits apoptosis, |
| Q12982 | BNIP2 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2 | 0.6 | ↓↓ | Suppresses cell death |
| Q9NR28 | Smac | Second mitochondria derived activator of caspases | 0.6 | ↓↓ | Promotes apoptosis |
| P21741 | – | Midkine | 0.9 | ↓↓ | Cell proliferation regulator, activates MAPK pathway |
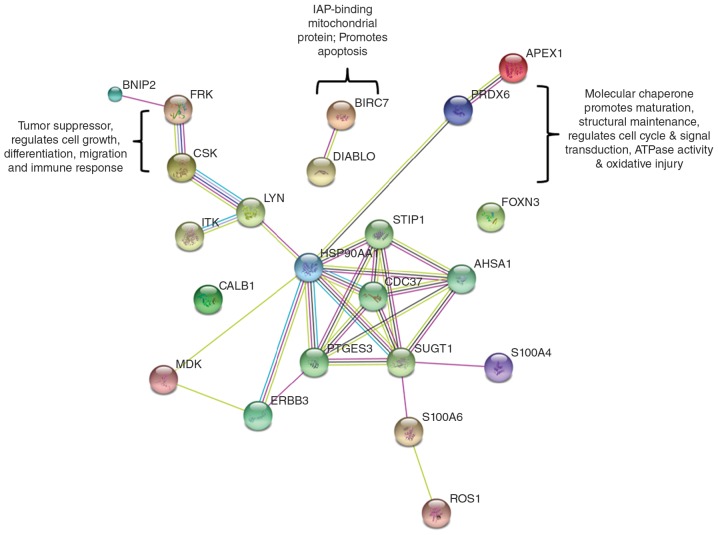
Protein-protein interaction
The STRING database was used to analyze the protein-protein interactions. The Fig. 2 indicates the protein-protein interaction generated by STRING with a medium confidence of 0.400. Prdx6 and FRK are directly or indirectly linked to Hsp90 and the combined score between Prdx6 and Hsp90 is 0.454, and that of FRK and Csk is 0.864. Another node of Diablo and Bric7 with a combined score of 0.993 was also observed. Table II shows the predicted functional partners and their respective functions. As presented in Fig. 2, Hsp90 is the primary functional interacting node in the STRING interaction.
Figure 2.
Protein-protein interaction of the identified proteins. STRING database, version 10 (http://string.embl.de) was used to determine the protein-protein interactions of the 18 proteins identified by antibody array analysis.
Table II.
List of predicted functional partners obtained from STRING database.
| Protein | Function |
|---|---|
| SUGT1 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae). May play role in ubiquitination and subsequent proteosomal degradation of target proteins |
| STIP1 | Stress-induced-phosphoprotein 1. Mediates the association of the molecular chaperones Hsc70 and Hsp90 |
| PTGES3 | Prostaglandin E synthase 3 (cytosolic). Molecular chaperone that localizes to genomic response elements in a hormone-dependent manner and disrupts receptor-mediated transcriptional activation, by promoting disassembly of transcriptional regulatory complexes |
| CDC37 | Cell division cycle 37 homolog. Co-chaperone that binds to numerous kinases and promotes their interaction with the Hsp90 |
| AHSA1 | AHA1, activator of heat shock 90 kDa protein ATPase homolog 1 |
Effect of hesperidin on protein expression
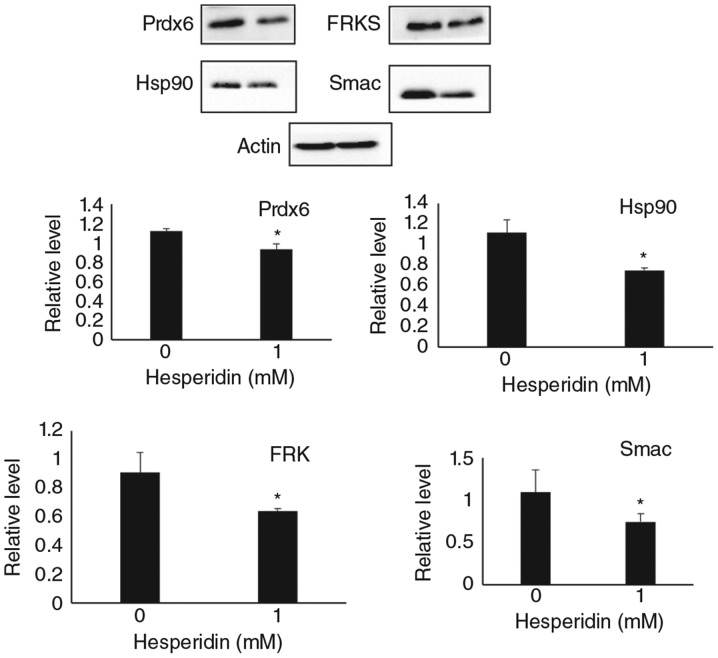
To validate the antibody array analysis results, western blot analysis was conducted using the same cell lysate that were used for antibody array analysis. Immunoblots of Hsp90, Prdx6, FRK and Smac were carried out. Hesperidin significantly reduced the protein expression of Hsp90, Prdx6, FRK and Smac (Fig. 3), which is consistent with the antibody array results.
Figure 3.
Expression of proteins altered during paraptosis. HepG2 cells were untreated or treated with hesperidin for 24 h and whole cell lysate prepared from these cells were subjected to western blot analysis. Protein level of Prdx6, Hsp90, FRK and Smac/DIABLO are shown relative to the value for untreated control cells. Data represent the mean ± standard deviation of three replicates independent experiments. *P<0.05 vs. control. Prdx6, peroxiredoxin 6; HSP, heat shock protein; FRK, Fyn related tyrosine kinase.
Discussion
In the present study, using antibody array techniques identified the proteins involved in hesperidin-induced cell death. Antibody array analysis has the advantaged of measuring multiple proteins in a parallel manner. A total of 19 proteins were identified and STRING analysis of these proteins indicated Hsp90 to be an important node within the entire network. Hsp90 is usually overexpressed in cancer cells and is considered as a potential target in cancer therapeutics. It is a chaperone protein required for the stability of various client proteins and many of these proteins are involved in cell signaling, proliferation and survival (10,11). Thus inhibition of Hsp90 has been considered as a promising way for cancer treatment. In the present study, the protein expression of Hsp90 was significantly reduced in hesperidin treated cells when compared with the untreated control cells. Many Hsp90 inhibitors like geldanamycin, 17-allylamino-17-demethoxygeldanamycin (17-AAG) and 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG) are currently being studied as anticancer agents (12,13). There are also studies that Hsp90 inhibition leads to ERK/MAPK activation in human cancer cells (14,15). In a previous study of the authors, it was reported that hesperidin-induced paraptosis occurs in HepG2 cells by activation of the ERK1/2 pathway (5). In addition, it was also observed that Hsp90 is an important node in the protein-protein interaction network. Thus, inhibition of Hsp90 may be an important upstream cascade in hesperidin-induced cell death in HepG2 cells.
Interestingly, protein expression of Smac/DIABLO (second mitochondria-derived activator of caspases/direct inhibitor of apoptosis-binding protein with low pI) was significantly reduced in hesperidin-treated HepG2 cells. It is a pro-apoptotic mitochondrial protein and negatively regulates the function of inhibitors of apoptosis proteins (IAPs). It promotes apoptosis by activating caspases (16). It was also observed that livin (BRIC7), a member of the IAP family, was upregulated in hesperidin-treated cells (Table I; Fig. 1). Previously, it was reported that livin promotes degradation of Smac/DIABLO through the proteasome ubiquitination pathway (17). As livin protein expression was upregulated with Smac/DIABLO inhibition by hesperidin treatment, the downstream caspase cascade may also be inhibited. As paraptosis is known to be a caspase independent cell death, downregulation of Smac/DIABLO supports the authors' previous results that hesperidin induced cell death in HepG2 cells is caspase independent and non-apoptotic (5).
In our previous study, it was observed that oxidative stress induced by hesperidin leads to paraptotic cell death in HepG2 cells (18). In the present study, it was observed that Prdx6, which is a member of the peroxiredoxins (Prdx) peroxidase family, protein expression was significantly reduced by hesperidin. As an anti-oxidant, Prdx serves a role in controlling intracellular reactive oxygen species expression. Thus, reduced protein expression of Prdx6 by hesperidin may cause the increase in oxidative stress, which further contributes to HepG2 cell death. Previously, it was reported that downregulation of Prdx6 expression has been reported to increase peroxide-induced cell death in liver cancer cells (19) and thiacremonone, an antioxidant which is generated in garlic after high pressure treatment inhibits lung cancer cell growth through inhibition of Prdx6 expression (20). Thus, Prdx6 is an important marker in hesperidin-induced cell death.
Among the proteins identified during antibody array analysis, FRK (Fyn-related tyrosine kinase) expression was also downregulated. FRK belongs to Src non-receptor tyrosine kinase family (21) and inhibition of Src family kinases is considered to be a potential target for cancer therapeutics. Src family kinases are usually overexpressed in a number of cancer cells-lung, colorectal, pancreatic and breast cancer cells (22). FRK is different from other Src family member and is predominantly expressed in epithelial tissues such as liver, kidney and lung (23). Chen et al (24) reported that FRK is overexpressed in liver cancer cells and knockdown of FRK reduces growth in HepG2 cells and cell proliferation in Hep3B cells. Thus, inhibition of FRK is a promising target in liver cancer. In the present study, antibody array analysis also identified many proteins regulating MAPK cascade, Ca2+ binding proteins, cell growth and proliferation regulatory proteins.
To the best of the authors' knowledge, the present study is the first study on using antibody array for protein profiling of paraptosis cell death induced by hesperidin. In the present study, many target proteins were identified and have also demonstrated their interactions with each other. From the protein-protein interaction generated by STRING Hsp90 was identified to be an important node in hesperidin-induced cell death. As a chaperon protein, Hsp90 is required for the stability of various client proteins and many of which are involved in cell signaling, proliferation and survival. As a result, inhibiting Hsp90 may play an important role in hesperidin-induced cell death. As in the authors' previous study, it has been determined that hesperidin induced cell death is paraptosis (5) and these proteins may be involved crucially in inducing cell death. However, further study on these proteins is required to determine their role in paraptosis. Overall, the present study provides a novel insight into the anticancer mechanism of hesperidin.
Acknowledgements
The present study was supported by a grant from the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (grant nos. 2012M3A9B8019303 and 2017R1A2B4003974).
References
- 1.Lee CJ, Wilson L, Jordan MA, Nguyen V, Tang J, Smiyun G. Hesperidin suppressed proliferations of both human breast cancer and androgen-dependent prostate cancer cells. Phytother Res. 2010;24(Suppl 1):S15–S19. doi: 10.1002/ptr.2856. [DOI] [PubMed] [Google Scholar]
- 2.Park HJ, Kim MJ, Ha E, Chung JH. Apoptotic effect of hesperidin through caspase3 activation in human colon cancer cells, SNU-C4. Phytomedicine. 2008;15:147–151. doi: 10.1016/j.phymed.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 3.Lee KH, Yeh MH, Kao ST, Hung CM, Liu CJ, Huang YY, Yeh CC. The inhibitory effect of hesperidin on tumor cell invasiveness occurs via suppression of activator protein 1 and nuclear factor-kappaB in human hepatocellular carcinoma cells. Toxicol Lett. 2010;194:42–49. doi: 10.1016/j.toxlet.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Yeh MH, Kao ST, Hung CM, Liu CJ, Lee KH, Yeh CC. Hesperidin inhibited acetaldehyde-induced matrix metalloproteinase-9 gene expression in human hepatocellular carcinoma cells. Toxicol Lett. 2009;184:204–210. doi: 10.1016/j.toxlet.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Yumnam S, Park HS, Kim MK, Nagappan A, Hong GE, Lee HJ, Lee WS, Kim EH, Cho JH, Shin SC, Kim GS. Hesperidin induces paraptosis like cell death in hepatoblatoma, HepG2 cells: Involvement of ERK1/2 MAPK [corrected] PLoS One. 2014;9:e101321. doi: 10.1371/journal.pone.0101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sperandio S, de Belle I, Bredesen DE. An alternative, nonapoptotic form of programmed cell death. Proc Natl Acad Sci USA. 2000;97:14376–14381. doi: 10.1073/pnas.97.26.14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siu KW, DeSouza LV, Scorilas A, Romaschin AD, Honey RJ, Stewart R, Pace K, Youssef Y, Chow TF, Yousef GM. Differential protein expressions in renal cell carcinoma: New biomarker discovery by mass spectrometry. J Proteome Res. 2009;8:3797–3807. doi: 10.1021/pr800389e. [DOI] [PubMed] [Google Scholar]
- 8.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang R, Jiang W, Yang J, Mao YQ, Zhang Y, Yang W, Yang D, Burkholder B, Huang RF, Huang RP. A biotin label-based antibody array for high-content profiling of protein expression. Cancer Genomics Proteomics. 2010;7:129–141. [PubMed] [Google Scholar]
- 10.Goetz MP, Toft DO, Ames MM, Erlichman C. The Hsp90 chaperone complex as a novel target for cancer therapy. Ann Oncol. 2003;14:1169–1176. doi: 10.1093/annonc/mdg316. [DOI] [PubMed] [Google Scholar]
- 11.Richter K, Buchner J. Hsp90: Chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 12.Chen TL, Gupta N, Lehman A, Ruppert AS, Yu L, Oakes CC, Claus R, Plass C, Maddocks KJ, Andritsos L, et al. Hsp90 inhibition increases SOCS3 transcript and regulates migration and cell death in chronic lymphocytic leukemia. Oncotarget. 2016;7:28684–28696. doi: 10.18632/oncotarget.8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, D'Andrea G, Dickler M, Moynahan ME, Sugarman S, et al. HSP90 inhibition is effective in breast cancer: A phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin Cancer Res. 2011;17:5132–5139. doi: 10.1158/1078-0432.CCR-11-0072. [DOI] [PubMed] [Google Scholar]
- 14.Donzé O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga F, Xu W, Karpova TS, McNally JG, Baron R, Neckers L. Hsp90 inhibition transiently activates src kinase and promotes src-dependent akt and erk activation. Proc Natl Acad Sci USA. 2006;103:11318–13322. doi: 10.1073/pnas.0604705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Ruiz G, Maldonado V, Ceballos-Cancino G, Grajeda JP, Melendez-Zajgla J. Role of smac/DIABLO in cancer progression. J Exp Clin Cancer Res. 2008;27:48. doi: 10.1186/1756-9966-27-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Huang Y, Song Z, Feng S, Tian X, Du W, Qiu X, Heese K, Wu M. Livin promotes smac/DIABLO degradation by ubiquitin-proteasome pathway. Cell Death Differ. 2006;13:2079–2088. doi: 10.1038/sj.cdd.4401959. [DOI] [PubMed] [Google Scholar]
- 18.Yumnam S, Hong GE, Raha S, Saralamma VV, Lee HJ, Lee WS, Kim EH, Kim GS. Mitochondrial dysfunction and Ca(2+) overload contributes to hesperidin induced paraptosis in hepatoblastoma cells, HepG2. J Cell Physiol. 2016;231:1261–1268. doi: 10.1002/jcp.25222. [DOI] [PubMed] [Google Scholar]
- 19.Walsh B, Pearl A, Suchy S, Tartaglio J, Visco K, Phelan SA. Overexpression of Prdx6 and resistance to peroxide-induced death in Hepa1-6 cells: Prdx suppression increases apoptosis. Redox Rep. 2009;14:275–284. doi: 10.1179/135100009X12525712409652. [DOI] [PubMed] [Google Scholar]
- 20.Jo M, Yun HM, Park KR, Park MH, Lee DH, Cho SH, Yoo HS, Lee YM, Jeong HS, Kim Y, et al. Anti-cancer effect of thiacremonone through down regulation of peroxiredoxin 6. PLoS One. 2014;9:e91508. doi: 10.1371/journal.pone.0091508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Wang Z, Luoh SM, Wood WI, Scadden DT. Cloning of FRK, a novel human intracellular SRC-like tyrosine kinase-encoding gene. Gene. 1994;138:247–251. doi: 10.1016/0378-1119(94)90817-6. [DOI] [PubMed] [Google Scholar]
- 22.Wheeler DL, Iida M, Dunn EF. The role of src in solid tumors. Oncologist. 2009;14:667–678. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irby RB, Yeatman TJ. Role of src expression and activation in human cancer. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- 24.Chen JS, Hung WS, Chan HH, Tsai SJ, Sun HS. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics. 2013;29:420–427. doi: 10.1093/bioinformatics/bts715. [DOI] [PubMed] [Google Scholar]