Abstract
PURPOSE
To develop and test a deep learning based computer-aided diagnosis (CAD) scheme of mammograms for classifying between malignant and benign masses.
METHODS
An image dataset involving 560 regions of interest (ROIs) extracted from digital mammograms was used. After down-sampling each ROI from 512×512 to 64×64 pixel size, we applied an 8 layer deep learning network that involves 3 pairs of convolution-max-pooling layers for automatic feature extraction and a multiple layer perceptron (MLP) classifier for feature categorization to process ROIs. The 3 pairs of convolution layers contain 20, 10, and 5 feature maps, respectively. Each convolution layer is connected with a max-pooling layer to improve the feature robustness. The output of the sixth layer is fully connected with a MLP classifier, which is composed of one hidden layer and one logistic regression layer. The network then generates a classification score to predict the likelihood of ROI depicting a malignant mass. A four-fold cross validation method was applied to train and test this deep learning network.
RESULTS
The results revealed that this CAD scheme yields an area under the receiver operation characteristic curve (AUC) of 0.696±0.044, 0.802±0.037, 0.836±0.036, and 0.822±0.035 for fold 1 to 4 testing datasets, respectively. The overall AUC of the entire dataset is 0.790±0.019.
CONCLUSIONS
This study demonstrates the feasibility of applying a deep learning based CAD scheme to classify between malignant and benign breast masses without a lesion segmentation, image feature computation and selection process.
Keywords: Computer aided diagnosis (CAD), Deep learning, Breast mass classification, Convolution neuron networks
1. INTRODUCTION
Breast cancer is the most prevalent cancer occurred in women, which accounts for 29% of all cancer cases and 13% of all cancer related deaths in women in USA [1]. In order to reduce breast cancer mortality rates, mammography screening is currently recommended and implemented as the only clinically acceptable imaging modality for the population-based breast cancer screening. However, breast lesions are very heterogeneous and the suspicious breast lesions are often overlapped with dense fibro-glandular tissue on the mammograms. It is quite difficult for radiologists to accurately distinguish between malignant and benign lesions [2], As a result, current mammography screening generates high false positive recall rates, which may potentially harm many cancer-free women routinely participating in the mammography screening [3]. Therefore, improving accuracy of classifying between malignant and benign lesions is an important and also difficult issue in current clinical practice.
For this purpose, in the last two decades, developing and testing computer-aided detection and diagnosis (CAD) schemes of mammograms have attracted wide research interest and efforts [4]. A typical CAD scheme processes each mammogram in three steps [5]. First, CAD searches for and segments the suspicious lesions from the image. Second, CAD computes a number of image features to extract characteristics of the segmented tumors. Third, CAD applies a machine learning based classifier based on a set of optimally selected features to predict the likelihood of the targeted lesion being associated with malignancy (“positive”). For the conventional CAD scheme, it is the critically important to accurately segment the lesion and identify the discriminatory image features, which directly determine the performance of the machine leaning classifier in the third step. However, accurate segmentation of breast masses from the mammograms is a major technical challenge in developing CAD schemes [6]. In addition, selecting effective image features heavily depends on the preference and/or prior experience of the CAD developers, as there are no commonly accepted methods or criteria for the feature selection. As a result, the existing CAD schemes generate high false-positive rates. It is controversial on whether using commercial CAD schemes can help radiologists improve cancer detection accuracy [7] or what the optimal approach is to use CAD cues in the clinical practice [8]. Despite the disappointment of using existing CAD schemes of mammograms in the clinical practice, researchers still believe that it is necessary to continuously explore and develop new CAD approaches [9], such as to cue more subtle lesions without increasing false-positive detections [10].
However, developing a computer-aided diagnosis scheme to classify between malignant and benign lesions is very different from detection of suspicious lesions. It has much higher requirement in lesion segmentation accuracy and identification of effective features [11]. Currently, no computer-aided diagnosis schemes are used in the clinical practice to help radiologists distinguish between malignant and benign lesions. In searching for new CAD concept and approaches, we noticed an emerging machine learning method of deep learning proposed in 2006 by Hinton et al [12], which has attracted extensive research effort and tested in many engineering application fields requiring image and pattern recognition, detection, parsing, registration, and estimation [13]. Due to the unique characteristics and advantages, deep learning method has also recently been tested and reported in medical image analysis field. The researchers have tested the feasibility of applying the deep learning networks to segment or recognize, isointense stage brain tissues [14], pancreas organ [15], and neuronal membranes [16] from MR images, CT images, and electron microscopy images, respectively. In addition, researchers have also utilized the deep convolution neural networks (CNN) to detect the chest pathology [17] or classify between the mild cognitive impairment (MCI) and Alzheimer’s disease (AD) [18].
Compared to the traditional classifiers, deep learning based classifiers have significantly different network architecture and learning approach. A deep learning network contains more image processing layers (i.e. 5–20 layers [19]) than the conventional image feature based machine learning classifiers. Each layer is a typical neural network such as convolution neural network (CNN) [20]. Instead of using a set of manually or automatically selected image features computed from the images, the deep learning network utilizes image itself as a single input. The effective image features are automatically learned and extracted by the lower layer networks. Accordingly, the higher layer networks use the extracted feature patterns and classify the images into different target categories. Previous studies have shown that using deep learning based classifiers might help reduce the gap between human vision and computer vision in pattern recognition [21] and also enabled to achieve a substantially higher classification performance [22] than the conventional classifiers.
Recently, researchers have also tested the feasibility of applying deep learning methods to CAD of mammograms, which include classification of mammographic density [23], prediction of ort-term breast cancer risk [24] and discrimination of different types of micro-calcifications [25]. In this study, we applied the deep learning method to develop and test a new CAD scheme for classifying between malignant and benign mass based breast lesions. The objective of this study is to investigate and demonstrate the feasibility of whether applying a deep leaning based CAD scheme can yield higher or comparable lesion classification performance without a lesion segmentation as well as image feature computation and selection. The details of this study including the scheme development and scheme testing results are presented in the following sections.
2. MATERIALS AND METHODS
2. 1 Image Dataset
In this study, the training and testing dataset was randomly collected from our previously established full-field digital mammography (FFDM) image database. The detailed description of the image database has been reported in a number of our previous studies [6, 26]. In brief, all FFDM images were acquired using Hologic Selenia FFDM machines and downloaded directly from the clinical PACS system after an image de-identification process. From our image database we created a dataset that includes 560 regions of interest (ROIs) for this study. The original size of all ROIs is 512×512 pixels (i.e. 38.4×38.4mm). Each ROI contains a verified breast soft-tissue mass based lesion. The center of each ROI overlaps with the center of each covered mass. No mass in this dataset has a maximum diameter greater than 38.4mm. All masses in the selected ROIs were biopsied. Based on the pathological reports of the biopsied examination, 280 masses were verified as benign and the others 280 were confirmed as malignant. Table 1 shows the distribution of mammographic density of each case based on the BIRADS rating and the boundary margin characteristic of each mass, which were provided by the radiologists in the original image reading and interpretation
Table 1.
Characteristic distribution of our testing dataset
| Breast density | 1 | 2 | 3 | 4 |
| 22 | 223 | 302 | 13 | |
| Breast margin | 1 | 2 | 3 | 4 |
| 85 | 235 | 138 | 102 |
Note: 1) BIRADS breast density score: 1-almost all fatty replaced, 2-scattered fibroglandular densities, 3-heterogeneously dense, 4-extremely dense; 2) BIRADS breast margin score: 1-smoothed, 2-irregular, 3-spiculated, 4-focal asymmetry.
2. 2 A Deep Learning based CAD Scheme
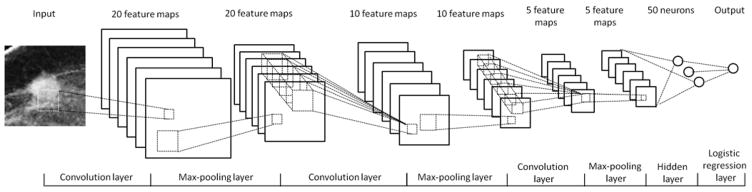
In order to classify between malignant and benign lesions, we designed a deep learning based network. As demonstrated in Figure 1, the network is composed of three pairs of convolution-max-pooling layers, one hidden layer, and one logistic regression layer. In this network, the first 3 pairs of the convolution-max-pooling layers are applied to automatically extract the features. The seventh hidden layer and the eighth logistic regression layer are used as a multiple layer perceptron (MLP) classifier to assign each testing case into malignant or benign case category. In order to effectively learn and extract the image features, we designed a convolution neural network (CNN), which is currently considered as the most effective neural network structure and has been widely used as the basic unit of the deep learning networks [20].
Figure 1.
Architecture of the deep learning networks used for the mass classification
Since a CNN applies different convolution kernels on input image, each kernel can be considered as one “feature”. In the deep learning networks, a number of CNNs are stacked to optimize the features [27]. In general, increasing the number of the CNN layers can increase the discriminatory power and may help improve the optimization effect. However, including more CNN layers requires a large volume training dataset and high computing power, which will significantly increase the training complexity [19, 28]. According to the previous studies, three convolution layers can achieve a satisfied performance in many different applications, and these architectures can also be accomplished under the single workstation environment [14]. Thus, a three convolution layer structure was selected and used to build a deep learning classifier for this new classification task.
From the previous CAD studies reported in the literature, we found that although a large number of image features [29, 30] could be computed, many of them are redundant and only a small set of features was finally selected to build a classifier [31]. The small number of features can typically improve robustness of the classifier with a limited training and testing dataset. Therefore, based on the limited dataset size, we designed a deep learning network that includes 3 layers to sequentially generate 20, 10, and 5 feature maps, respectively.
Using the 3 CNN layers, our deep learning network can automatically learn and define 5 optimal image features from the initial input of the training ROIs. Each layer applies a number of convolution kernels on the input images. The convolution kernel size of the feature map of these three layers are 9×9, 5×5, and 5×5, respectively, which were determined by the previous publication [14] and our own experimental experience. Given that the malignant (benign) breast lesions should be invariant in size (stretching) and rotation, a max-pooling layer is followed by each convolution layer to improve the network robustness. The pooling size is determined to be 2×2, which is most recommended in the previous studies [14]. The results generated by the 3 CNN layers are used as the input feature for the multiple layer perceptron based classifier in the seventh and eighth layers of the deep learning network.
Accordingly, the entire architecture of the deep learning network is as follows: In the first pair of convolution-max-pooling layers, the first convolution layer is connected with the input image. In order to reduce the computing complexity and accelerate the training progress, we down sampled each ROI from 512×512 to 64×64 pixels using a traditional averaging method. Thus, there are a total of 4096 (64×64) inputs in the first convolution layer. In this layer, 20 convolution kernels generate 20 feature maps, which are computed as follows [20, 32]:
| (1) |
In the above formula, is the pixel value at position (i, j) of the kth output feature map, and is the value on the pixel (i, j) of the lth input image. In the first layer, we have only one input image, thus l = 1. is the weight value at coordinate (u, v) of the kth convolution kernel, which applies on the lth input image. bl is the bias applied on the lth input image, and the convolution operation is defined as [20, 32]:
| (2) |
Considering the margin effect of the 9×9 convolution kernel, the size of each output feature map shrinks from 64×64 to 56×56. The 20 output feature maps are taken as the input of the second 2×2 max-pooling layer. In the second max-pooling layer, 20×56×56 inputs are divided into 20×28×28 units, each of which contains a 2×2 neighbor pixel pool. For each pool, the layer detects the maximal pixel value within the 2×2 neighborhood as the output. Thus, the second layer produces 20×28×28 outputs. Similarly, the third convolution layer applies 10 feature maps using a 5×5 filter stack on the 20×28×28 input values to generate 10×24×24 outputs, which are down sampled into 10×12×12 outputs by the fourth 2×2 max-pooling layer. The fifth and sixth layers in the third pair have 5 feature maps also using a 5×5 filter bank in the convolution layer and 2×2 down sampling in the sixth max-pooling layer, which finally produces 5×4×4 output values. These 5×4×4 output values are fully connected with the seventh hidden layer that contains 50 neurons. The seventh and eighth layers consist a MLP classifier, which is mathematically represented as [32, 33]:
| (3) |
Where matrices W(1), W(2), and vectors b(1), b(2) are the weight matrices and bias vectors for the seventh and eighth layer, respectively. In the formula, sigmoid is the logistic sigmoid function: sigmoid(x) = 1/(1 + exp(−x)). Finally, the output value of the network represents a classification score ranging from 0 to 1. The larger score value indicates the high probability of the testing ROI depicting a malignant mass. The detailed parameters used in our deep learning network are summarized and presented in Table 2.
Table 2.
Details of the structures of the each layer in the deep learning network
| 1st layer | 2nd layer | 3rd layer | 4th layer | 5th layer | 6th layer | 7th layer | 8th layer | |
|---|---|---|---|---|---|---|---|---|
| Layer type | Convolution | Max-pooling | Convolution | Max-pooling | Convolution | Max-pooling | Hidden | Logistic regression |
| Number of feature maps | 20 | N/A | 10 | N/A | 5 | N/A | N/A | N/A |
| Filter size | 9×9 | N/A | 5×5 | N/A | 5×5 | N/A | N/A | N/A |
| Input size | 64×64 | 20×56×56 | 20×28×28 | 10×24×24 | 10×12×12 | 5×8×8 | 5×4×4 | 50 |
| Output size | 20×56×56 | 20×28×28 | 10×24×24 | 10×12×12 | 5×8×8 | 5×4×4 | 50 | 2 |
2.3 Performance evaluation
In order to assess the performance of this new deep learning network, we applied a 4-fold cross validation based network training and testing method. The entire dataset was divided into four independent partitions. Each had 70 malignant ROIs and 70 benign ROIs. The distribution of mammographic density and boundary margin characteristics are illustrated in Table 3. Thus, the network was trained and tested four times. In each cycle three partitions with 420 cases were used to train the network and one with 140 cases was applied to test network.
Table 3.
Characteristic distribution of 4 fold testing dataset
| Breast density | Fold | 1 | 2 | 3 | 4 |
| 1 | 10 | 62 | 66 | 2 | |
| 2 | 3 | 57 | 80 | 0 | |
| 3 | 2 | 47 | 86 | 5 | |
| 4 | 7 | 57 | 70 | 6 | |
| Breast margin | 1 | 2 | 3 | 4 | |
| 1 | 37 | 35 | 40 | 28 | |
| 2 | 8 | 61 | 32 | 39 | |
| 3 | 33 | 64 | 26 | 17 | |
| 4 | 7 | 75 | 40 | 18 |
Note: 1) BIRADS breast density score: 1-almost all fatty replaced, 2-scattered fibroglandular densities, 3-heterogeneously dense, 4-extremely dense; 2) BIRADS breast margin score: 1-Smoothed, 2- Irregular, 3-Spiculated, 4 - Focal assymetry
During the optimization, a mini-batch statistic gradient descent method was also applied, which aims to help more effectively and efficiently converge the randomly selected initial parameters into the optimal parameters while reducing the computing complexity [34]. The selection of the number of the mini-batch is a trade-off between the computing efficiency and optimization stability. In this study, the training dataset was divided into 21 mini-batches, which was determined by the previous publication [32] and our own experimental experience. The learning rate of the CNN was selected as a fixed rate of 0.02 and the training epoch was selected as 40. For each epoch, the test dataset was applied to assess the classification performance of the network. The relationship between training epoch and network performance (for both training and testing datasets) was recorded and analyzed.
The entire training and testing process was conducted on a Dell T3600 workstation equipped with a Quadcore 2.80 GHz processor, 4GB RAM, and a Nvidia Quadro 2000D GPU card. Using the network-generated classification scores for all testing cases, the final classification performance of the deep learning algorithm based network was evaluated by using an evaluation index namely, the area under the receiving operation characteristic (ROC) curve (AUC), which is computed by using a publically available maximum likelihood based ROC fitting program (ROCKIT, http://xray.bsd.uchicago.edu/krl/roc_soft.htm, University of Chicago).
3. RESULTS
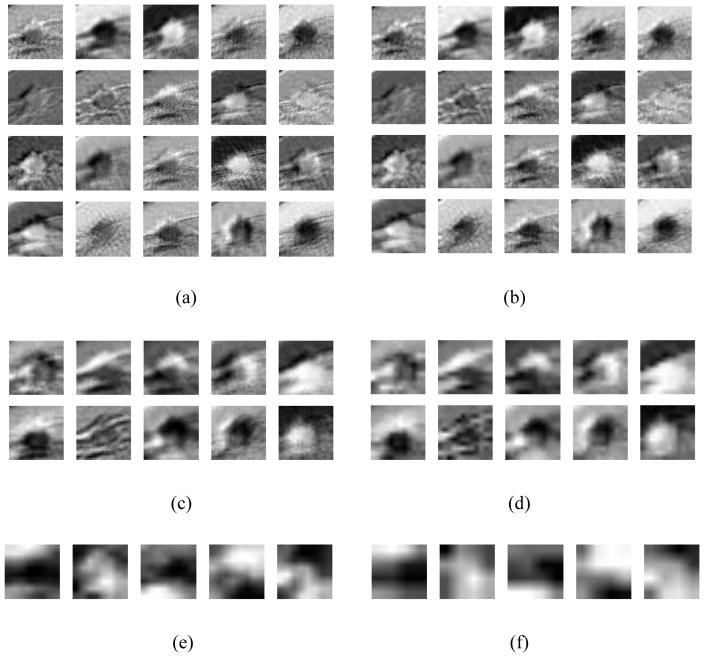
For an illustration purpose to demonstrate working concept and visual classification or comparison results, our trained/optimized deep learning network was applied to the input ROI demonstrated in Figure 1. Figure 2 shows all feature output images (ROIs) geenrated from the deep learning network layers 1 to 6, respectively. Figure 2 (a) demonstrates 20 output feature map images of the first convonlution layer (layer 1). These features illustrate the image characteristics in the following aspects: 1) Image edges, especially the breast mass edge; 2) Image textures, such as the rings of the surrounding tissues; and 3) The segmented mass from the background. These three types of features are the most critical characteristics learned and extracted by the CNN network to distinguish between the malignant and benign ROIs.
Figure 2.
The feature output map images generated by layers 1 – 6 of our deep learning classifier (a) – (f) when the network was applied on the input ROI indicated in Figure 1.
Figure 2(b) shows the results generated by layer 2. Comparing to Figure 2 (a), the output features remove a lot of details during the max-pooling process, which significantly improves the classifier’s robustness, as the classifier will be less sensitive to the subtle changes on the input details. The results in Figure 2(c) and (d) are more abstract comparing to the features shown in Figure 2(a) and (b). In Figure 2(c), the 20 input features are fused into 10 features to reduce the feature redundancy, while the details are further removed by layer 4 (the second max-pooling layer), as demonstrated in Figure 2(d). In Figure 2(e) and (f), the 10 features are finally combined into 5 features, with a size of 8×8 and 4×4, respectively. In this layer, the output feature are processed as highly abstracted, independent features, which are no longer similar to the originial input image. These highly abstracted features can then be used as input for the multiple perception classifiers in layers 7 and 8.
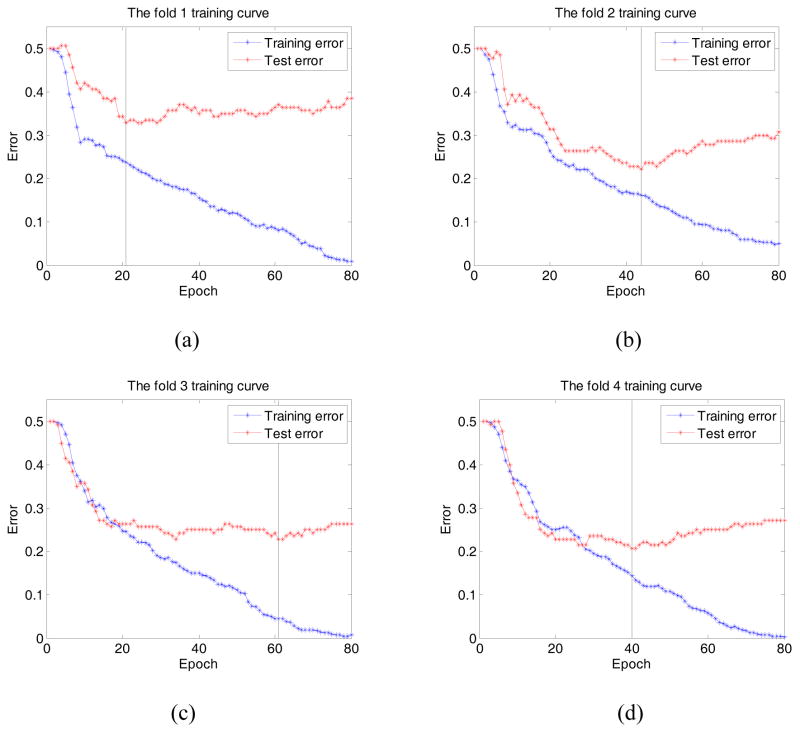
Figure 3 shows four figures of cross-validation folds 1 – 4 ((a) – (d)). Each figure demonstrates and compares two performance curves generated from the training and testing datasets, respectively. In general, the classification errors of both training and testing datasets monotonically reduce as the training epoch number increases initially. However, once the number of epoch reaches a critical point, the classificion errors of the testing dataset starts to increase. For example, as illustrated in Figure 3 (b), the classification error of the training dataset monotonically decreases during the entire training process, which reaches 5% at epoch 80. However, for the testing dataset, the error decreases at first, which yields the minimum of 22.14% at in epoch 44. Then, as the training process proceeds, the testing error gradually increases to 30.71% at epoch 80. For the other 3 folds, the minimum error was yielded at epoch 21, 61, and 40, with a value of 32.86%, 22.86%, and 20.71%, respectively. On the other hand, at epoch 40, the network achieves an error of 35%, 23.57%, 25%, and 20.71% for fold 1 – 4, respectively, which indicates the classifier was trained sufficiently in the experiment.
Figure 3.
The training curve of fold 1 – 4 (a) – (d). It shows that the testing errors yields 35%, 23.57%, 25%, and 20.71% at epoch 40 for the training fold 1, 2, 3, and 4, respectively. The vertical line indicates the epoch corresponding to the optimal training error.
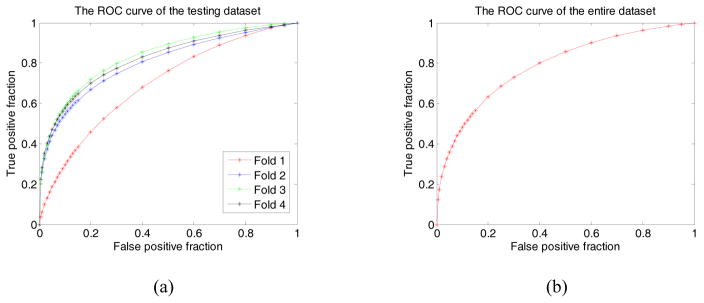
Figure 4 (a) and (b) illustrate the performance of the optimally trained networks for fold 1 – 4 and the entire dataset, respectively. In Figure 4(a), the computed AUC values are 0.696±0.044, 0.802±0.037, 0.836±0.036, and 0.822±0.035, for the testing dataset in fold 1 – 4 cross-validation cycles. At the specificity of 0.8 (or a false positive rate of 0.2), the CAD scheme yielded classification sensitivities of 45.8%, 66.9%, 71.8%, and 69.9%, respectively. When combining the classification scores of all 560 testing ROIs in all four partitions, Figure 4 (b) displays a ROC curve yieleded from the entire testing dataset, which achieves an AUC of 0.790±0.019. At specificity of 0.8, the classification sensitivity yields 63.1%.
Figure 4.
Illustration of 4 ROC curves of the 4 testing dataset (partitians) in 4 training folds 1 – 4(a), and the ROC curve of total testing dataset (b). In these ROC curves AUC values are 0.696±0.044, 0.802±0.037, 0.836±0.036, and 0.822±0.035 for 4 testing partitians in the training fold 1 – 4 respectively, while the AUC of the entire dataset is 0.790±0.019.
4. DISCUSSION
In this study we developed and tested a new deep learning network based CAD scheme to classify between malignant and benign breast lesions (masses) depicting on the digital mammograms. Our experimental results demonstrated that comparing to the existing conventional CAD schemes, this new deep learing based CAD scheme has a number of unique characteristics and/or potential advantages.
First, unlike a conventional hit-or-miss type CAD (detection) schemes that only detect the locations of the suspicious lesions, CAD (diagnosis) schemes for classifying between malignant and benign masses require much accurate lesion segmentation. The mass segmentation accuracy will directly affect or determine the accuracy of the computed mass-related image feature values. However, given the fact of that the morphology of the lesions and overlapping fibro-glandular tissue are highly complicated, previously studies have indicated that there were no “one-fit-all” segmenting methods that enable to successfully segment the diverse lesions depicting on mammograms [6]. In this study, 66.6% of masses as shown in Table 1 (373 / 560) were characterized by the radiologists as irregular or spiculated masses, which were often difficult to be accurately and/or reliably segmented by the conventional CAD schemes. Thus, one advantage of applying a deep learning network based CAD scheme is to avoid the image feature errors introduced from the error or inconsistency of lesion segmentation.
Second, although developing CAD schemes without lesion segmentation has also been previously developed and tested using a content-based image retrieval (CBIR) based CAD schemes [26, 35], the deep learning approach is different and may also have advantage. Unlike the CBIR based CAD schemes, which typically use a k-nearest neighborhood (KNN) based “lazy” learning concept and can be very computational intensive or inefficient in generating a classification score for each testing ROI [36], the deep learning network is pre-trained and its global optimization function (similar to a conventional artificial neural network) can be directly applied to all testing cases (ROIs). Thus, the deep learning based CAD scheme can be more efficiently used in the future clinical practice.
Third, due to the superior architecture of the deep learning networks, the deep learning based classifier do not need to human intervention to design or define the lesion-related image features that need to be computed by CAD schemes. Since there is a big gap between the human vision and computer vision, it is very difficult to subjectively define the effective image features used and implemented in the CAD schemes. In the past two decades, although a large number of morphological and texture based image features have been computed and used in CAD of mass classification, many of these features are highly correlated, which reduces the robustness of the CAD schemes. Thus, how to optimally selecting a small set of effective image features is always an important but difficult task in CAD development [37]. However, deep learning approach is designed to learn the intrinsic characteristics of the lesion images, which cover both a mass and its surround background tissue structure, without human intervention. In this study, we demonstrated that these parameters used in the network could be automatically optimized during the training process. As a result, the deep learning network automatically learned, computed and identified the relevant image features depicting on the training samples. Although due to the use of different testing datasets, we are unable to directly compare this performance level with many of previously reported CAD performance levels (in particular for those CBIR-based CAD schemes as shown in Table 4), our new deep learning network based CAD scheme also yielded a promising and comparable classification performance, which demonstrated the feasibility of using this new approach to provide a solid foundation for the future development and improvement.
Table 4.
Summary and comparison of previously reported CBIR-based CAD schemes of mammographic masses and the scheme performance
| Authors | Number of ROIs | Lesion Segmentation | Image Features | Reported Performance |
|---|---|---|---|---|
| Tourassi, et al, 2003 [38] | 809 masses and 656 manually selected negative ROIs | No | Mutual information | AUC = 0.87±0.01 |
| Alto, et al, 2005 [39] | 20 malignant and 37 benign masses | Yes | 14 gray level co-occurrence matrix (GLCM) based texture features | Classification Accuracy = 61% |
| Tao, et al, 2007 [40] | 121 malignant masses and 122 benign masses | Yes | 3 shape, texture and intensity features | Precision = 82.3% |
| Alolfe, et al, 2008 [41] | 57 malignant masses and 32 normal ROIs | No | 88 features computed from the 1st order statistics, GLCM and fractal dimension | Sensitivity = 68.42% at Specificity of 65.63% |
| Park, et al, 2009 [42] | 1500 masses and 1500 CAD-detected false-positive lesions | Yes | Fractal dimension and 14 morphological features | AUC = 0.851 (95% CI: 0.837 – 0.864) |
| Wang, et al, 2009 [36] | 200 masses and 200 CAD-detected false-positive lesions | No | Pearson’s correlation | AUC = 0.704±0.019 |
| Gundreddy, et al, 2015 [26] | 100 malignant and 100 benign Lesions | No | Two region heterogeneity related features | AUC = 0.832±0.040 |
AUC – Area under a receiver operating characteristic (ROC) curve.
This is our first CAD study that applied the deep learning concept and approach to classify between malignant and benign masses depicting on digital mammograms. The study also has several limitations and/or uninvestigated issues. First, although the deep learning method has demonstrated its substantial superior performance in many different computer vision applications, development of deep learning based CAD scheme is far behind maturity. As a result, performance of our deep learning based CAD scheme may not be statistically higher than the conventional CBIR schemes when we directly compare the AUC value of the ROC curve. It is an open question on how to sufficiently optimize the deep neural network under the specific medical imaging conditions, which is significantly different from the computer vision task. In this study, the dataset is relatively small in developing a deep learning network based CAD scheme. Although the size of our image dataset may be sufficient to develop and preliminarily evaluate the performance of a conventional CAD scheme, the most of well-performed deep learning classifiers are typically trained by very large datasets. Given that establishing a very large cancer image dataset is not an easy task, the alternative approaches need to be explored in the future studies. For example, previous study has shown that a well-trained deep learning network could be repurposed to a new computer vision task, which is substantially different from the original training dataset [43]. Using this concept, we can test the feasibility of combining both a large non-medical image dataset and a relatively small cancer image dataset to build the deep learning network. In this new scheme, non-medical image dataset is used to train the front layers with larger feature dimensions, and the cancer image dataset is used to train the last several layers with smaller feature dimensions, which may be able improve the classifying accuracy and robustness of the CAD scheme.
Second, similar to the conventional CAD schemes [5], the performance of the new deep learning network based CAD scheme also heavily depends on the distribution and/or diversity of the ROIs (or cases) in the specific training and testing datasets. In our 4-fold cross validation, the scheme yielded substantially lower classification performance with AUC = 0.696±0.044 on Fold 1, while the AUC > 0.8 were yielded from other three folds. The lower AUC in Fold 1 also affects the overall classification performance of the entire dataset with AUC = 0.790±0.019. The low performance of Fold 1 validation may be attributed by the fact that Fold 1 has a quite different image feature distribution as comparing to other 3 folds. As shown in Table 3, Fold 1 has substantially higher ratios of masses with smooth boundary and low density (Rated with BIRADS 1). Specifically, although Fold 1 only includes 25% of cases in the entire dataset, it includes 43.5% (37 / 85) of masses rated “smoothed” and 45.5% (10 / 22) of cases rated BIRADS 1 in mammographic density. For the mass classification task, the mammographic density and mass margin are two most important features extracted by the CAD scheme. When the deep learning network was sufficiently trained by the dataset with smaller prevalence of smoothed masses and lower mammographic density (BIRADS 1) cases, the scheme thus yielded a relatively lower classification performance applying to Fold 1. The results indicate the importance of applying the large and more balanced datasets to develop the highly performed and robust CAD schemes including those using the new deep learning networks.
Third, we only tested a deep learning network that is based on the convolution neuron network and multiple layer perceptron networks. Some other types of deep learning networks, such as restricted Boltzmann machine and/or stacked denoising auto-decoder [44] can also be investigated and tested in the future studies. The network architecture may be further optimized by using more convoluting layers or applying new structures such as joint deep learning methods [26, 45].
In summary, we investigated the feasibility of applying deep learning concept to automatically classify the malignant and benign breast masses using digital mammograms. Comparing to other conventional CAD architectures (including CBIR approach), our study results demonstrated that deep learning method can avoid the potential errors or biases introduced from the lesion segmentation and sub-optimal image feature extraction. Thus, deep learning approach has the potential to be a good alternative in developing CAD schemes. If our results can be verified in future studies, this paradigm change approach may have a significantly high clinical impact to help improve efficacy of mammography screening in the future clinical practice.
Acknowledgments
This work is supported in part by Grants of R01CA160205 and R01CA197150 from the National Cancer Institute, National Institutes of Health, USA. The authors would also like to acknowledge the support from the Peggy and Charles Stephenson Cancer Center, University of Oklahoma as well.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Fenton JJ, et al. Reality check: Perceived versus actual performance of community mammographers. Am J Roentgenol. 2006;187:42–46. doi: 10.2214/AJR.05.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodersen J, Siersma VD. Long-Term Psychosocial Consequences of False-Positive Screening Mammography. Ann Fam Med. 2013;11:106–115. doi: 10.1370/afm.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishikawa RA. Current status and future directions of computer-aided diagnosis in mammography. Comput Med Imaging Graph. 2007;31:224–235. doi: 10.1016/j.compmedimag.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Zheng B, et al. Computer-aided detection of breast masses depicted on full-field digital mammograms: a performance assessment. Br J Radiol. 2012;85:e153–161. doi: 10.1259/bjr/51461617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver A, et al. A review of automatic mass detection and segmentation in mammographic images. Med Image Anal. 2010;14:87–110. doi: 10.1016/j.media.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Fenton JJ, et al. Effectiveness of computer-aided detection in community mammography practice. J Natl Cancer Inst. 2011;103:1152–1161. doi: 10.1093/jnci/djr206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hupse R, et al. Computer-aided detection of masses at mammography: Interactive decision support versus prompts. Radiology. 2013;266:123–129. doi: 10.1148/radiol.12120218. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa RM, Gur D. CADe for early detection of breast cancer - current status and why we need to continue to explore new approaches. Acad Radiol. 2014;21:1320–1321. doi: 10.1016/j.acra.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Tan M, Aghaei F, Wang Y, Zheng B. Developing a new case based computer-aided detection scheme and an adaptive cueing method to improve performance in detecting mammographic lesions. Phys Med Biol. 2017;62:358–376. doi: 10.1088/1361-6560/aa5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Aghaei F, Zarafshani A, Qiu Y, Qian W, Zheng B. Computer-aided classification of mammographic masses using visually sensitive image features. J Xray Sci Technol. 2017;25:171–186. doi: 10.3233/XST-16212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinton GE, Salakhutdinov PR. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 13.Jaccard N, Rogers TW, Morton EJ, Griffin LD. Detection of concealed cars in complex cargo X-ray imagery using deep learning. J Xray Sci Tehcnol. 2016 doi: 10.3233/XST-16199. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Li R, Deng H, et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. 2015;108:214–224. doi: 10.1016/j.neuroimage.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth H, et al. DeepOrgan: Multi-level deep convolutional networks for automated pancreas segmentation. 18th International Conference on Medical Image Computing and Computer-Assisted Intervention – MICCAI; 2015; pp. 556–564. [Google Scholar]
- 16.Ciresan D, et al. Deep neural networks segment neuronal membranes in electron microscopy images. Proc NIPS. 2012 [Google Scholar]
- 17.Bar Y, et al. Deep learning with non-medical training used for chest pathology identification. Proc SPIE. 2015;9414:94140V. [Google Scholar]
- 18.Suk H-I, et al. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Structure & Function. 2015;220:841–859. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szegedy C, et al. Going deeper with convolutions. Computer Vision and Pattern Recognition. 2014:1–12. [Google Scholar]
- 20.Lecun Y, et al. Gradient-based learning applied to document recognition. Proc IEEE. 1998:2278–2324. [Google Scholar]
- 21.Taigman Y, et al. DeepFace: closing the gap to human-level performance in face verification. IEEE Conference in Computer Vision and Pattern Recognition (CVPR); 2014. pp. 1701–1708. [Google Scholar]
- 22.Tian Y, et al. Pedestrian Detection aided by Deep Learning Semantic Tasks. Computer Vision and Pattern Recognition. 2014:1–14. [Google Scholar]
- 23.Petersen K, et al. Breast tissue segmentation and mammographic risk scoring using deep learning. 12th International Workshop of igital Mammography (IWDM); 2014; pp. 88–94. [Google Scholar]
- 24.Qiu Y, et al. An initial investigation on developing a new method to predict short-term breast cancer risk based on deep learning technology. Proc SPIE. 2016;9785:978521. [Google Scholar]
- 25.Wang JH, et al. Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci Rep. 2016;6 doi: 10.1038/srep27327. Article number 27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gundreddy RR, et al. Assessment of performance and reproducibility of applying a content-based image retrieval scheme for classification of breast lesions. Med Phys. 2015;42:4241–4249. doi: 10.1118/1.4922681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang W, et al. Deepidnet: Multi - stage and deformable deep convolutional neural networks for object detection. 2014. arXiv:1409.3505 [cs.CV] [DOI] [PubMed] [Google Scholar]
- 28.Simonyan K, Zisseman A. Very deep convolutional networks for large-scale image recognition. 2015. arXiv:1409.1556[cs.CV] [Google Scholar]
- 29.Tan M, Pu J, Zheng B. A new and fast image feature selection method for developing an optimal mammographic mass detection scheme. Med Phys. 2014;41:313–324. doi: 10.1118/1.4890080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan M, Zheng B, Leader JK, Gur D. Association between changes in mammographic image features and risk for near-term breast cancer development. IEEE Trans Med Imaging. 2016;35:1719–1728. doi: 10.1109/TMI.2016.2527619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu YC, et al. Feature selection for the automated detection of metaphase chromosomes: performance comparison using a receiver operating characteristic method. Anal Cell Pathol. 2014 doi: 10.1155/2014/565392. Article ID: 565392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LISA Lab. Deep Learning Tutorial. University of Montreal; 2015. pp. 51–62. [Google Scholar]
- 33.Witten I, Frank E, Hall M. Data mining: Practical machine learning tools and techniques. 3. Elsevier; 2011. [Google Scholar]
- 34.Zhang T. Solving large scale linear prediction problems using stochastic gradient descent algorithms. Proceedings of the twenty-first international conference on Machine learning; 2004; pp. 1–8. [Google Scholar]
- 35.Zheng B. Computer-aided diagnosis in mammography using content-based image retrieval approaches: current status and future perspectives. Algorithms. 2009;2:828–849. doi: 10.3390/a2020828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Park SC, Zheng B. Improving performance of content-based image retrieval schemes in searching for similar breast mass regions: an assessment. Phys Med Biol. 2009;54:949–961. doi: 10.1088/0031-9155/54/4/009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan M, Pu J, Zheng B. Optimization of breast mass classification using sequential forward floating selection (SFFS) and a support vector machine (SVM) model. Int J Comput Assist Radiol Surg. 2014;9:1005–1020. doi: 10.1007/s11548-014-0992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tourassi GD, et al. Computer-assisted detection of mammographic masses: A template matching scheme based on mutual information. Med Phys. 2003;30:2123–2130. doi: 10.1118/1.1589494. [DOI] [PubMed] [Google Scholar]
- 39.Alto H, Rangayyan RM, Desautels JE. Content-based retrieval and analysis of mammographic masses. J Electron Imaging. 2005;14:023016. [Google Scholar]
- 40.Tao Y, et al. A preliminary study of content-based mammographic masses retrieval. Proc SPIE. 2007;6514:65141Z. [Google Scholar]
- 41.Alolfe MA, et al. Development of a computer-aided classification system for cancer detection from digital mammograms. Radio Science Conference, 2008 NRSC 2008 National; 2008; pp. 1–8. [Google Scholar]
- 42.Park SC, Wang XH, Zheng B. Assessment of performance improvement in content-based medical image retrieval schemes using fractal dimension. Acad Radiol. 2009;16:1171–1178. doi: 10.1016/j.acra.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donahue J, et al. DeCAF: A deep convolutional activation feature for generic visual recognition. 2013. arXiv:1310.1531 [cs.CV] [Google Scholar]
- 44.Vincent P, et al. Extracting and composing robust features with denoising autoencoders. Proc. of the 25th international conference on Machine learning; 2008; pp. 1096–1103. [Google Scholar]
- 45.Ouyang W, Wang WX. Joint deep learning for pedestrian detection. Proc. of 2013 IEEE International Conference on Computer Vision (ICCV); 2013; pp. 2056–2063. [Google Scholar]