Abstract
The reversible oxidation of protein cysteine residues (Cys-SH) is a key reaction in cellular redox signaling involving initial formation of sulfenic acids (Cys-SOH), which are commonly detected using selective dimedone-based probes. Here, we report that significant portions of dimedone-tagged proteins are susceptible to cleavage by DTT reflecting the presence of perthiosulfenic acid species (Cys-SSOH) due to similar oxidation of hydropersulfides (Cys-SSH), since Cys-S-dimedone adducts are stable toward DTT. Combined studies using molecular modeling, mass spectrometry, and cell-based experiments indicate that Cys-SSH are readily oxidized to Cys-SSOH, which forms stable adducts with dimedone-based probes. We additionally confirm the presence of Cys-SSH within protein tyrosine kinases such as EGFR, and their apparent oxidation to Cys-SSOH in response NADPH oxidase activation, suggesting that such Cys-SSH oxidation may represent a novel, as yet uncharacterized, event in redox-based signaling.
Abbreviations: Cys-SH, protein cysteine thiol; Cys-SOH, protein sulfenic acid; Cys-SSH, protein persulfide; Cys-SSOH, protein cysteine perthiosulfenic acid; EGFR, epidermal growth factor receptor; Dimedone, 5,5-dimethyl-1,3-cyclohexanedione; DTT, Dithiothreitol; RSB, reducing sample buffer
Keywords: Thiol oxidation, Sulfenic acid, Dimedone, Hydrogen peroxide, NADPH oxidase, Redox signaling
Graphical abstract
1. Introduction
Oxidation-reduction chemistry forms an increasingly appreciated feature of biological signaling pathways, which includes the reversible oxidation of protein cysteine residues (Cys-SH) to influence protein structures and functions [1], [2]. In such redox signaling mechanisms, protein Cys-SH are oxidized by H2O2 generated by activation of e.g. NOX family NADPH oxidases or mitochondria, to initially generate intermediate sulfenic acids (Cys-SOH) and subsequent disulfide species [3], [4], [5], [6]. While a reactive and transient species, development of selective molecular probes, such as the 1,3-diketone-containing 5,5-dimethyl-1,3-cyclohexanedione (dimedone), have allowed for the detection of Cys-SOH in a range of proteins during biological redox signaling [7], [8], [9]. Moreover, intermediate formation of Cys-SOH has been implicated in direct modulation of protein function, for example in the regulation of protein tyrosine kinases [10], [11], [12].
Although formation of Cys-SOH is commonly appreciated as an intermediate in protein cysteine oxidation, some questions have been raised with respect to the specificity of dimedone-based reagents to detect them (e.g. [6], [13]). In this regard, it is becoming apparent that biological thiols can also exist in the form of hydropersulfide or polysulfide species (Cys-SSnH), in both low-molecular weight thiols (e.g. glutathione persulfide; GSSH) and in various proteins [14], [15], [16], [17]. The origin of such Cys-SSnH species is debated [14], [17], [18], [19], but likely sources involve some particular transsulfurases and the chemical reaction of cysteines or their oxidized derivatives with hydrogen sulfide (H2S), an important small molecule mediator in biological systems [20], [21], [22], to generate hydropersulfide species (Cys-SSH) [23], [24], [25]. Moreover, studies towards the chemical biology of Cys-SSH indicate that they have enhanced nucleophilic properties and one-electron reductant activity compared to corresponding sulfhydryl species, in part due to markedly reduced pKa [14], [15], [25]. Because of the common presence of protein cysteine persulfide species (R-SSH) in biological systems, we speculated that they represent important targets for cellular oxidants such as H2O2, leading to intermediate formation of perthiosulfenic acid species (R-SSOH), which are also detectable with dimedone-based reagents.
The present studies indeed indicate that a significant fraction of dimedone-metabolized proteins in resting or stimulated cells is susceptible to cleavage by thiol-based reductants such as DTT, suggesting the presence of Cys-SSOH species. Moreover, increases in apparent Cys-SSOH in various proteins in response to NADPH oxidase activation suggest that such previously unrecognized intermediate might play novel and unique roles in redox-based signaling.
2. Materials and methods
2.1. Cell culture and treatments
Human embryonic kidney cells (HEK293) or human pulmonary mucoepidermoid epithelial cells (NCI-H292) were grown in Dulbecco's Modified Eagle Medium (DMEM: Life Technologies) or RPMI 1640 medium containing 10% fetal bovine serum and 1% penicillin/streptomycin, respectively, as previously described [26], [27]. For experimentation, cells were seeded at 100,000 cells/well in 24-well plates or 500,000 in 6-well plates (Corning). For studies involving cell stimulation with ATP (Sigma, St. Louis, MO; 100 µM) or EGF (100 ng/mL Calbiochem), confluent cell monolayers were previously cultured overnight in the absence of serum. Following treatments, cell lysates were collected by placing cells on ice in 100 µL Western solubilization lysis buffer (50 mM HEPES, 250 mM NaCl, 1.5 mM MgCl2, 1% Triton-X100, 10% glycerol, 1 mmol/L ethyleneglycol-bis-(β-aminoethylether)-N,N,N9,N9-tetraacetic acid, 1 mM phenylmethylsulfonyl fluoride, 2 mM Na3VO4, 10 mg/mL aprotinin, and 10 mg/mL leupeptin; pH 7.4) per well for 30 min. Lysates were collected by scraping, briefly sonicated, and cleared of insoluble material by centrifugation (14,000 rpm, 5 min) for analysis.
2.2. Analysis of protein Cys-SOH or Cys-SSOH
For analysis of protein sulfenylation (Cys-SOH) and perthiosulfenylation (Cys-SSOH), cells were lysed in Western solubilization buffer (WSB) containing 1 mM DCP-bio1 (Kerafast or EMD Millipore Sigma), 200 U/mL catalase (Worthington, Lakewood, NJ) and 10 mM N-ethylmaleimide (Sigma) and incubated for 1 h on ice. Equal amounts of cell lysates were mixed with Laemli sample buffer, either in the presence or absence of 25 mmol/L DTT for 30 min, and separated by 10% SDS-PAGE for Western blotting with streptavidin-peroxidase (see below). For analysis of specific proteins of interest, excess DCP-bio1 reagent was removed from DCP-Bio1-derivatized lysates by 6 successive washes with 20 mM Tris-HCl (pH 7.4) on Amicon Ultra-0.5 Centrifugal Filter Devices (Millipore). DCP-bio1-tagged proteins were subsequently collected with high capacity NeutrAvidin-agarose beads (Pierce) and washed successively with 1% SDS, 4 mol/L urea, and 1 mol/L NaCl [28]. Beads were then washed with 100 mmol/L ammonium bicarbonate either in the presence or absence of 10 mmol/L DTT for 30 min to assess the difference of sulfenic acid and perthiosulfenic acid tagged proteins.
2.3. Analysis of protein Cys-SSH
Analysis of protein hydropersulfides (Cys-SSH) was performed essentially as previously described [16]. Briefly, H292 cells were serum starved, purged for 30 min with nitrogen, and lysed in WSB containing 1.0 mM Iodoacetyl-PEG2-biotin (IAB; Thermo) and 2000 U/mL catalase (bovine liver, Worthington) and incubated for 1 h on ice. Lysates containing ~ 200 µg protein were added to high capacity Neutravidin-agarose beads (Pierce) and washed 5 times with WSB followed by three washes of Tris-buffered saline (50 mM Tris-Cl, 150 mM NaCl, pH 7.4; TBS) including 0.05% SDS. The beads, which contain alkylated Cys-SH or Cys-SSH species, were then resuspended for 30 min in TBS containing 0.5% Tween-20 (TBST) in the presence of 25 mM DTT, to elute persulfide-containing proteins [16]. Parallel samples were resuspended in the absence of DTT as a negative control. Both eluates were analyzed separately by Western blot, in comparison with residual proteins bound to these beads, which were eluted with reducing sample buffer (RSB) containing 125 mM Tris-Cl, 4% SDS, 20% w/v Glycerol, 0.5 mM β-mercaptoethanol, 0.02% bromophenol blue, pH 6.8). For appropriate comparison, equivalent volumes of each sample were analyzed.
2.4. Western blotting
Cell lysates containing equal amounts of protein (15–35 µg, measured using BCA protein assay kit; Pierce) or proteins eluted from Neutravidin beads were mixed with reducing or non-reducing sample buffer (see above) and loaded on 10% SDS-PAGE gels and transferred to nitrocellulose membranes, and probed using antibodies against Src (L4A1; Cell Signaling 1∶1000), EGFR (C74B9 Cell Signaling 1:1000), or anti-cysteine sulfenic acid (Millipore Sigma; 07–2139; which specifically detects dimedone-conjugated cysteine residues) or with streptavidin peroxidase polymer ultrasensitive (1:10,000; Sigma). Primary antibodies were probed with rabbit or mouse-specific secondary antibodies conjugated with HRP (Cell Signaling) and detected by enhanced chemiluminescence (Pierce) using an Amersham 600 SE Imaging System. Western blot band densities were quantified using ImageQuant TL (v8.1.0.0).
2.5. DFT calculations
Density functional theory (DFT) calculations were performed with Jaguar v8.4 [29]. Gas-phase geometry optimizations were performed with the M06-L functional [30] and 6–311++G** basis set. Geometries obtained for ethanepersulfide (CH3-CH2-S-S-H) are consistent with geometric parameters from structurally defined hydropersulfides (Table S1, Fig. S1). [31], [32] Thermodynamic corrections to enthalpy and entropy were obtained from frequency calculations with the M06-L functional, and geometry minima were confirmed on the basis of no imaginary frequencies. Gas-phase electronic energies were corrected using the SM8 [33], [34] solvation model for solvation in water solvent. Solvation single point calculations were performed with M06 [35], M06-2x [35], B3LYP-3D [36], PBE-D3 [37], and PBE0-D3 [38] to assess the variance in energies with a variety of functionals.
2.6. Identification of Cys-SOH and Cys-(S)nOH via dimedone labeling by liquid chromatography (LC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) (LC-ESI-MS/MS)
Cys-SOH and Cys-(S)nOH were identified as dimedone adducts by LC-ESI-MS/MS. Specifically, Cys-SOH/Cys-(S)nOH were formed in the reaction of 0.1 mM l-cysteine and 0.3 mM Na2S2 in 10 mM Tris-HCl buffer (pH 7.4) as described below, yielding a reaction mixture containing 19% Cys-SH, 33% Cys-SSH, and 35% Cys-SSSH, as determined by labeling with β-(4-hydroxyphenyl)ethyl iodoacetamide (HPE-IAM). [39] To the reaction mixture were added H2O2 (0–25 mM) and 5 mM dimedone, which were then were incubated at 37 °C for 3 h to form respective Cys-SOH/Cys-(S)nOH dimedone adducts. The reaction products treated with dimedone were diluted with 0.1% formic acid containing known amounts of isotope-labeled internal standards, which were then subjected to LC-ESI-MS/MS analyses as reported earlier [15]. Multiple reaction monitoring (MRM) parameters for each dimedone adduct are summarized in Table S4 in the Supplementary Information.
2.6.1. Preparation of Cys-S/Cys-(S)n-dimedone standards for LC-ESI-MS/MS analysis
Authentic Cys-S/CysS-(S)n-dimedone adducts were synthesized according to our method previously described with modifications, and were used as the standards for LC-ESI-MS/MS [15]. To form Cys-(S)nH, 0.5 mM l-cysteine was reacted with 0.5 mM NaHS with NO generated from 0.5 mM 1-hydroxy-2-oxo-3-(N-methyl-3-aminopropyl)−3-methyl-1-triazene (Dojindo Laboratories, Kumamoto, Japan) in 10 mM Tris-HCl buffer (pH 7.4) under aerobic conditions at room temperature for 30 min, after which 2 mM H2O2 and 10 mM dimedone (Wako Pure Chemical Industries, Osaka, Japan) were added to the reaction mixture, and the mixtures were incubated further at 37 °C for 3 h to produce respective Cys-SOH/Cys-(S)nOH adducts. Similarly, stable isotope-labeled dimedone adducts of CysS-(34S)nOH were synthesized by reacting dimedone with CysS-(34S)nOH formed with cysteine and polysulfides generated with NaH34S instead of NaH32S. Various dimedone adducts thus synthesized were purified by HPLC (Prominence; Shimadzu Corporation, Kyoto, Japan), on a reverse-phase column (YMC-Triart C18 column, 50 × 2.0 mm inner diameter; YMC, Kyoto, Japan) under the following elution conditions: mobile phases A (0.1% formic acid) and B (0.1% formic acid in methanol) with a linear gradient from 5% to 90% B for 15 min at a flow rate of 0.2 mL/min at 40 °C.
3. Results and discussion
3.1. Dimedone labeling of cellular proteins reveals the presence of DTT-reducible adducts
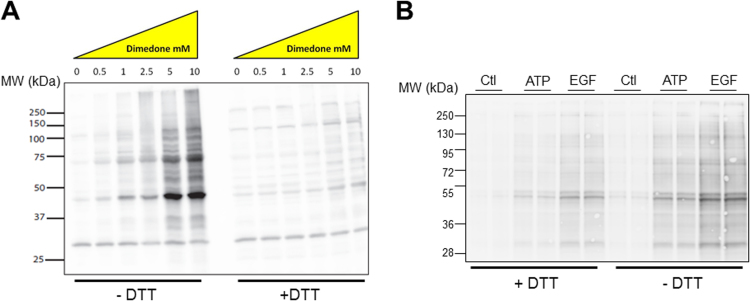
Detection of protein sulfenic acids (Cys-SOH) is typically performed using dimedone-based probes and analysis under reducing conditions (ie. in the presence of DTT). However, when HEK293 cells were incubated in the presence of dimedone for 2 h, and subsequently lysed and mixed with Laemlli sample buffer in either the absence or presence of DTT, subsequent SDS-PAGE and Western blot analysis with an α-dimedone antibody indicated that formation of dimedone-adducts was significantly more prominent when analyzed under non-reducing conditions (Fig. 1A), suggesting that a substantial fraction of detected adducts represents Cys-SSn-dimedone (n ≥ 1) conjugates. Similar findings were observed in NCI-H292 human pulmonary mucoepidermoid cells and analysis of cell lysates with the sulfenic acid probe DCP-bio1, which were subsequently mixed with SDS-PAGE sample buffer in the presence or absence of DTT, and analyzed by blotting with streptavidin-HRP to detect DCP-Bio1-tagged proteins (Fig. 1B) [7], [28]. In this case, we also addressed potential formation of Cys-SOH in response to cellular H2O2 production by cell stimulation with either exogenous ATP (100 µM) or epidermal growth factor (EGF; 100 ng/mL), which activate the NOX enzymes DUOX1 and NOX2, respectively [12], [27]. Indeed, cell stimulation with ATP and EGF promoted increased DCP-bio1-tagged proteins analyzed under reducing conditions, as expected, but more dramatically enhanced overall intensity of DCP-bio1-tagged protein bands when analyzed under non-reducing conditions (in the absence of DTT) (Fig. 1B). Although our findings indicate the presence of Cys-SSn-DCP-bio1 species, that would have gone undetected under reducing conditions, it is alternatively possible that reductants such as DTT may also be capable of reversing Cys-S-dimedone adducts, thus minimizing their detection [40], [41]. To address this, we performed LC-MS/MS analysis of preformed dimedone adducts of Cys-SH or Cys-SSH and followed their decomposition in the presence of various concentrations of DTT. As illustrated in Fig. 2, while DTT induced dose-dependent degradation of Cys-SS-dimedone, due to cleavage of its disulfide bond, it did not significantly degrade Cys-S-dimedone adducts, even at concentrations of 100 mM. Hence, our observation of DTT-reducible dimedone-tagged proteins reflects the presence of Cys-SSn-dimedone species, which most likely originated from dimedone labeling of oxidized persulfide species (Cys-SSnOH) species, by forming additional Cys-SSn-dimedone adducts, was also tested in several cell-based studies. Indeed, formation of such oxidized persulfides has been implicated previously during oxidation of mercaptopyruvate sulfurtransferase [42], but our findings indicate that such formation of thiosulfenate (Cys-S-SO-) or perthiosulfenic acid species occurs more widely in a large number of cellular proteins.
Fig. 1.
Analysis of dimedone labeling of protein sulfenic acids under reducing or non-reducing conditions. (A) HEK293 cells were incubated in the presence of indicated concentrations of dimedone, and cells were lysed and mixed with Laemlli sample buffer in the presence or absence of DTT before analysis by Western blot using α-dimedone antibody. (B) Analysis of DCP-Bio1-tagged proteins in H292 cells upon 10-min stimulation with either ATP (100 µM) or EGF (100 ng/mL) under reducing (+ DTT) or non-reducing (- DTT) conditions. Representative blots of 3 separate experiments are shown.
Fig. 2.
Analysis of decomposition of Cys-S-dimedone or Cys-SS-dimedone in the presence of DTT. Preformed Cys-S-dimedone and Cys-SS-dimedone adducts (10 nM each) were incubated in the presence of various concentrations of DTT in 30 mM HEPES buffer (pH 7.5) at 37 °C for 1 h, followed by quantification by LC-ESI-MS/MS analysis. Data are means ± s.d. (n = 4). *P < 0.05; **P < 0.01 (vs. 0 mM DTT).
3.2. DFT calculations and LC-MS/MS analysis predict facile oxidation of RSSH to RSSOH and labeling with dimedone
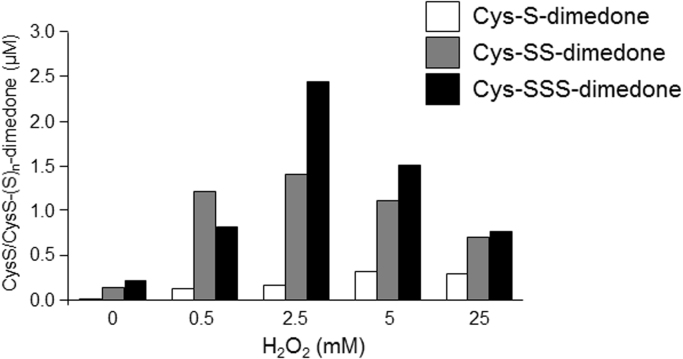
To support our proposed detection of Cys-SSn-dimedone species due to derivatization of Cys-SSnOH, we performed density functional theory (DFT) calculations on ethanethiol (Et-SH) and ethyldisulfane (Et-SSH) models, to predict the relative reactivity of H2O2 with R-SSH compared to R-SH with respect to generating corresponding sulfenic acids (R-SSOH versus R-SOH), as well as their subsequent reaction with dimedone. We first calibrated DFT geometries with X-ray crystal structures of hydropersulfide species [31], [32] (Fig. S1, Table S2) and determined that computed ΔG°’s are consistent with a set of diverse methods (Tables S3 and S4, Figs. S2 and S3). In line with our hypothesis, ΔG°’s for H2O2-dependent oxidation of Et-SSH to Et-SSOH were calculated to be ~ 6.2 kcal/mol more favorable compared to similar oxidation of Et-SH to Et-SOH (Fig. 3). Moreover, conjugation of either Et-SOH and Et-SSOH with the dimedone-like compound cyclohexane-1,3-dione is in both cases exergonic, although Et-SOH reacts with dimedone ~ 5.7 kcal/mol more favorably compared to Et-SSOH (Fig. 3). Hence, these calculations predict that R-SSH species are readily oxidized by H2O2 to R-SSOH, relative to comparable oxidation of R-SH to R-SOH, and that both R-SOH and R-SSOH are reactive towards dimedone to form stable adducts. Dimedone probes are not reactive towards R-SH, but could conceivably react with Cys-SSH to form Cys-S-dimedone with elimination of H2S. However, computed values of ΔG° indicate that dimedone adduction to Et-SSH is energetically uphill by ~ 7.6 kcal/mol (~ 15.1 kcal/mol less favorable compared to dimedone reacting with Et-SSOH), suggesting that dimedone is highly selective towards R-SOH or R-SSOH species compared to R-SSH species (Fig. S4). Overall, these calculations predict that R-SSH can be readily oxidized to R-SSOH and that commonly employed dimedone compounds used to trap R-SOH are also capable of trapping R-SSOH. In an attempt to experimentally confirm the ability of dimedone to detect R-SSOH, we prepared Cys-SnH species by reacting l-cysteine with Na2S2 and analyzed reaction products with H2O2 and dimedone with LC-MS/MS (Fig. 4; Table S4) [15]. As shown, H2O2 induced dose-dependent formation of Cys-S-dimedone and especially Cys-SS-dimedone and Cys-SSS-dimedone, reflecting intermediate formation of Cys-SnOH. The greater yield of Cys-SS-dimedone and Cys-SSS-dimedone species compared to Cys-S-dimedone is consistent with the greater susceptibility of perthiol species (Cys-SSH, Cys-SSSH) to oxidation by H2O2. Autoxidation of Cys-SSnH likely accounts for detection of dimedone adducts in the absence of H2O2, since direct reaction of dimedone with Cys-SSnH is deemed unlikely based on DFT calculations (Fig. S4). Finally, the observed decrease in dimedone adducts at [H2O2] > 5.0 mM most likely reflects further oxidation of Cys-SSnH to sulfinic and sulfonic acid species, which are unreactive with dimedone. In aggregate, these analyses support the formation of Cys-SSOH during oxidation of protein persulfides and its detection using dimedone-based probes.
Fig. 3.
DFT models and computed Gibbs Free Energies of the oxidation of (A) Et-SH to Et-SOH and (B) Et-SSH to Et-SSOH by H2O2 and subsequent reaction with dimedone to form (A) Et-S-dimedone and (B) Et-SS-dimedone.
Fig. 4.
Analysis of dimedone derivatization of Cys-(S)nOH by LC-MS/MS. Quantitative identification of Cys-(S)nOH formed during reaction of Cys-(Sn)H with H2O2 by derivatization with dimedone, as described in Materials and Methods. Data are representative of 2–3 separate analyses.
3.3. Protein tyrosine kinases contain persulfide species that are susceptible to H2O2
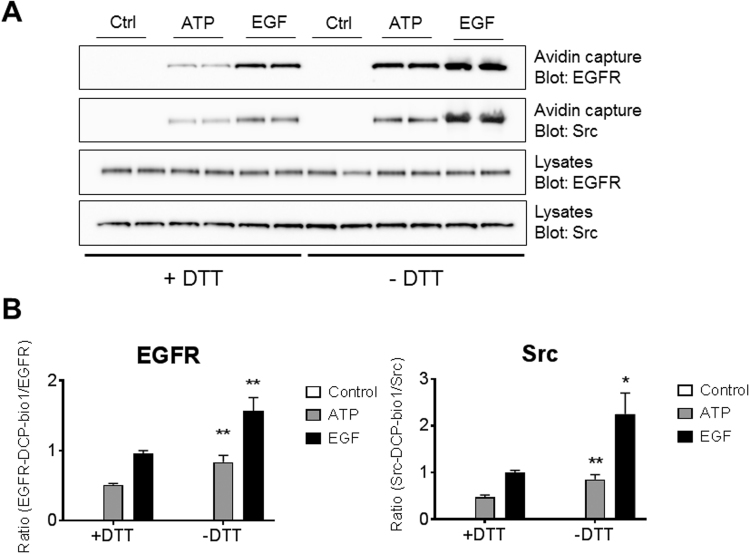
Previous studies have indicated that NOX-dependent H2O2 production, in response to stimulation with extracellular ATP or EGF regulates the activation of redox-sensitive tyrosine kinases such as epidermal growth factor receptor (EGFR) and Src, by sulfenylation of conserved cysteines within their kinase domains [12], [27], [43], [44]. Indeed, epithelial cell stimulation with ATP and EGF was found to generate H2O2 by activation of DUOX1 and NOX2, respectively [12], [45]. Since both ATP and EGF enhance formation of DTT-reducible dimedone conjugates (Fig. 1 B), we speculated whether Cys-SSOH may be formed in these kinases due to oxidation of pre-existing Cys-SSH species. To this end, DCP-Bio1-derivatized cell lysates from ATP- or EGF-stimulated H292 cells (Fig. 1) were subjected to neutravidin purification [7], [28], and neutravidin-purified proteins were washed in the presence or absence of DTT prior to elution in reducing sample buffer (RSB), and then analyzed by SDS-PAGE and Western blotting for EGFR or Src. As shown, greater amounts of DCP-Bio1-labeled EGFR and Src were recovered from ATP- or EGF-stimulated cells under non-reducing washing conditions when compared to washing conditions that included DTT (Fig. 5), indicating the formation of reducible DCP-Bio1 conjugates in these proteins in response to ATP and EGF that likely reflect the presence of Cys-SSnOH within these proteins. Quantification of relative band densities indicates that DCP-Bio1 tagging of EGFR and Src in cells treated with either stimulus was ~ 60–120% higher in the absence of DTT washing (Fig. 5B), suggesting that a significant portion of DCP-Bio1 reactivity within these proteins may actually reflect the presence of Cys-SSnOH intermediates, as potentially relevant post-translational modifications in redox signaling. One potential caveat in these measurements is that DTT reduction may also lead to recovery of proteins that are indirectly bound through intermolecular disulfides to DCP-bio1-conjugated proteins, which would be mistakenly attributed to Cys-SSOH-containing proteins. However, to our knowledge there is no evidence for intermolecular disulfide crosslinking of EGFR with other proteins, although this may be true for Src [46]. Therefore, we assume that our findings reflect the presence of Cys-SSnOH within EGFR and Src. Another issue worth considering is that alkylating agents, such as N-ethylmaleimide (NEM; which was used in lysis buffers in these studies), can react with Cys-SOH species [41] and potentially also with Cys-SSOH, although it would have to outcompete reactions of dimedone or DCP-Bio1 with these products. If anything, this would not invalidate our conclusions and would in fact result in underestimation of the amounts of Cys-SOH and Cys-SSOH species.
Fig. 5.
(A) Representative Western blot analysis of Cys-SSOH species in EGFR and Src in stimulated H292 cells by DCP-bio1 labeling and avidin capture and processing in the presence or absence of DTT. (B) Densitometry analysis of relative band density of Western blots from 8 replicates from 3 independent experiments. Ratios were arbitrarily set at 1 for EGF-stimulated samples analyzed with DTT. Data represents means ± s.e.m. * P < 0.05 and ** P < 0.01 based on Student's t-test.
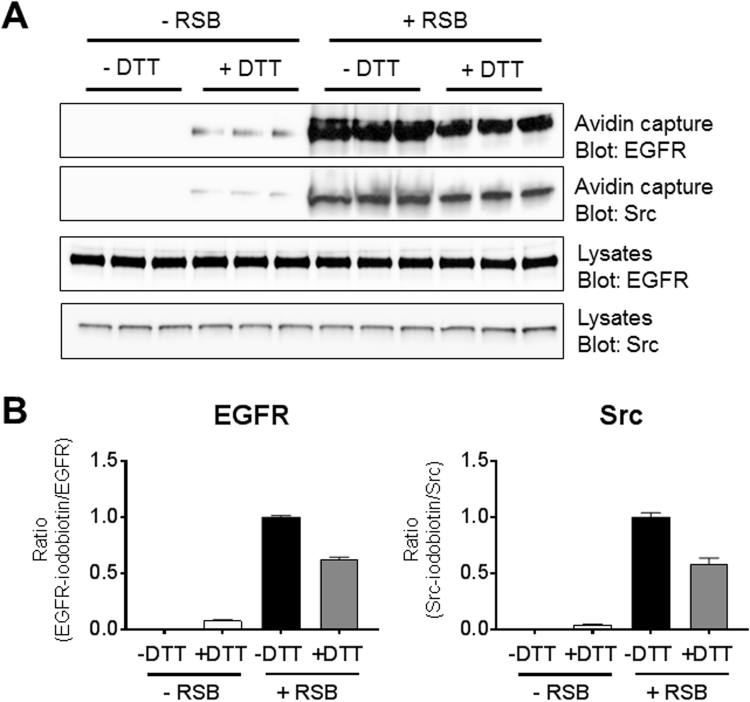
If our findings indeed reflect the formation of Cys-SSnOH species within EGFR and Src in response to H2O2 production, this would imply that a fraction of Cys residues within these kinases exists as Cys-SSnH. To address this, we utilized a recently developed procedure to detect the presence of protein Cys-SSnH, ProPerDP [16], based on alkylation of both Cys-SH and Cys-SSH with Iodoacetyl-PEG2-Biotin (IAB) and differential elution of avidin-purified proteins in the presence or absence of DTT. As shown in Fig. 6, analysis of DTT-eluted proteins from avidin-purified IAB-derivatized proteins from H292 cell lysates indeed revealed detectable amounts of EGFR and Src, indicating the presence of Cys-SSnH, in comparison to control elution in the absence of DTT (Fig. 6; left 6 lanes). Analysis of avidin-purified proteins using SDS-containing reducing sample buffer (RSB), which represents combined IAB-tagged Cys-SH and Cys-SSnH species, indicated slightly lower recovery of EGFR and Src from avidin beads that were initially prewashed with DTT (Fig. 6; right 6 lanes), thus indirectly demonstrating the presence of Cys-SSnH in both proteins. It is important to consider some technical limitations of this methodology [16]. First, EGFR and Src contain multiple cysteine residues, and potential IAB conjugation of both Cys-SH and Cys-SSH within the same protein would render Cys-SSH undetectable with this method. Additionally, as is also the case for the DCP-bio1 experiments in Fig. 5, DTT reduction could lead to an increase in the signal for EGFR and Src bound through intermolecular disulfides and would be mistakenly identified as Cys-SSH-containing proteins. However, and as mentioned previously, this is expected to be negligable. Since IAB can react with Cys-SOH and thus presumably also with Cys-SSOH, our analysis of Cys-SSH might also include Cys-SSOH. Lastly, our analysis of Cys-SSH may also be compromised by reaction of IAB with Cys-SOH to the corresponding Cys-SOR adduct, which could be reduced by DTT during the initial elution step. This would mean that this detection method for Cys-SSH might inadvertently also detect some Cys-SOH. However, since the reduction of Cys-SOR by DTT is rather slow (< 50% after 1 h [42]), we believe this is a relatively minor concern. Notwithstanding these limitations, the present findings strongly indicate the presence of Cys-SSnH within these protein kinases and quantification of relative Western blot band densities suggests that ~ 5–8% of all reduced Cys residues within EGFR and Src may exist as Cys-SSnH (Fig. 6B). The estimated proportion of Cys-SSn-dimedone in overall dimedone conjugates in these proteins in response to cellular H2O2 production (Fig. 5B) would suggest that Cys-SSnH indeed appears to be a preferred target compared to Cys-SH for oxidation to corresponding Cys-SSnOH intermediates, in agreement with our DFT calculations and LC-MS/MS analysis.
Fig. 6.
(A) Representative Western blot analysis of Cys-SSH within EGFR and Src in H292 cells, by lysis in the presence of IAB, following by avidin capture and initial elution in the presence or absence of DTT (-RSB) and subsequent elution with SDS-containing reducing sample buffer (RSB). (B) Relative band densities of Western blot analysis of avidin-captured proteins, normalized to input lysates (with ratios of –DTT; + RSB sample arbitrarily set at 1). Data represents means ± s.e.m from 3 replicates.
4. Conclusions
In summary, the present work indicates that dimedone trapping approaches that are commonly used to detect protein sulfenylation (Cys-SOH) can also detect Cys-SSnOH species, and implicates Cys-SSOH as a previously unrecognized species as a potential additional intermediate in thiol-based redox signaling. Importantly, while previous efforts to identify sulfenylated proteins, in so-called “sulfenomes”, commonly use biotin-tagging approaches and avidin capture under reducing conditions [7], [47], [48], alternative approaches have been reported that do not and would potentially also include Cys-SSOH in their sulfenome analysis [49]. Additionally, evidence is emerging for the biological significance of Cys-SOH in redox-dependent activation of e.g. protein kinases such as EGFR and Src [11], [50], [51], the functional importance of Cys-SSnH or Cys-SSnOH within these proteins is still unclear. Given the general importance of non-catalytic cysteine residues within protein kinases in redox regulation or as a target for irreversible kinase inhibitors [52], [53], our findings warrant future studies to assess the biological relevance of Cys-SSnH and Cys-SSnOH intermediates within these proteins. Identification of these and related novel intermediates further exemplifies the expanding pleiotropic nature of redox-dependent protein regulation by reversible cysteine modifications.
Acknowledgements
The authors gratefully acknowledge the following sources of research support: National Institutes of Health Grants R01HL085646 and ES021476 to AvdV; Fellowship F32 HL129706 to DEH; Grants-in-Aid for Scientific Research from the Ministry of Education, Sciences, Sports, and Technology (MEXT), Japan, to TA (26111008, 26111001, 15K21759, 25253020, 16K15208); and research grants from the Deutsche Forschungsgemeinschaft (DFG; grant SFB1036 to TPD) and the Hungarian National Science Foundation (OTKA; grant No.: K109843 to PN). Computational resources were provided to JL by Vermont Advanced Computing Core (VACC) and Stampede from the Extreme Science and Engineering Discovery Environment (XSEDE, NSF Grant No. ACI-1053575). The authors also thank Dr. Christopher J. Cramer for helpful discussions and comments.
Footnotes
The authors declare no competing financial interests.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.10.006.
Contributor Information
Takaaki Akaike, Email: takaike@med.tohoku.ac.jp.
Albert van der Vliet, Email: albert.van-der-vliet@med.uvm.edu.
Appendix A. Supplementary material
Supplementary material
References
- 1.Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 3.Claiborne A., Yeh J.I., Mallett T.C., Luba J., Crane E.J., Charrier V., Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38:15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- 4.Poole L.B., Karplus P.A., Claiborne A. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 5.Gupta V., Carroll K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta. 2013:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heppner D.E., Janssen-Heininger Y.M.W., van der Vliet A. The role of sulfenic acids in cellular redox signaling: reconciling chemical kinetics and molecular detection strategies. Arch. Biochem. Biophys. 2017;616:40–46. doi: 10.1016/j.abb.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson K.J., Klomsiri C., Codreanu S.G., Soito L., Liebler D.C., Rogers L.C., Daniel L.W., Poole L.B. Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods Enzymol. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta V., Carroll K.S. Profiling the reactivity of cyclic C-nucleophiles towards electrophilic sulfur in cysteine sulfenic acid. Chem. Sci. 2016;7:400–415. doi: 10.1039/c5sc02569a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta V., Yang J., Liebler D.C., Carroll K.S. Diverse Redoxome Reactivity profiles of carbon nucleophiles. J. Am. Chem. Soc. 2017;139:5588–5595. doi: 10.1021/jacs.7b01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulsen C.E., Truong T.H., Garcia F.J., Homann A., Gupta V., Leonard S.E., Carroll K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truong Thu H., Ung, Peter M.-U., Palde Prakash B., Paulsen Candice E., Schlessinger A., Carroll Kate S. Molecular basis for redox activation of epidermal growth factor receptor kinase. Cell Chem. Biol. 2016;23:837–848. doi: 10.1016/j.chembiol.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heppner D.E., Hristova M., Dustin C.M., Danyal K., Habibovic A., van der Vliet A. The NADPH oxidases DUOX1 and NOX2 play distinct roles in redox regulation of epidermal growth factor receptor signaling. J. Biol. Chem. 2016;291:23282–23293. doi: 10.1074/jbc.M116.749028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forman H.J., Davies M.J., Krämer A.C., Miotto G., Zaccarin M., Zhang H., Ursini F. Protein cysteine oxidation in redox signaling: caveats on sulfenic acid detection and quantification. Arch. Biochem. Biophys. 2017;617:26–37. doi: 10.1016/j.abb.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuevasanta E., Lange M., Bonanata J., Coitiño E.L., Ferrer-Sueta G., Filipovic M.R., Alvarez B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 2015;290:26866–26880. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., Yamamoto M., Ono K., Devarie-Baez N.O., Xian M., Fukuto J.M., Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dóka É., Pader I., Bíró A., Johansson K., Cheng Q., Ballagó K., Prigge J.R., Pastor-Flores D., Dick T.P., Schmidt E.E., Arnér E.S.J., Nagy P. A novel persulfide detection method reveals protein persulfide-and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2016;2:e1500968. doi: 10.1126/sciadv.1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akaike T., Ida T., Wei F., Nishida M., Muagai Y., Alam M.M., ihara H., Sawa T., Matsunaga T., Kasamatsu S., Nishimura A., Morita M., Tomizawa K., Nishimura A., Wantanabe S., Inaba K., Shima H., Tanuma N., Jung M., Fujii S., Wantanabe Y., Ohmuraya M., Nagy P., Feelisch M., Fukuto J.M., Motohashi H. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017 doi: 10.1038/s41467-017-01311-y. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul B.D., Snyder S.H. H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- 19.Toohey J.I. Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem. 2011;413:1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 20.Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol. Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 21.Kimura H. Hydrogen sulfide: its production, release and functions. Amino Acids. 2011;41:113–121. doi: 10.1007/s00726-010-0510-x. [DOI] [PubMed] [Google Scholar]
- 22.Kashfi K., Olson K.R. Biology and therapeutic potential of hydrogen sulfide and hydrogen sulfide-releasing chimeras. Biochem. Pharmacol. 2013;85:689–703. doi: 10.1016/j.bcp.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono K., Akaike T., Sawa T., Kumagai Y., Wink D.A., Tantillo D.J., Hobbs A.J., Nagy P., Xian M., Lin J. Redox chemistry and chemical biology of H 2 S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic. Biol. Med. 2014;77:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida M., Sawa T., Kitajima N., Ono K., Inoue H., Ihara H., Motohashi H., Yamamoto M., Suematsu M., Kurose H., van der Vliet A., Freeman B.A., Shibata T., Uchida K., Kumagai Y., Akaike T. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012;8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianco C.L., Chavez T.A., Sosa V., Saund S.S., Nguyen Q.N.N., Tantillo D.J., Ichimura A.S., Toscano J.P., Fukuto J.M. The chemical biology of the persulfide (RSSH)/perthiyl (RSS·) redox couple and possible role in biological redox signaling. Free Radic. Biol. Med. 2016;101:20–31. doi: 10.1016/j.freeradbiomed.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stottmeier B., Dick T.P. Redox sensitivity of the MyD88 immune signaling adapter. Free Radic. Biol. Med. 2016;101:93–101. doi: 10.1016/j.freeradbiomed.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Sham D., Wesley U.V., Hristova M., van der Vliet A. ATP-mediated transactivation of the epidermal growth factor receptor in airway epithelial cells involves DUOX1-dependent oxidation of Src and ADAM17. PLoS One. 2013;8:e54391. doi: 10.1371/journal.pone.0054391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klomsiri C., Nelson K.J., Bechtold E., Soito L., Johnson L.C., Lowther W.T., Ryu S.E., King S.B., Furdui C.M., Poole L.B. Use of dimedone-based chemical probes for sulfenic acid detection evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 2010;473:77–94. doi: 10.1016/S0076-6879(10)73003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochevarov A.D., Harder E., Hughes T.F., Greenwood J.R., Braden D.A., Philipp D.M., Rinaldo D., Halls M.D., Zhang J., Friesner R.A. Jaguar: a high‐performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013;113:2110–2142. [Google Scholar]
- 30.Zhao Y., Truhlar D.G. A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interactions. J. Chem. Phys. 2006;125:194101. doi: 10.1063/1.2370993. [DOI] [PubMed] [Google Scholar]
- 31.Franzi R., Geoffroy M., Bernardinelli G. Photolytic damage in triphenylmethyldisulphane single crystals: the crystal structure of Ph3CSSH and the ESR study of Ph3CSS radical pairs. Mol. Phys. 1984;52:947–954. [Google Scholar]
- 32.Bailey T.S., Zakharov L.N., Pluth M.D. Understanding hydrogen sulfide storage: probing conditions for sulfide release from hydrodisulfides. J. Am. Chem. Soc. 2014;136:10573–10576. doi: 10.1021/ja505371z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marenich A.V., Olson R.M., Kelly C.P., Cramer C.J., Truhlar D.G. Self-consistent reaction field model for aqueous and nonaqueous solutions based on accurate polarized partial charges. J. Chem. Theory Comput. 2007;3:2011–2033. doi: 10.1021/ct7001418. [DOI] [PubMed] [Google Scholar]
- 34.Olson R.M., Marenich A.V., Cramer C.J., Truhlar D.G. Charge model 4 and intramolecular charge polarization. J. Chem. Theory Comput. 2007;3:2046–2054. doi: 10.1021/ct7001607. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y., Truhlar D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]
- 36.Becke A.D. Density‐functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 37.Perdew J.P., Burke K., Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77:3865. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 38.Adamo C., Barone V. Toward reliable density functional methods without adjustable parameters: the PBE0 model. J. Chem. Phys. 1999;110:6158–6170. [Google Scholar]
- 39.Numakura T., Sugiura H., Akaike T., Ida T., Fujii S., Koarai A., Yamada M., Onodera K., Hashimoto Y., Tanaka R., Sato K., Shishikura Y., Hirano T., Yanagisawa S., Fujino N., Okazaki T., Tamada T., Hoshikawa Y., Okada Y., Ichinose M. Production of reactive persulfide species in chronic obstructive pulmonary disease. Thorax. 2017 doi: 10.1136/thoraxjnl-2016-209359. [DOI] [PubMed] [Google Scholar]
- 40.Gupta V., Paritala H., Carroll K.S. Reactivity, selectivity, and stability in sulfenic acid detection: a comparative study of nucleophilic and electrophilic probes. Bioconjugate Chem. 2016;27:1411–1418. doi: 10.1021/acs.bioconjchem.6b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisz J.A., Bechtold E., King S.B., Poole L.B., Furdui C.M. Thiol‐blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J. 2013;280:6150–6161. doi: 10.1111/febs.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagahara N., Nirasawa T., Yoshii T., Niimura Y. Is novel signal transducer sulfur oxide involved in the redox cycle of persulfide at the catalytic site cysteine in a stable reaction intermediate of mercaptopyruvate sulfurtransferase? Antioxid. Redox Signal. 2012;16:747–753. doi: 10.1089/ars.2011.4468. [DOI] [PubMed] [Google Scholar]
- 43.Hristova M., Habibovic A., Veith C., Janssen-Heininger Y., Dixon A.E., Geiszt M., van der Vliet A. Airway epithelial DUOX1 mediates allergen-induced IL-33 secretion and activation of type 2 immune responses. J. Allergy Clin. Immunol. 2016;137:1545–1556. doi: 10.1016/j.jaci.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habibovic A., Hristova M., Heppner D.E., Danyal K., Ather J.L., Janssen-Heininger Y.M.W., Irvin C.G., Poynter M.E., Lundblad L.K., Dixon A.E., Geiszt M., van der Vliet A. DUOX1 mediates persistent epithelial EGFR activation, mucous cell metaplasia, and airway remodeling during allergic asthma. JCI Insight. 2016;1:e88811. doi: 10.1172/jci.insight.88811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hristova M., Veith C., Habibovic A., Lam Y.-W., Deng B., Geiszt M., Janssen-Heininger Y.M., van der Vliet A. Identification of DUOX1-dependent redox signaling through protein S-glutathionylation in airway epithelial cells. Redox Biol. 2014;2:436–446. doi: 10.1016/j.redox.2013.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans J.V., Ammer A.G., Jett J.E., Bolcato C.A., Breaux J.C., Martin K.H., Culp M.V., Gannett P.M., Weed S.A. Src binds cortactin through an SH2 domain cystine-mediated linkage. J. Cell Sci. 2012;125:6185–6197. doi: 10.1242/jcs.121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leonard S.E., Reddie K.G., Carroll K.S. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 48.Yang J., Gupta V., Carroll K.S., Liebler D.C. Site-specific mapping and quantification of protein S-sulfenylation in cells. Nat. Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waszczak C., Akter S., Eeckhout D., Persiau G., Wahni K., Bodra N., Van Molle I., De Smet B., Vertommen D., Gevaert K. Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2014;111:11545–11550. doi: 10.1073/pnas.1411607111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Truong T.H., Carroll K.S. Redox regulation of EGFR signaling through cysteine oxidation. Biochemistry. 2012;51:9954–9965. doi: 10.1021/bi301441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heppner D.E., van der Vliet A. Redox-dependent regulation of epidermal growth factor receptor signaling. Redox Biol. 2016;8:24–27. doi: 10.1016/j.redox.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fry D.W., Bridges A.J., Denny W.A., Doherty A., Greis K.D., Hicks J.L., Hook K.E., Keller P.R., Leopold W.R., Loo J.A. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. Proc. Natl. Acad. Sci. USA. 1998;95:12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen M.S., Zhang C., Shokat K.M., Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material