Abstract
Circulating tumor cells (CTCs), a type of cancer cells that spreads from primary or metastatic tumors into the bloodstream, can lead to a new fatal metastasis. As a new type of liquid biopsy, CTCs have become a hot pursuit and detection of CTCs offers the possibility for early diagnosis of cancers, earlier evaluation of chemotherapeutic efficacy and cancer recurrence, and choice of individual sensitive anti-cancer drugs. The fundamental challenges of capturing and characterizing CTCs are the extremely low number of CTCs in the blood and the intrinsic heterogeneity of CTCs. A series of microfluidic devices have been proposed for the analysis of CTCs with automation capability, precise flow behaviors, and significant advantages over the conventional larger scale systems. This review aims to provide in-depth insights into CTCs analysis, including various nanomaterial-based microfluidic chips for the capture and detection of CTCs based on the specific biochemical and physical properties of CTCs. The current developmental trends and promising research directions in the establishment of microfluidic chips for the capture and detection of CTCs are also discussed.
Keywords: circulating tumor cells, nanomaterial, microfluidic chips, cancer.
Introduction
Cancer has become a leading cause of death and expected to grow worldwide with population aging and growth 1, 2. The major cause of death in patients with cancer is tumor metastasis, accounting for approximately 90% of the mortality. The mechanism of tumor metastasis has not been fully understood, but an important step of the metastatic process is the transport of cancer cells that spread through the blood from the primary tumor site 3-6.
Ashworth reported that tumor cells in human blood samples of patients with cancer were analogous to those in tumor tissue, standing for the first discovery of circulating tumor cells (CTCs). CTCs, defined as the small number of cancer cells spreading from primary tumor sites or metastatic lesions into the blood circulation, are responsible for the spread of cancer to distant sites 7. Numerous clinical studies has indicated that levels of CTCs are an indicator of survival in metastatic cancer patients, indicating CTCs can act as a biomarker in cancer diagnosis and therapy assessment 8-10. For example, if CTCs count is more than 5 CTCs per 7.5 mL of whole blood for patients with metastatic cancer, the patients have a lower survival rate 11. There are currently over 270 clinical trials that validate CTCs as biomarkers based on the registration at ClinicalTrials.gov, and numerous studies confirm that CTCs could be acted as prognostic markers for cancer (the survival time is longer when the CTC concentration is lower) 9-13.
In addition, capturing of CTCs directly from blood samples of cancer patients is a noninvasive method. As a new type of liquid biopsy, it does not require the extraction of tumor tissues from the cancer patients, but still offers some information about the cancer for early diagnosis of cancers and choice of individual sensitive anti-cancer drugs. Hence, it is highly essential for capturing CTCs directly from the patient's blood in cancer research.
However, capturing CTCs with high purity and efficiency is still a challenge 14, 15. Firstly, compared to a great deal of normal blood cells, CTCs are extremely rare in the patient blood stream (about one CTC in a billion blood cells can be found in patient's blood) 16. Secondly, as there is no unique biomarker for the identification of CTCs, the intrinsic heterogeneity of CTCs exhibits a challenge. And the last challenge is that the isolated CTCs should remain viable and pure, and be detachable for subsequent cellular characterization and functional analysis.
To date, various approaches have been developed for CTC capture and detection 17-21. However, there is only one product that has been approved by the US food and drug administration (FDA) for detecting CTCs in 2004. CellSearch, an immunomagnetic enrichment method, relies on targeting the epithelial cell adhesion molecule (EpCAM) specific to epithelial cells 22-24. It has been the most widely used approach and is still the only CTC detection approach with FDA clearance in clinical applications. In CellSearch, anti-EpCAM-coated magnetic beads capture CTCs of epithelial origin upon passing through the magnetic field. After capture, the retained cells will subsequently be treated with fluorescent antibody conjugates against epithelial markers (EpCAM+), a nuclear stain (DAPI+), cytokeratins (CK+), and a leukocyte marker (CD45-) for distinguishing CTCs from leukocytes, enabling the observation of the differences between CTCs and normal cells. CellSearch can reach an average recovery rate of blood samples by spiking with different numbers of tumor cells of no less than 80% 25-27.
Although many platforms have successfully established for the capture and detection CTC, the low sensitivity and selectivity, the high detection cost and the complicated detection process of these approaches are matters of concern 28, 29. Hence, it is necessary to perform advanced material interfaces to improve efficient capture and sensitive detection of rare CTCs for clinical cancer studies and applications. Moreover, microfluidic chip has become one of the mainstream technologies for CTC study due to some advantages including miniaturization, portability, cost-effectiveness and the abilities of single cell analysis and online isolation/detection 30-33. And a variety of microfluidic chip platforms have been developed for CTCs analysis 34-38.
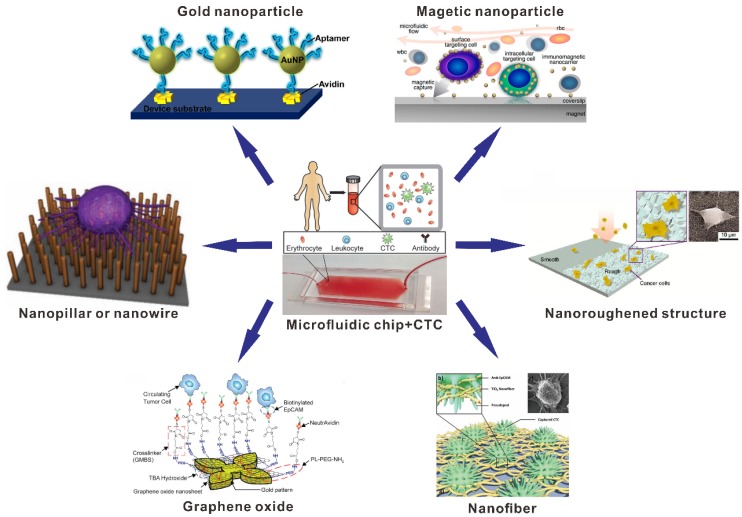
In this paper, we review recent significant progress in nanomaterial-based microfluidic chip for CTCs analysis. Firstly, we introduce the biochemical and physical properties of CTCs and the features were used for the design of microfluidic chip. Secondly, we will summarize the representative nanomaterial-based microfluidic chips performed for capture, detection and release of CTCs, with focus on the biochemical properties of CTCs (Figure 1). Lastly, the challenges and potential promising research directions regards microfluidic chips for CTCs capture and detection are also discussed.
Figure 1.
Schematic illustration of nanomaterial-based microfluidic chips (nanopillar, nanowire, gold nanoparticle, magnetic nanoparticle, graphene oxide, nanofiber and nanoroughened structure) for the capture and detection of circulating tumor cells (CTCs). Reprinted with permission from ref. 34, 67, 70, 75, 87, 88, 95, 96.
Physical and Biochemical Properties of CTCs
The key technical challenge in CTC study is isolation and detection and many CTC capture and detection platforms utilize the physical and biochemical features of CTCs 39. CTCs were first described in 1869 by Prof. Ashworth, when he microscopically observed the blood of a patient with cancer. CTCs identification was initially done by trained cytologists in term of fragmentation and elongated nuclei of the chromatin because of the similarity of CTC to the metastatic cancer cells. 40. Then many researchers investigated the gravity of CTCs. The specific gravity of CTCs and leukocytes in gravity gradient centrifugation was studies by Seal et al., and the results demonstrated that the specific gravity of CTCs was bigger than leukocytes and the method of gravity gradient centrifugation has become to be a potential method for CTCs capture and detection 41. In addition to the physical features of cell gravity, recent data suggested that CTC clusters may have 50 times greater metastatic potential than individual CTCs 42-44. With the development of fluorescent staining and microscopy technologies, more insight into the physical features of CTCs including cell deformability and cell size were studied recently. Many studies in breast cancer demonstrated that the size of CTCs was typically larger than normal blood cells, which was utilized as a criterion for CTCs detection 45, 46. Besides, high deformability is an anther significant mechanical feature for CTCs, especially for CTCs in order to go through the small-diameter capillaries and successfully metastasize 47. All the differences of physical features including size, gravity, and deformability between normal blood cells and CTCs, could be utilized for the enhanced capture of CTC from blood samples.
In addition, CTCs also express some unique biochemical properties that can be used for enhanced CTCs analysis. EpCAM is the most commonly used biochemical markers for the capture and detection of CTCs 48. EpCAM is a transmembrane glycoprotein mediating Ca2+-independent cell-cell adhesion in epithelia 49, 50. EpCAM is absent in blood cells, but it is overexpressed on the human adenocarcinoma cells. Therefore, EpCAM is one of the CTC-associated tumor markers, and CTC capture techniques are widely performed based on anti-EpCAM antibodies 51, 52. The CellSearch system employs a conjugation of EpCAM antibodies to magnetic beads for capturing CTCs through a magnetic field 25-27, 53.
For instance, one of the CTC affinity-based microfluidic chip platforms showed the remarkable gains in performance that could be achieved with a microfluidic chip approach. Toner's group reported a microfluidic chip platform based on anti-EpCAM antibody-coated microposts for CTCs capture and detection (Figure 2A) 54. Based on the specific interactions between the anti-EpCAM antibody-coated microposts and target CTCs, viable CTCs capture can be achieved from whole blood samples under precisely controlled laminar flow conditions. The results demonstrated that this microfluidic chip successfully captured CTCs from patients' blood samples with high sensitivity and without requisite pre-labelling or processing of samples. Toner's group also reported a herringbone-based microfluidic chip for CTCs analysis (Figure 2B) 55. The herringbone-based microchip consisted eight microchannels with patterned herringbone structures to generate microvortices for disrupting the laminar flow streamlines. The interactions between EpCAM antibody-coated surface and CTCs can be enhanced by providing passive mixing of blood cells in this microchip. Consequently, the high efficiency of CTCs capture was obtained in the artificial CTCs blood by spiking blood with different densities of tumor cells and clinical blood samples from cancer patients.
Figure 2.
Anti-EpCAM antibody-based representative microfluidic chips for CTC capture. (A) CTC-chip. Reprinted with permission from ref. 54. (B) Herringbone-chip. Reprinted with permission from ref. 55.
Many studies have also demonstrated that human epidermal growth factor receptor 2 (HER2) and epithelial growth factor receptor (EGFR) are expressed in CTCs of both early and metastatic cancer patients. The change of HER2 expression from low level to high level also occurred along with breast cancer progression and recurrence 48, 56, 57. Molecular analysis of CTCs from the blood sample of patients with lung cancer offers the possibility of monitoring changes in EGFR expression during the course of treatment 58. Therefore, HER2 and EGFR are considered to be CTCs-related tumor markers, and have also been widely developed for the capture and detection of CTCs. For example, Yu et al. have performed evidence of epithelial-mesenchymal transition (EMT) in human breast cancer specimens 48. Epithelial and mesenchymal markers were expressed in rare primary tumor cells, but mesenchymal cells were highly enriched in CTCs. And using a novel approach of three antibodies (EGFR, HER2, and EpCAM), breast cancer cells can be captured more efficiently.
Nanomaterial-Based Microfluidic Chip for CTC Capture
Nanomaterial-based nanostructured platforms have been performed to mimic the basement membrane and natural extracellular matrix 59, 60. The major advantage of the nanomaterial-based nanostructured surface is the enhanced local topographic interactions between nanoscale components of the cellular surface and nanostructures, resulting in improving the affinity of cell capture 61, 62. Therefore, nanostructured substrates can be combined with the affinity interactions-based CTC capture strategy in the microfluidic chip, which can enhance CTC capture efficiency and emerge as a promising approach for CTCs analysis. Different types of nanomaterial-based with microfluidic chips for CTCs capture and detection will be briefly introduced, including common nanopillar, nanowire, gold nanoparticle, magnetic nanoparticle, graphene oxide, nanofiber and nanoroughened structure.
Nanopillar and Nanowire
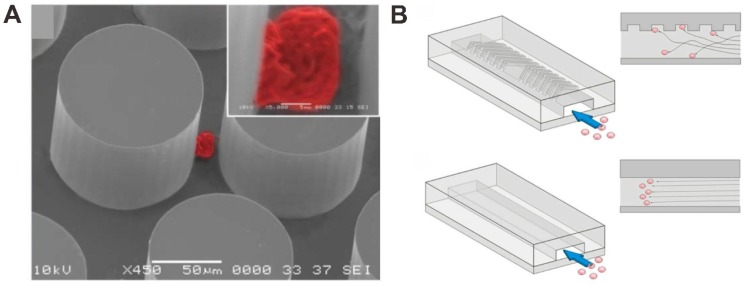
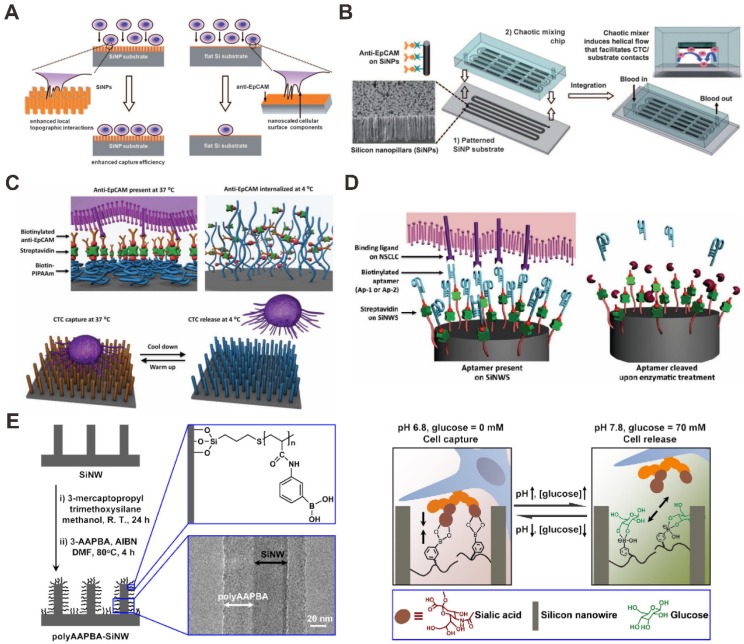
The nanopillar or nanowire-based substrates have been performed and utilized to enhance the CTCs capture in blood by using the surface adhesion of the cells. For instance, Wang et al. used anti-EpCAM antibody-modified silicon nanopillars (SiNPs) to enhance CTCs capture and detection (Figure 3A) 63. Firstly, nanopillars were prepared on silicon wafers by using a wet chemical etching approach. In addition, they can control the length of the nanopillars by changing the etching times. In order to test the performance of cell capture on the SiNPs, the MCF-7 cell line (an EpCAM-positive breast-cancer cell line) was introduced onto the SiNPs substrate and flat silicon substrates. The results demonstrated that the cell capture efficiency was 45-65% on SiNPs compared to only 4-14% on flat silicon substrates, showing that nanopillars with antibodies are available for improving cell capture. The performance of SiNPs on CTCs analysis was tested in the artificial CTCs blood samples by spiking different numbers of cancer cells into blood and the platform provides a convenient alternative for CTCs detection. Compared with SiNPs, quartz nanowires were also fabricated and developed for CTCs capture and detection in the artificial CTCs blood samples to evaluate the performance for clinical study 64. Besides, a uniform multiscale TiO2 nanorod array is fabricated to provide a “multi-scale interacting platform” for cell capture, which exhibits excellent capture specificity and sensitivity of the target cancer cells after modification with bovine serum albumin (BSA) and DNA aptamer 65. The capture yield of artificial blood samples on the BSA-aptamer TiO2 nanorod substrates is up to 85-95%, revealing the potential application of the TiO2 nanorods on efficient and sensitive capture of rare CTCs.
Figure 3.
Nanopillar (NP) and nanowire (NW) -based microfluidic chip for CTC capture and detection. (A) Anti-EpCAM antibody-coated SiNP substrate. Reprinted with permission from ref. 63. (B) Anti-EpCAM antibody -coating SiNP substrate with an overlaid microfluidic chaotic mixing chip. Reprinted with permission from ref. 66. (C) SiNW-based platform for CTC capture and release with temperature stimulation. Reprinted with permission from ref. 67. (D) SiNW-based platform for CTC capture and release with enzymatic treatment. Reprinted with permission from ref. 68. (E) SiNW-based platform for CTC capture and release with pH and glucose stimulation. Reprinted with permission from ref. 69
Furthermore, higher CTC capture efficiency could be achieved by increasing the contact frequency between nanopillar substrate and tumor cells in the microfluidic chip. Wang et al. also integrated SiNPs into a microfluidic chip with serpentine chaotic micromixers, obtaining a high capture efficiency of tumor cell (Figure 3B) 66. The CTCs capture microfluidic chip platform integrates two functional components: a long chaotic mixing channel in cell-substrate contact frequency and a patterned SiNPs substrate with anti-EpCAM antibody modifying for capturing EpCAM-expressing cells. In order to evaluate the CTCs capture performance of the integrated microfluidic chip platform, CTCs samples were prepared by spiking EpCAM-positive cancer cell lines into three kinds of solutions (PBS buffer, lysed blood, and whole blood). The results demonstrated that more than 95% of target tumor cell capture efficiency was performed in CTCs samples by the chip, providing an efficient approach for capturing and detecting of CTCs.
For nanowire surfaces-based CTCs capture methods, different approaches have been reported for CTCs capture and release. Hou et al. developed a microfluidic chip for CTC capture and release based on thermally responsive Poly(Nisopropylacrylamide) brushes-modified silicon nanowires (SiNWs) substrate (Figure 3C) 67. This approach showed perfect performances in capturing tumor cells at 37 °C with great efficiency, and releasing the captured tumor cells at 4 °C with high viability. At 37 °C, anti-EpCAM and hydrophobic domains of the polymer brushes are present on the surfaces of substrates, enabling CTC capture. When the temperature is reduced to 4 °C, the conformational change of the polymer brushes induces an internalization of anti-EpCAM, leading to CTC release. In another report, Shen et al. have developed an inexpensive and efficient CTC analysis capable of enriching, identifying and enumerating CTCs in blood samples from patients with prostate cancer (Figure 3D) 68. In the paper, by integrated CTC selective DNA aptamer with the SiNWs substrates, a novel integrated SiNWs microfluidic chaotic mixture was developed. This SiNWs-based microfluidic chip platform can not only improve CTC capture efficiency, but also realize controllable CTCs release via nuclease treatment. Recently, Liu et al. performed a glucose-responsive and pH strategy for CTCs capture and release based on poly(acrylamidephenylboronic acid)-grafted aligned SiNWs (Figure 3E) 69. Capturing and releasing of CTCs could be successfully achieved by precisely controlling the glucose and pH concentration in cell samples. With the increase of pH from 6.8 to 7.8 and the presence of glucose, the SiNWs substrate changed the state from cell-adhesive to cell-repulsive. Under the condition of pH 7.8, the substrate became glucose responsive, releasing targeted cells in the presence of glucose and capturing targeted cells in the absence of glucose. The SiNWs-based approach for capture and release of CTCs is available with higher cell viability, showing the potential for cancer diagnosis.
Nanoparticle
Gold Nanoparticle
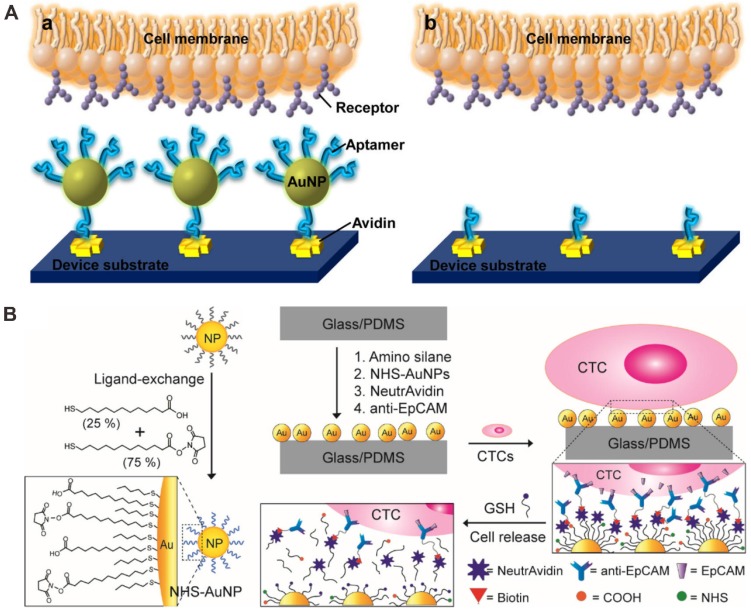
For the microfluidic chip deposited with by nanoparticles covalent binding or physical adsorption, the effects play a significant role in CTCs capture by enhancing ligand-cell interactions. Sheng et al. performed a novel microfluidic chip device combining multivalent nanoparticles for efficient capture and detection of CTCs (Figure 4A) 70. Up to 95 aptamer ligands were attached onto each gold nanoparticle (approximately 14 nm). When the microchannel of a microfluidic chip was modified by gold nanoparticles (AuNPs), an increase of 39-fold in binding affinity was developed compared with flat surface coated with aptamer alone. The results showed that efficiency of cell capture increased from 49% by using aptamer alone to 92% by using AuNPs-aptamer, indicating the strong potential of AuNPs-based microfluidic chip device for the analysis of CTCs.
Figure 4.
Gold nanoparticle (AuNP) -based microfluidic chip for CTC capture and detection. (A) Aptamer modified AuNP surface. Reprinted with permission from ref. 70. (B) Anti-EpCAM antibody modified AuNP surface for CTC capture and release by ligand exchange. Reprinted with permission from ref. 71.
Moreover, Park et al. utilized a thiolated ligand-exchange reaction with AuNPs on a herringbone chip to isolate and release cancer cells from whole blood (Figure 4B) 71. The AuNPs were composed of a mixed monolayer of 12-mercaptododecanoic acid N-hydroxysuccinimide ester and 11-mercaptoundecanoic acid. Then the functionalized AuNPs were immobilized within the herringbone channels through reactions with amine groups on the surface, and the chip with bound NeutrAvidin-AuNP assemblies was coated with anti-EpCAM antibodies via tetravalent biotin-NeutrAvidin binding to facilitate specific tumor cell binding. Using this microfluidic chip, the isolated cancer cells from mesenchymal and epithelial cancer cell lines as well as metastatic breast cancer patient samples can be recovered through simple thiol exchange reactions without any significant damage.
Magnetic nanoparticle
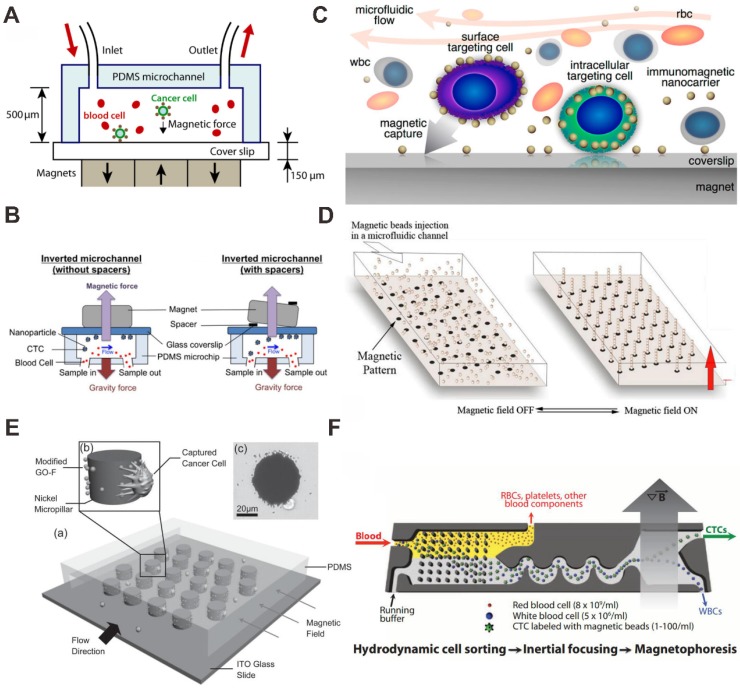
Magnetic nanoparticle (MNP) can be performed to make use of surface expression of biochemical properties of the tumor cell. In the presence of a magnetic field performed with permanent magnets under the chip, MNPs functionalized with anti-EpCAM antibody were utilized to capture target tumor cells. The sample throughput and capture efficiency can be precisely controlled through the design of the microfluidic chips, the magnetic field strength, and the flow rate.
As a breakthrough technology, micro-nuclear magnetic resonance technology detects target with MNPs, showing potential in highly sensitive and rapid detection. Lee et al. studied a miniaturized diagnostic magnetic resonance (DMR) platform for analysis of mammalian cells 72. The handheld DMR device can perform measurements on cancer cells by using magnetic particles as a proximity sensor for amplifying molecular interactions.
Zhang et al. have studied both theoretically and experimentally the effects of parameters on the capture efficiency of tumor cell (Figure 5A) 73. They demonstrated that more than 85% spiked tumor cells in blood samples can be captured with anti-EpCAM antibody modified Fe3O4 nanoparticles using block magnets at a flow rate of 10 mL/h. They also studied that inverting the microfluidic channel (magnet placed on top of the microfluidic chip) can improve the performance of the CTCs separation (Figure 5B) 74. In this method, the direction of the magnetic field force is opposite to that of the gravity. Hence, the effect of red blood cell sedimentation on the capture of CTC is greatly reduced.
Figure 5.
Magnetic nanoparticle (MNP) -based microfluidic chip for CTC capture and detection. (A) Anti-EpCAM antibody functionalized MNP bind to CTCs with the magnet placed on bottom of the channel. Reprinted with permission from ref. 73. (B) Anti-EpCAM antibody functionalized MNP bind to CTCs with the magnet placed on top of the channel. Reprinted with permission from ref. 74. (C) Antibody functionalized AuNP-Fe3O4 bind to CTCs. Reprinted with permission from ref. 75. (D) Antibody functionalized MNP self-assembly for CTC capture. Reprinted with permission from ref. 78. (E) Graphite oxide (GO) -coated MNPs with a micropillar device for CTC capture and release. Reprinted with permission from ref. 77. (F) Antibody functionalized MNPs bind to CTCs with a microstructured substrate for CTC sort, capture and detection. Reprinted with permission from ref. 80.
To improve the stability and biocompatibility of the MNPs, some modified methods are used, for example AuNPs modified MNP 75 and graphite-coated MNP 76, 77. Using Fe3O4-Au core-shell NPs with the microfluidic chip-based immunomagnetic separation, Wu et al. demonstrated that the capture efficiency of CTC can be highly improved by duplex targeting (Figure 5C) 75. CTCs were targeted from blood samples by anti-EpCAM antibody-functionalized MNPs and isolated by magnetic force in a microfluidic chamber with high capture efficiency. In order to reduce the need for macroscopic biopsies and generalize the use of fine needle aspirates. Saliba et al. proposed a novel method for cell sorting by using columns of biofunctionalized superparamagnetic beads self-assembled in a channel of microfluidic chip onto an array of magnetic traps prepared (Figure 5D) 78. On the mixtures of cells from culture cell lines (leukemia cell line and lymphoid cell line), the results demonstrated the cell capture efficiency was better than 94%, and the possibility to cultivate in situ the captured cells.
In some devices, the microstructures are designed to facilitate or enhance cell capture with MNPs in microfluidic chips 77, 79. For example, using graphite-coated MNP with the microchip-based immunomagnetic separation, Yu et al. reported a micropillar device for capturing and releasing of cancer cells (Figure 5E) 77. Graphite oxide-modified Fe3O4 MNPs were functionalized with a specific anti-EpCAM antibody. Under magnetic field manipulation, nickel micropillars attracted MNPs for increased interaction with cells and antibody presentation. Greater than 40% efficiency of cell capture in blood sample was achieved. By removing the magnetic field, 92.9% of captured cells were released and 78% of the released cancer cells are viable. A breakthrough in marker-free isolation of CTCs was enabled by an inertial focusing chip and a negative depletion strategy, also developed by Toner's group 80, 81. The first stage within the chip used hydrodynamic size-based sorting to retain white blood cells (WBCs) and CTCs (Figure 5F) 80. Firstly, the sample of WBCs or CTCs is incubated with antibody-functionalized MNPs. After labeling, the cells are loaded into the chip and separated into a bulk fraction. If WBCs are captured with MNPs, the CTCs can be isolated without the use of a specific marker for their identification. The combination of a high-throughput, broadly applicable and automatable rare CTCs sorting approach with cytology standards and accepted molecular assays will make the integration of CTC-based diagnostics into the clinical study of cancer.
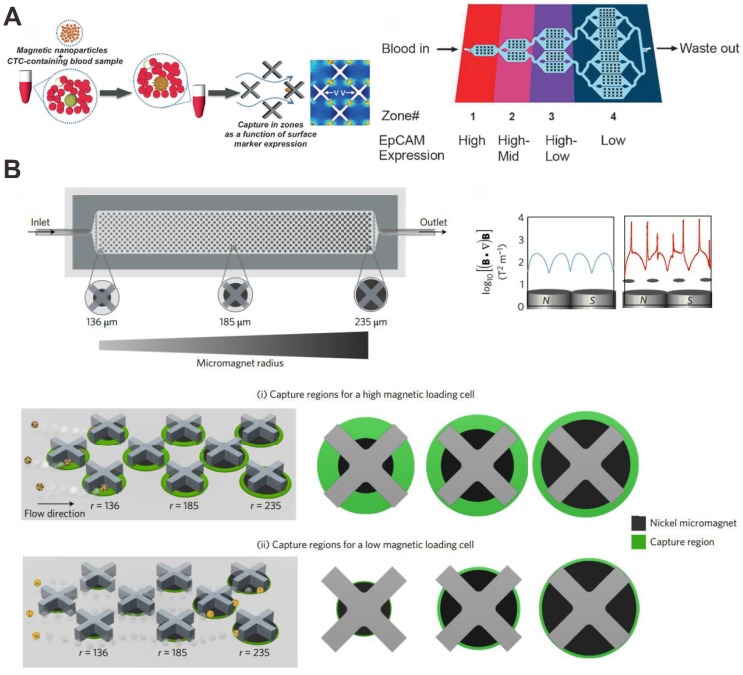
A recent advance in the separation of CTC subpopulations with a microfluidic device was realized by possessing different zones that would selectively capture cells with different levels of surface maker and MNPs. Kelley's group has performed a series of work about this 82-86. Mohamadi et al. analyzed the levels of surface protein expression with MNPs functionalized with specific antibodies using microfluidics, and used this type of phenotypic information to provide a profile of subpopulations of CTCs present in patient samples (Figure 6A) 84. CTCs with anti-EpCAM antibody functionalizing MNPs can be sorted corresponding to the levels of a surface marker. Many existing devices seek simply to isolate the set of all CTCs in a sample, and do not separate them into distinct subpopulations. To address this problem, microfabricated X-shaped structures were patterned within the device to create regions of low flow, allowing high EpCAM CTCs to be separated from low EpCAM CTCs. The microfluidic chip has been demonstrated to capture low numbers of CTCs from blood, and was proven to be effective in sorting tumor cells with varying levels of EpCAM expression.
Figure 6.
MNP and X-shaped microstructure-based microfluidic chip for CTC capture and detection. (A) Anti-EpCAM antibody functionalized MNP bind to CTCs with X-shaped velocity valleys. Reprinted with permission from ref. 84. (B) Anti-EpCAM antibody functionalized MNP bind to CTCs with an array of X-shaped structures varied magnetic capture zones. Reprinted with permission from ref. 85.
On the basis of this, Poudineh et al. report a new method for CTC characterization, called magnetic ranking cytometry, which allows us to profile the heterogeneous CTC subpopulations (Figure 6B) 85. The approach leverages immunomagnetic separation for profiling CTCs as a function of their surface marker expression. X-shaped structures within the microfluidic channel generate regions with slow flow and favorable capture dynamics, a requirement for the capture of cancer cells that are tagged with anti-EpCAM antibody functionalizing MNPs; whereas highly discretized sorting of subpopulations is achieved via the introduction of differently sized nickel micromagnets. The micromagnets are positioned concentrically within the X-shaped microstructures, creating regions with low flow and high magnetic field gradients that are ideal for capturing CTCs with even low levels of magnetic loading. The approach classifies CTCs with single-cell resolution in accordance with their expression of phenotypic surface markers, even in the presence of whole blood.
Besides, the ability to process whole blood, capture CTCs, lyse the cancer cells, and analyze mRNA expression by using microfluidic chips has been further developed 86. The CTCs bound to anti-EpCAM antibody-labeled MNPs are captured in the microfluidic device with microfabricated X-shaped structures, and analyzed for gene expression profiles by using nanostructured microelectrode biosensors. This entire workflow was carried out within a single integrated microfluidic device and was completed within 30 min. The results demonstrate that the gene expression module accurately profiles the expression of prostate-specific genes in CTCs captured from whole blood.
Graphene Oxide
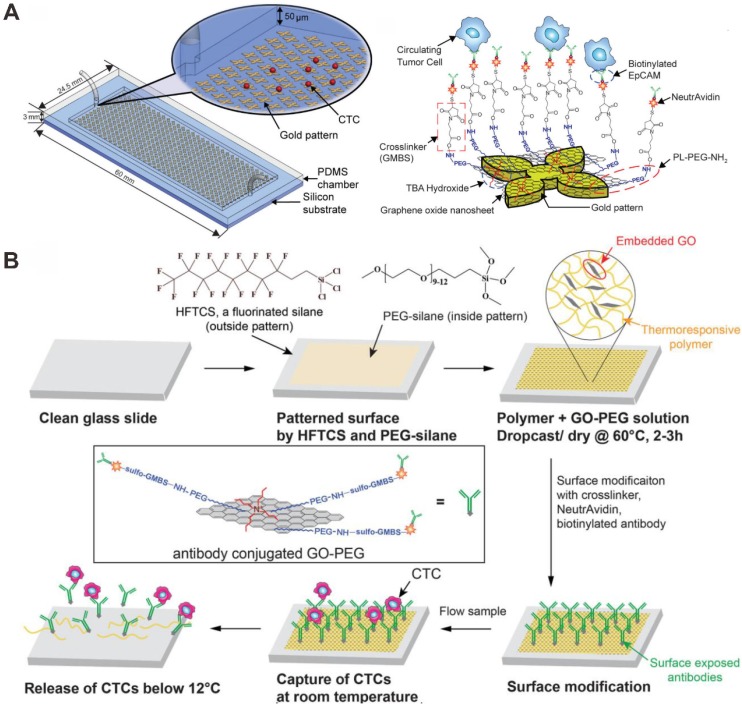
Graphene oxide (GO) is a promising nanomaterial as a component in applications, such as biosensors for cancer cell and DNA detection, polymer composites and paper-like materials. GO sheets have been shown for enhancing cell proliferation due to their biocompatibility with cells. Yoon et al. performed the GO-based microfluidic chip for CTCs capture and detection (Figure 7A) 87. GO nanosheets were able to self-assemble on the patterned gold surface by electrostatic attraction. A series of linker chemistries, such as cross-linker and biotin-avidin chemistry, were utilized to ultimately functionalize the substrate with an anti-EpCAM antibody. Human breast cancer cell lines (MCF-7 and Hs-578T) and a human prostate cancer cell line (PC-3) were spiked into buffer or blood and flowed through the microchip. The captured cells were cultured on the patterned gold surface with GO sheets. Due to the low number of CTCs, the culture of captured CTCs would allow for downstream analysis. The results demonstrated that the microchip can isolate CTCs from early stage cancer patients. However, this microfluidic chip device shares the common drawback across most immunoaffinity based technologies: the limitation of post-capture analysis because of difficulty in releasing viable cells from the capture microfluidic chip.
Figure 7.
Graphene oxide (GO) -based microfluidic chip for CTC capture and detection. (A) Anti-EpCAM antibody functionalized GO bind to CTCs. Reprinted with permission from ref. 87. (B) Anti-EpCAM antibody functionalized polymer-GO for the capture and release of CTCs. Reprinted with permission from ref. 88.
Subsequently, Yoon et al. designed a novel microfluidic chip, which was coated with a composite film of functionalized GO dispersed in a matrix of thermal-sensitive polymer with a lower critical solution temperature (Figure 7B) 88. The polymer matrix provided temperature dependent modulation of capture or release functionality. For EpCAM-positive cancer cells (MCF-7 breast cancer cells, LNCaP prostate cancer cells, and H1650 lung cancer cells), the anti-EpCAM antibody-modified GO-polymer microchip platform achieved high capture efficiency of CTCs (84.93-95.21%). After cell release, the remained cells exhibited 91.68% viability. The microfluidic chip can make it possible for various downstream analyses that require integrity of the targeted cell population.
Other Nanomaterial
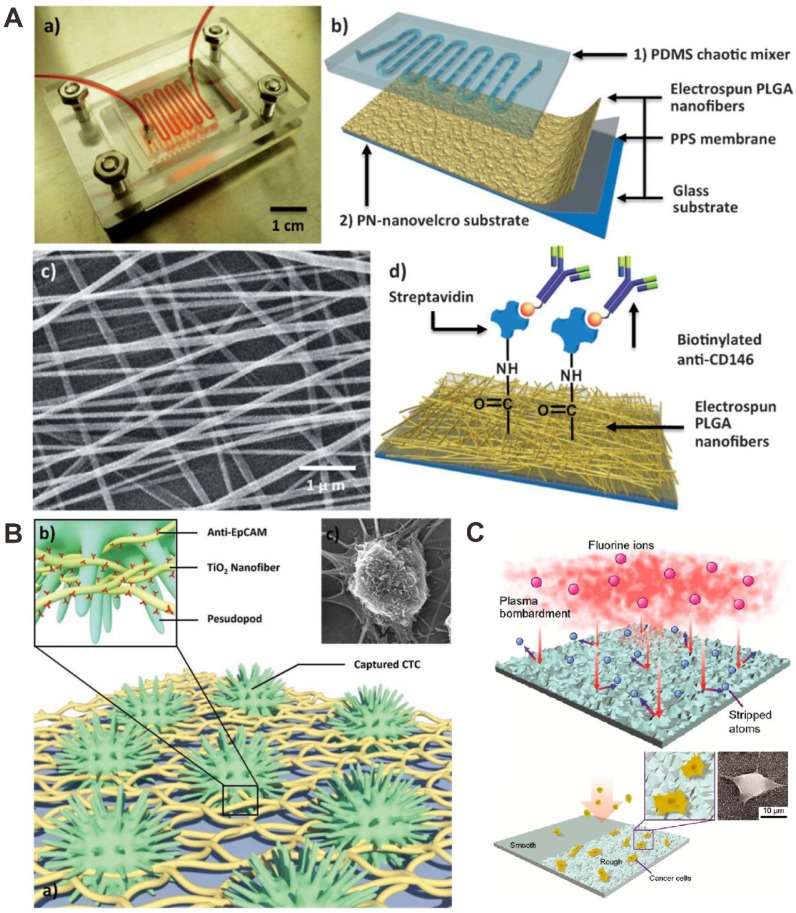
Hou et al. performed a polymer nanofiber-embedded microfluidic chips, abbreviated PN-nanovelcro chip, which were developed for the detection, specific isolation and molecular analysis of single CTC (Figure 8A) 89. The microchip was composed of two functional components: an overlaid poly(dimethylsiloxane) (PDMS) chaotic mixer and a nanovelcro substrate, which was fabricated by electrospining poly(lactic-co-glycolic acid) (PLGA) nanofibers onto the poly(phenylene sulfide) (PPS) membrane, followed by streptavidin-mediated conjugation of a cell-capture antibody. Target tumor cells were efficiently captured, and cell isolation was subsequently isolated by using the commercial laser microdissection technique with the enhanced local interaction between PLGA nanofibers and tumor cell.
Figure 8.
Other material-based microfluidic chip for CTC capture and detection. (A) Anti-CD146 antibody modified polymer poly(lactic-co-glycolic acid) (PLGA) nanofiber for the capture and isolation of CTCs. Reprinted with permission from ref. 89. (B) Anti-EpCAM antibody modified TiO2 nanofiber for improved CTC capture. Reprinted with permission from ref. 95. (C) Nanorough glass surfaces for label-free capture of CTCs. Reprinted with permission from ref. 96.
The electrospun nanofibers are easy to fabricate, have a large surface-to-volume ratio and high porosity. Recently, electrospun nanofibers have been used as substrates to capture CTCs 90-94. For example, Shi's group used hyaluronic acid-functionalized electrospun PLGA nanofibers embedded in a microfluidic chip for the capture and detection of CTCs 94. The hyaluronic acid was covalently conjugated onto polyethyleneimine-modified electrospun PLGA nanofibers. The results show that the CD44 overexpressing CTCs could be selectively captured by the nanofibers in the microfluidic chip platform and this method effectively captured cancer cells of different origins from blood samples.
Zhang et al. developed a new CTC detection assay based on an electrospun TiO2 nanofibers-deposited substrate (Figure 8B) 95. The electrospun TiO2 nanofibers better mimic these horizontally oriented nanostructures for improving cell affinity. The CTCs capture platform was prepared with cell capture agent conjugated onto the surface of TiO2 nanofibers. Using nanofibers modified nanosubstrates, tumor cells from artificial CTCs blood samples and blood samples of cancer patients were efficiently captured and detected.
Compared to normal blood cells, the differential adhesion preference of CTCs makes nanoroughened surfaces as an alternative approach for CTCs detection. The adhesion strength of cancer cells can be affected by nanotopographic sensing, while adhesion of normal blood cells cannot be sensitive to nanotopographic cues. Chen et al. reported an effective and simple strategy for CTCs capture using nanorough glass surfaces (Figure 8C) 96. The results demonstrated that the cell surface area can be increased with nanoroughened surfaces for cell adhesion, and reactions. When nanoroughened surfaces are performed, the use of capture antibodies for CTC is not necessary. This method is expected to lead to better enrichment and isolation strategies for accurate diagnosis and therapeutic choices.
Conclusions and Future Perspectives
Although CTC has become a hotspot research field and many CTC capture and detection technologies have been developed, translation of these technologies from laboratory to clinical practice is nontrivial. The CellSearch system is the first and only CTC product approved by the FDA for CTC analysis in cancer diagnosis more than a decade ago. In this review, we discussed the existing nanomaterial-based microfluidic chips for CTC detection. Without capture, CTCs cannot be isolated form human blood for subsequent phenotype identification and molecular analysis. With ligand capture, the process of CTC capture and detection is time-consuming and automation is not currently possible. With microfluidic chips and ligand capture, the CTC without expression (or very low expression) of the biomarker (e.g. EpCAM) cannot be captured. With nanomaterial-based microfluidic chips and ligand capture, the CTCs with very low expression of the biomarker (e.g. EpCAM) can be captured, but the slow flow rate of the microfluidic results in a long time for CTC capture and detection.
Nanomaterials, with high surface-area-to-volume ratio, can address the problems of low purity and insufficient cancer cell capture efficiency. While nanomaterial-based nanostructured platforms are promising for the CTC capture and detection, most of them performed perfect in laboratory study. High capture efficiency and purity are still problems for these approaches to be available to clinical practice. In addition, the nonspecific capture of blood cells is still a problem for further enhancing the purity of cell capture. The combination of nanomaterials with microfluidic chip platforms are suggested, and throw light on the capture and detection of CTC.
For future CTC study, microfluidic chip devices are designed to meet the standards: i) shorter analytical time for CTC capture and detection; ii) enhanced capture efficiency, detection specificity and sensitivity; iii) enhanced capability for sequential analysis after capturing and releasing; iv) enhanced capability in operating procedures for high-throughput. For future development, single cell resolution analysis, e.g. single cell sequencing, single cell proteomics, is essential for CTCs characterization, which has significance in biology and clinics. Furthermore, nanomaterial-based microfluidic chip devices will be performed in achieving point-of-care cancer diagnosis and play an important role in developing personalized therapeutics for cancer patients.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81601571, 31370868 and 21375152), the Medical Scientific Research Foundation of Guangdong Province (No. A2017033), the Fundamental Research Funds for the Central Universities (No. 16ykzd13) and a Start-up Grant from Sun Yat-Sen University.
Abbreviations
- AuNPs
gold nanoparticles
- BSA
bovine serum albumin
- CTCs
circulating tumor cells
- DMR
diagnostic magnetic resonance
- EGFR
epithelial growth factor receptor
- EMT
epithelial-mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- FDA
food and drug administration
- GO
graphene oxide
- HER2
human epidermal growth factor receptor 2
- MNP
magnetic nanoparticle
- PDMS
polydimethylsiloxane
- PLGA
polylactic-co-glycolic acid
- PPS
polyphenylene sulfide
- SiNPs
silicon nanopillars
- SiNWs
silicon nanowires
- WBCs
white blood cells.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. Ca-Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Ca-Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 6.Khan N, Mukhtar H. Cancer and metastasis: prevention and treatment by green tea. Cancer Metast Rev. 2010;29:435–45. doi: 10.1007/s10555-010-9236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaks V, Koopman CD, Werb Z. Circulating tumor cells. Science. 2013;341:1186–8. doi: 10.1126/science.1235226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng SD, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF. et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–62. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 9.Riethdorf S, Fritsche H, Mueller V, Rau T, Schindibeck C, Rack B. et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–8. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 10.Riethdorf S, Mueller V, Zhang L, Rau T, Loibl S, Komor M. et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16:2634–45. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 11.Dotan E, Cohen SJ, Alpaugh KR, Meropol NJ. Circulating tumor cells: evolving evidence and future challenges. Oncologist. 2009;14:1070–82. doi: 10.1634/theoncologist.2009-0094. [DOI] [PubMed] [Google Scholar]
- 12.Wong VCL, Ko JMY, Lam CT, Lung ML. Succinct workflows for circulating tumor cells after enrichment: From systematic counting to mutational profiling. Plos One. 2017;12:e0177276. doi: 10.1371/journal.pone.0177276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danila DC, Pantel K, Fleisher M, Scher HI. Circulating tumors cells as biomarkers progress toward biomarker qualification. Cancer J. 2011;17:438–50. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coumans FAW, Ligthart ST, Uhr JW, Terstappen LWMM. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res. 2012;18:5711–8. doi: 10.1158/1078-0432.CCR-12-1585. [DOI] [PubMed] [Google Scholar]
- 15.Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–31. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 17.Bhana S, Wang YM, Huang XH. Nanotechnology for enrichment and detection of circulating tumor cells. Nanomedicine-UK. 2015;10:1973–90. doi: 10.2217/nnm.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li YQ, Chandran BK, Lim CT, Chen XD. Rational design of materials interface for efficient capture of circulating tumor cells. Adv Sci. 2015;2:1500118. doi: 10.1002/advs.201500118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green BJ, Safaei TS, Mepham A, Labib M, Mohamadi RM, Kelley SO. Beyond the capture of circulating tumor cells: next-generation devices and materials. Angew Chem Int Edit. 2016;55:1252–65. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 20.Shen ZY, Wu AG, Chen XY. Current detection technologies for circulating tumor cells. Chem Soc Rev. 2017;46:2038–56. doi: 10.1039/c6cs00803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song YL, Tian T, Shi YZ, Liu WL, Zou Y, Khajvand T. et al. Enrichment and single-cell analysis of circulating tumor cells. Chem Sci. 2017;8:1736–51. doi: 10.1039/c6sc04671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H. et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 23.Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY. et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 24.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 25.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK. et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–32. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 26.Khan MS, Kirkwood A, Tsigani T, Garcia-Hernandez J, Hartley JA, Caplin ME. et al. Circulating tumor cells as prognostic markers in neuroendocrine tumors. J Clin Oncol. 2013;31:365–72. doi: 10.1200/JCO.2012.44.2905. [DOI] [PubMed] [Google Scholar]
- 27.Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A. et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–63. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 28.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F. et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Brit J Cancer. 2011;105:847–53. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M. et al. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer I. 2009;101:61–6. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YQ, Zhang WJ, Qin LD. Mesenchymal-mode migration assay and antimetastatic drug screening with high-throughput microfluidic channel networks. Angew Chem Int Edit. 2014;53:2344–8. doi: 10.1002/anie.201309885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang YQ, Zhou LD, Qin LD. High-throughput 3D cell invasion chip enables accurate cancer metastatic assays. J Am Chem Soc. 2014;136:15257–62. doi: 10.1021/ja5072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YQ, Wu MH, Han X, Wang P, Qin LD. High-throughput, label-free isolation of cancer stem cells on the basis of cell adhesion capacity. Angew Chem Int Edit. 2015;54:10838–42. doi: 10.1002/anie.201505294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun DP, Lu J, Chen ZG, Yu YY, Li YB. A novel three-dimensional microfluidic platform for on chip multicellular tumor spheroid formation and culture. Microfluid Nanofluid. 2014;17:831–42. [Google Scholar]
- 34.Qian WY, Zhang Y, Chen WQ. Capturing cancer: Emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small. 2015;11:3850–72. doi: 10.1002/smll.201403658. [DOI] [PubMed] [Google Scholar]
- 35.Yoon HJ, Kozminsky M, Nagrath S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. Acs Nano. 2014;8:1995–2017. doi: 10.1021/nn5004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myung JH, Hong S. Microfluidic devices to enrich and isolate circulating tumor cells. Lab Chip. 2015;15:4500–11. doi: 10.1039/c5lc00947b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong B, Zu YL. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377–94. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin M, Chen JF, Lu YT, Zhang Y, Song JZ, Hou S. et al. Nanostructure embedded microchips for detection, isolation, and characteriiation of circulating tumor cells. Accounts Chem Res. 2014;47:2941–50. doi: 10.1021/ar5001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HTK, Lee W. et al. Label-free cell separation and sorting in microfluidic systems. Anal Bioanal Chem. 2010;397:3249–67. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson O. Method for cytological detection of cancer cells in blood. Cancer. 1962;15:171–5. doi: 10.1002/1097-0142(196201/02)15:1<171::aid-cncr2820150123>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 41.Seal SH. Sieve for isolation of cancer cells and other large cells from blood. Cancer. 1964;17:637–42. doi: 10.1002/1097-0142(196405)17:5<637::aid-cncr2820170512>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 42.Seal SH. Silicone flotation: A simple quantitative method for the isolation of free-floating cancer cells from the blood. Cancer. 1959;12:590–5. doi: 10.1002/1097-0142(195905/06)12:3<590::aid-cncr2820120318>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 43.Alexander RF, Spriggs AI. The differential diagnosis of tumour cells in circulating blood. J Clin Pathol. 1960;13:414–24. doi: 10.1136/jcp.13.5.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheinin TM, Koivuniemi AP. Large benign cells in circulating blood and their significance in identification of cancer cells. Cancer. 1962;15:972–7. doi: 10.1002/1097-0142(196209/10)15:5<972::aid-cncr2820150512>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 45.Hsieh HB, Marrinucci D, Bethel K, Curry DN, Humphrey M, Krivacic RT. et al. High speed detection of circulating tumor cells. Biosens Bioelectron. 2006;21:1893–9. doi: 10.1016/j.bios.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 46.Marrinucci D, Bethel K, Bruce RH, Curry DN, Hsieh B, Humphrey M. et al. Case study of the morphologic variation of circulating tumor cells. Hum Pathol. 2007;38:514–9. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 47.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–3. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 48.Yu M, Bardia A, Wittner B, Stott SL, Smas ME, Ting DT. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomura T, Katunuma N. Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J Med Invest. 2005;52:1–9. doi: 10.2152/jmi.52.1. [DOI] [PubMed] [Google Scholar]
- 50.Pauli C, Munz M, Kieu C, Mack B, Breinl P, Wollenberg B. et al. Tumor-specific glycosylation of the carcinoma-associated epithelial cell adhesion molecule EpCAM in head and neck carcinomas. Cancer Lett. 2003;193:25–32. doi: 10.1016/s0304-3835(03)00003-x. [DOI] [PubMed] [Google Scholar]
- 51.Hughes AD, King MR. Use of naturally occurring halloysite nanotubes for enhanced capture of flowing cells. Langmuir. 2010;26:12155–64. doi: 10.1021/la101179y. [DOI] [PubMed] [Google Scholar]
- 52.Van der Auwera I, Peeters D, Benoy IH, Elst HJ, Van Laere SJ, Prove A. et al. Circulating tumour cell detection: a direct comparison between the CellSearch System, the AdnaTest and CK-19/mammaglobin RT-PCR in patients with metastatic breast cancer. Brit J Cancer. 2010;102:276–84. doi: 10.1038/sj.bjc.6605472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helo P, Cronin AM, Danila DC, Wenske S, Gonzalez-Espinoza R, Anand A. et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–73. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. P Natl Acad Sci USA. 2010;107:18392–7. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao S, Li Y, Li J, Li CF, Zhang W, Yang ZQ. et al. Quantitative determination of HER2 expression by confocal microscopy assay in CTCs of breast cancer. Oncol Rep. 2010;23:423–8. [PubMed] [Google Scholar]
- 57.Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T. et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. New Engl J Med. 2006;355:2733–43. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 58.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV. et al. Detection of mutations in EGFR in circulating lung-cancer cells. New Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacquemart I, Pamula E, De Cupere VM, Rouxhet P, Dupont-Gillain CC. Nanostructured collagen layers obtained by adsorption and drying. J Colloid Interf Sci. 2004;278:63–70. doi: 10.1016/j.jcis.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 60.Geckil H, Xu F, Zhang X, Moon S, Demirci U. Engineering hydrogels as extracellular matrix mimics. Nanomedicine-UK. 2010;5:469–84. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma ZW, Kotaki M, Inai R, Ramakrishna S. Potential of nanofiber matrix as tissue-engineering scaffolds. Tissue Eng. 2005;11:101–9. doi: 10.1089/ten.2005.11.101. [DOI] [PubMed] [Google Scholar]
- 62.Wang L, Asghar W, Demirci U, Wan Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today. 2013;8:347–87. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K-i. et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Edit. 2009;48:8970–3. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M. et al. Nanowire substrate-based laser scanning cytometry for quantitation of circulating tumor cells. Nano Lett. 2012;12:2697–704. doi: 10.1021/nl2041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun N, Li XP, Wang ZL, Zhang RH, Wang JE, Wang KW. et al. A multiscale TiO2 nanorod array for ultrasensitive capture of circulating tumor cells. Acs Appl Mater Inter. 2016;8:12638–43. doi: 10.1021/acsami.6b02178. [DOI] [PubMed] [Google Scholar]
- 66.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L. et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Edit. 2011;50:3084–8. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY. et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater. 2013;25:1547–51. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y. et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv Mater. 2013;25:2368–73. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu H, Li Y, Sun K, Fan J, Zhang P, Meng J. et al. Dual-responsive surfaces modified with phenylboronic acid-containing polymer brush to reversibly capture and release cancer cells. J Am Chem Soc. 2013;135:7603–9. doi: 10.1021/ja401000m. [DOI] [PubMed] [Google Scholar]
- 70.Sheng W, Chen T, Tan W, Fan ZH. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. Acs Nano. 2013;7:7067–76. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park MH, Reátegui E, Li W, Tessier SN, Wong KHK, Jensen AE. et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J Am Chem Soc. 2017;139:2741–9. doi: 10.1021/jacs.6b12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–74. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP. et al. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip. 2011;11:3449–57. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang YY, Hoshino K, Chen P, Wu CH, Lane N, Huebschman M. et al. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed Microdevices. 2013;15:673–81. doi: 10.1007/s10544-012-9718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu CH, Huang YY, Chen P, Hoshino K, Liu H, Frenkel EP. et al. Versatile immunomagnetic nanocarrier platform for capturing cancer cells. Acs Nano. 2013;7:8816–23. doi: 10.1021/nn403281e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Z, Hong G, Wang H, Welsher K, Tabakman SM, Sherlock SP. et al. Graphite-coated magnetic nanoparticle microarray for few-cells enrichment and detection. Acs Nano. 2012;6:1094–101. doi: 10.1021/nn2034692. [DOI] [PubMed] [Google Scholar]
- 77.Yu X, He R, Li S, Cai B, Zhao L, Liao L. et al. Magneto-controllable capture and release of cancer cells by using a micropillar device decorated with graphite oxide-coated magnetic nanoparticles. Small. 2013;9:3895–901. doi: 10.1002/smll.201300169. [DOI] [PubMed] [Google Scholar]
- 78.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC. et al. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. P Natl Acad Sci USA. 2010;107:14524–9. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim S, Han SI, Park MJ, Jeon CW, Joo YD, Choi IH. et al. Circulating tumor cell microseparator based on lateral magnetophoresis and immunomagnetic nanobeads. Anal Chem. 2013;85:2779–86. doi: 10.1021/ac303284u. [DOI] [PubMed] [Google Scholar]
- 80.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E. et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L, Gibbs T. et al. Nanoparticle-mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew Chem Int Edit. 2015;54:139–43. doi: 10.1002/anie.201409376. [DOI] [PubMed] [Google Scholar]
- 83.Labib M, Green B, Mohamadi RM, Mepham A, Ahmed SU, Mahmoudian L. et al. Aptamer and antisense-mediated two-dimensional isolation of specific cancer cell subpopulations. J Am Chem Soc. 2016;138:2476–9. doi: 10.1021/jacs.5b10939. [DOI] [PubMed] [Google Scholar]
- 84.Poudineh M, Labib M, Ahmed S, Nguyen LNM, Kermanshah L, Mohamadi RM. et al. Profiling functional and biochemical phenotypes of circulating tumor cells using a two-dimensional sorting device. Angew Chem Int Edit. 2017;56:163–8. doi: 10.1002/anie.201608983. [DOI] [PubMed] [Google Scholar]
- 85.Poudineh M, Aldridge PM, Ahmed S, Green BJ, Kermanshah L, Nguyen V. et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat Nanotechnol. 2017;12:274–281. doi: 10.1038/nnano.2016.239. [DOI] [PubMed] [Google Scholar]
- 86.Mohamadi RM, Ivanov I, Stojcic J, Nam RK, Sargent EH, Kelley SO. Sample-to-answer isolation and mRNA profiling of circulating tumor cells. Anal Chem. 2015;87:6258–64. doi: 10.1021/acs.analchem.5b01019. [DOI] [PubMed] [Google Scholar]
- 87.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol. 2013;8:735–41. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoon HJ, Shanker A, Wang Y, Kozminsky M, Jin Q, Palanisamy N. et al. Tunable Thermal-Sensitive Polymer-Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv Mater. 2016;28:4891–7. doi: 10.1002/adma.201600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X. et al. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Edit. 2013;52:3379–83. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao YL, Zhu XY, Liu H, Luo Y, Wang SG, Shen MW. et al. Dendrimer-functionalized electrospun cellulose acetate nanofibers for targeted cancer cell capture applications. J Mater Chem B. 2014;2:7384–93. doi: 10.1039/c4tb01278j. [DOI] [PubMed] [Google Scholar]
- 91.Zhao YL, Fan ZY, Shen MW, Shi XY. Capturing hepatocellular carcinoma cells using lactobionic acid-functionalized electrospun polyvinyl alcohol/polyethyleneimine nanofibers. Rsc Adv. 2015;5:70439–47. [Google Scholar]
- 92.Zhao YL, Fan ZY, Shen MW, Shi XY. Hyaluronic acid-functionalized electrospun polyvinyl alcohol/polyethyleneimine nanofibers for cancer cell capture applications. Adv Mater Interfaces. 2015;2:1500256. [Google Scholar]
- 93.Fan ZY, Zhao YL, Zhu XY, Luo Y, Shen MW, Shi XY. Folic acid modified electrospun poly(vinyl alcohol)/polyethyleneimine nanofibers for cancer cell capture applications. Chinese J Polym Sci. 2016;34:755–65. [Google Scholar]
- 94.Xu GW, Tan YL, Xu TG, Yin D, Wang MY, Shen MW. et al. Hyaluronic acid-functionalized electrospun PLGA nanofibers embedded in a microfluidic chip for cancer cell capture and culture. Biomater Sci-UK. 2017;5:752–61. doi: 10.1039/c6bm00933f. [DOI] [PubMed] [Google Scholar]
- 95.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q. et al. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24:2756–60. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 96.Chen WQ, Weng SN, Zhang F, Allen S, Li X, Bao LW. et al. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. Acs Nano. 2013;7:566–75. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]