Abstract
Adaptive responses to thermal stress in poikilotherms plays an important role in determining competitive ability and species distributions. Amino acid substitutions that affect protein stability and modify the thermal optima of orthologous proteins may be particularly important in this context. Here, we examine a set of 2,770 protein-coding genes to determine if proteins in a highly invasive heat tolerant blue mussel (Mytilus galloprovincialis) contain signals of adaptive increases in protein stability relative to orthologs in a more cold tolerant M. trossulus. Such thermal adaptations might help to explain, mechanistically, the success with which the invasive marine mussel M. galloprovincialis has displaced native species in contact zones in the eastern (California) and western (Japan) Pacific. We tested for stabilizing amino acid substitutions in warm tolerant M. galloprovincialis relative to cold tolerant M. trossulus with a generalized linear model that compares in silico estimates of recent changes in protein stability among closely related congeners. Fixed substitutions in M. galloprovincialis were 3,180.0 calories per mol per substitution more stabilizing at genes with both elevated dN/dS ratios and transcriptional responses to heat stress, and 705.8 calories per mol per substitution more stabilizing across all 2,770 loci investigated. Amino acid substitutions concentrated in a small number of genes were more stabilizing in M. galloprovincialis compared with cold tolerant M. trossulus. We also tested for, but did not find, enrichment of a priori GO terms in genes with elevated dN/dS ratios in M. galloprovincialis. This might indicate that selection for thermodynamic stability is generic across all lineages, and suggests that the high change in estimated protein stability that we observed in M. galloprovincialis is driven by selection for extra stabilizing substitutions, rather than by higher incidence of selection in a greater number of genes in this lineage. Nonetheless, our finding of more stabilizing amino acid changes in the warm adapted lineage is important because it suggests that adaption for thermal stability has contributed to M. galloprovincialis’ superior tolerance to heat stress, and that pairing tests for positive selection and tests for transcriptional response to heat stress can identify candidates of protein stability adaptation.
Keywords: allosteric function, Gibbs free energy, global warming, positive selection, protein stability, transcriptome
Introduction
Temperature plays a central role in governing rates of physiological activities and determining the stabilities of macromolecules such as proteins (Somero 1995; Hochachka and Somero 2002; Fields etal. 2015). Protein adaptation to temperature strongly influences an poikilothermic species’ competitive ability and plays an important role in determining where such species can survive and thrive (Sunday etal. 2011). There is growing concern that global climate change will have major effects on species’ distributions (Occhipinti-Ambrogi 2007; Rahel and Olden 2008; Somero 2010). One manifestation of this risk is the spread of invasive species. The past few decades have seen an increase in invasive species establishment and persistence in northern latitudes (Ruiz et al. 2000; Stachowicz et al. 2002; Hellmann et al. 2008). Therefore, it is important to understand the physiological and molecular basis of species-specific differences in temperature tolerance, especially when studying differences between closely related native and invasive species (Watt and Dean 2000; Petit 2004; Gu and Hilser 2009; Somero 2010; Lockwood and Somero 2011a; Fields etal. 2015).
Blue mussels of the genus Mytilus are a promising study system for the investigation of differences in physiological tolerance among closely related native and invasive species at the molecular level because of their recent divergence and their similar effective population sizes, levels of polymorphism, and rates of molecular evolution (Ahmed and Sparks 1970; Seed 1992; Sarver and Foltz 1993; Comesana et al. 1999; Bierne et al. 2003; Cruz et al. 2005; Saarman and Pogson 2015). It is known that Mytilus galloprovincialis evolved in the Mediterranean and is unique among blue mussels in its ability to invade and outcompete native congeners in new localities. For example, M. galloprovincialis has successfully colonized California, British Columbia, Japan, Chile, New Zealand, the Auckland Islands, southern Australia and South Africa (Grant and Cherry 1985; Seed 1992; Heath et al. 1995; Geller 1999; Rawson et al. 1999; Robinson et al. 2007; Westfall and Gardner 2010; Borsa et al. 2012). Physiological differences between M. galloprovincialis and other blue mussels have been documented for cardiac performance, metabolic rates, enzymatic activity levels, and mortality rates, especially at temperatures between 26 °C and 32 °C (Buckley et al. 2001; Braby and Somero 2006; Tomanek and Zuzow 2010; Dowd and Somero 2013). Since high temperatures in this thermal range are common in intertidal habitats during low tides (Dowd and Somero 2013), these studies suggest that superior thermal resistance might contribute to M. galloprovincialis’ capacity to outcompete the native M. trossulus in the southern extent of its distribution in Japan and California (Sarver and Foltz 1993; Gardner 1994; Suchanek etal.1997; Rawson et al. 1999; Braby and Somero 2006; Somero 2010).
It is widely known that temperature has widespread effects on the functional rates and structural stabilities of proteins (Somero 1995; Zavodszky etal. 1998; Fields etal. 2015). Orthologous proteins of organisms adapted to different temperatures typically show differences in thermal responses in ligand-binding abilities, catalytic rates, and stabilities of higher order structures (Hochachka and Somero 2002). Recent studies of several proteins involved in cellular energy metabolism have revealed that adaptive differences between orthologues of closely related species can result from small numbers of amino acid changes (Fields etal. 2006; Dong and Somero 2009; Lockwood and Somero 2012). These substitutions often modify the numbers and strengths of weak-bond interactions in the native folded protein, thereby modifying the protein’s net Gibbs free energy of stabilization (ΔGstab). However, the proteomic distribution of temperature-related adaptive substitutions such as those documented in metabolic proteins remains unknown (see Lockwood and Somero 2012). In the genus Mytilus, a study by Lockwood etal. (2010) used transcriptional response to investigate physiological traits involved in surviving acute heat stress, and found that 1,531 genes had responses to experimental change in temperature from 13 °C to 32 °C. However, only 96 of these had a response specific to heat tolerant M. galloprovincialis, suggesting that only a subset of genes involved in heat stress response are subject to species-specific thermal selection.
Here, we test predictions established from previous studies that compared 1) genome-wide patterns in thermophiles relative to mesophiles, 2) a small number of glycolytic enzymes invitro in species inhabiting narrow ranges of temperature, and 3) transcriptional responses in warm and cold tolerant Mytilus congeners. Comparisons among species with stark differences in thermal range have indicated there is a genome-wide influence of temperature on both protein stability and amino acid content (Fields and Somero 1998; Zeldovich et al. 2007; Cherry 2010), but that stabilizing amino acid changes in thermophiles are concentrated in proteins involved in metabolism (Gu and Hilser 2009). On the other hand, studies comparing glycolytic enzymes invitro have demonstrated that enzyme kinetic parameters (Km and/or Kcat) can adapt to subtle changes in temperature (e.g., Graves and Somero 1982; Lockwood and Somero 2012; reviewed in Fields etal. 2015). Studies of transcriptional responses to heat stress indicate that only a small portion of the genome might be subject to temperature-related selection unique to the warm tolerant lineage (Lockwood et al. 2010). These previous studies lead to the prediction that different thermal environments can cause adaptive divergence in protein structure and function among closely related species, but that these differences might be limited to a small subset of the genome.
In this study, we investigate the importance of temperature-related selection on protein stability in contributing to the evolution of the invasive M. galloprovincialis’ distinctive heat tolerance. We use in silico estimates of changes in Gibb’s free energy of amino acid substitutions to test for more stabilizing changes in heat tolerant M. galloprovincialis relative to cold tolerant M. trossulus. We apply a generalized linear model to test this prediction, thereby accounting for phylogenetic history, other possible conflicting selective forces, and transcriptional response. We also test for enrichment in heat tolerant M. galloprovincialis of a priori thermally sensitive GO terms identified in a comparative study of protein stability between thermophiles and mesophiles by Gu and Hilser (2009). Our findings contribute to understanding of the mechanisms of thermal adaptation, and have implications for the potential of species to track the narrow shifts in temperature predicted with global warming.
Materials and Methods
RNA Sequencing in Mytilus
For each species, we collected one young adult individual (7–10 mm shell length) from various worldwide localities for RNA extraction (table 1). Thrichomya hirsuta was used as the outgroup in all phylogenetic analysis. All mussels from abroad were dissected on the day of collection, preserved in RNAlater RNA Stabilization Solution (Life Technologies, Gaithersburg, MD), shipped on ice and stored in a −80 °C freezer for a maximum of 3 months before RNA extraction.
Table 1.
Summary of Transcriptomic RNA Sequence Data Collected
| Species | Date | Locality | Coordinates | Total Readsa | Average Coverage | % Missing |
|---|---|---|---|---|---|---|
| Mytilus galloprovincialis | Feb, 2011 | Oakland, CA | 37.805°, −122.257° | 17,845,567 | 68.9 | 1.67% |
| Mytilus edulis | Mar, 2012 | Darling Marine Center, ME | 43.935°, −69.581° | 9,957,638 | 50.3 | 4.19% |
| Mytilus trossulus | Dec, 2010 | Newport Harbor, OR | 44.631°, −124.049° | 17,285,811 | 70.1 | 1.51% |
| Mytilus californianus | N/A | CA | 36.955°, −122.025° | N/A | N/A | N/A (0%) |
| Trichomya hirsuta | Oct, 2012 | N Stradbroke Is, Australia | −27.503°, 153.401° | 27,983,710 | 73.6 | 5.39% |
Total number of reads that mapped to Mytilus californianus reference expressed sequence tags.
Total RNA was extracted using the Agencourt RNAdvance Tissue Kit (Beckman Coulter, Brea, CA) following the manufacturer’s instructions. We removed any gonads present in larger individuals to improve overlap in transcript capture, minced all remaining tissues using a razor blade on a glass slide, and used 15 μg for the RNA extraction. We measured RNA concentration on a Qubit 2.0 Fluorometer (Invitogen, Carlsbad, CA). Between 2,180 and 5,500 ng (table 1) were submitted for Illumina sequencing to Hudson Alpha Institute for Biotechnology (HAIB) in Huntsville, Alabama. HAIB performed mature mRNA isolation, complementary DNA library preparation, cluster generation, and Illumina sequencing. Each of six samples was barcoded with a multiplex identifier adaptor and sequenced under the Illumina HiSeq platform to yield ∼20 million 50-mer paired-end reads per individual.
Reference M. californianus EST Database Open-Reading-Frame Prediction and Annotation
We used a Sanger sequenced expressed sequence tag (EST) library from M. californianus as a reference for transcript alignments, prediction of protein-coding regions, and annotations (Gracey etal. 2008) [accession numbers: ES7325872–ES738966; ES408175–ES387463]. Dr Brent Lockwood and colleagues kindly provided the M. californianus 13,082 gene EST library in a refined format that included only unique single-copy protein-coding genes, as determined by the PartiGene bioinformatics pipeline (Parkinson etal. 2004; Lockwood et al. 2010). We predicted the single most likely protein-coding open reading frame (ORF) per transcript and performed annotation of each transcript with TRANSDECODER, a Trinity plugin (Grabherr etal. 2011). The output from TRANSDECODER was parsed with a custom python script, which selected the strand, sense or antisense, that we used for each transcript based on maximum length. This method produced 9,219 unique ORFs for the study, which we refer to here as “genes.” ORFs that were unlikely to be protein-coding were distinguished by two criteria: 1) elevation of dN/dS ratios >2 with the CODEML program of PAML v.4.5 (Yang 2007), and 2) no significant BLASTX, BLASTP, or protein domain search hit. These proteins were excluded from all further analyses.
Alignments to the Reference M. californianus EST Library
We trimmed raw reads and discarded Casava “filtered” flagged reads using the fastq_illumina_filter v1.0.1 using default parameters (http://cancan.cshl.edu/labmembers/gordon/fastq_illumina_filter/). We then applied a quality filter (Phred score >20, length > 40) with EU_UTIL’s FastqMcf toolkit (Aronesty 2011) and visualized these results with FastQC (Andrews 2010). Between 11,086,567 and 32,328,073 (table 1) high-quality (mean quality Phred score of 39) short-paired Illumina reads (mean 40 bp in length) from M. edulis, M. trossulus, M. galloprovincialis, and T. hirsuta were aligned to the M. californianus EST library using Sequence Search and Alignment by Hashing Algorithm (SSAHA2) (Ning et al. 2001). SSAHA2 is an appropriate tool for species with high polymorphisms such as Mytilus species because of its flexibility in the degree of mismatches allowed (Li and Durbin 2010). The reference assembly hash table was generated using a K-mer length of 13 and a skip step of six. Paired-end reads were aligned to this using a skip step of six and “solexa” defaults.
A consensus alignment for each species was generated for each gene following the methods described in Kober and Pogson (2017). Unless specified, we performed data processing using BioPerl (Stajich etal. 2002), BioPython (Cock etal. 2009), the Newick Utilities (Junier and Zdobnov 2010), and the R statistical package (Ihaka and Gentleman 1996), with the Open Grid Scheduler (available at http://gridscheduler.sourceforge.net/) for job scheduling. Read mapping qualities <30 were discarded and only the top highest MAPQ scored reads in the case of duplicates were retained. Pileups were created with the PySAM (available at http://code.google.com/p/pysam) interface to SAMTOOLS (Li et al. 2009) and sites with base quality scores <25 were discarded. Valid alleles were represented on >8 reads and a frequency >0.126. Insertion and deletions were ignored. Polymorphic sites were retained in the alignments with IUPAC ambiguity codes. Between 9,957,638 and 27,983,710 high-quality (mean Phred score 39) short paired-end Illumina reads (mean 40 bp in length) from M. edulis, M. trossulus, M. galloprovincialis, M. galloprovincialis, and T. hirsuta mapped successfully to the M. californianus EST reference sequences (table 1). Overall coverage and percentage of sites covered averaged 73.6× and 97.7%, respectively. We used a final alignment of 2,770 protein-coding genes > 60 codons in length with ambiguous sites removed for subsequent phylogenetic and protein stability analyses.
Phylogenetic Analysis and Tests for Positive Selection
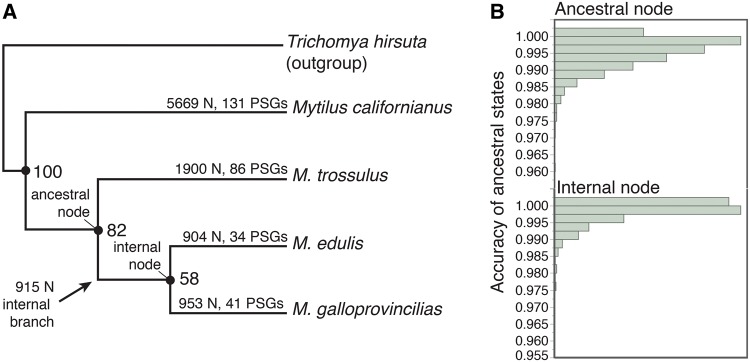
We applied a phylogenetic approach to overcome the challenge of phylogenetic nonindependence (Felsenstein 1985), an issue well recognized in the context of protein evolution (Graves and Somero 1982). PhyML v3.0 (Guindon etal. 2003) was used to create gene trees, with a GTR model of nucleotide evolution, “free rates” for each class from the data, optimized tree topologies, branch length, and rate parameters, and the best tree topology of nearest neighbor interchange and subtree pruning and regrafting search operations. The topology of the most commonly observed ML tree (fig. 1) was in concordance with the previously published phylogeny of this group (Vermeij 1991; Seed 1992; Samadi etal. 2007), and we use the best ML tree for each locus in subsequent analyses.
Fig. 1.
—Phylogenetic results and ancestral reconstructions, showing (A) the cladogram of the most frequent tree obtained from the maximum likelihood analysis in PhyML v3.0 (Guindon etal. 2003), and (B) the distribution of the accuracy of all ancestral states predicted in the CODEML program of the PAML v.4.5 package (Yang 2007). Node labels represent the percent gene trees that support the presented topology, branch labels represent number of amino acid changes detected (N), and the number of positively selected genes detected (PSGs) from the positive selection analysis in the CODEML program of the PAML v.4.5 package (Yang 2007).
We tested for significantly elevated dN/dS ratios among orthologs against the null expectation of 1 using the PAML v.4.5 package (Yang 1997, 2007) prior to testing for the stabilizing or destabilizing effects of amino acid substitutions on protein stability. There were three reasons for adopting this approach. First, genes with elevated dN/dS ratios are candidates for positive selection and thus may identify genes experiencing thermal adaptation (Dasmeh et al. 2013). Second, protein coding genes experience a variety of potentially conflicting selective forces, with the strongest forces being driven by antagonistic coevolution (Hughes and Nei 1988; Murphy 1993; Civetta and Singh 1995; Swanson and Vacquier 2002; Civetta 2003; Schlenke and Begun 2003; Clark et al. 2006; Drosophila 12 Genomes Consortium 2007; Haerty et al. 2007; Kober and Pogson 2013). Consequently, these loci might experience elevated rates of stabilizing or destabilizing substitutions irrespective of adaptation to temperature. Finally, there is evidence that clients of heat-shock proteins have elevated dN/dS ratios (Lachowiec et al. 2013, 2015) making it important to account for interactive effects between estimates of protein stability and dN/dS ratios. Maximum likelihood tests for elevated dN/dS ratios were performed using the CODEML program in PAML v.4.5 (Yang 1997, 2007). For each locus, ancestral states were inferred with model 0, and then used to test for elevated dN/dS ratios above the null expectation of 1 with the improved log-likelihood ratio test, “test 2,” of Zhang etal. (2005). In this test, the null hypothesis is the branch-site model A where dN/dS ratio varies among branches and codons, with a total of four site classes in the sequence: there are two site classes along the background lineages with ratios ω0 or ω1 fixed to 0 and 1, respectively, but along the lineage of interest (the foreground lineage), the null model has an dN/dS ratio ω2 fixed to 1, and the alternative model has a constrained dN/dS ratio at ω2 ≥ 1 (Zhang etal. 2005). Tests were performed for all four terminal branches leading to the extant Mytilus species. We corrected for multiple tests using a Bonferroni correction of the family-wise error rate for each branch tested for each protein (Lu and Guindon 2014).
Transcriptional Response to Heat Stress
Transcriptional response to heat stress was included in our analyses because genes with significant changes in expression level under experimental heat stress are candidates of physiological traits involved in thermal tolerance (Lockwood etal. 2010). Additionally, transcription patterns are correlated with both rates of molecular evolution and protein stability (Drummond et al. 2005; Drummond and Wilke 2008; Serohijos et al. 2012), which makes it important to account for these associations, irrespective of the direction of the association. For example, heat-shock proteins are predicted to have high transcriptional response to heat stress (Lockwood et al. 2010) but not elevated dN/dS ratios because of their high level of selective constraint. Clients of heat-shock proteins may be expected to have high dN/dS ratios (Lachowiec etal. 2013, 2015) but not a transcriptional response to heat stress. Proteins with thermally adaptive substitutions might be expected to have both a transcriptional response and elevated dN/dS ratios. We parsed transcriptional response data reported by (Lockwood etal. 2010) into two broad categories using the clustering algorithm of the “heatmap.2” function in the R package “ggplot2” (Wickham 2009). We scored all loci with significant differences in transcription level when held at 13 °C versus 32 °C as a response to heat stress, and any loci with no significant difference in transcription as showing no response to heat stress.
eScape Site-Specific Estimates of Protein Stability in Mytilus Species
We estimated changes in Gibbs free energy of stabilization (ΔΔGstab) of each amino acid substitution along each terminal branch of the 2,770 gene trees (fig. 1A) using eScape v2.1 (Gu and Hilser 2008). Adaptive divergence is predicted to increase protein stability at upper thermal extremes and increase protein flexibility (i.e., decrease stability) at lower thermal extremes. However, accurate description of protein thermodynamics is difficult because proteins are only marginally stable and exist as transient ensembles of conformational microstates (Wrabl etal. 2011). Thus, accounting for multiple subensembles is necessary but is computational difficult. eScape v2.1 (Gu and Hilser 2008) overcomes this difficulty in silico by using amino acid sequence data to model the contribution of each residue to the stability constant, a metric representing the equilibrium of the natively folded and the multiple unfolded states of a protein (D’Aquino etal. 1996). Estimates of the contribution of each residue to the stability constant (ΔGstab) are completed in a sliding window along the length of an amino acid sequence alignment. This sliding window approach allows for local estimates of ΔGstab across stretches <20 residues in length, and makes eScape an excellent tool for detecting subtle changes in protein thermodynamics such as those important in adaptive stabilizing substitutions (Fields etal. 2015). We estimated ΔGstab of each residue of the 2,770 gene alignments, and calculated ΔΔGstab as the difference between the ΔGstab of each amino substitution (fig. 1A) and the ΔGstab of the ancestral state inferred for the branch-sites tests from CODEML, which were all of high accuracy (fig. 1B): The overall mean posterior probability was 0.996 (±0.004 SD; fig. 1B).
Generalized Linear Model to Detect Differences in ΔΔGstab among Lineages
We tested the prediction of more stabilizing ΔΔGstab mutations in heat tolerant M. galloprovincialis relative to cold tolerant M. trossulus using a generalized linear likelihood model in JMP v11.0.0 (SAS Institute Inc 1989–2013) using ΔΔGstab as the dependent variable and a full factorial set of independent variables, including the lineage of the amino acid substitution, results from the tests for elevated dN/dS ratios, and the transcriptional responses described in Lockwood etal.’s (2010) study. We investigated genes of interest with searches using the Protein Homology/analogY Recognition Engine v2.0 (Kelley and Sternberg 2009; Kelley etal., 2015), and mapped amino acid substitutions and estimates of ΔGstab of M. galloprovincialis, M. trossulus, and the internal node onto the predicted protein structure.
Enrichment Analysis for A Priori GO Terms
We also tested for enrichment of GO terms at genes with elevated dN/dS ratios that showed shifts in protein stability in thermophiles relative to mesophiles in Gu and Hilser’s (2009) study. Gene ontology (GO) terms (Gene Ontology Consortium 2004) were identified with the “trinotate” pipeline (available at http://trinotate.sourceforge.net/) based on a homology searches with NCBI-BLASTx and NCBI-BLASTp against the SWISSPROT database (version “Trinotate.Oct2013”) with an e-value cutoff of 10e-5. We used a weighted algorithm that took the hierarchical structure of GO terms into account in the Bioconductor package topGO v2.14 (Gentleman et al. 2004; Alexa et al. 2006) and looked for overlap of these terms with a priori GO terms. The a priori GO terms included seven molecular function terms (enzyme regulator, translation regulator activity, protein transporter activity, ion transporter activity, helicase activity, catalytic activity, and structural molecule activity), a single biological process term (secretion), and two cellular component terms (nucleus and chromosome).
Results
Sequence-based estimates of protein energy landscapes in Mytilus spp. identified more stabilizing amino acid changes in heat tolerant M. galloprovincialis relative to cold tolerant M. trossulus, especially in genes with elevated dN/dS ratios and those with transcriptional responses to heat stress. This pattern was driven by ΔΔGstab outliers in M. galloprovincialis. We did not obtain evidence that selection for protein stability was strong enough to enrich a priori candidates of thermal sensitively in genes with elevated dN/dS ratios in M. galloprovincialis relative to other congeners.
Differences in ΔΔGstab among Lineages
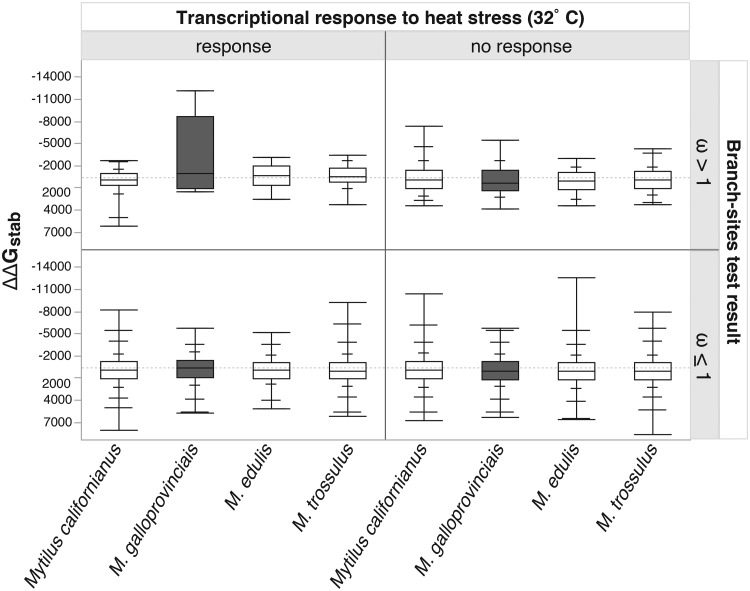
We identified 40 genes with significantly elevated dN/dS ratios in M. galloprovincialis, 34 genes in M. edulis, 87 genes in M. trossulus, and 130 genes in M. californianus (table 2;supplemental materials 1 and 2, Supplementary Material online). The greater number genes with elevated dN/dS ratios detected in M. trossulus and M. galloprovincialis is consistent with previous work showing positive selection is more likely detected in longer branches (Nielsen et al. 2005; Kosiol et al. 2008; Kober and Pogson 2017), and suggests limited power to detect positive selection in M. galloprovincialis and M. edulis (fig. 1) within this data. Our generalized linear model found that amino acid substitutions in M. galloprovincialis were 3,180.0 calories per mol more stabilizing in a subset of 65 loci with both elevated dN/dS ratios and transcriptional response to heat stress (fig. 2), and 705.8 calories per mol more
Table 2.
Results of Branch-Site Tests for Positive Selection in PAML v.4.5 (Yang 2007)
| Lineage | Total Sites Calleda | Number of Genes Tested | Mean Length of Genes | Mean ωb | Number of PSGsc | Mean LRTd of PSGs |
|---|---|---|---|---|---|---|
| Mytilus galloprovincialis | 1,639,598 | 1996 | 420.89 | 0.11912 | 40 | 7.2386 |
| M. edulis | 1,467,662 | 2046 | 435.88 | 0.1039 | 34 | 8.2249 |
| M. trossulus | 1,635,612 | 2117 | 442.50 | 0.1251 | 87 | 6.5140 |
| M. californianus | 1,897,333 | 2819 | 440.48 | 0.1277 | 130 | 6.6383 |
| Overall | 1,470,508 | 2770 | 434.94 | 0.1190 | 282 | 7.1539 |
In total, 631,194 of these sites that were called for all lineages were used in tests of positive selection.
Mean dN/dS ratio per gene for the lineage.
PSGs, positively selected genes.
Likelihood-ratio test score.
Fig. 2.
—Quantile box-plots of mean eScape v2.1 estimates of change in Gibbs free energy of stability (ΔΔGstab) in calories per mol for each amino acid substitution identified in the 2,770 orthologs analyzed, separated by branch-sites tests for elevated dN/dS ratios (ω) and transcriptional response to heat stress at 32 °C (Lockwood et al. 2010). ΔΔGstab was estimated as the mean difference between the derived ΔGstab and the ancestral ΔGstab for each lineage for each substitution identified in the CODEML program of the PAML v.4.5 package (Yang 2007). More negative ΔΔGstab values signify more stabilizing amino acid substitutions.
stabilizing across all 2,770 loci investigated (table 3). The full model was highly significant (P value = 0.0009), and many individual parameter estimates were significant, notably in the M. galloprovincialis lineage (table 3). Removal of 26 M. galloprovincialis outlier orthologs with ΔΔGstab outside of the 95% quantiles reduced the model to nonsignificance (P value = 0.1464).
Table 3.
Summary of Generalized Linear Model Effect Sizes of Change in Gibbs Free Energy Constant (ΔΔGstab) by Lineage
| ßa of ΔΔGstab | SE | LRCS | P Value | |
|---|---|---|---|---|
| By lineage | ||||
| Mytilus californianus | 26.14 | 198.23 | 0.02 | 0.895 |
| Internal branch | 693.65 | 608.12 | 1.30 | 0.254 |
| M. galloprovincialis | −705.80 | 263.95 | 7.15 | 0.008 |
| M. edulis | 36.89 | 68.28 | 0.29 | 0.589 |
| By lineage by positive selection (PS) | ||||
| M. californianus by PS | 79.74 | 199.80 | 0.16 | 0.690 |
| Internal branch by PS | 606.43 | 603.13 | 1.01 | 0.315 |
| M. galloprovincialis by PS | −616.26 | 266.83 | 5.33 | 0.021 |
| M. edulis by PS | 0.00 | N/A | N/A | N/A |
| By lineage by heat stress induced transcriptional response (HSITR) | ||||
| M. californianus by HSITR | 229.12 | 198.23 | 1.34 | 0.248 |
| Internal branch by HSITR | 555.74 | 608.12 | 0.84 | 0.361 |
| M. galloprovincialis by HSITR | −707.32 | 263.95 | 7.18 | 0.007 |
| M. edulis by HSITR | −58.21 | 68.28 | 0.73 | 0.394 |
| By lineage by PS by HSITR | ||||
| M. californianus by PS by HSITR | 187.39 | 199.80 | 0.88 | 0.348 |
| Internal branch by PS by HSITR | 438.10 | 603.13 | 0.53 | 0.468 |
| M. galloprovincialis by PS by HSITR | −615.77 | 266.83 | 5.32 | 0.021 |
| M. edulis by PS by HSITR | 0.00 | N/A | N/A | N/A |
Significant coefficients (ß) indicated in bold. SE, standard error of the coefficient (ß); LRCS, likelihood ratio χ2 of the coefficient (ß).
Coefficient of predicted ΔΔGstab against Mytilus trossulus (full model P value = 0.0009).
Energy Profiles of Orthologs with Elevated d N/dS Ratios and Transcriptional Response to Heat Stress
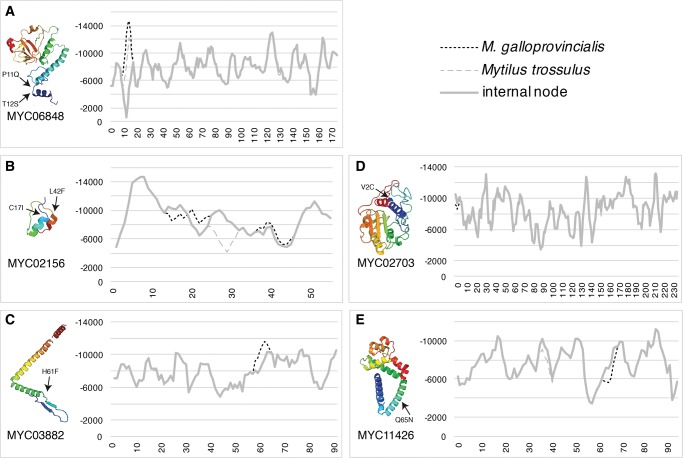
Genes with transcriptional response to heat stress and significantly elevated dN/dS ratios in M. galloprovincialis fell within five contigs; MYC06848, MYC02156, MYC03882, MYC07203, and MYC11426 (fig. 3). The identity and patterns of ΔΔGstab within these genes suggested that substitutions within coiled regions of down-regulated proteins had the most stabilizing effects (fig. 3 and supplemental material 1, Supplementary Material online).
Fig. 3.
—Protein structure predictions based on searches using the Protein Homology/analogY Recognition Engine v2.0 (Kelley and Sternberg 2009) and eScape v2.1 estimates of in Gibbs free energy of stability (ΔGstab), in calories per mol, for loci with elevated dN/dS ratios (ω) and transcriptional response to heat stress in Mytilus galloprovincialis. Three genes exhibited clear evidence of stabilizing substitutions in M. galloprovincialis: (A) MYC06848, (B) MYC03882, and (C) MYC02156. Two genes showed minimal changes in energy profiles: (D) MYC07203, and (E) MYC11426. More negative ΔGstab values signify higher protein stability. Protein structure image is colored by rainbow from red at the amine-terminus to blue at the carboxyl-terminus, with α-helices shown as helices, β-strands shown as arrows, and coils shown as narrow lines. See supplemental material 1, Supplementary Material online, for details.
Uniform Enrichment of A Priori GO Terms
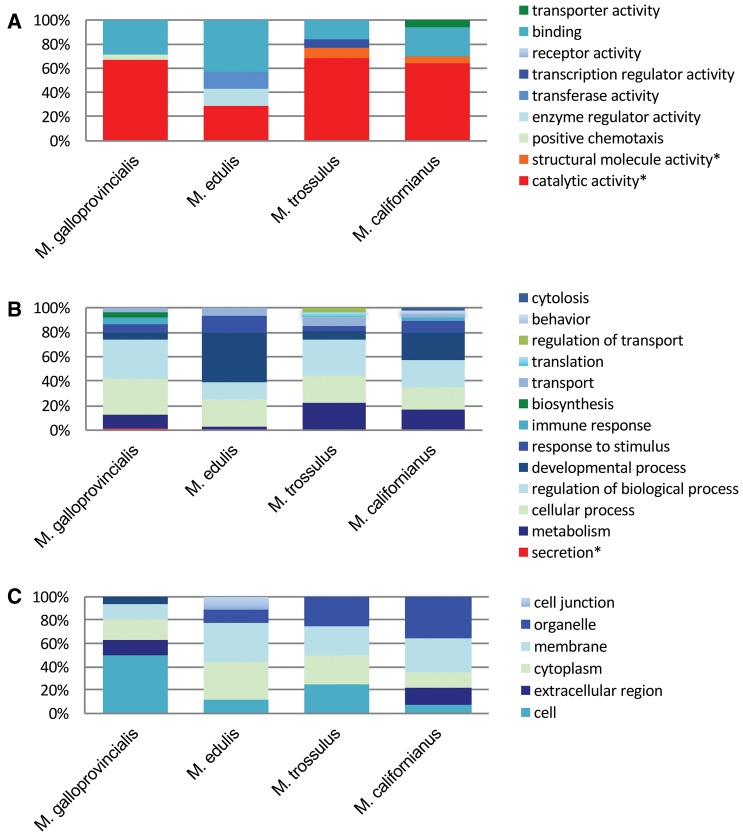
All a priori GO terms were equally common in all four Mytilus species analyzed. At candidate positively selected genes, we found 20 GO terms enriched in M. galloprovincialis, 86 in M. edulis, 66 in M. trossulus, and 51 in M. californianus (supplemental material 3, Supplementary Material online). Nearly all of these GO terms were low in the GO hierarchy (child terms) and only a single candidate GO term, “structural molecule activity,” was enriched in M. trossulus. When we compared the parent terms of this list, we found a much higher degree of overlap with those associated with thermal sensitivity in Gu and Hilser’s (2009) study, but that they were uniformly distributed across all four Mytilus species (fig. 4). Enriched a priori GO terms included “structural molecular activity,” “catalytic activity,” and “secretion” (fig. 4).
Fig. 4.
—Frequency of top level GO terms in the list of enriched (A) molecular function, (B) biological process, and (C) cellular component GO terms based on topGO v2.14 (Gentleman etal. 2004; Alexa etal. 2006) analysis of lineage-specific positively selected genes in Mytilus galloprovincialis, M. edulis, M. trossulus, and M. californianus. Terms that overlap with those showing a strong thermal response in the Gu and Hilser (2009) study are marked with * and shown in warm colors.
Discussion
Obtaining evidence for genome-wide selection on protein stability is particularly challenging, but recent computational approaches (Vertrees et al. 2005; Tehei etal. 2006; Gu and Hilser 2008; Li et al. 2012; reviewed in Somero et al. 2016) are a promising avenue forward (Benedix et al. 2009; Wrabl et al. 2011). The present study represents one of the first attempts to identify amino acid substitutions that stabilize protein structure in a species group known to have one lineage (M. galloprovincialis) that has undergone adaptation to warmer temperatures (Braby and Somero 2006; Lockwood et al. 2010; Somero 2010; Tomanek and Zuzow 2010; Lockwood and Somero 2011a). As expected, in silico estimates of ΔΔGstab suggested stronger genome-wide selection on proteins for thermodynamic stability in heat tolerant M. galloprovincialis than in cold tolerant M. trossulus. More stabilizing amino acid substitutions had fixed in M. galloprovincialis relative to M. trossulus (table 3), and genes with both elevated dN/dS ratios and transcriptional response to heat stress had more stabilizing changes in M. galloprovincialis by ∼3,180 calories per mol per substitution than in M. trossulus (fig. 2 and table 3). This level of ΔΔGstab per substitution is in the range of documented adaptive stabilizing mutations in Mytilus species and in other systems (D’Amico et al. 2001; Dasmeh et al. 2013; Fields et al. 2015) and is above the level expected by random mutation (Tokuriki et al. 2007). This suggests that amino acid substitutions that account for significantly higher ΔΔGstab in M. galloprovincialis represent an adaptive response to heat stress during its evolutionary history in the warm Mediterranean Sea (Thunell 1979).
A priori tests for GO term enrichment revealed that selection for thermodynamic stability was not restricted to M. galloprovincialis but was present across all lineages. It is possible that difference in average ΔΔGstab observed in M. galloprovincialis resulted from selection for stabilizing substitutions of greater effect, rather than selection acting on a greater number of genes. This pattern has been described previously for the evolution in myoglobin in cetaceans compared with terrestrial mammals (Dasmeh et al. 2013) and is supported by the importance of outliers in M. galloprovincialis in generating the signal of thermal adaptation. This hypothesis is also supported by the evolutionary and natural history of the group, which indicates significant overlap in thermal history (Seed 1992; Suchanek etal. 1997) and occasional exposure to thermal stress during tidal and weather extremes in all Mytilus species within their current distributions (Fields etal. 2006; Somero 2010; Dowd and Somero 2013).
The widespread signals of enrichment of a priori GO terms across all Mytilus species could, however, indicate nonrepresentative a priori terms, insensitivity of the tests for positive selection, or lack of statistical power. The a priori GO terms used were based on thermophiles (Gu and Hilser 2009) and may not be appropriate for small shifts in thermal tolerance from 26 °C to 32 °C. For example, there are biases in amino-acid usage in thermophiles (Zeldovich et al. 2007; Cherry 2010) such that mesophiles may experience different patterns of constraint and positive selection. In addition, the a priori GO terms could represent vulnerability to any environmental stressor—such as oxidative, salinity, and radiation stress—rather than to thermal stress per se (Regoli and Principato 1995; Pampanin etal. 2005; Fasulo etal. 2008; Hamer etal. 2008; Lockwood and Somero 2011b; AlAmri etal. 2012; Trudeau etal. 2016) rendering them a priori more likely to produce significant branch-sites tests regardless of thermal differences (Oliver etal. 2010). Another possibility is that elevated dN/dS ratios may have been insensitive to the relevant substitutions, either because of general insensitivity of branch-sites tests to single amino acid changes (Drosophila 12 Genomes Consortium 2007; Ellegren 2008; Gharib and Robinson-Rechavi 2013; Lu and Guindon 2014) or because of the action of stronger selective agents such as antagonistic coevolution (Hughes and Nei 1988; Civetta and Singh 1995; Haerty et al. 2007). Indeed, many of the enriched GO terms are candidates for involvement in the innate immune response terms (e.g., immune response, response to stimulus, fig. 4) or sexual conflict (e.g., female germline ring canal formation, ovarian fusome organization, germ-line cyst formation, supplemental material 3, Supplementary Material online). Finally, uniform enrichment could be due to lack of statistical power of our tests, which included only 34 to 130 genes with elevated dN/dS ratios, and only 2,770 genes total of the 13,759 ESTs available in the M. californianus EST library (Gracey etal. 2008). We suggest future research should aim to increase the number of genes analyzed and, when available in the literature, attention should be directed at a priori genes and GO terms known to be involved in thermal adaptation in eukaryotic mesophiles.
The energy profiles of orthologs with both elevated dN/dS ratios and transcriptional response to heat stress in M. galloprovincialis (fig. 3) indicated that the largest changes in energy profile occurred in the expected direction (i.e., increasing stability) in warm tolerant M. galloprovincialis orthologs relative to cold tolerant M. trossulus. These changes tended to occur in coiled regions of local high stability, thus increasing the top peaks of stability of the whole protein (fig. 3A–C). Additionally, the largest stabilizing ΔΔGstab estimates occurred in genes down-regulated when exposed to heat stress (fig. 3A and B). The changes in energy profiles of other orthologs with both elevated dN/dS ratios and transcriptional response to heat stress were very minor, occurred within an α-helix or in up-regulated genes, and were unreliable (fig. 3D) or exhibited a mix of difference in ΔGstab in the predicted and unpredicted directions (fig. 3E). These results suggest that adaptive stabilizing substitutions in M. galloprovincialis tended to occur within coiled regions of down-regulated genes with significantly elevated dN/dS ratios. However, a larger sample of genes with more comprehensive coverage of all open reading frames is needed to determine the generality of these patterns.
Conclusion
We observed significantly more stabilizing amino acid substitutions in M. galloprovincialis than in cold tolerant M. trossulus and suggest that these substitutions have contributed to its ability to tolerate heat stress several degrees warmer than its congeners. Our results provide confidence that it is possible to make in silico estimates of protein energy profiles using DNA sequence alone with the eScape method (Gu and Hilser 2008) in much smaller windows of thermal adaptation than originally tested (Gu and Hilser 2009). Adaptive shifts in protein energy landscapes appeared to be concentrated in a small subset of genes and tended to occur within coiled regions of heat stress induced down-regulated genes with significantly elevated dN/dS ratios.
Global warming has created concerns that poikilotherms will fail to evolve quickly enough to track rising temperatures. A key physiological response to a warming climate may be the fine-tuning of protein stability to maintain efficient allosteric function. Thermal adaptation in proteins is facilitated by the modulation of both local flexibility and protein-wide stability (Somero et al. 2016). However, the strength of the resulting selection for stability or flexibility is heretofore undocumented, and there are many other selection pressures important in sexually reproducing eukaryotes that may overpower any selection for protein stability. Our results suggest that adaptation of protein structure to the narrow shifts in temperature predicted with global warming may be an important component of species survival. However, our results also suggest that there may be some level of selection for thermodynamic stability across all Mytilus species, and that the higher level of stabilizing changes observed in M. galloprovincialis may have been caused by stabilizing substitutions of greater effect in the warm adapted lineage relative to the cold adapted lineage in a small subset of the genome. The remaining questions merit further study, and we suggest that future work targets a larger number of genes, especially those with both elevated dN/dS ratios and transcriptional response to heat stress, and should focus on experimental confirmation of the accuracy of in silico estimates of thermodynamic stability.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online at https://doi.org/10.1093/gbe/evx190.
Supplementary Material
Acknowledgments
We thank K. M. Kay, R. C. Vrijenhoek, and P. T. Raimondi for their comments and suggestions, and especially G. M. Somero for his extensive help in developing the methodology and for his inspirational research, which helped establish the framework of the manuscript. Samples from Maine and Australia were kindly collected by C. Riginos, for which we are grateful. We also thank B. E. Nobua-Behrmann for her help with statistical analysis. This work was funded by an Achievement Award for College Scientists Foundation, the University of California President’s Dissertation Year Fellowship, the Davis Memorial Scholarship, and the Myers Oceanographic and Marine Biology Trust. In addition to facilities at UC Santa Cruz and UC San Francisco, this work used the Center for Comparative Genomics at the California Academy of Sciences, and the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
Literature Cited
- Ahmed M, Sparks A.. 1970. Chromosome number, structure and autosomal polymorphism in the marine mussels Mytilus edulis and Mytilus californianus. Biol Bull. 138(1):1–13. [Google Scholar]
- AlAmri OD, Cundy AB, Di Y, Jha AN, Rotchell JM.. 2012. Ionizing radiation-induced DNA damage response identified in marine mussels, Mytilus sp. Environ Pollut. 168:107–112. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T.. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22(13):1600–1607. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FASTQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc, last accessed April 24, 2013.
- Aronesty E. 2011. Command-line tools for processing biological sequencing data. Expression analysis. Available from: http://code.google.com/p/ea-utils, last accessed June 6, 2013.
- Benedix A, Becker CM, de Groot BL, Caflisch A, Böckmann RA.. 2009. Predicting free energy changes using structural ensembles. Nat Methods 6(1):3–4. [DOI] [PubMed] [Google Scholar]
- Bierne N, et al. 2003. Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol Ecol. 12(2):447–461. [DOI] [PubMed] [Google Scholar]
- Borsa P, Rolland V, Daguin-Thiébaut C.. 2012. Genetics and taxonomy of Chilean smooth-shelled mussels, Mytilus spp. (Bivalvia: Mytilidae). C R Biol. 335(1):51–61. [DOI] [PubMed] [Google Scholar]
- Braby CE, Somero GN.. 2006. Following the heart: temperature and salinity effects on heart rate in native and invasive species of blue mussels (genus Mytilus). J Exp Biol. 209(Pt 13):2554–2566. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Owen ME, Hofmann GE.. 2001. Adjusting the thermostat: the threshold induction temperature for the heat-shock response in intertidal mussels (genus Mytilus) changes as a function of thermal history. J Exp Biol. 204(Pt 20):3571–3579. [DOI] [PubMed] [Google Scholar]
- Cherry JL. 2010. Highly expressed and slowly evolving proteins share compositional properties with thermophilic proteins. Mol Biol Evol. 27(3):735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A, Singh RS.. 1995. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J Mol Evol. 41(6):1085–1095. [DOI] [PubMed] [Google Scholar]
- Civetta A. 2003. Shall we dance or shall we fight? Using DNA sequence data to untangle controversies surrounding sexual selection. Genome 46(6):925–929. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ.. 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131(1):11–22. [DOI] [PubMed] [Google Scholar]
- Cock PA, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25(11):1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comesana AS, Toro JE, Innes DJ, Thompson RJ.. 1999. A molecular approach to the ecology of a mussel (Mytilus edulis–M. trossulus) hybrid zone on the east coast of Newfoundland, Canada. Mar Biol. 133:213–221. [Google Scholar]
- Cruz F, Perez M, Presa P.. 2005. Distribution and abundance of microsatellites in the genome of bivalves. Gene 346:241–247. [DOI] [PubMed] [Google Scholar]
- D’Amico S, Gerday C, Feller G.. 2001. Structural determinants of cold adaptation and stability in a large protein. J Biol Chem. 276:25791–25796. [DOI] [PubMed] [Google Scholar]
- D’Aquino JA, et al. 1996. The magnitude of the backbone conformational entropy change in protein folding. Proteins 25:143–156. [DOI] [PubMed] [Google Scholar]
- Dasmeh P, Serohijos AWR, Kepp KP, Shakhnovich EI, Ben-Tal N.. 2013. Positively selected sites in cetacean Myoglobins contribute to protein stability. PLoS Comput Biol. 9(3):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Somero GN.. 2009. Temperature adaptation of cytosolic malate dehydrogenases of limpets (genus Lottia): differences in stability and function due to minor changes in sequence correlate with biogeographic and vertical distributions. J Exp Biol. 212(Pt 2):169–177. [DOI] [PubMed] [Google Scholar]
- Dowd WW, Somero GN.. 2013. Behavior and survival of Mytilus congeners following episodes of elevated body temperature in air and seawater. J Exp Biol. 216(Pt 3):502–514. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 102:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO.. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134(2):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2008. Comparative genomics and the study of evolution by natural selection. Mol Ecol. 17(21):4586–4596. [DOI] [PubMed] [Google Scholar]
- Fasulo S, et al. 2008. Expression of metallothionein mRNAs by in situ hybridization in the gills of Mytilus galloprovincialis, from natural polluted environments. Aquat Toxicol. 88(1):62–68. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- Fields PA, Dong Y, Meng X, Somero GN.. 2015. Adaptations of protein structure and function to temperature: there is more than one way to ‘skin a cat’. J Exp Biol. 218:1801–1811. [DOI] [PubMed] [Google Scholar]
- Fields PA, Rudomin EL, Somero GN. 2006. Temperature sensitivities of cytosolic malate dehydrogenases from native and invasive species of marine mussels (genus Mytilus): sequence-function linkages and correlations with biogeographic distribution. J Exp Biol. 209:656–667. [DOI] [PubMed] [Google Scholar]
- Fields PA, Somero GN.. 1998. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci U S A. 95(19):11476–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JPA. 1994. The structure and dynamics of naturally occurring hybrid Mytilus edulis Linnaeus, 1758 and Mytilus galloprovincialis Lamarck, 1819 (Bivalvia, Mollusca) populations review and interpretation. Arch Hydrobiol. 1–2:37–71. [Google Scholar]
- Geller JB. 1999. Decline of a native mussel masked by sibling species invasion. Conserv Biol. 13(3):661–664. [Google Scholar]
- Gene Ontology Consortium. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5(10):R80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M.. 2013. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol Biol Evol. 30(7):1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracey AY, et al. 2008. Rhythms of gene expression in a fluctuating intertidal environment. Curr Biol. 18(19):1501–1507. [DOI] [PubMed] [Google Scholar]
- Grant WS, Cherry MI.. 1985. Mytilus galloprovincialis Lmk. in southern Africa. J Exp Mar Biol Ecol. 90(2):179–191. [Google Scholar]
- Graves JE, Somero GN.. 1982. Electrophoretic and functional enzymic evolution in four species of eastern Pacific barracudas from different thermal environments. Evolution 36(1):97–106. [DOI] [PubMed] [Google Scholar]
- Gu J, Hilser VJ.. 2008. Predicting the energetics of conformational fluctuations in proteins from sequence: a strategy for profiling the proteome. Structure 16(11):1627–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Hilser VJ.. 2009. Sequence-based analysis of protein energy landscapes reveals nonuniform thermal adaptation within the proteome. Mol Biol Evol. 26(10):2217–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Haerty W, et al. 2007. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177(3):1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer B. 2008. Effect of hypoosmotic stress by low salinity acclimation of Mediterranean mussels Mytilus galloprovincialis on biological parameters used for pollution assessment. Aquat Toxicol. 89(3):137–151. [DOI] [PubMed] [Google Scholar]
- Heath DD, Rawson PD, Hilbish TJ.. 1995. PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can J Fish Aquat Sci. 52(12):2621–2627. [Google Scholar]
- Hellmann JJ, Byers JE, Bierwagen BG, Dukes JS.. 2008. Five potential consequences of climate change for invasive species. Conserv Biol. 22(3):534–543. [DOI] [PubMed] [Google Scholar]
- Hochachka PW, Somero GN.. 2002. Biochemical adaptation: mechanism and process in physiological evolution. New York: Oxford University Press. [Google Scholar]
- Hughes AL, Nei M.. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335(6186):167–170. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman RC.. 1996. R: a language for data analysis and graphics. J Comput Graph Stat. 5(3):299–314. [Google Scholar]
- Junier T, Zdobnov E.. 2010. The Newick utilities: high-throughput phylogenetic tree processing in the Unix shell. Bioinformatics 26(13):1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE.. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 4(3):363–371. [DOI] [PubMed] [Google Scholar]
- Kober KM, Pogson GH.. 2013. Genome-wide patterns of codon bias are shaped by natural selection in the purple sea urchin, Strongylocentrotus purpuratus. G3 Genes Genomes Genet. 3(7):1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober KM, Pogson GH.. 2017. Genome-wide signals of positive selection in strongylocentrotid sea urchins. BMC Genomics. 8(1):555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, et al. 2008. Patterns of positive selection in six mammalian genomes. PLoS Genet. 4(8):e1000144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowiec J, et al. 2013. The protein chaperone HSP90 can facilitate the divergence of gene duplicates. Genetics 193(4):1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowiec J, Lemus T, Borenstein E, Queitsch C.. 2015. Hsp90 promotes kinase evolution. Mol Biol Evol. 32(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. 2012. Prots: a fragment based protein thermo‐stability potential. Proteins 80(1):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood BL, Sanders JG, Somero GN.. 2010. Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. J Exp Biol. 213(20):3548–3558. [DOI] [PubMed] [Google Scholar]
- Lockwood BL, Somero GN.. 2011a. Invasive and native blue mussels (genus Mytilus) on the California coast: the role of physiology in a biological invasion. J Exp Mar Biol Ecol. 400:167–174. [Google Scholar]
- Lockwood BL, Somero GN.. 2011b. Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol Ecol. 20:517–529. [DOI] [PubMed] [Google Scholar]
- Lockwood BL, Somero GN.. 2012. Functional determinants of temperature adaptation in enzymes of cold- versus warm-adapted mussels (genus Mytilus). Mol Biol Evol. 29(10):3061–3070. [DOI] [PubMed] [Google Scholar]
- Lu A, Guindon S.. 2014. Performance of standard and stochastic branch-site models for detecting positive selection among coding sequences. Mol Biol Evol. 31(2):484–495. [DOI] [PubMed] [Google Scholar]
- Murphy PM. 1993. Molecular mimicry and the generation of host defense protein diversity. Cell 72(6):823–826. [DOI] [PubMed] [Google Scholar]
- Nielsen R, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3(6):0976–0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC.. 2001. SSAHA: a Fast search method for large DNA databases. Genome Res. 11(10):1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhipinti-Ambrogi A. 2007. Global change and marine communities: alien species and climate change. Mar Pollut Bull. 55(7–9):342–352. [DOI] [PubMed] [Google Scholar]
- Oliver TA, et al. 2010. Whole-genome positive selection and habitat-driven evolution in a shallow and a deep-sea urchin. Genome Biol Evol. 2(0):800–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampanin DM, et al. 2005. Susceptibility to oxidative stress of mussels (Mytilus galloprovincialis) in the Venice Lagoon (Italy). Mar Pollut Bull. 50(12):1548–1557. [DOI] [PubMed] [Google Scholar]
- Parkinson J, et al. 2004. PartiGene – Constructing partial genomes. Bioinformatics 20(9):1398–1404. [DOI] [PubMed] [Google Scholar]
- Petit RJ. 2004. Biological invasions at the gene level. Divers Distributions 10(3):159–165. [Google Scholar]
- Rahel FJ, Olden JD.. 2008. Assessing the effects of climate change on aquatic invasive species. Conserv Biol. 22(3):521–533. [DOI] [PubMed] [Google Scholar]
- Rawson PD, Agrawal V, Hilbish TJ.. 1999. Hybridization between the blue mussels Mytilus galloprovincialis and M. trossulus along the Pacific coast of North America: evidence for limited introgression. Mar Biol. 134(1):201–211. [Google Scholar]
- Regoli F, Principato G.. 1995. Glutathione, glutathione-dependent and antioxidant enzymes in mussel, Mytilus galloprovincialis, exposed to metals under field and laboratory conditions: implications for the use of biochemical biomarkers. Aquat Toxicol. 31(2):143–164. [Google Scholar]
- Robinson TB, Branch GM, Griffiths CL, Govender A, Hockey PAR.. 2007. Changes in South African rocky intertidal invertebrate community structure associated with the invasion of the mussel Mytilus galloprovincialis. Mar Ecol Prog Ser. 340:163–171. [Google Scholar]
- Ruiz GM, Fofonoff PW, Carlton JT, Wonham MJ, Hines AH.. 2000. Invasion of coastal marine communities in North America: apparent patterns, processes, and biases. Annu Rev Ecol Syst. 31(1):481–531. [Google Scholar]
- Saarman NP, Pogson GH.. 2015. Introgression between invasive and native blue mussels (genus Mytilus) in the central California hybrid zone. Mol Ecol. 24(18):4723–4738. [DOI] [PubMed] [Google Scholar]
- Samadi S, et al. 2007. Molecular phylogeny in mytilids supports the wooden steps to deep-sea vents hypothesis. C R Biol. 330(5):446–456. [DOI] [PubMed] [Google Scholar]
- Sarver SK, Foltz DW.. 1993. Genetic population structure of a species’ complex of blue mussels (Mytilus spp.). Mar Biol. 117(1):105–112. [Google Scholar]
- Schlenke TA, Begun DJ.. 2003. Natural selection drives Drosophila immune system evolution. Genetics 164(4):1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed R. 1992. Systematics evolution and distribution of mussels belonging to the genus Mytilus – an overview. Am Malacol Bull. 9:123–137. [Google Scholar]
- Serohijos AWR, Rimas Z, Shakhnovich EI.. 2012. Protein biophysics explains why highly abundant proteins evolve slowly. Cell Rep. 2(2):249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero GN. 1995. Proteins and temperature. Ann Rev Physiol. 57:43–68. [DOI] [PubMed] [Google Scholar]
- Somero GN. 2010. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine “winners” and “losers”. J Exp Biol. 213(6):912–920. [DOI] [PubMed] [Google Scholar]
- Somero GN, Lockwood BL, Tomanek L.. 2016. Biochemical adaptation: response to environmental challenges, from life’s origins to the anthropocene. Sunderland, Massachusetts: Sinauer Associates, Inc. [Google Scholar]
- Stachowicz JJ, Terwin JR, Whitlatch RB, Osman RW.. 2002. Linking climate change and biological invasions: ocean warming facilitates nonindigenous species invasions. Proc Natl Acad Sci U S A. 99(24):15497–15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, et al. 2002. The Bioperl Toolkit: Perl modules for the life sciences. Genome Res. 12(10):1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek TH, Geller JB, Kreiser BR, Mitton JB.. 1997. Zoogeographic distributions of the sibling species Mytilus galloprovincialis and M. trossulus (Bivalvia: Mytilidae) and their hybrids in the North Pacific. Biol Bull. 193(2):187–194. [DOI] [PubMed] [Google Scholar]
- Sunday JM, Bates AE, Dulvy NK.. 2011. Global analysis of thermal tolerance and latitude in ectotherms. Proc R Soc B Biol Sci. 278(1713):1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD.. 2002. The rapid evolution of reproductive proteins. Nat Rev Genet. 3(2):137–144. [DOI] [PubMed] [Google Scholar]
- Tehei M, Daniel R, Zaccai G.. 2006. Fundamental and biotechnological applications of neutron scattering measurements for macromolecular dynamics. Eur Biophys J. 35(7):551–558. [DOI] [PubMed] [Google Scholar]
- Thunell RC. 1979. Pliocene – Pleistocene paleotemperature and paleosalinity history of the Mediterranean Sea: results from DSDP Sites 125 and 132. Mar Micropaleontol. 4:173–187. [Google Scholar]
- Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS.. 2007. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 369(5):1318–1332. [DOI] [PubMed] [Google Scholar]
- Tomanek L, Zuzow MJ.. 2010. The proteomic response of the mussel congeners Mytilus galloprovincialis and M. trossulus to acute heat stress: implications for thermal tolerance limits and metabolic costs of thermal stress. J Exp Biol. 213(20):3559–3574. [DOI] [PubMed] [Google Scholar]
- Trudeau DL, Kaltenbach M, Tawfik DS.. 2016. On the potential origins of the high stability of reconstructed ancestral proteins. Mol Biol Evol. 33(10):2633–2641. [DOI] [PubMed] [Google Scholar]
- Vermeij GJ. 1991. Anatomy of an invasion: the trans-Arctic interchange. Paleobiology 17(03):281–307. [Google Scholar]
- Vertrees J, Barritt P, Whitten S, Hilser VJ.. 2005. COREX/BEST server: a web browser-based program that calculates regional stability variations within protein structures. Bioinformatics 21(15):3318–3319. [DOI] [PubMed] [Google Scholar]
- Watt WB, Dean AM.. 2000. Molecular-functional studies of adaptive genetic variation in prokaryotes and eukaryotes. Annu Rev Genet. 34:593–622. [DOI] [PubMed] [Google Scholar]
- Westfall KM, Gardner JPA.. 2010. Genetic diversity of Southern hemisphere blue mussels (Bivalvia: Mytilidae) and the identification of non-indigenous taxa. Biol J Linn Soc. 101(4):898–909. [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Wrabl JO, et al. 2011. The role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem. 159(1):129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zavodszky P, Kardos J, Svingor A, Petsko GA.. 1998. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci U S A. 95(13):7406–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI.. 2007. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 3(1):e5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.