ABSTRACT
Streptococcus pneumoniae expresses capsular polysaccharides (CPSs) to protect itself from opsonophagocytic killing. The genes responsible for capsules synthesized by the Wzy-dependent mechanism, which accounts for 96 of the 98 known pneumococcal capsule types, are in a chromosomal region known as the cps locus. The nucleotide sequence in this region has been determined for all serotypes. In contrast, not all CPS structures have been defined. The structure of the serotype 35C polysaccharide was recently reported, but the presence of O-acetyltransferase genes in the serotype 35C cps locus suggested that it could be incomplete, as the reported structure contains no O-acetylation. In addition, the genetic distinction of serotype 35C from the closely related serotype 42 was unclear, as their reported cps loci are nearly identical. To clarify these discrepancies, we obtained serotype 35C and 42 clinical and reference isolates and studied their serological and genetic properties, as well as the structures of CPSs purified from reference isolates. We demonstrated that the O-acetyltransferase WciG was functional in serotype 35C but nonfunctional in serotype 42 due to a deletion in wciG. Serotype 35C was O-acetylated at the 5- and 6-positions of 3-β-galactofuranose, as well as the 2-position of 6-β-galactofuranose. However, serotype 42 has only O-acetylation at 3-β-galactofuranose, an observation consistent with its loss of WciG functionality, which is associated with O-acetylation at the 2-position and subsequent reaction with typing antiserum 35a. These findings provide a comprehensive view of the genetic, biochemical structural, and serological bases of serotypes 35C and 42.
KEYWORDS: O-acetyltransferase, Streptococcus pneumoniae, biochemical structure, capsular polysaccharide, genetic basis, serological profile, serotyping
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a major human pathogen frequently responsible for pneumonia, as well as sepsis and meningitis. Despite the availability of vaccines and antibiotics, pneumococcal infections remain a significant cause of morbidity and mortality: pneumococcal infections account for ∼11% of all worldwide deaths of children younger than 5 years old (1). As a species, pneumococci can express at least 98 chemically and serologically distinct capsule types (serotypes) (2, 3), but only a subset of these is frequently observed in sepsis and meningitis, which together are termed invasive pneumococcal diseases (IPDs). Pathogenic pneumococcal isolates from patients with IPDs almost always express polysaccharide capsules (4), which shield pneumococci from host immunity and dramatically increase their virulence (5).
The capsule is also the primary interface with the host, and antibodies against the capsule are protective. The highly successful pneumococcal conjugate vaccines (PCVs) are designed to elicit antibodies to the serotypes causing high rates of IPD. Widespread vaccination has removed the vaccine-targeted serotypes from both asymptomatic carriage (6) and IPD (7), but clearance of these serotypes from nasopharyngeal carriage has increased the relative prevalence of rarer serotypes such as serotype 35B (8, 9). Thus, serotypes that are currently rare may become more widespread after implementation of subsequent generations of conjugate vaccines.
Because vaccines must target serotypes that are prevalent in the populations that will receive the vaccine, it is essential to accurately monitor the distribution of pneumococcal serotypes in IPD. For example, after the 7-valent conjugate vaccine, which contains serotype 6B, was introduced, it appeared that serotype 6A rose in prevalence rather than being eliminated through cross-reactive immunity to serotype 6B. Through the use of monoclonal antibodies, it was determined that the increasing serotype was in actuality a previously unrecognized serotype, 6C and that serotype 6A had diminished as expected (10). Accurate epidemiology requires a comprehensive understanding of the structural and serologic bases of pneumococcal capsules in order to distinguish closely related serotypes. As indirect serotyping based on genetic sequences continues to replace traditional Quellung serotyping in popularity, it has become essential to understand the genetic bases differentiating serotypes as well.
While extensive studies of the capsule have led us to fully understand these relationships for most serotypes (2, 3), serotypes 35C and 42 are an exception. Antigenically, they are closely related, sharing most serological reactivities and differentiated by only factor serum 35a, which is reactive with serotype 35C but not serotype 42 (Table 1). Despite distinct immunological profiles, their reference cps loci are nearly identical (see Table S1 in the supplemental material) and contain the same genes in the same arrangement, with two intact O-acetyltransferase genes (wcjE and wciG [see Fig. 2A]) (11). However, biochemical studies showed that serotype 42 capsular polysaccharide (CPS) has two O-acetylations on its 3-linked β-galactofuranose (Galf) (12, 13), while a commercial serotype 35C capsule preparation contained no O-acetylation (13). In addition, both serotypes are known to bind ficolin-2, which targets O-acetyl groups in some pneumococcal serotypes (14). Taken together, these data suggest that our understanding of these serotypes is incomplete. Seeking to resolve these inconsistencies, we investigated the genetic, biochemical, and immunologic differences of the two serotypes.
TABLE 1.
Accepted reactivities of serotypes 35C, 42, and 4 with antisera and antigenic formulae of these serotypes
| Serotype | Reactivitya |
|||||
|---|---|---|---|---|---|---|
| Antiserum pool G | Factor serum |
|||||
| 35a | 35b | 35c | 29b | 42a | ||
| 35C | + | + | – | + | – | + |
| 42 | + | – | – | + | – | + |
| 4 | – | – | – | – | – | – |
+, Reaction; –, no reaction.
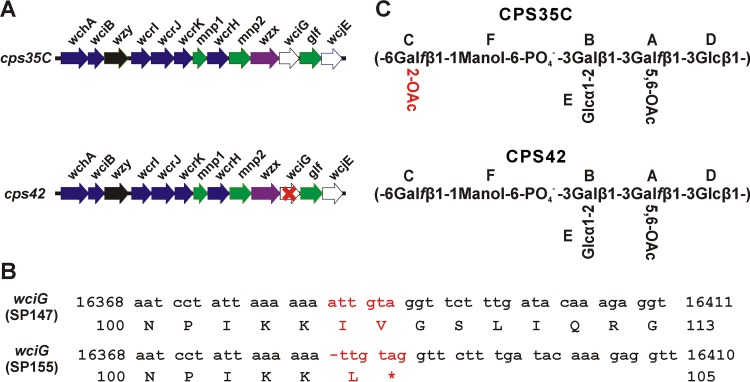
FIG 2.
Genetic and structural differences between serotypes 35C and 42. (A) Arrangement of structure-determining genes in the published cps loci of S. pneumoniae serotypes 35C (accession no. CR931706) and 42 (accession no. KY009533). The red “X” in wciG of cps42 reflects our finding in this study that wciG of serotype 42 is defective. (B) Comparison of nucleotide and amino acid sequences of wciG genes. The differences between the two sequences are highlighted in red. In cps42 of SP155, deletion of a single nucleotide “a” after nucleotide 16383 results in premature termination of WciG translation. Numbering is with respect to GenBank accession no. CR931706. (C) Complete structures of serotype 35C and 42 polysaccharides as determined by NMR spectroscopy. The uppercase letters in the structures indicate the residues whose 1H and 13C chemical shifts are given in Table 3 and in the text. The sole difference between the two polysaccharides is the O-acetylation at 2-position of residue C, which is highlighted in red.
RESULTS
Immunologic studies of isolates typed as serotypes 35C and 42.
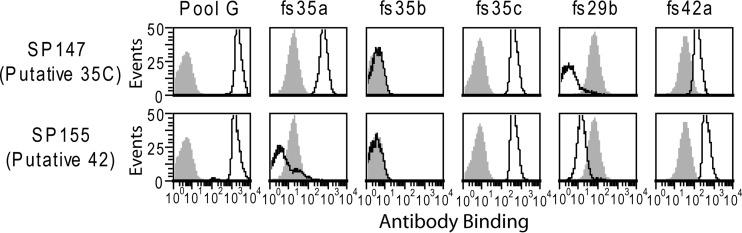
We obtained nine strains putatively assigned as serotype 35C and one putatively assigned as serotype 42 from the U.S. Centers for Disease Control and Prevention (CDC) (Table 2). This represents their entire inventory of these serotypes, as serotypes 35C and 42 are currently exceedingly rare in the United States (7, 15). We then characterized the serologic properties of these isolates with a flow cytometric serotyping assay using the commercial typing antisera listed in Table 1. Staining of the two reference isolates is shown in Fig. 1, which is representative for the remaining isolates. Pool G typing antiserum, which recognizes serotypes 35C and 42 (15), strongly stained all putative 35C and 42 isolates. Each isolate was positive for factor sera 35c and 42a, whereas binding by factor sera 35b and 29b was at or below the levels of the irrelevant serotype control (serotype 4 strain TIGR4), which represents binding by noncapsular antibodies in these reagents. Factor serum 35a is the sole reagent that distinguishes serotypes 35C and 42 (Table 1), and indeed it reacted with all putative serotype 35C isolates but not SP155, the serotype 42 reference isolate. Thus, SP155 was the sole isolate with the serotypic features associated with serotype 42.
TABLE 2.
Strains used in this study
| Straina | Putative serotype | FCSAb serotype | wciG status | GenBank accession no. | Sourcec |
|---|---|---|---|---|---|
| Reference isolates | |||||
| SP147 | 35C | 35C | Intact | KY091876 | CDC |
| SP155* | 42 | 42 | Disrupted | KY009533 | CDC |
| TIGR4 | 4 | 4 | NAd | NC_003028 | ATCC |
| Clinical isolates | |||||
| 5705-06† | 35C | 35C | Intact | KX470740 | CDC |
| PATH1895 | 35C | 35C | Intact | KY091869 | CDC |
| PATH1896 | 35C | 35C | Intact | KY091870 | CDC |
| PATH1901 | 35C | 35C | Intact | KY091871 | CDC |
| PATH2897 | 35C | 35C | Intact | KY091872 | CDC |
| PATH4040 | 35C | 35C | Intact | KY091873 | CDC |
| PATH4191 | 35C | 35C | Intact | KY091874 | CDC |
| PATH4348 | 35C | 35C | Intact | KY091875 | CDC |
| Laboratory-derived strains | |||||
| KAG1017†e | KY124245 | This study | |||
| KAG1021*f | This study |
*, The mnp2 gene in strains SP155 and KAG1021 encodes 14637A→G with respect to the reference serotype 35C cps locus (GenBank accession no. CR931706); †, the mnp2 genes in strains 5705-06 and KAG1017 correspond to CR931706.
FCSA, flow cytometric serotyping assay.
CDC, Centers for Disease Control and Prevention, Active Bacterial Core surveillance, Atlanta, GA; ATCC, American Type Culture Collection, Manassas, VA.
NA, not applicable.
5705-06ΔwciG.
SP155 aliA::pKAG2008; aliA+ wciG+.
FIG 1.
Serological analysis of putative serotype 35C and 42 reference isolates. Indicated strains were assayed by flow cytometric staining for binding to the indicated typing antisera. Gray shading represents binding to TIGR4, which does not contain the indicated antigens (see Table 1) and represents binding by antibodies in these reagents directed against epitopes other than the capsule.
Genetic basis of serotype 42.
We recently reported that functionality of the putative O-acetyltransferase WciG is associated with factor serum 35a recognition in serogroup 35 (3). Since serotype 42 is serologically identical to serotype 35C except for recognition by factor serum 35a, we hypothesized that wciG of serotype 42 may have inactivating mutations. Indeed, upon sequencing serotype 42 isolate SP155, we found that the cps locus contains a single nucleotide deletion in wciG that results in frameshift and premature termination at residue 105 of 333 (Fig. 2B). Inactivation of wciG is consistent with the presence of only a single acetylated residue of the serotype 42 polysaccharide repeat unit, putatively mediated by WcjE, in the other reported serotype 42 structures (12, 13). The wciG genes of all isolates serologically classified as serotype 35C encoded an intact WciG (see Table 2 for accession numbers).
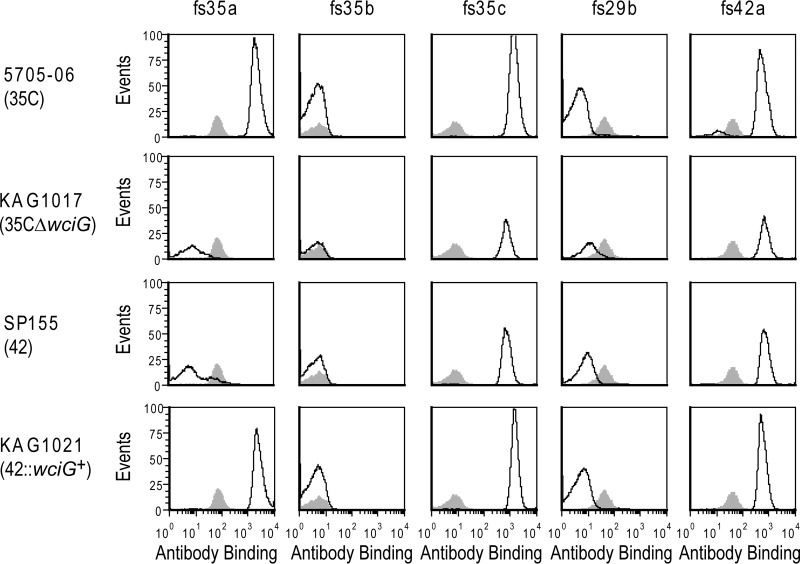
Across the coding sequence of the cps locus, SP155 encodes only one other deviation from the serotype 35C reference sequence (GenBank accession no. CR931706), a polymorphism in mnp2, which results in a missense substitution at amino acid 358 of 359 (G358R with respect to GenBank accession no. CR931706). Although it seemed unlikely that the mnp2 polymorphism accounted for the phenotype, to confirm that wciG functionality was responsible for the observed serologic differences, we created a wciG-null mutant in the serotype 35C strain 5705-06 (Fig. 3). This strain, KAG1017, did not react with factor serum 35a and was serologically typed as serotype 42. To perform the complementary experiment, we introduced a functional wciG into the SP155 genome; the resulting strain, KAG1021, reacted with factor serum 35a and thus became serologically 35C (Fig. 3). Therefore, WciG functionality is the determinant distinguishing serotype 35C (functional WciG) from serotype 42 (nonfunctional WciG). Since WciG mediates O-acetylation in serotype 35B (3), this result suggested that CPS35C was O-acetylated and that the reported structure of CPS35C (13) was incomplete.
FIG 3.
Serological panel demonstrating that WciG functionality differentiates serotypes 35C and 42. The indicated strains were assayed by flow cytometric staining for binding to the indicated typing antisera. Gray shading represents binding to TIGR4, which does not contain the indicated antigens (see Table 1) and represents binding by antibodies to noncapsular epitopes.
Structural differences between CPS35C and CPS42.
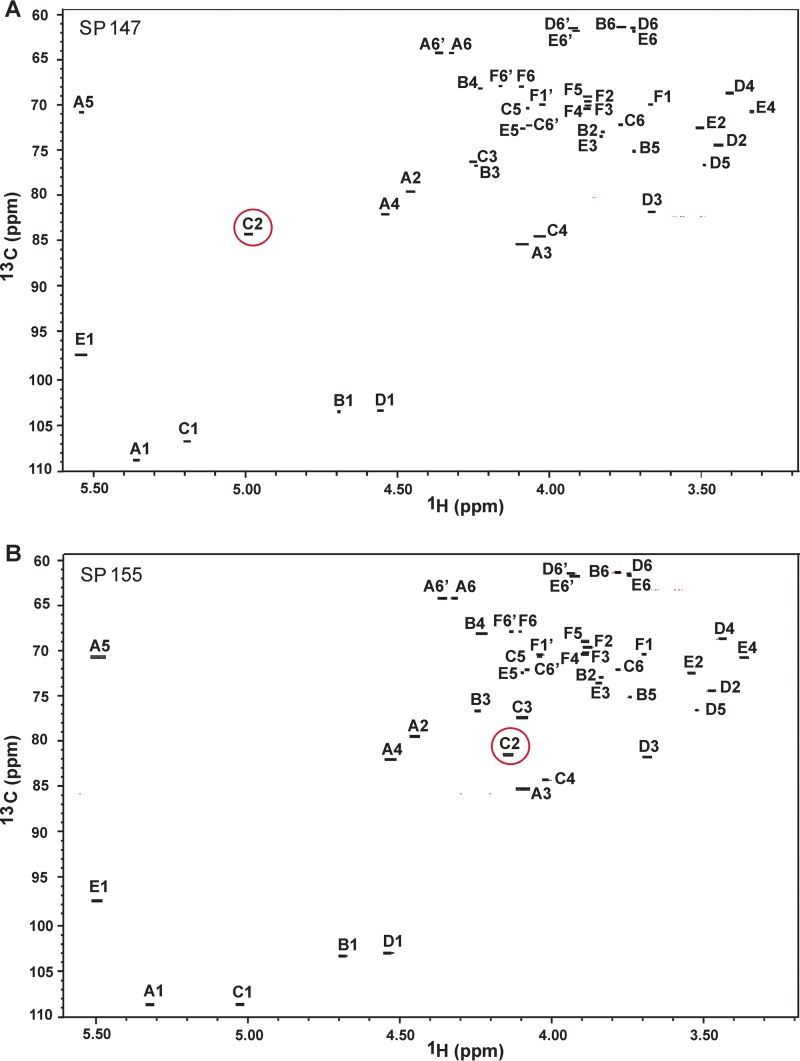
To characterize the O-acetylation of the serotype 35C and 42 capsules, capsular polysaccharides were purified from isolates SP147 (serotype 35C) and SP155 (serotype 42) (Table 2), and their structures were determined by high-resolution nuclear magnetic resonance (NMR) spectroscopy. The NMR spectra and complete chemical shift assignments are presented in Fig. 4 and 5 and Tables 3 and 4. The 1H-13C heteronuclear single quantum coherence (HSQC) spectra of these polysaccharides matched well with those of a commercial CPS42 preparation (13) with the exception of peaks corresponding to residue C (6-β-Galf) in serotype 35C, which were shifted and suggested differences at this residue in serotype 35C (capital letters, e.g., “C,” represent the residues in capsular polysaccharides and are underlined to distinguish these from carbon atoms).
FIG 4.
Anomeric and central regions of multiplicity-edited HSQC spectra of serotype 35C (A) and serotype 42 (B) polysaccharides. The chemical shift of 1H in residue C at the 2-position (C2, red circle) is affected by O-acetylation.
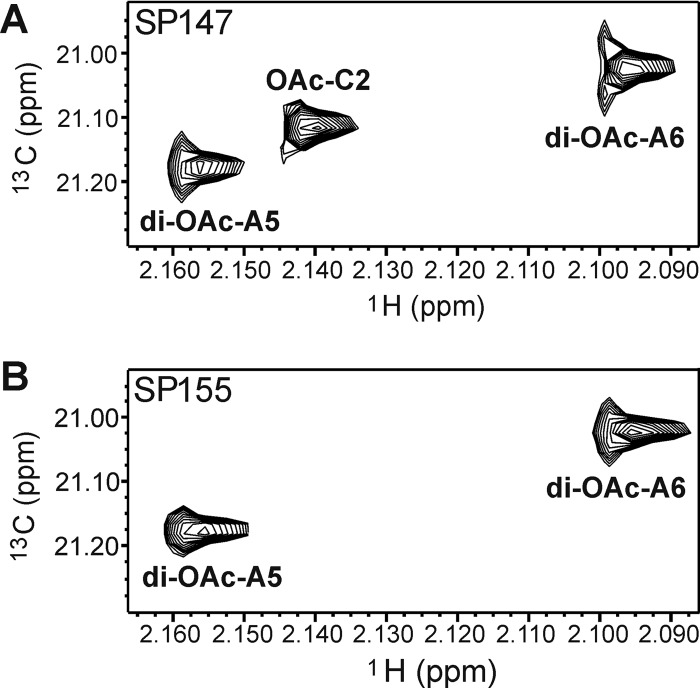
FIG 5.
1H-13C HSQC spectra in the O-acetyl methyl regions of serotype 35C (A) and serotype 42 (B) polysaccharides. Chemical shifts of these peaks are listed in Table 4. This peak at 2.140/21.12 ppm is assigned O-acetylation at 2-position of residue C (OAc-C2), which is clearly present in serotype 35C but completely lost in serotype 42. Both isolates contain full O-acetylation at the 5- and 6-positions of residue A (di-OAc-A5 and di-OAc-A6).
TABLE 3.
Residue-by-residue comparison of HSQC 1H and 13C chemical shift of S. pneumoniae serotype 35C and 42 polysaccharides
| Polysaccharide (isolate source) | Residue/structure | Chemical shift (ppm)a |
|||||
|---|---|---|---|---|---|---|---|
| 1-H, 1-C | 2-H, 2-C | 3-H, 3-C | 4-H, 4-C | 5-H, 5-C | 6-H, 6-C | ||
| 35C (SP147) | A/3-β-Galf | 5.360 | 4.457 | 4.109 | 4.540 | 5.540 | 4.322, 4.362 |
| 108.80 | 79.64 | 85.50 | 82.17 | 70.87 | 64.29 | ||
| 42 (SP155) | 5.361 | 4.457 | 4.108 | 4.540 | 5.542 | 4.322, 4.362 | |
| 108.80 | 79.63 | 85.49 | 82.18 | 70.87 | 64.29 | ||
| 35C (SP147) | B/3-β-Galp | 4.692 | 3.824 | 4.241 | 4.231 | 3.721 | 3.763 |
| 103.55 | 73.06 | 76.76 | 68.21 | 75.25 | 61.41 | ||
| 42 (SP155) | 4.703 | 3.820 | 4.243 | 4.229 | 3.722 | 3.765 | |
| 103.57 | 73.07 | 76.80 | 68.20 | 75.25 | 61.41 | ||
| 35C (SP147) | C/6-β-Galf | 5.192 | 4.990 | 4.250 | 4.072 | 4.071 | 3.769, 4.073 |
| 106.70 | 84.40 | 76.41 | 84.66 | 70.48 | 72.31 | ||
| 42 (SP155) | 5.054 | 4.138 | 4.091 | 4.019 | 4.031 | 3.763, 4.074 | |
| 108.78 | 81.70 | 77.57 | 83.91 | 70.57 | 72.25 | ||
| 35C (SP147) | D/3-β-Glcp | 4.556 | 3.443 | 3.663 | 3.406 | 3.489 | 3.724, 3.922 |
| 103.37 | 74.54 | 81.92 | 68.77 | 76.72 | 61.52 | ||
| 42 (SP155) | 4.551 | 3.443 | 3.664 | 3.406 | 3.494 | 3.726, 3.924 | |
| 103.35 | 74.54 | 81.92 | 68.77 | 76.71 | 61.54 | ||
| 35C (SP147) | E/3-α-Glcp | 5.542 | 3.504 | 3.829 | 3.335 | 4.087 | 3.721, 3.912 |
| 97.82 | 72.63 | 73.69 | 70.85 | 72.58 | 61.75 | ||
| 42 (SP155) | 5.542 | 3.513 | 3.829 | 3.333 | 4.090 | 3.726, 3.913 | |
| 97.82 | 72.62 | 73.69 | 70.85 | 72.58 | 61.76 | ||
| 35C (SP147) | F/1-Manol-6 | 3.669, 4.020 | 3.873 | 3.873 | 3.876 | 3.876 | 4.102, 4.161 |
| 70.02 | 69.67 | 70.19 | 70.38 | 69.14 | 68.00 | ||
| 42 (SP155) | 3.764, 4.031 | 3.868 | 3.878 | 3.877 | 3.875 | 4.097, 4.158 | |
| 70.52 | 69.72 | 70.26 | 70.39 | 69.12 | 67.97 | ||
Chemical shifts of polysaccharides were recorded at 25°C. Pairs of values separated by a comma are H, H′ values. Underlined numbers indicate O-acetylated positions, and numbers in boldface indicate di-O-acetylated positions.
TABLE 4.
O-acetyl groups identified in the serotype 35C polysaccharide
| Position on residuea | Chemical shift (ppm) |
|||
|---|---|---|---|---|
| Methyl 1H | Methyl 13C | Carbonyl 13C | Link 1H | |
| 6-di-OAc on residue A | 2.096 | 21.03 | 174.70 | 4.322, 4.362 |
| 5-di-OAc on residue A | 2.156 | 21.18 | 174.42 | 5.540 |
| 2-OAc on residue C | 2.140 | 21.12 | 174.13 | 4.990 |
The number indicates position of the carbon atom on sugar residues. Residue A is 3-β-galactofuranose, and residue C is 6-β-galactofuranose in the serotype 35C repeat unit. OAc, O-acetylation.
The acetyl/methyl regions of HSQC (Fig. 5) clearly demonstrated the distinct difference between the two polysaccharides. CPS from SP147 had two O-acetyl peaks at the same chemical shifts as those of the SP155 CPS but contained an additional peak at 2.140 ppm in 1H/21.12 ppm in 13C absent in SP155 (Fig. 5A). The site of this unique O-acetyl/methyl group was determined using standard NMR scalar coupling correlations. As noted above, the chemical shifts observed for other residues were identical in these polysaccharides (Table 3 and Fig. 4); thus, the unique feature of serotype 35C must arise from O-acetylation of residue C. In all resonances of this residue, the peak at 5.192 ppm in 1H/106.70 ppm in 13C was characteristic of an anomeric center. HSQC-total correlation spectroscopy (TOCSY) showed cross peaks between 5.192 ppm (C1-H) and 84.40 ppm (C2-C) and 76.41 ppm (C3-C). The former peak was correlated with the resonance at 4.990 ppm (C2-H), and the latter peak was correlated with the resonance at 4.250 ppm (C3-H) by double-quantum-filtered coherence spectroscopy (DQF-COSY). The assignments of the other resonances overlapped with those of the unacetylated residue C (13). In addition, this O-acetyl/methyl group resonance was correlated with the carbonyl carbon resonance at 174.13 ppm, which in turn showed a cross peak to 4.990 ppm (C2-H) (Table 4). These results suggested that the unique spectrum of serotype 35C represented an O-acetyl group at the 2-position of residue C (C2). This position was 100% O-acetylated since only one peak appeared at 4.990 ppm of C2-H in CPS35C of SP147 in the HSQC spectrum (Fig. 4A). We found no evidence of this O-acetylation in serotype 42, where the peak at 4.138 ppm of C2-H represented unacetylated form (Fig. 4B).
Both polysaccharides were O-acetylated at residue A (Fig. 5). The two O-acetyl peaks at 2.156/21.18 and 2.096/21.03 in 1H/13C represented di-O-acetyl groups at 5- and 6-positions of residue A (3-β-Galf). The relative heights of these peaks in the HSQC spectra suggested 100% di-O-acetylation forms in both; however, in our previously reported structure of commercial serotype 42 polysaccharide, there was one form with 54% O-acetylation at both A5 and A6, a second form with 30% acetylation at A6, and a third form without acetylation (13).
As shown in Fig. 2C, the completed structure of serotype 35C contains O-acetylation at the 2-position of 6-β-Galf and 5- and 6-positions of 3-β-Galf. Serotype 42 lacks O-acetylation on 3-β-Galf because of its nonfunctional WciG.
DISCUSSION
We demonstrated here the genetic, biochemical, and serological bases for serotypes 35C and 42 and revealed that WciG functionality distinguishes these closely related serotypes. Serotype 35C encodes two functional O-acetyltransferases, WciG and WcjE, whereas serotype 42 encodes functional WcjE but a nonfunctional WciG. The chemical structures suggest that WcjE acetylates the 5- and 6-positions of 3-β-Galf in the polysaccharides of both serotypes. This is consistent with other reports of O-acetylation in serotypes 20, 31, 33A, 35A, and 47A (2), which also carry wcjE (11, 16). Thus, WciG acetylates the 2-position of the 6-β-Galf in the serotype 35C repeat unit, and 2-OAc-βGalf (“OAc” represents O-acetylation) is essential for factor serum 35a recognition. This is supported by the recent report of serotype 35D, which is a WciG-deficient variant of serotype 35B and also fails to bind factor serum 35a (3). The presence of wciG in the cps locus is associated with 2-OAc-βGalf in many serotypes for which the structures are known (2, 11).
The completed structure of the serotype 35C polysaccharide explains and reconciles the genetic, biochemical and serologic characteristics of serotypes 35C and 42. We previously reported the structure of a commercial preparation of CPS35C, which had no O-acetylation (i.e., <10%) (13). We speculated that the commercial preparation was purified under different conditions and may not have preserved the capsular O-acetylation. However, it is unclear whether these differences were due to strain-to-strain variation or to differences in cultivation or capsular purification since O-acetyl groups are somewhat labile. In addition, our results suggest that the previously reported DNA sequence of serotype 42 cps locus (GenBank accession no. CR931715), which contains an intact wciG, appears to have been obtained from a serotype 35C isolate that was mistyped as serotype 42. Because serological discrimination of the two closely related serotypes relies on a single reagent (factor serum 35a) targeting a labile and possibly variable modification, serotype 35C may be easily mistyped as serotype 42 or vice versa (11). Indeed, serotype 42 was the most frequent incorrectly assigned serotype in a study of serology-based serotyping in Europe, assigned as serotype 35C by half the laboratories tested (17); serotype 35C was not included in the study. In addition, some serotype 35C isolates might inherently express reduced amounts of O-acetylation due, presumably, to the influence of elements outside the cps locus resulting in weak factor serum 35a recognition and the recording of a negative result. Further studies of serotype 35C isolates may provide additional insights into this common case of mistaken identity.
Elucidation of the genetic basis for serotype 42 will directly assist studies of pneumococcal epidemiology. Continuous monitoring of pneumococcal serotype epidemiology is vital to understanding the impact of PCVs since their efficacy depends on pneumococcal serotype distributions, and PCVs have altered distributions of pneumococcal serotypes. Serotypes are increasingly determined indirectly by genotyping; indeed, the CDC currently identifies pneumococcal serotypes of isolates obtained through their Active Bacterial Core surveillance (ABCs) program by analysis of genetic sequences obtained by whole-genome sequencing. Our information should enable the CDC and others to easily distinguish serotypes 35C and 42, which are difficult to distinguish even with typing antisera.
Historically, serotype 42 was excluded from serogroup 35 because it did not react with factor serum 35a and, according to the tradition of the Danish naming system, a member of any serogroup should react with factor serum “a” of the serogroup. Given the close relationship between serotypes 35C and 42, it could be reasonable to consider serotype 42 as a member of serogroup 35. However, as our understanding of the genetic, structural, and serological relationships between serotypes evolves, relationships of the serotypes seem likely to become more muddied still, and it is more important to be aware of the limitations of the classical nomenclature than it is to ensure that a given serotype has the “correct” name.
Acetyl groups are often the epitopes of antibodies. They are the targets of many serotyping reagents, innate immune molecules (e.g., ficolin-2 [14, 18] and siglecs [19]), and vaccine-elicited antibodies (20). Pneumococcal cps loci harbor many acetyltransferase genes (11), and several serogroups contain serotype pairs: one with a functional transferase and the other with nonfunctional transferase. For example, WcjE-positive and -negative pairs have been observed in serogroups 9, 11, and 33 (11, 21). wciG is carried by 13 serotypes, including serotype 42 and every member of serogroup 35 (11), and wciG functionality differentiates serotype 35B from the recently identified serotype 35D (3, 22). Thus, wciG functionality may be an as-yet-unexplored mechanism of significantly increasing capsule diversity among pneumococci. Since O-acetylation of polysaccharides is an important modification in many bacteria, such as Shigella spp. (23, 24), Salmonella spp. (25), Neisseria meningitidis (26–28), and Streptococcus agalactiae (29), it would be beneficial to systematically study variation in O-acetyltransferase functionality in many bacterial species.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are indicated in Table 2. Pneumococci were grown on blood agar plates overnight at 37°C with 5% CO2. Plated bacteria were inoculated into Todd-Hewitt broth (Difco) with 5% yeast extract (THY). Glycerol stocks were prepared by mixing two parts culture with two parts fresh THY and one part 80% glycerol and freezing at −70°C in aliquots. Transformations were plated on THY agar plates and overlaid with THY agar containing 25 μg/ml 2-,3-,5-triphenyltetrazolium chloride to aid visualization of colonies. When specified, THY broth and THY overlay were supplemented with spectinomycin (100 μg/ml) or erythromycin (0.3 μg/ml).
FCSAs.
Flow cytometric serotyping assays (FCSAs) were performed as previously described (3) using frozen glycerol stocks as prepared above by staining bacteria with the indicated reagents, washing by centrifugation, and staining with secondary antibodies. Commercial typing antisera and factor sera (Table 2) were obtained from Statens Serum Institut (SSI, Denmark) and used at a 1:10,000 final dilution prior to detection with a phycoerythrin-conjugated anti-rabbit secondary antibody (product 4010-09; Southern Biotech, Birmingham, AL) at a final dilution of 1:1,000. After washing, stained bacteria were read on a BD FACSCalibur.
DNA sequencing.
DNA sequencing was performed by the Heflin Center for Genomic Science at the University of Alabama at Birmingham using Sanger sequencing, except for strain SP155. SP155 was sequenced and assembled by the Centers for Disease Control and Prevention Active Bacterial Core surveillance program using Illumina sequencing and VelvetOptimiser.
Genetic manipulations.
Primers used in this study are presented in Table S2 in the supplemental material. wciG was deleted in serotype 35C strain 5705-06 through allelic exchange with the aad9 spectinomycin resistance gene. Briefly, fragments of upstream and downstream flanking homology of the cps locus were amplified from prepared chromosomal DNA using primer sets 5163/3184 and 5230/3160 with homology at the 3′ and 5′ ends, respectively, to aad9. aad9 was amplified from pCLT1242 (30) using the primer set 5229/3184. Fragments were connected by overlap extension PCR and transformed into pneumococci as previously described (3) with selection for spectinomycin resistance, yielding strain KAG1017. The surrounding region was sequenced to verify that only the desired sequence was affected. Strain SP155 was complemented for wciG by insertion of 5705-06 wciG downstream of aliA using a pneumococcal suicide vector as previously described (3). The resulting strain, KAG1021, was sequenced in the affected region to verify correct insertion of the plasmid.
Biochemical studies.
Capsular polysaccharides of SP155 and SP147 were purified by ethanol precipitation and anion-exchange chromatography as previously described (31) with the exception that dialysis was performed against 5 mM Tris-HCl at pH ∼7 rather than at pH 7.4, with a subsequent purification by size exclusion chromatography over Sephacryl S300 resin. After each purification, polysaccharide was detected by an inhibition enzyme-linked immunosorbent assay (ELISA) in which polysaccharide in purification fractions competed against plate-bound serotype 35C polysaccharide (SSI) for binding of factor serum 42a (used at 1:2,700 dilution). Polysaccharide-containing fractions were pooled, dialyzed against water, and lyophilized.
NMR spectroscopy.
NMR studies were performed as previously described (13) with native polysaccharides. Prior to NMR, polysaccharides (3 to 10 mg) were lyophilized twice from 99.8% D2O and taken up in 99.996% D2O. Spectra were recorded at 25°C in Bruker Avance spectrometers running Topspin 3 software at 500 and 600 MHz. All proton and carbon chemical shifts were referenced relative to internal acetone using δ 1H = 2.225 ppm and δ 13C = 31.07 ppm.
Multiplicity-edited HSQC was used to distinguish methylene from methine groups, which was useful for identification of C6 groups of hexoses, as well as C1 and C6 groups of alditols in polysaccharides. The common homonuclear two-dimensional NMR methods of DQF-COSY, TOCSY, and nuclear overhauser effect spectroscopy (NOESY) were augmented by the hybrid method HSQC-TOCSY, which, in the crowded carbohydrate spectra, was enhanced by high digital resolution in the indirect dimension (13C). A second hybrid pulse sequence, HSQC-NOESY, also recorded at high digital resolution in 13C, was especially valuable for correlating C5 of β-Galp with H1 and H4. Heteronuclear multiple bond correlation (HMBC) spectra were used for identifying linkage positions and assignments of residues. All NMR data were processed by NMRpipe and NMRDraw (NMRScience) with analysis by NMRview (One Moon Scientific).
Accession number(s).
All relevant nucleotide sequences were submitted to GenBank under the accession numbers presented in Table 2.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Centers for Disease Control and Prevention Streptococcus lab for providing isolates and their sequence data for SP155. In particular, we thank Bernard Beall, Sopio Chochua, Robert E. Gertz, Jr., Lesley McGee, and Benjamin J. Metcalf.
This study was supported by the National Institutes of Health (NIH) grants HL105346 (K.A.G.) and AG050607 (M.H.N.), NIH grant HHSN272201200005D (M.H.N.), NIH P01-HL-107153 (C.A.B.), and the National Natural Science Foundation of China grant 31470003 (J.Y.). The University of Alabama at Birmingham (UAB) has intellectual property rights on some reagents developed by M.H.N., and M.H.N. and K.A.G. are UAB employees. The authors declare no additional conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00822-17.
REFERENCES
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geno KA, Saad JS, Nahm MH. 2017. Discovery of novel pneumococcal serotype, 35D: a natural WciG-deficient variant of serotype 35B. J Clin Microbiol 55:1416–1425. doi: 10.1128/JCM.00054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park IH, Geno KA, Sherwood LK, Nahm MH, Beall B. 2014. Population-based analysis of invasive nontypeable pneumococci reveals that most have defective capsule synthesis genes. PLoS One 9:e97825. doi: 10.1371/journal.pone.0097825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery OT, Dubos R. 1931. The protective action of a specific enzyme against type III pneumococcus infection in mice. J Exp Med 54:73–89. doi: 10.1084/jem.54.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Brien KL, Dagan R. 2003. The potential indirect effect of conjugate pneumococcal vaccines. Vaccine 21:1815–1825. doi: 10.1016/S0264-410X(02)00807-1. [DOI] [PubMed] [Google Scholar]
- 7.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, Reingold A, Thomas A, Schaffner W, Craig AS, Smith PJ, Beall BW, Whitney CG, Moore MR. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis 201:32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 8.Miyazaki H, Shibuya R, Midorikawa N, Chang B, Ohnishi M, Matsumoto T. 2017. Serotype distribution and antimicrobial susceptibility of Streptococcus pneumoniae strains isolated in Japan after introduction of the routine immunization program. J Infect Chemother 23:234–240. doi: 10.1016/j.jiac.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 9.Olarte L, Kaplan SL, Barson WJ, Romero JR, Lin PL, Tan TQ, Hoffman JA, Bradley JS, Givner LB, Mason EO, Hulten KG. 2017. Emergence of multidrug-resistant pneumococcal serotype 35B among children in the United States. J Clin Microbiol 55:724–734. doi: 10.1128/JCM.01778-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MC, Nahm MH. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol 45:1225–1233. doi: 10.1128/JCM.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen BO, Meier S, Paulsen BS, Redondo AR, Skovsted IC. 2014. Determination of native capsular polysaccharide structures of Streptococcus pneumoniae serotypes 39, 42, and 47F and comparison to genetically or serologically related strains. Carbohydr Res 395:38–46. doi: 10.1016/j.carres.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Bush CA, Cisar JO, Yang J. 2015. Structures of capsular polysaccharide serotypes 35F and 35C of Streptococcus pneumoniae determined by nuclear magnetic resonance and their relation to other cross-reactive serotypes. J Bacteriol 197:2762–2769. doi: 10.1128/JB.00207-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brady AM, Calix JJ, Yu J, Geno KA, Cutter GR, Nahm MH. 2014. Low invasiveness of pneumococcal serotype 11A is linked to ficolin-2 recognition of O-acetylated capsule epitopes and lectin complement pathway activation. J Infect Dis 210:1155–1165. doi: 10.1093/infdis/jiu195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Statens Serum Institut. 2013. Streptococcus pneumoniae: textbook in serotyping, virulence factors, and enzyme-linked immunosorbent assay (ELISA) for measuring pneumococcal antibodies. Statens Serum Institut, Hillerød, Denmark. [Google Scholar]
- 16.Beynon LM, Richards JC, Perry MB. 1997. Identification of the common antigenic determinant shared by Streptococcus pneumoniae serotypes 35A and 20 capsular polysaccharides: structural analysis of the Streptococcus pneumoniae serotype 35A capsular polysaccharide. Eur J Biochem 250:163–167. doi: 10.1111/j.1432-1033.1997.00163.x. [DOI] [PubMed] [Google Scholar]
- 17.Konradsen HB. 2005. Validation of serotyping of Streptococcus pneumoniae in Europe. Vaccine 23:1368–1373. doi: 10.1016/j.vaccine.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Krarup A, Thiel S, Hansen A, Fujita T, Jensenius JC. 2004. L-ficolin is a pattern recognition molecule specific for acetyl groups. J Biol Chem 279:47513–47519. doi: 10.1074/jbc.M407161200. [DOI] [PubMed] [Google Scholar]
- 19.Carlin AF, Lewis AL, Varki A, Nizet V. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol 189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajam G, Carlone GM, Romero-Steiner S. 2007. Functional antibodies to the O-acetylated pneumococcal serotype 15B capsular polysaccharide have low cross-reactivities with serotype 15C. Clin Vaccine Immunol 14:1223–1227. doi: 10.1128/CVI.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calix JJ, Nahm MH. 2010. A new pneumococcal serotype, 11E, has a variably inactivated wcjE gene. J Infect Dis 202:29–38. doi: 10.1086/653123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staples M, Graham RM, Hicks V, Strachan J, Goncalves Da Silva A, Peverall J, Wicks V, Jennison AV. 2017. Discovery of Streptococcus pneumoniae serogroup 35 variants in Australian patients. Clin Microbiol Infect doi: 10.1016/j.cmi.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 23.Clark CA, Beltrame J, Manning PA. 1991. The oac gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene 107:43–52. doi: 10.1016/0378-1119(91)90295-M. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Knirel YA, Lan R, Senchenkova SN, Luo X, Perepelov AV, Wang Y, Shashkov AS, Xu J, Sun Q. 2014. Identification of an O-acyltransferase gene (oacB) that mediates 3- and 4-O-acetylation of rhamnose III in Shigella flexneri O antigens. J Bacteriol 196:1525–1531. doi: 10.1128/JB.01393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slauch JM, Mahan MJ, Michetti P, Neutra MR, Mekalanos JJ. 1995. Acetylation (O-factor 5) affects the structural and immunological properties of Salmonella typhimurium lipopolysaccharide O antigen. Infect Immun 63:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry DS, Lynn F, Lee CH, Frasch CE, Bash MC. 2002. Effect of O acetylation of Neisseria meningitidis serogroup A capsular polysaccharide on development of functional immune responses. Infect Immun 70:3707–3713. doi: 10.1128/IAI.70.7.3707-3713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borrow R, Longworth E, Gray SJ, Kaczmarski EB. 2000. Prevalence of de-O-acetylated serogroup C meningococci before the introduction of meningococcal serogroup C conjugate vaccines in the United Kingdom. FEMS Immunol Med Microbiol 28:189–191. doi: 10.1111/j.1574-695X.2000.tb01475.x. [DOI] [PubMed] [Google Scholar]
- 28.Claus H, Borrow R, Achtman M, Morelli G, Kantelberg C, Longworth E, Frosch M, Vogel U. 2004. Genetics of capsule O-acetylation in serogroup C, W-135, and Y meningococci. Mol Microbiol 51:227–239. doi: 10.1046/j.1365-2958.2003.03819.x. [DOI] [PubMed] [Google Scholar]
- 29.Lewis AL, Nizet V, Varki A. 2004. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc Natl Acad Sci U S A 101:11123–11128. doi: 10.1073/pnas.0403010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong S, McPherson SA, Tan L, Chesnokova ON, Turnbough CL Jr, Pritchard DG. 2008. Anthrose biosynthetic operon of Bacillus anthracis. J Bacteriol 190:2350–2359. doi: 10.1128/JB.01899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton RL, Geno KA, Saad JS, Nahm MH. 2016. Pneumococcus with the “6E” cps locus produces serotype 6B capsular polysaccharide. J Clin Microbiol 54:967–971. doi: 10.1128/JCM.03194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.