ABSTRACT
In Pseudomonas aeruginosa, the ferric uptake regulator (Fur) protein controls both metabolism and virulence in response to iron availability. Differently from other bacteria, attempts to obtain fur deletion mutants of P. aeruginosa failed, leading to the assumption that Fur is an essential protein in this bacterium. By investigating a P. aeruginosa conditional fur mutant, we demonstrate that Fur is not essential for P. aeruginosa growth in liquid media, biofilm formation, and pathogenicity in an insect model of infection. Conversely, Fur is essential for growth on solid media since Fur-depleted cells are severely impaired in colony formation. Transposon-mediated random mutagenesis experiments identified pyochelin siderophore biosynthesis as a major cause of the colony growth defect of the conditional fur mutant, and deletion mutagenesis confirmed this evidence. Impaired colony growth of pyochelin-proficient Fur-depleted cells does not depend on oxidative stress, since Fur-depleted cells do not accumulate higher levels of reactive oxygen species (ROS) and are not rescued by antioxidant agents or overexpression of ROS-detoxifying enzymes. Ectopic expression of pch genes revealed that pyochelin production has no inhibitory effects on a fur deletion mutant of Pseudomonas syringae pv. tabaci, suggesting that the toxicity of the pch locus in Fur-depleted cells involves a P. aeruginosa-specific pathway(s).
IMPORTANCE Members of the ferric uptake regulator (Fur) protein family are bacterial transcriptional repressors that control iron uptake and storage in response to iron availability, thereby playing a crucial role in the maintenance of iron homeostasis. While fur null mutants of many bacteria have been obtained, Fur appears to be essential in Pseudomonas aeruginosa for still unknown reasons. We obtained Fur-depleted P. aeruginosa cells by conditional mutagenesis and showed that Fur is dispensable for planktonic growth, while it is required for colony formation. This is because Fur protects P. aeruginosa colonies from toxicity exerted by the pyochelin siderophore. This work provides a functional basis to the essentiality of Fur in P. aeruginosa and highlights unique properties of the Fur regulon in this species.
KEYWORDS: biofilm, colony formation, infection, iron uptake, oxidative stress, Pseudomonas aeruginosa, pyochelin, siderophores
INTRODUCTION
Iron is a critical nutrient for almost all forms of life, because of its fundamental role as a redox cofactor of important enzymes involved in different cellular functions, such as respiration, catabolism, amino acid and nucleoside synthesis, and stress response. Although essential for nutrition, iron can also have deleterious effects if supplied in excess to living organisms. Indeed, intracellular Fe2+ catalyzes the formation of reactive oxygen species (ROS) through the Fenton reaction, which can damage biological macromolecules, such as proteins and nucleic acids, and lipids (1).
To reconcile their nutritional iron requirement with the need to maintain the intracellular iron content below hazardous levels, bacteria have evolved sophisticated strategies to control iron homeostasis. While most bacteria have the genetic potential to express different mechanisms for iron acquisition, as a general rule, the expression of iron uptake genes is repressed when sufficient amounts of intracellular iron are available. This negative control occurs through the activity of a transcriptional repressor of the Fur (or DtxR) family (2). When loaded with Fe2+, this repressor forms homodimers that bind the operator sequence (Fur box) within target promoters, thereby blocking the transcription of iron uptake genes (3). In addition to this pivotal regulatory mechanism, which is responsible for the restricted expression of iron uptake genes under conditions of iron starvation, iron acquisition is also regulated by additional mechanisms that overall allow optimal expression of a given iron uptake system only when it is effective in delivering iron to the cell (4). In many bacteria, Fur also positively controls the expression of nonessential iron-using proteins or ROS-detoxifying enzymes by repressing the transcription of small RNAs, which in turn inhibits the translation of target mRNAs (3, 5). As a result, genes that are involved in iron storage and/or detoxification are induced in the presence of high intracellular iron levels, thus counteracting free-iron toxicity, as well as decreasing the amount of sequestered (protein-bound) iron under low-iron conditions.
In spite of the crucial role of Fur in preserving iron homeostasis in bacterial cells, fur gene knockout mutants of several bacterial species have been obtained. These mutants showed variable and species-specific phenotypes, ranging from constitutive expression of iron uptake systems and/or virulence factors to impaired acid, serum, or oxidative stress resistance and defective motility and/or biofilm formation (3). In general, fur deletion has minor effects on bacterial growth in vitro though it can affect pathogenicity, since fur mutants of some pathogenic species appear less virulent and/or invasive than their wild-type counterparts (3, 6). However, this is not a rule, given that some bacterial pathogens, such as pathogenic Escherichia coli strains and Vibrio vulnificus, showed no differences in infectivity between wild-type and fur mutant strains in animal models (7–9), suggesting that the relevance of Fur during bacterial infections is species specific and likely infection model specific.
Notably, fur (or its analogous gene ideR) has been described as an essential gene in few bacterial species, such as Pseudomonas aeruginosa and Mycobacterium tuberculosis, because of the inability to obtain viable fur knockout mutants (10–12). The relevance of fur for the fitness of the opportunistic human pathogen P. aeruginosa was recently corroborated by two independent transposon sequencing (Tn-seq) studies reporting that fur insertion mutants were outcompeted in transposon mutant pools under several different growth conditions (13, 14). In silico predictions and in vitro experiments, including microarray comparison of gene expression in iron-replete and iron-depleted cells and Fur DNA pulldown experiments, were used to define the Fur regulon in P. aeruginosa (15–18). Overall, these and further confirmatory studies revealed that Fur, directly or indirectly via Fur-regulated alternative sigma factors or AraC-like transcriptional regulators, basically represses all iron uptake genes, including those for biosynthesis and transport of the endogenous siderophores pyoverdine and pyochelin, heme uptake and transport of exogenous iron chelators, and some virulence factors. Moreover, Fur positively regulates iron storage proteins (bacterioferritins), some enzymes involved in carbon catabolism and respiration, and ROS-scavenging enzymes through Fur-mediated repression of the two small RNAs PrrF1 and PrrF2 (reviewed in references 19 and 20). This led to the proposal that Fur essentiality could be related to the inability of P. aeruginosa to grow through fermentation and thus its strict requirement to respire, which generates considerable potential for the generation of ROS that would not be efficiently detoxified in Fur-deficient cells (20, 21). However, while the importance of Fur for resistance to oxidative stress is overall conserved in bacteria (3), it should be noted that fur knockout mutants of other nonfermenting Gram-negative bacteria (e.g., Burkholderia multivorans, Moraxella catarrhalis, and some Pseudomonas species) have been obtained (22-26), suggesting that an alternative, more specific harmful effect(s) could account for the critical role of Fur in P. aeruginosa physiology.
In this work, we used a conditional fur mutant of P. aeruginosa reference strain PAO1 to investigate the effect of Fur depletion both in vitro and in vivo and employed a transposon mutagenesis approach to identify genetic determinants affecting the growth of Fur-depleted cells. Our results revealed that Fur-depleted P. aeruginosa cells are similar to wild type with regard to planktonic growth, biofilm formation, and pathogenicity in an insect model of infection. In contrast, we found that Fur is essential for colony growth on agar plates and that biosynthesis of the pyochelin siderophore is partially responsible for this defect, through a mechanism that apparently does not depend on ROS generation.
RESULTS
Validation of the conditional fur mutant to investigate the effect of Fur depletion in P. aeruginosa.
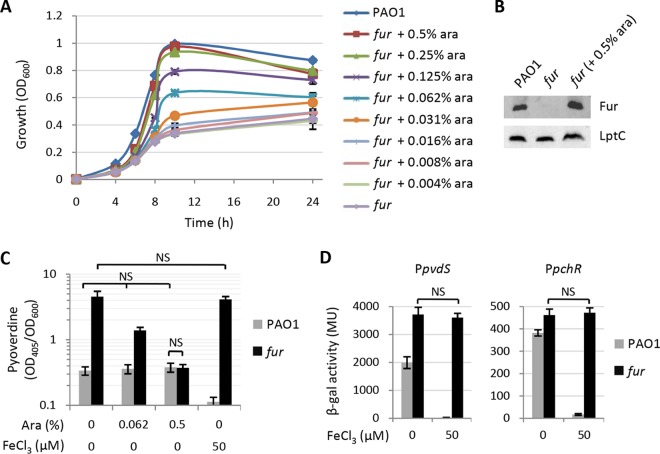
To investigate the possible involvement of Fur in cell aggregation-mediated induction of iron uptake and virulence genes in P. aeruginosa, we recently generated a conditional fur mutant of P. aeruginosa PAO1. This mutant carries an arabinose-inducible copy of the fur coding sequence inserted into a neutral site (attB) of the PAO1 genome and an in-frame deletion in the endogenous fur gene (27). With the aim of corroborating the suitability of the conditional fur mutant to study the effect of Fur depletion on P. aeruginosa cells, we analyzed this mutant for (i) the effect of arabinose on growth, (ii) intracellular Fur levels, and (iii) expression of Fur-repressed genes in response to iron. As shown in Fig. 1A, the growth of the conditional fur mutant in microtiter plates in Mueller-Hinton (MH) broth was impaired with respect to that of parental strain PAO1 and was gradually restored by arabinose in a dose-dependent manner. In the absence of arabinose, Fur was undetectable by Western blotting in cells of the conditional fur mutant, while it was detected at wild-type levels in mutant cells cultured in the presence of an arabinose concentration (0.5%) (Fig. 1B) that completely restored the growth of the conditional fur mutant. Since the inability to immunodetect a given protein is not sufficient to rule out the possibility that basal protein levels potentially present in the conditional mutant still have an effect on Fur-dependent genes, we also monitored pyoverdine production, an easily detectable phenotype that is promptly repressed by Fur in response to iron (28). Pyoverdine production was about 10-fold higher in the fur mutant than in PAO1, but it decreased to wild-type levels in the presence of 0.5% arabinose (Fig. 1C). More importantly, while the addition of 50 μM FeCl3 to the medium shut down pyoverdine production by wild-type cells, it had no effect on the pyoverdine levels of the fur mutant grown in the absence of arabinose (Fig. 1C), indicating that iron-mediated repression of Fur-regulated genes is abrogated in the conditional mutant. This was confirmed by the observation that exogenously added iron was unable to repress transcription from promoters of genes directly repressed by Fur (pvdS and pchR) in the conditional fur mutant (Fig. 1D). Taken together, these preliminary experiments provide evidence that our conditional fur mutant can be used as a tool to investigate the effect of Fur depletion in P. aeruginosa.
FIG 1.
Validation of the arabinose-dependent P. aeruginosa PAO1 conditional fur mutant. (A) Growth (OD600) of P. aeruginosa PAO1 and the conditional fur mutant in MH broth supplemented with increasing concentrations of arabinose (ara) in microtiter plates at 37°C with shaking at 200 rpm. The growth of wild-type PAO1 was not significantly affected by any arabinose concentration (data not shown). (B) Intracellular levels of Fur in P. aeruginosa PAO1 and conditional fur mutant cells grown for 10 h in MH broth supplemented or not with 0.5% arabinose, determined by Western blotting of whole-cell lysates (25 μg of protein) with an anti-Fur polyclonal antibody. The constitutive LptC protein (involved in LPS transport) was used as a loading control. (C) Pyoverdine production by P. aeruginosa PAO1 and conditional fur mutant cells grown at 37°C for 20 h in MH broth supplemented or not with different arabinose concentrations or 50 μM FeCl3. (D) β-Galactosidase activity (in Miller units [MU]) expressed by PAO1 and the conditional fur mutant carrying promoter probe plasmid pMP220 PpvdS (left) or pMP220 PpchR (right) after 14 h of growth in MH broth supplemented or not with 50 μM FeCl3. Values in panels A, C, and D are the mean (±standard deviation) from at least three independent assays, while the image in panel B is representative of three assays showing similar results. Unless otherwise stated, values in panels C and D are significantly different (P < 0.01). NS, not significant (P > 0.05).
Fur-depleted P. aeruginosa cells show severe growth defects on solid media.
To investigate the effect of Fur depletion on P. aeruginosa in greater depth, we compared the planktonic growth of wild-type strain PAO1 with that of the conditional fur mutant in flask cultures by using different media containing different iron concentrations. The conditional fur mutant was able to grow in the complex medium MH broth (about 5 to 10 μM iron concentration) (29), although with lower growth rates and an about 50% reduction in growth yields with respect to wild-type PAO1 (Fig. 2A). The differences in growth rates and yields between the two strains were markedly reduced in the iron-deprived complex medium TSBD (ca. 1.5 μM iron concentration) (30) (Fig. 2B), while no differences were observed in the iron-depleted minimal medium DCAA, where the iron concentration is very low (<0.1 μM) (31) (Fig. 2C; note the different y axis scales in Fig. 2). Supplementation of the iron-poor media TSBD and DCAA with iron (50 μM FeCl3) exacerbated the growth defects of the fur mutant in both media, resulting in growth profiles comparable to those observed in MH broth (Fig. 2). Notably, the growth-promoting effect of exogenously added iron on the wild type was much stronger in the minimal medium DCAA than in the complex medium TSBD (Fig. 2B and C), confirming that iron is a major limiting factor for P. aeruginosa growth in DCAA (31).
FIG 2.
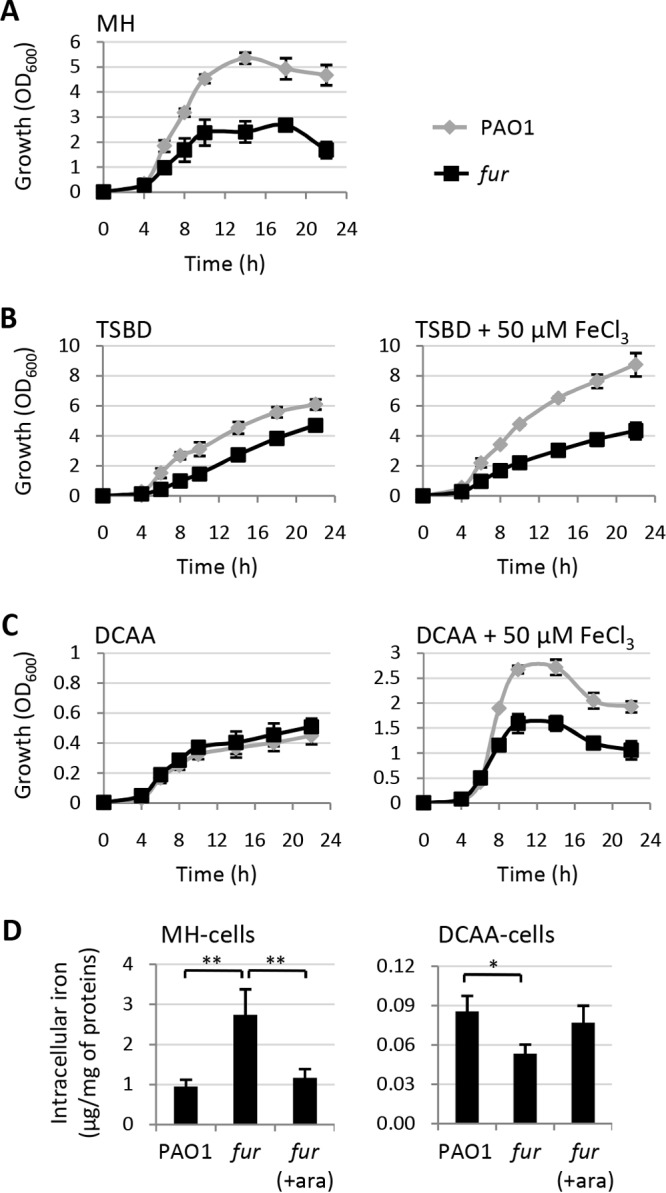
Planktonic growth of Fur-depleted cells is impaired under high-iron culture conditions. (A to C) Growth (OD600) of P. aeruginosa wild-type PAO1 and the conditional fur mutant in MH broth (A), TSBD (B), or DCAA (C) in flasks at 37°C with vigorous agitation (200 rpm) in the absence (left) or presence (right) of exogenously added iron (50 μM FeCl3). (D) Intracellular iron levels (μg/mg of protein) in wild-type and fur mutant PAO1 cells grown in MH broth (left) or DCAA (right) supplemented or not with 0.5% arabinose (ara) for 14 h at 37°C with shaking at 200 rpm. Values are the mean (±standard deviation) from at least three independent assays. Asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01).
To directly correlate growth capabilities with intracellular iron levels, we determined the total iron content of PAO1 and conditional fur mutant cells cultured in MH broth or in DCAA in the presence or absence of arabinose. In line with the relevant role of Fur as a repressor of iron uptake genes under iron-replete conditions (3), Fur-depleted cells cultured in MH broth showed levels of iron about 3-fold higher than those of the wild type or the fur mutant grown in the presence of arabinose (Fig. 2D). Intracellular iron levels were strongly reduced (about 10-fold) in PAO1 cells cultured in the iron-depleted medium DCAA and were about 35% lower in Fur-depleted cells than in Fur-replete ones (Fig. 2D), demonstrating that under strong iron deprivation (DCAA), Fur-depleted cells are unable to accumulate more iron than Fur-replete ones.
In parallel, we also assessed the colony growth of the wild type and the conditional fur mutant on the above-mentioned media solidified with 1.5% agar. Surprisingly, growth of the fur mutant was almost completely abrogated in MH broth plates, while it was restored to wild-type levels in the presence of arabinose (Fig. 3). Such a defect in colony growth was also evident in DCAA (Fig. 3), in spite of the evidence that the conditional fur mutant did not show any defect in planktonic growth in liquid DCAA (Fig. 2C). The colony growth defect was, however, much stronger in DCAA supplemented with 50 μM FeCl3 (Fig. 3), confirming that Fur depletion causes an increase in iron sensitivity also during colony development. Similar results were obtained in TSBD agar (data not shown). Accordingly, addition of exogenous iron (50 μM FeCl3) to MH agar plates completely prevented the growth of the conditional fur mutant in the absence of arabinose (Fig. 3).
FIG 3.
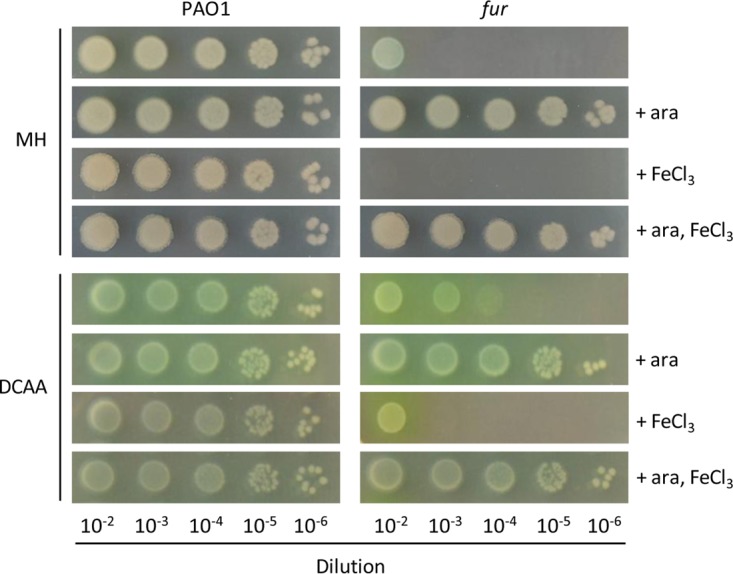
Fur is essential for P. aeruginosa growth on solid media. Shown is the colony growth of P. aeruginosa wild-type PAO1 and the conditional fur mutant on MH or DCAA agar plates supplemented or not with 0.5% arabinose (ara) or 50 μM FeCl3 as indicated. Exponential-phase cultures in MH broth or DCAA with 0.5% arabinose were normalized to an OD600 of 1, and 5 μl of the 10−2 to 10−6 dilutions, corresponding to ca. 105 to 10 viable cells, respectively, were spotted onto the plates, which were then incubated for 20 h (MH agar) or 24 h (DCAA agar) at 37°C. The images are representative of at least three independent experiments with similar results.
As a whole, these experiments demonstrate that Fur-depleted cells are proficient in planktonic growth, although they display growth defects that are proportional to the iron availability in the culture medium and, consequently, to the iron content of the cells. In contrast, Fur depletion strongly impairs the ability to grow as colonies on solid media, which is only partially affected by iron availability, indicating that the lack of Fur in P. aeruginosa cells has specific detrimental consequences during colony growth.
Fur is not required for biofilm formation and acute infection of Galleria mellonella.
To investigate whether the colony growth defects of the conditional fur mutant could be ascribed to a higher sensitivity of Fur-depleted cells to cell-cell contacts, we verified the behavior of the conditional fur mutant during biofilm growth, which involves extensive contact between bacterial cells. For this purpose, both microtiter plate and flow cell assays were used. Notably, no significant differences in biofilm biomass were observed between the conditional fur mutant and the wild-type PAO1 strain in the microtiter plate assay (Fig. 4A), and this was further confirmed by using the flow cell assay, in which the fur mutant formed mature biofilms overall comparable to those of Fur-replete cells (Fig. 4B). As a control, we also assessed flow cell biofilm formation by a P. aeruginosa strain with a conditional mutation in a strictly essential protein (LptH) (32), which indeed was only able to adhere to the surface without developing mature (mushroom-like) biofilms (Fig. 4B). These data demonstrate that Fur is dispensable for P. aeruginosa biofilm formation, in agreement with previous results obtained with a strain carrying a point mutation in fur (33), likely ruling out the possibility that the observed defect of Fur-depleted cells in colony formation is due to a cell contact-dependent inhibitory mechanism(s).
FIG 4.
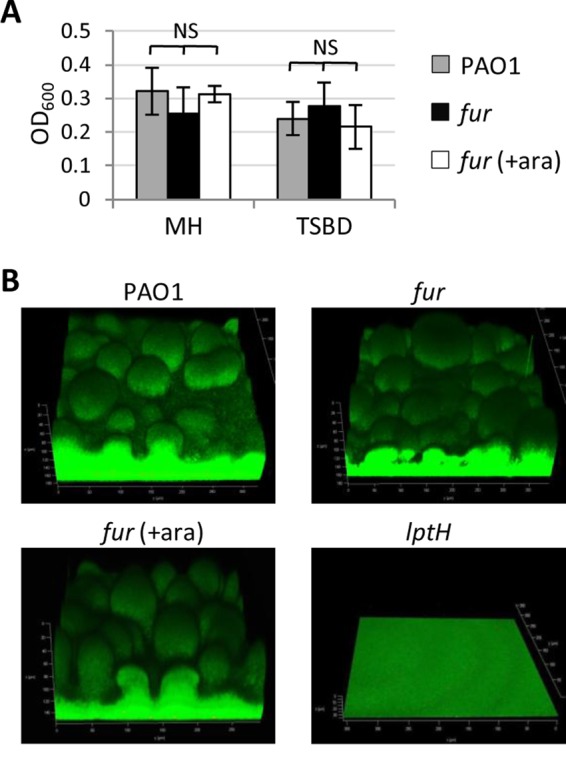
P. aeruginosa Fur is not required for biofilm formation. (A) Biofilm formation in 96-well polystyrene microtiter plates of wild-type PAO1 and the conditional fur mutant in MH broth or TSBD (supplemented or not with 0.5% arabinose [ara]) after 24 h at 37°C under static conditions. Values are the mean (±standard deviation) from three independent assays. NS, not significant (P > 0.05). (B) Biofilms formed in flow cells at 48 h by PAO1, the conditional fur mutant, and the lptH conditional mutant in 1% TSB supplemented or not with 0.5% arabinose (ara). The images are representative of two independent experiments with similar results.
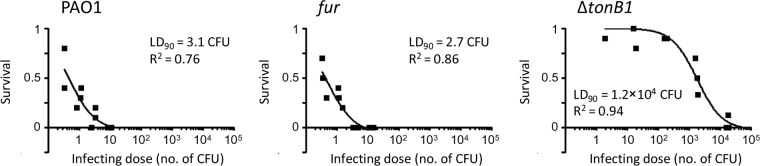
The above-described results suggest that Fur should not be considered an essential protein sensu stricto in P. aeruginosa, since Fur-depleted cells are able to grow planktonically in both iron-replete and -depleted media, as well as to form mature biofilms (Fig. 1, 2, and 4). To further corroborate this evidence, we investigated the effect of Fur depletion on P. aeruginosa pathogenicity by using an acute-infection model based on the larvae of the insect G. mellonella (34). Larvae were infected with different doses of either wild-type or conditional fur mutant cells, and lethality was monitored for 5 days. The number of bacterial cells theoretically required to kill 90% of the larvae (LD90) inferred from survival curves was 3.1 cells for wild-type PAO1, in line with previous reports (32, 35), and 2.7 cells for the conditional fur mutant (Fig. 5), implying that there are no significant differences in infectivity between Fur-replete and -depleted cells in this model of infection. In contrast, a PAO1 ΔtonB1 mutant that is impaired in ferric iron uptake across the outer membrane and has recently been demonstrated to be strongly defective in pathogenicity in a mouse pulmonary model (36) had an LD90 of 1.2 × 104 cells (Fig. 5), about 4,000-fold higher than that of PAO1 or the conditional fur mutant, thus confirming that G. mellonella is a suitable model for investigation of the infectivity of iron metabolism mutants. Overall, these results demonstrate that P. aeruginosa Fur is not required to cause lethal infection in G. mellonella. While obvious differences among animal models of infection exist, these results provide the first demonstration of the nonessential role of Fur also in an in vivo model.
FIG 5.
Fur is dispensable for acute P. aeruginosa infection of G. mellonella larvae. Shown are survival curves of G. mellonella larvae infected with different doses of P. aeruginosa PAO1, the conditional fur mutant, or the PAO1 ΔtonB1 mutant. The LD90 and R2 values of each curve are also shown.
The pch locus is involved in the colony growth defects of Fur-depleted cells.
While Fur does not appear to be required for planktonic growth, biofilm formation, and infection (Fig. 2, 4, and 5), Fur depletion makes P. aeruginosa cells severely defective in colony growth on solid media (Fig. 3). With the aim of investigating the mechanism(s) underlying the inhibitory or toxic effect of the lack of Fur-mediated repression on colony development, transposon mutagenesis screening was performed to select transposon insertion derivatives of the conditional fur mutant able to grow on MH agar plates in the absence of arabinose. The screening of almost 30,000 transposon insertion mutants in two independent experiments obtained 30 clones that developed mature colonies on MH agar plates within 24 h of growth at 37°C. Direct comparison of the colony growth kinetics of wild-type PAO1, the conditional fur mutant (used as positive and negative controls, respectively), and the 30 transposon insertion mutants selected during the first screening highlighted six clones whose colony growth phenotype resembled that of the wild-type strain (see Fig. S1 in the supplemental material). Mapping of the transposon insertion sites revealed that in three mutants, the transposon was inserted in the araC-PBAD regulatory region, which controls the expression of the exogenous copy of fur in the conditional mutant (Table S1) (35), while in the other three clones, the transposon was inserted in the regulatory gene pchR or in the biosynthetic operon pchEFGHI involved in the biogenesis of the pyochelin siderophore (Fig. S1 and Table S1) (37). The insertion of the transposon in the araC-PBAD regulatory element led us to speculate that the corresponding mutants could grow because of dysregulated AraC-mediated repression of fur expression. This hypothesis was confirmed by the observation that these mutants showed iron-dependent inhibition of a Fur-repressed phenotype (i.e., pyoverdine production) that was comparable to that observed in the wild type (Fig. S2). In contrast, the pchE transposon insertion mutant (used as a control) was fully insensitive to iron repression, just like its parental conditional fur mutant (Fig. S2). Therefore, the former mutants were no longer considered in this work.
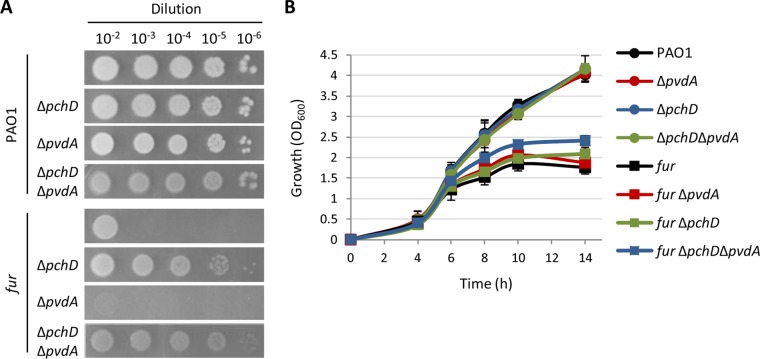
The involvement of the pyochelin locus in the growth defect of Fur-depleted cells on agar plates was verified by generating an in-frame deletion in the pyochelin biosynthetic gene pchD by using a mutagenesis construct already available in our laboratory (Table S1). In agreement with the results of the transposon mutagenesis analysis, pchD deletion in the conditional fur mutant restored growth on MH agar plates, although at levels lower than that of the wild type (Fig. 6A). Notably, loss of pyoverdine production because of deletion of the pyoverdine biosynthetic gene pvdA did not rescue the growth of the conditional fur mutant (Fig. 6A), indicating that the growth-promoting effect associated with the deficiency in pyochelin biosynthesis cannot be ascribed to a general defect in siderophore-mediated iron uptake. Accordingly, colony growth of the conditional fur mutant with both pchD and pvdA deleted was overall comparable to that of the isogenic ΔpchD mutant and still impaired with respect to that of wild-type PAO1 or its isogenic pvdA and pchD single and double mutants (Fig. 6A). In contrast, the pchD deletion in the fur mutant did not affect planktonic growth in MH broth, which was only marginally restored in the siderophore-null fur ΔpchD ΔpvdA mutant (Fig. 6B). Overall, these data strongly suggest that the toxic effect associated with the pch locus in Fur-depleted P. aeruginosa cells is specific to colony growth.
FIG 6.
The pch locus is involved in the colony growth defect of Fur-depleted cells. (A) Colony growth of P. aeruginosa wild-type PAO1, the conditional fur mutant, and isogenic pyoverdine-defective (ΔpvdA) and/or pyochelin-defective (ΔpchD) mutants on MH agar plates inoculated as described in the legend to Fig. 3. The images are representative of at least three independent experiments. (B) Planktonic growth of the strains tested in panel A in MH broth in flasks at 37°C with shaking at 200 rpm. Values are the mean (±standard deviation) from three independent assays.
Colony growth defects of Fur-depleted cells are not related to oxidative stress.
An early report on P. aeruginosa PAO1 mutants carrying different point mutations in the fur gene highlighted that cells expressing defective variants of Fur were more sensitive to oxidative stress (38), in agreement with the indirect role of Fur as a positive regulator of important ROS-detoxifying enzymes, such as the catalase KatA and the superoxide dismutase SodB (39). Moreover, it has recently been demonstrated that pyochelin can act as an antibiotic against some bacterial species by inducing the generation of ROS (40, 41). The growth-inhibitory effect of pyochelin is not related to its iron-chelating (siderophore) activity and is suppressed by ROS-scavenging compounds such as catecholate siderophores (e.g., enterobactin) and ascorbic acid (40).
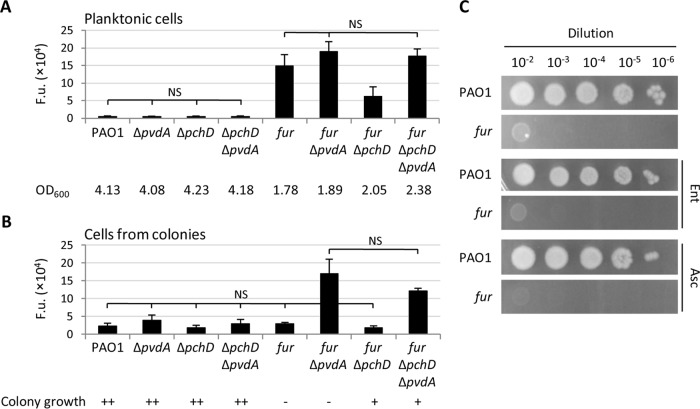
To determine whether pch-associated toxicity in Fur-depleted cells is dependent on pyochelin-induced oxidative stress, we monitored oxidative stress in wild-type and fur mutant PAO1 cells proficient or deficient in siderophore production by using a fluorescence assay based on the ROS probe 2′,7′-dichlorodihydrofluorescein (42). As expected, planktonic cells of the fur mutant cultured in MH broth displayed a strong increase in intracellular ROS levels compared to those of wild-type cells (Fig. 7A). Pyochelin deficiency significantly reduced ROS in Fur-depleted cells, while pyoverdine deficiency slightly increased them (Fig. 7A), in line with the protective effect of catecholate siderophores such as pyoverdine against oxidative stress (40). Thus, data from liquid cultures corroborate the notion that Fur and pyoverdine protect from oxidative stress, as opposed to pyochelin, which promotes ROS production.
FIG 7.
Oxidative stress does not account for the colony growth defect of Fur-depleted cells. ROS levels in cells of wild-type P. aeruginosa PAO1, the conditional fur mutant, and isogenic pyoverdine-defective (ΔpvdA) and/or pyochelin-defective (ΔpchD) mutants cultured in MH broth (A) or on MH agar plates (B). Values are the mean (±standard deviation) from three independent experiments and, unless otherwise stated, are significantly different (P < 0.05). NS, not significant. Mean OD600 values and colony growth phenotypes (++, wild-type growth; +, slightly impaired growth; −, strongly defective growth) are shown below the graphs in panels A and B, respectively. (C) Colony growth of PAO1 and the conditional fur mutant on MH agar plates supplemented or not with 5 mM ascorbic acid (Asc) or 5 μM enterobactin (Ent). The images are representative of three independent experiments. Identical results were obtained with 1 mM Asc and 1 μM Ent (data not shown).
Intriguingly, the contribution of Fur to protection from oxidative stress was negligible during colony growth. Indeed, ROS levels were comparable in wild-type PAO1 and the fur mutant cultured on MH agar plates, irrespective of pyochelin production, while a strong boost in oxidative stress was observed in pyoverdine-deficient Fur-depleted cells (Fig. 7B). By combining growth data (Fig. 6) with intracellular ROS levels (Fig. 7), it can be argued that there is no direct correlation between the growth of Fur-depleted cells and ROS accumulation in both the planktonic and colony modes. Indeed, the planktonic growth of fur ΔpvdA and fur ΔpchD mutants was comparable (Fig. 6B), even if fur ΔpvdA mutant cells accumulate almost 4-fold larger amounts of ROS (Fig. 7A). Moreover, the ΔpchD mutation restored the growth of the conditional fur mutant on agar plates (Fig. 6A) without significantly affecting intracellular ROS levels (Fig. 7A). Finally, both fur and fur ΔpvdA mutants showed marked defects in colony growth (Fig. 6A) but only the latter overproduced ROS on agar plates (Fig. 7B). Thus, the rescue effect of pch mutations on the colony growth of Fur-depleted cells does not appear to be related to the oxidative stress induced by pyochelin. This is consistent with the observations that (i) exogenously provided enterobactin or ascorbic acid, which was previously shown to counteract the growth-inhibitory activity of pyochelin on susceptible strains (40), did not stimulate the growth of the conditional fur mutant on agar plates (Fig. 7C) and (ii) the overexpression of a ROS-detoxifying enzyme (i.e., catalase KatA or superoxide dismutase SodB) partially restored the planktonic growth of Fur-depleted cells without exerting any rescue effects on colony growth (Fig. S3).
Pyochelin toxicity during colony growth depends on intracellular iron availability.
The results so far obtained highlight the involvement of pyochelin biosynthesis in the growth defect of colonies of Fur-depleted cells. However, we did not observe any inhibitory effect of exogenously provided pyochelin (up to 250 μM) on the colony growth of both fur ΔpchD and fur ΔpchD ΔpvdA mutants on MH agar plates (data not shown), suggesting that pyochelin biosynthesis, rather than pyochelin-mediated iron import, may be responsible for toxicity.
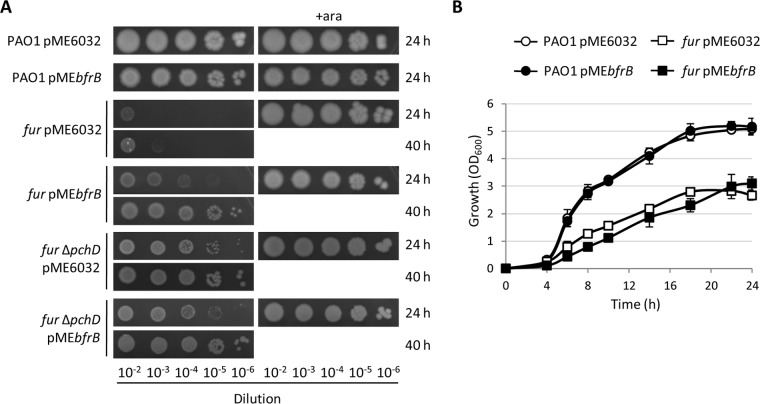
To determine whether the availability of intracellular iron may affect pyochelin-related toxicity, we overexpressed the main intracellular iron storage protein of P. aeruginosa (the bacterioferritin BfrB) (43) in wild-type and fur mutant cells. Notably, BfrB overexpression partially restored the colony growth of Fur-depleted cells (Fig. 8A) without stimulating planktonic growth (Fig. 8B). In the presence of arabinose, the colony growth of both the wild type and the conditional fur mutant was not affected by BfrB overexpression (Fig. 8A), indicating that the rescue effect of BfrB is specific to the colony growth of Fur-depleted cells. To tentatively correlate BfrB-mediated rescue of fur mutant colony growth with pyochelin-related toxicity, we also overexpressed BfrB in the pyochelin-deficient fur ΔpchD conditional mutant. Interestingly, no differences in colony growth were observed between fur ΔpchD cells overexpressing or not overexpressing BfrB, both in the presence and in the absence of arabinose (Fig. 8A), suggesting that the rescue effect of BfrB in the fur-depleted background is dependent on the synthesis/presence of pyochelin and that the availability of intracellular iron is a prerequisite for pyochelin toxicity during colony growth.
FIG 8.
Overexpression of bacterioferritin partly restores colony growth in Fur-depleted cells. Shown is the colony (A) and planktonic (B) growth of PAO1 and conditional fur mutant cells carrying the empty vector pME6032 or the bacterioferritin-expressing vector pMEbfrB in MH broth (or on MH agar) supplemented with 0.5 mM IPTG at 37°C. Images in panel A were taken after 24 or 40 h of growth and are representative of three experiments with similar results. Values in panel B are the mean (±standard deviation) from two independent experiments performed in duplicate. ara, arabinose.
Pyochelin-related toxicity in Fur-depleted cells is not a common feature of all Pseudomonas species.
While several attempts to obtain fur deletion mutants of P. aeruginosa failed (10, 11; this work), fur knockout mutants (obtained by deletion or insertional mutagenesis) of other Pseudomonas species, namely, P. syringae pv. tabaci, P. fluorescens, and P. pseudoalcaligenes, were generated and characterized (24–26). While the fur knockout mutant of P. pseudoalcaligenes was generated in a siderophore-null strain (26) and P. syringae pathovars do not synthesize pyochelin (44), some strains of P. fluorescens produce enantio-pyochelin, the enantiomer of P. aeruginosa pyochelin (44, 45). Unfortunately, the ability of the P. fluorescens strain (TSS) used for fur mutagenesis to produce pyochelin was not investigated (25, 46), thus hampering attempts to correlate Fur essentiality with pyochelin biosynthesis in other Pseudomonas species.
To determine experimentally whether pyochelin production has deleterious effects on the growth of Fur-depleted cells in a different Pseudomonas species, we introduced the cosmid pME3300, which carries the whole pyochelin locus from P. aeruginosa PAO1 (47), or the empty cosmid pLAFR3 as a control into P. syringae pv. tabaci Δfur mutant BL33 (24). As expected, pyochelin was efficiently produced in P. syringae pv. tabaci Δfur cells harboring pME3300 (Fig. 9A). However, differently from what was observed in P. aeruginosa, pyochelin biosynthesis did not influence the colony growth of the P. syringae pv. tabaci Δfur mutant on MH agar (Fig. 9B) and King's B agar (data not shown), implying that the toxicity associated with the pch locus in Fur-depleted cells is not shared by all pseudomonads.
FIG 9.
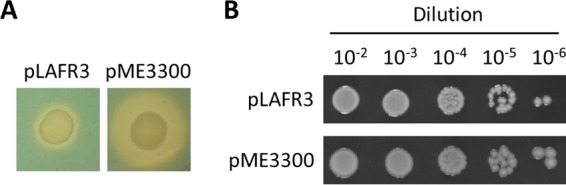
Pyochelin production does not affect the colony growth of a P. syringae pv. tabaci fur deletion mutant. (A) CAS agar assay showing siderophore production after 48 h of growth at 37°C by P. syringae pv. tabaci Δfur mutant BL33 carrying a cosmid containing the entire pch genomic region from P. aeruginosa PAO1 (pME3300) or the empty control (pLAFR3). (B) Colony growth on MH agar plates supplemented with 12 μg/ml tetracycline after 48 h at 37°C of P. syringae pv. tabaci Δfur mutant BL33 carrying pME3300 or pLAFR3. Images are representative of two experiments with similar results. Comparable results were obtained with King's B agar plates (data not shown).
DISCUSSION
By using a conditional fur mutant of P. aeruginosa PAO1, we have demonstrated that the proposed essentiality of Fur in this species is likely related to the inability of Fur-depleted cells to grow efficiently and develop colonies on agar plates. In agreement with previous reports based on mutant strains harboring point mutations in the fur gene (10, 33, 38), we observed that Fur-depleted cells were still able to (i) grow planktonically, although their growth was defective under iron-rich conditions, likely because of dysregulated iron acquisition (Fig. 2) and a consequent increase in oxidative stress (Fig. 7), and (ii) form mature biofilms on abiotic surfaces (Fig. 4). Moreover, we demonstrated that, differently from other P. aeruginosa essential gene conditional mutants generated by the same mutagenesis strategy (31, 34), the conditional fur mutant is not impaired in infectivity and virulence in an insect model (Fig. 5). This result implies that the growth defects and high oxidative stress observed in vitro do not affect the ability of the conditional fur mutant to cause infection, at least in a simple model of acute infection. The retained virulence of Fur-depleted cells, however, needs to be corroborated in more complex infection models (e.g., vertebrate and/or chronic infection models) in which intracellular iron homeostasis could play a more relevant role during the infection process. It is worth noting that in vivo Tn-seq experiments did not detect fur mutants among transposon mutants impaired in mouse gastrointestinal tract colonization (48). While this result suggests that the nonessentiality of Fur for P. aeruginosa pathogenicity may extend to other types of infection, experimental validation by using the P. aeruginosa conditional fur mutant in more complex animal models of infection is needed to confirm this hypothesis.
The main findings of this work are that (i) Fur-depleted cells are severely impaired in colony growth on agar plates (Fig. 3) and (ii) colony growth of Fur-depleted cells can be partially rescued by insertion or deletion mutations in the pyochelin locus (Fig. 6; also see Fig. S1 in the supplemental material). While pyochelin has been demonstrated to exert a growth-inhibitory effect on some bacterial species by inducing ROS (40, 41), we observed that pyochelin-related toxicity does not correlate with intracellular levels of ROS during colony growth and is not counteracted by antioxidant agents or by overexpression of ROS-detoxifying enzymes (Fig. 7 and S3) that were previously reported to protect sensitive cells from pyochelin-induced oxidative stress (40). Thus, mechanisms other than ROS generation are likely involved in pyochelin toxicity to Fur-depleted cells during colony growth.
Since colony growth defects of Fur-depleted cells are exacerbated in the presence of high iron levels (Fig. 3) and partially relieved in pyochelin-defective mutants (Fig. 6), one could hypothesize that pyochelin exerts its toxicity by loading the cells with too much iron, which cannot be properly managed in Fur-depleted cells with impaired iron homeostasis. However, at least two lines of evidence argue against this hypothesis. First, while many studies have demonstrated that pyoverdine represents the primary P. aeruginosa siderophore for iron uptake under all of the in vitro and in vivo conditions investigated so far (36, 49, 50), colony growth is not restored in pyoverdine-deficient Fur-depleted cells (Fig. 6). Second, exogenously provided pyochelin is not able to inhibit the colony growth of pyochelin-deficient fur mutant cells, suggesting that pyochelin biosynthesis, rather than its uptake, poisons Fur-depleted cells. This result also reasonably rules out the possibility that pyochelin toxicity in the absence of Fur is related to its ability to chelate and mediate the acquisition of other metals (51). However, the inability to measure intracellular and extracellular pyochelin levels during colony growth does not allow experimental verification of whether pyochelin toxicity arises from cytoplasmic and/or extracellular accumulation of the siderophore. The finding that pyochelin production has no effect on the viability of Fur-depleted P. aeruginosa cells during planktonic growth (Fig. 6), and of a fur deletion mutant of a different Pseudomonas species (Fig. 9), implies that toxicity likely arises from a specific effect of pyochelin biosynthesis on a pathway(s) important for growth on agar and/or colony formation that is not conserved in all pseudomonads. Notably, the inhibitory effect of pyochelin biosynthesis is relieved by reducing cytoplasmic levels of free iron (Fig. 8), indicating that intracellular iron contributes to pyochelin-dependent poisoning of Fur-depleted cells during colony growth. However, whether the observed toxic effect of pyochelin biosynthesis relies on the intracellular toxicity of pyochelin itself, any of its metabolic precursors (37), or a specific biosynthetic enzyme remains to be determined.
A limitation of the present study is the failure to obtain a clear fur null mutant. Indeed, although we demonstrated that Fur-depleted cells can grow planktonically and show partially restored growth on agar plates upon inactivation of the pyochelin biosynthetic pathway (Fig. 2 and 6), our many attempts to generate an unmarked fur deletion mutant failed, even in a pyochelin-deficient (ΔpchD) background (data not shown). This could be ascribed to growth defects of pyochelin-deficient Fur-depleted cells (fur ΔpchD) compared with wild-type cells (Fig. 6), which are expected to favor reversion to the wild type during mutant generation and selection procedures. However, it cannot be ruled out that some phenotypes of the conditional fur mutant could be affected by residual expression of Fur in this mutant. Indeed, although gene expression assays excluded the presence of Fur at levels sufficient to exert iron-mediated repression on selected Fur-regulated genes and Fur was undetectable by Western blot analysis (Fig. 1), it is tempting to speculate that very low levels of Fur in the conditional mutant could still play some crucial role in P. aeruginosa physiology, possibly independent of its well-known activity as a transcriptional repressor. Irrespective of this margin of uncertainty, our results clearly indicate that Fur-depleted P. aeruginosa cells are still able to grow, form biofilms, and cause infection, whereas they are strongly impaired in colony growth because of some toxic effect related to pyochelin biosynthesis. This work thus provides new evidence of the complex interplay between iron metabolism and multicellular behaviors in P. aeruginosa (27, 33, 52–54) and provides the basis for future investigations of a still unexplored role of the pyochelin locus in the physiology of this pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. Bacteria were routinely cultured in Luria-Bertani broth for genetic manipulation, while growth assays were performed with MH broth, King's B medium, and the iron-depleted media TSBD (30) and DCAA (31), to which FeCl3 was added at the concentrations indicated when required. Chrome azurol S (CAS) agar plates (55) were used to detect siderophore production. When specified, growth media were supplemented with l-arabinose, enterobactin, or ascorbic acid (Sigma-Aldrich) at the concentrations indicated. Growth assays in liquid cultures were performed with 96-well microtiter plates (200 μl of medium in each well) or with 50-ml flasks containing 10 ml of medium at 37°C with vigorous shaking (200 rpm). Growth assays on solid media were performed by plating 5 μl of serial 10-fold dilutions of bacterial suspensions in saline normalized to an optical density at 600 nm (OD600) of 1 (from late-exponential-phase cultures grown in the presence of 0.5% arabinose) on media solidified with 1.5% agar. When required, antibiotics were added at the following concentrations for E. coli (the concentrations used for P. aeruginosa are shown in parentheses): ampicillin, 100 μg/ml; nalidixic acid, 20 μg/ml; carbenicillin, 0 μg/ml (250 μg/ml); chloramphenicol, 30 μg/ml (350 μg/ml); gentamicin, 20 μg/ml (50 μg/ml); tetracycline, 12 μg/ml (50 to 100 μg/ml). Pyochelin was purified from a pyoverdine-deficient P. aeruginosa mutant as described previously (56).
Growth and pyoverdine measurements.
Growth was measured as the OD600 of bacterial cultures in a Victor3V microplate reader (Perkin-Elmer). Flask culture samples were appropriately diluted in sterile growth medium, and the OD600 was determined spectrophotometrically. Pyoverdine production was measured as the OD405 of culture supernatants appropriately diluted in 0.1 M Tris-HCl (pH 8), normalized to the OD600 of the corresponding cultures (57).
Generation of plasmids and deletion mutants.
Recombinant DNA was manipulated as described elsewhere (58). The pMEkatA, pMEsodB, and pMEbfrB constructs were generated by PCR amplification and directional cloning of the DNA fragment encompassing the coding sequence of each gene into isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible shuttle vector pME6032 (59) by using the primers and restriction enzymes described in Table S2.
pvdA or pchD deletion mutants were generated by using a specific derivative of the pEX18Tc or pDM4 suicide plasmid (Table S1) as previously described (56, 60). Gene deletions were verified by PCR and DNA sequencing.
Transposon mutagenesis and identification of transposon insertion sites.
For random transposon mutagenesis, biparental matings were set up between E. coli S17.1 λpir carrying suicide plasmid pLM1, containing a mini-Tn5 transposon (61), and the P. aeruginosa conditional fur mutant. The transposon insertion mutants were selected on MH agar plates containing 50 μg/ml gentamicin and 20 μg/ml nalidixic acid to directly select transposon insertion revertants of the conditional fur mutant able to grow on plates in the absence of arabinose. A 1/100 dilution of each conjugation mixture was also plated on plates supplemented with 0.5% arabinose to determine transposon mutagenesis efficiency. Plates were incubated at 37°C, and colonies appearing 24 h after inoculation were reisolated on MH agar plates (without arabinose) together with wild-type strain PAO1 and the conditional fur mutant as controls. Clones showing growth on MH agar plates comparable to that of the wild-type strain were considered for further analysis.
To map transposon insertions, genomic DNA was isolated from selected transposon mutants and digested with BamHI. The transposon does not contain a BamHI restriction site and has an R6K origin of replication. The digested DNA was self-ligated with T4 DNA ligase and introduced by transformation into E. coli S17.1 λpir. Plasmid DNA was isolated from gentamicin-resistant colonies and sequenced with Tn5-specific primers TnpRL17-1 and TnpRL13-2 (62) (Table S2). Transposon insertions were mapped by using the BLAST program of the Pseudomonas Genome Database (http://www.pseudomonas.com).
β-Galactosidase assays.
The β-galactosidase activity from P. aeruginosa cells carrying the different reporter plasmids (Table S1) was determined spectrophotometrically with o-nitrophenyl-β-d-galactopyranoside as the substrate, normalized to the OD600 of the bacterial culture, and expressed in Miller units (63).
Measurement of intracellular ROS.
Bacteria from overnight cultures in MH broth supplemented with 0.5% arabinose were either subcultured in MH broth or streaked onto MH agar plates in the presence or absence of 0.5% arabinose. After 14 h (planktonic cultures) or 24 h (agar plate cultures) of growth, cells were collected by centrifugation or scraped off the plates, washed twice with phosphate-buffered saline (PBS), and resuspended in PBS at an OD600 of about 2. 2′,7′-Dichlorodihydrofluorescein diacetate (Sigma-Aldrich) was added to bacterial suspensions at a final concentration of 10 mM. Bacterial cells were incubated for 20 min at 37°C in the dark, washed twice with PBS, and then suspended in the same volume of PBS. Two hundred microliters of each sample was then transferred to a 96-well microtiter plate. Fluorescence was measured at 485-nm excitation and 535-nm emission wavelengths with a Victor3V microplate reader (Perkin-Elmer) and normalized to the OD600 of each sample.
Measurement of intracellular iron.
Bacteria were cultured in 50 ml of MH broth or 300 ml of DCAA, supplemented or not with 0.5% arabinose, in flasks at 37°C with shaking at 200 rpm for 14 h. Cells were collected by centrifugation, washed with saline, lysed in concentrated HNO3, and analyzed by inductively coupled plasma optical emission spectrometry (ICP-OES) with an ICP-OES 710 Varian spectrometer (Agilent Technologies). Cells were also collected from a 1-ml aliquot of each culture to determine the whole protein concentration by using the DC protein assay kit (Bio-Rad) with bovine serum albumin as the standard. Iron levels determined by ICP-OES were normalized to the protein content of each sample.
Western blot analysis.
Appropriate volumes of exponentially growing bacterial cultures in MH broth in flasks were centrifuged, and pellets were suspended in SDS-PAGE loading buffer (0.25 M Tris-HCl [pH 6.8], 2% SDS, 10% β-mercaptoethanol, 20% glycerol) for SDS-PAGE analysis of whole-cell extracts. Pellets from identical culture volumes were also collected to determine the cellular protein concentration of each sample by using the DC protein assay kit (Bio-Rad) with bovine serum albumin as the standard. Volumes of SDS-PAGE samples corresponding to 25 μg of protein were loaded onto the gels. Proteins resolved by SDS-PAGE were electrotransferred onto a nitrocellulose filter (Hybond-C extra; Amersham) and probed for Fur or LptC with custom rabbit polyclonal antibodies and a goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich). Anti-Fur and anti-LptC antibodies were generated at GenScript (http://www.genscript.com/custom-polyclonal-antibody-production-services.html) with keyhole limpet hemocyanin-conjugated peptides as antigens (Fur epitope, EIVRERGFELVDHN; LptC epitope, NAHSLQYQEDGSLD). The epitopes were selected with the OptimumAntigen design tool (GenScript). Filters were developed with ECL chemiluminescent reagents (Amersham) and visualized on a ChemiDoc XRS+ system (Bio-Rad).
Biofilm assays.
For the microtiter plate biofilm assay, bacteria from overnight cultures in MH broth or TSBD containing 50 μM FeCl3 and 0.5% arabinose were inoculated into fresh medium to an OD600 of 0.002 and dispensed into 96-well polystyrene plates (150 μl per well). After 24 h of incubation at 37°C under static conditions, the wells were washed several times with distilled water. Attached cells were stained with 0.1% crystal violet (175 μl) at room temperature (RT) for 15 min and washed several times with distilled water to remove unbound dye. Biofilm-bound crystal violet was eluted in absolute ethanol (200 μl) at RT for 15 min, and for each well, 100 μl of the resulting solution was aliquoted into a sterile microtiter plate. The released crystal violet was measured as OD600 in a Victor3V microplate reader (Perkin-Elmer). Triplicate independent experiments with at least three wells per condition were performed.
The flow cell biofilm assay was performed as previously described (64). Briefly, biofilm flow chambers were inoculated with overnight cultures (in tryptic soy broth [TSB] containing 0.5% arabinose) of P. aeruginosa strains constitutively expressing green fluorescent protein (GFP) from pMRP9-1 (Table S1) that were diluted to an OD600 of 0.15 in 1% TSB. A flow of 1% TSB (supplemented with 0.5% arabinose when required) was initiated after 2 h with a peristaltic pump at a rate of ∼10 ml/h. An upright Leica SPE TCS laser scanning confocal microscope was used to image biofilms. GFP was excited at 488 nm, and fluorescence emission was collected in the range of 504 to 530 nm. Z-stacks of two-dimensional confocal images were rendered in three dimensions with Imaris (Bitplane).
G. mellonella infection assay.
P. aeruginosa strains were grown in MH broth with 0.5% arabinose, and serial dilutions of bacterial cell suspensions in saline were injected into G. mellonella larvae as described previously (34). Ten larvae were infected with each infecting dose, and infected larvae were placed separately in a 3.5-cm petri dish. Larvae were incubated at 30°C for up to 5 days and monitored for death. Survival curves, LD90s, and R2 values were determined with the GraphPad Prism software as previously described (65).
Statistical analysis.
Statistical analysis was performed with the GraphPad Instat software by using one-way analysis of variance, followed by the Tukey-Kramer multiple-comparison test.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Carlo Bonchi for assistance in pyochelin purification and to Alessia Falsetti and Renato Baciocchi for performing ICP-OES measurements. We also thank Hyang Suk Baik for kindly providing P. syringae pv. tabaci Δfur mutant BL33.
This work was supported by grants from the Italian Cystic Fibrosis Research Foundation (FFC10/2013), the Sapienza University of Rome (Ateneo 2015), and the Pasteur Institute-Cenci Bolognetti Foundation to F.I. E.B. was supported by a grant from the Dyna and Fala Weinstock Foundation.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00472-17.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Fillat MF. 2014. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llamas MA, Imperi F, Visca P, Lamont IL. 2014. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev 38:569–597. doi: 10.1111/1574-6976.12078. [DOI] [PubMed] [Google Scholar]
- 5.Oglesby-Sherrouse AG, Murphy ER. 2013. Iron-responsive bacterial small RNAs: variations on a theme. Metallomics 5:276–286. doi: 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porcheron G, Dozois CM. 2015. Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet Microbiol 179:2–14. doi: 10.1016/j.vetmic.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 7.Porcheron G, Habib R, Houle S, Caza M, Lépine F, Daigle F, Massé E, Dozois CM. 2014. The small RNA RyhB contributes to siderophore production and virulence of uropathogenic Escherichia coli. Infect Immun 82:5056–5068. doi: 10.1128/IAI.02287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Ngeleka M, Potter AA, Allan BJ. 2002. Effect of fur mutation on acid-tolerance response and in vivo virulence of avian septicemic Escherichia coli. Can J Microbiol 48:458–462. doi: 10.1139/w02-042. [DOI] [PubMed] [Google Scholar]
- 9.Alice AF, Naka H, Crosa JH. 2008. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect Immun 76:4019–4037. doi: 10.1128/IAI.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton HA, Johnson Z, Cox CD, Vasil AI, Vasil ML. 1996. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol 21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 11.Prince RW, Cox CD, Vasil ML. 1993. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol 175:2589–2598. doi: 10.1128/jb.175.9.2589-2598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun 70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C. 2015. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol 45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 16.Palma M, Worgall S, Quadri LE. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch Microbiol 180:374–379. doi: 10.1007/s00203-003-0602-z. [DOI] [PubMed] [Google Scholar]
- 17.van Oeffelen L, Cornelis P, Van Delm W, De Ridder F, De Moor B, Moreau Y. 2008. Detecting cis-regulatory binding sites for cooperatively binding proteins. Nucleic Acids Res 36:e46. doi: 10.1093/nar/gkn140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochsner UA, Vasil ML. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci U S A 93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornelis P, Matthijs S, Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 20.Vasil ML. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20:587–601. doi: 10.1007/s10534-006-9067-2. [DOI] [PubMed] [Google Scholar]
- 21.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 22.Furano K, Campagnari AA. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect Immun 71:1843–1848. doi: 10.1128/IAI.71.4.1843-1848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuhara S, Komatsu H, Goto H, Ohtsubo Y, Nagata Y, Tsuda M. 2008. Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 154:1763–1774. doi: 10.1099/mic.0.2007/015537-0. [DOI] [PubMed] [Google Scholar]
- 24.Cha JY, Lee JS, Oh JI, Choi JW, Baik HS. 2008. Functional analysis of the role of Fur in the virulence of Pseudomonas syringae pv. tabaci 11528: Fur controls expression of genes involved in quorum-sensing. Biochem Biophys Res Commun 366:281–287. doi: 10.1016/j.bbrc.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Wang HR, Hu YH, Zhang WW, Sun L. 2009. Construction of an attenuated Pseudomonas fluorescens strain and evaluation of its potential as a cross-protective vaccine. Vaccine 27:4047–4055. doi: 10.1016/j.vaccine.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 26.Becerra G, Merchán F, Blasco R, Igeño MI. 2014. Characterization of a ferric uptake regulator (Fur)-mutant of the cyanotrophic bacterium Pseudomonas pseudoalcaligenes CECT5344. J Biotechnol 190:2–10. doi: 10.1016/j.jbiotec.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Visaggio D, Pasqua M, Bonchi C, Kaever V, Visca P, Imperi F. 2015. Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front Microbiol 6:902. doi: 10.3389/fmicb.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiburzi F, Imperi F, Visca P. 2008. Intracellular levels and activity of PvdS, the major iron starvation sigma factor of Pseudomonas aeruginosa. Mol Microbiol 67:213–227. doi: 10.1111/j.1365-2958.2007.06051.x. [DOI] [PubMed] [Google Scholar]
- 29.Girardello R, Bispo PJ, Yamanaka TM, Gales AC. 2012. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J Clin Microbiol 50:2414–2418. doi: 10.1128/JCM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohman DE, Sadoff JC, Iglewski BH. 1980. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect Immun 28:899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visca P, Ciervo A, Sanfilippo V, Orsi N. 1993. Iron-regulated salicylate synthesis by Pseudomonas spp. J Gen Microbiol 139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]
- 32.Fernández-Piñar R, Lo Sciuto A, Rossi A, Ranucci S, Bragonzi A, Imperi F. 2015. In vitro and in vivo screening for novel essential cell-envelope proteins in Pseudomonas aeruginosa. Sci Rep 5:17593. doi: 10.1038/srep17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A 102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 182:3843–3845. doi: 10.1128/JB.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo Sciuto A, Fernández-Piñar R, Bertuccini L, Iosi F, Superti F, Imperi F. 2014. The periplasmic protein TolB as a potential drug target in Pseudomonas aeruginosa. PLoS One 9:e103784. doi: 10.1371/journal.pone.0103784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minandri F, Imperi F, Frangipani E, Bonchi C, Visaggio D, Facchini M, Pasquali P, Bragonzi A, Visca P. 2016. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect Immun 84:2324–2335. doi: 10.1128/IAI.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youard ZA, Wenner N, Reimmann C. 2011. Iron acquisition with the natural siderophore enantiomers pyochelin and enantio-pyochelin in Pseudomonas species. Biometals 24:513–522. doi: 10.1007/s10534-010-9399-9. [DOI] [PubMed] [Google Scholar]
- 38.Hassett DJ, Sokol PA, Howell ML, Ma JF, Schweizer HT, Ochsner U, Vasil ML. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J Bacteriol 178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A 101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adler C, Corbalán NS, Seyedsayamdost MR, Pomares MF, de Cristóbal RE, Clardy J, Kolter R, Vincent PA. 2012. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS One 7:e46754. doi: 10.1371/journal.pone.0046754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong KS, Cheow YL, Lee SM. 2017. The role of reactive oxygen species in the antimicrobial activity of pyochelin. J Adv Res 8:393–398. doi: 10.1016/j.jare.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Zhong Z, Xu Z, Chen L, Wang Y. 2010. 2′,7′-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: forty years of application and controversy. Free Radic Res 44:587–604. doi: 10.3109/10715761003709802. [DOI] [PubMed] [Google Scholar]
- 43.Eshelman K, Yao H, Punchi Hewage AN, Deay JJ, Chandler JR, Rivera M. 2017. Inhibiting the BfrB:Bfd interaction in Pseudomonas aeruginosa causes irreversible iron accumulation in bacterioferritin and iron deficiency in the bacterial cytosol. Metallomics 9:646–659. doi: 10.1039/C7MT00042A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 45.Youard ZA, Mislin GL, Majcherczyk PA, Schalk IJ, Reimmann C. 2007. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J Biol Chem 282:35546–35553. doi: 10.1074/jbc.M707039200. [DOI] [PubMed] [Google Scholar]
- 46.Zhou ZJ, Zhang L, Sun L. 2015. Pseudomonas fluorescens: fur is required for multiple biological properties associated with pathogenesis. Vet Microbiol 175:145–149. doi: 10.1016/j.vetmic.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Serino L, Reimmann C, Baur H, Beyeler M, Visca P, Haas D. 1995. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol Gen Genet 249:217–228. doi: 10.1007/BF00290369. [DOI] [PubMed] [Google Scholar]
- 48.Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, Lory S, Pier GB. 2013. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog 9:e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takase H, Nitanai H, Hoshino K, Otani T. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect Immun 68:1834–1839. doi: 10.1128/IAI.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonchi C, Frangipani E, Imperi F, Visca P. 2015. Pyoverdine and proteases affect the response of Pseudomonas aeruginosa to gallium in human serum. Antimicrob Agents Chemother 59:5641–5646. doi: 10.1128/AAC.01097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braud A, Hannauer M, Mislin GL, Schalk IJ. 2009. The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J Bacteriol 191:3517–3525. doi: 10.1128/JB.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Overhage J, Bains M, Brazas MD, Hancock RE. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patriquin GM, Banin E, Gilmour C, Tuchman R, Greenberg EP, Poole K. 2008. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol 190:662–671. doi: 10.1128/JB.01473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiens JR, Vasil AI, Schurr MJ, Vasil ML. 2014. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio 5:e01010-13. doi: 10.1128/mBio.01010-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 56.Frangipani E, Bonchi C, Minandri F, Imperi F, Visca P. 2014. Pyochelin potentiates the inhibitory activity of gallium on Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:5572–5575. doi: 10.1128/AAC.03154-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imperi F, Tiburzi F, Fimia GM, Visca P. 2010. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ Microbiol 12:1630–1642. doi: 10.1111/j.1462-2920.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 59.Heeb S, Blumer C, Haas D. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol 184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Imperi F, Putignani L, Tiburzi F, Ambrosi C, Cipollone R, Ascenzi P, Visca P. 2008. Membrane-association determinants of the omega-amino acid monooxygenase PvdA, a pyoverdine biosynthetic enzyme from Pseudomonas aeruginosa. Microbiology 154:2804–2813. doi: 10.1099/mic.0.2008/018804-0. [DOI] [PubMed] [Google Scholar]
- 61.Dubern JF, Cigana C, De Simone M, Lazenby J, Juhas M, Schwager S, Bianconi I, Döring G, Eberl L, Williams P, Bragonzi A, Cámara M. 2015. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ Microbiol 17:4379–4393. doi: 10.1111/1462-2920.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 63.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 64.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, Rich JD, Parsek MR, Kaever V, Harrison JJ, Banin E. 2015. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:11359–11364. doi: 10.1073/pnas.1421450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. doi: 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.