ABSTRACT
The study of the minimum set of genes required to sustain life is a fundamental question in biological research. Recent studies on bacterial essential genes suggested that between 350 and 700 genes are essential to support autonomous bacterial cell growth. Essential genes are of interest as potential new antimicrobial drug targets; hence, our aim was to identify the essential genome of the cystic fibrosis (CF) isolate Burkholderia cenocepacia H111. Using a transposon sequencing (Tn-Seq) approach, we identified essential genes required for growth in rich medium under aerobic and microoxic conditions as well as in a defined minimal medium with citrate as a sole carbon source. Our analysis suggests that 398 genes are required for autonomous growth in rich medium, a number that represents only around 5% of the predicted genes of this bacterium. Five hundred twenty-six genes were required to support growth in minimal medium, and 434 genes were essential under microoxic conditions (0.5% O2). A comparison of these data sets identified 339 genes that represent the minimal set of essential genes required for growth under all conditions tested and can be considered the core essential genome of B. cenocepacia H111. The majority of essential genes were found to be located on chromosome 1, and few such genes were located on chromosome 2, where most of them were clustered in one region. This gene cluster is fully conserved in all Burkholderia species but is present on chromosome 1 in members of the closely related genus Ralstonia, suggesting that the transfer of these essential genes to chromosome 2 in a common ancestor contributed toward the separation of the two genera.
IMPORTANCE Transposon sequencing (Tn-Seq) is a powerful method used to identify genes that are essential for autonomous growth under various conditions. In this study, we have identified a set of “core essential genes” that are required for growth under multiple conditions, and these genes represent potential antimicrobial targets. We also identified genes specifically required for growth under low-oxygen and nutrient-limited environments. We generated conditional mutants to verify the results of our Tn-Seq analysis and demonstrate that one of the identified genes was not essential per se but was an artifact of the construction of the mutant library. We also present verified examples of genes that were not truly essential but, when inactivated, showed a growth defect. These examples have identified so-far-underestimated shortcomings of this powerful method.
KEYWORDS: Burkholderia, Tn-Seq, essential genes, minimal genome
INTRODUCTION
The study of the essential genome is fundamental to further our understanding of basic biological processes and how these interact to support life. Furthermore, due to increased antibiotic resistance of many pathogens, researchers are examining essential genes in order to identify novel antimicrobial drug targets (1–4). Burkholderia cenocepacia is an opportunistic pathogen that is inherently resistant to a number of antimicrobials, notably, polymyxin B and other cationic antimicrobial peptides, due to amino arabinose being incorporated naturally into its lipopolysaccharide (LPS) (5–8).
The B. cenocepacia genome consists of three large replicons and an optional plasmid that are predicted to encode around 7,500 proteins. Recent work has shown that it is possible to delete the third replicon, leading to the reclassification of this chromosome as a megaplasmid, termed pC3 (9, 10). B. cenocepacia is a member of a metabolically diverse group of bacteria termed the Burkholderia cepacia complex (BCC), whose members can cause serious infections in patients who suffer from cystic fibrosis (CF). The majority of BCC CF infections are caused by either B. cenocepacia or Burkholderia multivorans, and BCC infection of the CF lung is usually associated with a poor prognosis for the patient (11, 12). The fact that B. cenocepacia is able to form biofilms, resist oxidative stress, persist within macrophages, and produce a wide range of virulence factors, including toxins, proteases, and type six secretion systems, greatly contributes to the clinical relevance of this organism. BCC infections are difficult to treat due to their inherent antibiotic resistance and a high rate of patient-to-patient transmission. This group of bacteria are a major problem for CF clinics; hence, new antimicrobials that target BCC strains are urgently needed (13–17).
Early studies into essential genes in B. cenocepacia have involved either computational predictions or the creation of conditional mutants that cause growth defects (8, 18, 19). While the bioinformatics approach does not provide a bona fide proof of the essentiality of a gene, the latter method is very laborious, which makes global studies of essential genes difficult to undertake. Saturation level transposon mutagenesis combined with next-generation sequencing, commonly known as Tn-Seq, has provided a quick and technically efficient method to analyze a bacterium's entire essential genome in a single experiment (20, 21). More recently, this approach was applied to B. cenocepacia J2315, creating a list of predicted essential genes for this strain that was not further validated (22).
In this study, we ascertained the essential genome of B. cenocepacia strain H111 in rich medium under aerobic and microoxic conditions as well as in minimal medium. A subset of the predicted essential genes were verified using a conditional mutant approach. Although the identified essential genes of H111 overlap those of J2315, there are considerable differences that not only identified genes that are only essential in one of the strains but also shed light on the pitfalls of this method.
RESULTS
Construction and sequencing of a B. cenocepacia H111 transposon (Tn) library.
The initial Tn library was taken from LB plates with antibiotic markers to select for the transposon postconjugation and stored in glycerol at −80°C. An aliquot of the library was grown through one round of batch culture in LB until early stationary phase (approximately 8 h) without antibiotics to reduce false-positive results for genes that help to deal with the stress caused by the selection. Sequencing of this mutant pool resulted in 7,839,743 reads being mapped to 856,501 unique insertions within protein- and RNA-coding regions of the genome. We determined that the average insertion frequencies within the coding regions were 6 bp, 9 bp, and 12 bp for chromosome 1, chromosome 2, and pC3, respectively. Data analysis was performed using the unique insertion density (UID) approach of the Tn-Seq Explorer software as detailed in Materials and Methods (23).
Three hundred ninety-eight genes were predicted to be essential using the suggested cutoff value of 0.01 (see Table S1 in the supplemental material). The majority of essential genes appear to be on chromosome 1, with only 40 genes being identified as essential on chromosome 2 and pC3 (Table 1). Most of the essential genes are related to core metabolic processes, particularly carbon metabolism and amino acid production. In addition, genes involved in protein synthesis, energy generation, cell cycle, and cell wall and membrane structure were overrepresented (Fig. 1).
TABLE 1.
Numbers of unique insertions, average insertion frequencies, and numbers of essential genes of the three B. cenocepacia H111 replicons in rich medium
| Chromosome | Size of coding regions (bp) | No. of unique insertions | Insertion frequency (bp) | No. of essential genes |
|---|---|---|---|---|
| 1 | 3,099,313 | 496,269 | 6 | 358 |
| 2 | 2,657,050 | 288,110 | 9 | 30 |
| 3 | 904,321 | 72,122 | 12 | 10 |
FIG 1.
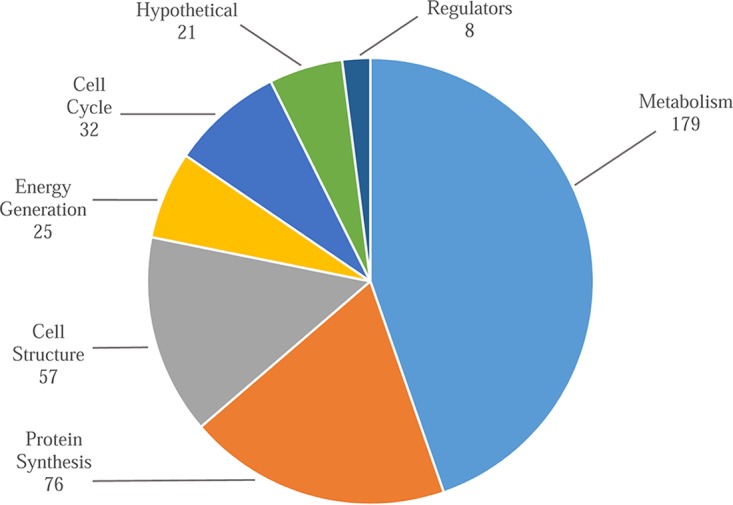
Functional categories of the 398 genes identified as being required for growth in rich medium.
Comparison of Tn-Seq data with published essential genes of B. cenocepacia.
To assess the accuracy of our Tn-Seq prediction, we cross-checked our data with previously reported essential genes. Wong et al. (22) conducted a TraDIS study of essential genes in B. cenocepacia J2315 and reported 383 genes as essential in rich medium. One hundred twenty-seven genes identified as essential in B. cenocepacia H111 were not reported as essential in J2315. Eighteen of these were tRNAs, which were not examined in that study, with the sequences of another 3 genes present but not annotated in the J2315 genome. Nineteen genes were in duplicated regions of J2315 and were likely for this reason not identified as essential in the J2315 study (22). Multiple rounds of batch culture were used in the J2315 study, and essential genes were investigated after each passage (22). Investigation of these data sets revealed that 44 H111 essential genes appeared in at least 1 of these “output pools,” with a further 22 genes being immediately upstream or downstream of genes identified in those data sets. This leaves 22 H111 essential genes that were not identified in J2315 (see Table S2 in the supplemental material). Three of these genes were within unique phage regions of H111, one of which was annotated as a DNA methylase and the other two likely encoding phage repressors. Information from the KEGG database revealed that 5 of the H111 essential genes, which were apparently nonessential in J2315, were related to the biosynthesis of arginine (24, 25). To investigate the essentiality of arginine biosynthesis, we constructed conditional BCAM0746 (arginosuccinate synthase gene) mutants in both strain backgrounds (Table 2). While the J2315 mutant grew well in the presence of glucose (nonpermissive condition), indicating that this gene is not essential in this strain, growth of the corresponding H111 mutant was strongly impaired. Importantly, supplementation of the medium with l-arginine rescued the growth defect of the H111 mutant (Fig. 2).
TABLE 2.
Validation of H111 candidate essential genesa
| Conditional mutant J2315 identifier | H111 RefSeq identifier | Function | Essentialb |
|---|---|---|---|
| B. cenocepacia | |||
| BCAM0632 | I35_RS19210 | N-Acetyltransferase | N |
| BCAM0746-strain H111 | I35_RS19780 | Arginosuccinate synthase | Y |
| BCAM0746-strain J2315 | I35_RS19780 | Arginosuccinate synthase | N |
| BCAM0911 | I35_RS20630 | 1-Deoxy-d-xylulose-5-phosphate synthase | Y |
| BCAM0913 | I35_RS20640 | tRNA threonylcarbamoyladenosine modification protein TsaD | Y |
| BCAM0918 | I35_RS20665 | RNA polymerase sigma factor RpoD | Y |
| BCAM0967 | I35_RS20915 | Succinate dehydrogenase | Y |
| BCAM0972 | I35_RS20940 | Type II citrate synthase | Y |
| BCAM0986 | I35_RS21020 | Aspartate-semialdehyde dehydrogenase | N |
| BCAM0994 | I35_RS21060 | Acetyl-CoAc carboxylase carboxyltransferase subunit beta | Y |
| BCAM0995 | I35_RS21065 | Bifunctional folylpolyglutamate synthase/dihydrofolate synthase | Y |
| BCAM0998 | I35_RS21080 | Amidophosphoribosyltransferase | N |
| BCAM1362 | I35_RS22520 | Cell division protein | Y |
| BCAM1931 | I35_RS25190 | Porin | N |
| BCAM2650 | I35_RS29140 | trans-2-Enoyl-CoA reductase | Y |
| BCAM2688 | I35_RS29330 | N-Acylglucosamine 2-epimerase | N |
| B. thailandensis E264/J2315 orthologue | |||
| BTH_I0484/BCAM0545 | I35_RS18760 | PTS fructose transporter subunit IIA | N |
| BTH_I1642/BCAM0855 | I35_RS20355 | UDP-glucose 6-dehydrogenase | N |
| BTH_II0656/BCAM0963 | I35_RS20895 | Hypothetical protein | N |
| BTH_II1941/BCAM1812 | I35_RS24870 | Agmatinase | N |
| BTH_II1863/BCAM2321 | I35_RS27455 | Electron transfer flavoprotein subunit alpha | N |
| BTH_II0780/BCAM2358A | I35_RS27645 | Hypothetical protein | N |
The conditional rhamnose-inducible mutants were spotted on LB plates supplemented with either 0.5% rhamnose (permissive condition) or 0.5% glucose (nonpermissive condition) as described in Materials and Methods. Growth was recorded after 24 h of incubation at 37°C.
Y, essential; N, nonessential.
CoA, coenzyme A.
FIG 2.
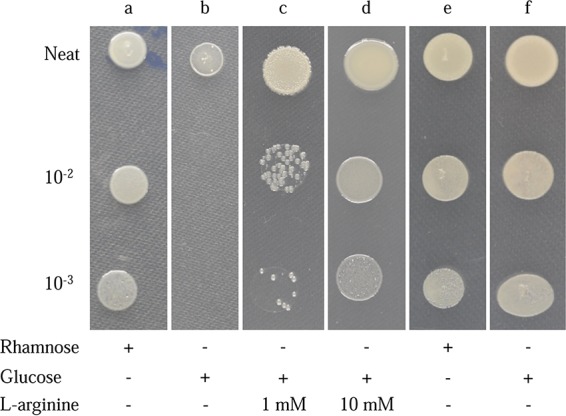
Conditional lethal phenotype of the rhamnose-inducible BCAM0746 mutant. Undiluted cultures of mutants grew visibly on plates supplemented with either glucose or rhamnose prior to depletion of the existing protein; however, at 100- and 1,000-fold dilutions, the mutant grew well in the presence of rhamnose (a) but was unable to grow on plates supplemented with glucose (b). Addition of 1 mM l-arginine partially restored growth of the BCAM0746 conditional mutant in the presence of glucose (c), and addition of 10 mM l-arginine restored growth to the level of the induced mutant (d). The J2315 BCAM0746 conditional mutant grew equally well in the presence of rhamnose (e) and glucose (f).
Bloodworth et al. (18) created 106 conditional mutants via transposon mutagenesis in strain B. cenocepacia K56-2. Many of these mutants were in operons, taking the total number of predicted essential genes in that study to 179. Ninety of these genes matched our Tn-Seq prediction, and 63 genes that were missing from our prediction were in operons with one or more of these genes (see Table S3 in the supplemental material). Of the 19 genes that did not match our Tn-Seq prediction, 12 were within 1 kb of an essential gene. Seven genes appeared to be nonessential in our Tn-Seq data set, and 7 genes identified as essential by Bloodworth et al. (18) had no orthologue in B. cenocepacia H111. In conclusion, these data show that although there is a considerable overlap of essential genes between B. cenocepacia strains H111, J2315, and K56-2, sets of essential genes can vary even between closely related strains, which suggests that cell viability can be achieved via different routes.
Data verification using conditional mutants.
To experimentally verify the Tn-Seq results, conditional mutants in which the native promoters of candidate genes were exchanged against the Escherichia coli rhamnose-inducible promoter PrhaB were created as described in Materials and Methods. Given that chromosome 1 carries mostly housekeeping genes, which match with previously published data sets of essential genes, we chose to verify 15 candidate genes identified on chromosome 2. Of these genes, 5 appeared to be nonessential under nonpermissive conditions (LB supplemented with 0.5% glucose): those encoding N-acetyltransferase (BCAM0632), aspartate-semialdehyde dehydrogenase (BCAM0986), amidophosphoribosyltransferase (BCAM0998), N-acylglucosamine 2-epimerase (BCAM2688), and a porin (BCAM1931). To assess the quality of our Tn-Seq approach, we also tested the essentiality of 6 orthologues from Burkholderia thailandensis E264 in LB that were reported as being essential in this organism (26) but did not appear in the H111 Tn-Seq data set. The 6 B. thailandensis orthologues were confirmed to be nonessential in B. cenocepacia H111, demonstrating that it was unlikely that our Tn-Seq methodology missed any essential genes (Table 2).
We noticed that one of the genes that could not be verified to be essential, BCAM2688, was previously found to be required for resistance of H111 to polymyxin (data not shown). As polymyxin targets the bacterial cell membrane, we hypothesized that the use of Irgasan to select against the E. coli donor and helper strains during transposon library construction counterselected mutants in this gene. Similar to polymyxin, Irgasan at high concentrations causes membrane damage (27). While the conditional BCAM2688 mutant grew well on LB plates supplemented with either polymyxin or Irgasan in the presence of rhamnose, growth was strongly impaired under nonpermissive conditions. This suggests that BCAM2688 is not an essential gene for growth in LB and that the respective mutants are not present in our library because of the use of Irgasan as a selective agent for library construction. BCAM1931 was also predicted to be essential in the Tn-Seq data set, with only one Tn insertion within the coding sequence, but we were unable to confirm this experimentally using a conditional mutant. Unlike the BCAM2688 mutant, this mutant was not susceptible to Irgasan or trimethoprim. Interestingly, this gene was also found to be essential in J2315 (22) and B. thailandensis E264 (26). Genes BCAM0632, BCAM0986, and BCAM0998, which could not be confirmed as essential, were close to the theoretical cutoff suggested by the Tn-Seq Explorer software as being essential.
Analysis of intergenic regions.
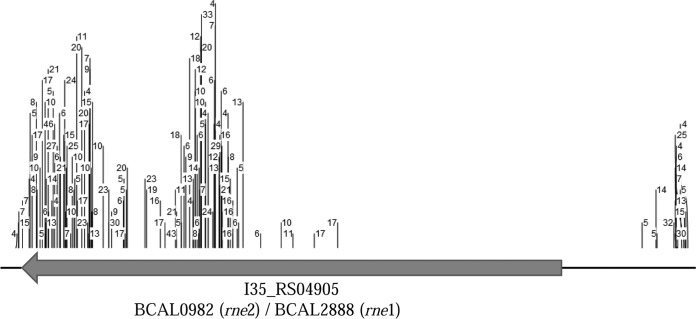
The analysis of intergenic regions is not currently supported by Tn-Seq Explorer. To circumvent this problem, the .ptt input file was adjusted to allow the software to examine the coordinates between genes rather than the coding regions. Two hundred sixteen intergenic regions of over 150 bp were identified as essential (Table S1). Some of these regions appeared to contain promoters of essential genes, but some were located between two nonessential genes. Four large intergenic regions were identified, 3 of which were annotated as pseudogenes in the RefSeq genome assembly (GCF_000236215.2). Another intergenic region spanning 720 bp appeared to be a promoter for RNase E (I35_RS04905). This gene had Tn insertions only within the second half of the gene, suggesting either that the N terminus of the resulting protein is sufficient for function or that the gene was not annotated correctly. Another intergenic region spanning 205 bp on chromosome 2 has a predicted noncoding small RNA in strain J2315. This small RNA (ncS62) is toxic when expressed in E. coli (29), and its function is at present unknown, although our Tn-Seq data set suggests that this small RNA is essential in strain H111.
Essential genes in ABC minimal medium.
Members of the genus Burkholderia are typically isolated from soil or the plant rhizosphere, where nutrients are limited. For this reason, we determined the essential genes required for growth in AB minimal medium with 0.2% citrate as the sole carbon source (ABC). Using a UID cutoff value of 0.007, 522 genes were identified as essential (see Table S4 in the supplemental material).
Of the 522 genes identified, 156 genes were specific for autonomous growth in minimal medium compared with LB. Most of these essential genes are involved in anabolic processes such as amino acid, fatty acid, purine, pyrimidine, and sugar biosynthesis. Twenty of the identified genes code for ribosomal proteins and chaperones. Conditionally essential ribosomal proteins have previously been described in E. coli, which requires the L21 ribosomal protein for growth at low temperatures, whereas L24 and L27 are essential at higher temperatures (30). Twenty-two of the identified genes appear to affect the cell structure, the majority of which are transporters or membrane proteins. There were few genes identified as important for energy generation and cell cycle changes, with the second-largest number of essential genes in ABC medium being hypothetical genes.
Genes related to the relief of oxidative stress were identified as essential in ABC minimal medium. Superoxide dismutase, pyrroloquinoline quinone (PQQ), and the pigment production phytoene synthase all reduce oxidative stress, and their genes appear to be essential in ABC minimal medium but not in LB.
In a previous study, we isolated 14 transposon insertion mutants in H111 that were not only attenuated in virulence in a wax moth model but also unable to grow in minimal medium (31). Ten of these were predicted to be essential using our Tn-Seq analysis in ABC medium, with the other 4 being found between essential genes (see Table S5 in the supplemental material). We also constructed BCAM0965 and BCAM2833 conditional mutants and confirmed that these genes are essential for growth in minimal medium but not in LB (data not shown).
The genes BCAM0986 and BCAM0998, which were borderline essential in LB, were clearly essential in ABC minimum medium. Testing of the respective conditional mutants confirmed that the BCAM0986 mutant was nonviable in ABC and the BCAM0998 mutant had a growth impairment compared with the H111 wild type. BCAM0986 is involved in threonine metabolism, and we were able to restore growth of the BCAM0986 conditional mutant by supplementation of ABC medium with 1 mM l-threonine (data not shown). In contrast, BCAM0632 was close to the cutoff for essentiality in both media, and the BCAM0632 conditional mutant grew as well as the wild type in ABC minimal medium (Fig. 3). This result suggests that growth of the BCAM0986 and BCAM0998 Tn5 mutants is dependent on certain nutritional components of LB that are depleted during growth. While these mutants grow well in fresh LB medium, they are unable to grow once these nutritional components are consumed, resulting in an underrepresentation of these mutants in LB medium.
FIG 3.
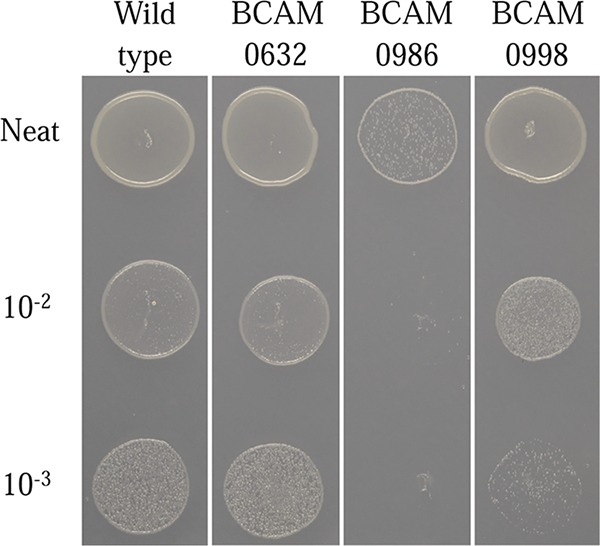
Conditional lethal phenotype of rhamnose-inducible mutants of BCAM0632, BCAM0986, and BCAM0998 on ABC minimal medium. The BCAM0632 conditional mutant had levels of growth comparable to those of the B. cenocepacia H111 wild type. The BCAM0986 conditional mutant was unable to grow on ABC medium, and the BCAM0998 conditional mutant had reduced growth at the 10−2 and 10−3 dilutions compared with the wild-type strain H111.
In conclusion, the ABC-specific essential genes encode mainly proteins required for the biosynthesis of core metabolites. A subset of genes related to oxidative stress were also identified, suggesting that growth in minimal medium is more stressful than growth in rich medium.
Essential genes under microoxic conditions.
Given that in the CF lung bacteria are limited for oxygen, we were interested in identifying essential genes required for microoxic growth in LB. Four hundred thirty-eight genes were identified as essential for growth under microoxic conditions (Table S6). Of the 93 genes that were specific for this condition compared with LB, 33 encode metabolic functions and 8 encode ribosomal proteins. Interestingly, under microoxic conditions, a number of modifications to the LPS structure appear to be essential along with a subset of genes encoding flagellar components. The L-ring, basal body modification, P-ring, and flagellar biosynthesis genes had very small numbers of transposon insertions compared with what was observed in LB and ABC minimal medium. In contrast, the flagellar brake protein had many more insertions under microoxic conditions than in aerobic growth in LB and minimal medium, suggesting that mutants with a defective brake protein have a growth advantage under microoxic conditions (Table 3). Interestingly, under low-oxygen conditions oxidative stress also appears to be a problem. The superoxide dismutase, PQQ, and phytoene synthase genes identified in the ABC experiment were also identified as essential for the microoxic environment. In addition, two exodeoxyribonuclease V subunit genes, which are involved in double-stranded DNA break repair, appear to be essential. This suggests that genotoxic stress prevails under microoxic conditions.
TABLE 3.
Comparison of the numbers of transposon insertions found in the flagellar biosynthesis genes in rich medium, minimal medium and microoxiaa
| Gene | Function | No. of unique insertions |
||
|---|---|---|---|---|
| LB | ABC | Microoxia | ||
| flgL | Flagellar hook-associated protein FlgL | 303 | 184 | 21 |
| flgK | Flagellar hook protein FlgK | 522 | 325 | 47 |
| —b | Flagellar brake protein YcgR | 158 | 77 | 776 |
| flgJ | Flagellar rod assembly protein FlgJ | 213 | 130 | 27 |
| — | Flagellar P-ring protein FlgI | 281 | 153 | 29 |
| — | Flagellar L-ring protein | 49 | 24 | 3 |
| flgG | Flagellar basal body rod protein FlgG | 231 | 108 | 25 |
| flgF | Flagellar basal body rod protein FlgF | 174 | 57 | 10 |
| flgE | Flagellar hook protein FlgE | 328 | 160 | 18 |
| — | Flagellar basal body rod modification protein FlgD | 106 | 56 | 8 |
| flgC | Flagellar basal body rod protein FlgC | 106 | 60 | 2 |
| flgB | Flagellar biosynthesis protein FlgB | 91 | 40 | 3 |
| — | Flagellar basal body P-ring biosynthesis protein FlgA | 164 | 58 | 6 |
| — | Flagellar biosynthesis protein FlgM | 87 | 69 | 31 |
| — | Flagellar synthesis chaperone protein FlgN | 113 | 58 | 52 |
Genes whose function is in bold were predicted as essential under microoxic conditions but not under normal growth conditions in LB and ABC medium.
—, no gene name assigned.
DISCUSSION
B. cenocepacia H111 has a large multireplicon genome (7.7 Mb) encoding approximately 7,500 proteins (32), giving this bacterium huge metabolic versatility. Yet, our analysis suggests that only 398 genes are essential under typical laboratory culture conditions. While the set of essential genes identified in this study of B. cenocepacia strain H111 are in good agreement with those identified in strain J2315, we noticed some interesting differences. We identified a leucine transporter gene, two genes involved in LPS biosynthesis, three genes involved in the cell cycle, and five genes related to arginine biosynthesis as essential in strain H111 but not J2315. Viability of the BCAM0746 (arginosuccinate synthase) conditional mutant was restored by the addition of 10 mM l-arginine to both rich and minimal media but not by the addition of 2× yeast extract or 2× tryptone.
It is unlikely that the arginine content of the media used in this study was vastly different from that used in the J2315 study, and we confirmed that in strain J2315 BCAM0746 was nonessential under the conditions used in this study. The high levels of externally added arginine, which rescued the growth defect of the BCAM0746 conditional mutant, suggest that H111 has a higher requirement for arginine than J2315 and even in rich medium de novo arginine biosynthesis is required for growth. This may indicate that H111 is impaired in arginine uptake or is draining the cellular arginine pool for the biosynthesis of an unknown metabolite.
The antibiotics used for selection of the transposon in our study were different from those used in the TraDIS study of J2315, which could contribute to variation between the data sets. Postconjugation, the H111 mutant library was selected using trimethoprim and Irgasan. During the verification of essential genes, BCAM2688 was identified as essential but only in the presence of Irgasan. BCAM2688 is annotated as encoding an N-acylglucosamine 2-epimerase, which catalyzes the reaction N-acyl-d-glucosamine ⇌ N-acyl-d-mannosamine (33) and is likely involved in sialic acid biosynthesis. Interestingly, it has been demonstrated that defects in sialic acid production sensitize Campylobacter jejuni to polymyxin (34). BCAM2688 was found to be essential for resistance to polymyxin as well as to colistin and SDS, demonstrating that this gene aids resistance to antimicrobials and disinfectants that target the cell membrane. This is an interesting observation, as Burkholderia species are inherently resistant to cationic antimicrobial peptides, which has been demonstrated to be achieved by incorporating amino arabinose naturally into the LPS (5, 6). Our results make BCAM2688 a promising target for antiresistance drugs, as inhibiting the encoded enzyme would render B. cenocepacia susceptible to existing antibiotics to which Burkholderia species are normally resistant.
Chromosome 2 essential region.
More than one-half of the essential genes on chromosome 2 are located within a large gene cluster between BCAM0911 and BCAM0995. Within this cluster are genes that are expected to be essential. Genes involved in the first steps of B vitamin, fatty acid, and folate biosynthesis are found within the cluster along with the housekeeping sigma factor gene rpoD and genes for parts of the tricarboxylic acid (TCA) cycle. For growth in ABC minimal medium, additional essential genes involved in amino acid and tRNA biosynthesis and the TCA cycle were identified. This cluster always appears on chromosome 2 in members of the genus Burkholderia, but these genes are found on the main chromosome in strains of the closely related genus Ralstonia. Chromosome 2, which is found in all Burkholderia species, has a typical plasmid type replication machinery, similar to pC3 (35), with the latter now being reclassified as a megaplasmid (10). It is tempting to speculate that the transfer of this essential region from chromosome 1 to an ancestral plasmid made this plasmid indispensable and contributed toward the separation of the Ralstonia genus from the Burkholderia genus.
Accessory essential genomes.
When examining ABC minimal medium assays, it was not anticipated that genes involved with the relief of oxidative stress would be identified. Our data suggest that oxidative stress is a problem in both minimal and microoxic environments, with PQQ, superoxide dismutase, and pigment production being identified as essential. While the global oxidative stress response regulator OxyR was not identified as essential in these experiments, the multiple antibiotic response regulator (MarR), which has been demonstrated to control both antibiotic and oxidative stress in E. coli, was (36). There are 28 MarR annotations in the B. cenocepacia H111 genome, and two different MarR orthologues were identified for ABC minimal medium (BCAM0731) and microoxic (BCAM0588) environments. It is conceivable that one of the MarR regulators could be part of the low-oxygen regulon, with the other being active in minimal media. BCAM0588 has been shown to be upregulated in B. cenocepacia microoxic RNA-Seq data sets (37, 38), which adds weight to this hypothesis. Further research is required to elucidate which processes are controlled by the MarR orthologues in B. cenocepacia H111 and why growth under minimal medium or microoxic conditions causes oxidative stress.
Interestingly, the flagellar L- and P-ring genes appeared to be essential under microoxic conditions. The flagellar biosynthesis genes have been demonstrated to be upregulated in microoxia (37, 38), but this cannot explain why only a subset of flagellar genes are essential in this environment. It could be due to the flagellar hook and filament not being correctly inserted into the membrane in these particular mutants, which causes structural damage due to the higher number of flagella. The flagellar brake protein appears to be strongly nonessential, with 776 insertions observed in the microoxic environment. This is a large number and was the 29th-most-hit gene in that experiment, strongly suggesting that inactivation of the flagellar brake gene provides a growth advantage in microoxia.
Tn-Seq error in identifying essential genes.
As pC3 is nonessential (9, 10), this replicon allowed an estimation of the error within our Tn-Seq experiments. There were 10 genes predicted to be essential that are carried by pC3 with 5 genes being expected as false positives. The repB and parAB genes were anticipated to lack Tn5 insertions, as an interruption of these replication genes results in loss of pC3 (9, 10), and thus these genes will appear to be void of insertions in the Tn-Seq analysis. The other two genes anticipated to lack Tn5 insertions were pilin biosynthesis genes, as it is likely that inactivation of certain genes of the pilus biosynthesis pathway may lead to the accumulation of toxic unassembled pilin (39, 40). This leaves 5 genes, all of which encode hypothetical proteins that are nonessential in this strain (10) but are predicted to be essential by Tn-Seq. Given that pC3 represents approximately a 7th of the size of the entire genome, we assume that there could be about 37 falsely identified genes, representing 7.5% of the results.
The use of antibiotic markers to select for transposon insertions during library construction is one reason why false-positive results appear in Tn-Seq data sets, as we exemplified in this study for BCAM2688 and its role in resistance to Irgasan. Uneven Tn distribution across the Burkholderia chromosomes could contribute to Tn-Seq error in these organisms. We noted 50% more Tn insertions in chromosome 1 than in chromosome 2 and twice as many insertions in chromosome 1 as in pC3. This phenomenon was also reported in the J2315 study (22), and it was noted that almost twice as many insertions were present on chromosome 1 as on chromosome 2 in a study of Burkholderia pseudomallei (4). At present, the reason for the observed uneven Tn distribution is unclear.
The average Tn coverage in the LB experiment was one insertion every 6, 9, and 12 bp for chromosome 1, chromosome 2, and pC3, respectively. This is very high coverage, and it is unlikely that essential genes were underestimated on chromosome 2 and pC3. A number of chromosome 2 essential genes were confirmed using conditional mutants, with BCAM0986 and BCAM0998 predicted to provide a fitness benefit in LB rather than being essential per se, further supporting that our cutoff for essentiality was accurate.
The relative fitness of each mutant would also play a role in Tn-Seq error. Genes that are found close to the predicted cutoff for essentiality may be viable but had a growth defect at some point during library selection. Two examples of this were BCAM0986 and BCAM0998, which appeared close to the cutoff for essentiality in LB and whose mutants likely encountered a growth defect, as specific nutrients were depleted during growth. BCAM0986, which encodes an aspartate-semialdehyde dehydrogenase, is involved in the conversion of l-aspartate to l-threonine (24, 25). All the genes that encode enzymes required for this conversion (BCAL1925, BCAL1926, BCAL2146, BCAM0899, and BCAM0986) were essential in the ABC minimal medium data set. BCAL1925, encoding a threonine synthase, was essential in LB, but this gene is also involved in vitamin B6 metabolism (24, 25). BCAL2146, which encodes the 1st step in the conversion of l-aspartate to l-threonine (24, 25), was borderline essential in LB. It is likely that mutants in BCAL2146 and BCAM0986 encountered a growth defect as threonine was depleted from LB, resulting in the underrepresentation of these mutants in the LB data set. In support of this hypothesis, we were able to restore growth of the BCAM0986 conditional mutant in ABC minimal medium by the addition of 1 mM l-threonine.
Gene BCAM0998 (purF) is annotated as a gene for amidophosphoribosyltransferase, which converts 5-phosphoribosyl-1-pyrophosphate (PRPP) to 5-phosphoribosyl-1-amine (PRA) using the ammonia group from l-glutamine (24, 25). This enzyme links the endpoint of the pentose phosphate pathway (PRPP) to purine biosynthesis, and this is another example of a mutant that likely had a growth impairment due to the depletion of certain nutrients during growth in LB. Interestingly, a BCAM0998 mutant was identified in a screen for reduced pathogenicity in a number of nonmammalian pathogenicity models (31). This demonstrates that in certain infection models, purines cannot be obtained from the host and de novo biosynthesis is required. Both BCAM0986 and BCAM0998 were confirmed to be essential in ABC (Fig. 3) but not LB medium.
Another issue is Tn-Seq data interpretation. It is assumed that the central part of a gene is required for function. After examining intergenic regions, we discovered a large intergenic region (720 bp) devoid of transposons. Directly downstream of this intergenic region was a gene annotated as RNase E, which had insertions only after approximately 1,200 bp of its coding sequence, suggesting that this is an essential gene (Fig. 4).
FIG 4.
Visualization of the numbers and positions of transposon insertions within the coding sequence and the upstream region of RNase E (I35_RS04905). Note that no insertions were found within a large part of the upstream intergenic region of this gene and that the first insertions are found only approximately 1,200 bp downstream of the annotated start of the reading frame. This gene is duplicated in B. cenocepacia J2315, with each functional copy being designated BCAL0982 and BCAL2888. Image taken from the LB data set.
The RNase E gene is an essential gene in E. coli, and the protein's functional domains are found in the N terminus (41), which could explain why this region is devoid of insertions. It could also be that the gene was not annotated correctly. This annotation issue was further exemplified when 3 pseudogenes (BCAL0904, BCAL1005, and BCAL2189) were identified in all experiments during the intergenic region analysis. The RefSeq assembly files from NCBI were used in this analysis. Postanalysis, it was discovered that these 3 pseudogenes had full annotations in the GenBank assembly files, also available from NCBI, and had full annotations in strain J2315 (42). We also identified 3 genes for which the sequence is present but not annotated on the J2315 genome. These examples demonstrate that inconsistencies in genome annotations contribute to error in Tn-Seq data analysis.
The core essential genome.
Comparison of the LB, ABC, and microoxic data sets revealed that there were 339 genes identified as essential in all 3 experiments, which can be considered the core essential genome of B. cenocepacia H111 (see Table S7 in the supplemental material). This accounts for only 5% of the coding capacity of this bacterium but is well within the range of essential genes reported in other bacteria. Three hundred thirty-six genes were suggested to be essential in Pseudomonas aeruginosa (43), 505 in Burkholderia pseudomallei (4), and 406 in Burkholderia thailandensis (26). The smallest experimentally verified genome shown to support autonomous growth was that of the redesigned synthetic cell Mycoplasma mycoides JCVI-syn3.0, which contained 473 genes (44).
Although there are drawbacks to identifying essential genes using Tn-Seq, a highly conserved set of 339 genes were identified in all experiments. Essential genes have been proposed as good targets for novel antimicrobials (3, 4), and hence, this set of common essential genes would make a good starting point for the development of novel antimicrobials targeting B. cenocepacia (Table S7).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are detailed in Table S8 in the supplemental material. Bacterial strains were routinely grown in Difco Lennox broth (LB) or AB medium (45) supplemented with 0.2% citrate (ABC) as the sole carbon source at 37°C with shaking at 200 rpm or LB or ABC agar plates (addition of 1.8% agar) at 37°C. The medium was supplemented with relevant antibiotics when appropriate, using the following concentrations: (i) for Escherichia coli, kanamycin (Kan), 25 μg ml−1, and trimethoprim (Tp), 50 μg ml−1; (ii) for B. cenocepacia, Tp, 50 μg ml−1. To select for B. cenocepacia after mating experiments, LB agar plates supplemented with Tp (50 μg ml−1) for strain H111 or Tp (500 μg ml−1) for strain J2315, polymyxin B (Pol) (50 μg ml−1), and 0.5% l-rhamnose were used. Media were supplemented with 0.5% l-rhamnose or 0.5% glucose for inducible strains as required.
Tn-Seq methodology.
Transposon mutagenesis was carried out by conjugation. Overnight cultures of the recipient strain (B. cenocepacia H111), the donor strain, E. coli SM10 λ-pir containing pLG99 (carrying a Tn5 transposon), and a helper strain, E. coli DH5α pRK2013 (to increase efficiency of transfer), were grown with the appropriate antibiotics. The optical density (OD) at 600 nm was measured, cultures were washed twice in fresh LB, and the OD was adjusted to 1. Recipient, donor, and helper strains were mixed in a 2:1:1 ratio (1 ml to 0.5 ml to 0.5 ml), centrifuged, resuspended in 100 μl LB, spotted onto a nitrocellulose filter (0.45-μm pore size) placed on an LB plate, and incubated for 5 h at 37°C. Cells were washed from the filter with 1 ml saline solution and plated on LB agar containing trimethoprim (50 μg/ml) and Irgasan (25 μg/ml) to counterselect E. coli. Plates were incubated at 37°C overnight, and the resulting colonies were washed from the plates with 1 ml LB (25% glycerol). Multiple 1-ml washes were stored in aliquots at −80°C. An estimated total of 1,000,000 mutants were generated from 44 parallel matings.
For Tn-Seq experiments, the mutant population was grown in liquid LB and ABC minimal medium under oxygenated and microoxic conditions (0.5% O2) as described previously (37). Cultures were grown to late exponential phase, and genomic DNA was extracted using the Qiagen DNeasy blood and tissue kit.
Sequencing of transposon junctions was undertaken using the circle method (46) with the following amendments. Final library amplification by PCR was carried out using Phusion polymerase (NEB) with the GC buffer in the place of Kapa polymerase. Postamplification, a size selection step was included to select for molecules between 230 and 380 bp, which were recovered from a 1% agarose gel using a Qiagen gel recovery kit. Samples were pair end sequenced using the 2 × 250-bp kit and the custom sequencing setup on the Illumina MiSeq platform.
Construction of conditional mutants and screening for essentiality.
The construction of conditional mutants in Burkholderia species using pSC200 has been well described. Briefly, this plasmid has an R6K origin of replication and contains a multiple cloning site (MCS) upstream of a rhaR-rhaS-PrhaB module. A portion of the coding sequence of the gene of interest is cloned into the MCS, and the plasmid is transferred to Burkholderia, where it integrates into the target sequence via homologous recombination, switching the native promoter to the rhamnose-inducible promoter (PrhaB) (4, 8, 18, 19).
To construct mutagenesis plasmids, 500- to 600-bp fragments of the 5′ end of selected genes were amplified by PCR using the primers detailed in Table S9 in the supplemental material. Primers were designed to remove the start codon and add an NdeI restriction site to the 5′ end and an XbaI site to the 3′ end of the resulting PCR products. The NdeI restriction site has an ATG, which negates the need for a start codon postcloning. PCR products were cleaned using a Qiagen PCR cleanup kit, restricted, and cloned into similarly digested pSC200 using standard molecular biology techniques (47). The resulting plasmids were maintained in E. coli CC118 λ-pir and sequenced prior to use. Plasmids were introduced into B. cenocepacia H111 by triparental mating as described above. Chromosomal integrants were selected on LB agar containing 50 μg ml−1 Tp for strain H111 or 500 μg ml−1 Tp for strain J2315, 50 μg ml−1 polymyxin B (Pol), and 0.5% (wt/vol) l-rhamnose, and the resulting mutants were stored at −80°C in 25% (wt/vol) glycerol (final).
To assess if a target gene was essential, overnight cultures were set up in LB broth containing 0.2% (wt/vol) l-rhamnose. Cultures were diluted to 10−2 and 10−3 in fresh LB and 2.5 μl of the neat overnight culture and each dilution culture was spotted onto both LB with 0.5% (wt/vol) l-rhamnose (to induce PrhaB) and LB with 0.5% (wt/vol) glucose (to repress PrhaB) plates. For minimal medium assays, an ABC plate with 0.5% (wt/vol) glucose was used. Cultures were grown at 37°C, and growth was assessed after 24 and 48 h.
Tn-Seq data analysis and bioinformatics.
Illumina sequence FASTQ files were trimmed for quality using trimmomatic-0.32 and the settings Leading:30, Trailing:30, Slidingwindow:4:20, Minlen:60 (48). Adapter sequences were then removed using Cutadapt (v1.9) (49).
Tn-Seq data were analyzed using an open-licensed software platform called Tn-Seq Explorer (23). Prior to utilizing Tn-Seq Explorer, .ptt and .rnt input file types had to be created. The Tn-Seq Explorer software was designed to utilize these file types to input the locations of the protein and RNA coding regions to the software, but these file types are no longer accessible via NCBI (23). Tn-Seq Explorer can access the old .ptt and .rnt files from an archive, but B. cenocepacia H111 is not in that archive. To circumvent this problem and allow future use of Tn-Seq Explorer, a custom Python script was written to extract protein and RNA annotations from the current RefSeq .gbff file and format the data as .ptt and .rnt files, which were loaded into Tn-Seq Explorer. The .ptt and .rnt files are available upon request.
Trimmed reads were mapped to the chromosome using the Bowtie 2 plugin of Tn-Seq Explorer; the default “–very-sensitive” command for mapping pair end reads was used (50). Reads were mapped to all three chromosomes simultaneously instead of mapping to the individual chromosomes to prevent reads mapping to orthologues rather than to their true locations. A sequence alignment map (SAM) file containing the positions of each read was produced. Each SAM file was extracted into Tn-Seq Explorer to examine essentiality. Transposons mapping within 5% of the start codon and 20% of the stop codon were excluded from the analysis. Unique insertion density (UID) and unique insertion counts (UIC) were calculated using the software. A prediction for the cutoff UID value was carried out for each experiment to separate the essential from the nonessential genes (23).
In this approach, the number of unique insertions of a given gene is divided by the gene length, returning the UID for that gene. The frequency of the UID values is calculated, and a plot of frequency versus UID is created. Typically, a bimodal distribution is observed, whereby genes that have very few or no insertions appear in high frequency on the left side of the plot. The plot quickly decreases as genes appear with higher numbers of transposons until the plot rises again for genes that can tolerate transposon insertions (4, 26, 46). It is at this crossover point that the predicted “cutoff” for essential genes is taken. Past this point, genes are considered to be able to tolerate high numbers of transposon mutations without affecting growth and are considered nonessential; the plot for the LB data set can be viewed in Fig. S10 in the supplemental material.
The locations and frequency of each Tn insertion were added to our RefSeq GenBank file using a custom Python script for visualization in a bioinformatics genome browser; these files are available upon request. COG and EggNOG functions were added to each of the essential genes using Mauve.
Availability of data.
Raw FASTQ files generated from the Illumina MiSeq platform are publicly available from the NCBI short reads archive (SRA) and can be found using the following link: https://www.ncbi.nlm.nih.gov/sra/?term=SRP115200. The accession numbers for individual data sets are as follows: LB data set, SRX3085715; ABC data set, SRX3085716; LB microoxic (0.5% O2) data set, SRX3085714.
Supplementary Material
ACKNOWLEDGMENTS
We thank Larry Gallagher (Department of Genome Sciences, University of Washington, Seattle, WA) for supplying pLG99 and a detailed protocol for the Tn-Seq circle method, Hans-Martin Fischer (Institute of Microbiology, ETH Zürich), who allowed us to conduct the microoxic experiments in his laboratory, and Lucy Poveda (Functional Genomics Center Zürich) for sequencing of our transposon libraries on the Illumina MiSeq platform.
This work was supported by the Swiss National Science Foundation (Project 31003A-169307/1).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00260-17.
REFERENCES
- 1.Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, Edwards TE, Armour B, Begley DW, Dieterich SH, Dranow DM, Abendroth J, Fairman JW, Fox D III, Staker BL, Phan I, Gillespie A, Choi R, Nakazawa-Hewitt S, Nguyen MT, Napuli A, Barrett L, Buchko GW, Stacy R, Myler PJ, Stewart LJ, Manoil C, Van Voorhis WC. 2013. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS One 8:e53851. doi: 10.1371/journal.pone.0053851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juhas M, Eberl L, Church GM. 2012. Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends Biotechnol 30:601–607. doi: 10.1016/j.tibtech.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C. 2015. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moule MG, Hemsley CM, Seet Q, Guerra-Assuncao JA, Lim J, Sarkar-Tyson M, Clark TG, Tan PB, Titball RW, Cuccui J, Wren BW. 2014. Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5:e00926-13. doi: 10.1128/mBio.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamad MA, Di Lorenzo F, Molinaro A, Valvano MA. 2012. Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia. Mol Microbiol 85:962–974. doi: 10.1111/j.1365-2958.2012.08154.x. [DOI] [PubMed] [Google Scholar]
- 6.Loutet SA, Valvano MA. 2011. Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Microbiol 2:159. doi: 10.3389/fmicb.2011.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortega X, Silipo A, Saldias MS, Bates CC, Molinaro A, Valvano MA. 2009. Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: truncation of the core oligosaccharide leads to increased binding and sensitivity to polymyxin B. J Biol Chem 284:21738–21751. doi: 10.1074/jbc.M109.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega XP, Cardona ST, Brown AR, Loutet SA, Flannagan RS, Campopiano DJ, Govan JR, Valvano MA. 2007. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J Bacteriol 189:3639–3644. doi: 10.1128/JB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agnoli K, Frauenknecht C, Freitag R, Schwager S, Jenul C, Vergunst A, Carlier A, Eberl L. 2014. The third replicon of members of the Burkholderia cepacia complex, plasmid pC3, plays a role in stress tolerance. Appl Environ Microbiol 80:1340–1348. doi: 10.1128/AEM.03330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, Sokol PA, Carlier A, Eberl L. 2012. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol Microbiol 83:362–378. doi: 10.1111/j.1365-2958.2011.07937.x. [DOI] [PubMed] [Google Scholar]
- 11.Jones AM, Dodd ME, Govan JR, Barcus V, Doherty CJ, Morris J, Webb AK. 2004. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59:948–951. doi: 10.1136/thx.2003.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartell JA, Yen P, Varga JJ, Goldberg JB, Papin JA. 2014. Comparative metabolic systems analysis of pathogenic Burkholderia. J Bacteriol 196:210–226. doi: 10.1128/JB.00997-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang K, Zhao H, Sun C, Lam CM, Chang S, Zhang K, Panda G, Godinho M, Martins dos Santos VA, Wang J. 2011. Exploring the metabolic network of the epidemic pathogen Burkholderia cenocepacia J2315 via genome-scale reconstruction. BMC Syst Biol 5:83. doi: 10.1186/1752-0509-5-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loutet SA, Valvano MA. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect Immun 78:4088–4100. doi: 10.1128/IAI.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDowell A, Mahenthiralingam E, Dunbar KE, Moore JE, Crowe M, Elborn JS. 2004. Epidemiology of Burkholderia cepacia complex species recovered from cystic fibrosis patients: issues related to patient segregation. J Med Microbiol 53:663–668. doi: 10.1099/jmm.0.45557-0. [DOI] [PubMed] [Google Scholar]
- 17.Varga JJ, Losada L, Zelazny AM, Kim M, McCorrison J, Brinkac L, Sampaio EP, Greenberg DE, Singh I, Heiner C, Ashby M, Nierman WC, Holland SM, Goldberg JB. 2013. Draft genome sequences of Burkholderia cenocepacia ET12 lineage strains K56-2 and BC7. Genome Announc 1(5):e00841-13. doi: 10.1128/genomeA.00841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloodworth RAM, Gislason AS, Cardona ST. 2013. Burkholderia cenocepacia conditional growth mutant library created by random promoter replacement of essential genes. Microbiologyopen 2:243–258. doi: 10.1002/mbo3.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhas M, Stark M, von Mering C, Lumjiaktase P, Crook DW, Valvano MA, Eberl L. 2012. High confidence prediction of essential genes in Burkholderia cenocepacia. PLoS One 7:e40064. doi: 10.1371/journal.pone.0040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brutinel ED, Gralnick JA. 2012. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol Microbiol 86:273–283. doi: 10.1111/j.1365-2958.2012.08196.x. [DOI] [PubMed] [Google Scholar]
- 21.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong YC, Abd El Ghany M, Naeem R, Lee KW, Tan YC, Pain A, Nathan S. 2016. Candidate essential genes in Burkholderia cenocepacia J2315 identified by genome-wide TraDIS. Front Microbiol 7:1288. doi: 10.3389/fmicb.2016.01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solaimanpour S, Sarmiento F, Mrazek J. 2015. Tn-seq explorer: a tool for analysis of high-throughput sequencing data of transposon mutant libraries. PLoS One 10:e0126070. doi: 10.1371/journal.pone.0126070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. 2013. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio 4:e00604-13. doi: 10.1128/mBio.00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villalain J, Mateo CR, Aranda FJ, Shapiro S, Micol V. 2001. Membranotropic effects of the antibacterial agent Triclosan. Arch Biochem Biophys 390:128–136. doi: 10.1006/abbi.2001.2356. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Sass AM, Van Acker H, Forstner KU, Van Nieuwerburgh F, Deforce D, Vogel J, Coenye T. 2015. Genome-wide transcription start site profiling in biofilm-grown Burkholderia cenocepacia J2315. BMC Genomics 16:775. doi: 10.1186/s12864-015-1993-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG. 2011. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol 413:751–761. doi: 10.1016/j.jmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwager S, Agnoli K, Kothe M, Feldmann F, Givskov M, Carlier A, Eberl L. 2013. Identification of Burkholderia cenocepacia strain H111 virulence factors using nonmammalian infection hosts. Infect Immun 81:143–153. doi: 10.1128/IAI.00768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlier A, Agnoli K, Pessi G, Suppiger A, Jenul C, Schmid N, Tummler B, Pinto-Carbo M, Eberl L. 2014. Genome sequence of Burkholderia cenocepacia H111, a cystic fibrosis airway isolate. Genome Announc 2(2):e00298-14. doi: 10.1128/genomeA.00298-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 34.Naito M, Frirdich E, Fields JA, Pryjma M, Li J, Cameron A, Gilbert M, Thompson SA, Gaynor EC. 2010. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J Bacteriol 192:2182–2192. doi: 10.1128/JB.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubarry N, Pasta F, Lane D. 2006. ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J Bacteriol 188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol 8:51–62. [PubMed] [Google Scholar]
- 37.Pessi G, Braunwalder R, Grunau A, Omasits U, Ahrens CH, Eberl L. 2013. Response of Burkholderia cenocepacia H111 to micro-oxia. PLoS One 8:e72939. doi: 10.1371/journal.pone.0072939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sass AM, Schmerk C, Agnoli K, Norville PJ, Eberl L, Valvano MA, Mahenthiralingam E. 2013. The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. ISME J 7:1568–1581. doi: 10.1038/ismej.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linhartova M, Bucinska L, Halada P, Jecmen T, Setlik J, Komenda J, Sobotka R. 2014. Accumulation of the type IV prepilin triggers degradation of SecY and YidC and inhibits synthesis of Photosystem II proteins in the cyanobacterium Synechocystis PCC 6803. Mol Microbiol 93:1207–1223. doi: 10.1111/mmi.12730. [DOI] [PubMed] [Google Scholar]
- 40.Hultgren SJ, Normark S, Abraham SN. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol 45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 41.Mackie GA. 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 42.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutchison CA III, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 45.Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J Mol Biol 23:99–112. doi: 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- 46.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 50.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.