Abstract
With early life likely to have existed in a hot environment, enzymes had to cope with an inherent drop in catalytic speed caused by lowered temperature. Here we characterize the molecular mechanisms underlying thermoadaptation of enzyme catalysis in adenylate kinase using ancestral sequence reconstruction spanning 3 billion years of evolution. We show that evolution solved the enzyme's key kinetic obstacle—how to maintain catalytic speed on a cooler Earth—by exploiting transition-state heat capacity. Tracing the evolution of enzyme activity and stability from the hot-start toward modern hyperthermophilic, mesophilic, and psychrophilic organisms illustrates active pressure versus passive drift in evolution on a molecular level, refutes the debated activity/stability trade-off, and suggests that the catalytic speed of adenylate kinase is an evolutionary driver for organismal fitness.
As a direct manifestation of molecular kinetic energy, temperature is a fundamental evolutionary driver for chemical reactions. However, it is currently not understood how the natural evolution of catalytic efficiency responds to marked changes in environmental temperatures. Theoretically, temperatures in biological processes could range from −50°C, where cells vitrify (1), to +140°C, when nucleic acids spontaneously hydrolyze (2). Life has entrenched itself in nearly every point along this scale, including extreme psychrophiles such as the bacteria Psychrobacter articus living in Arctic ice veins at −20°C (3) and the hyperthermophile Archean Pyrlobus fumarii that grows at 121°C (4).
It is likely that early life existed in a hot environment. The hot-start hypothesis is supported by several lines of evidence, including the presence of thermophilic organisms on the branches closest to the roots of the universal tree of life (5), the ratio of oxygen and silica isotopes in sedimentary chert and surrounding seawater (6), lower-viscosity seawater during the Archean eon (7), and reconstruction of ancestral proteins showing that the oldest nodes in phylogenetic trees are the most thermophilic (8, 9). The hot-start hypothesis implies that life had to adapt to cooler temperatures due to cooling of the Earth. What does this temperature adaptation mean for enzymes? Enzymes had to contend with the evolutionary pressures on stability and activity. Chemical reaction rates, whether catalyzed or uncatalyzed, inherently scale with temperature. Hence, a hypothetical thermophilic enzyme encountering cooler environments must evolve increased catalytic activity at lower temperatures (Fig. 1A), while accommodating relaxed selection on thermostability. However, if this mesophilic ancestor, once evolved to cooler conditions, returned to a hot environment, then selection would favor an increase in melting temperature, with no pressure on its already optimized catalytic activity (Fig. 1A).
Fig. 1. Evolutionary pressures on enzymes during adaptation to environmental temperature changes.
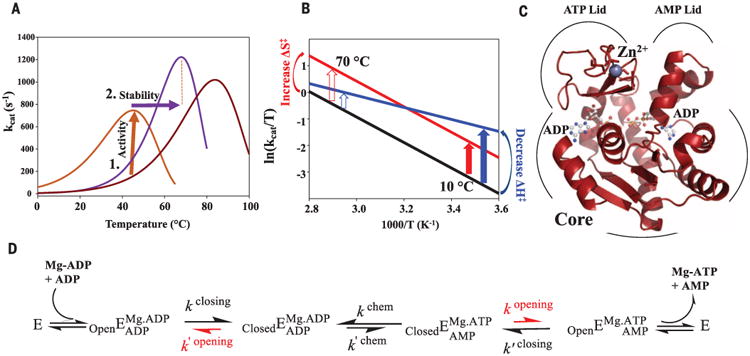
(A) Hypothetical ancestral protein (dark red) needs to increase activity at lower temperatures (brown arrow), resulting in a second ancestral enzyme (brown). Evolution from cold to hot imposes pressure on increased stability (purple arrow), resulting in a modern thermophile (purple). (B) Eyring plot illustrating proposed mechanisms for increasing enzymatic activity at colder temperatures by increasing ΔS‡ (red) versus decreasing ΔH‡ (blue) relative to the uncatalyzed reaction (black) (12, 13, 34) with larger rate acceleration at low temperatures by ΔH‡ (blue arrow). (C) The 1.2 Å x-ray structure of ANC1 Adk with two ADPs bound. (D) Reaction scheme for Adk catalysis, highlighting lid opening as the rate-limiting step (red) [from (22)].
Although intense experimental effort has gone into understanding protein thermostability (8, 10, 11), the challenge of evolving efficient enzymatic turnover at lower temperatures has not been addressed. Indeed, Wolfenden and colleagues raised this question as a crucial evolutionary obstacle (12, 13). The authors reasoned that in an early hot environment, primordial chemical reactions could occur readily, but would become slow at lower temperatures because of their steep temperature dependencies (12, 13). They suggested that enzymes solved the thermal kinetic obstacle at lower temperatures by decreasing the enthalpic activation barrier (ΔH‡), resulting in a shallower temperature dependence (Fig. 1B) (12, 13). Here we investigate the molecular mechanisms underlying thermoadaptation of enzyme catalysis using ancestral sequence reconstruction (ASR) spanning about 3 billion years of evolution.
ASR [reviewed in (14)] is a valuable tool for experimentally testing molecular evolutionary questions. ASR uses genetic patterns stored in the sequences of extant proteins, and the phylogenetic relationships between them, to infer sequences of ancestors along an evolutionary trajectory. We focused on the enzyme adenylate kinase (Adk) because it is an essential enzyme found in nearly every life form, it has a strong correlation between organismal growth temperature and Adk stability (15, 16), and its catalytic mechanism is well understood (Fig. 1, C and D). Adk catalyzes the reversible conversion of Mg/ adenosine 5′-triphosphate (Mg/ATP) and adenosine 3′,5′-monophosphate (AMP) into two adenosine 5-diphosphate (ADP) molecules, maintaining nucleotide concentrations in the cell.
The phylum Firmicutes is an ancient bacterial lineage [∼3 billion years ago (bya)], and its contemporary species inhabit a wide range of environments (17), including aerobic deep polar sediment (Bacillus marinus), hot thermal springs (B. stearothermophilus), and common soil (B. subtilis), as well as hot anaerobic environments such as seafloor hydrothermal vents (C. subterraneus) (Fig. 2A). In addition, modern Firmicutes have a particularly well-conserved core genome and slow mutational rates (5, 17). A robust coestimated phylogeny and alignment of modern Fir-micute Adk sequences from the National Center for Biotechnology Information database was constructed using BAli-Phy (18) (Fig. 2A, figs. S1 and S2, and data file S1). The strength of the reconstructed phylogeny and alignment is illustrated by strong posterior probabilities along branch sites (fig. S1) and its topological resemblance with previously reported trees based on 16 genes found in all three domains of life (5).
Fig. 2. Evolution of thermostability through the reconstructed Adk phylogeny spanning 2.5 to 3 bya.
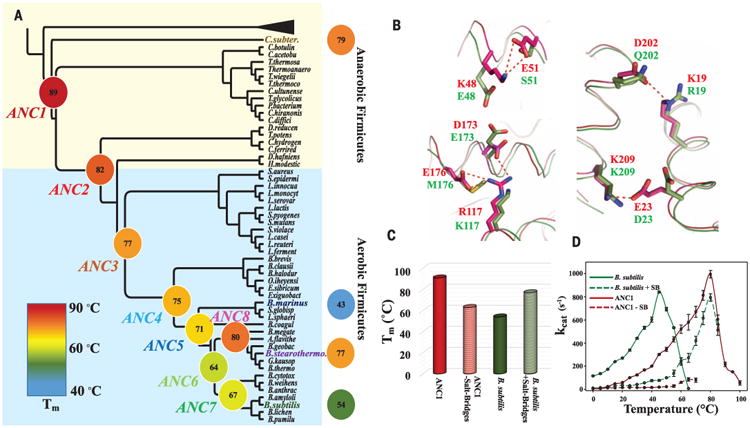
(A) Collapsed cladogram of the tree (see fig. S1) used to resurrect ancestral Adks (nomenclature and colors for the 12 Adks are used throughout the manuscript). Measured Tms are indicated and illustrated by a continuous color scale. (B) Superposition of ANC1 (red, 5G3Y) and B. subtilis Adk (green, 1P3J) structures suggests salt bridges (dotted red lines) in ANC1 responsible for high Tm. (C) Removing these salt bridges from ANC1 or adding them into B. subtilis results in a drastic decrease or increase in Tm, respectively. (D) Corresponding activity changes of these mutant proteins. Errors are standard errors from the linear fit based on eight time points for each temperature.
We reconstructed eight nodes along the lineage from the oldest ancestor corresponding to the divergence of aerobic and anaerobic Firmicutes, estimated divergence between 3 and 2.6 bya (17, 19), toward modern-day thermophilic (B. stearothermophilus, C. subterraneus), mesophilic (B. subtilis), and psychrophilic (B. marinus) Adks (Fig. 2A and figs. S1 and S2). The great majority of sites in the reconstructed sequences have >0.95 posterior probability (fig. S3). As an independent control for phylogenetic uncertainty and proposed biases with maximum likelihood-based reconstructions (20), three of the nodes (ANC1, ANC3, ANC4) were also predicted using sequences sampled from the posterior distribution inferred during the BAli-phy run. Those nodes had properties nearly identical to those predicted from maximum likelihood (table S1), thus confirming that the reconstructed sequences are robust to the uncertainty inherent in the methods. Eight ancestral and four modern Adk enzymes were overexpressed in Escherichia coli and purified. All ancestors are active enzymes with kcat values comparable to those of their modern descendants (table S2), even though they contained up to 47 amino acid differences (ANC1) relative to any modern Adk.
First, we characterized the evolutionary trajectories of melting temperatures using differential scanning calorimetry (DSC) (Fig. 2A). ANC1 starts with the highest melting temperature (Tm) of 89°C. Melting temperatures gradually decrease along the tree from the oldest reconstructed enzyme toward modern-day mesophilic and psychrophilic enzymes. In one branch, the modern thermophile B. stearothermophilus reevolved thermostability from an ancestor that had adopted to cooler climates (ANC6, Tm 64°C).
To view this evolutionary path through a structural lens, we solved high-resolution crystal structures of ANC1, ANC3, and ANC4 bound to ADP or the substrate analog Ap5A [P1, P5-di(adenosine-5′) pentaphosphate] (table S3). A global comparison of our ancestor structures among each other and with crystal structures of the modern mesophilic, psychrophilic, and thermophilic Adk enzymes (1P3J, 1S3G, 1ZIN) hinted at salt bridges as the primary source for differential stabilities (Fig. 2B, fig. S4, and table S4). Several unique salt bridges sequentially disappear during evolution toward colder environments and then reappear in species that subsequently adapt to hot niches (fig. S5 and table S5). To experimentally test this mechanistic interpretation, we mutated residues responsible for the salt bridge differences in ANC1 to the corresponding residues found in Adk from B. subtilis, and vice versa (Fig. 2, B and C). The changes in Tm from interchanging only these five salt bridges indicate that they are indeed the primary contributors to thermostability. Both contrived “swap mutant enzymes” are catalytically impaired (Fig. 2D), indicating epistasis, meaning that the backgrounds in which these salt bridges are placed influence the effects of the mutations (14).
The thermostability data provided the starting point to address the question of how catalytic power evolved in response to environmental temperature change. For this purpose, catalytic rates were measured between 0° and 100°C (Fig. 3) for all 13 enzymes. First, all eight ancestors display catalytic activities at their optimal temperatures that differ by less than a factor of 3 from those of modern Adk enzymes, providing an additional internal control for accurately reconstructed sequences (11). Second, ancestors and modern Adk enzymes display the typical trend of increased activity with temperature followed by a steep decline due to thermal denaturation (fig. S6). Third, below 25°C, ANC1 and ANC2 have a strong temperature dependence with very low catalytic rates, agreeing with the prediction that ancient enzymes are adapted exclusively to hot environments.
Fig. 3. Evolution of enzyme activity over about 2.5 to 3 bya.
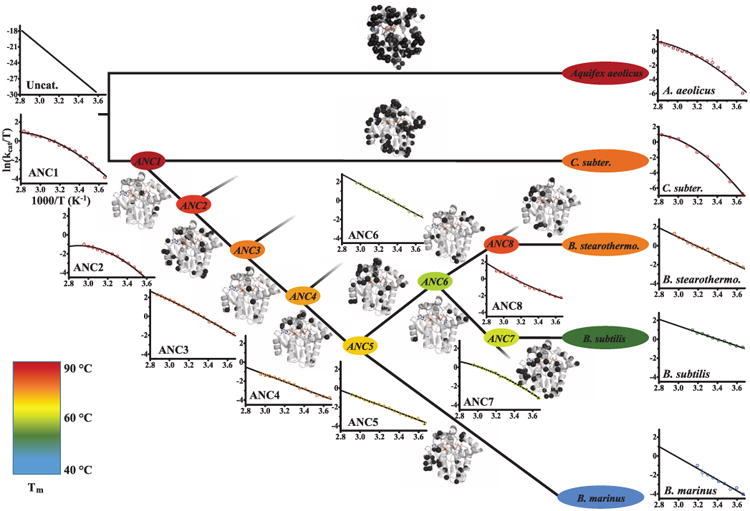
Temperature dependence of kcat is shown as Eyring plots including the fits to Eq. 1, and is compared to the uncatalyzed reaction reported (34). Tms are shown using color code of Fig. 2. For fitting, only data for temperatures before unfolding set in are used, with a nonzero term when validated by an F test (see fig. S6 for complete activity–temperature profiles). Along the evolutionary trajectory from ANC1 via mesophilic ANC6 to modern Adks, positive curvature evolved to straight Eyring plots. In contrast, C. subterraneus and A. aeolicus Adks retain positive curvature. Sites of mutations between the nodes are plotted as black spheres on the structure of ANC1. Error bars as in Fig. 2D.
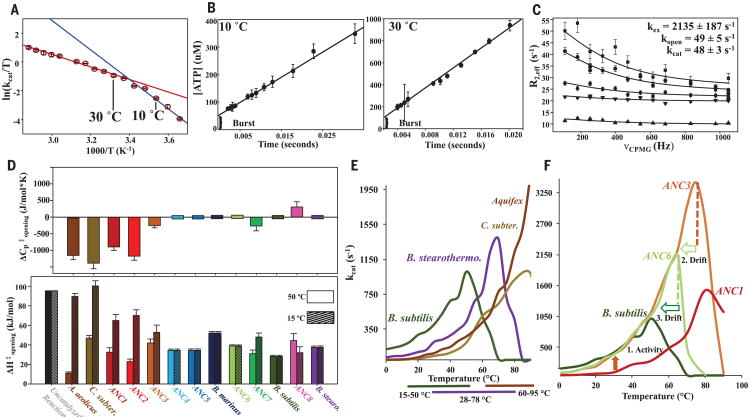
To elucidate how enzymatic turnover evolved in response to temperature changes, we generated Eyring plots for each modern and ancestral Adk [see Fig. 1B as an illustration of the hypothesis put forward in (12, 13)]. Unexpectedly, the oldest ancestors have positively curved Eyring plots. The curvature is less pronounced for the more recent ancestors (Fig. 3). Nonlinear Eyring plots can be trivially caused either by protein denaturation at higher temperatures or a temperature-dependent reversible inactivation equilibrium. Both possibilities were ruled out by nuclear magnetic resonance (NMR) spectra for this temperature range (fig. S7). Also, subsaturating substrate concentration due to a large increase in Km at elevated temperatures was excluded as a potential explanation (fig. S8). Another potential source of a curved Eyring plot is a change in the rate-limiting step at different temperatures (Fig. 4A) (21). Because lid opening has been shown to be rate-limiting for modern Adk from various organisms, we considered that the chemical step of phosphoryl transfer might become rate-limiting at a certain temperature. However, quench flow experiments show a product burst both at low and high temperatures for ANC1, indicating a conventional rate-limiting conformational change at both temperatures following a faster phosphoryl-transfer step (Fig. 4B), in agreement with earlier work on E. coli and Aquifex aeolicus Adk (22). We confirmed this result by NMR relaxation experiments on ANC1 during catalysis (22), determining a lid-opening rate of 49 s−1 at 15°C that is within experimental error of the corresponding kcat (Fig. 4C).
Fig. 4. Change in as evolutionary driver for thermoadaptation of catalysis and implications for organismal fitness.
(A) Eyring plot of ANC1 activity with theoretical explanation of curvature caused by a change in rate-limiting step between 10° and 30°C between two steps with different temperature dependencies (red and blue lines). (B) Quench-flow experiment of 100 μM ANC1 and 5 mM Mg/ADP shows a burst for fast phosphoryl transfer, followed by the rate-limiting lid opening (linear part) (mean ± SEM; N = 3 experiments). (C) 15N CPMG relaxation (22) data of representative residues of ANC1 during catalysis at saturating concentrations of Mg/ADP at 15°C. Error bars denote uncertainty in the ratio of cross-peaks, estimated as root mean square deviation from duplicates. (D) Heat capacity of activation for lid-opening dynamics ( ) for all Adk enzymes determined from the data of Fig. 3 fitted to Eq. 1 (top) and corresponding ΔH‡ values at 50° and 15°C (bottom) (standard errors in the fitted parameters). (E) Correlation between activity–temperature profiles of modern Adks with their known growth temperatures (35–38) (bottom bars). (F) Activity–temperature profiles along the evolutionary path from ANC1 to modern B. subtilis labeling active pressure for increased activity at lower temperatures (orange arrow from ANC1 to ANC3), creating a hyperactive and hyperstable enzyme in ANC3 and subsequently a slower passive drift to decreased stabilities via ANC6 to B. subtilis (light green and green open arrow).
Having established that kcat reports on lid opening over the relevant temperature range, we asked whether the curved Eyring plots arise from a nonzero change in heat capacity of activation ( ; Eq. 1 is repeated in supplementary materials with all terms defined):
| (1) |
reflects the difference in heat capacity between the transition state and the closed state, and a nonzero value necessarily imparts temperature dependence to the activation enthalpy. Fitting the activity data to such an extended model including a term (Eq. 1) indeed rationalizes temperature adaptation during evolution of enzyme catalysis over 3 billion years (Fig. 3). The strong negative of the oldest ancestors results in extremely slow catalysis at low temperatures (Figs. 3 and 4D), a tolerable situation for these hot-Earth ancestors. Notably, this kinetic obstacle to catalysis during thermoadaptation to colder environmental temperatures was removed by progressively reducing toward zero in evolution toward modern enzymes (Fig. 3). Tracing the changes along the evolutionary pathways exposes a gradual trend (Fig. 4D) of greatly increasing the catalytic rates at lower temperatures, the parameter under direct evolutionary pressure. Negative values produce a larger enthalpy of activation at lower temperatures, causing the severe drop in catalytic rate constants (Fig. 4D and Eq. 1).
Intriguingly, values of found here are comparable in magnitude to the ones measured for protein-folding kinetics (23). Moreover, Fersht and colleagues had speculated that could in principle influence enzyme catalysis for cases where conformational transitions are rate-limiting (23), as is shown here. Although it would be desirable to identify whether solvation of hydrophobic residues or charged residues, or differences in dynamics, are responsible for the measured , this is unachievable because (i) the heat capacity of the transition state cannot be estimated, (ii) the values are small, and (iii) the molecular character of the transition state is unknown (23).
In summary, this thermoadaptation mechanism for catalysis differs from the original hypothesis of a uniform decrease in ΔH‡ in the evolution of enzyme catalysis (Fig. 1B) (12, 13) and creates an even larger rate acceleration at lower temperatures relative to the uncatalyzed rate. The source of rate acceleration by the oldest ancestral Adk relative to the nonenzymatic rate is indeed a decrease in ΔH‡ (Figs. 3 and 4), as postulated by Wolfenden (12, 13), but only at the high primordial temperatures. Adk evolution over the next 3 billion years encountered a decrease in the environmental temperatures and was driven by a corresponding change in the heat capacity of activation. This process resulted in comparable ΔH‡ values for all ancestors and modern extremophiles at their corresponding optimal temperatures (Fig. 4D).
To further test our model, we characterized two additional modern hyperthermophiles, C. subterraneus and A. aeolicus Adk. These organisms have apparently remained thermophilic throughout their evolutionary history (Fig. 3 and S9) and therefore never experienced sustained selection for low-temperature catalytic efficiency. As predicted by our model, both modern enzymes behave like our oldest ancestor, showing large negative values, in sharp contrast to hyper-thermophilic B. stearothermophilus (Figs. 3 and 4D). The negligible of B. stearothermophilus seems to be a biophysical-vestigial leftover from its evolutionary history in a cooler environment. These results illustrate elements of “evolutionary memory,” thereby resolving long-standing controversies in interpreting differences in modern extremophiles within the same enzyme class (24).
Starting from modern Bacillus Adk enzymes, Shamoo's lab demonstrated a direct link between Adk stability and organismal fitness (15, 16). Comparing our in vitro Adk activity temperature profiles with known growth temperature ranges of the organisms leads to the hypothesis that a minimal catalytic rate of Adk is needed for survival (Fig. 4E). We propose that differential fitness at low temperatures is directly linked to the enzymatic rate: The survival of B. stearothermophilus at mesophilic temperatures is a holdover from the mesophilic sojourn taken by its ancestor, explaining its perplexingly broad organismal fitness (25) (Fig. 4E).
According to the view that protein stability must be sacrificed to support the conformational flexibility necessary for enzymatic activity, the marginal melting temperatures in mesophiles and psychrophiles would be an adaptive trait (26). We do not observe this correlation, as ANC3 is both thermostable and catalytically active at low temperatures, resulting in a “super-enzyme” with the unprecedented optimal kcat among Adks of ∼3500 s−1 (Fig. 4F). Adk is especially relevant to the flexibility/stability trade-off debate, as the rate-limiting step is a dynamic lid-opening process (22). With no pressure on either maintaining or losing stability, the melting temperatures of Adk enzymes likely decreased by genetic drift toward modern mesophiles and psychrophiles while the activity was maintained by purifying selection (Fig. 4F) (26, 27). Our evidence against the activity/stability trade-off is immediately pertinent to biotechnology for producing very active enzymes that are also hyperstable.
How do our results compare to previous studies on the evolution of thermostability? Thermostabilities of ANC1 and ANC2 show excellent agreement with melting temperatures of ancestral beta-lactamases (89° to 90°C) at ∼ 3 bya (28) and the thermostability of ancestors of isopropylmalate dehydrogenase along the Bacilli clade of Firmicutes over 1 bya (11). The evolution of Adk's Tms over 3 bya correlates with the change in Earth's temperature as first reported for the elongation factor Tu (8), although those measured Tms are in general below the estimated Earth temperatures whereas Adk's Tm values are above. These results, using different enzymes and techniques for phylogenetic inference, demonstrate the robustness and reproducibility of ASR. Notably, our results deliver the structural mechanisms for the evolution of thermostability from ancestors to modern enzymes that could not be achieved for other reconstructed systems (8, 11). A comparison of only modern B. subtilis and B. stearothermophilus Adk identified salt bridges as being crucial for the observed differences in thermostability (29). These salt bridges differ from the ones we identified in the evolutionary trajectory, highlighting that similar strategies can be used by evolution in different ways to increase the protein stability in response to environmental pressure.
Tracing the evolution of catalytic power under thermal pressure revealed as the unexpected evolutionary driver for enzymatic speed. In protein chemistry, heat capacities have almost entirely been studied for the temperature dependence of thermodynamic equilibria (ΔCp), such as between the folded and unfolded states (30). as a source of nonlinear Eyring plots had only been measured for nonenzymatic reactions (31) and protein folding (23). A nonzero in enzyme catalysis where conformational transitions are rate-limiting is analogous to a nonzero in protein folding (23), an idea that suggests a unified view of protein folding that includes conformational transitions within the folded ensemble (32, 33). The quantitative characterization of the evolution of enzyme activity and stability exposes the action of active selection versus passive drift in evolution on a molecular level, reveals a surprising mechanism of thermoadaptation for catalysis, opposes the commonly assumed activity/stability trade-off concept, and suggests the catalytic speed of Adk as an evolutionary driver for organismal fitness.
Supplementary Material
Fig. S1: Same phylogeny as in Fig. 2 with full clades shown here. Values indicate posterior probabilities of important nodes. All nodes not labeled have probabilities above 0.65 except four nodes denoted with a dotted circle.
Fig. S2: Multiple sequence alignment of recreated ancestral Adk enzymes from PAML (46) and BAli-Phy (18) along with modern sequences characterized in this study, using Adk from B. subtilis as reference sequence. Beta sheets are shown as yellow arrows and alpha helices as pink cylinders.
Fig. S3: Histogram of reconstructed residues binned according to posterior probability of the predicted residue. The color code defined in Fig. 2 is used for the ancestors
Fig. S4: (A) Maximum likelihood structural superposition of Adk crystal structures from ANC1, ANC3, ANC4, B. subtilis and B. stearothermophilus using the THESEUS package (63, 64). (B) Putty representation of deviation among all structures.
Fig. S5: Structural analysis of the evolution of thermal stability from (A) ANC1 to ANC4, (B) ANC4 to B. stearothermophilus, (C) ANC4 to B. subtilis and (D) ANC4 to B. marinus using published crystal structures (29, 65) and the ones solved in this manuscript (5G3Y, 5G3Z, 5G40). (A) Four clusters of ion pairs (shown as black dotted lines in ANC1) are lost during the evolutionary pathway from ANC1 to ANC4, potentially explaining the drop of Tm of ANC4 relative to ANC1 by 12 °C (see also experiments in Fig. 2C, D). (B) B. stearothermophilus evolved from ANC4 via the mesophilic ancestor ANC6. During its evolutionary pathway, the ion-pair D75-K78 as well as the H-bond E119-Q199 are lost, while K19-E202 is reformed and new ion-pairs R116-E198 and D114-K180 evolve. This rebalancing of salt bridges might lead to a similar Tm. (C) Four ion pairs are lost during the evolutionary pathway from ANC4 to B. subtilis resulting in a drop of Tm by 21 °C. (D) Two ion pairs and one H-bond interaction (R128-Q159) are lost during the evolutionary pathway from ANC4 to B. marinus possibly playing an important role in the drop of Tm by 31 °C.
Fig. S6: Full Eyring plots of all studied Adk enzymes showing the decrease in activity at higher temperatures due to denaturation. Note that this decline (or strong curvature) is there for all enzymes due to general unfolding at temperatures close to Tm, and is much steeper than the shallow positive curvature for ANC1, ANC2, C. subterraneus and A. aeolicus due to negative ΔCp‡ of kcat. For fitting of activity data to Eq.1, only data before any unfolding sets in were used (See Fig. S7). Error bars are described in Fig. 3. Black line shows fit with ΔCp‡ = 0. Red line shows fit including ΔCp‡ as a parameter. Asterisks indicate significant fit enhancements from the inclusion of ΔCp‡ as a parameter (p < 0.05 from F-tests). The temperature dependence of activity shown here unambiguously demonstrates how to differentiate between a decrease in activity with increased temperature caused by denaturation from a true ΔCp‡ effect on the catalytic step.
We note that in three publications to date from the Arcus lab, a decline in activity at temperatures close to the melting temperatures has been incorrectly interpreted as large negative heat capacity of activation for enzymatic reactions (ΔCp‡ in the order between 4 to 11 kJ/mol*K) (66-68). As shown (11, 69), the measured decline in activity at higher temperatures in (66-68) is due to unfolding of the enzyme, in full agreement with our results of a steep decline of activity close to the corresponding Tm.
Fig. S7: Curved Eyring plots in the temperature range fitted are not due to protein denaturation, or temperature dependent inactivation equilibrium. (A) ANC1 is fully folded up to 45 °C as measured by NMR 1H 15N HSQC spectra of 1 mM ANC1 at pH 7.0. Chemical shift changes are in agreement with expected linear amide chemical shift changes due to increasing temperature with no sign of denaturation setting in. Eyring plots in the temperature range between 0 °C and 45 °C are used to fit Eq. 1 to determine of kcat. (B-F) The equilibrium model of an active-inactive interconversion with a strong temperature dependence of its equilibrium can be ruled out as mechanism for the curved Eyring plots. (B) Eyring equation that includes an active-inactive equilibrium of the enzyme with a temperature dependent Keq. (C) Fits of Eyring plots of activity data from ANC1 to the equation above for a model of Hot inactivation and Cold inactivation. (D) Table of ΔG for this equilibrium at different temperatures. (E) Percentage in the inactive and active states at each temperatures illustrating the exponential change with temperature. (F) 1H 15N HSQC spectra during enzymatic turnover of ANC1 at different temperatures shows chemical shift changes that are linear with respect to temperature for all residues in the protein, shown here are some representative examples. This is inconsistent with the equilibrium model for which the shifts would be proportional to the population change, hence an exponential dependence on temperature (see E).
Fig. S8: Verification that in the temperature range that is used to determine of kcat, the enzyme is indeed under kcat conditions (saturating concentrations for Mg/ADP). Michealis-Menten curve for ANC1 for Mg/ADP shows that while Km increases with temperature as expected, the enzyme is always at saturating conditions for the performed assays with 4 mM Mg/ADP. Uncertainties are ± SEM from three experiments.
Fig. S9: Tree adapted from (70) that was calculated based on a continuously updated set of 16SRNA sequences. The clades of Firmicutes and A. aeolicus used in our study are circled, highlighting that A. aeolicus sits near the root of life suggesting it has not moved through a mesophilic ancestor.
Table S1: Comparison of biophysical parameters for ancestral enzymes constructed by maximum likelihood (ML) and Bayesian methods. Corresponding sequences are shown in Fig. S2. Strikingly, enzymes from the same nodes resurrected from ML or Bayesian methods display very similar biophysical properties within the error of the methods. This result strengthens the validity or our ASR tree and consequently the evolutionary conclusions drawn. This is in contrast to the resurrected ancestors for IPMDH in (11) where the authors describe a big error. Uncertainties of Tm are the standard error in the fitted parameter to a 2-state model. Uncertainties of maximum activity are standard errors based on 8 data points at Topt of the ancestral enzymes. Uncertainties of ΔCp‡opening are the standard error in fitted parameter of Eq. 1.
Table S2: Biophysical parameters for thermal stability and catalytic activity for studied Adk enzymes. values cause the temperature dependence of ΔH‡opening and ΔS‡opening according to Eq. 1, and we list here the calculated values at a low and high temperature for better visualization of the magnitude of change in these parameters as a function of temperature. Uncertainties are described in Table S1.
Table S3: Data collection and refinement statistics for the 4 solved x-ray structures of ANC1, ANC3 and ANC4. *Values in parentheses correspond to the highest-resolution shell.
Table S4: What-if analysis (71) of solved crystal structures. The only significant differences are in the number of ion pairs.
Table S5: Comparison of ion-pairs identified from the x-ray structures that possibly are responsible for differences in thermal stability between the studied Adks. Green shaded are cluster forming ion-pairs, while white cells cannot form the ion-pair, either due to loss of acceptor or donor or due to a greater distance than 4 Å. Grey shaded is a tight H-bond cluster found in all Adks but the psychrophile B. marinus.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (HHMI); the Office of Basic Energy Sciences, Catalysis Science Program; U.S. Department of Energy award DE-FG02-05ER15699; and NIH grant GM100966 (to D.K) and NIH grants GM096053 and GM094468 (to D.L.T.). We thank the Advanced Light Source (ALS), Berkeley, CA, USA, for access to beamlines BL8.2.1 and BL8.2.2. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the HHMI. The ALS is supported by the director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract DE-AC02-05CH11231. The Protein Data Bank accession codes are 5G3Y (ANC1*Zn*ADP*ADP), 5G3Z (ANC3*Zn*Mg*Ap5A), 5G40 (ANC4*Zn*AMP*ADP), and 5G41 (ANC4*Zn*Mg*Ap5A).
References and Notes
- 1.Clarke A, et al. PLOS ONE. 2013;8:e66207. doi: 10.1371/journal.pone.0066207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowan DA. Trends Microbiol. 2004;12:58–60. doi: 10.1016/j.tim.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Deming JW. Curr Opin Microbiol. 2002;5:301–309. doi: 10.1016/s1369-5274(02)00329-6. [DOI] [PubMed] [Google Scholar]
- 4.Blöchl E, et al. Extremophiles. 1997;1:14–21. doi: 10.1007/s007920050010. [DOI] [PubMed] [Google Scholar]
- 5.Ciccarelli FD, et al. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 6.Knauth LP, Lowe DR. Geol Soc Am Bull. 2003;115:566–580. [Google Scholar]
- 7.Fralick P, Carter JE. Precambrian Res. 2011;191:78–84. [Google Scholar]
- 8.Gaucher EA, Govindarajan S, Ganesh OK. Nature. 2008;451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 9.Akanuma S, et al. Proc Natl Acad Sci USA. 2013;110:11067–11072. doi: 10.1073/pnas.1308215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 11.Hobbs JK, et al. Mol Biol Evol. 2012;29:825–835. doi: 10.1093/molbev/msr253. [DOI] [PubMed] [Google Scholar]
- 12.Stockbridge RB, Lewis CA, Jr, Yuan Y, Wolfenden R. Proc Natl Acad Sci USA. 2010;107:22102–22105. doi: 10.1073/pnas.1013647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfenden R. Cell Mol Life Sci. 2014;71:2909–2915. doi: 10.1007/s00018-014-1587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms MJ, Thornton JW. Nat Rev Genet. 2013;14:559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couñago R, Shamoo Y. Extremophiles. 2005;9:135–144. doi: 10.1007/s00792-004-0428-x. [DOI] [PubMed] [Google Scholar]
- 16.Couñago R, Chen S, Shamoo Y. Mol Cell. 2006;22:441–449. doi: 10.1016/j.molcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Letelier A, Olmedo-Alvarez G, Eguiarte LE, Souza V. Astrobiology. 2012;12:674–684. doi: 10.1089/ast.2011.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redelings BD, Suchard MA. Syst Biol. 2005;54:401–418. doi: 10.1080/10635150590947041. [DOI] [PubMed] [Google Scholar]
- 19.Battistuzzi FU, Feijao A, Hedges SB. BMC Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams PD, Pollock DD, Blackburne BP, Goldstein RA. PLOS Comput Biol. 2006;2:e69. doi: 10.1371/journal.pcbi.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Case A, Stein RL. Biochemistry. 2003;42:3335–3348. doi: 10.1021/bi020668l. [DOI] [PubMed] [Google Scholar]
- 22.Kerns SJ, et al. Nat Struct Mol Biol. 2015;22:124–131. doi: 10.1038/nsmb.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveberg M, Tan YJ, Fersht AR. Proc Natl Acad Sci USA. 1995;92:8926–8929. doi: 10.1073/pnas.92.19.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petsko GA. Methods Enzymol. 2001;334:469–478. doi: 10.1016/s0076-6879(01)34486-5. [DOI] [PubMed] [Google Scholar]
- 25.Zeigler DR. Microbiology. 2014;160:1–11. doi: 10.1099/mic.0.071696-0. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein RA. Proteins. 2011;79:1396–1407. doi: 10.1002/prot.22964. [DOI] [PubMed] [Google Scholar]
- 27.Bornberg-Bauer E, Chan HS. Proc Natl Acad Sci USA. 1999;96:10689–10694. doi: 10.1073/pnas.96.19.10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risso VA, et al. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 29.Bae E, Phillips GN., Jr J Biol Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 30.Prabhu NV, Sharp KA. Annu Rev Phys Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 31.Canepa C. Chem Cent J. 2011;5:22. doi: 10.1186/1752-153X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nussinov R, Wolynes PG. Phys Chem Chem Phys. 2014;16:6321–6322. doi: 10.1039/c4cp90027h. [DOI] [PubMed] [Google Scholar]
- 33.Wolynes PG, Eaton WA, Fersht AR. Proc Natl Acad Sci USA. 2012;109:17770–17771. doi: 10.1073/pnas.1215733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockbridge RB, Wolfenden R. J Biol Chem. 2009;284:22747–22757. doi: 10.1074/jbc.M109.017806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtmann G, Bremer E. J Bacteriol. 2004;186:1683–1693. doi: 10.1128/JB.186.6.1683-1693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols DS, Nichols PD, McMeekin TA. Sci Prog. 1995;78:311–348. [Google Scholar]
- 37.Nazina TN, et al. Int J Syst Evol Microbiol. 2001;51:433–446. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- 38.Huber R, Stetter KO. Methods Enzymol. 2001;330:11–24. doi: 10.1016/s0076-6879(01)30367-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Same phylogeny as in Fig. 2 with full clades shown here. Values indicate posterior probabilities of important nodes. All nodes not labeled have probabilities above 0.65 except four nodes denoted with a dotted circle.
Fig. S2: Multiple sequence alignment of recreated ancestral Adk enzymes from PAML (46) and BAli-Phy (18) along with modern sequences characterized in this study, using Adk from B. subtilis as reference sequence. Beta sheets are shown as yellow arrows and alpha helices as pink cylinders.
Fig. S3: Histogram of reconstructed residues binned according to posterior probability of the predicted residue. The color code defined in Fig. 2 is used for the ancestors
Fig. S4: (A) Maximum likelihood structural superposition of Adk crystal structures from ANC1, ANC3, ANC4, B. subtilis and B. stearothermophilus using the THESEUS package (63, 64). (B) Putty representation of deviation among all structures.
Fig. S5: Structural analysis of the evolution of thermal stability from (A) ANC1 to ANC4, (B) ANC4 to B. stearothermophilus, (C) ANC4 to B. subtilis and (D) ANC4 to B. marinus using published crystal structures (29, 65) and the ones solved in this manuscript (5G3Y, 5G3Z, 5G40). (A) Four clusters of ion pairs (shown as black dotted lines in ANC1) are lost during the evolutionary pathway from ANC1 to ANC4, potentially explaining the drop of Tm of ANC4 relative to ANC1 by 12 °C (see also experiments in Fig. 2C, D). (B) B. stearothermophilus evolved from ANC4 via the mesophilic ancestor ANC6. During its evolutionary pathway, the ion-pair D75-K78 as well as the H-bond E119-Q199 are lost, while K19-E202 is reformed and new ion-pairs R116-E198 and D114-K180 evolve. This rebalancing of salt bridges might lead to a similar Tm. (C) Four ion pairs are lost during the evolutionary pathway from ANC4 to B. subtilis resulting in a drop of Tm by 21 °C. (D) Two ion pairs and one H-bond interaction (R128-Q159) are lost during the evolutionary pathway from ANC4 to B. marinus possibly playing an important role in the drop of Tm by 31 °C.
Fig. S6: Full Eyring plots of all studied Adk enzymes showing the decrease in activity at higher temperatures due to denaturation. Note that this decline (or strong curvature) is there for all enzymes due to general unfolding at temperatures close to Tm, and is much steeper than the shallow positive curvature for ANC1, ANC2, C. subterraneus and A. aeolicus due to negative ΔCp‡ of kcat. For fitting of activity data to Eq.1, only data before any unfolding sets in were used (See Fig. S7). Error bars are described in Fig. 3. Black line shows fit with ΔCp‡ = 0. Red line shows fit including ΔCp‡ as a parameter. Asterisks indicate significant fit enhancements from the inclusion of ΔCp‡ as a parameter (p < 0.05 from F-tests). The temperature dependence of activity shown here unambiguously demonstrates how to differentiate between a decrease in activity with increased temperature caused by denaturation from a true ΔCp‡ effect on the catalytic step.
We note that in three publications to date from the Arcus lab, a decline in activity at temperatures close to the melting temperatures has been incorrectly interpreted as large negative heat capacity of activation for enzymatic reactions (ΔCp‡ in the order between 4 to 11 kJ/mol*K) (66-68). As shown (11, 69), the measured decline in activity at higher temperatures in (66-68) is due to unfolding of the enzyme, in full agreement with our results of a steep decline of activity close to the corresponding Tm.
Fig. S7: Curved Eyring plots in the temperature range fitted are not due to protein denaturation, or temperature dependent inactivation equilibrium. (A) ANC1 is fully folded up to 45 °C as measured by NMR 1H 15N HSQC spectra of 1 mM ANC1 at pH 7.0. Chemical shift changes are in agreement with expected linear amide chemical shift changes due to increasing temperature with no sign of denaturation setting in. Eyring plots in the temperature range between 0 °C and 45 °C are used to fit Eq. 1 to determine of kcat. (B-F) The equilibrium model of an active-inactive interconversion with a strong temperature dependence of its equilibrium can be ruled out as mechanism for the curved Eyring plots. (B) Eyring equation that includes an active-inactive equilibrium of the enzyme with a temperature dependent Keq. (C) Fits of Eyring plots of activity data from ANC1 to the equation above for a model of Hot inactivation and Cold inactivation. (D) Table of ΔG for this equilibrium at different temperatures. (E) Percentage in the inactive and active states at each temperatures illustrating the exponential change with temperature. (F) 1H 15N HSQC spectra during enzymatic turnover of ANC1 at different temperatures shows chemical shift changes that are linear with respect to temperature for all residues in the protein, shown here are some representative examples. This is inconsistent with the equilibrium model for which the shifts would be proportional to the population change, hence an exponential dependence on temperature (see E).
Fig. S8: Verification that in the temperature range that is used to determine of kcat, the enzyme is indeed under kcat conditions (saturating concentrations for Mg/ADP). Michealis-Menten curve for ANC1 for Mg/ADP shows that while Km increases with temperature as expected, the enzyme is always at saturating conditions for the performed assays with 4 mM Mg/ADP. Uncertainties are ± SEM from three experiments.
Fig. S9: Tree adapted from (70) that was calculated based on a continuously updated set of 16SRNA sequences. The clades of Firmicutes and A. aeolicus used in our study are circled, highlighting that A. aeolicus sits near the root of life suggesting it has not moved through a mesophilic ancestor.
Table S1: Comparison of biophysical parameters for ancestral enzymes constructed by maximum likelihood (ML) and Bayesian methods. Corresponding sequences are shown in Fig. S2. Strikingly, enzymes from the same nodes resurrected from ML or Bayesian methods display very similar biophysical properties within the error of the methods. This result strengthens the validity or our ASR tree and consequently the evolutionary conclusions drawn. This is in contrast to the resurrected ancestors for IPMDH in (11) where the authors describe a big error. Uncertainties of Tm are the standard error in the fitted parameter to a 2-state model. Uncertainties of maximum activity are standard errors based on 8 data points at Topt of the ancestral enzymes. Uncertainties of ΔCp‡opening are the standard error in fitted parameter of Eq. 1.
Table S2: Biophysical parameters for thermal stability and catalytic activity for studied Adk enzymes. values cause the temperature dependence of ΔH‡opening and ΔS‡opening according to Eq. 1, and we list here the calculated values at a low and high temperature for better visualization of the magnitude of change in these parameters as a function of temperature. Uncertainties are described in Table S1.
Table S3: Data collection and refinement statistics for the 4 solved x-ray structures of ANC1, ANC3 and ANC4. *Values in parentheses correspond to the highest-resolution shell.
Table S4: What-if analysis (71) of solved crystal structures. The only significant differences are in the number of ion pairs.
Table S5: Comparison of ion-pairs identified from the x-ray structures that possibly are responsible for differences in thermal stability between the studied Adks. Green shaded are cluster forming ion-pairs, while white cells cannot form the ion-pair, either due to loss of acceptor or donor or due to a greater distance than 4 Å. Grey shaded is a tight H-bond cluster found in all Adks but the psychrophile B. marinus.