Abstract
Purpose
This study intends to investigate the feasibility of using hyperspectral imaging (HSI) to detect and delineate cancers in fresh, surgical specimens of patients with head and neck cancers.
Experimental Design
A clinical study was conducted in order to collect and image fresh, surgical specimens from patients (N = 36) with head and neck cancers undergoing surgical resection. A set of machine-learning tools were developed to quantify hyperspectral images of the resected tissue in order to detect and delineate cancerous regions which were validated by histopathologic diagnosis. More than two million reflectance spectral signatures were obtained by HSI and analyzed using machine-learning methods. The detection results of HSI were compared with autofluorescence imaging and fluorescence imaging of two vital-dyes of the same specimens.
Results
Quantitative HSI differentiated cancerous tissue from normal tissue in ex vivo surgical specimens with a sensitivity and specificity of 91% and 91%, respectively, and which was more accurate than autofluorescence imaging (P < 0.05) or fluorescence imaging of 2-NBDG (P < 0.05) and proflavine (P < 0.05). The proposed quantification tools also generated cancer probability maps with the tumor border demarcated and which could provide real-time guidance for surgeons regarding optimal tumor resection.
Conclusions
This study highlights the feasibility of using quantitative HSI as a diagnostic tool to delineate the cancer boundaries in surgical specimens, and which could be translated into the clinic application with the hope of improving clinical outcomes in the future.
Introduction
Worldwide, more than 500,000 patients are diagnosed with head and neck squamous cell carcinoma (HNSCC) each year, and thus posing substantial economic burdens (1, 2). Head and neck cancers comprise a heterogeneous group of tumors arising from the oral cavity, pharynx, larynx, paranasal sinuses, and salivary and thyroid glands (3). Surgery remains one of the major treatment options for this disease, and with the primary objectives of maximizing tumor removal while minimizing the damage to healthy tissue. Extensive resection with unnecessary removal of normal tissue can leave patients with serious functional and aesthetic deficits, and thus compromising their ability to perform vital daily functions, such as chewing, swallowing, or speaking. However, if the tumor margins are not accurately defined and the diseased tissue is not completely removed, cancer is likely to persist or recur. A positive surgical margin is associated with a poor prognosis in terms of increased local recurrence and decreased overall patient survival (4). Therefore, the ability to define the tumor margins with a high degree of accuracy is critical for maximizing the efficacy of surgical treatment and the patient’s subsequent quality of life, both of which might produce significant cost savings.
Achieving resection margin adequacy is highly “operator dependent” with respect to surgeons and pathologists (5). Visual inspection and palpation are routinely used by a surgeon to differentiate between tumor and normal tissue during surgery (6), and which is known to be subjective and not easily quantifiable. Intraoperative frozen section assessment of surgical margins is widely used to assist in complete tumor extirpation in head and neck surgery. However, frozen section histopathology requires multiple, well-trained professionals, increases the cost and the duration of procedures, extends anesthesia-related risks, and provides diagnostic information at only a few discrete locations of the resection margins (7). Freezing artifacts, such as distortion of tissue architecture, uneven sectioning or poor staining, increases the likelihood of interpretive errors of histologic diagnosis. The overall frozen section accuracy has been reported to be only 71.3% and 77.9% in the evaluation of close (<5 mm) or positive final margins in two comprehensive studies (7, 8). Therefore, intraoperative imaging techniques that can help the surgeon to visualize and guide the tumor excision could improve complete surgical resection while preserving vital anatomic structures and functions of patients with head and neck cancer.
To address the issue, a variety of optical imaging approaches utilizing intrinsic contrast, such as autofluorescence imaging (9, 10) and narrow band imaging (11), or extrinsic contrast, such as near-infrared (NIR) fluorescence imaging (12, 13), have been studied to guide surgical resection in head and neck cancers (14). Despite the great potential of these optical modalities, objective and quantitative delineation of the surgical margin is still lacking and surgical resections were subjected to the qualitative observations of the optical images by the surgeons. Here, we propose to investigate the utility of hyperspectral imaging (HSI) in conjunction with quantitative machine-learning techniques as a diagnostic tool to demarcate tumor from the normal tissue during surgery. The advantage of HSI is that it is a hybrid optical modality that combines wide-field imaging and spectroscopy to simultaneously attain both spatial and spectral information, and which makes it possible to spatially demarcate the tumor boundaries without using an exogenous contrast agent (15). Our group has previously reported the use of quantitative HSI for the detection and delineation of head and neck cancer in several preclinical animal studies (16–19).
To this end, we conducted a proof-of-concept study in order to determine the feasibility of using quantitative HSI to distinguish tumor from normal tissue in fresh, surgical specimens from patients with head and neck cancers obtained from a variety of anatomic sites, including the oral cavity, larynx, pharynx, thyroid gland, paranasal sinus, and nasal cavity. To obtain relevant diagnostic information from hypercubes, we developed two, machine-learning frameworks to quantify hyperspectral images, and taking into the account the intra-and interpatient spectral variabilities. Pseudo-color maps were generated to indicate cancer probabilities and demarcate tumor borders, and which could be valuable for assisting the surgeon to accurately pinpoint the tumor margins and thus facilitate radical resection. The proposed quantification methods were validated by the pathology diagnosis. Furthermore, the diagnostic performance of label-free HSI was compared with other optical imaging methods, including autofluorescence imaging and fluorescence imaging of two topical dyes, that is, 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose (2-NBDG) and proflavine, that were previously used for the detection of head and neck cancers (20–22) as well as breast cancer (23, 24). To the best of our knowledge, this is the first time that HSI and machine learning–based quantification tools were tested and validated in a wide variety of head and neck cancer tissue from human patients.
Materials and Methods
Hyperspectral imaging instrumentation
A description of the HSI instrument is given in ref. 19. Briefly, an HSI system called Maestro (Perkin Elmer Inc.) was used for acquiring the hyperspectral dataset. This system mainly consists of a flexible, fiber-optic lighting system, a solid-state liquid crystal tunable filter (LCTF, bandwidth 20 nm), a spectrally optimized lens, and a 12-bit, high-resolution, charge-coupled device (CCD). A Cermax-type, 300 W, Xenon light source is used as white light excitation. The reflected light from the surface is split into a series of narrow spectral bands by the LCTF and then collected by the CCD camera. This instrument is capable of acquiring images from 450 to 900 nm with 5-nm increments.
Patient recruitment and specimen collection
Patients with head and neck cancers scheduled to undergo surgical resection with curative intent at Emory University Midtown Hospital were consented for our study. All tissue was collected under the Head and Neck Satellite Tissue Bank (HNSB, IRB00003208) protocol approved by the Emory University Institutional Review Board. The samples were de-identified and coded by a Clinical Research Coordinator before being released to our laboratory. During the surgery, resected specimens were sent to the pathology room for margin assessment. Selected surgical margins were processed for frozen-section, histopathologic evaluation, as routinely performed independently of our research. When specimen handling related to direct patient care was complete, three, fresh, tissue samples, including clinically visible tumor, clinically normal tissue, and tumor–normal interface (tumor with adjacent normal tissue), were procured from the main specimen of each consented patient. Tissue regions with pathology ink were excluded. Specimen collection and imaging did not affect the procedure time in the operating room (OR) or the content and verification of the final pathology report.
Fresh surgical specimen imaging
Figure 1 shows an overview of the clinical study design. Briefly, fresh surgical specimens collected in the hospital were kept in cold PBS and were immediately transported to our imaging center. The resected specimen was washed carefully with PBS in order to eliminate the blood contamination on the tissue surface. A series of imaging steps were performed as follows:
Step 1: White and dark reference hypercubes were acquired before tissue imaging. White reference image cubes are acquired by placing a standard white reference board in the field of view. The dark reference cubes are acquired by keeping the camera shutter closed in absence of light (25, 26).
Step 2: The specimens were placed on a nonreflective blackboard on the imaging stage. Reflectance hyperspectral images of the specimen were obtained from 450–900 nm with 5-nm intervals.
Step 3: Autofluorescence images were acquired with blue light excitation at 455- and a 490-nm long-pass emission filter.
Step 4: Fluorescence images of 2-NBDG and proflavine were acquired. By topically apply 2-NBDG and proflavine to the surgical specimen, we may identify cancerous regions from fluorescence images and compare the fluorescence imaging with label-free, wide-field HSI. Detailed procedures for vital dye fluorescence imaging methods were previously described in ref. 27. Here is a brief summary of the imaging procedures:
2-NBDG imaging: tissue specimens were first incubated in a 160 μmol/L solution of 2-NBDG (Cayman Chemical) in 1 × PBS for 20 minutes at 37°C The specimens were then washed once with PBS, after which fluorescence images of the tissue were obtained using the blue excitation and 490-nm long-pass emission.
Proflavine imaging: tissue samples were first incubated in a 0.01% (w/v) solution of proflavine (Sigma Aldrich) in 1 × PBS for 2 minutes at room temperature. Next the specimens were washed once with PBS, after which fluorescence images were obtained using blue excitation and 490 nm emission. The fluorescence signal from proflavine staining is much brighter than that of 2-NBDG, and thus allowing for imaging of proflavine-stained tissue after 2-NBDG staining.
Figure 1.
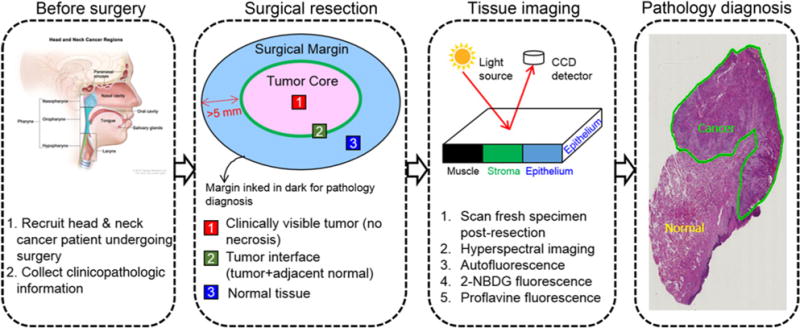
Overview of the clinical study design for fresh surgical specimen imaging.
Histologic processing and annotation
After all the imaging procedures were finished, we stained three tissue samples with red, blue, and yellow color ink in order to preserve its orientation during histologic processing. The inked specimens were fixed in 10% buffered formalin overnight and then sent to the Pathology Department for standard histologic processing. Two to three serial sections with 5 μm thickness from the imaging surface were cut and stained with hematoxylin and eosin (H&E) staining. These tissue sections were then digitally scanned for the pathology diagnosis. A clinically experienced pathologist examined the histology slides and outlined the tumor boundary on the digitized slides as the gold standard to validate the quantification method.
Pre-processing of hypercube
The method for data preprocessing consists of reflectance calibration and glare removal:
- Reflectance normalization: The purpose of this step is to remove the spectral non-uniformity of the illumination device and the influence of the dark current. The raw radiance data can be converted into normalized reflectance using the following equation:
where Iref(x, y, λ) is the normalized reflectance value at the pixel location (x,y) and the wavelength band λ. Iraw(x, y, λ) is the raw intensity value of a sample pixel (x,y). Iwhite(x,y, λ) and Idark(x,y, λ) are the corresponding pixels from the white and dark reference images at the same wavelength as the sample image.(1) Glare removal: Glare spots are frequently observed in optical images, and which are attributed to the specular reflection from the moist tissue surface. Glare pixels do not contain diagnostic information from under the tissue surface and can introduce artifacts in feature extraction and deteriorate classification results. Therefore, we propose to detect and remove glare pixels from the normalized hypercube as described in ref. 19.
Feature extraction
Hyperspectral imaging
To reduce the computational time without reducing the accuracy, spectral curves were averaged in nonoverlapping blocks of m × m in order to yield a spectral signature per block. All of the spectral information available in the hyperspectral data was utilized. Blocks containing glare pixels were excluded from the classification process. Each block was assigned a label as tumor or normal.
Multispectral imaging
For comparison, we evaluated the multi-spectral imaging (MSI) features utilizing the spectral peak bands of oxygenated hemoglobin (540 nm, 575 nm) and deoxygenated hemoglobin (555 nm), and which have been shown to aid in visualization increased vasculature in the oral cavity during malignant transformation (28). The average multispectral intensities were used as features for MSI classification.
Conventional RGB imaging
We simulated the conventional RGB imaging by converting the illumination wavelength for each band in the range of 450 to 900 nm into the constituent RGB values as perceived by humans, and then averaging the contribution to R, G, and B for each band. The average RGB intensity values of each block were used as features for the RGB imaging classification.
Autofluorescence imaging
Average fluorescence intensity from 500 to 650 nm with 10 nm increments within each block was extracted as features for classification.
Fluorescence imaging
The mean fluorescence intensity of 2-NBDG or proflavine images from 500 to 650 nm with 10-nm increments within each block was extracted as features for classification.
Classification
Pathological validation and ROI selection
The gold standard for cancer detection is pathological diagnosis. As the tumor profile was outlined in the digitized H&E stained histologic images by a pathologist, we need to register the pathological image with the hypercube of the tissue specimen in order to delineate the tumor region in hyperspectral images, and which would be the gold standard for validating our quantification method. To do this, we first synthesized an RGB color image from each hypercube, and then manually registered the histologic image with the synthesized RGB image using affine registration with software Analyze 10.0 (Mayo Clinic). Region of interests (ROI) that were pathologically confirmed to be tumor or normal were chosen for quantitative analysis.
Predictive modeling
A total of 36 research subjects were included in this study (Table 1) for quantitative analysis. To determine the optimal classifier for hypercube analysis, we evaluated and compared a variety of machine learning classifiers in order to analyze the spectra and determine the classification criteria, allowing samples from all cancer tissue categories to be separated from samples corresponding to normal tissue. Classifiers including linear discriminant analysis (LDA; ref. 29), quadratic discriminant analysis (QDA), ensemble LDA (30), linear support vector machine (SVM), and kernel SVM with radial basis function (RBF) function (31), as well as random forest (RF; ref. 32) were used for supervised learning. All of the data were processed and analyzed with software developed in-house that operates in a MATLAB environment (MATLAB 2015b, MathWorks). The LIBSVM software package (31) was used for both linear and kernel SVMs. To account for intra- and interpatient spectral variations, two predictive analysis frameworks were developed and validated in this study. Details of the methods were described below.
Table 1.
Summary of the surgical specimens and patient information for quantitative analysis
| Primary tumor site | Specimen origin | ID | Age | Gendera | Racea | Histologic typea |
|---|---|---|---|---|---|---|
| Oral cavity | Tongue | 1 | 55 | F | W | SCC |
| Tongue | 2 | 43 | M | W | SCC | |
| Tongue | 3 | 67 | F | W | SCC | |
| Tongue | 4 | 75 | F | W | SCC | |
| Tongue | 5 | 60 | M | I | SCC | |
| Tongue | 6 | 86 | F | W | SCC | |
| Tongue | 7 | 58 | F | W | SCC | |
| FOM | 8 | 50 | M | W | SCC | |
| FOM | 9 | 51 | F | W | SCC | |
| FOM | 10 | 57 | M | W | SCC | |
| FOM | 11 | 62 | M | W | SCSC | |
| Soft palate | 12 | 61 | F | AA | SCC | |
| Mandibule | 13 | 53 | M | W | SCC | |
| Mandibule | 14 | 44 | M | W | SCC | |
| Gingiva | 15 | 76 | M | I | SCC | |
| Alveolar ridge | 16 | 81 | F | AA | SCC | |
| Larynx | Supraglottis | 17 | 44 | M | AA | SCC |
| Supraglottis | 18 | 54 | F | W | SCC | |
| Glottis | 19 | 57 | M | AA | SCC | |
| Glottis | 20 | 69 | M | AA | SCC | |
| Pharynx | Hypopharynx | 21 | 66 | M | W | SCC |
| Paranasal and nasal | Maxillary sinus | 22 | 73 | F | W | SCC |
| Maxillary sinus | 23 | 65 | M | W | SCC | |
| Nose | 24 | 74 | M | W | ASC | |
| Gland | Thyroid | 25 | 69 | M | AA | PTC |
| Thyroid | 26 | 59 | M | A | PTC | |
| Thyroid | 27 | 24 | F | I | PTC | |
| Thyroid | 28 | 37 | M | I | PTC | |
| Thyroid | 29 | 22 | M | H | PTC | |
| Thyroid | 30 | 30 | F | AA | PTC | |
| Thyroid | 31 | 18 | M | W | FTC | |
| Thyroid | 32 | 37 | F | AA | PTC | |
| Thyroid | 33 | 35 | M | W | PTC | |
| Thyroid | 34 | 71 | M | W | PTC | |
| Thyroid | 35 | 35 | F | W | MTC | |
| Parotid | 36 | 39 | M | AA | PA |
Gender: F, Female; M, Male.
Race: A, Asian; AA, African American; H, Hispanic; I, Indian; W, White.
Histologic type: PA, pleomorphic adenoma.
Intrapatient classification
For each of the enrolled research subjects, tissue samples were taken at the clinically visible tumor center without necrosis, surrounding normal tissue, and at the tumor–normal tissue interface when available. So we proposed to conduct intrapatient classification, which used the tumor and normal sample of each patient, as training data to build a predictive model and then evaluated the model performance on the tumor–normal interface tissue sample of the same patient. The sensitivity and specificity of the classifier for each patient was calculated based on how many tumor pixels or blocks were correctly classified and how many normal pixels were correctly classified within the selected ROIs on the tumor– normal interface tissue. The advantage of this method is to detect and delineate the tumor regions in the tumor–normal interface sample quickly and to eliminate the influence of interpatient heterogeneity by utilizing spectra from patient-self. As listed in Table 1, 25 patients with all three pieces of tissue available were selected for intrapatient classification, including cancers from the oral cavity (N= 12), larynx and pharynx (N = 3), maxillary sinus (N = 2), nasal cavity (N = 1), thyroid (N = 6), and parotid (N = 1). The number in the brackets refers to the number of patients.
Interpatient classification
To account for interpatient heterogeneity, we proposed another framework called interpatient classification. Tissue samples from 31 research subjects, as listed in Table 1, were analyzed using this method. Each patient included in this analysis had at least two tissue samples (tumor and normal) available. Leave-one-patient out cross-validation was performed to account for variability in classification performance. We separated 31 patients into two cohorts, that is, nonthyroid and thyroid cancer, in order to reduce spectral variability due to the cancer site. In the first cohort, 20 patients with squamous cell carcinoma (SCC), spindle cell squamous carcinoma (SCSC) or adenosquamous carcinoma (ASC), including cancers from the oral cavity (N = 13), larynx and pharynx (N = 4), and paranasal and nasal cavity (N = 3) were grouped together for leave-one-out analysis. During each run, 19 out of 20 were randomly selected and all of their tissue specimens were used as training data, while all of the tissue specimens from the remaining patient were used as testing data. This process was repeated 20 times for the first cohort.
Similarly, 11 patients with thyroid cancers, including nine papillary thyroid carcinomas (PTC), one follicular thyroid carcinoma (FTC), and one medullary thyroid carcinoma (MTC), were considered as the second cohort for this analysis. During each run, 10 out of 11 patients were randomly chosen for training and the remaining patient for testing. This procedure was repeated 11 times for this cohort.
For each patient, we calculated how many normal pixels/blocks were correctly classified for a normal specimen, how many tumor pixels/blocks were correctly classified for a tumor specimen, as well as the sensitivity and specificity on a tumor-normal interface specimen.
Performance metric
We evaluated the performance of classifiers with receiver operating characteristic (ROC) curves, the areas under the curve (AUC), accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Classification accuracy, sensitivity, specificity, PPV, and NPV were determined using the following equations (TN, true negative; TP, true positive; FP, false positive; FN, false negative):
Results
Reflectance spectrum of head and neck tissue
Figure 2 shows the spectra of tumor and normal tissue samples from a variety of head and neck cancer sites in human patients. We found that the measured average spectra for all cancerous tissue differ significantly (P < 0.0001) from that of normal tissue in all the cancer sites (33). However, the spectra also showed large within-class variations, and which may be primarily attributed to the heterogeneity of head and neck tissue. The characteristic dip of hemoglobin can be observed in the reflectance spectra of all cancer sites. In general, the mean reflectance intensity of tumor tissue is higher than that of normal tissue in most of these anatomical sites.
Figure 2.
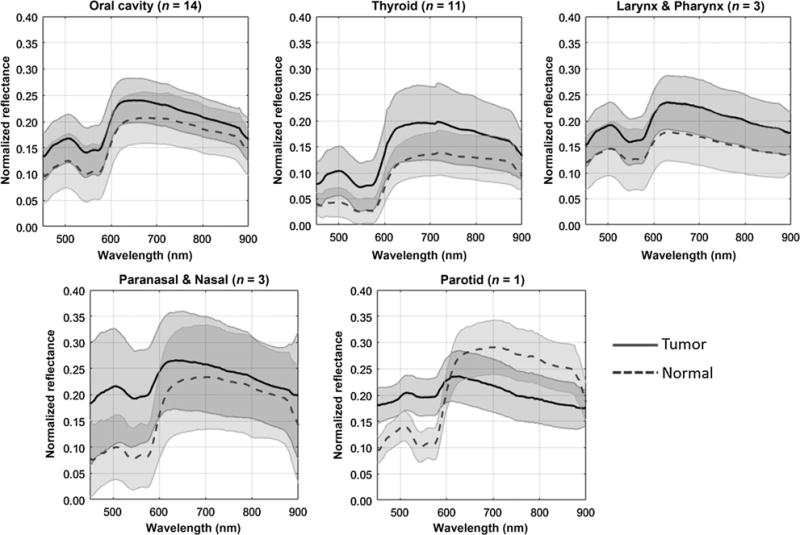
Average spectral curve of tumor and normal tissue samples from various head and neck cancer sites, including the oral cavity, thyroid, larynx, pharynx, parotid, paranasal sinus, and nasal cavity of human patients. The solid line and dash line represent the mean spectra of cancer and normal tissue, and the shaded area centered on the two lines represents the standard deviation.
Cancer prediction with intrapatient classification
Tissue from different anatomic sites may have a different structure and composition, so the optimal block size for averaging spectra could differ in order to best characterize the tissue. As shown in Fig. 3A, the diagnostic performances of spectral features extracted from block sizes of 1 × 1, 3 × 3, 5 × 5, 7 × 7, 9 × 9, and 11 × 11 were compared with specimens from the oral cavity, thyroid, larynx, and paranasal sinus. The optimal block size for the oral cavity is 7 × 7, for the thyroid and larynx is 5 × 5, and for the paranasal sinus is 1 × 1. Among all the classifiers tested for intrapatient classification, ensemble LDA was chosen as the optimal classifier due to its superior overall performance (Fig. 3B). To use HSI toward real-time guidance of surgical resection, we generated a cancer probability map with the tumor border outlined in green, as shown in Fig. 3C–F. This color-coded map may assist a surgeon to better visualize and assess tumor borders during surgery.
Figure 3.
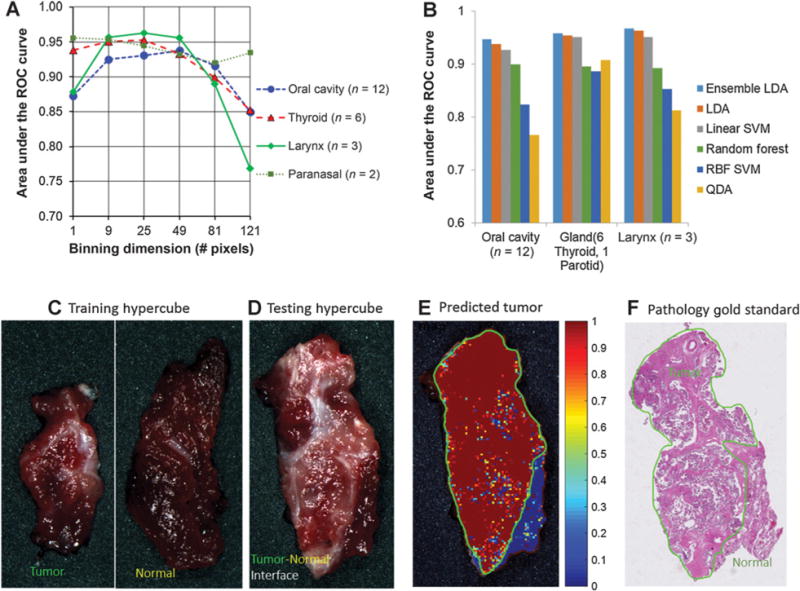
Diagnostic performance of HSI with the intrapatient classification method. A, Different block sizes and classifiers for the distinction of tumor from normal tissue in multiple, anatomic sites in head and neck cancer patients. C–F, Shows an example of a thyroid cancer detection result. C, Training hypercube with a tumor specimen and a normal specimen. D, Testing a hypercube with tumor and normal interface tissue. E, Cancer probability map generated by the ensemble LDA classifier. The green line is the tumor border generated by thresholding on the probability map. The color bar shows the likelihood of it being cancerous tissue. F, Pathology gold standard with the tumor region outlined within the green region by a clinically experienced pathologist.
Diagnostic performance of HSI, autofluorescence imaging and fluoresecence imaging
As shown in Table 2, using reflectance spectra from HSI, we were able to distinguish between tumor and normal tissue with an average accuracy, sensitivity, and specificity of 89%, 90%, and 90% from the oral cavity, 91%, 91%, and 93% from glandular tissue (thyroid and parotid), 94%, 95%, and 90% from the larynx and pharynx, and 90%, 90% and 90% from paranasal and nasal tissue, respectively. Overall, HSI outperformed autofluorescence imaging and 2-NBDG and proflavine fluorescence imaging for tumor detection in all of these anatomical sites.
Table 2.
Diagnostic performance of HSI, autofluorescence imaging, 2-NBDG fluorescence imaging, and proflavine fluorescence imaging using the intra-patient classification method
| Cancer site | Imaging method | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Oral cavity (n = 12) | HSI | 0.95 ± 0.05 | 89% ± 8% | 90% ± 7% | 90% ± 8% |
| Autofluorescence | 0.82 ± 0.20 | 81% ± 16% | 80% ± 16% | 80% ± 19% | |
| 2-NBDG | 0.83 ± 0.14 | 79% ± 14% | 79% ± 15% | 79% ± 14% | |
| Proflavine | 0.68 ± 0.18 | 66% ± 14% | 64% ± 16% | 69% ± 15% | |
| Gland (6 thyroid, 1 parotid) | HSI | 0.96 ± 0.04 | 91% ± 7% | 91% ± 8% | 93% ± 6% |
| Autofluorescence | 0.72 ± 0.31 | 67% ± 33% | 80% ± 20% | 71% ± 35% | |
| 2-NBDG | 0.84 ± 0.18 | 80% ± 17% | 78% ± 19% | 82% ± 16% | |
| Proflavine | 0.80 ± 0.23 | 78% ± 21% | 73% ± 26% | 82% ± 15% | |
| Larynx and pharynx (n = 3) | HSI | 0.97 ± 0.03 | 94% ± 4% | 95% ± 4% | 90% ± 7% |
| Autofluorescence | 0.74 ± 0.26 | 74% ± 21% | 74% ± 28% | 79% ± 22% | |
| 2-NBDG | 0.97 ± 0.04 | 92% ± 7% | 92% ± 8% | 92% ± 5% | |
| Proflavine | 0.79 ± 0.23 | 77% ± 20% | 77% ± 20% | 74% ± 23% | |
| Paranasal and nasal (n = 3) | HSI | 0.96 ± 0.02 | 90% ± 5% | 90% ± 5% | 90% ± 4% |
| Autofluorescence | 0.81 ± 0.11 | 76% ± 10% | 79% ± 8% | 72% ± 14% | |
| 2-NBDG | 0.76 ± 0.18 | 68% ± 19% | 64% ± 20% | 79% ± 7% | |
| Proflavine | 0.80 ± 0.08 | 73% ± 6% | 69% ± 11% | 79% ± 2% |
Figure 4 shows an example of tongue cancer detection using label-free HSI, autofluorscence imaging, and fluorescence imaging of 2-NBDG and proflavine. A predictive model was trained on the cancer and normal tongue tissue, as shown in the first column, and was tested on the cancer interface tissue of the same patient, as shown in the second column. The tongue cancer prediction maps by HSI and autofluorescence imaging were highly accurate, whereas 2-NBDG and proflavine was not able to correctly classify the normal tissue in the cancer interface specimen. This is likely due to the nonspecific uptake of the dye by the normal head and neck tissue.
Figure 4.
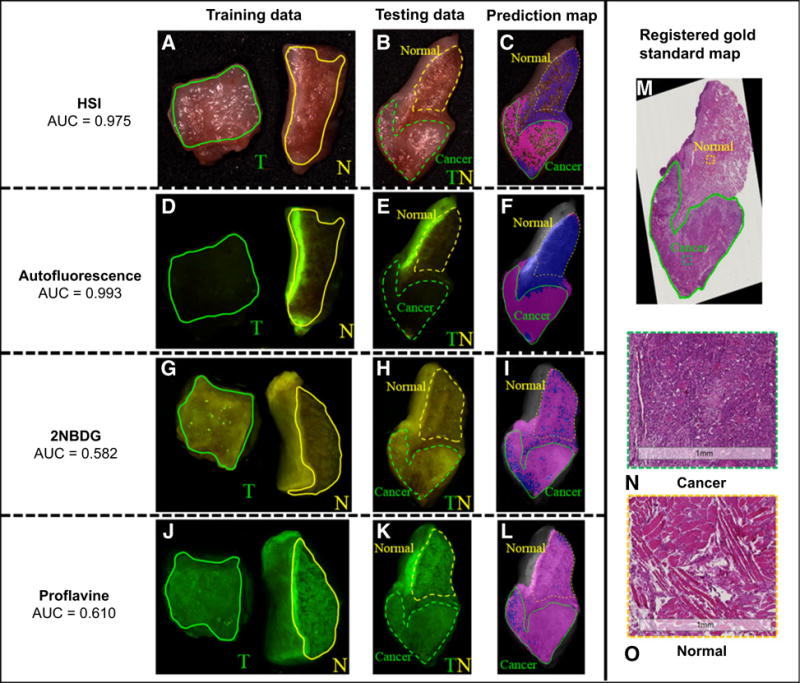
Tongue cancer detection using the intrapatient classification method. A, D, G, and J are the RGB composite image from hypercube, autofluorecence imaging, 2-NBDG, and proflavine fluorescence imaging, with the green and yellow solid lines outlining the cancer and normal tissue regions for training predictive models. B, E, H, and K are the corresponding RGB composite image of cancer–normal interface tissue for testing model performance, with the green and yellow dashed line outlining the region that we are certain to be tumor and normal for quantitative evaluation. C, F, I, and L are the predicted cancer map for HSI, autofluorescence, 2-NBDG, and proflavine imaging, with magenta and blue color denoting predicted malignant and normal tissue. Only regions within the green and yellow curve are used for quantitative evaluation. Glare pixels identified from hyperspectral images were excluded from classification, and, therefore, not labeled in the prediction map. M is the registered histology gold standard image, with the cancerous region outlined inside the green line by an clinically experienced pathologist. N and O are the enlarged cancer and normal histology image from the selected region of the image in M.
Cancer prediction with interpatient classification
Figure 5 plotted the ROC curves of individual cancer patients with intrapatient classification (A) and interpatient classification (B). With reflectance spectra from HSI, we obtained an average AUC of 0.88 and 0.91 for the two cohorts of patients previously described. The diagnostic performance of HSI for cancer detection was slightly lower than the results of the intrapatient classification, probably due to the influence of interpatient heterogeneity. Supplementary Figure S1 shows an example of thyroid cancer detection from hyperspectral images with interpatient classification. We noticed a much lower reflectance spectrum from normal thyroid tissue than from thyroid cancer tissue. We also discovered that cancer and normal tissue were accurately detected in all three tissue samples (cancer, normal, cancer interface).
Figure 5.
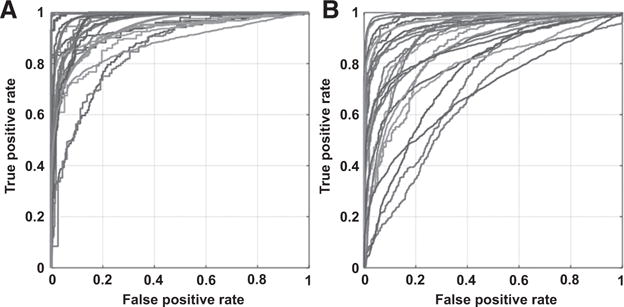
ROC curves of intrapatient classification (A) and interpatient classification (B) with HSI for individual patients.
Diagnostic performances of HSI, MSI, and RGB
To evaluate the diagnostic value of different wavelength regions, we grouped the wavelengths from 450 to 900 nm into different subregions, including 450 to 600 nm, 605 to 850 nm, and 855 to 900 nm and compared their classification performances. As shown in Table 3, we found that oral cavity cancer, the whole spectrum including both visible and NIR light, was able to distinguish cancer from normal tissue from the oral cavity with a higher sensitivity and specificity than the visible wavelength region alone. Although for thyroid cancer, the visible wavelength region from 450 to 600 nm yielded the highest average classification accuracy for thyroid cancers. Furthermore, we found that HSI was superior to MSI and conventional RGB imaging in the detection of oral cavity cancer (Supplementary Table S1).
Table 3.
Classification performance of wavelength subregions of HSI for the detection of head and neck cancer with intrapatient classification and interpatient classification
| Primary site | Classification method | Wavelength range (nm) | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Oral cavity | Intrapatient classification | 450–600 | 0.87 ± 0.11 | 82% ± 11% | 81% ± 13% | 82% ± 10% |
| 605–850 | 0.89 ± 0.12 | 84% ± 12% | 83% ± 12% | 85% ± 12% | ||
| 855–900 | 0.76 ± 0.14 | 72% ± 11% | 73% ± 11% | 71% ± 12% | ||
| 450–900 | 0.95 ± 0.05 | 89% ± 8% | 90% ± 7% | 90% ± 8% | ||
| Interpatient classification | 450–600 | 0.88 ± 0.12 | 82% ± 11% | 82% ± 11% | 81% ± 12% | |
| 605–850 | 0.82 ± 0.11 | 77% ± 11% | 78% ± 10% | 76% ± 12% | ||
| 855–900 | 0.71 ± 0.11 | 67% ± 9% | 67% ± 9% | 66% ± 11% | ||
| 450–900 | 0.91 ± 0.10 | 85% ± 10% | 85% ± 9% | 84% ± 12% | ||
| Thyroid | Intrapatient classification | 450–600 | 0.97 ± 0.06 | 93% ± 8% | 93% ± 9% | 95% ± 6% |
| 605–850 | 0.95 ± 0.06 | 91% ± 9% | 90% ± 10% | 92% ± 7% | ||
| 855–900 | 0.93 ± 0.09 | 89% ± 10% | 88% ± 12% | 90% ± 8% | ||
| 450–900 | 0.96 ± 0.05 | 92% ± 7% | 92% ± 9% | 94% ± 6% | ||
| Interpatient classification | 450–600 | 0.92 ± 0.07 | 87% ± 9% | 86% ± 9% | 88% ± 9% | |
| 605–850 | 0.94 ± 0.05 | 88% ± 6% | 88% ± 6% | 88% ± 6% | ||
| 855–900 | 0.84 ± 0.11 | 78% ± 11% | 77% ± 10% | 79% ± 10% | ||
| 450–900 | 0.91 ± 0.09 | 86% ± 10% | 85% ± 11% | 87% ± 9% |
Next, we looked at the cancer prediction accuracy for individual surgical specimens, including purely normal, purely cancer, and cancer–normal interface. As shown in Supplementary Table S2, for patient within the first cohort (oral cavity, larynx, paranasal, and nasal), HSI achieved an accuracy of 82% for classifying cancerous regions in cancer tissue and an accuracy of 88% in classifying normal regions in normal tissue and. In addition, HSI was able to distinguish cancer from normal tissue with an average accuracy, sensitivity, and specificity of 82%, 66%, and 76% in tumor–normal interface tissue. For patients with thyroid carcinoma, HSI was able to accurately detect 86% of the cancerous tissue from cancer specimens and 87% of the normal tissue from normal specimens (Supplementary Table S3). For cancer interface tissue, HSI achieved an accuracy, sensitivity, and specificity of 85%, 86%, and 87%, respectively, for the detection of thyroid carcinoma. As the specimen from only one patient of the thyroid cohort was MTC, the classification results for this patient (P35) were relatively poor.
Discussion
HSI is a wide-field modality able to sense tumors in varying depth using VIS and NIR light illumination (at clinically relevant sensing depths). The scanning time is on the order of seconds. It is an objective, fast, and cost-effective tool that may provide real-time assessment of complete resection margins. This technique is able to quantify not only surface mucosa but also different faces of the resection margins. This study for the first time evaluated the diagnostic potential of HSI for the detection and delineation of head and neck cancer in fresh surgical specimens from a variety of anatomic sites, including the oral cavity, thyroid, larynx, pharynx, parotid, paranasal sinus, and nasal cavity. We demonstrated that HSI combined with machine-learning techniques enabled accurate discrimination between normal and cancerous tissue from fresh surgical specimens of a variety of head and neck cancer sites. Although this study was designed only for imaging the ex vivo surgical specimen, label-free HSI could be used for rapid and objective assessment of head and neck cancer margins during surgery and in turn, improve clinical decision-making and patient outcomes.
To facilitate the clinical application of HSI, it is of paramount importance to develop fast and accurate quantification tools for the large amounts of hyperspectral data. HSI is able to acquire a stack of two-dimensional images over a wide range of spectral bands, and thus generates a three-dimensional hypercube containing rich spectral-spatial information. Intensities over all of the spectral bands form a spectral signature for each pixel of the hypercube, and these spectral signatures are associated with the biochemical and morphological changes in tissue. Therefore, hyperspectral images which contain a spectral signature at each image pixel, can be analyzed to potentially identify various pathologic conditions. The challenges of hypercube analysis lie in the large size of the spectra-spatial dataset and the intra- and interpatient spectral variability.
To efficiently analyze hyperspectral data for intraoperative applications, two quantification tools were developed using intra- and interpatient classification. In this feasibility study, the quantification results were validated by the pathologic diagnosis in formalin-fixed paraffin-embedded tissue, which was reported to be more accurate than the frozen pathology (7, 8). In the future, we would like to further assess the value of HIS by comparing its performance with surgeons’ assessment in the OR, and also with frozen sections diagnosis in the pathology room. Both intra- and interpatient classification methods are very promising for evaluating the surgical margins of an ex vivo specimen and for intraoperative surgical guidance. In the intrapatient analysis, a series of images of the surgical bed could be collected before, during, and after tumor resection of the same patient based on preoperative imaging and surgeons’ assessment. Clinically visible tumor and normal tissue spectra may be used to build predictive models, and residual tumors in images of the surgical bed after resection may be detected with the model in ~0.3 s. This method is not affected by interpatient spectral heterogeneities, even with much smaller training dataset. As for the interpatient analysis, it would be desirable to establish a spectral database of cancer and normal tissue from a large number of patients and a variety of tissue types and anatomic sites. A predictive model could be built and optimized in advance based on these datasets. The model could be directly utilized to predict the presence of cancerous tissue during the surgical resection. Furthermore, the model could be utilized to predict the margin status of the resected, fresh surgical specimen in the pathology room, which could save the time and cost associated with frozen-section diagnosis. Because the large heterogeneity of cancer patients, in our study interpatient classification was not as accurate as intrapatient classification for the detection of cancerous tissue. Future work will continue to improve the diagnostic performance for interpatient classification from two aspects: (1) enlarge the training database by incorporating more hyperspectral dataset of a wide variety of surgical tissue samples; and (2) try deep learning-based algorithm to further boost the classification accuracy.
We also demonstrated that for oral cancer detection, it would be preferable to use the whole spectrum of 450 to 900 nm containing both visible and NIR light in order to provide a more accurate diagnostic measure, and which indicated that visible and NIR light contained complementary information for diagnosis. Furthermore, HSI was also more accurate than HSI, MSI, and RGB imaging for oral cancer detection. Because the reflectance spectra captured the alteration of absorption and scattering properties of tissue associated with malignant transformations, molecular fingerprinting based on inverse modeling of reflectance spectra obtained by HSI may shed new light on our understanding of human head and neck cancer biology.
We previously demonstrated the utility of HSI for head and neck cancer detection in a subcutaneous cancer animal model (17, 19, 34) and a chemically induced, oral cancer model (35). Uptake of the 2-NBDG fluorescent deoxyglucose derivative was associated with increased metabolic activity. A recent study also reported that wide-field optical imaging with 2-NBDG could accurately distinguish the pathologically normal and abnormal biopsies of patients with head and neck cancers (20). Furthermore, proflavine was also applied for distinguishing between benign and neoplastic mucosa in the head and neck (22). Here, we showed that label-free HSI was superior to autofluorescence imaging and fluorescence imaging of two dyes, that is, 2-NBDG and proflavine, in the detection of head and neck cancers in fresh surgical specimens. The fact that HSI does not require the use of an exogenous contrast agent to provide optical contrast favors its clinical translation.
As proof of the concept, this study included a limited number of patients in each anatomic site at the head and neck region. We are attempting to include more patients and build a large spectral database with various type of tissue components including mucosa, connective tissue, muscle, gland, etc. from different anatomic sites. Such a database would be helpful to predict cancer presence in the surgical margin in the future. Although larynx and thyroid lesions may be taken out as whole organs in many cases, there are also partial resections depending on the extent of the tumors and it is not unusual to take margins from the main specimen or surgical bed for intraoperative pathology assessment.
As HSI is relatively new to the field of cancer imaging, there has not been an intraoperative HSI commercially available. In this feasibility study, the closed field imaging system represents the first study of using HSI to identify head and neck cancers in surgical specimens of human patients. Interference from exogenous light is a common problem for intraoperative optical imaging techniques. To address these problems, we may either turn off the OR light as adopted by the NIR fluorescence imaging (12), or use spatially modulated illumination as used in spatial frequency domain imaging (36). In the future, a system combining the HSI instrument with the quantification tools needs to be designed to produce high-quality images. We will further evaluate this system on the mucosal and deep surgical margins as well as more challenging cases like highly invasive/cohesive patterns of invasion in both the OR and the pathology laboratory. HSI could be utilized to quickly scan a large field of view and then highlight those highly suspicious pixels or regions after quantitative image classification. These suspicious tumor foci could be taken out for pathology validation to determine the sensitivity of the technology and to correlate with the local recurrence rate. Furthermore, HSI could also be integrated with a microscope to provide more detailed information for microscopic tumor detection. The long-term goal of this work is to provide an adjunct tool for real-time margin assessment during surgery, and thus potentially improving cancer resection while preserving vital anatomic structures and functions.
Supplementary Material
Translational Relevance.
Surgical resection is the backbone of the treatment options for head and neck cancers. The complex anatomy and vital physiologic role of the tumor-involved structures dictate that the treatment goals are not only to improve survival outcomes but also to preserve organ function. Here we present for the first time the application of quantitative hyperspectral imaging as a sensitive and specific diagnostic tool for the detection and delineation of head and neck cancers in a wide variety of anatomic sites. Hyperspectral imaging combined with machine learning-based quantification methods provides an objective, fast, and cost-effective tool that may allow real-time assessment of complete resection margins.
Acknowledgments
We thank all of the surgeons and surgical nurses from the Department of Otolaryngology, as well as the pathologists and pathologist assistants from the Department of Pathology and Laboratory Medicine at the Emory University Hospital Midtown, for their help with surgical specimen collection and pathology diagnosis. We also thank our lab members: Xulei Qin, James Dormer, Rongrong Guo, Ling Ma, Zhiqiang Tian, and Yun Zhang, for their assistance with imaging during the experiment.
Grant Support
This research is supported in part by the NIH grants (CA176684, R01CA156775, and CA204254) and by Developmental Funds from the Win-ship Cancer Institute of Emory University under award number P30CA138292.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
M.R. Patel is a consultant/advisory board member for AstraZeneca and Intuitive Surgical. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: G. Lu, J.V. Little, A.Y. Chen, B. Fei
Development of methodology: G. Lu, B. Fei
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): G. Lu, J.V. Little, X. Wang, H. Zhang, M.R. Patel, M.W. El-Deiry, A.Y. Chen, B. Fei
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): G. Lu, J.V. Little, B. Fei
Writing, review, and/or revision of the manuscript: G.Lu, J.V. Little, M.R. Patel, C.C. Griffith, M.W. El-Deiry, A.Y. Chen, B. Fei
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): G. Lu, J.V. Little, C.C. Griffith, B. Fei Study supervision: M.W. El-Deiry, B. Fei
References
- 1.Wissinger E, Griebsch I, Lungershausen J, Foster T, Pashos C. The economic burden of head and neck cancer: a systematic literature review. Pharmaco Economics. 2014;32:865–82. doi: 10.1007/s40273-014-0169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386–96. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Haddad RI, Shin DM. Recent advances in head and neck cancer. Indian J Plast Surg. 2008;359:1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 4.Sutton DN, Brown JS, Rogers SN, Vaughan ED, Woolgar JA. The prognostic implications of the surgical margin in oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2003;32:30–4. doi: 10.1054/ijom.2002.0313. [DOI] [PubMed] [Google Scholar]
- 5.Hinni ML, Ferlito A, Brandwein-Gensler MS, Takes RP, Silver CE, Westra WH, et al. Surgical margins in head and neck cancer: a contemporary review. Head Neck. 2013;35:1362–70. doi: 10.1002/hed.23110. [DOI] [PubMed] [Google Scholar]
- 6.Keereweer S, Kerrebijn JF, Driel PAA, Xie B, Kaijzel E, Snoeks TA, et al. Optical image-guided surgery—where do we stand? Mol Imaging Biol. 2011;13:199–207. doi: 10.1007/s11307-010-0373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du E, Ow TJ, Lo YT, Gersten A, Schiff BA, Tassler AB, et al. Refining the utility and role of frozen section in head and neck squamous cell carcinoma resection. Laryngoscope. 2016;126:1768–75. doi: 10.1002/lary.25899. [DOI] [PubMed] [Google Scholar]
- 8.DiNardo LJ, Lin J, Karageorge LS, Powers CN. Accuracy, utility, and cost of frozen section margins in head and neck cancer surgery. Laryngoscope. 2000;110:1773–6. doi: 10.1097/00005537-200010000-00039. [DOI] [PubMed] [Google Scholar]
- 9.Poh CF, Zhang L, Anderson DW, Durham JS, Williams PM, Priddy RW, et al. Fluorescence visualization detection of field alterations in tumor margins of oral cancer patients. Clin Cancer Res. 2006;12:6716–22. doi: 10.1158/1078-0432.CCR-06-1317. [DOI] [PubMed] [Google Scholar]
- 10.Poh CF, Anderson DW, Durham J, et al. Fluorescence visualization–guided surgery for early-stage oral cancer. JAMA Otolaryngol Head Neck Surg. 2016;142:209–16. doi: 10.1001/jamaoto.2015.3211. [DOI] [PubMed] [Google Scholar]
- 11.Farah CS, Dalley AJ, Nguyen P, Batstone M, Kordbacheh F, Perry-Keene J, et al. Improved surgical margin definition by narrow band imaging for resection of oral squamous cell carcinoma: a prospective gene expression profiling study. Head Neck. 2014:832–9. doi: 10.1002/hed.23989. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal EL, Warram JM, de Boer E, Chung TK, Korb ML, Brandwein-Gensler M, et al. Safety and tumor specificity of cetuximab-IRDye800 for surgical navigation in head and neck cancer. Clin Cancer Res. 2015;21:3658–66. doi: 10.1158/1078-0432.CCR-14-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iqbal H, Pan Q. Image guided surgery in the management of head and neck cancer. Oral Oncol. 2016;57:32–9. doi: 10.1016/j.oraloncology.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Keereweer S, Van Driel PBAA, Snoeks TJA, Kerrebijn JDF, Baatenburg de Jong RJ, Vahrmeijer AL, et al. Optical image-guided cancer surgery: challenges and limitations. Clin Cancer Res. 2013;19:3745–54. doi: 10.1158/1078-0432.CCR-12-3598. [DOI] [PubMed] [Google Scholar]
- 15.Lu G, Fei B. Medical hyperspectral imaging: a review. J Biomed Opt. 2014;19:10901. doi: 10.1117/1.JBO.19.1.010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akbari H, Halig LV, Schuster DM, Osunkoya A, Master V, Nieh PT, et al. Hyperspectral imaging and quantitative analysis for prostate cancer detection. J Biomed Opt. 2012;17:076005. doi: 10.1117/1.JBO.17.7.076005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu G, Halig L, Wang D, Qin X, Chen ZG, Fei B. Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging. J Biomed Opt. 2014;19:106004. doi: 10.1117/1.JBO.19.10.106004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pike R, Lu G, Wang D, Chen ZG, Fei B. A minimum spanning Forest based method for noninvasive cancer detection with hyperspectral imaging. IEEE Trans Biomed Eng. 2015;PP:1. doi: 10.1109/TBME.2015.2468578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu G, Wang D, Qin X, Halig L, Muller S, Zhang H, et al. Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery. J Biomed Opt. 2015;20:126012. doi: 10.1117/1.JBO.20.12.126012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Z, Loja MN, Farwell DG, Luu QC, Donald PJ, Amott D, et al. Widefield optical imaging of changes in uptake of glucose and tissue extracellular pH in head and neck cancer. Cancer Prev Res (Phila) 2014;7:1035–44. doi: 10.1158/1940-6207.CAPR-14-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muldoon TJ, Roblyer D, Williams MD, Stepanek VMT, Richards Kortum R, Gillenwater AM. Noninvasive imaging of oral neoplasia with a highresolution fiber-optic microendoscope. Head Neck. 2011;9999:1–8. doi: 10.1002/hed.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vila P, Park C, Pierce M, Goldstein G, Levy L, Gurudutt V, et al. Discrimination of benign and neoplastic mucosa with a high-resolution microendoscope (HRME) in head and neck cancer. Ann Surg Oncol. 2012;19:3534–9. doi: 10.1245/s10434-012-2351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langsner RJ, Middleton LP, Sun J, Meric-Bernstam F, Hunt KK, Drezek RA, et al. Wide-field imaging of fluorescent deoxy-glucose in ex vivo malignant and normal breast tissue. Biomed Opt Express. 2011;2:1514–23. doi: 10.1364/BOE.2.001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobbs JL, Ding H, Benveniste AP, Kuerer HM, Krishnamurthy S, Yang W, et al. Feasibility of confocal fluorescence microscopy for real-time evaluation of neoplasia in fresh human breast tissue. J Biomed Opt. 2013;18:106016. doi: 10.1117/1.JBO.18.10.106016. [DOI] [PubMed] [Google Scholar]
- 25.Sun DW. Hyperspectral imaging for food quality analysis and control. San Diego, CA: Academic Press; 2010. Preprocessing of hyperspectral images; p. 303. [Google Scholar]
- 26.Park B, Lu R. Hyperspectral imaging technology in food and agriculture. New York: Springer; 2015. Procedures for acquiring hyperspectral image cubes; p. 321. [Google Scholar]
- 27.Hellebust A, Rosbach K, Wu JK, Nguyen J, Gillenwater A, Vigneswaran N, et al. Vital-dye-enhanced multimodal imaging of neoplastic progression in a mouse model of oral carcinogenesis. J Biomed Opt. 2013;18:126017. doi: 10.1117/1.JBO.18.12.126017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subhash N, Mallia JR, Thomas SS, Mathews A, Sebastian P, Madhavan J. Oral cancer detection using diffuse reflectance spectral ratio R540/R575 of oxygenated hemoglobin bands. J Biomed Opt. 2006;11:014018. doi: 10.1117/1.2165184. [DOI] [PubMed] [Google Scholar]
- 29.Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936;7:179–88. [Google Scholar]
- 30.Tin Kam H. The random subspace method for constructing decision forests. IEEE Trans Pattern Anal Mach Intell. 1998;20:832–44. [Google Scholar]
- 31.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- 32.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 33.Good P. Permutation tests: a practical guide to resampling methods for testing hypotheses. New York: Springer-Verlag; 2013. [Google Scholar]
- 34.Baowei F, Akbari H, Halig LV. Hyperspectral imaging and spectral-spatial classification for cancer detection. Biomedical Engineering and Informatics (BMEI), 2012 5th International Conference on. 2012:62–4. [Google Scholar]
- 35.Lu G, Qin X, Wang D, Muller S, Zhang H, Chen A, et al. Hyperspectral imaging of neoplastic progression in a mouse model of oral carcinogenesis. Proceedings of SPIE. 2016:978812. doi: 10.1117/12.2216553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laughney AM, Krishnaswamy V, Rizzo EJ, Schwab MC, Barth RJ, Cuccia DJ, et al. Spectral discrimination of breast pathologies in situusing spatial frequency domain imaging. Breast Cancer Res. 2013;15:R61. doi: 10.1186/bcr3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


