Abstract
Boswellia sacra, a frankincense producing endemic tree, has been well known for its cultural, religious and economic values. However, the tree has been least explored for the associated microsymbiota in the rhizosphere. The current study elucidates the fungal and bacterial communities of the rhizospheric regions of the wild and cultivated B. sacra tree populations through next generation sequencing. The sequence analysis showed the existence of 1006±8.9 and 60.6±3.1 operational taxonomic unit (OTUs) for bacterial and fungal communities respectively. In fungal communities, five major phyla were found with significantly higher abundance of Ascomycota (60.3%) in wild population and Basidiomycota (52%) in cultivated tree rhizospheres. Among bacterial communities, 31 major phyla were found, with significant distribution of Actinobacteria in wild tree rhizospheres, whereas Proteobacteria and Acidobacteria were highly abundant in cultivated trees. The diversity and abundance of microbiome varied significantly depending upon soil characteristics of the three different populations. In addition, significantly higher glucosidases, cellulases and indole-3-acetic acid were found in cultivated tree’s rhizospheres as compared to wild tree populations. for these plants to survive the harsh arid-land environmental conditions. The current study is a first comprehensive work and advances our knowledge about the core fungal and bacterial microbial microbiome associated with this economically important tree.
Introduction
Symbioses of microbial communities have been of special interest to ecologists to understand their role in plant growth and development under extreme living conditions such as desert conditions [1]. Marginalize conditions often provides new insights to elucidate the basis of survival in water deficient and sever heat stress environments. Besides, plant’s unique genetic makeup, symbiosis of native microflora plays an essential role in plant life. These microorganisms, predominantly occupying the rhizosphere, enable processes involved in nutrient transport, essential secondary metabolites secretions, and abiotic and biotic factors mitigations [2–6]. The plant in return provides a favorable niche for specific kinds of microbes to grow and reproduce whilst also sharing some of the beneficial exudates and nutrients [7].
During the association with host, fungi and bacteria produces various kinds of extracellular enzymes or exozymes, which target various macromolecules such as carbohydrates, lignin, organic phosphate, proteins, and sugar-base polymers to breakdown into transportable product throughout the cells [8]. A similar prospect has been noted for indole acetic acid (IAA), a phytohormones, which help plant root and growth development during normal and stress conditions [9]. Besides initiating the host-symbiosis process, some of these exozymes and IAA counteract plant pathogenic infections and extend abiotic stress tolerance [9]. However, such interaction has been very little known about the microbial symbionts of trees growing sub-tropical deserts, especially on Boswellia sacra [10].
B. sacra is an economically important frankincense producing tree of the Sultanate of Oman [11]. Resin from Boswellia has been traded as incense from the southern coast of Arabia to the Mediterranean region for more than a millennium [12]. There are more than twenty Boswellia species, and B. sacra is an endemic species that grows specifically in Dhofar region of Oman. It is a keystone species that is known to provide an important oleo gum resin, which has long-standing cultural and medicinal history. The essential oil and boswellic acids have been known to possess potent anticancer activities [13, 14]. The local population obtains solid and semi-solid resin (commonly known as Luban) by making a series of wounds/incisions in the bark of the tree (Fig 1). The annual production of Omani frankincense ranges between 80 to 100 tons from nearly 500,000 trees [15, 16]. In some areas, the collection of resin is an economically favorable use of land than crop production and accounts for the majority of a rural household’s income [17].
Fig 1. Plant habitats and their location.
B. scara growing in wild and in cultivated conserved areas.
Very least is known about the microbial community profile of B. sacra. Recently, El-Nagerabi et al. [18] and Khan et al. [9] have shown the endophytic microbial communities of the tree. However, looking at the tree life and its habitats, they are unique as these often experience wide array of stresses such as heat, UV incidence, drought and strong wind [1]. Moreover, there is a significant difference between the wild and cultivated trees growth, development and resin production ability. In case of economically important plants growing in such a marginalize ecosystem, very few studies have been carried out on their microflora, where some are restricted to only bacterial communities. Some of the examples includes Rehmannia glutinosa [19], Rumex patientia [20], Polygonum cuspidatum [21], Aloe vera [22] Rhododendron arboretum [23], Agave species [24], Cacti [25] and Thymus zygis [26]. In current study, we aimed to elucidate the core fungal and prokaryotic communities, and their structure in the three population of B. sacra tree through detailed metagenomics and bioinformatics approaches.
Materials and methods
Sampling area and collection
Rhizospheric soil samples of Boswellia sacra were collected from three major location at Adonab (BSA; N17°20.47’ E054°04.51), Dowkah National Park (BSD; N19°4.89’ E054°22.81) and Dowkah valley (BSW; N19°07.76’ E054°25.43) respectively in dry summer season. These locations are famous for B. sacra tree population. Rhizospheric soil samples adhering to root surface (upto 60 inches deep) were collected from these regions (Table 1). The soil analysis were performed according to Adhikari [27]. A total of fifty tree’s root regions were selected for rhizospheric soil collection from three respective sites approximately 50 km away from each other (Fig 1). The rhizospheric soil collected from each tree community was pooled into two replicates.
Table 1. Physical and chemical properties of soil samples collected from the rhizospheres of three B. sacra populations.
| Clay (%) | Sand (%) | Silt (%) | Bulk density (%) | Organic matter (%) | Texture | EC (dS m−1) | pH | Nitrates (mg/L) | |
|---|---|---|---|---|---|---|---|---|---|
| BSA | 13.2±0.92a | 64±1.9c | 17.3±0.84a | 2.9±0.23a | 2.6±0.39a | sandy loam | 18.5±0.29a | 7.82±0.43a | 2.1±0.2a |
| BSD | 8.2±0.12b | 75.5±1.2ab | 12.8±0.92ab | 1.4±0.85b | 2.1±0.9a | sandy loam | 18.1±0.68a | 6.43±0.23b | 1.1±0.01b |
| BSW | 4.1±0.38c | 88.8±1.8a | 6.1±0.43c | 0.2±0.81c | 0.8±0.2b | sandy | 5.87±0.42b | 5.49±0.1c | 0.45±0.1c |
Values in each column are the mean of five replications and presented with standard deviation. The different letter(s) with the values in each column showed the values are significantly different among three sampling population of B. sacra.
Microbial products analysis in rhizospheric soil
To quantify extracellular enzymes, the method of Marx et al. [28] and Khan et al. [9] was adopted with some modifications. Briefly, all the substrates (S1 Table) were obtained from Sigma-Aldrich Co. Ltd in crystalline form. Ten milliliters of a 10 mM stock solution of each 4-methylumbelliferone (MUB) substrate was prepared, while the assay procedures were the same for each substrate. Depending on the substrate, a 7-MUB standard was used. A 10 mM stock solution of pure MUB was prepared in methanol (0.1762 g of 4-methylumbelliferone in 100 mL). This stock solution was diluted in sodium acetate (pH 5.2) buffer to 1 μM and stored at 4°C (S1 Table). The soil samples were processed for exozymes analysis using the method of Marx et al. [28] through Shimadzo (Tokyo Japan) fluorescence spectrophotometer (S1 Methods). The Indole acetic acid quantification of soil sample was performed using the method of Khan et al. [9], S1 Methods).
Sample preparation, DNA extraction, and sequencing
The mixture of rhizospheric soil samples were multiplexed and subject to total DNA extraction through combined manual and kit based methods (Supporting Information Methods S1). PCR free libraries of each DNA sample were made by amplifying the internal transcribe spacer (3F/4R (ITS3‐4di) and 16S (V3-V4) for fungal and prokaryotic communities respectively (S1 Methods; Coleman-Derr et al. [24]. For 16S, peptide nucleic acid (PNA) clamps were used to reduce the chloroplast and mitochondrial contamination. A Paired-end 250 bp sequencing approach was performed on an Illumina MiSeq instrument (Illumina Inc., San Diego, CA, USA) operating with v2 chemistry (User Guide Part # 15027617 Rev. L). All quality sequences related to this project are available in the NCBI Sequence Read Archive (SRA) under project ID RA337739, BioProject PRJNA337739, 16S accessions (KY694695 –K694751), and ITS accessions (KY694662 –KY694694).
Data processing and analyses
The raw Fastq reads were processed through a custom pipeline developed at the Macrogen Inc. (Seoul, South Korea; S1 Methods). Raw reads were contaminant-filtered, quality trimmed, merged and clustered to pro- taxonomic units (OTUs), respectively, at 95% and 97% identity using the UPARSE pipeline. FLASH was also used for short reads length adjustment to improve genome assemblies [29]. Taxonomies were assigned to each OTU using the RDP Naıve Bayesian Classifier [30] with custom reference databases. We used the CD-HIT-OTU package [31] and its variant tailored for Illumina reads. Another heuristic, the UCLUST greedy algorithm [32] (included in the free 64-bit version 6.0.307 of USEARCH) as implemented in the QIIME [32] script “pick_otus.py” (v1.8.0) was also used with default parameters. OTUs whose RDP classifications did not match their expected taxonomic kingdoms (Fungi and Bacteria/Archaea, respectively) were removed. Average read counts varied by sample type for different data sets. To reduce low-abundance and spurious OTUs, technical reproducibility thresholds determined empirically from technical replicates as in Lundberg et al. [33] were set and OTUs kept only if they had at least two reads in at least three samples (ITS data) or at least seven reads in at least three samples (16S data). To check for chimeric sequences amongst the different categories of sequences, the UCHIME algorithm [34] included in the free version 6.0.307 of USEARCH was used. Two variations of the program were run and compared. First the de novo mode in which the varying abundances of sequences in the input were exploited. Secondly, we used the reference mode in which decisions are made using a database of chimera-free sequences. Thirdly, the latest product from Robert C. Edgar titled UPARSE [35] was applied to our data (included in the free version 9.0.2132 of USEARCH). Mothur analysis were performed for community richness and diversity analysis [36].
Statistical analysis
All samples were analyzed in triplicate. The data are presented as the mean ± standard error of the mean (SEM). Differences were evaluated using one-way analysis of variance (ANOVA). Differences were considered significant at P < 0.05 and were calculated by GraphPad Prism Version 6.01 (GraphPad Software, San Diego, CA, USA). The mean values were compared using Duncan’s multiple range tests at P < 0.05 (SAS 9.1, Cary, NC, USA). The R statistical framework version 2.11 was used for NMDS ordination plots (metaMDS), beta-dispersion (betadis), PERMANOVA (adonis), permutational ANOVA (aovp) and the estimation of diversity indices and PAST (v3.01).
Results
Microbial community diversity associated with the three populations of B. sacra
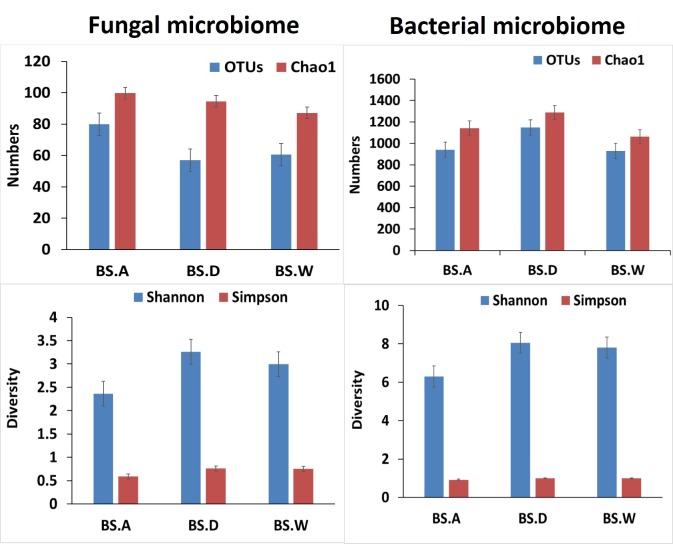
We assessed the prokaryotic and fungal communities of the rhizosphere of three distinctive populations of B. sacra viz. (i) BSA, (ii) BSD and their comparison with wildly grown tree population (BSW). The profiles of microbial communities were elucidated through MiSeq sequencing platform. A total of 678Mbp and 1.24Gbp of high quality read data (S2 Table) was generated for fungal and bacterial microbial communities respectively (S2 Table). The average bases counted were 1,219,536±19.1 and 546,307±14.5 for fungi and bacterial microorganisms respectively (S2 Table). The mean GC content for ITS was 49.44%; whereas it was 58.32% for 16S. The mean read count of fungal communities was 127±2.1, 995±43.2 (BSA), 98,088±5.1 (BSD) and 75,979±103.9 (BSW), whereas, for bacterial communities, it was 146,650±21.5 (BSA), 199,516±34.9 (BSD) and 188,036±44.9 (BSW). This suggests that the wild population of B. sacra tree showed a lower read count than the cultivated population (S2 Table).
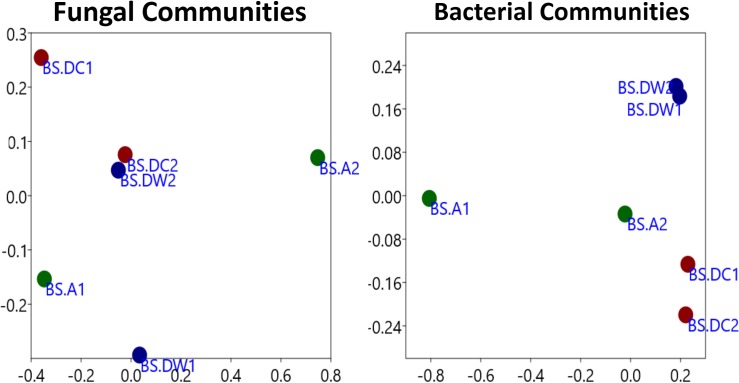
In case of OTU analysis, significantly higher (P<0.0019; average 1006±9.9) OTUs were revealed in bacterial communities, whereas, in fungi, it was significantly lower (P<0.0019; average 60.6±3.1). This suggests a higher microbial diversity of bacterial species. In case of population specific fungal OTUs, BSA showed significantly higher (P<0.02; 102±3.9 OTUs as compared to BSD and BSW population. However, in case of diversity indices, the Shannon was significantly higher (ANOSIM; R2 = 0.718, p< 0.00071) in BSD as compared to BSA and BSW populations of B. sacra (Fig 2). In case of bacterial communities, BSD population was found with significantly higher OTUs (P<0.001; 1178±13.5) as compared with other two populations. The Shannon diversity indices was also higher for BSD (Fig 2). The bacterial communities were the highest in abundance and OTUs number as compared to fungi (Fig 2). In case of nMDS of fungal communities, all the three populations and their replications were significantly distant a part from each other, however, BSW and BSD were found closely associated (R2 = 0.78; P<0.031; Fig 2).
Fig 2. Distribution of fungal and bacterial communities operational taxonomic units (OTUs) and Chao-1 of each replica from data generated through MiSeq sequencing (16S and ITS) of the rhizospheric samples from wild and cultivated rhizosphere of B. sacra tree.
In case of nMDS analysis of bacterial communities (R2 = 0.937; P<0.0092), BSW formed a separate combined group suggesting not much difference in the OTUs of the replicates. The BSA replications were significantly distant a part from each other, showing a heterogeneous diversity of fungal OTUs across same replication. The distance among replicates of BSD was not higher as compared to BSA population (Fig 3). The replicates of BSD population were comparatively not distance a part. The rarefaction curve of the three populations also revealed similar conjoining properties (S1 Fig), where fungal communities are distant apart than bacterial ones in the three samples. This was also similar to the results of hierarchal clustering (P<0.05 of taxa and sample), in which BSA formed a distinctive clade with BSD and BSW, instead of forming same clade with its own replication (S1 Fig). However, this was different in bacterial communities as each sample formed a cladogram with each other (S1 Fig).
Fig 3. Nonmetric Multidimensional Scaling (NMDS) plots for Bray–Curtis distances of different rhizospheric populations of B. sacra.
The nMDS plot was made in PAST v3.0 (New Zeeland).
Fungal microbial communities in the Rhizosphere of B. sacra
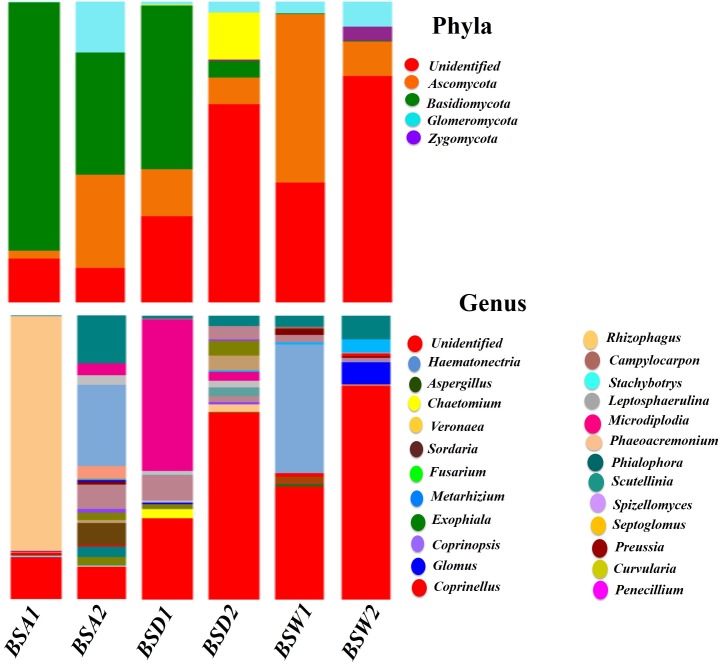
Rhizospheric microbial diversity among the three populations varied greatly. In the fungal phylum, Ascomycota, Basidiomycota, Chytridiomycota, Glomeromycota and Zygomycota were the abundant in BSA, BSD and BSW populations. Among populations, Ascomycota was significantly higher (P<0.001; 60%) in wild population (BSW), which was followed by BSA (47%) of B. sacra. Basidiomycota, on the other hand, was significantly higher (P<0.001; 52%) in BSA rhizosphere (Fig 4; S3 Table). The unidentified fungal communities were sharing a major proportion of data, which was about 76% in BSW and 65% in BSD populations. Glomeromycota and Zygomycota were only abundant in BSD. The diversity of fungal genus was encompassed around unidentified microbes.
Fig 4. Relative abundances and shared core OTUs of fungal phyla and genus found in the rhizosphere of wild and cultivated populations of B. sacra tree.
Taxa with abundance < 5% are presented as “Others/Unidentified”.
In correlation with phyla, the genus too was varyingly distributed among the three populations of B. sacra (Fig 4; S3 Table). The results showed that Haematonectria was highly abundant (1.1 to 8.1%) in all the three populations. Where, Aspergillus, Exophiala, Coprinopsis and Veronaea were significantly higher (P<0.001; 4 to 7.6%) only in BSA samples as compared to BSD and BSW samples. Glomus and Rhizophagus were solely abundant in BSD population. Contrarily, Chaetomium and Spizellomyces were highly abundant (P<0.0001; 4 to 7.8%) in BSW population as compared to other two samples (Fig 4; S3 Table).
Among fungal species, the unidentified category showed higher proportion in the sample. The BSA, BSD and BSW population shared 45.4%, 53.1% and 62.3% respectively (S2 Fig). Pezizaceae sp, Haematonectria haematococca, and Sebacinales sp. were the most abundant across the three populations. Among these three species, Pezizaceae sp was significantly (P<0.00001; 22.6%) abundant in BSW population (S2 Fig) as compared to other two populations. H. haematococca, on the other hand, was 3.7, 5.2 and 1.5% in BSA, BSD and BSW populations. However, Sebacinales sp. Exophiala xenobiotica, Veronaea botryose, Coprinopsis nivea, and Aspergillus niger were significantly higher (P<0.00001; ranging from 1 to 3.8%) in OTUs abundance in BSA population only (S2 Fig).
Bacterial diversity and the key players
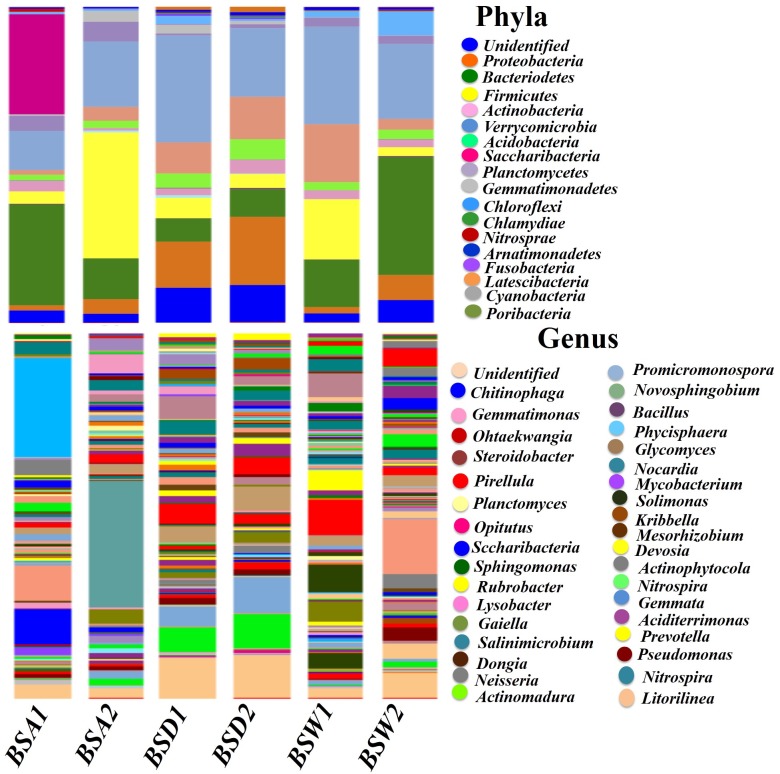
Acidobacteria, Actinobacteria, Bacteroidetes, Proteobacteria, Planctomycetes, and Gemmatimonadetes were the most highly (P<0.0001) abundant bacterial phyla in the rhizosphere of the three populations. However, these varied greatly in distribution across different samples (Fig 5; S4 Table). For example, Acidobacteria, Planctomycetes. Gemmatimonadetes and Proteobacteria were significantly higher (P<0.0001; from 5 to 29%) in BSD rhizosphere, whereas, Actinobacteria was significantly higher in BSW (P<0.0001; 27%). Bacteroidetes and Verrucomicrobia were significantly higher (P<0.01; from 4 to 28%) in BSA. Firmicutes was significantly higher (P<0.0032; 5.2%) in BSW as compared to other populations. Chlamydiae, Armatimonadetes, Chloroflexi, Candidatus, Latescibacteria, and Nitrospirae were not significantly different among three populations (Fig 5; S4 Table).
Fig 5. Abundances and shared core OTUs of bacterial phyla and genus from the rhizosphere of wild and cultivated populations of B. sacra tree.
Taxa with abundance <5% are presented as “Others/Unidentified”.
The abundances of genus among different population also varied greatly as around 191 different genera were observed throughout the data. For example, Chitinophaga, Steroidobacter, Opitutus, Ohtaekwangia, Streptomyces, Promicromonospora, Glycomyces, and Novosphingobium were significantly higher (P<0.0001; from 1.2 to 20.1%) in BSA rhizospheric samples as compared to BSD and BSW (Fig 5; S4 Table). Gemmatimonas, Pirellula, and Lysobacter were significantly abundant (P<0.0001; from 1 to 6%) in BSD rhizospheric samples as compared to BSA and BSW. Phenylobacterium, Planctomyces, and Sphingomonas (P<0.008; from 1 to 8.6%) were significantly abundant in BSW rhizosphere as compared to BSA and BSD samples (Fig 5; S4 Table). Whereas, Gemmatimonas, Ohtaekwangia, Gaiella, Mycobacterium, and Dongia were commonly distributed across the three rhizospheric samples from BSA, BSD and BSW (Fig 5; S4 Table). In case of bacterial species, the abundance of uncultured bacteria was high across all the three samples. However, Gamma-proteobacterium, Acidobacteria sp., Planctomycete sp., Firmicutes sp., Actinomycete sp., Escherichia coli, Prevotella nanceiensis, Chloroflexi sp., Rubrobacter sp., Lactobacillus reuteri, Bacillus niabensis, and Actinoplanes sp. were identified and distributed across the rhizosphere of BSA, BSD and BSW ranging from 0.5 to 6.28%.
Soil, exozymes and IAA analysis of the rhizosphere of three populations
The wild population are growing in extreme water deficiency–a desert environment, whereas, the cultivated are growing the same climate but facilitated with required water. The soil structure in these three regions (BSA, BSD and BSW) varied with respect to soil texture, pH, organic matter and nutrient concentration (Table 1), which may act as potential selection factors affecting rhizosphere communities and plant growth. Additionally, the physical structure of these three soils varied with BSA soil having a higher clay percentage (13.2%), as compared to BSD and BSW. Similarly, the highest sand percentage (88.8%) was found in BSW soil (Table 1). The Pearson’s correlation coefficients between the soil samples suggest that significantly higher (P<0.05) correlation exist for bulk density with clay, pH and nitrates; where the pH significantly (P<0.001) varies with nitrates content in the three soil samples (S5 Table).
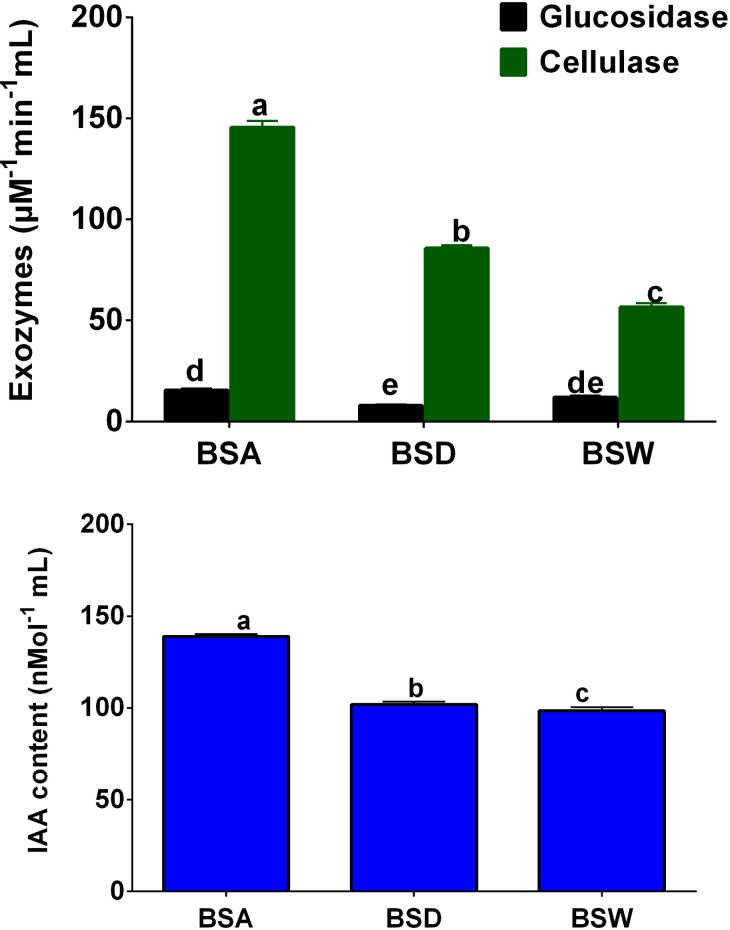
The glucosidases and cellulases were quantified in the rhizospheric samples of BSA, BSD and BSW populations. The results showed that cellulases was significantly higher (P<0.0001; 148.4±1.8μM/min/mL) in BSA rhizospheric samples, followed by BSD samples (91.8±2.3μM/min/mL; Fig 6). In the rhizosphere of wild B. sacra population, very significantly lower amount of cellulases was found. The glucosidases were found in very low concentrations in all the rhizospheric samples, however, BSA had significantly higher (P<0.018; 17.1±0.9μM/min/mL) amount as compared to other two populations. In case of IAA content in the rhizospheric samples of the three populations, a significantly higher amount was detected in BSA (P<0.001; 137.8±2.1nM/mL) samples as compared to other populations (Fig 6).
Fig 6. Exozymes (glucosidases and cellulases) and indole acetic acid quantification in rhizosphere of wild and cultivated populations of B. sacra tree.
Discussion
The current study investigated the microbiomes associated with the rhizosphere of three populations of Boswellia sacra tree. The tree is endemic to Oman and possess rich affiliation culturally and economically [9]. However, its various populations both cultivated and wild are frequently exposed to regenerative threats [37]. In this regard, rhizospheric microbial communities and their diversity has been shown to be retrospective of the host plant health, growth and development [38, 39]. Results showed varying distribution of microorganisms in the rhizosphere of the three populations. Many of the current insights into the interactions and processes of rhizospheric microbiome have emerged from studies on agricultural or horticultural crop plants and model species such as Arabidopsis thaliana and Medicago truncatula [40, 41]. However, a considerable progress is also being made in understanding the microbial ecology of the rhizosphere of non-cultivated plant species in natural ecosystems [42, 43] and how microorganisms influence resource allocation, biodiversity and above-ground interactions with herbivores and their natural enemies [44, 45].
Current study elucidated the fungal and bacterial microbial communities of three B. sacra tree populations, which showed a varying response in the metagenomic data output as well as the number of OTUs. This was also demonstrated in recent studies that host-specific traits, including broad morphological characteristics and specific genetic pathways and gene products [39, 44, 45], can have significant effects on microbiome composition and diversity [24, 38, 43]. Though the climatic and soil conditions are quite similar, however, the cultivated populations (BSA and BSD) are different due to regular supply of water as compared to BSW. The cultivated and non-cultivated plants and their rhizosphere have shown to possess a varying niche of symbionts. This was previously reported by Coleman-Derr et al. [24], who showed a significant difference among different populations of Agave plant, which share some similarity in growth conditions with B. sacra. In conformity, Peiffer et al. [38] also showed that the structure and diversity of bacterial communities vary significantly across the rhizosphere of the same plant species collected from different location. Though, we collected rhizospheric samples from the tree with the same age and height, however, in addition to varying conditions of soil, the plant itself can influence the distribution of microbiome through different phases of development. A similar conclusion was also drawn by Chaparro et al., [40], where the bacterial community diversity changed with the seedling, vegetative, bolting and flowering stages of model plant Arabidopsis. [46] also showed a similar conclusion from analyzing the rhizospheric microbiota of Solanum tuberosum growing in different climatic conditions. [47] also showed that the location and age of the host plant can drastically impact bacterial community abundances.
Our results showed that OTUs composition varied greatly among representative replica as well as the different rhizospheric units. This too can be co-related with the plant exudation, which in turn depends on the environmental conditions where it grows. In current case, since, the rhizosphere of wild population was much deprived from the essential growth resources, therefore, it was showing lesser abundance on OTUs. Climate on the other hand, implicates the abundance of different microbial communities, thus effecting on the total OTUs of the rhizosphere. Peiffer et al. [46] also reported that different climatic results in the variation of OTUs across potato rhizosphere. A similar conclusion was also drawn for the rhizosphere of Maize and Soybean plants [48]. However, such information is scarce for the wild and cultivated economical important trees. Though, there are a few studies on the tree rhizosphere and its microbiome such as Taxus [41], Quercus robur [47], Avicennia marina [49] and Populus [50, 51]]. However, current study elucidates the microbiomes of this economically important tree for the first time. In addition, most of the studies are restricted to bacterial communities, whereas, less emphasis has been made on the fungal communities, suggesting a future need to consider the holistic approach to understand the microbiome in the rhizosphere.
Besides the abundance, the distribution of microbial communities also differed across the three population of rhizosphere. Basidiomycota, Ascomycota, Glomeromycota and Zygomycota were abundant fungal phyla in the rhizosphere of the three populations. However, the unidentified proportion in the population varied greatly specially across BSW, suggesting that it could be much higher than the available in currently available data at the NCBI. In addition, the highest composition of Glomeromycota and Zygomycota in BSW suggest a novel niche of the microbiome to support plant growth in such a harsh climatic and nutrient deficient environment. The presence of these two phyla suggest greater deterrence against pathogenic infection to the host as was shown for the rhizospheric fungi of Panax notoginseng, where Zygomycota was 46.2% in abundance [52]. This was also suggested by Shakya et al. [50] and Li et al. [53] where they have observed higher abundance of Glomeromycota and Zygomycota in Populus deltoides and Pipper nigrum L rhizospheres. This finding is consistent with previous studies on arid land plants [24, 25]. In current results, the Haematonectria, Veronaea, Aspergillus and Coprinopsis were significantly abundant in BSA and BSD, suggesting a better arsenal of the host plant to protect from harmful pathogenic fungal attack to the root structure. This was also shown by Xiong et al. [54], who compared the fungal community composition of black pepper plants and their ability to counteract against Fusarium wilt disease. BSW, on the other hand, showed the greater abundance of Chaetomium and Pezizaceae as compared to BSD and BSA population, which are very least reported from the arid land ecosystem. Previously, these were also found in the root zones of date palms [55], and Agave [6].
In case of bacterial communities, Actinobacteria, Acidobacteria, Proteobacteria and Bacteroidete were significantly abundant bacterial phyla from the three rhizospheric populations. These are some of the predominantly abundant bacterial species found in the metagenomic dataset obtained from various plants and rhizosphere [22, 45]. These have also been reported in some of the important medicinal plants such as Agave species [6], Ginseng [56], Polygonum cuspidatum [41], Thymus zygis [26], Rhododendron arboreum [23], Sapindus saponaria [45], Taxus baccata and Aloe vera [22] etc. However, the distribution of core phyla such as Proteobacteria and Actinobacteria in BSW suggest higher potential of the host plant to tackle harsh environmental conditions. A similar conclusion was also shown by Kaplan et al. [57] and Marasco et al. [58], suggesting the higher abundance Proteobacteria and Actinobacteria in desert rhizosphere can help well. The significantly higher number of “Unidentified” sequences in bacteria could be due to (i) greater number of sequences of uncultured microbes, (ii) less sequenced microbial genomes, and/or (iii) absence of homologues sequence in NCBI [43].
Similarly, various abiotic factors, including soil depth, availability of soil nutrient, water content, temperature, aeration and soil management practices may affect the structure and activity of soil microbial community and abundances [59–62]. In current study, our results noticeably demonstrated that soil textural differences significantly affected bacterial population and that smaller size fractions (silt and clay) host higher bacterial community than larger size particles (sand). Among these soil textures we found that bacterial population varied greatly across different sample. The highest bacterial populations among different genus were significantly higher (1.2 to 20.1%) in BSA rhizospheric sample having higher percentage of clay and silt as compared to BSD and BSW (S1 Table). However, some genus like Sphingomonas and Planctomycetes were significantly abundant in BSW rhizosphere having highest percentage of sand (88.8%) as compared to BSD and BSA. Our results agree with previous studies that soil texture is one of the most important factors contributing soil microbial population [63–68]. Similarly, various studies reported that finer particle fractions were suitable for bacterial survival because smaller size particles provide a protective habitat for microorganism through pore size exclusion of predators [64, 69, 70]. Furthermore, higher bacterial diversity in clay and silty soil may be due to higher water contents, organic matter and nutrient availability [71, 72]. Differences in fungal community structure were observed in these samples too. Previously researchers reported effects of soil texture on fungal community and their abundances [73–75]. We found in our results that fungal communities were positively correlated with the soil sand contents and negatively with silt and clay. Similar results were reported by Wubet et al. [76] that fungal community and diversity were positively correlated to the first NMDS axis with the soil sand content and C:N ratio, while negative correlations were found with pH, silt, and clay content.
Besides, the distribution and diversity of specific microbiota, the ability of microbes in producing bioactive and potent substance in the rhizosphere have been considered essentially important for the host plant growth [77, 78]. In this mutualistic interaction, the production of bioactive metabolites such as phytohormones and extra-cellular enzymes etc can further pave a way for sustainable growth of the host [8]. As reported earlier that plant cell wall, predominantly composed of lignocellulose, serves as the main barrier to endophytic microbes. These microbes’ secrets numerous cell wall degrading enzymes, such as cellulase to break the plant cell wall to enter the plant [79]. Similarly Solomon and Matthews [80], demonstrated that the colonization of endophytes in plant internal tissues involved the production of cellulases and glucosidases [81, 82] such indicating the cell wall degrading enzymes were most likely a key determinant for the bacteria to initially enter and colonize the plant host to promote plant growth [44]. In our study, we found significantly higher amounts of cellulases as compared to glucosidases in BSA rhizosphere than BSD and BSW. A previous study showed that the endophytic symbionts with B. sacra can increase plant growth by producing IAA and extra-cellular enzymes [9]. These extracellular enzymes such as cellulases are key to maintain high organic matter for the host plant and terrestrial carbon cycle [83]. Increased amount of cellulases in BSA followed by BSD populations, suggest an active system of maintaining soil carbon influx, which on the other hand also shows a healthy soil life support system for the host. However, since, in the BSW rhizosphere, the tree populations are often confronted with extreme growth conditions, showing a minimal microbial activity, which is also in correlation with our MiSeq data for lower abundance of both fungal and bacterial communities. Ofek-Lalzar et al. [84] and Lopez-Mondejar et al. López-Mondéjar, Zühlke [83] also suggested some similar dynamics of rhizospheric communities. This is also in conformity with IAA content in BSA population. Where, IAA production has been shown a major feature of plant growth promoting activity of both fungal and bacterial rhizospheric communities as suggested by Adriaenssens [1], Cipriano et al.[85] and Khan et al.[9].
Conclusion
The current results provide a genomic baseline to further deepen our understanding of the complex microbe interactions with plants growing in arid land ecosystem especially with B. sacra. To a certain degree our results are in correlation with recent metagenomic data on the diversity of microbiomes associated with arid/semi-arid tree populations. These plants were studied for the first time. The identification of specific taxa particularly at genus levels can provide a new insight for future research work on the associated functions and inter-play of enriched species in rhizosphere of Boswellia species. We also predict that secretion of exozymes, essential metabolites and soil characteristics are the key to variation in the microbial community. Current report offer an opportunities and detailed insight of core microbiome in the rhizosphere of this economically and ecologically important frankincense producing tree species.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by The Oman Research Council (TRC) through ORG/EBR/15/007. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adriaenssens EM, Van Zyl L, De Maayer P, Rubagotti E, Rybicki E, Tuffin M, et al. Metagenomic analysis of the viral community in Namib Desert hypoliths. Environ Microbiol. 2015;17(2):480–95. doi: 10.1111/1462-2920.12528 [DOI] [PubMed] [Google Scholar]
- 2.Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Annual review of phytopathology. 2000;38(1):145–80. [DOI] [PubMed] [Google Scholar]
- 3.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microb. 2003;69(4):1875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–38. doi: 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- 5.Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux JP, L'Haridon F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210(3):1033–43. doi: 10.1111/nph.13808 [DOI] [PubMed] [Google Scholar]
- 6.Coleman‐Derr D, Desgarennes D, Fonseca‐Garcia C, Gross S, Clingenpeel S, Woyke T, et al. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016;209(2):798–811. doi: 10.1111/nph.13697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17(8):478–86. doi: 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 8.Allison SD, Vitousek PM. Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biology and Biochemistry. 2005;37(5):937–44. [Google Scholar]
- 9.Khan AL, Al-Harrasi A, Al-Rawahi A, Al-Farsi Z, Al-Mamari A, Waqas M, et al. Endophytic fungi from Frankincense tree improves host growth and produces extracellular enzymes and indole acetic acid. Plos One. 2016;11(6):e0158207 doi: 10.1371/journal.pone.0158207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. The ISME journal. 2012;6(7):1378 doi: 10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffaelli M, Mosti S, Tardelli M. The Frankincense Tree (Boswellia sacra Flueck., Burseraceae) in Dhofar, southern Oman: field-investigations on the natural populations. Webbia. 2003;58(1):133–49. [Google Scholar]
- 12.Gebrehiwot K, Muys B, Haile M, Mitloehner R. Introducing Boswellia papyrifera (Del.) Hochst and its non-timber forest product, frankincense. International forestry review. 2003;5(4):348–53. [Google Scholar]
- 13.Al-Harrasi A, Al-Saidi S. Phytochemical analysis of the essential oil from botanically certified oleogum resin of Boswellia sacra (Omani Luban). Molecules. 2008;13(9):2181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi M, Sung B, Shen Y, Hur K, Link A, Boland CR, et al. Boswellic acid exerts antitumor effects in colorectal cancer cells by modulating expression of the let-7 and miR-200 microRNA family. Carcinogenesis. 2012;33(12):2441–9. doi: 10.1093/carcin/bgs286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farah MH. Non-timber forest product (NTFP) extraction in arid environments: land-use change, frankincense production and the sustainability of Boswellia sacra in Dhofar (Oman): The University of Arizona; 2008. [Google Scholar]
- 16.Eshete A, Sterck FJ, Bongers F. Frankincense production is determined by tree size and tapping frequency and intensity. Forest ecology and management. 2012;274:136–42. [Google Scholar]
- 17.Tolera M, Sass-Klaassen U, Eshete A, Bongers F, Sterck FJ. Frankincense tree recruitment failed over the past half century. Forest Ecology and Management. 2013;304:65–72. [Google Scholar]
- 18.El-Nagerabi SA, Elshafie AE, Alkhanjari SS. Endophytic fungi associated with endogenous Boswellia sacra. Biodiversitas Journal of Biological Diversity. 2013;15(1). [Google Scholar]
- 19.Wu L, Wang H, Zhang Z, Lin R, Zhang Z, Lin W. Comparative metaproteomic analysis on consecutively Rehmannia glutinosa-monocultured rhizosphere soil. Plos One. 2011;6(5):e20611 doi: 10.1371/journal.pone.0020611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi X, Wang E, Xing M, Zhao W, Chen X. Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World Journal of Microbiology and Biotechnology. 2012;28(5):2257–65. doi: 10.1007/s11274-012-1033-2 [DOI] [PubMed] [Google Scholar]
- 21.Song SM, Mu J, Hu WL, Xiao PG. Unearthing microbial diversity of Taxus rhizosphere via MiSeq high-throughput amplicon sequencing and isolate characterization. Sci Rep-Uk. 2016;6:22006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akinsanya MA, Goh JK, Lim SP, Ting ASY. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genomics data. 2015;6:159–63. doi: 10.1016/j.gdata.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debnath R, Yadav A, Gupta VK, Singh BP, Handique PJ, Saikia R. Rhizospheric Bacterial Community of Endemic Rhododendron arboreum Sm. Ssp. delavayi along Eastern Himalayan Slope in Tawang. Front Plant Sci. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, et al. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016;209(2):798–811. Epub 2015/10/16. doi: 10.1111/nph.13697 ; PubMed Central PMCID: PMCPMC5057366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fonseca-García C, Coleman-Derr D, Garrido E, Visel A, Tringe SG, Partida-Martínez LP. The cacti microbiome: interplay between habitat-filtering and host-specificity. Front Microbiol. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pascual J, Blanco S, García-López M, García-Salamanca A, Bursakov SA, Genilloud O, et al. Assessing bacterial diversity in the rhizosphere of Thymus zygis growing in the Sierra Nevada National Park (Spain) through culture-dependent and independent approaches. Plos One. 2016;11(1):e0146558 doi: 10.1371/journal.pone.0146558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adhikari P, Shukla MK, Mexal JG. Spatial variability of electrical conductivity of desert soil irrigated with treated wastewater: implications for irrigation management. Applied and Environmental Soil Science. 2011;2011. [Google Scholar]
- 28.Marx M-C, Wood M, Jarvis S. A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil biology and biochemistry. 2001;33(12):1633–40. [Google Scholar]
- 29.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. doi: 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73(16):5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Fu L, Niu B, Wu S, Wooley J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief Bioinform. 2012;13(6):656–68. doi: 10.1093/bib/bbs035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 33.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86 doi: 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. doi: 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu P, Xiong X, Xu Z, Lu C, Cheng H, Lyu X, et al. Bacterial Communities in the Rhizospheres of Three Mangrove Tree Species from Beilun Estuary, China. Plos One. 2016;11(10):e0164082 doi: 10.1371/journal.pone.0164082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeiffer S, Mitter B, Oswald A, Schloter-Hai B, Schloter M, Declerck S, et al. Rhizosphere microbiomes of potato cultivated in the High Andes show stable and dynamic core microbiomes with different responses to plant development. Fems Microbiol Ecol. 2017;93(2). [DOI] [PubMed] [Google Scholar]
- 39.Taketani RG, Lançoni MD, Kavamura VN, Durrer A, Andreote FD, Melo IS. Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microbial ecology. 2017;73(1):153–61. doi: 10.1007/s00248-016-0835-4 [DOI] [PubMed] [Google Scholar]
- 40.Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. The ISME journal. 2014;8(4):790 doi: 10.1038/ismej.2013.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Da Cheng Hao SMS, Mu J, Hu WL, Xiao PG. Unearthing microbial diversity of Taxus rhizosphere via MiSeq high-throughput amplicon sequencing and isolate characterization. Sci Rep-Uk. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17(8):478–86. Epub 2012/05/09. doi: 10.1016/j.tplants.2012.04.001 . [DOI] [PubMed] [Google Scholar]
- 43.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206(4):1196–206. doi: 10.1111/nph.13312 [DOI] [PubMed] [Google Scholar]
- 44.Tian B-Y, Cao Y, Zhang K-Q. Metagenomic insights into communities, functions of endophytes, and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci Rep-Uk. 2015;5:srep17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia A, Polonio J, Polli A, Santos C, Rhoden S, Quecine M, et al. Rhizosphere bacteriome of the medicinal plant Sapindus saponaria L. revealed by pyrosequencing. Genetics and molecular research: GMR. 2016;15(4). [DOI] [PubMed] [Google Scholar]
- 46.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proceedings of the National Academy of Sciences. 2013;110(16):6548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meaden S, Metcalf C, Koskella B. The effects of host age and spatial location on bacterial community composition in the English Oak tree (Quercus robur). Env Microbiol Rep. 2016;8(5):649–58. [DOI] [PubMed] [Google Scholar]
- 48.Guo M, Gong Z, Miao R, Su D, Li X, Jia C, et al. The influence of root exudates of maize and soybean on polycyclic aromatic hydrocarbons degradation and soil bacterial community structure. Ecological Engineering. 2017;99:22–30. [Google Scholar]
- 49.Alzubaidy H, Essack M, Malas TB, Bokhari A, Motwalli O, Kamanu FK, et al. Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene. 2016;576(2):626–36. [DOI] [PubMed] [Google Scholar]
- 50.Shakya M, Gottel N, Castro H, Yang ZK, Gunter L, Labbé J, et al. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. Plos One. 2013;8(10):e76382 doi: 10.1371/journal.pone.0076382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hacquard S, Schadt CW. Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 2015;205(4):1424–30. doi: 10.1111/nph.13133 [DOI] [PubMed] [Google Scholar]
- 52.Miao C-P, Mi Q-L, Qiao X-G, Zheng Y-K, Chen Y-W, Xu L-H, et al. Rhizospheric fungi of Panax notoginseng: diversity and antagonism to host phytopathogens. Journal of ginseng research. 2016;40(2):127–34. doi: 10.1016/j.jgr.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Zu C, Wang C, Yang J, Yu H, Wu H. Different responses of rhizosphere and non-rhizosphere soil microbial communities to consecutive Piper nigrum L. monoculture. Sci Rep-Uk. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong W, Zhao Q, Xue C, Xun W, Zhao J, Wu H, et al. Comparison of fungal community in black pepper-vanilla and vanilla monoculture systems associated with vanilla Fusarium wilt disease. Front Microbiol. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Khayri JM, Jain SM, Johnson DV. Date Palm Genetic Resourse and Utilization: Springer; 2015. [Google Scholar]
- 56.Tan Y, Cui Y, Li H, Kuang A, Li X, Wei Y, et al. Diversity and composition of rhizospheric soil and root endogenous bacteria in Panax notoginseng during continuous cropping practices. J Basic Microb. 2017;57(4):337–44. [DOI] [PubMed] [Google Scholar]
- 57.Kaplan D, Maymon M, Agapakis CM, Lee A, Wang A, Prigge BA, et al. A survey of the microbial community in the rhizosphere of two dominant shrubs of the Negev Desert highlands, Zygophyllum dumosum (Zygophyllaceae) and Atriplex halimus (Amaranthaceae), using cultivation-dependent and cultivation-independent methods. Am J Bot. 2013;100(9):1713–25. doi: 10.3732/ajb.1200615 [DOI] [PubMed] [Google Scholar]
- 58.Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, et al. A drought resistance-promoting microbiome is selected by root system under desert farming. Plos One. 2012;7(10):e48479 doi: 10.1371/journal.pone.0048479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marschner P, Yang C-H, Lieberei R, Crowley D. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biology and Biochemistry. 2001;33(11):1437–45. [Google Scholar]
- 60.Griffiths RI, Whiteley AS, O'donnell AG, Bailey MJ. Influence of depth and sampling time on bacterial community structure in an upland grassland soil. Fems Microbiol Ecol. 2003;43(1):35–43. doi: 10.1111/j.1574-6941.2003.tb01043.x [DOI] [PubMed] [Google Scholar]
- 61.Lauber CL, Strickland MS, Bradford MA, Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biology and Biochemistry. 2008;40(9):2407–15. [Google Scholar]
- 62.Andrew DR, Fitak RR, Munguia-Vega A, Racolta A, Martinson VG, Dontsova K. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl Environ Microb. 2012;78(21):7527–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microb. 2003;69(3):1800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, Zheng J, Pan G, Zhang X, Li L, Rolf T. Changes in microbial community structure and function within particle size fractions of a paddy soil under different long-term fertilization treatments from the Tai Lake region, China. Colloids and Surfaces B: Biointerfaces. 2007;58(2):264–70. doi: 10.1016/j.colsurfb.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 65.Fang M, Motavalli PP, Kremer RJ, Nelson KA. Assessing changes in soil microbial communities and carbon mineralization in Bt and non-Bt corn residue-amended soils. Appl Soil Ecol. 2007;37(1):150–60. [Google Scholar]
- 66.Kanazawa S, Filip Z. Distribution of microorganisms, total biomass, and enzyme activities in different particles of brown soil. Microbial Ecology. 1986;12(2):205–15. doi: 10.1007/BF02011205 [DOI] [PubMed] [Google Scholar]
- 67.Buyer JS, Roberts DP, Russek-Cohen E. Microbial community structure and function in the spermosphere as affected by soil and seed type. Can J Microbiol. 1999;45(2):138–44. [Google Scholar]
- 68.Hamarashid NH, Othman MA, Hussain M-AH. Effects of soil texture on chemical compositions, microbial populations and carbon mineralization in soil. Egypt J Exp Biol(Bot). 2010;6(1):59–64. [Google Scholar]
- 69.Postma J, Van Veen J. Habitable pore space and survival ofRhizobium leguminosarum biovartrifolii introduced into soil. Microbial Ecology. 1990;19(2):149–61. doi: 10.1007/BF02012096 [DOI] [PubMed] [Google Scholar]
- 70.Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microb. 2001;67(9):4215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grayston S, Campbell C, Bardgett R, Mawdsley J, Clegg C, Ritz K, et al. Assessing shifts in microbial community structure across a range of grasslands of differing management intensity using CLPP, PLFA and community DNA techniques. Appl Soil Ecol. 2004;25(1):63–84. [Google Scholar]
- 72.Cookson WR, Abaye DA, Marschner P, Murphy DV, Stockdale EA, Goulding KW. The contribution of soil organic matter fractions to carbon and nitrogen mineralization and microbial community size and structure. Soil Biology and Biochemistry. 2005;37(9):1726–37. [Google Scholar]
- 73.Burke DJ, López-Gutiérrez JC, Smemo KA, Chan CR. Vegetation and soil environment influence the spatial distribution of root-associated fungi in a mature beech-maple forest. Appl Environ Microb. 2009;75(24):7639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jumpponen A, Jones KL, David Mattox J, Yaege C. Massively parallel 454‐sequencing of fungal communities in Quercus spp. ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol Ecol. 2010;19(s1):41–53. [DOI] [PubMed] [Google Scholar]
- 75.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microb. 2009;75(15):5111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wubet T, Christ S, Schöning I, Boch S, Gawlich M, Schnabel B, et al. Differences in soil fungal communities between European beech (Fagus sylvatica L.) dominated forests are related to soil and understory vegetation. Plos One. 2012;7(10):e47500 doi: 10.1371/journal.pone.0047500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan AL, Hamayun M, Kang S-M, Kim Y-H, Jung H-Y, Lee J-H, et al. Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC microbiology. 2012;12(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khan AL, Hussain J, Al-Harrasi A, Al-Rawahi A, Lee I-J. Endophytic fungi: resource for gibberellins and crop abiotic stress resistance. Critical reviews in biotechnology. 2015;35(1):62–74. doi: 10.3109/07388551.2013.800018 [DOI] [PubMed] [Google Scholar]
- 79.Lagaert S, Beliën T, Volckaert G, editors. Plant cell walls: Protecting the barrier from degradation by microbial enzymes. Semin Cell Dev Biol; 2009: Elsevier. [DOI] [PubMed] [Google Scholar]
- 80.Solomon EB, Matthews KR. Use of fluorescent microspheres as a tool to investigate bacterial interactions with growing plants. Journal of food protection. 2005;68(4):870–3. [DOI] [PubMed] [Google Scholar]
- 81.Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe In. 2006;19(8):827–37. [DOI] [PubMed] [Google Scholar]
- 82.Reinhold-Hurek B, Maes T, Gemmer S, Van Montagu M, Hurek T. An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol Plant Microbe In. 2006;19(2):181–8. [DOI] [PubMed] [Google Scholar]
- 83.López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep-Uk. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ofek-Lalzar M, Sela N, Goldman-Voronov M, Green SJ, Hadar Y, Minz D. Niche and host-associated functional signatures of the root surface microbiome. Nat Commun. 2014;5:4950 doi: 10.1038/ncomms5950 [DOI] [PubMed] [Google Scholar]
- 85.Cipriano MA, Lupatini M, Lopes-Santos L, da Silva MJ, Roesch LF, Destéfano SA, et al. Lettuce and rhizosphere microbiome responses to growth promoting Pseudomonas species under field conditions. Fems Microbiol Ecol. 2016;92(12). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.