Abstract
Silver nanoparticles (AgNP) are promising candidates for fighting drug-resistant infections because of their intrinsic antimicrobial effect. The design of high-yield antimicrobial molecules may inadvertently cause variation in host cells’ biological responses. While many factors affect AgNPs’ efficacy, their surface is exposed to the biological environment and thus plays a critical role in both the preservation of antimicrobial efficacy against pathogens and the modulation of host cells cytotoxicity. This work investigated an engineered biomimetic interface approach to controlling AgNP surface properties to provide them a competitive advantage in a biological environment. Here, a fusion protein featuring a silver-binding peptide (AgBP) domain was engineered to enable self-assembly and track assembly by a green fluorescent protein (GFP) reporter. Following AgNP functionalisation with GFP–AgBP, their antimicrobial and cytotoxic properties were evaluated. GFP–AgBP binding affinity to AgNPs was evaluated using localized surface plasmon resonance sensing. The GFP–AgBP biomimetic interface on AgNPs’ surfaces provided sustained antibacterial efficacy at low concentrations based on bacterial growth inhibition assays. Viability and cytotoxicity measurements in fibroblast cells exposed to GFP–AgBP protein-functionalised AgNPs showed significant improvement compared to controls. Biointerface engineering offers promise towards tailoring AgNP antimicrobial efficacy while addressing safety concerns to maintain optimum cellular interactions.
Keywords: anti-microbial, biointerface, nanoparticles
1. Introduction
The high rates of biomaterial-associated infections and the global healthcare burden associated with the increase in drug-resistant bacteria have prompted significant efforts to develop effective antimicrobial agents.1,2 For the oral cavity, infections can occur in approximately 5% of implanted dental devices, with these infections having a broad adverse systemic impact.3–5 In the USA, nearly 300 000 dental implants are placed each year to replace or restore craniofacial defects or missing teeth.6 The high number of craniofacial infection is primarily attributed to poor material compatibility, the volatile environment with numerous oral bacteria and the formation of an impenetrable biofilm.7 These biofilms shield the bacteria from shear stresses, traditional antibiotics and native immune responses, while simultaneously allowing some of the bacteria to escape and invade new surfaces at remote sites.1,8,9 Furthermore, antibiotic drugs relied on to fight local and systemic infections are being undermined by bacterial resistance – so much so that numerous authoritative health institutions have fervently informed the global population about the burgeoning threat and imminent health crisis regarding antibiotic-resistant bacterial strains.10,11 There has been an increasingly significant effort to develop antimicrobial agents to combat drug-resistant bacteria while protecting patients from systemic infections and their consequences. Innovative approaches to biomaterial design for dental implants have focused on mitigating the risk of infection at the implant–tissue interface by coating or functionalising the implant surface with various antimicrobial moieties.9
Numerous antimicrobial moieties and device coatings have been clinically approved to combat the persistent and challenging burden of infections by either limiting bacterial adhesion or actively killing the bacteria upon direct contact.12 Silver (Ag) species,13–24 metal oxides,25–29 various proteins and peptides,30–36 charged polymers,37–39 quaternary ammonium salts40,41 and selenium species42,43 are among the various agents which have been identified as promising weapons in this combat. Silver species, and specifically silver nanoparticles (NPs), have received significant attention due to their distinctive physico-chemical and antimicrobial properties. Prior to the introduction of antibiotics in the 1940s, silver was the most important antimicrobial agent in medicine, utilised for over six millennia to combat infection.13 Even though silver has been used without any documented cases of bacterial resistance, cellular toxicity reports have triggered major safety concerns about silver NPs compromising healthy tissue in humans. The antimicrobial efficacy and the potential cytotoxicity of silver NPs in culture have been shown to be highly dependent on many factors, including, but not limited to, NP size, concentration, stabilisation, charge or surface functionalisation properties. The ability to tune and control the competing properties of biological and biocidal effects is critical in determining an optimal compromise between maintaining antimicrobial properties while mitigating risk of adverse host cytotoxic impacts.
Many studies have shown that the overall bactericidal efficacy is highly dependent on the size and the shape of the silver NPs used.20,22,44–46 The general trend observed indicates that smaller NPs exhibit a more potent antimicrobial effect.23,45 Martinez-Castanon et al.22 showed that minimum inhibitory concentration (MIC) of silver NPs on Staphylococcus aureus strains changes from 7·5 to 16·67 μg/ml and eventually to 33·71 μg/ml as the NPs’ size changes from 7 to 29 nm and to 89 nm respectively. Most prior work performed on size-dependent antimicrobial effect of silver NPs was against Escherichia coli as a test bacterial strain. However, with rising interest in the oral microbiome and bacteria-related caries destruction of biomineralised tooth tissue, many groups started to focus on oral pathogens, since dental caries remains the most prevalent infectious disease of mankind.47 For one of the most abundant oral pathogens, Streptococcus mutans, the size-dependent effect was shown for NPs with sizes of 8·4, 16·1 and 98 nm, where the MIC changed to 102, 146 and 320 μg/ml respectively.18 The same trend was also recently shown on a different oral pathogen strain, where the smallest NPs (5 nm) were found to have the largest antimicrobial effect.20 For example, for S. mutans and S. sanguis, an MIC of 100 μg/ml was observed for 5-nm silver NPs compared to an MIC of 200 μg/ml in the case of 55-nm silver NPs.23 The size dependence of the activity may be explained by one mechanism of silver NP action against bacteria, namely the interaction of the particle with the bacterial cell membrane with the consequential reduction in cell membrane permeability and respiration. In this case, the smaller particles have a greater ability to cover a larger area of the membrane, thus having a more potent effect on altering vital cellular functions.
Silver NPs have also been evaluated for their cytotoxic effect on local tissue and host cell integrity. Size-dependent cytotoxic effects have also been observed by several groups. Hernandez-Sierra et al.19 investigated the cytotoxic effects of silver NPs, sized 10, 20, and 80 nm, on local dental tissues, evaluated following exposure times of 1, 3 and 7 d. Results indicated that silver NPs 20 nm or smaller increased cytotoxicity in both a time-and dose-dependent matter when compared to 80–100-nm silver NPs, which did not significantly modify the viability of cultured primary human cells.19 Silver NPs with a size range of 10–25 nm were reported to be more likely to promote cellular apoptosis, as well as increased reactive oxygen species production, when compared to larger silver NPs with a size of 80 nm.48 Results measuring the cytotoxic effects of 10-nm silver NPs on lung epithelial cells were shown to be surface coating independent when compared to those for silver NPs sized 40 and 75 nm.44 Another group reported that 200 μM of 20 nm silver NPs did not induce cytotoxic effects in human dermal fibroblast cells after incubation for up to 8 h.49 Interestingly, another study reported a subsequent apoptosis of a fibroblast cell line treated with 1–100-nm silver NPs.50 Overall, an optimal silver NP size may be dependent on its intended use and application, where a balance must be established to select a silver particle size at which the antimicrobial properties are maintained while cytotoxic effects to host cells are mitigated.
These contradictory reports support the fact that it is critical to investigate surface functionalisation techniques as a way to mediate antimicrobial properties and cytotoxicity. Coating or functionalising silver NPs’ surfaces with polymers or biomolecules, including proteins, alters the silver NPs’ size and properties due to the formation of a coronal structure formed upon subsequent contact with biological fluids from host tissues.3,30,33,36,51 Hard and soft coronal structures and compositions alter the silver NP structure and size and therefore can affect both their innate antimicrobial properties and cellular cytotoxicity.33,35 The effects of the transient, soft corona layer on silver NP function and cellular uptake have not yet been studied in detail due to the inability to distinguish accurately the rapidly exchanging, weakly bound proteins in various biological environments.51 However, the sustained complex protein corona properties have been linked to NP aggregation and cellular association.36,51 Agglomeration substantially reduces the antimicrobial capacity of silver NPs, so maintaining a dispersed and stable silver NP population is critical. There is strong evidence that different protein components existing in biological environments may compete for the silver NP surface and create a secondary protein corona signature. The complex protein corona composition of silver NPs in the host cell environment may trigger different biological responses affecting cellular and bio-physico-chemical mechanisms in the host cells, which are of utmost importance for deciphering and addressing the safety concerns associated with the use of silver NPs as antimicrobial agents.
In this work, the authors explored a biomimetic interface as a surface functionalisation approach on silver NPs using engineered fusion proteins able to self-assemble at the biomaterial interface. Over the past decade, inorganic binding peptides (BPs) have been proven as robust surface functionalisation agents on different metal and metal oxide surfaces.32,52–54 In the authors’ previous work, a multifunctional fusion protein was designed, which included a silver BP, identified using phage display technology, coupled to a green fluorescent protein (GFP).34 It was demonstrated that the silver BP tag directs the fusion protein to self-immobilise selectively onto silver NP arrays on ferroelectric materials.55 Herein, the authors explored the bactericidal and cytotoxic effects of their fusion protein-stabilised silver NPs. The binding and stability of the fusion protein to the NPs was first investigated using localised surface plasmon resonance (LSPR) sensing. With the notable selective binding obtained at the NP interfacial surface, the antimicrobial effect of the fusion protein-coated particles on the S. mutans bacterial strain was explored next. Encouraged with the minimum inhibition concentration obtained with relatively low silver NP concentration, the authors next demonstrated a significant reduction in cytotoxicity effects on fibroblast cells and, in addition, observed an improved metabolic activity for treated cells compared to control silver NPs. The biomimetic interface approach as a surface functionalisation technique could be expanded to other nanoscale material surfaces where specific inorganic BPs could be engineered into other functional proteins and peptides to provide a competitive advantage by building on biological self-assembly.
2. Materials and methods
2.1 Production of GFP–silver BP fusion protein
Maltose-binding peptide (MBP)-tagged proteins were expressed and purified as previously described.34,56 Two MBP-tagged proteins were expressed in this work to include GFP or GFP with silver BP and denoted as MBP–GFP and MBP–GFP–silver BP respectively. Briefly, following protein expression, cells were harvested by way of centrifugation at 4000 relative centrifugal force (rcf) for 30 min. Cell pellets were resuspended in column buffer (20 mM Tris–hydrochloric acid (HCl), 200 mM sodium chloride (NaCl), 1 mM ethylenediaminetetraacetic acid (Edta; pH 7·4)). Cells were disrupted by way of sonication, and cell debris was removed by centrifugation. Supernatant was loaded onto a buffer-equilibrated amylose resin column (New England Biolabs) and washed to remove unbound proteins. MBP-tagged proteins were eluted using column buffer supplemented with 10 mM maltose. Pure protein fractions were concentrated to 1 mg/ml by using Amicon Ultra-15 centrifugal filters (Millipore) and transferred to a cleavage buffer (20 mM Tris-hydrochloric acid, 100 mM sodium chloride and 2 mM calcium chloride (CaCl2), pH 8·0). The MBP tag was enzymatically removed using 30 μl of 1 mg/ml factor Xa (New England Biolabs) added to a 2·5 mg/ml fusion protein substrate. The reaction was incubated at 16°C overnight under gentle agitation. Each fusion protein cleavage mixture was loaded onto a hydroxyapatite (Bio-Rad) column equilibrated in 20 mM sodium phosphate and 200 mM sodium chloride (pH 7·2) buffer. After maltose molecules were washed away, protein samples were eluted with 0·5 M sodium phosphate (pH 7·2). Hydroxyapatite-eluted protein samples were next loaded onto a regenerated amylose resin column. MBP-free protein samples were collected as the flow-through. Protein purity was assessed using sodium dodecyl sulfate polyacrylamide gel electrophoresis.
2.2 Silver NP functionalisation
Stock, citrate-coated silver NPs sized 20, 40 and 80 nm (Ted Pella) were diluted to desired concentrations. Forty-nanometre silver NPs were selected as the ideal size for detailed analysis, reported herein, following initial experimental results on cellular cytotoxicity and bactericidal efficacy. The stock concentration for the 40-nm silver NPs was provided as 0·024 mg/ml of silver. Silver NPs were incubated with a range of protein concentrations for both GFP and GFP–silver BP. Silver NPs were diluted from the stock in purified deionised (DI) water (18·2 MΩ cm, pH ≅ 7·0) and allowed to mix with protein for 24 h at room temperature (25°C) with gentle agitation (final pH 7·0).
The binding affinity and stability of the engineered bifunctional protein, GFP–silver BP, onto silver NPs was studied using LSPR. Silver NPs were diluted in purified DI water from the stock to achieve a final concentration of 40 μM. A range of protein concentrations of either GFP alone or GFP–silver BP were compared to silver NPs diluted in pure DI water only. LSPR was performed on silver NPs prior to exposure to proteins or water control. Following 24 h of incubation time, absorbance spectra were taken to study spectral peak shifts due to changes in silver NP size or shape and again following a wash step. Washes were done for all silver NP samples by pelleting by way of centrifugation for 30 min at 3000 rcf. The supernatant was carefully collected to remove unbound proteins, and silver NPs were then reconstituted in purified DI water and subjected to final LSPR analysis.
Silver NP functionalisation for antibacterial and cellular viability studies were performed under similar conditions. Stock, citrate-coated 40-nm silver NPs were diluted to 2× final concentrations and incubated with 40 μM GFP–silver BP functional peptide or were diluted in DI water for only 2 h at room temperature with gentle mixing (the 2 h of incubation time was selected following time-dependent binding analysis, not reported herein). Each sample was pelleted by way of centrifugation and resuspended in either brain–heart infusion (BHI) medium or Dulbecco’s modified Eagle’s medium (DMEM) for use in bacterial assays or fibroblast cell viability assays, respectively.
2.3 Antimicrobial activity assays
The antimicrobial activity of the GFP–silver BP-functionalised and citrate-coated silver NPs was studied in the S. mutans (American Type Culture Collection (ATCC) 25175) bacterial strain. The lyophilised bacteria were reconstituted in BHI medium (BD Difco), consistent with the ATCC standard protocol, and streaked onto a BHI agar (BD Difco) plate. The S. mutans plate was incubated for 24 h at 37°C and in the presence of 5% carbon dioxide (CO2). Overnight cultures were prepared prior to each experiment by inoculating 10 ml of fresh BHI medium with a single colony of bacteria and incubating for 16 h. The S. mutans cultures were then diluted, and growth was monitored by way of optical density measurements at 600 nm on a Cytation 3 imaging multimode plate reader (BioTek) until a final concentration of 107 colony-forming units (CFUs)/ml was reached.
Bacterial disc diffusion assay was carried to test the antimicrobial effect of NPs. BHI agar plates were prepared and stored at 4°C and allowed to acclimate to room temperature 30 min prior to experiments. Spread onto a pre-prepared BHI agar plate were 106 CFUs of S. mutans through the aseptic technique. After 5 min, loaded discs were placed into each labelled quadrant of the plate. Porous, 30-μm polyethylene filter discs were utilised to allow for uninhibited silver NP diffusion. Each disc was soaked in desired concentrations of protein-stabilised silver NPs and compared to the corresponding concentrations of the stock, citrate-coated silver NPs. A positive control of 10 μg/ml ampicillin (Sigma) in DI water was employed, and DI water was used as a negative control. Following placement of the discs, S. mutans plates were incubated for 24 h at 37°C with 5% carbon dioxide. The average diameter of the inhibition zone around each disc was measured using a digital calliper. The measured disc diameter was subtracted from the average zone of inhibition calculated for each treatment to normalise measurements for data analysis and representation. Duplicate samples were prepared for each silver NP concentration studied. Experiments were repeated three times, each assay using an S. mutans grown from a different individual colony.
2.4 Cellular cytotoxicity assays
The murine-derived fibroblast cell line NIH/3T3 (ATCC CRL-1658) was used as a preliminary model to determine any effects that the functionalised silver NPs had on cellular viability and cytotoxicity. Culture conditions followed the ATCC protocol and previously established methods. Briefly, cells were grown at 37°C, supplemented with 5% carbon dioxide in air in DMEM (ATCC 30-2002) containing 10% foetal bovine serum (Gibco) and 1% penicillin–streptomycin (Gibco). The cell culture medium was changed the day after plating, and complete media changes were conducted every 48 h. Cells were subcultured at 80% confluency or biweekly with 0·25% trypsin–Edta (Gibco).
To follow cellular viability, NIH/3T3 cells were first seeded into 96-well plates with black side/clear (BrandTech) at 104 cells/well. Following complete cell attachment to plate, the medium was changed and various concentrations of untreated (stock, citrate-coated) and GFP–silver BP protein-functionalised silver NPs were added to each well and allowed to incubate for 24 h at 37°C with 5% carbon dioxide. To evaluate cellular viability, 10% v/v of AlamarBlue reagent (Life Technologies) was added to each treated well at the 22 h time point, allowing cells to metabolise the reagent for about 2 h prior to the time point of 24 h. Both absorbance and fluorescence measurements were recorded, according to company protocol, to assess quantitatively cellular viability following 24 h of incubation with silver NP treatments.
Cellular viability and cytotoxicity was also quantified using the Live/Dead viability/cytotoxicity kit (Life Technologies) to indicate intracellular esterase activity and plasma membrane integrity. Cells were plated and treated in 96-well plates following the same protocol mentioned earlier. Following a 24-h incubation, cells were imaged using both bright-field and fluorescence imaging (BioTek Cytation 3) to observe cell morphology and quantify cellular viability/cytotoxicity respectively. The calcein acetoxymethyl ester (green fluorescent) stain identified healthy, viable cells, whereas ethidium homodimer-1 (red fluorescent) labelled the loss of the membrane integrity of dead cells. Images were taken for each well to determine the fluorescent contribution of the GFP-containing protein prior to Live/Dead staining and were subtracted for accurate staining analysis. Plates were treated such that eight repeats of each silver NP concentration were averaged and compared to respective cell-only controls. A minimum of three plates were used for each concentration and treatment for analysis and normalised to respective plate controls. All results are presented as a mean ± standard deviation.
3. Results and discussion
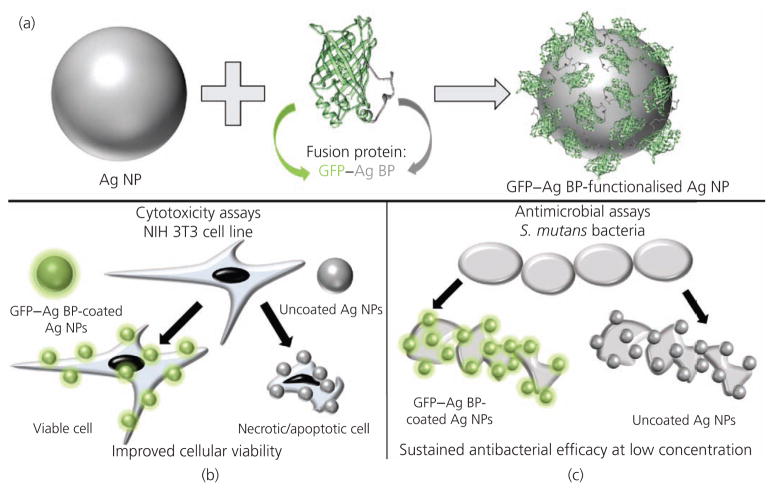
The schematic drawing shown in Figure 1 demonstrates the multifunctional fusion protein approach described in this work. The authors developed a robust NP biointerface by using the GFP engineered to include a silver BP to produce a multifunctional fusion protein that self-assembles on the surface of a silver NP. Silver NPs functionalised with the GFP–silver BP fusion proteins were tested for antimicrobial activity over a range of concentrations. The same concentrations were used to conduct cytotoxicity and cell viability assays to determine any differences in the response between cells treated with stock NPs compared to silver NPs with a biomimetic protein interface.
Figure 1.
Schematic diagrams demonstrating (a) the GFP–silver BP functionalisation of silver NPs and (b) the observed improvement in cellular viability in cell culture while (c) maintaining the inherent antibacterial efficacy at relatively low concentrations
3.1 Characterisation of silver NPs
Prior to characterisation of silver NPs, preliminary data (not shown) was collected to determine an optimal silver NP size to conduct experiments based on cytotoxicity studies using NIH/3T3 cells as well as antimicrobial diffusion assays conducted against S. mutans bacteria. Silver NPs sized 20, 40 and 80 nm were selected for preliminary analysis to determine an optimised NP size to explore in greater detail herein. Consistent with literature, NIH/3T3 cells exposed to 20-nm silver NPs were observed to develop cytotoxic effects compared to 40- and 80-nm silver NPs prepared at the same concentrations.19,20,44 The authors also observed a limited antimicrobial activity against S. mutans using 80-nm silver NPs.23,45 Therefore, the authors selected 40-nm silver NPs to conduct their studies.
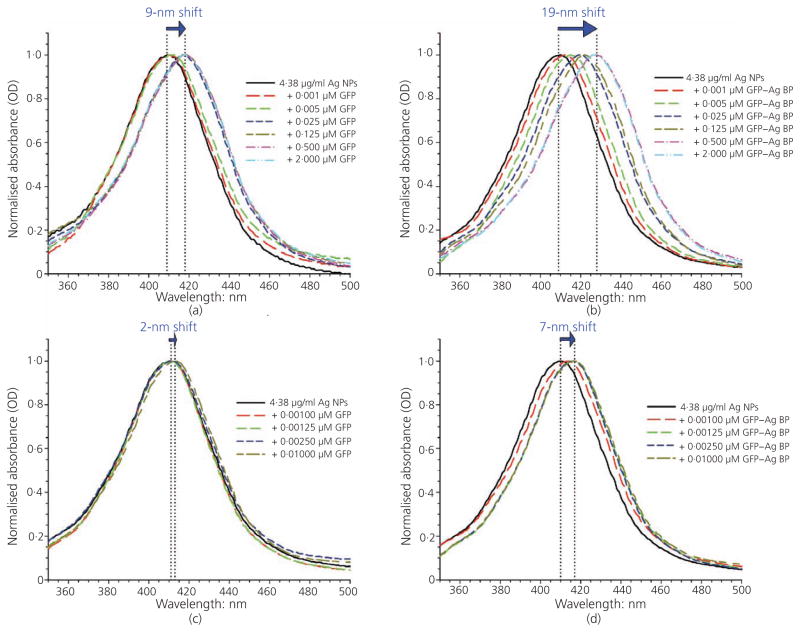
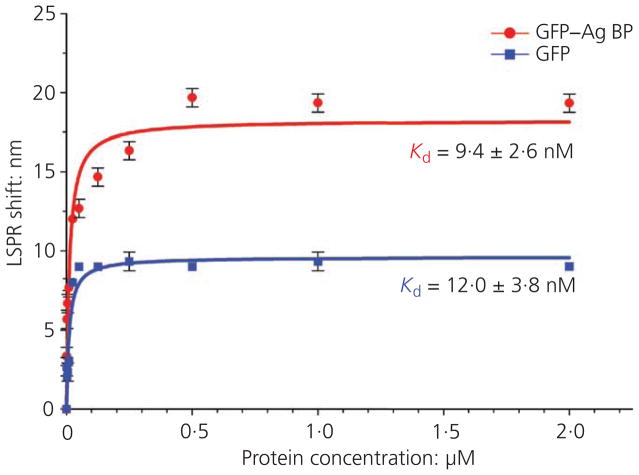
The binding affinity and stability of the engineered bifunctional protein, GFP–silver BP, onto silver NPs was studied using LSPR and compared against silver NPs incubated with GFP protein alone (no silver affinity tag) (Figure 2). The LSPR spectra, formed as a result of the collective oscillations of conduction band electrons for metals such as silver, present a characteristic wavelength of maximum absorption dependent on silver NP size, shape and environmental conditions. In general, a red shift (longer wavelength) in the peak LSPR band corresponds to an analyte binding onto the NP surface, altering the silver NP surface structure.57 Distinct, narrow spectral peaks indicate that NPs remained discrete in suspension and did not agglomerate. The stock, citrate-coated 40-nm silver NPs used presented a strong characteristic absorption peak at about 410 nm, following LSPR spectral scanning range from 300 to 700 nm at 2-nm increments. The LSPR spectra were then taken to determine shifts in the characteristic spectra for 40-nm silver NPs functionalised with the authors’ GFP–silver BP affinity protein compared to GFP protein alone. Following a 24-h incubation, 40-nm silver NPs coated with either GFP or GFP–silver BP proteins revealed maximum red shifts in the spectra of 9 and 19 nm respectively, compared to untreated (stock, citrate-coated) controls with an average spectral peak of 409 nm (Figures 2(a) and 2(b)). A final spectral scan was collected for each silver NP treatment following extensive washing of the NPs incubated with the proteins to remove weakly bound and/or excess protein. After the wash step, the silver NPs displayed a spectra peak red shift of only 2 and 7 nm for silver NPs coated with GFP and GFP–silver BP respectively compared to untreated control samples (Figures 2(c) and 2(d)). The observed red shifts in the spectra could be associated with the increased silver NP sizes due to robust surface functionalisation – that is, the maintained 7-nm shift observed for the GFP–silver BP-functionalised silver NPs can be attributed to the presence of the silver-binding affinity tag (silver BP) of the authors’ engineered fusion protein forming a stable (following washing) protein layer around the silver NPs compared to the non-specific binding using GFP protein alone. These observations are consistent with recent literature findings for silver NP functionalisation monitored by ultraviolet–visible absorbance red shift due to dampened surface plasmon resonance.24,33,58 LSPR experiments run at different protein concentration were used to calculate the Kd values (dissociation constant) to compare quantitatively the protein binding affinity onto the NP surface (Figure 3). The GFP–silver BP protein resulted in significantly higher binding (Kd of ~9·4 nM) compared to GFP protein without silver BP tag (Kd of ~12·0 nM).
Figure 2.
LSPR spectra to confirm protein functionalisation on the silver NPs. LSPR spectral peaks collected after 24 h of incubation with (a) GFP wild-type protein and (b) engineered bifunctional protein GFP–silver BP. A red shift is observed as protein concentration increases. LSPR shifts following the 24-h incubation and wash step to remove unbound (or weakly-bound) protein indicate (c) a sustained 2-nm shift for GFP and (d) a 7-nm shift for the GFP–silver BP spectra. GFP–silver BP is shown to provide a more stable and robust affinity silver NP coating when compared to the wild-type protein alone
Figure 3.
Specific binding fit analysis for averaged LSPR shifts obtained at different protein concentrations for GFP–silver BP and GFP proteins. Dissociation constant (Kd) values were calculated as about 9·4 and 12·0 nM for silver NPs coated with either GFP–silver BP or GFP wild-type protein respectively
3.2 Antimicrobial efficacies of the silver NPs
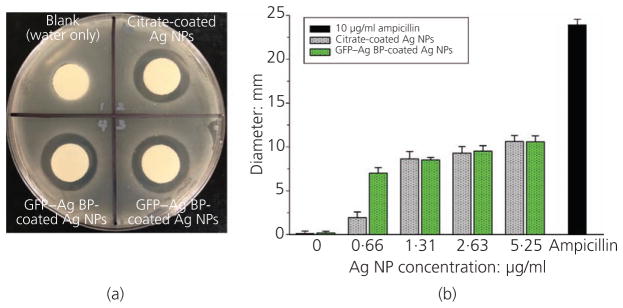
The antimicrobial efficiency of the functionalised silver NPs was studied using growth inhibition models and disc diffusion assays in an S. mutans bacterial strain. Figure 4(a) shows a representative plate from the disc diffusion assay experiments. Clearly observed zones of bacterial growth inhibition are observed for sections exposed to the 40-nm silver NPs-loaded discs compared to a control (water-only) disc. Figure 4(b) indicates a trend suggesting that silver BP–GFP-functionalised NPs may have an enhanced antimicrobial efficacy, although not statistically significant across all concentrations tested. The zones of inhibition were statistically similar for concentrations of 1·31, 2·63 and 5·25 μg/ml for the citrate-coated control and protein-functionalised samples (Figure 4(b)). At the lowest concentration, 0·66 μg/ml, the GFP–silver BP-functionalised silver NPs showed a significantly larger zone of inhibition compared to control NPs. This difference can be attributed to the fact that the fusion protein-coated NPs boasted a more stable biomimetic interface compared to the citrate-coated controls, allowing them to diffuse more easily from the loaded disc. At higher concentrations, silver NPs have a greater potential to agglomerate, based on spatial constraints in the treatment wells (in which the discs were pre-soaked for loading). Agglomerated silver NPs may be trapped or stuck to the polyethylene discs; therefore, the authors attribute the inhibition zone plateau at increasing silver NP concentrations to reduced diffusion capabilities.
Figure 4.
S. mutans agar diffusion assay. (a) Representative image for observed zones of inhibition for the 2·63 μg/ml silver NP concentration comparing GFP–silver BP-coated silver NP loaded discs (duplicate samples shown) and citrate-coated silver NPs discs with respect to a blank control. (b) Graphical representation of measured inhibition zone diameters across four different concentrations studied to compare silver NPs coated with GFP–silver BP or citrate (stock silver NPs). Ampicillin (10 μg/ml) was loaded onto a disc as a positive control. Data trend indicates enhanced antimicrobial function with increasing silver NP concentration for both coatings. GFP–silver BP-coated silver NPs have a significantly greater antimicrobial effect than citrate-coated silver NPs at the lowest concentration studied (0·66 μg/ml)
It is important to note that the engineered protein coating does not limit the natural antimicrobial efficacy of silver NPs; on the contrary, it may be providing a robust biomimetic interface which can enhance silver NP stability (non-agglomeration) in a biologically relevant environment. Numerous reports in the literature utilising a variety of chemically and covalently attached polymers or biomolecular moieties to functionalise and stabilise silver NPs also showed changes and an overall reduction in antimicrobial efficacy due to the functionalisation technique.58,59 However, in light of recent findings regarding protein corona formation and altered size effects of silver NPs, the thickness and chemical properties imparted by the coatings can lead to significant changes in cellular cytotoxicity. The authors’ protein self-assembly technique provides a tailored approach to functionalising NPs with tunable and predictable surface properties while still maintaining the inherent antimicrobial functionality of silver NPs.
3.3 Cell culture fibroblast cellular viability model
The effects of the protein-functionalised silver NPs were also studied in cell culture by using a fibroblast cell line to determine changes in cytotoxicity to mammalian cells subjected to those concentrations that demonstrated antimicrobial efficacy. Conflicting reports have shown a significant range of cytotoxic effects on cells in the presence of silver NPs in numerous cell line cultures and animal models.16,19,44,46,48,49,59–62 Recent review articles describe these findings in detail with respect to silver NP size, shape, surface functionalisation, concentration and stability, among other properties.14,16,21,24,33,35,61 The authors’ studies were conducted with 40-nm silver NPs, a size considered to be within the range to mitigate cytotoxic effects in cells.44 The silver NP concentration range selected for the authors’ study was based on recent studies and the antimicrobial assays that they performed which had demonstrated a sustained antimicrobial efficacy. The maximum silver NP concentration studied, 5·25 μg/ml, is notably lower compared to those in many literature reports on cellular cytotxicity.14,18,44,49,58 The authors attribute this favourable outcome to their novel protein self-assembly silver NP functionalisation technique, which offers enhanced stability without significantly altering silver NP size. Greater stability at the surface prevents agglomeration and allows for maintaining or increasing the NPs’ antimicrobial properties while providing the added benefit of reducing their cytotoxic effects.
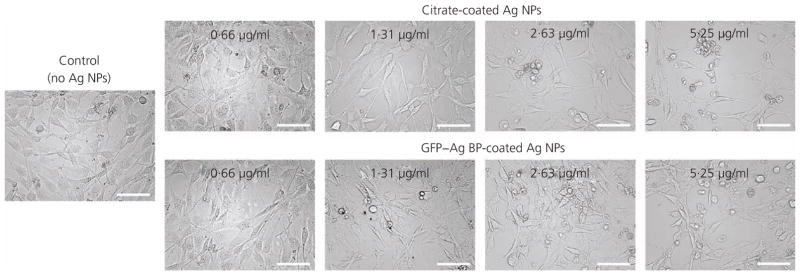
The authors followed the morphological changes for the NIH/3T3 fibroblast cells by bright-field imaging following a 24-h treatment with functionalised and control (citrate-coated) silver NPs. Adherent fibroblast cells were imaged prior to treatment to ensure uniform confluency and morphology across all plates. Representative images are provided in Figure 5, which displays the morphological changes in the cells following a 24-h incubation with various silver NP treatments, compared to control (sham-treated) cells. In all concentrations, cells subjected to the stock, citrate-coated NPs appeared to have a less healthy cell morphology compared to cells treated with respective concentrations of GFP–silver BP protein-coated silver NPs. Concurrent to the literature, cells appeared more viable and healthy at the lower silver NP concentrations, whereas the number of adherent cells remaining in each well suggested that the higher concentrations were more cytotoxic due to an increase in cellular detachment owing to reduced viability.
Figure 5.
Bright-field imaging of the NIH/3T3 fibroblast cells treated with GFP–silver BP- or citrate-coated silver NPs at designated concentrations. Following a 24-h incubation, the cells appear less elongated and are shown to detach as silver NP concentration is increased. The morphological effects appear less severe for GFP–silver BP-coated silver NPs compared to those for stock silver NPs at the same concentrations. Images are shown at ×20 magnification; scale bar represents 100 μm
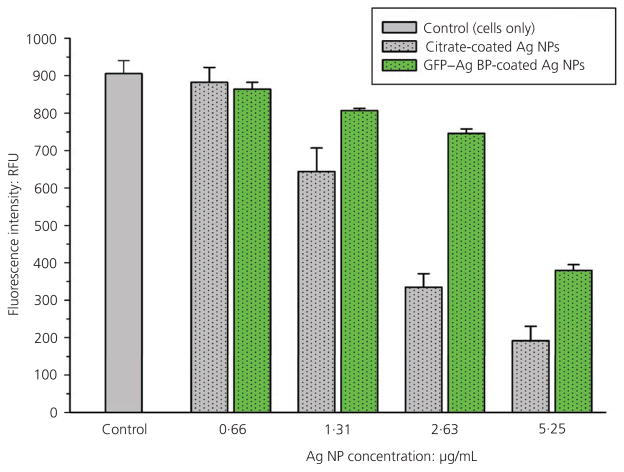
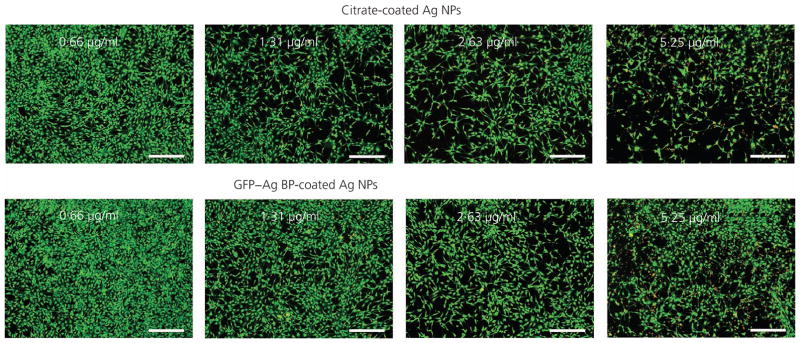
In parallel, cellular viability was quantitatively studied using two independent methods. First, cells were subjected to the AlamarBlue metabolic viability assay. Figure 6 shows average fluorescence intensity measurements from AlamarBlue-treated wells. Overall, a concentration-dependent decrease in cellular viability was observed with increasing silver NP concentrations for both the citrate-coated and GFP–silver BP functionalised NP treatments. However, a statistically significant improvement was observed in cellular viability for cells treated with protein-functionalised silver NPs compared to stock, citrate-coated silver NPs at concentrations of 1·31, 2·63 and 5·25 μg/ml. Comparable results were also achieved using Live/Dead staining methods, as shown in representative images in Figure 7. The Live/Dead staining technique labels healthy, viable cells in green and cytotoxic cells (compromised membrane integrity) are stained with red fluorescent dye. Because the authors’ engineered fusion protein featured GFP, each plate was imaged prior to Live/Dead staining and the fluorescent contribution of the GFP protein subtracted from the Live/Dead assay. The data confirmed an improved cellular viability (green stain) and reduced cytotoxicity for NIH/3T3 cells treated with protein-functionalised silver NPs compared to citrate-coated controls at all silver NP concentrations. The enhanced viability of cells exposed to GFP–silver BP-stabilised NPs demonstrates a clear dependence on silver NP surface functionalisation for silver NPs of the same size and concentration, a finding reflected within the vast number of related publications. The authors’ results were in agreement with the relevant literature, showing that cellular cytotoxicity increased with greater silver NP concentrations. However, cells treated with the protein-stabilised silver NPs show a significant reduction in cytotoxic activity (as a ratio to viable cells remaining) at each concentration as compared to citrate-coated controls.
Figure 6.
Cell viability of NIH/3T3 fibroblast cells exposed to increasing concentrations of silver NPs that are coated with either GFP–silver BP or citrate (control). There is a decreasing trend for cellular viability, measured by AlamarBlue fluorescence activity, as silver NP concentrations increase. Cells incubated with silver NPs functionalised with GFP–silver BP maintain a significantly higher cellular viability compared to cells exposed to citrate-coated silver NPs at corresponding concentrations (for concentrations of 1·31 μg/ml and higher). RFU, relative fluorescence units
Figure 7.
Representative images of the NIH/3T3 fibroblast cells treated with GFP–silver BP- or citrate-coated silver NPs at designated concentrations by Live/Dead fluorescence. Following a 24-h incubation with silver NP treatments, viable cells are identified by their green stain; increased cellular cytotoxicity (red stain) is noted at increasing concentrations for citrate-coated compared to GFP–silver BP-coated silver NPs. Protein-functionalised silver NPs maintain a greater viability throughout increased concentrations compared to citrate-coated silver NPs alone. Images are shown at ×4 magnification; scale bar represents 300 μm
4. Conclusions
While studying biological interactions with materials at the nanoscale is extremely important to address their safety concern, these analyses are often still deficient for studies on silver NPs. Silver NPs offer an exceptional opportunity to combat infections and treat antibiotic-resistant bacteria, but their widespread use has been limited because of unanswered questions regarding toxicity to host cells and the related protein complexes that create a corona on their surfaces. The protein corona confounds quantitative and qualitative analysis. One way to overcome these complex interactions may be to engineer the silver NP surface by using biological self-assembly to provide a competitive advantage over other competitive absorption processes at the interface. Combinatorial-selected inorganic BPs may offer an opportunity to provide a relatively stable interface at the silver NP surfaces that better serves a biomedical application. Herein, the authors genetically engineered a silver BP and used it to form a biointerface to stabilise the surface of silver NPs. By providing the peptide tag to bind silver within the multifunctional protein complex, the authors succeeded in creating a protein cage around the NPs where the resulting interface may have a better chance to stand the complex ongoing biological interactions. The GFP–silver BP fusion protein demonstrates its ability to bio-self-assemble onto silver NPs in a tunable, concentration-dependent manner and displays a strong binding stability. The results showed that a low concentration of 40-nm silver NPs, stabilised with the engineered fusion protein, provides a local antibacterial efficacy at a suitably low concentration while mitigating the cytotoxicity and improving viability in a fibroblast cell model in culture. These results offer a one-step, environmentally friendly and biomimetic approach to NP functionalisation to overcome infection-associated implant failure in medical and dental devices. Furthermore, by substituting the GFP moiety with a protein or peptide tailored to reduce the local inflammatory response, the functionalisation approach may help to promote wound healing while maintaining enhanced antimicrobial activity at the biomaterial interface.
Acknowledgments
This research was supported by National Institutes of Health (NIH)–National Institute of Arthritis and Musculoskeletal and Skin Diseases AR062249-03 and NIH–National Institute of Dental and Craniofacial Research R01DE025476-01 and through New Faculty General Research Fund (NFGRF) support provided through the University of Kansas. The authors would like to thank Marcos Paulo Simone Barbosa for his help with bacterial cell culture and data analysis.
Contributor Information
Sarah Kay VanOosten, PhD Student, Bioengineering Research Center, Department of Bioengineering, University of Kansas, Lawrence, KS, USA.
Esra Yuca, Postdoctoral Research Fellow, Bioengineering Research Center, Department of Mechanical Engineering, University of Kansas, Lawrence, KS, USA; Research Associate, Department of Molecular Biology, Yildiz Technical University, Istanbul, Turkey.
Banu Taktak Karaca, Postdoctoral Research Fellow, Bioengineering Research Center, Department of Mechanical Engineering, University of Kansas, Lawrence, KS, USA.
Kyle Boone, PhD Student, Bioengineering Research Center, Department of Bioengineering, University of Kansas, Lawrence, KS, USA.
Malcolm L. Snead, Professor and Chair, Division of Biomedical Sciences, Center for Craniofacial Molecular Biology, Herman Ostrow School of Dentistry, The University of Southern California, Los Angeles, CA, USA.
Paulette Spencer, Ackers Distinguished Professor and Director, Bioengineering Research Center, Department of Mechanical Engineering, University of Kansas, Lawrence, KS, USA.
Candan Tamerler, Wesley G. Cramer Professor, Bioengineering Research Center, Department of Mechanical Engineering, University of Kansas, Lawrence, KS, USA.
References
- 1.Darouiche RO. Device-associated infections: a macroproblem that starts with microadherence. Clinical Infectious Diseases. 2001;33(9):1567–1572. doi: 10.1086/323130. [DOI] [PubMed] [Google Scholar]
- 2.Ventola CL. Challenges in evaluating and standardizing medical devices in health care facilities. Pharmacy and Therapeutics. 2008;33(6):348–359. [PMC free article] [PubMed] [Google Scholar]
- 3.Besinis A, De Peralta T, Tredwin CJ, Handy RD. Review of nanomaterials in dentistry: interactions with the oral microenvironment, clinical applications, hazards, and benefits. ACS Nano. 2015;9(3):2255–2289. doi: 10.1021/nn505015e. [DOI] [PubMed] [Google Scholar]
- 4.Xuedong Z. Dental Caries: Principles and Management. Springer; Berlin, Germany: 2015. [Google Scholar]
- 5.Fernandes JMFA, Menezes VA, Albuquerque AJR, et al. Improving antimicrobial activity of dental restorative materials. In: Virdi MS, editor. Emerging Trends in Oral Health Sciences and Dentistry. InTech; Rijeka, Croatia: 2015. pp. 65–82. [Google Scholar]
- 6.Gaviria L, Salcido JP, Guda T, Ong JL. Current trends in dental implants. Journal of the Korean Association of Oral and Maxillofacial Surgeons. 2014;40(2):50–60. doi: 10.5125/jkaoms.2014.40.2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh C, Lee K, Cheong Y, et al. Comparison of the oral microbiomes of canines and their owners using next-generation sequencing. PLoS One. 2015;10(7):e0131468. doi: 10.1371/journal.pone.0131468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallo J, Holinka M, Moucha CS. Antibacterial surface treatment for orthopaedic implants. International Journal of Molecular Sciences. 2014;15(8):13849–13880. doi: 10.3390/ijms150813849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerli W, Trampuz A. Biomaterial-associated infection: a perspective from the clinic. In: Moriarty FT, Zaat AJS, Busscher JH, editors. Biomaterials Associated Infection: Immunological Aspects and Antimicrobial Strategies. Springer; New York, NY, USA: 2013. pp. 3–24. [Google Scholar]
- 10.WHO (World Health Organization) Report: Antimicrobial Resistance: Global Report on Surveillance. WHO Press; Geneva, Switzerland: 2014. [Google Scholar]
- 11.Ventola CL. The antibiotic resistance crisis: part 1: Causes and threats. Pharmacy and Therapeutics. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks BD, Brooks AE, Grainger DW. Antimicrobial medical devices in preclinical development and clinical use. In: Moriarty FT, Zaat AJS, Busscher JH, editors. Biomaterials Associated Infection: Immunological Aspects and Antimicrobial Strategies. Springer; New York, NY, USA: 2013. pp. 307–354. [Google Scholar]
- 13.Alexander JW. History of the medical use of silver. Surgical Infections. 2009;10(3):289–292. doi: 10.1089/sur.2008.9941. [DOI] [PubMed] [Google Scholar]
- 14.Corrêa JM, Mori M, Sanches HL, et al. Silver nanoparticles in dental biomaterials. International Journal of Biomaterials. 2015;2015:485275. doi: 10.1155/2015/485275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dallas P, Sharma VK, Zboril R. Silver polymeric nanocomposites as advanced antimicrobial agents: classification, synthetic paths, applications, and perspectives. Advances in Colloid and Interface Science. 2011;166(1–2):119–135. doi: 10.1016/j.cis.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 16.dos Santos CA, Seckler MM, Ingle AP, et al. Silver nanoparticles: therapeutical uses, toxicity, and safety issues. Journal of Pharmaceutical Sciences. 2014;103(7):1931–1944. doi: 10.1002/jps.24001. [DOI] [PubMed] [Google Scholar]
- 17.Duran N, Duran M, de Jesus MB, et al. Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomedicine: Nanotechnology, Biology, and Medicine. 2016;12(3):789–799. doi: 10.1016/j.nano.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Espinosa-Cristobal LF, Martinez-Castanon GA, Martinez-Martinez RE, et al. Antibacterial effect of silver nanoparticles against Streptococcus mutans. Materials Letters. 2009;63(29):2603–2606. [Google Scholar]
- 19.Hernandez-Sierra JF, Galicia-Cruz O, Angelica SA, et al. In vitro cytotoxicity of silver nanoparticles on human periodontal fibroblasts. Journal of Clinical Pediatric Dentistry. 2011;36(1):37–41. doi: 10.17796/jcpd.36.1.d677647166398886. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z, Rong K, Li J, Yang H, Chen R. Size-dependent antibacterial activities of silver nanoparticles against oral anaerobic pathogenic bacteria. Journal of Materials Science: Materials in Medicine. 2013;24(6):1465–1471. doi: 10.1007/s10856-013-4894-5. [DOI] [PubMed] [Google Scholar]
- 21.Marin S, Vlasceanu GM, Tiplea RE, et al. Applications and toxicity of silver nanoparticles: a recent review. Current Topics in Medicinal Chemistry. 2015;15(16):1596–1604. doi: 10.2174/1568026615666150414142209. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Castanon GA, Nino-Martinez N, Martinez-Gutierrez F, Martinez-Mendoza JR, Ruiz F. Synthesis and antibacterial activity of silver nanoparticles with different sizes. Journal of Nanoparticle Research. 2008;10(8):1343–1348. [Google Scholar]
- 23.Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 24.Wei L, Lu J, Xu H, et al. Silver nanoparticles: synthesis, properties, and therapeutic applications. Drug Discovery Today. 2015;20(5):595–601. doi: 10.1016/j.drudis.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allaker RP, Memarzadeh K. Nanoparticles and the control of oral infections. International Journal of Antimicrobial Agents. 2014;43(2):95–104. doi: 10.1016/j.ijantimicag.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Khan ST, Musarrat J, Al-Khedhairy AA. Countering drug resistance, infectious diseases, and sepsis using metal and metal oxides nanoparticles: current status. Colloids and Surfaces B: Biointerfaces. 2016;146:70–83. doi: 10.1016/j.colsurfb.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 27.Durán N, Nakazato G, Seabra AB. Antimicrobial activity of biogenic silver nanoparticles, and silver chloride nanoparticles: an overview and comments. Applied Microbiology and Biotechnology. 2016;100(15):6555–6570. doi: 10.1007/s00253-016-7657-7. [DOI] [PubMed] [Google Scholar]
- 28.Mftah A, Alhassan FH, Al-Qubaisi MS, et al. Physicochemical properties, cytotoxicity, and antimicrobial activity of sulphated zirconia nanoparticles. International Journal of Nanomedicine. 2015;10:765–774. doi: 10.2147/IJN.S66058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Li J, Qian S, et al. Antibacterial surface design of titanium-based biomaterials for enhanced bacteria-killing and cell-assisting functions against periprosthetic joint infection. ACS Applied Materials & Interfaces. 2016;8(17):11162–11178. doi: 10.1021/acsami.6b02803. [DOI] [PubMed] [Google Scholar]
- 30.Lynch I, Dawson KA. Protein–nanoparticle interactions. Nano Today. 2008;3(1–2):40–47. [Google Scholar]
- 31.Yucesoy DT, Hnilova M, Boone K, et al. Chimeric peptides as implant functionalization agents for titanium alloy implants with antimicrobial properties. JOM (1989) 2015;67(4):754–766. doi: 10.1007/s11837-015-1350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazici H, O’Neill MB, Kacar T, et al. Engineered chimeric peptides as antimicrobial surface coating agents toward infection-free implants. ACS Applied Materials & Interfaces. 2016;8(8):5070–5081. doi: 10.1021/acsami.5b03697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durán N, Silveira CP, Durán M, Martinez DST. Silver nanoparticle protein corona and toxicity: a mini-review. Journal of Nanobiotechnology. 2015;13(55):1–17. doi: 10.1186/s12951-015-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall Sedlak R, Hnilova M, Grosh C, et al. Engineered Escherichia coli silver-binding periplasmic protein that promotes silver tolerance. Applied and Environmental Microbiology. 2012;78(7):2289–2296. doi: 10.1128/AEM.06823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YK, Choi EJ, Webster TJ, Kim SH, Khang D. Effect of the protein corona on nanoparticles for modulating cytotoxicity and immunotoxicity. International Journal of Nanomedicine. 2015;10:97–113. doi: 10.2147/IJN.S72998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nature Nanotechnology. 2012;7(12):779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 37.Carmona-Ribeiro AM, de Melo Carrasco LD. Cationic antimicrobial polymers and their assemblies. International Journal of Molecular Sciences. 2013;14(5):9906–9946. doi: 10.3390/ijms14059906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siedenbiedel F, Tiller JC. Antimicrobial polymers in solution and on surfaces: overview and functional principles. Polymers. 2012;4(1):46–71. [Google Scholar]
- 39.Mi L, Jiang S. Integrated antimicrobial and nonfouling zwitterionic polymers. Angewandte Chemie International Edition. 2014;53(7):1746–1754. doi: 10.1002/anie.201304060. [DOI] [PubMed] [Google Scholar]
- 40.Jennings MC, Minbiole KPC, Wuest WM. Quaternary ammonium compounds: an antimicrobial mainstay and platform for innovation to address bacterial resistance. ACS Infectious Diseases. 2015;1(7):288–303. doi: 10.1021/acsinfecdis.5b00047. [DOI] [PubMed] [Google Scholar]
- 41.Buffet-Bataillon S, Tattevin P, Bonnaure-Mallet M, Jolivet-Gougeon A. Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds – a critical review. International Journal of Antimicrobial Agents. 2012;39(5):381–389. doi: 10.1016/j.ijantimicag.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Alberto EE, Rossato LL, Alves SH, Alves D, Braga AL. Imidazolium ionic liquids containing selenium: synthesis and antimicrobial activity. Organic & Biomolecular Chemistry. 2011;9(4):1001–1003. doi: 10.1039/c0ob01010c. [DOI] [PubMed] [Google Scholar]
- 43.Tran PA, Webster TJ. Antimicrobial selenium nanoparticle coatings on polymeric medical devices. Nanotechnology. 2013;24(15):155101. doi: 10.1088/0957-4484/24/15/155101. [DOI] [PubMed] [Google Scholar]
- 44.Gliga AR, Skoglund S, Wallinder IO, Fadeel B, Karlsson HL. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release. Particle and Fibre Toxicology. 2014;11:11. doi: 10.1186/1743-8977-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Applied and Environmental Microbiology. 2007;73(6):1712–1720. doi: 10.1128/AEM.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yen HJ, Hsu SH, Tsai CL. Cytotoxicity and immunological response of gold and silver nanoparticles of different sizes. Small. 2009;5(13):1553–1561. doi: 10.1002/smll.200900126. [DOI] [PubMed] [Google Scholar]
- 47.Caufield PW, Li Y, Dasanayake A. Dental caries: an infectious and transmissible disease. Compendium of Continuing Education in Dentistry. 2005;26(5):10–16. [PubMed] [Google Scholar]
- 48.Braydich-Stolle LK, Lucas B, Schrand A, et al. Silver nanoparticles disrupt GDNF/Fyn kinase signaling in spermatogonial stem cells. Toxicological Sciences. 2010;116(2):577–589. doi: 10.1093/toxsci/kfq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Lu X, Ma J. Toxicity of silver nanoparticles to human dermal fibroblasts on microRNA level. Journal of Biomedical Nanotechnology. 2014;10(11):3304–3317. doi: 10.1166/jbn.2014.1974. [DOI] [PubMed] [Google Scholar]
- 50.Hsin YH, Chena CF, Huang S, et al. The apoptotic effect of nanosilver is mediated by a ROS- and JNK-dependent mechanism involving the mitochondrial pathway in NIH3T3 cells. Toxicology Letters. 2008;179(3):130–139. doi: 10.1016/j.toxlet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Miclaus T, Beer C, Chevallier J, et al. Dynamic protein coronas revealed as a modulator of silver nanoparticle sulphidation in vitro. Nature Communications. 2016;7:11770. doi: 10.1038/ncomms11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yucesoy DT, Karaca BT, Cetinel S, et al. Direct bioelectrocatalysis at the interfaces by genetically engineered dehydrogenase. Bioinspired, Biomimetic and Nanobiomaterials. 2015;4(1):79–89. [Google Scholar]
- 53.Balcioglu BK, Ozdemir-Bahadir A, Hinc D, Tamerler C, Erdag B. Cost effective filamentous phage based immunization nanoparticles displaying a full-length hepatitis B virus surface antigen. Advances in Bioscience and Biotechnology. 2014;5(1):46–53. [Google Scholar]
- 54.Zin MT, Munro AM, Gungormus M, et al. Peptide-mediated surface-immobilized quantum dot hybrid nanoassemblies with controlled photoluminescence. Journal of Materials Chemistry. 2007;17(9):866–872. [Google Scholar]
- 55.Hnilova M, Liu X, Yuca E, et al. Multifunctional protein-enabled patterning on arrayed ferroelectric materials. ACS Applied Materials & Interfaces. 2012;4(4):1865–1871. doi: 10.1021/am300177t. [DOI] [PubMed] [Google Scholar]
- 56.Yuca E, Karatas AY, Seker UO, et al. In vitro labeling of hydroxyapatite minerals by an engineered protein. Biotechnology and Bioengineering. 2011;108(5):1021–1030. doi: 10.1002/bit.23041. [DOI] [PubMed] [Google Scholar]
- 57.Podila R, Chen R, Ke PC, Brown JM, Rao AM. Effects of surface functional groups on the formation of nanoparticle-protein corona. Applied Physics Letters. 2012;101(26):263701. doi: 10.1063/1.4772509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kujda M, Ocwieja M, Adamczyk Z, et al. Charge stabilized silver nanoparticles applied as antibacterial agents. Journal of Nanoscience and Nanotechnology. 2015;15(5):3574–3583. doi: 10.1166/jnn.2015.9727. [DOI] [PubMed] [Google Scholar]
- 59.Jiang-Jen L, Wen-Chun L, Rui-Xuan D, Shan-hui H. The cellular responses and antibacterial activities of silver nanoparticles stabilized by different polymers. Nanotechnology. 2012;23(6):065102. doi: 10.1088/0957-4484/23/6/065102. [DOI] [PubMed] [Google Scholar]
- 60.Arora S, Jain J, Rajwade JM, Paknikar KM. Interactions of silver nanoparticles with primary mouse fibroblasts and liver cells. Toxicology and Applied Pharmacology. 2009;236(3):310–318. doi: 10.1016/j.taap.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 61.Sharma VK, Siskova KM, Zboril R, Gardea-Torresdey JL. Organic-coated silver nanoparticles in biological and environmental conditions: fate, stability and toxicity. Advances in Colloid and Interface Science. 2014;204:15–34. doi: 10.1016/j.cis.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Yu X, Hong F, Zhang YQ. Bio-effect of nanoparticles in the cardiovascular system. Journal of Biomedical Materials Research Part A. 2016;104(11):2881–2897. doi: 10.1002/jbm.a.35804. [DOI] [PubMed] [Google Scholar]