Abstract
IFITM3 is involved in cell adhesion, apoptosis, immune, and antivirus activity. Furthermore, IFITM3 gene has been considered as a preferential marker for inflammatory diseases, and positive correlation to pathological grades. Therefore, we assumed that IFITM3 was regulated by different signal pathways. To better understand IFITM3 function in inflammatory response, we cloned swine IFITM3 gene, and detected IFITM3 distribution in tissues, as well as characterized this gene. Results indicated that the length of swine IFITM3 gene was 438 bp, encoding 145 amino acids. IFITM3 gene expression abundance was higher in spleen and lungs. Moreover, we next constructed the eukaryotic expression vector PBIFM3 and transfected into PK15 cells, finally obtained swine IFITM3 gene stable expression cell line. Meanwhile, we explored the effects of LPS on swine IFITM3 expression. Results showed that LPS increased IFITM3 mRNA abundance and exhibited time-dependent effect for LPS treatment. To further demonstrate the mechanism that IFITM3 regulated type I IFNs production, we also detected the important molecules expression of TLR4 signaling pathway. In transfected and non-transfected IFITM3 PK15 cells, LPS exacerbated the relative expression of TLR4-NFκB signaling molecules. However, the IFITM3 overexpression suppressed the inflammatory development of PK15 cells. In conclusion, these data indicated that the overexpression of swine IFITM3 could decrease the inflammatory response through TLR4 signaling pathway, and participate in type I interferon production. These findings may lead to an improved understanding of the biological function of IFITM3 in inflammation.
Keywords: IFITM3, characterization, anti-inflammation, TLR4 signaling pathway, lipopolysaccharide, Immunology and Microbiology Section, Immune response, Immunity
INTRODUCTION
Swine diseases have disturbed many intensive farms heavily in economic and social losses, and aroused extensive attention to the prevention and treatment of swine diseases. Due to the complex genetic diversity, current vaccination and antiviral strategies are not valid [1]. The innate immune is the first line for host defense against infections. As one type of potent innate immunomodulators, interferons (IFNs) exert an important role in the host against infection and pathogen invasion via several diverse mechanisms. IFNs can inhibit a variety of viruses, bacteria, and parasites infections [2].
Interferon inducible trans-membrane protein 3 (IFITM3) is a double trans-membrane protein that belongs to the IFITM family members. IFITM3 is mainly stimulated by IFNs participating in various biological processes. Hence, IFITM3 is involved in interferon-triggered processes, such as anti-proliferative activities of different pathogenesis, antivirus infections in innate immune response, cell adhesion, cell apoptosis, and germ cell homing [2-10]. More importantly, IFITM3 is also a potent antiviral and anti-inflammatory effector in the host innate immune system [7, 8, 11]. At present, the characteristics and antiviral activity of human IFITM3 have been broadly investigated, and mainly focused on human-associated pathogens.
Currently, studies on swine IFITM3, especially regarding its function, are very few. Although previous studies on IFITMs mainly focused on their roles in embryonic development, their functions as host antiviral factors were only recently discovered by RNA interference genomic screening for host factors involved in influenza virus infection [7]. It was subsequently revealed that IFITMs can restrict the early stages of replication for a wide variety of viruses, including Influenza virus, West Nile viruses, Ebola viruses, and Corona virus [7, 8, 12]. Researchers found that IFITM3 gene was mainly isolated from severely inflamed mucosa and considered as a biomarker for ulcerative colitis [6, 13]. Furthermore, IFITM3 expression increased in gastric cancer, and colorectal tumors [14-16]. There exists a positive correlation between the IFITM3 expression levels and the pathological glioma grades [17]. Previous studies have shown that toll like receptor 4 (TLR4) may play a major role in various inflammatory diseases. Literatures demonstrated that TLR4 expression up-regulated in rheumatoid arthritis, systemic lupus erythematosus, and multiple sclerosis [18-23]. Bacterial lipopolysaccharide (LPS), which is recognized by host innate immune receptor TLR4 and subsequently triggers inflammation by activation of a transcriptional factor IFN regulatory factor and nuclear factor-κB (NFκB), resulting in up-regulation of various inflammatory mediators [24].
However, the role of swine IFITM3 in inflammation and the regulatory signal pathways is unclear. In this study, we first cloned swine IFITM3 gene from spleen tissues and examined swine IFITM3 mRNA location in tissues. To better understand IFITM3 function in inflammation, we next constructed the eukaryotic expression vector PBIFM3 and transfected into PK15 cells, as well as obtained swine IFITM3 gene stable expression cell line. Meanwhile, we explored the effects of LPS on swine IFITM3 expression. We further also detected the important molecules expression of TLR4 signaling pathway. Together, these data indicated that overexpression of swine IFITM3 could decrease the inflammatory response through TLR4 signaling pathway, and participate in type I interferon production. These conclusions may be helpful to understand deeply the biological function of IFITM3 in inflammation.
RESULTS
Characterization and expression pattern of IFITM3 gene
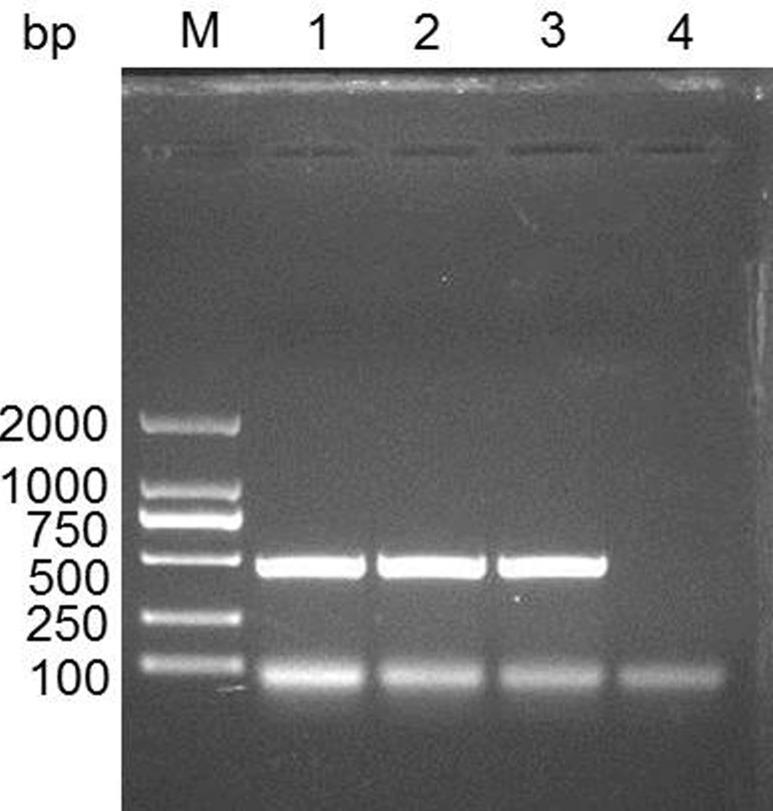
We searched the predicted IFITM3 CDS (NM_001201382.1) in the NCBI database. The length of IFITM3 CDS is 438 bp, and encodes 145 amino acids. We cloned IFITM3 CDS and identified it using DNA sequencing. The data demonstrated the expected nucleotide sequence of IFITM3 as NCBI database described (Figure 1).
Figure 1. Cloning the IFITM3 gene in swine spleen tissue.
M, DL2000 DNA marker; 1-3, PCR product for swine IFITM3; 4, negative control.
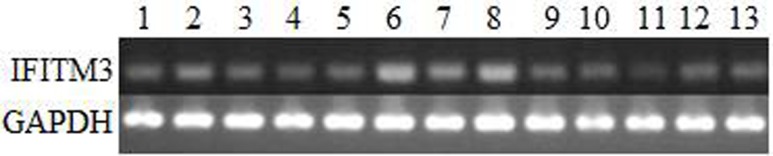
To illustrate the expression patterns of IFITM3 mRNA in tissues, we detected IFITM3 mRNA expression in 13 healthy swine tissues (Figure 2). Results showed that IFITM3 mRNA was expressed in all 13 tissues. Moreover, the IFITM3 mRNA abundance was relatively higher in spleen and lung, while the IFITM3 mRNA in heart and ileum was hardly detected (Figure 2). These differential expression implicated that IFITM3 might be involved in the functional regulation of immunity.
Figure 2. Expression patterns of the IFITM3 mRNA in various swine tissues.
1, skin; 2, muscle; 3, adipose; 4, heart; 5, liver; 6, spleen; 7, kidney; 8, lung; 9, duodenum; 10, jejunum; 11, ileum; 12, cecum; 13, rectum.
Transfection efficiency and IFITM3 overexpression
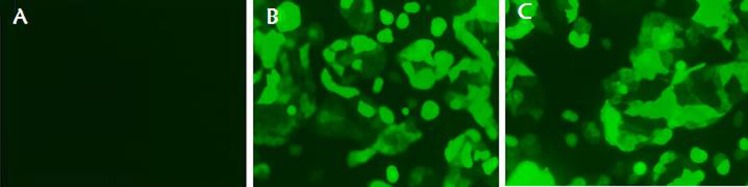
To further evaluate the IFITM3 function in vitro, we next constructed the eukaryotic expression vector PBIFM3 and transfected into PK15 cells, as well as obtained swine IFITM3 stable expression cell line. Based on Piggyback (PB) vector carries the green fluorescent protein (GFP) gene. When the PB vector is integrated into the host cell genome, GFP protein is expressed in the host cells, which can be seen under a fluorescence microscope. We clearly observed the green fluorescence in the transfected PBIFM3 and PBv, but not in the non-transfected PK15 cells (Figure 3). These results indicated that vectors with the GFP gene were successfully transfected into cells and integrated into the genome. Through screening, we obtained stable transfected cell lines. To evaluate IFITM3 expression efficiency, we further detected the expression of IFITM3 using real-time RT-PCR, combined with observation under microscope (Figure 3). The IFITM3 mRNA expression up-regulated significantly in PBIFM3 cells, compared with that in PBv cells (p < 0.0001) (Figure 4). Thus, IFITM3 was successfully transfected into PK15 cells and efficiently overexpressed.
Figure 3. Observation of green fluorescence in PK15 cells transfected with IFITM3 expression vectors.
Green fluorescence was observed clearly using fluorescence microscopy in the PBv (B), and PBIFM3 (C) cells, but not in non-transfected PK15 cells (A).
Figure 4. Overexpression of the swine IFITM3 gene in PBIFM3 cells.
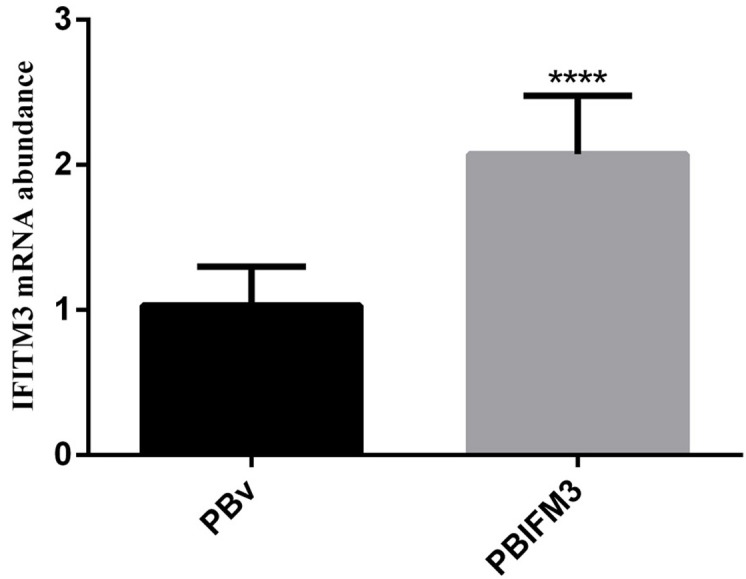
GAPDH was used as an internal housekeeping gene. **** P < 0.0001 vs. PBv.
IFITM3 expression in inflammatory response
LPS is the most commonly used experimental inflammatory factor. We treated the PBIFM3 and PBv cells with or without LPS to detect IFITM3 expression in inflammatory response. As shown in Figure 5, compared with 0 h cells, LPS increased IFITM3 mRNA abundance in PBIFM3 and PBv, and exhibited the time-dependent effect for LPS treatment. However, IFITM3 mRNA abundance in PBIFM3 cells was notably higher than that in PBv cells (p < 0.0001). These data noted that the IFITM3 expression can be up-regulated in inflammatory response.
Figure 5. Effect of LPS on swine IFITM3 expression.
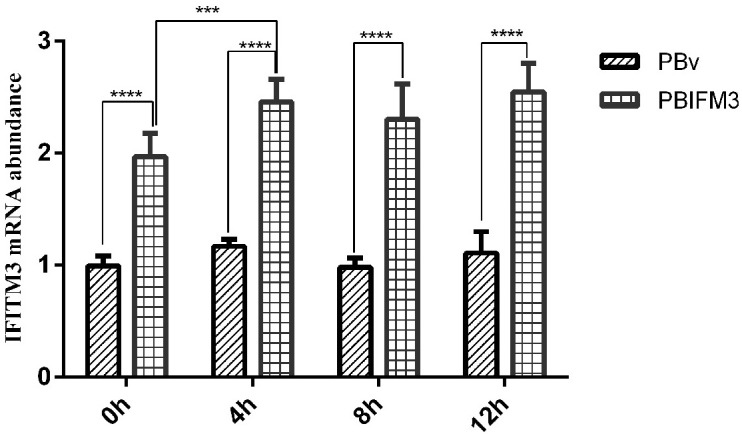
Graphs demonstrate the expression of IFITM3 in PBIFM3 and PBv cells stimulated with or without LPS (100 μg/ml). In LPS groups, cells were incubated correspondingly with LPS (100 μg/mL) for 4, 8, 12 h. Cells in 0 h (control) group were cultured in basal medium. ****P < 0.0001 vs. PBv or 0 h group.
IFITM3 inhibits type I interferons production
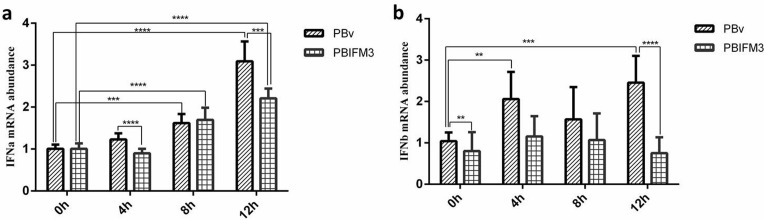
Type I interferons are critical for blocking pathogens infection by promoting hundreds of IFN-stimulated genes production [25]. IFITM3 gene has been identified in several cellular processes mediated by IFNs, such as antivirus, cell adhesion and anti-proliferative activities [3]. We detected type I interferons production in PBIFM3 cells with or without LPS treatment. As shown in Figure 6, IFNɑ was significantly up-regulated in the PBIFM3 and PBv cells, as well as dependent on the time of LPS treatment. However, IFNɑ abundance in PBIFM3 cells was lower than that in PBv cells. Similar results were also observed in the change of IFNβ expression, IFNβ expression was not entirely dependent on the time of LPS treatment. Results implicated that IFITM3 decreased the IFNs production during inflammatory activity.
Figure 6. Effect of swine IFITM3 on the expression of IFNα and IFNβ.
Graphs demonstrate the expression of IFNα (a) and IFNβ (b) in PBIFM3 and PBv cells stimulated with or without LPS (100 μg/ml). In LPS groups, cells were incubated correspondingly with LPS (100 μg/mL) for 4, 8, 12 h. Cells in 0 h (control) group were cultured in basal medium. **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. PBv or 0 h group.
IFITM3 suppresses TLR4 signaling pathway
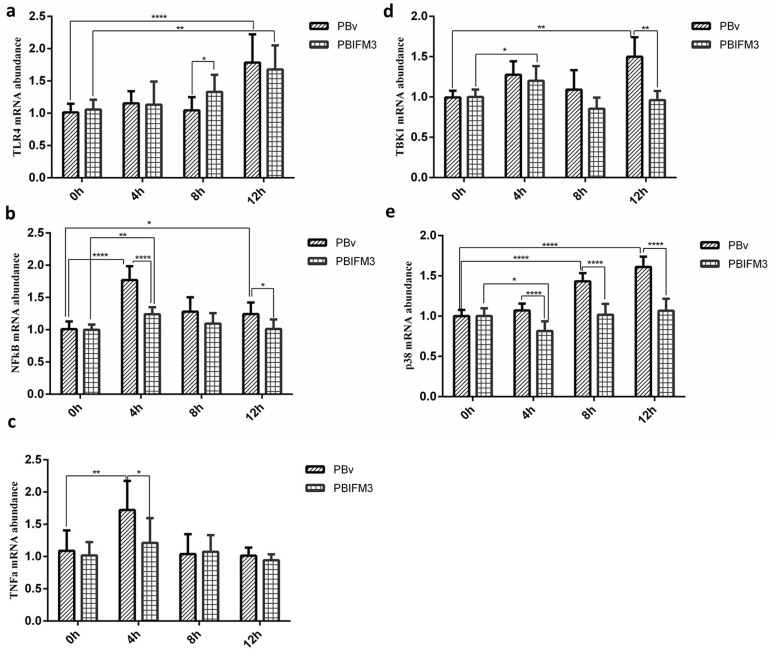
To further clarify the mechanism that IFITM3 regulated type I IFNs production, we also detected the important molecules expression of TLR4 signaling pathway, which is the classical pathway of LPS. In transfected and non-transfected IFITM3 PK15 cells, LPS exacerbated the relative expression of TLR4-NFκB signaling key molecules (Figure 7), and up-regulated the expression of TLR4, NFκB, p38 mitogen-activated protein kinases (p38 MAPK), and tumor necrosis factor alpha (TNFα), compared with 0 h treatment cultured with basal medium only (Figure 7). However, the IFITM3 overexpression suppressed the inflammatory development of PK15 cells (Figure 7), and down-regulated the expression of TLR4, NFκB, p38 MAPK, and TNFα, compared with the PBv cells treated with LPS (Figure 7). Consistently, the levels of TNFα and NFκB reached at peak 4 h after LPS treatment both of the PBIFM3 and PBv cells, then decreased over time. The relative expression of TLR4 and p38 MAPK were dependent on the time of LPS treatment. However, the change of TANK-binding kinase 1 (TBK1) expression had no significance correlation with LPS treatment time. Collectively, these results indicated that IFITM3 was involved in the TLR4-NFκB signaling pathway and played a pivotal role in inflammatory response.
Figure 7. Effect of swine IFITM3 on the expression of components of the TLR4 signaling pathway.
Graphs demonstrate the expression of TLR4 (a), NFκB (b), TNFα (c), TBK1 (d) and p38 (e) in PBIFM3 and PBv cells stimulated with or without LPS (100 μg/ml). In LPS groups, cells were incubated correspondingly with LPS (100 μg/mL) for 4, 8, 12 h. Cells in 0 h (control) group were cultured in basal medium. *P < 0.05,**P < 0.01, ****P < 0.0001 vs. PBv or 0 h group.
DISCUSSION
IFITM proteins are approximately 130 amino acids in length and are conserved in most vertebrate species [3]. Xu et al reported that the full-length cDNA of swine IFITM3 from lymph node tissue is 438 base pairs (bp), and encodes 145 amino acids residues [26]. In this study, we cloned swine IFITM3 gene, and detected the IFITM3 distribution in tissues, as well as functional characterized this gene. The results similar with Xu et al [26] were also observed. The length of swine IFITM3 gene was also 438 bp, encoding 145 amino acids. According to the gene distribution in tissues, IFITM3 gene expression abundance is higher in spleen and lungs.
IFITM3 has been reported to participate in adhesion, apoptosis, immune response, growth and development, and germ cell homing [3]. More importantly, IFITM3 exhibits the greatest protection against the broadest range of viruses, including influenza A virus, flaviviruses, hepaciviruses, and reoviruses, etc [11, 12, 25, 27]. IFITM3 blocked human influenza virus infection during the early stages of virus entry, precisely, after hemifusion, likely due to the relatively high constitutive expression in the endosomal and lysosomal of host [28]. Furthermore, the single-nucleotide polymorphism of IFITM3 gene resulted in decreasing IFITM3 protein expression, which has been linked with a higher risk of virus infections [29, 30]. IFITM3 knockout mice increased morbidity and mortality associated with seasonal or pandemic influenza virus infection [29, 31]. Recent studies identified that IFITM3 gene was a powerful biomarker for ulcerative colitis [6, 13]. Moreover, IFITM3 up-regulated in gastric cancer, and colorectal tumors [14-16]. Furthermore, study showed that there existed a positive correlation between the IFITM3 expression levels and pathological grades [17]. In this study, the IFITM3 mRNA abundance was hardly detected in ileum (Figure 2). We considered that different intestine and pathological grades may be the major reason resulting in the differential result. However, some studies indicated that IFITM3 was necessary to regulate the complex IFN-α response [32, 33]. Therefore, we assumed that IFITM3 could be regulated by several signal pathways.
As we all know, TLR4-NFκB pathway participates in various inflammatory cytokines production, which is classical pathway of LPS, and subsequently triggers inflammation by activation of a transcriptional factor IFN regulatory factor and NFκB, resulting in the up-regulation of various inflammatory mediators [24]. To better understand IFITM3 function in inflammatory response, we transfected PBIFM3 vector into PK15 cells to achieve IFITM3 overexpression. PB is a DNA transposon which was originally isolated from genomes of baculoviruses that infect cabbage looper moth Trichoplusiani [34]. PB vector uses the TTAA sequence to insert into target sites, and displays little selectivity for particular regions of the genome other than a modest preference for DNase I sensitivity regions [35, 36]. Others have shown that PB is distinguished by its ability to remain active when fused to a DNA binding domain and that such fusions can bias insertion toward cognate sites [37, 38, 39]. Hence, we used PB vector and constructed the eukaryotic expression vector PBIFM3, and then transfected into PK15 cells, as well as obtained swine IFITM3 stable expression cell line. Meanwhile, we explored the effects of LPS on swine IFITM3 expression. Results showed that LPS increased IFITM3 mRNA abundance and exhibited the time-dependent effect for LPS treatment.
TLR4-NFκB signaling has been reported to play a critical role in the LPS-induced expression of inflammatory cytokines such as TNFα and interleukin 1 beta (IL-1β) in macrophages and gingival fibroblasts [40, 41]. p38 MAPK belongs to a class of serine/threonine kinases and expresses in most tissues [42]. p38 MAPK activates a wide range of transcription factors, protein kinases, cytosolic and nuclear proteins, all of which lead to diverse responses such as inflammation, cell differentiation, apoptosis, and cytokine production [43]. TBK1 plays important roles in innate immunity. TBK1 mediates the activation of interferon regulatory factor, leading to the induction of type I IFNs [44]. Furthermore, Nakajima et al has shown that the TBK1, p38 MAPK, and NFκB pathways are involved in the IFITM3 expression induced by LPS [45]. We detected type I interferon production in PBIFM3 cells with or without LPS treatment. As shown in Figure 6, IFITM3 down-regulated IFNs production during inflammatory activity. To further illustrate the mechanism that IFITM3 regulated type I IFNs production, we next detected the important molecules expression of TLR4 signaling pathway. In transfected and non-transfected IFITM3 PK15 cells, LPS aggravated the relative expression of TLR4-NFκB signaling molecules (Figure 7). However, the IFITM3 overexpression suppressed the inflammatory development of PK15 cells (Figure 7).
In summary, these data indicated that the overexpression of swine IFITM3 could decrease the inflammatory response through TLR4 signaling pathway, and participate in type I interferon production. These findings may lead to an improved understanding of the biological function of IFITM3 in inflammation.
MATERIALS AND METHODS
Reagents
LPS (E. coli O55:B5), and puromycin were bought from Sigma (St Louis, MO, USA). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were obtained from Gibco (Life Technologies, Carlsbad, CA, USA). Trizol reagent, Prime Script RT reagent kit, SYBR Premix Ex Taq, and pMD19-T were purchased from TaKaRa Bio Inc. (Shiga, Japan). PB vector and PB transposes were obtained from SBI Ltd. (Palo Alto, CA, US). Other reagents were purchased from Sino Pharm Chemical Reagent Ltd. (Shanghai, China).
Tissues samples
A total of 13 tissues samples including skin, muscle, fat, heart, liver, spleen, kidney, lung, dudens, jejunum, ileum, cecum, rectum from 5 healthy swines (Crossbred: Duroc × Landrace × Yorkshire) were collected, washed three times in phosphate buffered saline (PBS, pH 7.2) and immediately snap-frozen in liquid nitrogen before being stored at -80◦C for further use.
Cloning IFITM3 gene
Swine IFITM3 genomic sequence (NM_001201382.1) was searched in the GenBank database. Using this sequence, a pair of primers (F: 5’-ACTGTCGACAT GAACTGCGCTTCCCAGCCCTTC-3’; R: 5’-ACTGCGGCCGCGTAGCCTCTGT AATCCTTTATG-3’) containing the Sal I and Not I (TaKaRa, Shiga, Japan) restriction sites were designed. Total RNA was extracted from swine tissues using Trizol reagent (TaKaRa, Shiga, Japan), and RNA was then used for cDNA synthesis. IFITM3 fragment was amplified; the final PCR product was isolated by electrophoresis using 1 % polyacrylamide gels, and purified using an agarose gel extraction kit. The purified target fragment was ligated into the plasmid pMD19-T and then transformed into competent E.coli DH5a cells. Recombinant plasmid was extracted from bacterial colonies and plasmid solution was subjected to agarose gel electrophoresis to confirm the presence of the correct sequence of IFITM3. The length of the amplification segment was 438 bp.
Expression pattern of IFITM3 mRNA in tissues
Total RNA was prepared from the snap-frozen tissue using Trizol reagent and real-time RT-PCR was adopted to detect the expression of IFITM3 mRNA in different tissues. The relative abundance of IFITM3 mRNA was evaluated by the melt cycle threshold method and standarded with swine GAPDH mRNA level as an inter reference with specific primers for GAPDH (listed in Table 1).
Table 1. Primers used for RT-PCR amplification.
| Gene | Primers | Length (bp) | Access No. |
|---|---|---|---|
| IFITM3 | F: 5’- TGGTGGGAGACATCATTGGG-3’ R: 5’- GAAAATTACCAGGGAGCCAGTG-3’ | 134 | NM_001201382.1 |
| IFNα | F: 5’- TGGTGCATGAGATGCTCCA-3’ R: 5’- GCCGAGCCCTCTGTGCT-3’ | 55 | XM_021062981.1 |
| IFNβ | F: 5’- TCGCTCTCCTGATGTGTTTC-3’ R: 5’- TTCTGACATGCCAAATTGCT-3’ | 94 | NM_001003923.1 |
| p38 | F: 5’- CTTACGGATGACCACGTTCAGT-3’ R: 5’- GCTCACAGTCTTCATTCACAGC-3’ | 127 | XM_005318767.3 |
| TNFα | F: 5’- AACCCTCTGGCCCAAGGA-3’ R: 5’- GGCGACGGGCTTATCTGA-3’ | 57 | NM_214022.1 |
| TBK1 | F: 5’- ACAGATTTTGGTGCAGCCAG-3’ R: 5’- CCTTATTCCTACGTGGCCCT-3’ | 229 | XM_021090854.1 |
| NFκB | F: 5’- CCCATGTAGACAGCACCACCTATGAT-3’ R: 5’- ACAGAGGCTCAAAGTTCTCCACCA-3’ | 132 | NM_001048232.1 |
| TLR4 | F: 5’- TCAGTTCTCACCTTCCTCCTG-3’ R: 5’- GTTCATTCCTCACCCAGTCTTC-3’ | 166 | NM_001293316.1 |
| GAPDH | F: 5’- TGGTGAAGGTCGGAGTGAAC-3’ R: 5’- GGAAGATGGTGATGGCCTTTC-3’ | 369 | XM_021091114.1 |
Vectors constructions
Construction of the eukaryotic expression vector PB-IFITM3 was as follows. IFITM3 cDNA (438 bp length) was subcloned from pMD19-T-IFITM3 into the PB vector (that contained GFP and puromycin resistance genes) after double restriction enzyme digestion. The obtained recombinant expression vector was called PB-IFITM3. Nhe I and EcoR I were used to digest PB-IFITM3, and then identified them through agarose gel electrophoresis. Sequencing using both forward and reverse primers was performed to confirm the above recombinant vectors.
Cell culture, transfection, and photograph
PK15 porcine kidney epithelial cells were maintained in DMEM medium including 10 % FBS (v/v) and 1 % penicillin/streptomycin (v/v), and cultured at 37°C with 5 % CO2 (MCO-5AC CO2 Incubator, Sanyo, Tokyo, Japan). Prior to transfection experiments, PK15 cells were plated at the density 2×105 cells per well in six-well plates. The PK15 cells were transfected with PB-IFITM3 and correspondingly in parallel with empty vector, using lipofectamine 2000 reagent (Life Technologies), according the manufacturer’s protocol. Based on transfecting plasmids, cells were divided into three groups: PB-IFITM3, PB-vector and non-transfection. After 12 h transfecting, the images were captured using DAS microscope Leitz DM RB with a dual mode cooled charged coupled device (CCD) camera (C4880; Hamamatsu, Japan) to observe GFP expression. The transfected cells were selected for on 5 μg/mL puromycin for 2 weeks. Cells that were not transfected with the target gene were killed, and we achieved stable transfection cell lines, named PK15-PB-IFITM3 (PBIFM3), and PK15-PB vector (PBv).
Transfection efficiency and overexpression detection
The PB vector carries both the purine gene and the GFP gene. Therefore, when the PB vector was transfected into cells and integrated into the genome, GFP protein was expressed in the host cells. After 12 h of transfection, the cells were observed under the fluorescence microscope to evaluate transfection efficiency. Overexpression of target gene (IFITM3) was detected using RT-PCR.
Cell treatment
The stably transfected cell lines, PBIFM3 and PBv, were divided respectively into 2 groups: control and LPS. In LPS groups, cells were incubated correspondingly with LPS (100 μg/mL) for 4, 8, 12 h. Cells in 0 h group were cultured in basal medium. After treatment, cells were collected to carry out target genes analysis using real-time PCR.
cDNA synthesis and PCR
Cells were seeded in a 6 well plates at 2×105 cells/well and treated with LPS at the concentration of 100 μg/mL. After 4, 8 and 12 h incubation, total RNA was extracted from cultured cells using Trizol reagent, and cDNA was prepared using a reverse transcriptase kit. PCR analysis was performed with a SYBR Green PCR Kit using a real-time fluorescence quantitative PCR instrument (Eppendorf Mastercycler eprealplex, Hamburg, Germany), with a reaction volume of 20 μL. The PCR program was as follows: 95 °C 15 s, 60 °C 20 s, and 72 °C 20 s, for 40 cycles.
Swine target gene mRNA specific primers are listed in Table 1. The synthesis of all primers was carried out by Shanghai Sangon Co. Ltd. (Shanghai, China). Each sample was analyzed in triplicate, and RT-PCR results were analyzed and evaluated using the relative quantity Ct method. Melting curve analysis was used to confirm the specificity of primers. The expression of the target genes were normalized as the ratio of target gene/GAPDH mRNA.
Statistical analysis
All results are presented as the mean ± standard deviation. SPSS (Statistical Package for the Social Sciences) statistical software (version 13.0) was used to analyze data (IBM SPSS Inc., Armonk, NY, USA). One-way analysis of variance (ANOVA) was employed to analyze the differences among groups. P-values less than 0.05 were taken to indicate statistically significant differences.
Acknowledgments
These authors are grateful to Yue-Ting Zheng and Jiang Wang for their suggestions in vector construction, and Chao Zhang, Bei-Bei Chu support in preparing this paper.
Abbreviations
- IFNs
interferons
- IFITM3
Interferon inducible trans-membrane protein 3
- TLR4
toll like receptor 4
- LPS
lipopolysaccharide
- PB
Piggyback
- GFP
green fluorescent protein
- MyD88
myeloid differentiation 88
- NFκB
Nuclear factor-κB
- p38 MAPK
p38 mitogen-activated protein kinases
- TNFα
tumor necrosis factor alpha
- TBK1
TANK-binding kinase 1
- bp
base pairs
- IL-1β
interleukin 1 beta
- CDS
coding sequence
Author contributions
Guo-Yu Yang and Yue-Ying Wang designed the experiments. He-Ping Li, Pei-Ge Chen and Yue-Ying Wang wrote manuscript. Fu-Tao Liu, Guang-Ming Zha, Yu-Jie Guo, Li-Qiang Han, and Xian-Qin Jiao conrtibuted to the experiments and writing. He-Shui Zhu, Kai Zhong and Wei-Fei Lu analyzed the data. All authors have read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This work was supported by the National Key Research and Development Program of China (2016YFD0500503), the National Natural Science Fund (31470120), Genetical modified Grant from Agricultural Ministry (2014ZX0801015B) and Grant from the Ministry of Agriculture (2011-G35). This project was partially supported by the Natural Science Fund of Henan Province (17A230016, 112102310705, 15A230022, 14A230010, 152300410070, 15A230017) and Zhengzhou City Natural Science Fund (141PPTGG407). The funding sources had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
REFERENCES
- 1.Kimman TG, Cornelissen LA, Moormann RJ, Rebel JM, Stockhofe-Zurwieden N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine. 2009;27:3704–18. doi: 10.1016/j.vaccine.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi T, Takaoka A. The interferon-/ system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr Opin Immunol. 2002;14:111–116. doi: 10.1016/S0952-7915(01)00305-3. [DOI] [PubMed] [Google Scholar]
- 3.Siegrist F, Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res. 2011;31:183–97. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- 4.Seyfried NT, Huysentruyt LC, Atwood JA, 3rd, Xia Q, Seyfried TN, Orlando R. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: a membrane proteomics approach. Cancer Lett. 2008;263:243–52. doi: 10.1016/j.canlet.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan J, Peng Z, Zhou C, Qiu G, Tang H, Sun Y, Wang X, Li Q, Le X, Xie K. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer. 2008;113:266–75. doi: 10.1002/cncr.23551. [DOI] [PubMed] [Google Scholar]
- 6.Hisamatsu T, Watanabe M, Ogata H, Ezaki T, Hozawa S, Ishii H, Kanai T, Hibi T. Interferon-inducible gene family 1-8U expression in colitis-associated colon cancer and severely inflamed mucosa in ulcerative colitis. Cancer Res. 1999;59:5927–31. [PubMed] [Google Scholar]
- 7.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–54. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol. 2010;84:8332–41. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidner JM, Jiang D, Pan XB, Chang J, Block TM, Guo JT. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84:12646–57. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J Biol Chem. 2013;288:17261–71. doi: 10.1074/jbc.M112.438515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–21. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 14.Seo GS, Lee JK, Yu JI, Yun KJ, Chae SC, Choi SC. Identification of the polymorphisms in IFITM3 gene and their association in a Korean population with ulcerative colitis. Exp Mol Med. 2010;42:99–104. doi: 10.3858/emm.2010.42.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, Perret C, Romagnolo B. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66:1949–55. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- 16.Li D, Peng Z, Tang H, Wei P, Kong X, Yan D, Huang F, Li Q, Le X, Li Q, Xie K. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 2011;17:3558–68. doi: 10.1158/1078-0432.CCR-10-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, Wang H, Zong G, Li P. The role of IFITM3 in the growth and migration of human glioma cells. BMC Neurol. 2013;13:210. doi: 10.1186/1471-2377-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spachidou MP, Bourazopoulou E, Maratheftis CI, Kapsogeorgou EK, Moutsopoulos HM, Tzioufas AG, Manoussakis MN. Expression of functional Toll-like receptors by salivary gland epithelial cells: increased mRNA expression in cells derived from patients with primary Sjögren’s syndrome. Clin Exp Immunol. 2007;147:497–503. doi: 10.1111/j.1365-2249.2006.03311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olofsson KM, Hjertner B, Fossum C, Press CM, Lindberg R. Expression of T helper type 17 (Th17)-associated cytokines and toll-like receptor 4 and their correlation with Foxp3 positive cells in rectal biopsies of horses with clinical signs of inflammatory bowel disease. Vet J. 2015;206:97–104. doi: 10.1016/j.tvjl.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Ferro M, Serrano Del Castillo C, Sánchez-Pernaute O. Cell membrane-bound TLR2 and TLR4: potential mredictors of active systemic lupus erythematosus and lupus nephritis. J Rheumatol. 2016;43:1444–5. doi: 10.3899/jrheum.151386. [DOI] [PubMed] [Google Scholar]
- 21.Menghini R, Campia U, Tesauro M, Marino A, Rovella V, Rodia G, Schinzari F, Tolusso B, di Daniele N, Federici M, Zoli A, Ferraccioli G, Cardillo C. Toll-like receptor 4 mediates endothelial cell activation through NF-B but is not associated with endothelial dysfunction in patients with rheumatoid arthritis. PLoS One. 2014;9:e99053. doi: 10.1371/journal.pone.0099053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duperray A, Barbe D, Raguenez G, Weksler BB, Romero IA, Couraud PO, Perron H, Marche PN. Inflammatory response of endothelial cells to a human endogenous retrovirus associated with multiple sclerosis is mediated by TLR4. Int Immunol. 2015;27:545–53. doi: 10.1093/intimm/dxv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordiglieri C, Marolda R, Franzi S, Cappelletti C, Giardina C, Motta T, Baggi F, Bernasconi P, Mantegazza R, Cavalcante P. Innate immunity in myasthenia gravis thymus: pathogenic effects of Toll-like receptor 4 signaling on autoimmunity. J Autoimmun. 2014;52:74–89. doi: 10.1016/j.jaut.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Mudhasani R, Tran JP, Retterer C, Radoshitzky SR, Kota KP, Altamura LA, Smith JM, Packard BZ, Kuhn JH, Costantino J, Garrison AR, Schmaljohn CS, Huang IC, et al. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J Virol. 2013;87:8451–64. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Qian P, Wu Q, Liu S, Fan W, Zhang K, Wang R, Zhang H, Chen H, Li X. Swine interferon-induced transmembrane protein, sIFITM3, inhibits foot-and- mouth disease virus infection in vitro and in vivo. Antiviral Res. 2014;109:22–9. doi: 10.1016/j.antiviral.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayana SK, Helbig KJ, McCartney EM, Eyre NS, Bull RA, Eltahla A, Lloyd AR, Beard MR. The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J Biol Chem. 2015;290:25946–59. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Zeng H, Kumar A, Belser JA, Maines TR, Tumpey TM. Constitutively expressed IFITM3 protein in human endothelial cells poses an early infection block to human influenza viruses. J Virol. 2016;90:11157–11167. doi: 10.1128/JVI.01254-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D. GenISIS Investigators; MOSAIC Investigators. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–23. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci USA. 2014;111:769–74. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Zhang A, Wan Y, Liu X, Qiu C, Xi X, Ren Y, Wang J, Dong Y, Bao M, Li L, Zhou M, Yuan S, et al. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brem R, Oraszlan-Szovik K, Foser S, Bohrmann B, Certa U. Inhibition of proliferation by 1-8U in interferon-alpha-responsive and non-responsive cell lines. Cell Mol Life Sci. 2003;60:1235–48. doi: 10.1007/s00018-003-3016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott R, Siegrist F, Foser S, Certa U. Interferon-alpha induces reversible DNA demethylation of the interferon-induced transmembrane protein-3 core promoter in human melanoma cells. J Interferon Cytokine Res. 2011;31:601–8. doi: 10.1089/jir.2010.0134. [DOI] [PubMed] [Google Scholar]
- 34.Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–69. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Mayhew D, Chen X, Johnston M, Mitra RD. “Calling cards” for DNA-binding proteins in mammalian cells. Genetics. 2012;190:941–9. doi: 10.1534/genetics.111.137315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Ewis H, Hice RH, Malani N, Parker N, Zhou L, Feschotte C, Bushman FD, Atkinson PW, Craig NL. A resurrected mammalian hAT transposable element and a closely related insect element are highly active in human cell culture. Proc Natl Acad Sci USA. 2013;110:E478–87. doi: 10.1073/pnas.1121543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–55. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 38.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–58. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 39.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 40.Kubota K, Sakaki H, Imaizumi T, Nakagawa H, Kusumi A, Kobayashi W, Satoh K, Kimura H. Retinoic acid-inducible gene-I is induced in gingival fibroblasts by lipopolysaccharide or poly IC: possible roles in interleukin-1beta, -6 and -8 expression. Oral Microbiol Immunol. 2006;21:399–406. doi: 10.1111/j.1399-302X.2006.00326.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Wu S, Jin X, Li M, Chen S, Teeling JL, Perry VH, Gu J. Retinoic acid-inducible gene-I mediates late phase induction of TNF-alpha by lipopolysaccharide. J Immunol. 2008;180:8011–9. doi: 10.4049/jimmunol.180.12.8011. [DOI] [PubMed] [Google Scholar]
- 42.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–17. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 43.Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773:1358–75. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Zhao W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 2013;587:542–8. doi: 10.1016/j.febslet.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakajima A, Ibi D, Nagai T, Yamada S, Nabeshima T, Yamada K. Induction of interferon-induced transmembrane protein 3 gene expression by lipopolys- accharide in astrocytes. Eur J Pharmacol. 2014;745:166–75. doi: 10.1016/j.ejphar.2014.08.034. [DOI] [PubMed] [Google Scholar]