Abstract
Reduced expression of Cadherin-Related Family Member 5 (CDHR5) was recently found implied in carcinogenesis of colon cancer, but its role in other tumors is unknown. We aimed to analyze the expression of CDHR5 in different subtypes of renal cell carcinoma. CDHR5 expression was immunohistochemically examined using tissue micro arrays (TMAs) covering 279 patients with primary renal cell carcinoma. Additionally, expression data from the TCGA (The Cancer Genome Atlas) of an independent cohort of 489 clear-cell RCC cases was evaluated. CDHR5 protein expression was found in 74.9% of cases, with higher levels seen in clear cell and papillary RCC. In the univariate analysis CDHR5 expression was significantly associated with a longer overall survival of RCC patients at the protein (p = 0.026, HR = 0.56) and transcript levels (TCGA-cohort: p = 0.0002, HR = 0.55). Importantly, differences in survival times were confirmed independently in multivariate analyses in a model with common clinicopathological variables at the transcript level (p = 0.0097, HR = 0.65). Investigation of the putative functional role of CDHR5 using TCGA data and Enrichment analysis for Gene Ontology and Pathways revealed associations with many metabolic and some tumor growth-associated processes and pathways. CDHR5 expression appears to be a promising and new independent prognostic biomarker in renal cell carcinoma.
Keywords: CDHR5, renal cell carcinoma, immunohistochemistry, prognostic marker
INTRODUCTION
In Germany, renal cell carcinoma (RCC) is the 8th most common cancer type in men and 10th most in women with 30,400 new cases and 10,500 deaths estimated for 2012 in Germany referring to a mortality of about 34% [1]. From 2000 to 2010 the cancer incidence increased by almost 10%. The majority of patients present at the time of initial diagnosis with relatively small tumors that are treated by radical nephrectomy or, as Van Poppel et al. [2] showed for low-stage RCC, by nephron-sparing surgery. However, 33% of the patients show symptomatic metastases at the time of diagnosis [3]. Furthermore, about 40% of the patients will develop metastasis or recurrences after nephrectomy [4, 5]. The 5-year relative survival rates range from 75% for men to 77% for women [6].
Metastatic RCC shows low response rates to standard chemotherapy. The management of advanced RCC has dramatically changed over the past decade. New multitargeted cancer therapies demonstrated improved response rates and a survival benefit for the patient, however, most patients with advanced RCC still have limited overall survival [7]. The large number of pathways, which are changed in tumor cells, open up the possibility for targeted new treatment options [8, 9].
Concerning this field of research, cell adhesion molecules, such as cadherin superfamily might be key players in developmental processes regarding to cell segregation and tumorigenesis [10, 11]. For some members of cadherin superfamily like E-cadherin and N-cadherin numerous clinical studies have shown that decreased expression of E-cadherin and thus overexpression of N-Cadherin promote motility and invasion [12]. The influence of cadherin’s could be shown for various tumor entities, including hereditary gastric cancer [13], breast cancer [14, 15] and renal cell carcinoma [16].
We decided to perform a further evaluation of the Cadherin-Related Family Member 5 (CDHR5), which is a protocadherin genomically located in the 11p15.5 chromosome region. Aberrations in this region are also associated with the Beckwith-Wiedmann syndrome and show as a second locus, a loss of heterozygosity in Wilms tumors [17-19]. Moreover CDHR5 is a protocadherin with a characteristic mucin-type repeat structure and two isoforms [20]. It is expressed in the basolateral membrane of epithelial structures during kidney and lung development [21]. CDHR5 plays an important role in brush border assembly in intestinal epithelium [22]. As it is reported for the cadherin superfamily, CDHR5 is a Ca2+-dependent cell-cell adhesion molecule including a possible role as a receptor for extracellular signals [23].
We aimed to assess the expression of CDHR5 in benign renal tissues and RCC and correlate these findings to clinicopathologic parameters including patient survival in two independent cohorts (Charité-Universitätsmedizin Berlin, Germany and the RCC cohort of The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/).
RESULTS
CDHR5 expression in renal and cancerous tissues
In normal renal tissues, CDHR5 was immunohistochemically detected in the proximal tubule with a pronunciation at the luminal brush border. The glomeruli, the distal tubuli, the collecting ducts and the stromal cells were all negative.
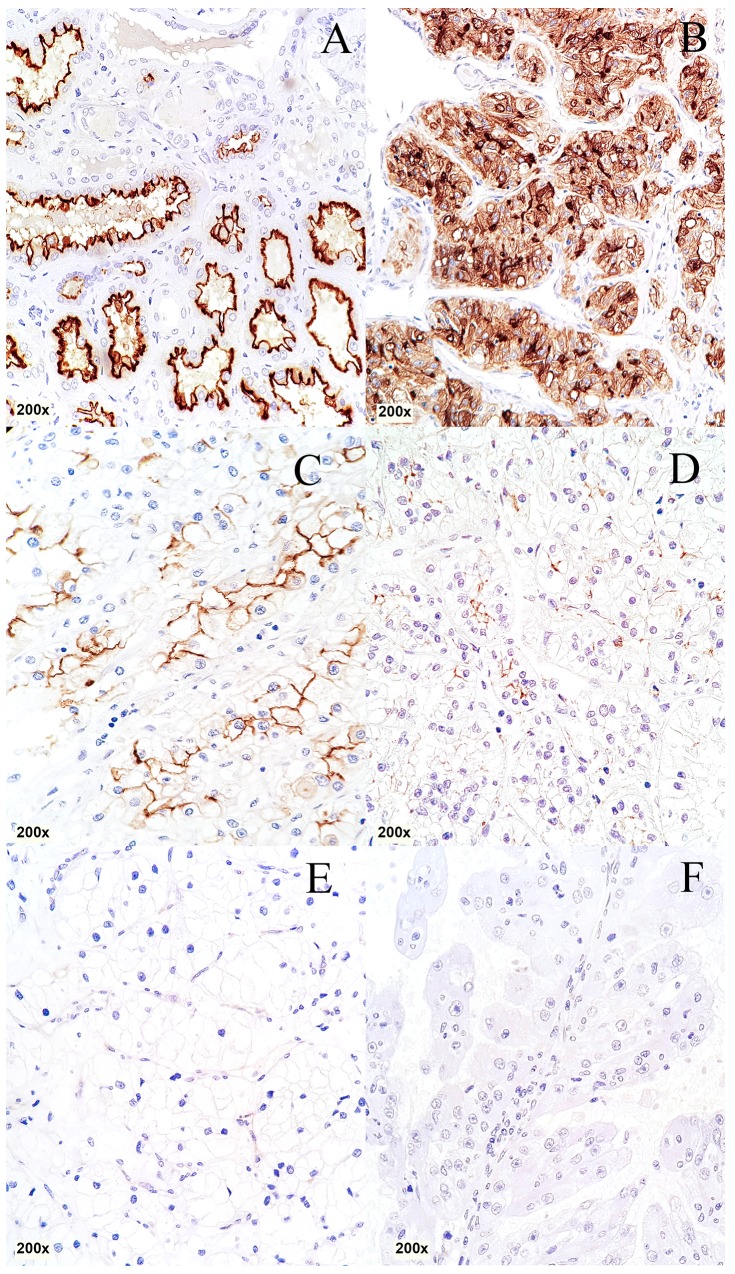
Among the cancerous tissue no CDHR5 expression could be observed in 70 of 279 evaluable cases (25.1%), whereas 125/279 (44.8%) showed a weak, 66/279 (23.7%) revealed a moderate and 18/279 (6.5%) showed a strong CDHR5 immunoreactivity. Representative immunostainings of CDHR5 in RCC are given in Figure 1.
Figure 1. Immunohistochemical staining of CDHR5 in different histological subtypes of RCC.
(A) Normal kidney, (B) clear cell RCC: positive “3+”, (C) clear cell RCC: positive “2+”, (D) clear cell RCC: positive “1+”, (E) clear cell RCC: negative “0”, (F): papillary RCC: negative “0”. (total magnification for every image 200x)
A stratified analysis of CDHR5 expression according to histological tumor subtype showed, that CDHR5 is mostly expressed in clear cell and papillary RCC (77.3% and 78.1% positive cases, respectively) whereas chromophobe RCC (n=9) showed no expression at all (Table 1).
Table 1. Associations of CDHR5 expression with clinicopathological parameters of RCC patients.
| All patients (n=279) | CDHR5-negative | CDHR5-positive | p-Value | |
|---|---|---|---|---|
| Median follow-up [months] | 87 (0-177) |
|||
| Evaluable cases | 279 | 70 (25.1%) | 209 (74.9%) | |
| Age, mean (range/SD) | 62 (30-86) | 61.0±11.4 | 61.2±9.4 | 0.89 |
| Gender | 0.049* | |||
| Men | 190 (68.1%) | 41 (21.6%) | 149 (78.4%) | |
| Women | 89 (31.9%) | 29 (32.6%) | 60 (67.4%) | |
| Histology | 0.918** | |||
| Clear cell | 238 (85.3%) | 54 (22.7%) | 184 (77.3%) | |
| Papillary | 32 (11.5%) | 7 (21.9%) | 25 (78.1%) | |
| Chromophobe | 9 (3.2%) | 9 (100.0%) | 0 (0%) | |
| pT status | 0.003$ | |||
| pT1 | 158 (56.6%) | 26 (16.5%) | 132 (83.5%) | |
| pT2 | 25 (9.0%) | 10 (40.0%) | 15 (60.0%) | |
| pT3a | 37 (13.2%) | 11 (29.7%) | 26 (70.3%) | |
| pT3b | 53 (19.0%) | 20 (37.7%) | 33 (62.3%) | |
| pT3c/pT4 | 6 (2.2%) | 3 (50.0%) | 3 (50.0%) | |
| pN status | 0.305* | |||
| pN0 | 144 (51.6%) | 37 (25.7%) | 107 (74.3%) | |
| pN1 | 19 (6.8%) | 7 (36.8%) | 12 (63.2%) | |
| pNx | 116 (41.6%) | |||
| ISUP grade | 0.419$ | |||
| G1 | 44 (16.3%) | 10 (22.7%) | 34 (77.3%) | |
| G2 | 152 (56.3%) | 33 (21.7%) | 119 (78.3%) | |
| G3 | 59 (21.9%) | 12 (20.3%) | 47 (79.7%) | |
| G4 | 15 (5.6%) | 6 (40.0%) | 9 (60.0%) | |
| No grade (chromophobe) | 9 | |||
| Metastasis | - | |||
| M0/x | 243 (87.1%) | 59 (24.3%) | 184 (75.7%) | |
| M1 synchronic | 25 (9.0%) | 7 (28.0%) | 18 (72.0%) | |
| M1 asynchronic | 11 (3.9%) | 4 (36.4%) | 7 (63.6%) | |
| ECOG performance status | 0.166$ | |||
| 0 | 189 (67.7%) | 43 (22.8%) | 146 (77.2%) | |
| 1 | 81 (29.0%) | 26 (32.1%) | 55 (67.9%) | |
| 2 | 9 (3.2%) | 1 (11.1%) | 8 (88.9%) |
* Mann-Whitney, ** Mann-Whitney for Clear cell vs Papillary histology, $ - Kruskall-Wallis-Test, SD – standard deviation.
In 100 (35.8%) of the 279 RCC tumor samples CDHR5 was expressed solely luminal. In the remaining cases the CDHR5 expression was diffuse or lost. In two thirds (157 of 238) of the clear-cell RCC cases, CDHR5 was expressed not only luminally, whereas, in 59.4% of the papillary RCCs (19 of 32) CDHR5 was exclusively expressed at the luminal brush border.
Association of CDHR5 expression with clinicopathological parameters and overall survival
Tissue samples from 279 RCC patients were analyzed (immunohistochemistry). A weak correlation between CDHR5 expression and the pT-stage (r = -0.143, p = 0.005) was found. A higher percentage of men showed CDHR5 expression with on average higher CDHR5 levels compared to women (p = 0.049). No significant association could be drawn between CDHR5 expression and (I) the pN-, M- and R-staging (II) patient age and (III) the ISUP Grade (Table 1).
For survival analyses we have used only the patients with clear cell histology (ccRCC) of RCC (n=238) to enable further comparisons to TCGA-data. CDHR5 expression was dichotomized into CDHR5-negative (n = 54) vs. CDHR5-positive (n = 184) cases. In a univariate Cox proportional Hazard analysis CDHR5-positive ccRCC patients showed a significantly lower risk of death compared to the CDHR5-negative cases (p = 0.027, HR = 0.58, CI 95% [0.358-0.939]). This finding could be confirmed in a Kaplan-Meier analysis (p = 0.025, Figure 2A). In a multivariate survival analysis with inclusion of the pT- and pN-stage, R-status and ISUP grading CDHR5 expression (immunohistochemistry) demonstrated no independent prognostic value (Table 2). Similar results for univariate and multivariate analyses were received in analysis with inclusion of clear cell and papillary histological types of RCC together (data not shown).
Figure 2. Kaplan-Meier analysis of overall survival.
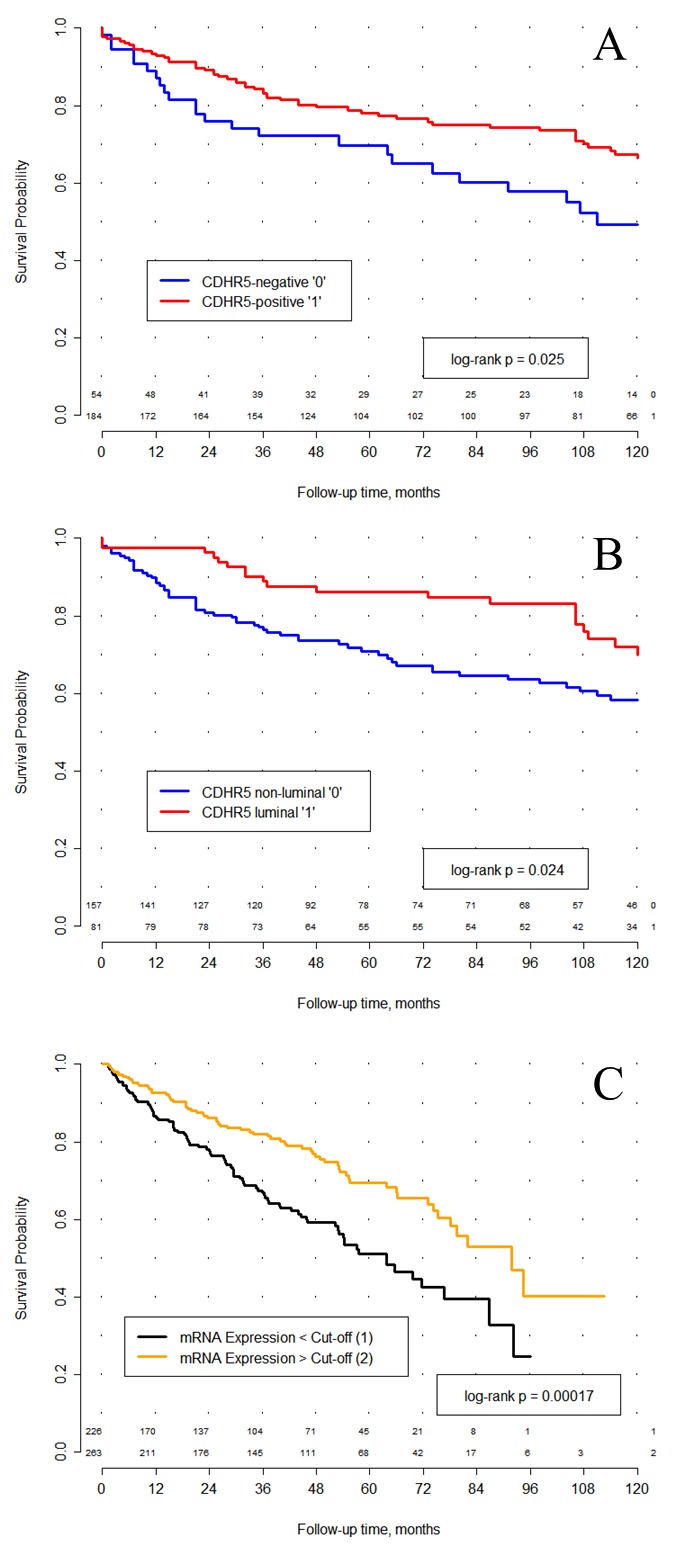
(A) In a cohort comprised of 238 renal cell carcinoma (RCC) patients with clear cell histology a significant association between CDHR5 expression and overall survival after surgery was observed. CDHR5 expression was dichotomized into CDHR5-negative (n = 54) vs. CDHR5-positive (n = 184), based on the complete absence or presence of any immunohistochemical protein expression. (B) CDHR5 expression was dichotomized into luminal CDHR5 expression (n = 81) vs. diffuse or no CDHR5 expression (n = 157). In a cohort comprised of 238 clear-cell RCC cases, patients with luminal CDHR5 expression showed a significant longer overall survival after surgery compared to those with no or diffuse CDHR5 expression. (C) In 489 RCC patient samples from the TCGA cohort the prognostic value of CDHR5 expression could be confirmed (log rank p = 0.00017). CDHR5 expression values (RNASeq) were dichotomized based on the best cut-off.
Table 2. Univariate and multivariate Cox proportional hazard analyses on overall survival in a sub-cohort of clear-cell renal cell carcinoma patients (n=238).
| Clinicopathological parameter / biomarker | Univariate Cox | Multivariate Cox | ||
|---|---|---|---|---|
| Hazard ratio [95% CI] | p-Value | Hazard ratio [95% CI] | p-Value | |
| Tumor stage: | ||||
| pT1 | 1.00 | - | 1.00 | - |
| pT2 | 2.69 [1.14-6-36] | 0.024 | 1.72 [0.68-4.38] | 0.255 |
| pT3+pT4 | 5.77 [3.43-9.72] | <0.001 | 4.07 [2.29-7.21] | <0.001 |
| ISUP grade: | ||||
| Grade 1 | 1.00 | - | 1.00 | - |
| Grade 2 | 3.05 [1.21-7.71] | 0.018 | 2.35 [0.92-6.04] | 0.075 |
| Grade 3 | 4.36 [1.64-11.56] | 0.003 | 3.15 [1.16-8.54] | 0.024 |
| Grade 4 | 9.48 [3.17-28.34] | <0.001 | 4.34 [1.36-13.85] | 0.013 |
| Surgical margin (R1 vs. R0) | 7.64 [3.96-14.75] | <0.001 | 3.25 [1.63-6.45] | 0.001 |
| Nodal status (pN1 vs. pN0/x) | 3.35 [1.72-6.52] | <0.001 | 1.25 [0.58-2.68] | 0.558 |
| CDHR5 immunohistochemistry (negative vs positive) | 1.73 [1.07-2.80] | 0.027 | 1.36 [0.81-2.26] | 0.243 |
| CDHR5 immunohistochemistry (luminal vs non luminal) | 0.56 [0.34-0.93] | 0.026 | # | # |
#CDHR5 luminal vs non luminal was not significant in the multivariate Cox proportional hazard analysis in the model with the same list of parameters.
Furthermore, the effect of the luminal limited CDHR5-expression on the prognosis of the RCC-patients (clear-cell and papillary) and on the correlation with clinicopathological parameters was examined. As described for the CDHR5-expression-strength a correlation between the luminal localization of the CDHR5-expression and the T-stage (r = -0.209, p = 0.001) could be found. No correlation between luminal expression of CDHR5 and the tumor grade (ISUP grading) was observed. Patients with tumors that at least partially comprise a tubulus like CDRH5-positivity had a favorable prognosis compared to those patients where the CDHR5-expression in the tumors is lost or not luminal restricted (p = 0.009, HR = 0.53, CI 95% [0.33-0.85]). This finding could be confirmed in a Kaplan-Meier analysis (p = 0.007, Figure 2B)
Analysis of prognostic role and investigation of putative biological role (TCGA-data)
Results from survival analysis were validated in an independent testing cohort comprised of clear cell RCC patients included in TCGA. A best cut-off for transcript number, based on the cut-off analysis (2580.166), classified patients approximately in the middle of the cohort into mCDHR5-low (n = 226), and mCDHR5-high (n = 263) groups. In univariate Cox proportional hazards analysis high CDHR5 mRNA expression (as assessed by RNASeq) showed a significantly lower risk of earlier death compared to patients with low CDHR5 mRNA expression (p = 0.0002, HR = 0.55, 95% CI [0.40-0.75]; Table 3). This finding was confirmed in a Kaplan-Meier analysis of overall survival (log rank p = 0.0002, Figure 2C).
Table 3. Univariate and multivariate Cox proportional hazard analyses on overall survival of clear-cell renal cell carcinoma patients from the TCGA cohort (n=489).
| Clinicopathological parameter / biomarker# | Univariate Cox | Multivariate Cox | ||
|---|---|---|---|---|
| Hazard ratio [95% CI] | p-Value | Hazard ratio [95% CI] | p-Value | |
| Tumor stage | ||||
| pT1 | 1.00 | - | 1.00 | - |
| pT2 | 1.44 [0.82-2.52] | 0.202 | 1.28 [0.72-2.26] | 0.401 |
| pT3a | 3.61 [2.45-5.33] | 9.68e-11 | 2.41 [1.58-3.69] | 4.68e-05 |
| pT3b | 3.46 [2.10-5.70] | 1.14e-06 | 2.57 [1.53-4.31] | 0.0004 |
| pT3c | 8.22 [1.98-34.13] | 0.004 | 4.55 [1.07-19.37] | 0.040 |
| pT4 | 12.55 [6.29-25.07] | 7.55e-13 | 4.64 [1.83-11.78] | 0.001 |
| ISUP grade | ||||
| Grade 2 | 1.00 | - | 1.00 | - |
| Grade 3 | 1.88 [1.26-2.81] | 0.002 | 1.48 [0.98-2.24] | 0.060 |
| Grade 4 | 5.67 [3.72-8.66] | 8.88e-16 | 2.83 [1.73-4.62] | 3.33e-05 |
| Nodal status (pN1 vs. pN0/x) | 3.75 [1.97-7.13] | 5.72e-05 | 1.12 [0.49-2.78] | 0.729 |
| CDHR5 mRNA Expression (high vs low*) | 0.55 [0.40-0.75] | 0.0002 | 0.65 [0.47-0.90] | 0.0097 |
# Surgical margin status was not available. * “high” means > cut-off, “low” means < cut-off.
A multivariate Cox proportional hazards of the CDHR5 mRNA expression using selected cut-off in the model including ISUP grade, pT-, pN-stage and R-Status showed an independent significant prognostic value (p = 0.0097, HR = 0.65, 95% CI [0.47-0.90]; Table 3).
The enrichment analysis for Gene Ontology and Pathways as an investigation tool for putative biological role of CDHR5 have shown the associations of this gene with important metabolic pathways (including many interactions with genes related to transmembrane and intracellular transport of different substances) and also with pathways related to cell motion, cell migration, apoptosis induction and signal transduction (Supplementary Figures 1 and 2).
DISCUSSION
In this study we showed for the first time CDHR5 expression in a population based RCC patient cohort. CDHR5 positivity is significantly associated with a longer overall survival time. The association between CDHR5 expression and RCC patient survival at the transcript level (TCGA RNAseq data) remained significant after multivariable adjustment for other clinicopathological parameters, including pT-, pN- categories, surgical margin status and grade. These results were confirmed in an independent RCC patient cohort from TCGA. Thus, the association appeared to be independent of current prognostic factors and could offer additional relevant prognostic information.
In the kidney, CDHR5 is primarily located at the luminal brush border in proximal tubules. So far, the physiological functions of CDHR5 are not sufficiently characterized. Similar to classical cadherins it is calcium ion binding and also β-catenin binding [24]. As a member of cadherin superfamily it can be assumed that CDHR5 mediates cell-cell adhesion and cell-signalling. In fact, specific function in normal cells is not yet described. In relation to pathogenesis CDHR5 influence is described for gallstone disease [25] and systemic sclerosis [26]. In addition to that, CDHR5 promoter methylation is a possible prognostic biomarker for cyst growth in polycystic kidney disease [27].
In tumors CDHR5 expression is altered. Recent studies showed that CDHR5 downregulation is a common event in colorectal carcinoma cells [28, 29]. In contrast, mesalazin therapy leads to a growth arrest due to higher CDHR5 expression in colorectal carcinoma (CRC) by inducing an increased transcription of the cyclin-dependent kinase inhibitor (p21waf-1) gene [24]. Besides the Cdx2 Homeobox, which regulates the balance between proliferation and differentiation in the adult intestinal epithelium, it also regulates CDHR5 expression by binding to two consensus Cdx2-binding sites. In CRC cell lines, expression of CDHR5 leads to a reduction of colonies and a disbanding of established colonies [30]. A possible mechanism is the ability of CDHR5 to retain β-catenin on the plasmatic membrane in CRC tumor cells. β-catenin acts as a transcription factor and is normally controlled by degradation through APC, which in turn can be inhibited by Wnt signalling. Therefore, CDHR5 has a direct influence on Wnt/β-catenin pathway, which is considered important in CRC tumorigenesis [31]. It can be assumed that catenins regulate gene expression and that these proteins form a functional network in the nucleus [32]. Former studies have indicated that the Wnt/β-catenin pathway may also be relevant in renal carcinogenesis [33]. Especially, the dysregulation of β-catenin in ccRCC is a predictor of a lower tumor mortality [34] and a potential target for therapy [35].
The ability of CDHR5 to retain β-catenin at the plasma membrane and to influence Wnt signaling is therefore consistent with the results of our study, as CDHR5 expression correlates with longer overall survival times of RCC patients in general. This was particularly pronounced in cases with a strict luminal localization of CDHR5 on the luminal brush border. CDHR5 expression was especially observed in clear cell and papillary RCCs which supports the notion that these subtypes share molecular associations with epithelia from proximal tubules [36] and that a correct expression of CDHR5 is important for the microvilli and epithelial brush border differentiation [22]. CDHR5 localization showed, in contrast to expression, significantly more reliable judgment about the overall patient survival.
Our analysis of the putative functional role of the CDHR5 based on the Enrichment analysis for Gene Ontology and Pathways provided some interesting findings, which certainly need to be proved in further functional studies, namely, that CDHR5 seems to exercise its main functions in the sphere of metabolic processes, although some important tumor growth-associated processes / pathways could be also linked to this gene.
A few limitations of the study need to be mentioned. A major limitation of this retrospective study is the relatively low number of patients with a non-clear cell histology, which are not sufficiently explored by our study. The role of cell adhesion molecules such as the cadherins in renal carcinogenesis ought to be explored further. This study is only descriptive and does not provide any functional analyses of CDHR5 in RCC cells. Further studies need to provide information whether CDHR5 has a comparable function in renal cell carcinogenesis like in CRC carcinogenesis.
In summary, this is the first study, which describes CDHR5 expression in RCC. CDHR5 loss appears to be a promising prognostic biomarker for clear cell renal cell carcinoma patients. The functional role of CDHR5 and its expression patterns in other renal tumor entities need to be unraveled in future studies.
MATERIALS AND METHODS
Ethics statement
The studies were approved by the Institutional Review Board (IRB) Charite - Universitätsmedizin Berlin (EA1/06/2004).
Patients and tissue microarray construction
The patient cohort and the construction of the tissue microarray were described in former studies [37]. Our cohort consists of two hundred seventy-nine primary RCC cases which were staged with regard to International Union against Cancer 2002 criteria at the Institute of Pathology, Charité-Universitätsmedizin Berlin. Patients underwent surgery at the Department of Urology, Charité-Universitätsmedizin Berlin, between 1993 and 2004. 190 (68.1%) of the patients were male and 89 were female (31.9%). The mean age at the time of surgery was 62 years (range 30-86). The dominating histological subtype was clear cell RCC (n = 238, 85.3%), papillary RCC were less common (n=32, 11.5%), as well as chromophobe RCC were rare (n = 9, 3.2%). All case were subjected to a central review to adjust the tumor grade according to the recommendations of ISUP 2012/WHO 2016 (YT, GK). Clinical characteristics are outlines in the Table 1.
For all 279 cases follow-up data were available. The median follow-up time was 87 months (range 0-177 months). In order to avoid contamination of progression data by age-related death we truncated follow-up times after 120 months. During this time 87 patients died (31.2%) after a median survival time of 31 months.
Immunohistochemistry
The TMAs were stained in the immunohistochemistry laboratory at the Institute of Pathology, Bonn. We used the Lab Vision Autostainer 480S system (Thermo Scientific, Waltham, MA, USA) together with the Thermo Scientific Reagents and the Medac C-DPVB 500 HRP detection system. The PT-Module was used for dewaxing and epitope retrieval (pH 6.0 at 99°C for 20 min). The polyclonal anti-CDHR5 antibody HPA009173 (Sigma Aldrich) was used (dilution 1:250), which was specificity-validated by the Human Protein Atlas (www.proteinatlas.org/). Slides were counterstained with hematoxylin, dehydrated and aqueously mounted.
CDHR5 immunoreactivity was semi-quantitatively scored and categorised as follows: negative (0), weak (1), moderate (2) and strong (3). Furthermore, the pattern of CDHR5 immunoreactivity was recorded (luminal vs. diffuse or negative).
TCGA-data mining
For validation of the results from the training study, an independent series of 489 ccRCC patients with follow-up (more than 30 days) and clinicopathological data was evaluated. The patients with ISUP Grade 1 (n=8) were not used in the univariate and multivariate analysis, as they showed a uniform excellent overall survival, which distorts the analysis when those patients are used as comparator. The results from the validation cohort shown here are in whole based upon RNASeq data generated by the TCGA Research Network: (http://cancergenome.nih.gov/). Normalized results of the mRNA expression were used for analysis. Overall survival was used as the end-point. The number of death events in the cohort was 158/489 (32.3%).
TCGA data set was also used for investigations of the functional role of CDHR5. Briefly, we have taken mRNA expression data of all genes evaluated using RNAseq (20501 genes) in tumor samples of patients with clear-cell RCC. Enrichment analysis for Gene Ontology and Pathways have been carried out using TCGABiolinks package for R to reveal the functions and to integrate the CDHR5 into certain pathways based on the analysis of associations with other genes. Two modifications of this analysis were performed. Firstly, we have analysed top 100 of genes, the mRNA expression of which was highly correlated with CDHR5 (Supplementary Tables 1 and 2), among them 50 genes with positive correlation and 50 genes with negative correlation. Secondly, we have performed the additional investigations for top 100 genes with differential expression (50 upregulated and 50 downregulated) between samples with high and low CDHR5 mRNA expression using the cut-off for transcript number from our prognostic studies (Supplementary Table 3), which proved to have the highest prognostic value with regard to survival and statistical significance (Cut-off = 2580.166).
Statistics
Statistical analysis was performed with SPSS, Version 21 (IBM SPSS Statistic) and R (version 3.2.2). Fisher’s exact test and chi-square test were used to evaluate statistical significance of the associations between CDHR5 expression and clinicopathological parameters. Bivariate correlation analysis was carried out using Spearman’s Rho. Survival analyses were conducted according to uni- and multivariate Cox Proportional Hazards and Kaplan-Meier analyses. P values refer to Wald test and Log-rank test. An error value of 5% (α = 0.05) was defined to infer statistical significance. As we do not consider such long follow-up relevant for RCC-specific survival, follow-up was truncated after 10 years. Extraction of the TCGA mRNA expression data was done in R using TCGA-Biolinks package. The best cut-off for the mRNA expression on the TCGA data was selected in R using the survMisc package (automatized systematic univariate Cox regression-based analysis of all available cut-offs for mRNA expression of CDHR5 using overall survival as end-point).
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
We thank Britta Beyer (TMA construction) and Susanne Steiner (Immunohistochemistry) for excellent technical support.
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- HR
hazard ratio
- ISUP
Internation Society of Urological Pathology
- RCC
renal cell carcinoma
- TCGA
The Cancer Genome Atlas
- WHO
World Health Organization
Author contributions
Study design: GK, KJ, CS, MOG, HJT
Data acquisition: FMB, SM, YT, CL, KJ, CS, GK
Data analysis: YT, FMB, SM, JE
Manuscript writing: FMB, SM, YT, GK
CONFLICTS OF INTEREST
All authors report no conflicts of interest.
FUNDING
Supported by EU grant FP7-PEOPLE-2010-IRSES (Marie Curie Actions–International Research Staff Exchange Scheme), project number 269186 “KRAB-ZNF–KRAB zinc finger gene biology in evolution and disease”.
REFERENCES
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Van Poppel H, Da Pozzo L, Albrecht W, Matveev V, Bono A, Borkowski A, Marechal JM, Klotz L, Skinner E, Keane T, Claessens I, Sylvester R, European Organization for Research and Treatment of Cancer (EORTC) A prospective randomized EORTC intergroup phase 3 study comparing the complications of elective nephron-sparing surgery and radical nephrectomy for low-stage renal cell carcinoma. Eur Urol. 2007;51:1606–15. doi: 10.1016/j.eururo.2006.11.013. https://doi.org/10.1016/j.eururo.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Pascual D, Borque A. Epidemiology of kidney cancer. Adv Urol. 2008;2008:782381. doi: 10.1155/2008/782381. https://doi.org/10.1155/2008/782381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam JS, Leppert JT, Belldegrun AS, Figlin RA. Novel approaches in the therapy of metastatic renal cell carcinoma. World J Urol. 2005;23:202–12. doi: 10.1007/s00345-004-0466-0. https://doi.org/10.1007/s00345-004-0466-0. [DOI] [PubMed] [Google Scholar]
- 5.Montie JE. Follow-up after partial or total nephrectomy for renal cell carcinoma. Urol Clin North Am. 1994;21:589–92. [PubMed] [Google Scholar]
- 6.German Centre for Cancer Registry Data Cancer in Germany 2009-2010. 2013.
- 7.Bedke J, Gauler T, Grünwald V, Hegele A, Herrmann E, Hinz S, Janssen J, Schmitz S, Schostak M, Tesch H, Zastrow S, Miller K. Systemic therapy in metastatic renal cell carcinoma. World J Urol. 2016;35:179–88. doi: 10.1007/s00345-016-1868-5. https://doi.org/10.1007/s00345-016-1868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oudard S, George D, Medioni J, Motzer R. Treatment options in renal cell carcinoma: past, present and future. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2007;18 Suppl 10:x25–31. doi: 10.1093/annonc/mdm411. https://doi.org/10.1093/annonc/mdm411. [DOI] [PubMed] [Google Scholar]
- 9.Bielecka ZF, Czarnecka AM, Szczylik C. Genomic analysis as the first step toward personalized treatment in renal cell carcinoma. Front Oncol. 2014;4:194. doi: 10.3389/fonc.2014.00194. https://doi.org/10.3389/fonc.2014.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–35. doi: 10.1242/jcs.000455. https://doi.org/10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 11.Paul R, Ewing CM, Jarrard DF, Isaacs WB. The cadherin cell-cell adhesion pathway in prostate cancer progression. Br J Urol. 1997;79 Suppl 1:37–43. doi: 10.1111/j.1464-410x.1997.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 12.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–88. doi: 10.1007/s00018-008-8281-1. https://doi.org/10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbier A, Guilford P. Hereditary diffuse gastric cancer. Adv Cancer Res. 2001;83:55–65. doi: 10.1016/s0065-230x(01)83002-5. [DOI] [PubMed] [Google Scholar]
- 14.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–44. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3:289–93. doi: 10.1186/bcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stassar MJ, Devitt G, Brosius M, Rinnab L, Prang J, Schradin T, Simon J, Petersen S, Kopp-Schneider A, Zöller M. Identification of human renal cell carcinoma associated genes by suppression subtractive hybridization. Br J Cancer. 2001;85:1372–82. doi: 10.1054/bjoc.2001.2074. https://doi.org/10.1054/bjoc.2001.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koufos A, Grundy P, Morgan K, Aleck KA, Hadro T, Lampkin BC, Kalbakji A, Cavenee WK. Familial Wiedemann-Beckwith syndrome and a second Wilms tumor locus both map to 11p15.5. Am J Hum Genet. 1989;44:711–9. [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg M, Wei M, Tycko B, Falikovich I, Warburton D. Identification and expression analysis of the human mu-protocadherin gene in fetal and adult kidneys. Am J Physiol Renal Physiol. 2002;283:F454–63. doi: 10.1152/ajprenal.00012.2002. https://doi.org/10.1152/ajprenal.00012.2002. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg M, Wei M, Yuan L, Murty VV, Tycko B. Biallelic expression of HRAS and MUCDHL in human and mouse. Hum Genet. 2003;112:334–42. doi: 10.1007/s00439-003-0907-7. https://doi.org/10.1007/s00439-003-0907-7. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adhes Migr. 2011;5:97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moulton DE, Crandall W, Lakhani R, Lowe ME. Expression of a novel cadherin in the mouse and human intestine. Pediatr Res. 2004;55:927–34. doi: 10.1203/01.PDR.0000125260.46861.32. https://doi.org/10.1203/01.PDR.0000125260.46861.32. [DOI] [PubMed] [Google Scholar]
- 22.Crawley SW, Shifrin DA, Grega-Larson NE, McConnell RE, Benesh AE, Mao S, Zheng Y, Zheng QY, Nam KT, Millis BA, Kachar B, Tyska MJ. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell. 2014;157:433–46. doi: 10.1016/j.cell.2014.01.067. https://doi.org/10.1016/j.cell.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg M, Peshkovsky C, Shifteh A, Al-Awqati Q. mu-Protocadherin, a novel developmentally regulated protocadherin with mucin-like domains. J Biol Chem. 2000;275:24622–9. doi: 10.1074/jbc.M000234200. https://doi.org/10.1074/jbc.M000234200. [DOI] [PubMed] [Google Scholar]
- 24.Parenti S, Ferrarini F, Zini R, Montanari M, Losi L, Canovi B, Ferrari S, Grande A. Mesalazine inhibits the beta-catenin signalling pathway acting through the upregulation of mu-protocadherin gene in colo-rectal cancer cells. Aliment Pharmacol Ther. 2010;31:108–19. doi: 10.1111/j.1365-2036.2009.04149.x. https://doi.org/10.1111/j.1365-2036.2009.04149.x. [DOI] [PubMed] [Google Scholar]
- 25.Chuang SC, Hsi E, Wang SN, Yu ML, Lee KT, Juo SH. Polymorphism at the mucin-like protocadherin gene influences susceptibility to gallstone disease. Clin Chim Acta Int J Clin Chem. 2011;412:2089–93. doi: 10.1016/j.cca.2011.07.015. https://doi.org/10.1016/j.cca.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Carmona FD, Gutala R, Simeón CP, Carreira P, Ortego-Centeno N, Vicente-Rabaneda E, García-Hernández FJ, García de la Peña P, Fernández-Castro M, Martínez-Estupiñán L, Egurbide MV, Tsao BP, Gourh P, et al. Novel identification of the IRF7 region as an anticentromere autoantibody propensity locus in systemic sclerosis. Ann Rheum Dis. 2012;71:114–9. doi: 10.1136/annrheumdis-2011-200275. https://doi.org/10.1136/annrheumdis-2011-200275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo YM, Shin Y, Hwang JA, Hwang YH, Lee S, Park EY, Kong HK, Park HC, Lee YS, Park JH. Epigenetic silencing of the MUPCDH gene as a possible prognostic biomarker for cyst growth in ADPKD. Sci Rep. 2015;5:15238. doi: 10.1038/srep15238. https://doi.org/10.1038/srep15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losi L, Parenti S, Ferrarini F, Rivasi F, Gavioli M, Natalini G, Ferrari S, Grande A. Down-regulation of μ-protocadherin expression is a common event in colorectal carcinogenesis. Hum Pathol. 2011;42:960–71. doi: 10.1016/j.humpath.2010.10.009. https://doi.org/10.1016/j.humpath.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Losi L, Grande A. Involvement of μ-protocadherin in colorectal carcinogenesis: a promise for clinico-pathological evaluation. Histol Histopathol. 2014;29:11–9. doi: 10.14670/HH-29.11. [DOI] [PubMed] [Google Scholar]
- 30.Hinkel I, Duluc I, Martin E, Guenot D, Freund JN, Gross I. Cdx2 controls expression of the protocadherin Mucdhl, an inhibitor of growth and β-catenin activity in colon cancer cells. Gastroenterology. 2012;142:875–85.e3. doi: 10.1053/j.gastro.2011.12.037. https://doi.org/10.1053/j.gastro.2011.12.037. [DOI] [PubMed] [Google Scholar]
- 31.Novellasdemunt L, Antas P, Li VS. Targeting wnt signaling in colorectal cancer. A review in the theme: cell signaling: proteins, pathways and mechanisms. Am J Physiol Cell Physiol. 2015;309:C511–21. doi: 10.1152/ajpcell.00117.2015. https://doi.org/10.1152/ajpcell.00117.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCrea PD, Gottardi CJ. Beyond β-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. https://doi.org/10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VON Schulz-Hausmann SA, Schmeel LC, Schmeel FC, Schmidt-Wolf IGH. Targeting the Wnt/beta-catenin pathway in renal cell carcinoma. Anticancer Res. 2014;34:4101–8. [PubMed] [Google Scholar]
- 34.Krabbe LM, Westerman ME, Bagrodia A, Gayed BA, Darwish OM, Haddad AQ, Khalil D, Kapur P, Sagalowsky AI, Lotan Y, Margulis V. Dysregulation of β-catenin is an independent predictor of oncologic outcomes in patients with clear cell renal cell carcinoma. J Urol. 2014;191:1671–7. doi: 10.1016/j.juro.2013.11.052. https://doi.org/10.1016/j.juro.2013.11.052. [DOI] [PubMed] [Google Scholar]
- 35.Fan W, Huang J, Xiao H. Histone deacetylase 10 suppresses proliferation and invasion by inhibiting the phosphorylation of β-catenin and serves as an independent prognostic factor for human clear cell renal cell carcinoma. Int J Clin Exp Med. 2015;8:3734–42. [PMC free article] [PubMed] [Google Scholar]
- 36.Cairns P. Renal cell carcinoma. Cancer Biomark Sect Dis Markers. 2010;9:461–73. doi: 10.3233/CBM-2011-0176. https://doi.org/10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fritzsche FR, Oelrich B, Johannsen M, Kristiansen I, Moch H, Jung K, Kristiansen G. Claudin-1 protein expression is a prognostic marker of patient survival in renal cell carcinomas. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:7035–42. doi: 10.1158/1078-0432.CCR-08-0855. https://doi.org/10.1158/1078-0432.CCR-08-0855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.