Significance
Understanding food web structure—networks of species and their feeding interactions—is central to predicting how ecosystems may respond to climate change. Feeding interactions are often determined by the traits individuals have, but how the effects of such traits and their intraspecific variation scale up from individuals to food webs is not known. Here, we use mathematical models to show that trait variation influences predator connectivity and trophic level within food webs. By doing so, we predict the occurrence of important structural features in food webs, and we confirm these predictions using a large global dataset of the best-resolved available food webs.
Keywords: intraspecific variability, individual variability, food webs, complex networks, consumer–resource interactions
Abstract
Food webs (i.e., networks of species and their feeding interactions) share multiple structural features across ecosystems. The factors explaining such similarities are still debated, and the role played by most organismal traits and their intraspecific variation is unknown. Here, we assess how variation in traits controlling predator–prey interactions (e.g., body size) affects food web structure. We show that larger phenotypic variation increases connectivity among predators and their prey as well as total food intake rate. For predators able to eat only a few species (i.e., specialists), low phenotypic variation maximizes intake rates, while the opposite is true for consumers with broader diets (i.e., generalists). We also show that variation sets predator trophic level by determining interaction strengths with prey at different trophic levels. Merging these results, we make two general predictions about the structure of food webs: (i) trophic level should increase with predator connectivity, and (ii) interaction strengths should decrease with prey trophic level. We confirm these predictions empirically using a global dataset of well-resolved food webs. Our results provide understanding of the processes structuring food webs that include functional traits and their naturally occurring variation.
Food webs often share structural features such as the existence of an upper limit to the number of trophic levels (1, 2), the relationship between the number of species and the number of feeding interactions (3–5), and the occurrence of highly repeated structural modules (6–8). These similarities occur despite enormous variation in constituent species and abiotic settings, which is rather surprising and suggests the existence of common fundamental organizing processes (1, 9, 10). It is still an open question, however, what these processes are and what their relative importance is for the structure of food webs (9, 11–14).
For example, simple food web models that take into account one or two niche dimensions can account for a surprisingly large number of structural features found in empirical food webs (5, 9, 10). However, it is still debated which dimensions of niche space are actually responsible for food web structure (15). Evidence suggests that the number of dimensions is low (16) and that some traits, such as body size, play a major role in structuring food webs (17–20). This makes sense, because body size and other key traits often determine the strength of predator–prey interactions (21–23). However, these traits are not fixed within populations. Intraspecific variation in phenotypes is pervasive, and such variation affects predator–prey dynamics and ecological processes through nonlinear relationships between traits and their ecological functions (24–30). However, while both genetic and phenotypic variation are commonplace within populations, how this variation scales up from individuals and populations to communities and food webs is largely unknown.
Here, we build a mechanistic understanding of how intraspecific phenotypic variation structures food webs by integrating processes across levels of biological organization. We make general theoretical predictions about food web structure that we test against a global dataset of well-resolved food webs. We show that phenotypic variation determines diet breadth, which generates common patterns in species connectivity and trophic level in most food webs. Our approach fosters a deeper understanding of the underlying processes leading to the emergence of widely pervasive food web structural patterns.
Phenotypic Variation and Predator Connectivity
We first explored the relationship between intake (foraging) rates, predator connectivity (i.e., the number of prey items), interaction strengths, and phenotypic variation. We assumed that predator–prey interactions are controlled by one or a few quantitative, and hence normally distributed, functional traits (e.g., body mass, SI Appendix, Appendix A) (24, 27, 31, 32). We also assumed a multispecies type II functional response, and the existence of an optimal predator trait value () at which its attack rate is maximal and its handling time is minimal, in such a way that the attack rate decreases and the handling time increases away from that optimum (22, 24, 27, 31–33) (SI Appendix, Appendix A). Under these assumptions, the mean intake rate of a predator for a given prey i, , can be written as follows:
| [1] |
where the functions and are the attack rate and handling time of the predator, respectively, is the underlying trait distribution, is the density of the ith prey, and is the predator density (SI Appendix, Appendix A).
This model thus incorporates the effect of intraspecific trait variation on foraging rates through its effect on attack rate and handling time. The squared distance between the mean trait value and the optimal trait value, , or phenotypic mismatch, can be seen as a measure of maladaptation. Indeed, when the phenotypic mismatch is small ( ; thus, ), small phenotypic variation leads to larger attack rates and smaller handling times, which results in larger foraging rates. The opposite is true when the phenotypic mismatch is large ( or ; thus, ). In this latter case, lower mismatch indicates greater potential for specialization on a given prey item.
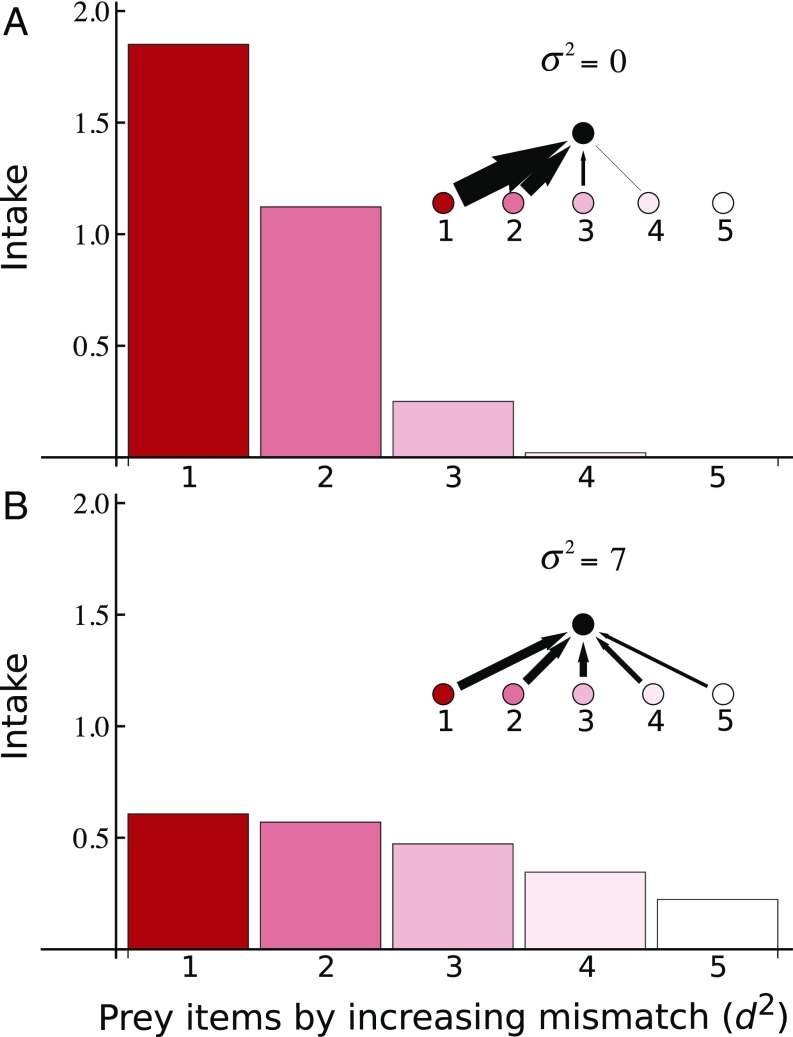
Many predators can consume a diverse array of prey types (34, 35), most of which will have different sets of traits, evolutionary histories, and, consequently, different levels of phenotypic mismatch with a particular predator (36, 37). Using Eq. 1, we calculated the predator’s intake rate for sets of potential hypothetical prey that vary in traits and mismatch (SI Appendix, Appendix A). By assessing how large the foraging rates of a predator are with each of its potential prey items, it is possible to assess how many connections that predator has and how strong those connections are (Fig. 1). Two patterns emerge from this analysis: (i) increasing phenotypic mismatch decreases overall intake rate, and (ii) increasing phenotypic variation increases predator connectivity by allowing the predator to prey upon an increasingly large set of prey items (Fig. 1). Large levels of phenotypic variation thus result in larger potential sets of edible prey. Moreover, total intake rate (i.e., the sum of all intake rates across all edible species) increases with phenotypic variation up to a certain point (Fig. 2; analytic details in SI Appendix, Appendix B), further suggesting that larger levels of phenotypic variation may benefit predators by increasing connectivity and gross energy gain. Of course, our results assume similar prey densities across levels of phenotypic mismatch. We notice that predators may be able to obtain adequate energy intake for prey with which they are highly mismatched if those prey are abundant, in which case the reported effects of phenotypic variation may be less important.
Fig. 1.
Intake rate for a predator with each of the species consumed or potentially consumed ordered by increasing phenotypic mismatch (d2 = 1 to d2 = 25) for increasing levels of phenotypic variation. (A) and (B) . We assumed interactions did not occur if foraging rates were smaller than 0.001 (e.g., Fig. 1A). For details on all other parameters, see SI Appendix, Appendix A.
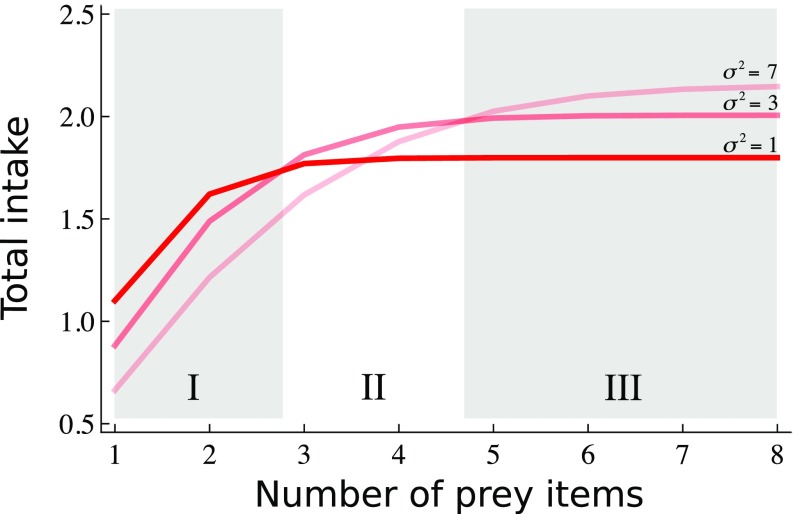
Fig. 2.
Total intake rate (sum of intake rates over all species consumed for a given predator) for three levels of phenotypic variation. Gray zones (I–III) represent the range of possible prey items at which low variation maximizes total intake (I), intermediate levels of variation maximize intake (II), or large levels of variation do (III) (details in SI Appendix, Appendix B).
Predators with small prey sets (specialists) do exist in nature (38), suggesting that low phenotypic variation may confer benefits in some settings. Indeed, our model indicates that, for predators that are constrained to prey upon smaller sets of species, smaller phenotypic variation leads to higher total intake rates (Fig. 2, zone I). In contrast, species with larger levels of phenotypic variation obtain larger total intake rates when they prey upon larger number of prey items (Fig. 2, zone III). Finally, intermediate phenotypic variation maximizes total intake rates when the potential number of prey is also intermediate (Fig. 2, zone II; see SI Appendix, Appendix B, for further exploration of parameter space). These results are driven by variation in the attack rate and they can also be obtained assuming no variation in the handling time or no handling time at all (SI Appendix, Appendix B).
These results provide a mechanistic explanation for the occurrence of two important ecological patterns: weak interaction strengths and diet specialization. First, we provide a simple mechanism by which predators can achieve large numbers of weak interactions without decreasing total intake rate, as phenotypic variation may be traded off with specialization to maintain adequate energy intake. Thus, large amounts of phenotypic variation can lead to weaker interaction strengths (27), which mostly stabilize food webs (7). Second, generalists and specialists can be defined in terms of how much intraspecific variation they have. Predators that prey upon few prey items (specialists) may maintain large enough total intake rates through low levels of phenotypic variation (large attack rates and low handling times, and thus large foraging rates with few prey items), while predators that prey upon large numbers of species (generalists) may maintain sufficient intake rates through greater phenotypic variation, resulting from smaller intakes from many different species (through small attack rates but large handling times with more prey items).
Variation and Predator Trophic Level
We assessed how phenotypic variation can determine predator trophic level and, through that, impact the structure of whole food webs. In the classic definition of trophic level (), TLi is the trophic level of prey i, and pij is the proportional contribution of prey i to the diet of predator j (2). Noting that pij can be written as the intake rate of species j when consuming species i divided by the sum of the intake rates across all species consumed, we rewrote the trophic level of species j (here C) as a function of intake rates (SI Appendix, Appendix C):
| [2] |
where the intake rates () are as in Eq. 1.
To understand the effect of phenotypic variation on trophic level, we analyzed a three-species system with a basal resource, a consumer, and an intraguild predator (Fig. 3 and SI Appendix, Appendix C). For simplicity, we focused on the case where predators only differ in whom they eat and their levels of phenotypic variation and mismatch, but different assumptions do not change our results (SI Appendix, Appendix C). In such case, it is possible to numerically solve the model for different levels of phenotypic variation for a focal species, and then find the equilibrium trophic level of that species and plot it against the amount of phenotypic variation considered. We separate the analyses into two distinct scenarios: (i) a scenario where the phenotypic mismatch of the focal predator, , is smaller for relatively low trophic level species than for relatively high trophic level species (i.e., a situation where predators in the food web rely more heavily on basal organisms but also can prey upon relatively high trophic level organisms whenever their phenotypic variation is large enough, Fig. 3A), and (ii) the reverse scenario where the phenotypic mismatch is smaller for relatively high trophic level species than for relatively low trophic level species (Fig. 3B).
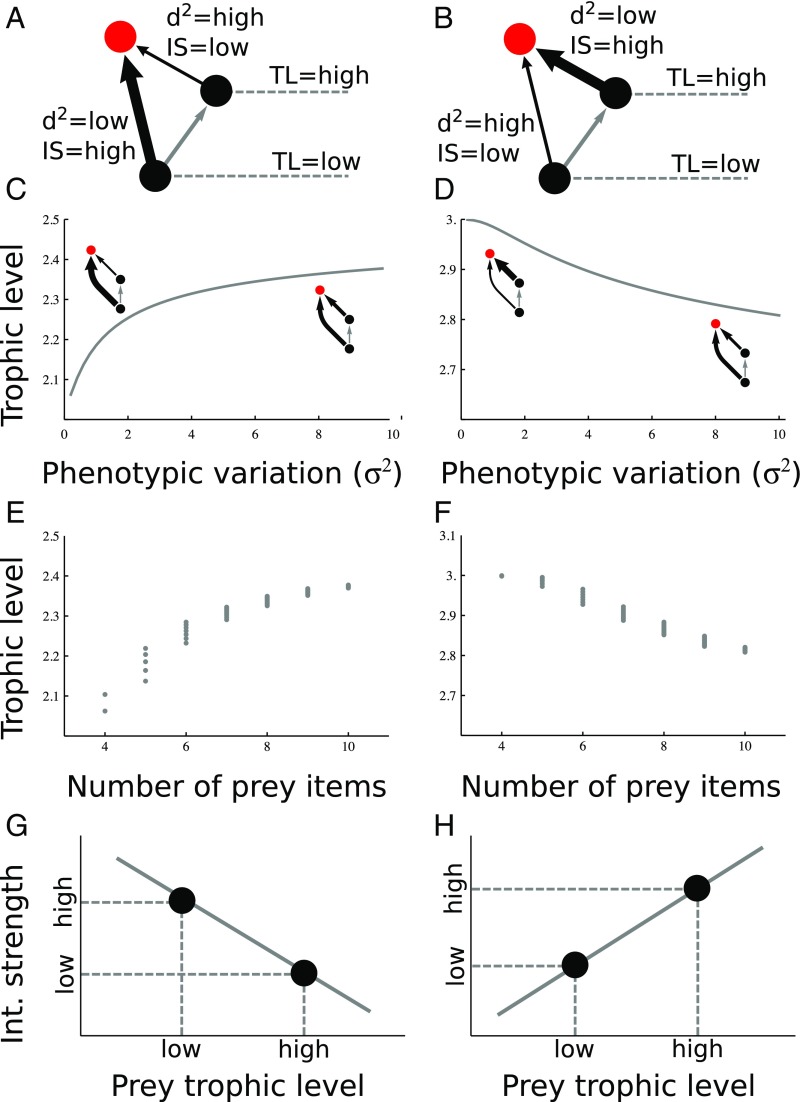
Fig. 3.
(Top row) Diagrams depicting the cases analyzed (case 1: A; case 2: B), with black balls representing prey, red balls representing the focal top predator, and arrows representing feeding interactions (thickness = strength). (Second row) How trophic level of the top species (red) changes with phenotypic variation whenever the predator relies more heavily on the bottom species (d2bottom = 0) than on the intermediate species (d2interm = 2) (C), and vice versa (D). (Third row) Predicted relationship between trophic level and predator connectivity for the first case (E), and the second case (F). (Bottom row) Predicted relationship between the trophic level of a prey item and the interaction strength of that particular interaction for the first case (G), and the second case (H). All parameters are as in the second row, and we used the same threshold for interactions as in Fig. 1.
In this three-species omnivory module, we found that the trophic level of the focal (top) predator increases with phenotypic variation when the predator relies more heavily on the relatively low trophic-level species (Fig. 3C), and decreases with variation when the predator relies more on the relatively high trophic-level species (Fig. 3D). When the top predator relies more heavily on the low trophic-level species, phenotypic variation increases the predator’s trophic level because its intake increasingly depends on preying upon the intermediate consumer (Fig. 3C). In contrast, phenotypic variation decreases trophic level when the predator is more heavily dependent on relatively high trophic-level species because the predator’s intake of the more basal species increases with variation (Fig. 3D).
Predicting the Effects of Trait Variation on Food Web Structural Patterns
Considering how variation jointly affects species connectivity and trophic level, our approach makes two general predictions as to how variation may determine food web structural properties. The first prediction involves the expected relationship between trophic level and connectivity in food webs: because both trophic level and the number of prey items change with phenotypic variation, we can plot them against each other (Fig. 3 E and F; see SI Appendix, Appendix C, for details). When predators can hunt at multiple trophic levels but rely more heavily on relatively low trophic-level species (case 1, Fig. 3A), our approach predicts a saturating, concave down relationship between trophic level and the number of prey items (Fig. 3E and SI Appendix, Appendix C). When predators are mostly dependent on relatively high trophic-level prey (case 2, Fig. 3B), our approach predicts a decreasing sigmoidal relationship between trophic level and number of prey items (Fig. 3F).
For interaction strengths, our approach predicts in case 1 (i.e., predators that rely more heavily on basal resources) a decrease in interaction strengths with an increase in the trophic level of the prey items consumed (Fig. 3G), and the opposite would happen for predators that depend on more higher trophic-level species (case 2, Fig. 3H). These last predictions are a consequence of the relationship between trophic level and phenotypic variation: when predators rely more heavily on basal resources, their interaction strength is large with the basal resource (because of their smaller phenotypic mismatch; Fig. 3A) but small with high trophic-level species (because of larger mismatch), leading to decreasing interaction strengths across trophic levels (Fig. 3G). The opposite happens when top consumers are more dependent on relatively high trophic-level prey than more basal resources (Fig. 3 B and H).
Testing the Predicted Food Web Structural Patterns
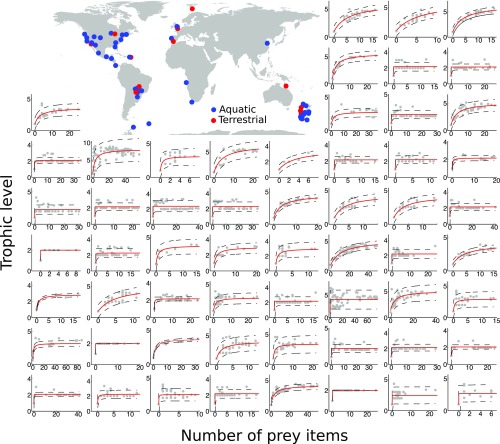
We used 58 of the best-resolved aquatic and terrestrial food webs currently available from around the globe to test the predictions of our theoretical approach (Fig. 4, map, and SI Appendix, Appendix E, Table S1). Some of these food webs are well known and have been used previously to understand both food web structure (4, 8, 9) and dynamics (39), but most have not (taken from Interaction Web DataBase and GlobalWeb food web database). To test our first prediction (Fig. 3 E and F), we used binary food webs, which record only the presence or absence of an interaction, rather than its strength. In such cases, we calculated the trophic level of all species in the food webs using . For binary food webs, we followed previous studies (4, 8, 9, 39) and assumed all pij values to be uniformly equal for all of the prey items of a given predator (pij = 1/number of prey items). To test our second prediction, we used a subset of these food webs for which there is an empirical measure of how prey impact their predators, our proxy for interaction strengths, as estimated by the researchers that originally collected the data (SI Appendix, Appendix D and Appendix E, Table S1).
Fig. 4.
As predicted, the trophic level of species in 58 empirical food webs show a saturating increase with the number of prey items, although food webs vary in how fast that saturation occurs. Gray dots are the values for both quantities in each species of the food web. Red lines are best fit lines with 95% prediction lines in dashed black. Food web names in order of appearance in SI Appendix, Appendix E.
Consistent with our first scenario, we found that all food webs analyzed show a saturating increase in trophic level with the number of prey items consumed per species, although food webs vary in how fast saturation occurs (Fig. 4). In some cases, the increase occurs very slowly (e.g., Fig. 4, Top Left), while in some others it occurs very quickly (e.g., Fig. 4, Bottom Left), immediately saturating from trophic level 1 to trophic level 2. This result suggests that most if not all consumers in these food webs may rely more heavily on basal species or species with lower trophic levels than on upper trophic level species, as per our model (Fig. 3 A and E).
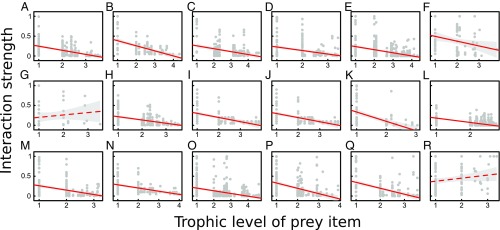
Because of this result, we would expect interaction strengths, or the impact of prey on their predators, to decrease with the trophic level of the prey (Fig. 3G). We regressed the interaction strength of each pairwise interaction against the trophic level of the prey for all predator–prey pairs in each food web for which such information was available (Fig. 5). Our results show that interaction strengths do decrease with the trophic level of the prey species in most (16 of 18) cases, consistent with our prediction.
Fig. 5.
As predicted by theory, the trophic level of a given prey type is generally negatively related to the interaction strength with that prey across predators in the food web (gray dots). Red lines represent predictions of a linear regression with 95% confidence intervals (gray shaded regions). We observed that, in all but two cases (G and R), the relationship between the two variables is significantly negative (P < 0.001). (A) Alvarado, slope = −0.10 ± 0.01 SE; (B) Angola, slope = −0.13 ± 0.02 SE; (C) Braço Morto, slope = −0.07 ± 0.01 SE; (D) Cádiz, slope = −0.09 ± 0.02 SE; (E) Chesapeake, slope = −0.08 ± 0.12 SE; (F) Corrente, slope = −0.12 ± 0.05 SE; (G) Huizache, slope = +0.06 ± 0.07 SE, P = 0.376; (H) Itaipú (83–87), slope = −0.08 ± 0.01 SE; (I) Itaipú (88–92), slope = −0.10 ± 0.02 SE; (J) Mondego, slope = −0.10 ± 0.02 SE; (K) Northern California, slope = −0.23 ± 0.04 SE; (L) Onca, slope = −0.11 ± 0.01 SE; (M) Paraná, slope = −0.11 ± 0.02 SE; (N) Reef, slope = −0.08 ± 0.01 SE; (O) Scotia Sea, slope = −0.07 ± 0.01 SE; (P) St. Marks, slope = −0.13 ± 0.02 SE; (Q) St. Martin, slope = −0.15 ± 0.02 SE; (R) UK Grassland, slope = +0.08 ± 0.05 SE, P = 0.097.
Discussion
Our results provide a mechanistic understanding of how traits and trait variation determines food web structure, thus addressing long-standing questions regarding the processes underpinning such structure. Our study connects ecological processes across multiple levels of biological organization, linking individuals and their traits to communities, and generates predictions about food web structure that are consistent with real food webs from around the world.
We showed that phenotypic variation plays a key role in determining the number and strength of predator–prey interactions a given predator can have. The relationship between variation and diet breadth is not necessarily new. In fact, the classic niche variation hypothesis (NVH) (40) posits that higher morphological variation arises in species with larger diet breadths as a consequence of a release in interspecific competition. The NVH thus also suggests the existence of a positive relationship between diet breath and variation. Our approach, however, assumes that larger variation is the cause, not the consequence, of larger diet breaths, as it allows species to successfully consume larger sets of potential prey. We also show that this variation can be maintained in the population, even though it reduces the intake the species could have from any given prey, because it maximizes the total intake rate under certain conditions (Fig. 2). A recent empirical study shows that increasing morphological variation in gill rake size and other consumption-related traits lead to larger isotopic variation in the three-spined stickleback (Gasterosteus aculeatus), indicative of wider diet breaths (41), which supports our results in a specific predator–prey system.
We also showed that phenotypic variation can affect the trophic level of a consumer and, through this, the architecture of the entire network of interacting species. This finding is consistent with a recent empirical study in perch (Perca fluviatilis) that demonstrated that trophic level is tightly related to individual dietary variation within populations (42). Along these lines, trait matching has been empirically observed to predict important structural patterns in mutualistic networks (43), predator–prey body size ratios influence food web structure and stability (44–47), and genetic variation can increase food web complexity in systems consisting of insect herbivores and different genotypes of plants (30). Together, these results show how traits, their variation, and their genetic basis can thus play an important role in determining food web structure and assembly.
A potential implication of our results is that changes in phenotypic variation may have profound effects on both the structure and the dynamics of food webs. Such changes can occur due to several factors, both biotic and abiotic. For example, natural selection can reduce variation in a trait, with effects on the feeding behavior of entire species (48). Also, increasing temperature can lead to a reduction in mean body size in ectotherms, a pattern known as the temperature–size rule (49), and these reductions in mean size can be accompanied by reductions in size variation (50). Both mean body size (17, 19, 51) and phenotypic variation can affect food web structure, so our results imply that temperature shifts may lead to rearrangements in food web structure through their effect on both the mean and variance of body size.
Overall, phenotypic variation in traits associated with foraging can explain important food web structural patterns by influencing predator connectivity, trophic level, and the relationship between these two with interaction strengths. Our results have important consequences for food web structure and dynamics in the context of increasing global temperatures, as both body size and its variation can change with temperature, potentially leading to structural rearrangements in food webs.
Supplementary Material
Acknowledgments
We thank Richard Williams, Jennifer Dunne, and Ulrich Brose for generously sharing data with us; two anonymous reviewers for their insightful suggestions; Justin Yeakel for insightful comments on an early version of this paper; and the Interaction Web Database and the GlobalWeb food web database for their data as well. J.P.G. was supported by a James S. McDonnell Postdoctoral Fellowship in Complex Systems, and J.P.D. was supported by United States–Israel Binational Science Foundation Grant 2014295.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703864114/-/DCSupplemental.
References
- 1.Pimm SL, Lawton JH, Cohen JE. Food web patterns and their consequences. Nature. 1991;350:669–674. [Google Scholar]
- 2.Williams RJ, Martinez ND. Limits to trophic levels and omnivory in complex food webs: Theory and data. Am Nat. 2004;163:458–468. doi: 10.1086/381964. [DOI] [PubMed] [Google Scholar]
- 3.Martinez ND. Constant connectance in community food webs. Am Nat. 1992;139:1208–1218. [Google Scholar]
- 4.Dunne JA, Williams RJ, Martinez ND. Network structure and biodiversity loss in food webs: Robustness increases with connectance. Ecol Lett. 2002;5:558–567. [Google Scholar]
- 5.Gravel D, Poisot T, Albouy C, Velez L, Mouillot D. Inferring food web structure from predator-prey body size relationships. Methods Ecol Evol. 2013;4:1083–1090. [Google Scholar]
- 6.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 7.McCann KS, Hastings A, Huxel GR. Weak trophic interactions and the balance of nature. Nature. 1998;395:794–798. [Google Scholar]
- 8.Williams RJ, Berlow EL, Dunne JA, Barabási A-L, Martinez ND. Two degrees of separation in complex food webs. Proc Natl Acad Sci USA. 2002;99:12913–12916. doi: 10.1073/pnas.192448799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RJ, Martinez ND. Simple rules yield complex food webs. Nature. 2000;404:180–183. doi: 10.1038/35004572. [DOI] [PubMed] [Google Scholar]
- 10.Beckerman AP, Petchey OL, Warren PH. Foraging biology predicts food web complexity. Proc Natl Acad Sci USA. 2006;103:13745–13749. doi: 10.1073/pnas.0603039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May RM. Will a large complex system be stable? Nature. 1972;238:413–414. doi: 10.1038/238413a0. [DOI] [PubMed] [Google Scholar]
- 12.McCann KS. The diversity-stability debate. Nature. 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 13.Neutel A-M, et al. Reconciling complexity with stability in naturally assembling food webs. Nature. 2007;449:599–602. doi: 10.1038/nature06154. [DOI] [PubMed] [Google Scholar]
- 14.Allesina S, et al. Predicting the stability of large structured food webs. Nat Commun. 2015;6:7842. doi: 10.1038/ncomms8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunne JA. The network structure of food webs. In: Pascual M, Dunne JA, editors. Ecological Networks. Linking Structure to Dynamics in Food Webs. Oxford Univ Press; New York: 2006. pp. 27–86. [Google Scholar]
- 16.Williams RJ, Purves DW. The probabilistic niche model reveals substantial variation in the niche structure of empirical food webs. Ecology. 2011;92:1849–1857. doi: 10.1890/11-0200.1. [DOI] [PubMed] [Google Scholar]
- 17.Petchey OL, Beckerman AP, Riede JO, Warren PH. Size, foraging, and food web structure. Proc Natl Acad Sci USA. 2008;105:4191–4196. doi: 10.1073/pnas.0710672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stouffer DB, Rezende EL, Amaral LA. The role of body mass in diet contiguity and food-web structure. J Anim Ecol. 2011;80:632–639. doi: 10.1111/j.1365-2656.2011.01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arim M, Abades SR, Laufer G, Loureiro M, Marquet PA. Food web structure and body size: Trophic position and resource acquisition. Oikos. 2010;119:147–153. [Google Scholar]
- 20.DeLong JP, et al. The body size dependence of trophic cascades. Am Nat. 2015;185:354–366. doi: 10.1086/679735. [DOI] [PubMed] [Google Scholar]
- 21.Ozgul A, et al. Coupled dynamics of body mass and population growth in response to environmental change. Nature. 2010;466:482–485. doi: 10.1038/nature09210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rall BC, et al. Universal temperature and body-mass scaling of feeding rates. Philos Trans R Soc Lond B Biol Sci. 2012;367:2923–2934. doi: 10.1098/rstb.2012.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLong JP. The body-size dependence of mutual interference. Biol Lett. 2014;10:20140261. doi: 10.1098/rsbl.2014.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiber SJ, Bürger R, Bolnick DI. The community effects of phenotypic and genetic variation within a predator population. Ecology. 2011;92:1582–1593. doi: 10.1890/10-2071.1. [DOI] [PubMed] [Google Scholar]
- 25.Bolnick DI, et al. Why intraspecific trait variation matters in community ecology. Trends Ecol Evol. 2011;26:183–192. doi: 10.1016/j.tree.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Violle C, et al. The return of the variance: Intraspecific variability in community ecology. Trends Ecol Evol. 2012;27:244–252. doi: 10.1016/j.tree.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Gibert JP, Brassil CE. Individual phenotypic variation reduces interaction strengths in a consumer-resource system. Ecol Evol. 2014;4:3703–3713. doi: 10.1002/ece3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibert JP, Dell AI, DeLong JP, Pawar S. Scaling-up trait variation from individuals to ecosystems. Adv Ecol Res. 2015;52:1–17. [Google Scholar]
- 29.Barabás G, D’Andrea R. The effect of intraspecific variation and heritability on community pattern and robustness. Ecol Lett. 2016;19:977–986. doi: 10.1111/ele.12636. [DOI] [PubMed] [Google Scholar]
- 30.Barbour MA, et al. Genetic specificity of a plant-insect food web: Implications for linking genetic variation to network complexity. Proc Natl Acad Sci USA. 2016;113:2128–2133. doi: 10.1073/pnas.1513633113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasseur DA, Amarasekare P, Rudolf VHW, Levine JM. Eco-evolutionary dynamics enable coexistence via neighbor-dependent selection. Am Nat. 2011;178:E96–E109. doi: 10.1086/662161. [DOI] [PubMed] [Google Scholar]
- 32.Gibert JP, DeLong JP. Individual variation decreases interference competition but increases species persistence. Adv Ecol Res. 2015;52:45–64. [Google Scholar]
- 33.Barrios-O’Neill D, et al. On the context-dependent scaling of consumer feeding rates. Ecol Lett. 2016;19:668–678. doi: 10.1111/ele.12605. [DOI] [PubMed] [Google Scholar]
- 34.Estes JA, Riedman ML, Staedler MM, Tinker MT, Lyon BE. Individual variation in prey selection by sea otters: Patterns, causes and implications. J Anim Ecol. 2003;72:144–155. [Google Scholar]
- 35.Araújo MS, Bolnick DI, Martinelli LA, Giaretta AA, Dos Reis SF. Individual-level diet variation in four species of Brazilian frogs. J Anim Ecol. 2009;78:848–856. doi: 10.1111/j.1365-2656.2009.01546.x. [DOI] [PubMed] [Google Scholar]
- 36.Brodie ED, Jr, Ridenhour BJ, Brodie ED., 3rd The evolutionary response of predators to dangerous prey: Hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution. 2002;56:2067–2082. doi: 10.1111/j.0014-3820.2002.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 37.Toju H, Sota T. Imbalance of predator and prey armament: Geographic clines in phenotypic interface and natural selection. Am Nat. 2006;167:105–117. doi: 10.1086/498277. [DOI] [PubMed] [Google Scholar]
- 38.Kofron CP. Foods and habitats of aquatic snakes (Reptilia, Serpentes) in a Louisiana swamp. J Herpetol. 1978;12:543–554. [Google Scholar]
- 39.Allesina S, Pascual M. Network structure, predator–prey modules, and stability in large food webs. Theor Ecol. 2007;1:55–64. [Google Scholar]
- 40.Van Valen L. Morphological variation and width of ecological niche. Am Nat. 1965;99:377–390. [Google Scholar]
- 41.Snowberg LK, Hendrix KM, Bolnick DI. Covarying variances: More morphologically variable populations also exhibit more diet variation. Oecologia. 2015;178:89–101. doi: 10.1007/s00442-014-3200-7. [DOI] [PubMed] [Google Scholar]
- 42.Svanbäck R, Quevedo M, Olsson J, Eklöv P. Individuals in food webs: The relationships between trophic position, omnivory and among-individual diet variation. Oecologia. 2015;178:103–114. doi: 10.1007/s00442-014-3203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehling DM, Jordano P, Schaefer HM, Böhning-Gaese K, Schleuning M. Morphology predicts species’ functional roles and their degree of specialization in plant-frugivore interactions. Proc Biol Sci. 2016;283:20152444. doi: 10.1098/rspb.2015.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emmerson MC, Raffaelli D. Body size, patterns of interaction strength and the stability of a real food web. J Anim Ecol. 2004;73:399–409. [Google Scholar]
- 45.Brose U, et al. Consumer-resource body-size relationships in natural food webs. Ecology. 2006;87:2411–2417. doi: 10.1890/0012-9658(2006)87[2411:cbrinf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 46.Jonsson T. Trophic links and the relationship between predator and prey body sizes in food webs. Community Ecol. 2014;15:54–64. [Google Scholar]
- 47.Gibert JP, DeLong JP. Temperature alters food web body-size structure. Biol Lett. 2014;10:20140473. doi: 10.1098/rsbl.2014.0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin’s finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 49.Atkinson D. Temperature and organism size—A biological law for ectotherms? Adv Ecol Res. 1994;25:1–58. [Google Scholar]
- 50.DeLong JP. Experimental demonstration of a “rate–size” trade-off governing body size optimization. Evol Ecol Res. 2012;14:343–352. [Google Scholar]
- 51.Stouffer DB, Bascompte J. Compartmentalization increases food-web persistence. Proc Natl Acad Sci USA. 2011;108:3648–3652. doi: 10.1073/pnas.1014353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.