Abstract
Atrazine is an herbicide with several known toxicologically relevant effects, including interactions with other chemicals. Atrazine increases the toxicity of several organophosphates and has been shown to reduce the toxicity of triclosan to D. magna in a concentration dependent manner. Atrazine is a potent activator in vitro of the xenobiotic-sensing nuclear receptor, HR96, related to vertebrate constitutive androstane receptor (CAR) and pregnane X-receptor (PXR). RNA sequencing (RNAseq) was performed to determine if atrazine is inducing phase I–III detoxification enzymes in vivo, and estimate its potential for mixture interactions. RNAseq analysis demonstrates induction of glutathione S-transferases (GSTs), cytochrome P450s (CYPs), glucosyltransferases (UDPGTs), and xenobiotic transporters, of which several are verified by qPCR. Pathway analysis demonstrates changes in drug, glutathione, and sphingolipid metabolism, indicative of HR96 activation. Based on our RNAseq data, we hypothesized as to which environmentally relevant chemicals may show altered toxicity with co-exposure to atrazine. Acute toxicity tests were performed to determine individual LC50 and Hillslope values as were toxicity tests with binary mixtures containing atrazine. The observed mixture toxicity was compared with modeled mixture toxicity using the Computational Approach to the Toxicity Assessment of Mixtures (CATAM) to assess whether atrazine is exerting antagonism, additivity, or synergistic toxicity in accordance with our hypothesis. Atrazine-triclosan mixtures showed decreased toxicity as expected; atrazine-parathion, atrazine-endosulfan, and to a lesser extent atrazine-p-nonylphenol mixtures showed increased toxicity. In summary, exposure to atrazine activates HR96, and induces phase I–III detoxification genes that are likely responsible for mixture interactions.
Keywords: gene expression, mixture model, nuclear receptor, aquatic toxicology, induction, concentration-dependent
1. Introduction
Daphnia species are aquatic crustaceans commonly used in ecotoxicology. D. pulex was the first crustacean to have its genome sequenced (Colbourne et al., 2011), and the D. magna genome is currently available online (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA298946). Subsequent phylogenetic analysis of the D. magna and D. pulex nuclear receptors (NRs) found many were strongly conserved between these Daphnia species, including HR96 (Litoff et al., 2014). HR96 is an arthropod homolog of the xenobiotic-sensing NRs, CAR and PXR found in vertebrates (King-Jones et al., 2006; Litoff et al., 2014). In D. magna, it is expressed at similar levels at all ages tested from less than 2-days old to reproductive adults at 14-days old (Sengupta et al., 2015). Daphnia HR96 is activated and sometimes inhibited by a number of different chemicals, including pesticides, plasticizers, pharmaceuticals, steroids, and fatty acids (Karimullina et al., 2012). In turn it regulates the expression of phase I–III detoxification enzymes and transporters (Karimullina et al., 2012; King-Jones et al., 2006). The chemical with the greatest potency for activating Daphnia HR96 is atrazine (Karimullina et al., 2012) making it a reference compound for the activation of HR96 similar to phenobarbital for CAR or hyperforin for PXR (Kawamoto et al., 1999; Moore et al., 2000). Therefore, atrazine can be used to estimate the potential effects of other potent HR96 activators or mixtures of HR96 activators.
Atrazine has been banned in Europe (EUROPA, 2004). However, worldwide it is still one of the most commonly used herbicides, the second most used herbicide in the United States as of 2014, and an environmentally relevant chemical found in ground and surface waters around the world (Baker, 2016; Belden and Lydy, 2000). Atrazine induces anti-oxidant responses, including GST activity in D. magna, probably due to HR96 activation (Sengupta et al., 2015). Surprisingly, atrazine co-treatment provided protection from triclosan and docosahexaenoic acid (DHA) toxicity, presumably due to the induction of GSTs (Sengupta et al., 2015). In contrast, the fungicide, chlorothanlonil that works through glutathione depletion, decreased the population growth rate of Dunaliella tertiolecta 1.83 times more when combined with atrazine then when either compound was provided individually (DeLorenzo and Serrano, 2003). Atrazine has also been shown to have interactions with other compounds, including organophosphates, as atrazine is associated with increased toxicity of chlorpyrifos in midge larvae and vertebrates (Belden & Lydy, 2000; Wacksman et al., 2006). Another organophosphate, malathion, caused reduced maturation time of oocytes in Xenopus laevis when co-exposed with atrazine, which ultimately led to a decrease in quality of the cells and abnormal cell division post-fertilization (Ji et al., 2016). Overall, atrazine interacts with a variety of compounds in several species.
Mixtures are generally described as either having additive, synergistic, or antagonistic activity. Additivity occurs when the effects of the individual compounds found in the mixture add to each other in a predictable manner. When the two compounds have the same mode of toxicity their additive toxicity is called concentration addition; when the mode of toxicity is different but the mixture effect is still predictable, it is known as independent joint action (Bliss, 1939; Olmstead & LeBlanc, 2005). When the toxicity of two compounds is greater than additive, it is called potentiation or synergism. Antagonism occurs when the toxicity of a mixture is less than expected by additivity (Feron and Groten, 2002; Metcalf, 1967). Some chemicals that cause synergism or antagonism do so through concentration-dependent mechanisms, causing induction or inhibition of detoxification enzymes that alter the metabolism of the toxic chemical. The Computational Approach to the Toxicity Assessment of Mixtures (CATAM) is a calculator that uses the EC50 and sigmoidal dose-response curve of individual chemicals to predict the additive toxicity of mixtures using either concentration addition or independent joint action (Rider and LeBlanc, 2005). Deviations in the experimental data from the predictive model indicate either antagonism or synergism depending on whether the toxicity results are above or below the model.
Concentration-dependent interactions often occur because of increased or decreased activity of detoxification enzymes (Baldwin & Roling, 2009; Rider & LeBlanc, 2005; Sengupta et al., 2015). For example, organophosphates like parathion and malathion require activation by cytochrome P450s (CYPs) to inhibit acetylcholinesterase and exert toxicity, but co-treatment with piperonyl butoxide, a CYP inhibitor, negates the toxicity of organophosphates (Rider and LeBlanc, 2005). The activation of PXR and subsequent induction of CYP3A4 increases the clearance of multiple chemotherapy drugs, immunosuppressants, and oral contraceptives (Harmsen et al., 2007; Moore et al., 2000; Vogel, 2001). Induction of drug transporters such as MDR1 and MRP2 increase clearance of chemotherapy drugs and ezetimibe, a cholesterol lowering drug (Oswald et al., 2006; Watkins et al., 2003). Not all drug-drug interactions due to NR activation result in decreased medication efficacy. For example, cyclophosphamide is activated by CYP2B6 to form 4-hydroxycyclophosphamide, and this process is increased in the presence of CAR activation leading to increased cytotoxicity in CAR-containing hepatocytes, but not CAR-null hepatocytes (Hedrich et al., 2016). We hypothesize that activation of HR96 will cause similar interactions as CAR/PXR due to enzyme or transporter induction.
Previous work has shown concentration-dependent protection by atrazine (20 or 40 μM) in D. magna exposed to the HR96 inhibitors triclosan or DHA (Sengupta et al., 2015). Given the potential for the HR96 activator, atrazine to alter the toxic response of D. magna, we examined its effects on gene expression via RNAseq. We hypothesized that atrazine would induce detoxification enzyme genes, such as GSTs, UDPGTs, and CYPs, and in turn allow us to predict classes of chemicals susceptible to mixture effects following HR96 activation.
2. Materials and Methods
2.1. Daphnia culture
D. magna were cultured in moderately hard water at 21°C in a Goldline SP-300 environmental chamber under a 16:8 light:dark cycle. The daphnids were fed laboratory cultured Raphidocelis subcapitata (Aquatic Biosystems, Fort Collins, CO USA) supplemented with TetraFin fish food (Masterpet Corp., New South Wales, Australia) as described previously (Ginjupalli and Baldwin, 2013).
2.2. Chemicals
Chemical exposures were performed with p-nonylphenol (99.9%) (Honeywell Riedel-de Haën, Morris Plains, NJ USA), bisphenol A (97%), triclosan (97%) (Sigma-Aldrich, St. Louis, MO USA), parathion (99.5%), chlorpyrifos (99.1%), endosulfan (99.5%), and atrazine (98.6%) (Chem Service, West Chester, PA USA). Stock solutions of each chemical were dissolved in dimethyl sulfoxide (DMSO) (99.7%) (Fisher Scientific, Fair Lawn, NJ).
2.3. RNA sequencing and qPCR confirmation
RNA was extracted from seven-day old daphnids exposed for 96h to 0, 20, or 40 μM atrazine with DMSO used as the solvent carrier under normal feeding conditions. Each treatment consisted of five beakers containing three daphnids each (n = 5). Following exposure, the samples were placed in RNAlater Stabilization Solution (Invitrogen, Carlsbad, CA USA) and stored at −80°C. RNA was extracted from the adult daphnids using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD USA) according to the manufacturer’s instructions, quantified with a NanoDrop Lite Spectrophotometer and purity of the samples were analyzed with a NanoDrop 8000 Spectrophotometer (Thermo Scientific, Waltham, MA USA). An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA USA) was used to assess the quality of the samples by assigning an RNA integrity number (RIN), and samples with RIN > 8 were determined to be of high enough quality for next generation sequencing.
Libraries were then prepared from the RNA and TruSeq Total RNA Library Prep kit (Illumina, San Diego, CA USA). Samples were sequenced to an average sequencing depth of 20,000,000 read pairs on an Illumina HiSeq2500, with a 2 × 125 paired-end module at Hollings Cancer Center at the Medical University of South Carolina. FastQC was used to check the quality metrics of the samples, and then Trimmomatic was used to trim low quality bases. After the reads were trimmed, 83.8% of the trimmed reads aligned using GSNAP to the D. magna reference genome (https://www.ncbi.nlm.nih.gov/assembly/GCA_001632505.1/). Subread feature counts software found reads that aligned with known genes. EdgeR (Huber et al., 2015; Robinson et al., 2009) and the raw reads counts were used to determine differential gene expression (DGE) analysis.
Low coverage genes, determined to be genes in which the lowest replicate number of samples had less than one count per million of expression, were filtered out of the analysis and samples were then normalized to scale their library sizes. Genes were determined to be differentially expressed if their adjusted p-value, false discovery rate, was less than 0.05. Differentially expressed genes were annotated with the Trinotate pipeline (Haas et al., 2013). The GO terms were analyzed for enrichment using the GOSeq method (Young et al., 2010) to adjust for gene length and expression bias, and enriched GO terms were visualized using Revigo and Cytoscape (Smoot et al., 2011; Supek et al., 2011). Similarly, the K numbers from the KEGG database were extracted from the genes with log2FC > 1 and entered into the KEGG Mapper (http://www.genome.ad.jp/kegg/) to determine biochemical pathways affected by atrazine exposure (Kanehisa and Goto, 2000). The normalized counts for genes of interest were averaged for the control and treated replicates and log transformed, then expression was compared in a heatmap constructed using Heatmap2 (R Core Team, 2017).
qPCR was performed to confirm accuracy of the RNAseq-based differential expression of specific genes in the samples following exposure to 0, 20 or 40 μM atrazine. Two μg RNA was used to synthesize cDNA as previously described (Sengupta et al., 2015) using MMLV reverse transcriptase (Promega, Madison, WI USA). qPCR was performed on an iCycler (Bio-Rad Laboratories, Hercules, CA USA) with 0.25X SyBR Green Master Mix (Qiagen, Germantown, MD USA) with a set of dilutions (1:1, 1:4, 1:16, 1:64, 1:256, and 1:1024) from a mix of untreated and treated samples as the standard curve to determine the efficiency of each qPCR reaction. Primer sets and annealing temperatures are available in Suppl. Table 1. Triplicate, 1:15 diluted samples were quantified by taking the efficiency of each curve raised to the power of the average threshold cycle (Ct) for each sample and then normalized to β-actin as described previously by us and others (Hernandez et al., 2009; Hernandez et al., 2007; Muller et al., 2002). Differences in gene expression were determined by One-way ANOVA followed by Fisher’s LSD post-hoc test using GraphPad Prism 7.0 software (Graphpad Software Inc, San Diego, CA USA).
2.4. Toxicity tests
Neonatal daphnids (age less than 24 h) were exposed to varying concentrations of a single chemical for 48 h in acute toxicity tests as described previously following guidelines by the USEPA (USEPA, 2002). Each treatment group consisted of five 50 ml beakers containing 40 ml of media with four neonates. After chemical concentrations were log10 transformed and survival normalized to 100%, sigmoidal dose response curves, LC50s, and Hillslopes for each chemical were generated using GraphPad Prism 7.0 (Baldwin & Roling, 2009; Rider & LeBlanc, 2005). Toxicity data were fit with a variable slope model Y=Bottom + (Top-Bottom)/(1+10^((LogIC50-X)*HillSlope) and fitted with least squares ordinary fit. Confidence intervals were produced assuming asymmetrical distribution, as recommended by Graphpad Prism.
Subsequent binary mixture exposures were performed with each chemical and 40 μM atrazine as this sublethal concentration of atrazine provided the largest GST induction and greatest protection from DHA and triclosan (Sengupta et al., 2015) and therefore has potential for interactions with other chemicals. For binary mixtures, each treatment group consisted of eight to ten 50 ml beakers each containing four neonates. A larger replicate number is required with the binary mixtures than single chemical toxicity tests in order to determine statistical differences between exposures. After 48 h, survival was plotted using Graphpad Prism and compared against the CATAM model used to predict mixture toxicity using the independent joint action model based on their Hillslope, individual LC50 and 95% CI (Baldwin & Roling, 2009; Rider & LeBlanc, 2005). Area under the curve (AUC) was calculated for each chemical’s individual dose-response, atrazine mixture, and CATAM dose-response curve using Graphpad Prism 7.0. The CATAM model’s AUC and standard error was calculated from the LC50 and the the upper and lower limits of the 95% CI of the LC50 and the Hillslope was used to determine the dose-response curve of the model as described. AUCs were compared using one-way ANOVA followed by Fisher’s LSD.
3. Results
3.1 RNA sequencing
RNAseq was performed on D. magna exposed to 0 or 40 μM atrazine to determine global differential gene expression. To illustrate the differences between samples, a Volcano plot was generated to show the logFC against the −log10(p-value) per gene. A total of 26,646 genes were identified through alignment to the D. magna genome; of these, 675 were significantly up-regulated and 441 were significantly down-regulated (Suppl. Fig. 1). A list of all the genes differentially expressed is provided as an Excel file (Suppl. Table 2), and is separated into genes with less than and greater than 2-fold changes.
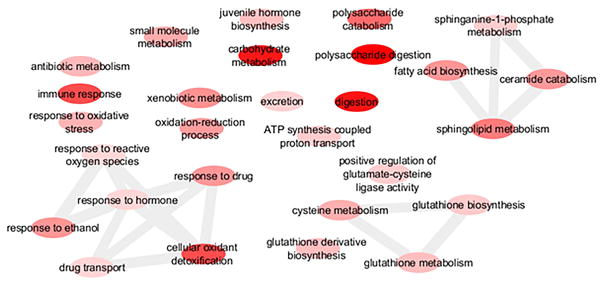
A total of 178 biological processes GO terms were enriched among the up-regulated genes (Suppl. Table 3). Enriched GO terms among the up-regulated genes included xenobiotic metabolism, response to oxidative stress, cellular oxidant detoxification, response to drug, and drug transport, consistent with an HR96-mediated Phase I–III detoxification response measured in Drosophila (King-Jones et al., 2006). Several enriched GO terms were also clustered by similarity and relate to GST activity, including glutathione biosynthesis and glutathione metabolism. Terms describing nutrient digestion further support an HR96 response as several terms associated with carbohydrate metabolism were induced. A fourth cluster of GO terms included sphingolipid metabolism, ceramide catabolism, fatty acid biosynthesis, and sphinganine-1-phosphate metabolism, consistent with HR96 activation and its role in mediating sphingomyelin, cholesterol, and triacylglycerol homeostasis (Bujold et al., 2010; Sengupta et al., 2017; Sieber & Thummel, 2009). Twenty-eight of the enriched GO terms were selected for their association with HR96 activation and detoxification and were visualized using Revigo and Cytoscape (Smoot et al., 2011; Supek et al., 2011) (Fig. 1). In addition processes such as rRNA catabolic process, apoptotic processes, and cellular response to nitric oxide, among others, were up-regulated (Suppl. Table 3) and these processes are not known to be mediated by HR96.
Fig. 1. GO term enrichment among the significantly up-regulated differentially expressed genes.
A sample of biological processes GO terms that were of relevance and significantly over-represented among the up-regulated genes were visualized using Revigo and Cytoscape. Each node represents a GO term. Darker colored nodes have lower p-values, and edges connect GO terms that have pairwise similarities that are in the strongest 3%. A full list of enriched biological processes GO terms is in Supplementary Table 3.
Only one biological processes GO term, DNA integration, was enriched among the significantly down-regulated genes. However, there are several terms associated with significantly down-regulated genes include galactosylceramide biosynthesis, oogenesis, and pupal chitin-based cuticle development, suggesting effects on genes that regulate development and reproduction (Suppl. Table 2). Atrazine at 40 μM reduces reproduction nearly 90% in D. magna (Sengupta et al., 2016). Therefore, perturbations in these biological processes is consistent with previous studies on atrazine exposure.
The KEGG database was used to confirm GO enrichment results and determine biochemical pathways perturbed by atrazine. A total of 101 biochemical pathways contained at least one gene product with significant induction, and a total of 47 biochemical pathways contained at least one gene with significant down-regulation. Eight biochemical pathways contained at least 5 induced genes, and two biochemical pathways contained at least 5 down-regulated genes. (Suppl. Table 4; Suppl. Table 5). Up-regulated pathways included the sphingolipid signaling and sphingolipid metabolism, glutathione metabolism, and metabolism of xenobiotics and drug metabolism, consistent with GO term enrichment analysis (Suppl. Table 3). An example pathway is the glutathione metabolism pathway, which had several enzymes significantly up-regulated, including glutathione synthetase (logFC=1.66), glutamate-cysteine ligase catalytic subunit (logFC = 2.19), aminopeptidase (logFC = 1.13), dipeptidase (logFC = 1.67), GST E10 (logFC = 3.1.), GST A4 (logFC = 2.16), and GST 1 (logFC = 1.33). Enzymes involved in the synthesis, conjugation, and metabolism of glutathione were all up-regulated due to atrazine exposure, supporting previous work which found increased GST activity in D. magna exposed to atrazine (Sengupta et al., 2015).
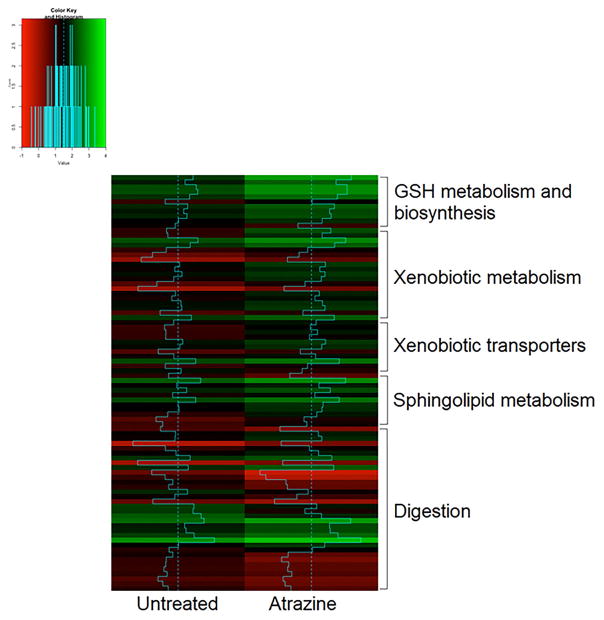
A heatmap was constructed to compare the expression of genes in glutathione metabolism and biosynthesis, xenobiotic metabolism, xenobiotic transporters, sphingolipid metabolism, and digestion pathways, including protein digestion (primarily down-regulated) and carbohydrate digestion (primarily up-regulated) (Fig. 2). Overall, expression was typically greater in the atrazine exposed group compared to the control group for the known HR96-mediated pathways such as detoxification and sphingolipid metabolism except digestion. Of the 147 genes with log fold change of 1.5 or greater, 96 were annotated, including 17 known detoxification genes such as ADH, esterases, peroxidases, GSTs, CYPs, ABC transporters, and a sulfotransferase. (Table 1; Suppl. Table 2). Overall, a large percentage (11.6%) of differentially expressed genes showing greater than 1.5-fold change are phase I–III detoxification genes; indicating preferential induction of chemical detoxification pathways. GO term enrichment and pathway analysis demonstrate significant induction of xenobiotic metabolism pathways by atrazine. Taken together with transcriptomics data indicating perturbations in sphingomyelin metabolism and digestion, the atrazine transcriptomics data is consistent with HR96 activation in vivo.
Fig. 2. Heatmap of relative expression of genes in pathways of interest.
Genes involved in glutathione metabolism and biosynthesis, xenobiotic metabolism, xenobiotic transporters, and sphingolipid metabolism generally had higher expression in atrazine exposed daphnids compared to the control. Digestion-related genes varied in expression in the atrazine group compared to the control.
Table 1.
Detoxification genes up-regulated due to 40 μM atrazine exposure.
| Gene | log2Fold Changea | FDRb | p-value | Locus tag/IDc |
|---|---|---|---|---|
| Aldo-keto reductase family 1 member B10/ADH A | 3.831 | 7.58E-11 | 3.18E-13 | APZ42_033983 |
| GST E10/1-1 | 3.103 | 4.07E-15 | 6.73E-18 | APZ42_014224 |
| Aldo-keto reductase family 1 member C23-like protein | 2.605 | 1.08E-15 | 1.64E-18 | APZ42_018563 |
| Organic cation transporter protein | 2.270 | 2.11E-12 | 6.39E-15 | APZ42_025994 |
| Glutamate-cysteine ligase catalytic subunit | 2.193 | 1.28E-12 | 3.6E-15 | APZ42_028324 |
| GST A4 | 2.167 | 1.65E-06 | 2.33E-08 | APZ42_020015 |
| UDPGT 2B7/Ugt36B | 1.843 | 5.74E-07 | 6.75E-09 | APZ42_014348 |
| Esterase FE4 | 1.779 | 1.98E-06 | 2.89E-08 | APZ42_031913 |
| CYP370A13 | 1.767 | 1.02E-06 | 1.32E-08 | APZ42_028687 |
| Dipeptidase 1 | 1.678 | 2.57E-08 | 8.07E-10 | APZ42_016051 |
| Glutathione synthetase | 1.657 | 3.42E-05 | 7.62E-07 | APZ42_019059 |
| CYP4C55 | 1.634 | 1.46E-05 | 2.76E-07 | APZ42_019414 |
| Cyp4C58 | 1.580 | 1.11E-04 | 2.97E-06 | APZ42_028083 |
| UDPGT 2B31 | 1.571 | 9.34E-04 | 3.45E-05 | APZ42_013190 |
| Glutamate--cysteine ligase regulatory subunit | 1.557 | 1.51E-06 | 2.09E-08 | APZ42_028089 |
| Solute carrier family 15 member 2 | 1.547 | 8.25E-04 | 2.97E-05 | APZ42_024423 |
| CYP370A9 | 1.541 | 1.34E-12 | 3.96E-15 | APZ42_028691 |
Log2Fold Change determined by EdgeR
False Discovery Rate
Locus tag/ID is the accession number for the NCBI database
3.2 qPCR
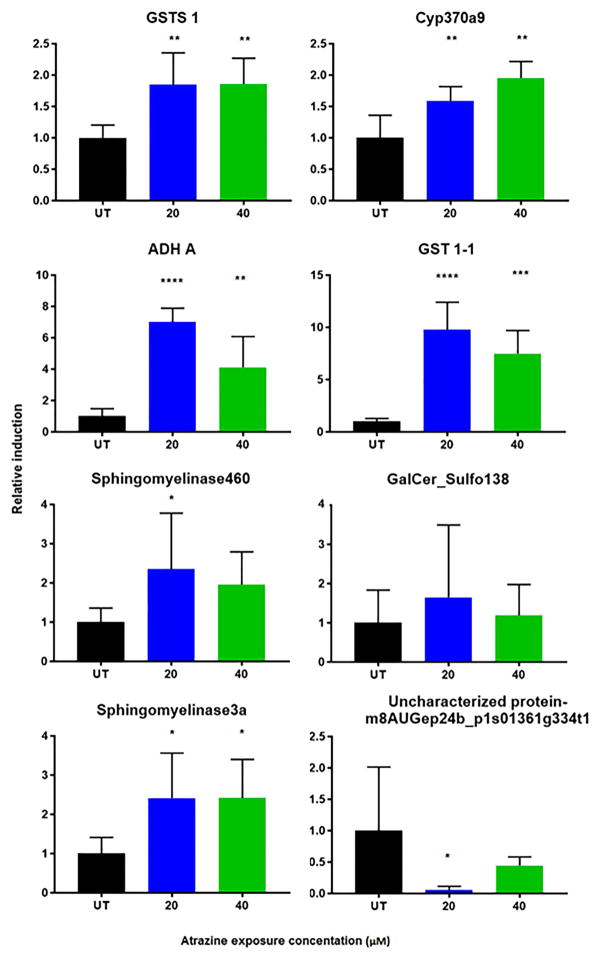
qPCR was used to confirm differential expression of several genes, especially those related to detoxification or sphingolipid homeostasis. Two GSTs, cyp370a9, ADH-A, and two sphingomyelinases were significantly up-regulated in the daphnids exposed to at least one of the atrazine concentrations (Fig. 3). An uncharacterized protein was confirmed to be down-regulated in the 20 μM treated daphnids; a galactosylceramide sulfotransferase gene identified to be down-regulated was not confirmed by qPCR (Fig. 3). qPCR validation of down-regulated genes can be difficult as genes with already low copy numbers may not respond until later qPCR cycles, which are associated with decreased reaction efficiencies (Morey et al., 2006). Overall, qPCR confirmed that several detoxification genes are significantly induced by atrazine.
Fig. 3. qPCR confirmed changes in gene expression measured by RNAseq.
Changes in gene expression were confirmed for several genes by qPCR at 0, 20 and 40 μM atrazine. Data are presented as mean ± SD. Asterisks denote p-values: * ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001; **** < 0.0001.
3.3 Toxicity tests
Standard 48h acute toxicity tests demonstrated that chlorpyrifos is the most toxic (LC50 = 2.9 pM) and bisphenol A (BPA) is the least toxic (LC50 = 66.53 μM) of the chemicals tested (Table 2; Suppl. Fig. 2, Suppl. Table 6). All six compounds tested were used at varying concentrations in binary mixture assays with 40 μM atrazine, as this concentration is not toxic, induces detoxification genes (Table 1; Fig. 3), and has been previously shown to provide concentration-dependent protection from triclosan (Sengupta et al., 2015). Measured toxicity of individual chemicals was compared to toxicity in the presence of atrazine, and to the independent joint action model using CATAM at each chemical’s LC50 value as well as the upper and lower 95% confidence intervals of the LC50 (Fig. 4).
Table 2. LC50 and Hillslope values for the chemicals of interest.
Values determined from neonates exposed to chemicals individually for 48 hours. Atrazine values from Sengupta et al., 2015.
| Chemical | LC50 (μM)(95% CI) | Hillslope |
|---|---|---|
| p-nonylphenol | 1.944 (1.722 – 2.194) | −5.895 (−8.000 to −3.790) |
| BPA | 66.53 (57.27 – 77.27) | −6.891 (−10.34 to −3.438) |
| parathion | 0.0122 (0.0103 – 0.0144) | −4.831 (−8.368 to −1.294) |
| chlorpyrifos | 0.002921 (0.002416 to 0.003449) | −2.498 (−3.795 to −1.496) |
| endosulfan | 4.644 (4.415 to 4.885) | −8.310 (−10.52 to −6.098) |
| triclosan | 1.083 (0.9951 to 1.186) | −4.365 (−6.091 to −3.083) |
| atrazine | 78.8 | −36.56 (−2.69e07 to 2.69e07) |
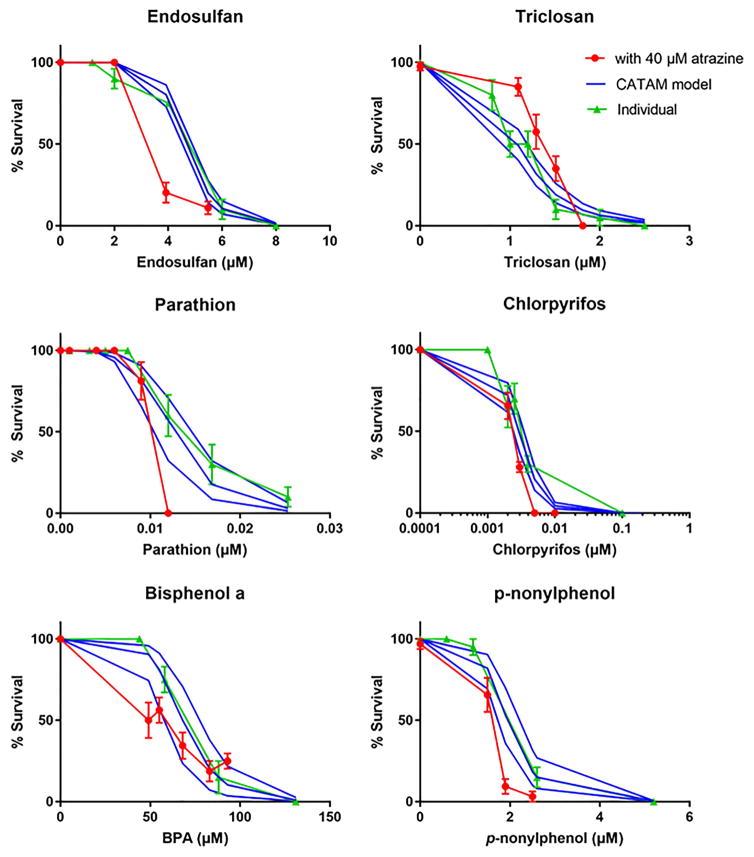
Fig. 4. Toxicity of individual chemicals and binary atrazine mixtures compared to the CATAM model.
Neonates were exposed to 40μM atrazine and one other chemical for 48 hours and survival measured. The measured toxicity of the individual chemicals and chemical mixture are compared to the estimated toxicity of the CATAM model. The CATAM model is shown as solid lines with the top and bottom lines indicative of the upper and lower 95% confidence intervals of the model based on the chemical’s Hillslope and LC50. Data are expressed as mean + SEM.
Percent survival and compliance with the independent joint action model varied depending on the chemical. Comparison of the dose-response curves from the standard toxicity tests to CATAM indicate that CATAM correctly estimated the toxicity of chlorpyrifos and BPA in the presence of atrazine (Fig. 4). To confirm these results, the LC50’s of the atrazine binary mixtures were determined and compared to the LC50 of the individual chemicals. The LC50 of atrazine + chlorpyrifos (0.00239 μM) and atrazine + BPA (67.75 μM) either fit well within the 95% CI of the individual chemicals or is extremely close. Thus, toxicity is considered additive.
Comparison of the curves produced from the toxicity tests to CATAM overestimated survival of daphnids exposed to endosulfan, high concentrations of parathion, and to a lesser extent p-nonylphenol, indicating synergism between these chemicals (Fig. 4). The LC50 values produced from the atrazine-containing binary mixtures confirmed synergistic toxicity for p-nonylphenol (1.449 μM) and endosulfan (1.825 μM), as LC50’s dropped 25% and 60%, respectively (Table 2). The LC50’s are well below the 95% CI’s for these two chemicals. The 95% CI of parathion is very close to parathion’s LC50 during atrazine co-treatment (0.0010 μM) and this may be due to atrazine exerting is synergistic effects only at high concentrations of parathion (Fig. 4). In addition, we determined AUC with GraphPad Prism 7.0 for the individual compounds, mixtures, and CATAM model. The AUCs were significantly less for the mixtures curves of atrazine with endosulfan, parathion, or p-nonylphenol compared to AUC for the compound individually, but also for the CATAM model. All three analyses used (dose-response curves, LC50 values, and AUC) indicate synergistic toxicity for atrazine-p-nonylphenol or atrazine-endosulfan mixtures, and two out of three analyses used indicate synergistic toxicity for atrazine-parathion mixture, further confirming that atrazine enhanced toxicity within all three of these mixtures (Fig 4; Suppl. Fig. 3, Suppl. Table 6).
Comparison of the curves produced from the toxicity tests to CATAM underestimated survival of daphnids exposed to triclosan, indicating concentration-dependent protection (antagonism) by atrazine (Fig. 4). The LC50 value produced from atrazine + triclosan (1.5 μM) is significantly higher than for triclosan alone (0.9951 μM to 1.186 μM) (Table 2), indicating that triclosan is the only chemical of those we tested with decreased toxicity in the presence of atrazine consistent with previous work (Sengupta et al., 2015). Furthermore, the AUC for atrazine in combination with triclosan was significantly greater than the CATAM model’s AUC or triclosan’s AUC, confirming survival was greater within the mixture (Suppl. Fig. 3).
4. Discussion
Atrazine, an efficacious HR96 activator, significantly up-regulates detoxification genes in D. magna (Suppl. Table 2–4; Table 1) consistent with HR96 activation (King-Jones et al., 2006; Sengupta et al., 2015; Sieber and Thummel, 2009). RNAseq demonstrated that GSTs, CYPs, ADH-A, and UDPGTs were significantly up-regulated at 40 μM atrazine and several of these were confirmed by qPCR at 20 and 40 μM atrazine (Fig. 3). The use of GO term enrichment and pathway analysis further demonstrated that terms and pathways associated with xenobiotic metabolism such as drug metabolism, drug metabolism by P450s, glutathione metabolism, oxidation-reduction reactions, and response to oxidative stress were all increased (Fig. 1–2; Table 1). The RNAseq data confirms previous research that demonstrates increased GST activity and antioxidant responses in D. magna following 20 and 40 μM atrazine exposure (Sengupta et al., 2015). Overall, these results support and confirm that atrazine is a potent inducer of detoxification genes most likely mediated by HR96 activation (Karimullina et al., 2012; King-Jones et al., 2006; Liu et al., 2017; Sengupta et al., 2015).
In addition, digestion, sphingolipid metabolism, cellular response to nitric oxide, and apoptotic processes were induced biological processes, among others (Fig. 1; Suppl. Table 3). Several of these pathways are known to be regulated by HR96 in Drosophila and potentially Daphnia such as xenobiotic metabolism, sphingolipid metabolism, and GST activity (Bujold et al., 2010; Horner et al., 2009; King-Jones et al., 2006; Sengupta et al., 2017). In Drosophila, HR96 regulates cholesterol and triacylglycerol homeostasis through the regulation of sterol metabolism genes, such as npc1b and magro, respectively (Bujold et al., 2010; Horner et al., 2009; Sieber and Thummel, 2009). Interestingly, atrazine did not induce magro in D. magna, in previous studies and the current one, although other enzymes involved in lipid metabolism such as ceramidase and sphingomyelinase 3A were increased in a concentration dependent manner (Suppl. Table 2; Fig. 3) (Sengupta et al., 2017). Other digestive pathways were increased including starch and sucrose metabolism (Suppl. Table 4). Last, pathways such as cellular response to nitric oxide or various apoptotic processes have not previously been shown to be HR96-mediated, and likely are mediated through activation of other pathways by atrazine.
During chemical stress, energy allocation is directed toward detoxification and survival and away from maintenance, reproduction and growth (Du et al., 2015; Roling et al., 2006; Scanlan et al., 2015)(Knops et al., 2001). Down-regulated GO terms such as oogenesis and pupal chitin-based cuticle development support this basic premise. Atrazine reduces reproduction significantly with 40 μM atrazine reducing fecundity nearly 90% (Sengupta et al., 2016). Furthermore, in Tribolium castaneum, HR96 mRNA levels are high in the ovaries, and the knock-down of HR96 through RNA interference (RNAi) resulted in the loss of reproduction, likely due to the interruption of development of embryos (Xu et al., 2010). Interestingly, galactosylceramide biosynthesis was a biological process associated with down-regulated genes and sphingolipid metabolism was associated with up-regulated genes (Suppl. Table 2). This indicates an increase in sphingolipids and sphingomyelin with an associated decrease in ceramides (Delgado et al., 2007). Sphingomyelin is primarily produced in neonates and lost as neonates develop into reproductive adults, further supporting a role for sphingomyelin/sphingolipid metabolism in reproduction (Sengupta et al., 2017). Previous research has shown that atrazine alters lipid profiles primarily by perturbing sphingomyelin/ceramide pathways, increasing sphingomyelin concentrations, and this is associated with repressed reproductive development (Sengupta et al., 2017, 2016). While Daphnia are unlikely to encounter a concentration as high as 40 μM in a natural system, atrazine has the potential to perturb both detoxification and reproduction and sphingolipid metabolism is associated with the decrease in reproduction.
Given the induction of GSTs, UDPGTs, CYPs, and multidrug transporters, among other detoxification enzymes, we predicted concentration-dependent protective antagonistic mixture interactions with atrazine and triclosan and atrazine and the phenols. Triclosan generates ROS and is primarily detoxified via glucuronidation with some sulfation (Ashrap et al., 2017; Binelli et al., 2009; Wu et al., 2010). RNAseq confirmed that several GSTs and UDPGTs were induced, which may detoxify triclosan or mitigate its toxic effects from ROS generation. Similar to previous studies, we saw an increase in survival in daphnids exposed to an atrazine and triclosan mixture when compared to triclosan alone and the CATAM model (Fig. 4). Previous work also demonstrated protective effects in 40 μM atrazine and 1–1.5 μM triclosan mixtures (Sengupta et al., 2015).
For BPA and p-nonylphenol, the CATAM model tended to underestimate toxicity and overestimate survival, though the extent by which it underestimated toxicity varied by concentration. Like triclosan, BPA and p-nonylphenol are hypothesized to be detoxified through glucuronide conjugation (Fay et al., 2015; Green et al., 2003). Despite the induction of several UDPGTs, antagonistic effects were not observed (Fig. 4; Suppl Fig. 3). At higher concentrations, p-nonylphenol weakly increased toxicity, while BPA’s trend did not deviate from the CATAM model. Previous work has demonstrated an additive effect with atrazine-BPA mixtures (Juhel et al., 2017). We did observe differences in the AUC for the atrazine-BPA mixtures (Suppl. Fig. 3); however this was the only one of three analysis methods (dose-response curves, LC50 values, AUC) that showed synergism and also appears to be the least stringent method.
For mixtures containing organochlorines or organophosphates, we expected that atrazine would increase toxicity because CYP induction would increase the formation of the toxic –sulfated and –oxon metabolites of endosulfan and parathion/chlorpyrifos. The CATAM model significantly underestimated endosulfan toxicity at a concentration of 4 μM (Fig. 4). Endosulfan is metabolized by CYPs to another toxic form, endosulfan sulfate, and generates ROS (Barata et al., 2005; Sebastian & Raghavan, 2017). Atrazine exposure induced several CYPs that could have contributed to increased toxicity through the production of endosulfan sulfate. Also induced were antioxidants enzymes like GSTs, but superoxide dismutase and catalase were not, potentially weakening D. magna’s ability to respond to ROS. The lack of superoxide dismutase and catalase induction also suggests that Nrf2 was not activated following atrazine exposure (Zhu et al., 2005). Overall, atrazine-endosulfan binary mixtures showed increased toxicity relative to the CATAM model for all three analyses, consistent with increased endosulfan sulfate production by CYPs.
The CATAM model underestimated the toxicity of parathion-atrazine mixtures as predicted, primarily at high concentrations of parathion. In atrazine-parathion mixtures, mortality was 100% at parathion’s LC50 (0.012 μM) (Fig. 4), presumably because of the increased paraoxon produced by CYPs (Mota et al., 2010). For chlorpyrifos-atrazine mixtures, the CATAM model was a fairly accurate predictive model, suggesting additive effects were occurring but not synergism. The CYP enzymes induced by atrazine may metabolize parathion, but not chlorpyrifos, potentially accounting for the difference in response to the atrazine mixture. In human microsomes, CYP2B6 was found to be important metabolic enzymes of parathion and chlorpyrifos (Foxenberg et al., 2007; Mutch & Williams, 2006). Chlorpyrifos metabolism by CYP2B6 primarily produced the toxic chlorpyrifos-oxon metabolite (Foxenberg et al., 2007), while other studies found CYP2B6 metabolism of parathion preferentially produced the non-toxic metabolite p-nitrophenol (Mutch & Williams, 2006). In D. magna, two CYP2 clan members, cyp370a13 and cyp370a9 (Baldwin et al., 2009, were induced by atrazine. However, as in humans, one of the up-regulated CYP enzymes induced by atrazine exposure may preferentially produce a toxic metabolite for one organophosphate (parathion) and the non-toxic metabolite for another (chlorpyrifos). Therefore, it is certainly possible that differences observed between chlorpyrifos and parathion toxicity in the presence of atrazine are caused by differences in the metabolism of these organophosphates by the corresponding induced CYPs.
4.2 Conclusions
RNAseq demonstrates that a sub-lethal concentration of the HR96 activator, atrazine up-regulates several detoxification enzymes such as GSTs, CYPs, and glucosyltransferases, and this response may aid D. magna response to chemical stressors. This work indicates that HR96 has similar functions in toxicology to its homologs, CAR and PXR. Similar to CAR and PXR, this response may cause adverse reactions with other chemicals (parathion, endosulfan, p-nonylphenol), or protective (triclosan) responses most likely depending on whether the induction of these chemicals leads to detoxification or activation. While the contaminated environment is far more complex than binary exposures, these findings provide a possible mechanistic explanation for atrazine mixture interactions in Daphnia that can be used for extrapolation in other mixture scenarios. Furthermore, it demonstrates that Daphnia possess the biochemical machinery necessary to respond and acclimate to chemical stressors.
Supplementary Material
Highlights.
Atrazine induces phase I–III detoxification consistent with HR96 activation
Atrazine-triclosan mixtures caused antagonism consistent with GST induction.
Atrazine-parathion and atrazine-endosulfan mixtures caused synergistic toxicity.
Most but not all interactions could be predicted based on enzyme induction.
Acknowledgments
This work was supported by NIEHS grant R15ES017321.
Abbreviations
- ADH
alcohol dehydrogenase
- AUC
area under the curve
- ABC transporter
ATP-binding cassette transporter
- BPA
bisphenol A
- CATAM
Computational Approach to the Toxicity Assessment of Mixtures
- CAR
constitutive androstane receptor
- CYP
cytochrome P450
- DHA
docosahexaenoic acid
- GST
glutathione S-transferase
- GO
gene ontology
- HR96
hormone receptor 96
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MRP
multi-drug resistance protein
- NR
nuclear receptor
- PXR
pregnane x receptor
- qPCR
quantitative real-time polymerase chain reaction
- ROS
reactive oxygen species
- RNAseq
RNA sequencing
- UDPGT
uridine 5′-diphospho-glucuronosyltransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashrap P, Zheng G, Wan Y, Li T, Hu W, Li W, Zhang H, Zhang Z, Hu J. Discovery of a widespread metabolic pathway within and among phenolic xenobiotics. Proc Natl Acad Sci U S A. 2017;114:6062–6067. doi: 10.1073/pnas.1700558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NT. Agricultural pesticide use estimates for the USGS National Water Quality Network, 1992–2014. 2016 doi: 10.5066/F7FF3QHP. [DOI] [Google Scholar]
- Baldwin WS, Roling JA. A concentration addition model for the activation of the constitutive androstane receptor by xenobiotic mixtures. Toxicol Sci. 2009;107:93–105. doi: 10.1093/toxsci/kfn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barata C, Varo I, Navarro JC, Arun S, Porte C. Antioxidant enzyme activities and lipid peroxidation in the freshwater cladoceran Daphnia magna exposed to redox cycling compounds. Comp Biochem Physiol - C Toxicol Pharmacol. 2005;140:175–186. doi: 10.1016/j.cca.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Belden JB, Lydy MJ. Impact of atrazine on organophosphate toxicity. Environ Toxicol Chem. 2000;19:2266–2274. doi: 10.1002/etc.5620190917. [DOI] [Google Scholar]
- Binelli A, Cogni D, Parolini M, Riva C, Provini A. In vivo experiments for the evaluation of genotoxic and cytotoxic effects of Triclosan in Zebra mussel hemocytes. Aquat Toxicol. 2009;91:238–244. doi: 10.1016/j.aquatox.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Bliss CI. The Toxicity of Poisons Applied Jointly. Ann Appl Biol. 1939;26:585–615. doi: 10.1111/j.1744-7348.1939.tb06990.x. [DOI] [Google Scholar]
- Bujold M, Gopalakrishnan A, Nally E, King-Jones K. Nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila melanogaster. Mol Cell Biol. 2010;30:793–805. doi: 10.1128/MCB.01327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Cáceres CE, Carmel L, Choi J, Detter JC, Dong Q, Dusheyko S, Eads D, Fröhlich T, Geiler-samerotte Ka, Gerlach D, Schaack S, Shapiro H, Shiga Y, Skalitzky C. The ecoresponsive genome of D. pulex. Science (80-) 2011;331:555–561. doi: 10.1126/science.1197761.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A, Casas J, Llebaria A, Abad JL, Fabriás G. Chemical tools to investigate sphingolipid metabolism and functions. ChemMedChem. 2007;2:580–606. doi: 10.1002/cmdc.200600195. [DOI] [PubMed] [Google Scholar]
- DeLorenzo ME, Serrano L. Individual and mixture toxicity of three pesticides; atrazine, chlorpyrifos, and chlorothalonil to the marine phytoplankton species Dunaliella tertiolecta. J Environ Sci Health B. 2003;38:529–538. doi: 10.1081/PFC-120023511. [DOI] [PubMed] [Google Scholar]
- Du X, Crawford DL, Oleksiak MF. Effects of Anthropogenic Pollution on the Oxidative Phosphorylation Pathway of Hepatocytes from Natural Populations of Fundulus heteroclitus. Aquat Toxicol. 2015;165:231–240. doi: 10.1016/j.aquatox.2015.06.009. [DOI] [PubMed] [Google Scholar]
- European Union. 2004. 32004D0248. 2004/248/EC: Commission Decision of 10 March 2004 concerning the non-inclusion of atrazine in Annex I to Council Directive 91/414/EEC and the withdrawal of authorisations for plant protection products containing this active substance (Text with EEA relevance) (notified under document number C(2004) 731)
- Fay MJ, Nguyen MT, Snouwaert JN, Dye R, Grant DJ, Bodnar WM, Koller BH. Xenobiotic metabolism in mice lacking the UDP-Glucuronosyltransferase 2 family. Drug Metab Dispos. 2015;43:1838–1846. doi: 10.1124/dmd.115.065482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron VJ, Groten JP. Toxicological evaluation of chemical mixtures. Food Chem Toxicol. 2002;40:825–839. doi: 10.1016/S0278-6915(02)00021-2. [DOI] [PubMed] [Google Scholar]
- Foxenberg RJ, McGarrigle BP, Knaak JB, Kostyniak PJ, Olson JR. Human hepatic cytochrome P450-specific metabolism of parathion and chlorpyrifos. Drug Metab Dispos. 2007;35:189–193. doi: 10.1124/dmd.106.012427. [DOI] [PubMed] [Google Scholar]
- Ginjupalli GK, Baldwin WS. The Time- and Age-dependent Effects of the Juvenile Hormone Analog Pesticide, Pyriproxyfen on Daphnia magna Reproduction. Chemosphere. 2013;92:1260–1266. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T, Swain C, Van Miller JP, Joiner RL. Absorption, bioavailability, and metabolism of para-nonylphenol in the rat. Regul Toxicol Pharmacol. 2003;38:43–51. doi: 10.1016/S0273-2300(03)00048-5. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Philip D, Bowden J, Couger MB, Eccles D, Li B, Macmanes MD, Ott M, Orvis J, Pochet N. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 2013;8:1–43. doi: 10.1038/nprot.2013.084.De. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen S, Meijerman I, Beijnen JH, Schellens JHM. The role of nuclear receptors in pharmacokinetic drug-drug interactions in oncology. Cancer Treat Rev. 2007;33:369–380. doi: 10.1016/j.ctrv.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Hedrich WD, Xiao J, Heyward S, Zhang Y, Zhang J, Baer MR, Hassan HE, Wang H. Activation of the constitutive androstane receptor increases the therapeutic index of CHOP in lymphoma treatment. Mol Cancer Ther. 2016;15:392–401. doi: 10.1021/acsnano.5b07425.Molecular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, Huang W, Chapman LM, Chua S, Moore DD, Baldwin WS. The Environmental Estrogen, Nonylphenol, Activates the Constitutive Androstane Receptor. Toxicol Sci. 2007;98:416–426. doi: 10.1017/S1368980009991996.Validation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Huang W, Moore DD, Baldwin WS. Sexually dimorphic regulation and induction of P450s by the constitutive androstane receptor (CAR) Toxicology. 2009;256:53–64. doi: 10.1016/j.tox.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Lee J, Lin YH, Jing G, Tsai LJ, Chen A, Hetrick L, Jocoy D, Liu J. Atrazine and malathion shorten the maturation process of Xenopus laevis oocytes and have an adverse effect on early embryo development. Toxicol Vitr. 2016;32:63–69. doi: 10.1016/j.tiv.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Juhel G, Bayen S, Goh C, Lee WK, Kelly BC. Use of a suite of biomarkers to assess the effects of carbamazepine, bisphenol A, atrazine, and their mixtures on green mussels, Perna viridis. Environ Toxicol Chem. 2017;36:429–441. doi: 10.1002/etc.3556. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimullina E, Li Y, Ginjupalli GK, Baldwin WS. Daphnia HR96 is a promiscuous xenobiotic and endobiotic nuclear receptor. Aquat Toxicol. 2012;116–117:69–78. doi: 10.1016/j.aquatox.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Horner MA, Lam G, Thummel CS. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Knops M, Altenburger R, Segner H. Alterations of physiological energetics, growth and reproduction of Daphnia magna under toxicant stress. Aquat Toxicol. 2001;53:79–90. doi: 10.1016/S0166-445X(00)00170-3. [DOI] [PubMed] [Google Scholar]
- Litoff EJ, Garriott TE, Ginjupalli GK, Butler L, Gay C, Scott K, Baldwin WS. Annotation of the Daphnia magna nuclear receptors: Comparison to Daphnia pulex. Gene. 2014;552:116–125. doi: 10.1016/j.immuni.2010.12.017.Two-stage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang L, Pan B, Wang C, Bao S, Nie X. Toxic effects of diclofenac on life history parameters and the expression of detoxification-related genes in Daphnia magna. Aquat Toxicol. 2017;183:104–113. doi: 10.1016/j.aquatox.2016.12.020. [DOI] [PubMed] [Google Scholar]
- Metcalf RL. Mode of Action of Insecticide Synergists. Annu Rev Entomol. 1967;12:229–256. doi: 10.1146/annurev.en.12.010167.001305. [DOI] [PubMed] [Google Scholar]
- Moore LB, Goodwin B, Jones Sa, Wisely GB, Serabjit-Singh CJ, Willson TM, Collins JL, Kliewer Sa. St. John’s wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc Natl Acad Sci U S A. 2000;97:7500–7502. doi: 10.1073/pnas.130155097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–93. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota LC, Hernandez JP, Baldwin WS. Constitutive androgen receptor-null mice are sensitive to the toxic effects of parathion: Association with reduced cytochrome P450-mediated parathion metabolism. Drug Metab Dispos. 2010;38:1582–1588. doi: 10.1124/dmd.110.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. [PubMed] [Google Scholar]
- Mutch E, Williams FM. Diazinon, chlorpyrifos and parathion are metabolised by multiple cytochromes P450 in human liver. Toxicology. 2006;224:22–32. doi: 10.1016/j.tox.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Olmstead AW, LeBlanc GA. Toxicity assessment of environmentally relevant pollutant mixtures using a heuristic model. Integr Env Assess Manag. 2005;1:114–122. doi: 10.1897/IEAM_2004-005R.1. [DOI] [PubMed] [Google Scholar]
- Oswald S, Haenisch S, Fricke C, Sudhop T, Remmler C, Giessmann T, Jedlitschky G, Adam U, Dazert E, Warzok R, Wacke W, Cascorbi I, Kroemer HK, Weitschies W, von Bergmann K, Siegmund W. Intestinal expression of P-glycoprotein (ABCB1), multidrug resistance associated protein 2 (ABCC2), and uridine diphosphate-glucuronosyltransferase 1A1 predicts the disposition and modulates the effects of the cholesterol absorption inhibitor ezetimibe in. Clin Pharmacol Ther. 2006;79:206–217. doi: 10.1016/j.clpt.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Rider CV, LeBlanc GA. An integrated addition and interaction model for assessing toxicity of chemical mixtures. Toxicol Sci. 2005;87:520–528. doi: 10.1093/toxsci/kfi247. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roling JA, Bain LJ, Gardea-Torresdey J, Bader J, Baldwin WS. Hexavalent chromium reduces larval growth and alters gene expression in mummichog (Fundulus heteroclitus) Environ Toxicol Chem. 2006;25:2725–33. doi: 10.1897/05-659r.1. [DOI] [PubMed] [Google Scholar]
- Scanlan LD, Loguinov AV, Teng Q, Antczak P, Dailey KP, Nowinski DT, Kornbluh J, Lin XX, Lachenauer E, Arai A, Douglas NK, Falciani F, Stapleton HM, Vulpe CD. Gene Transcription, Metabolite and Lipid Profiling in Eco-Indicator Daphnia magna Indicate Diverse Mechanisms of Toxicity by Legacy and Emerging Flame-Retardants. Env Sci Technol. 2015;49:7400–7410. doi: 10.1002/nbm.3369.Three. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian R, Raghavan SC. Molecular Mechanism of Endosulfan Action in Mammals. J Biosci. 2017;42:149–153. doi: 10.1007/s12038-016-9655-4. [DOI] [PubMed] [Google Scholar]
- Sengupta N, Gerard PD, Baldwin WS. Perturbations in Polar Lipids, Starvation Survival and Reproduction Following Exposure to Unsaturated Fatty Acids or Environmental Toxicants in Daphnia magna. Chemosphere. 2016;144:2302–2311. doi: 10.1038/nbt.3121.ChIP-nexus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Litoff EJ, Baldwin WS. The HR96 activator, atrazine, reduces sensitivity of D. magna to triclosan and DHA. Chemosphere. 2015;128:299–306. doi: 10.1002/dev.21214.Developmental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Reardon DC, Gerard PD, Baldwin WS. Exchange of polar lipids from adults to neonates in Daphnia magna: Perturbations in sphingomyelin allocation by dietary lipids and environmental toxicants. PLOS. 2017;12:1–25. doi: 10.1371/journal.pone.0178131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 Nuclear Receptor Controls Triacylglycerol Homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011:6. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Environmental Protection Agency. Environ Prot 266. 5. 2002. Oct, Methods for Measuring the Acute Toxicity of Effluents and Receiving Waters to Freshwater and Marine Organisms. EPA-821-R-02-012. [Google Scholar]
- Vogel G. A Worrisome Side Effect of an Antianxiety Remedy. Science (80-) 2001;291:37. doi: 10.1126/science.291.5501.37. [DOI] [PubMed] [Google Scholar]
- Wacksman MN, Maul JD, Lydy MJ. Impact of atrazine on chlorpyrifos toxicity in four aquatic vertebrates. Arch Environ Contam Toxicol. 2006;51:681–689. doi: 10.1007/s00244-005-0264-8. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Maglich JM, Moore LB, Wisely GB, Noble SM, Davis-Searles PR, Lambert MH, Kliewer SA, Redinbo MR. 2.1 Å crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry. 2003;42:1430–1438. doi: 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- Wu J, Liu J, Cai Z. Determination of triclosan metabolites by using in-source fragmentation from high-performance liquid chromatography/negative atmospheric pressure chemical ionization ion trap mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:1828–1834. doi: 10.1002/rcm.4558. [DOI] [PubMed] [Google Scholar]
- Xu J, Tan A, Palli SR. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J Insect Physiol. 2010;56:1471–1480. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Itoh K, Yamamoto M, Zweier JL, Li Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: Protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005;579:3029–3036. doi: 10.1016/j.febslet.2005.04.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.