Abstract
Antagonists of MDM2-p53 interaction are emerging anti-cancer drugs utilized in clinical trials for malignancies that rarely mutate p53, including melanoma. We discovered that MDM2-p53 antagonists protect DNA from drug-induced damage in melanoma cells and patient-derived xenografts. Among the tested DNA damaging drugs were various inhibitors of Aurora and Polo-like mitotic kinases, as well as traditional chemotherapy. Mitotic kinase inhibition causes mitotic slippage, DNA re-replication, and polyploidy. Here we show that re-replication of the polyploid genome generates replicative stress which leads to DNA damage. MDM2-p53 antagonists relieve replicative stress via the p53-dependent activation of p21 which inhibits DNA replication. Loss of p21 promoted drug-induced DNA damage in melanoma cells and enhanced anti-tumor activity of therapy combining MDM2 antagonist with mitotic kinase inhibitor in mice. In summary, MDM2 antagonists may reduce DNA damaging effects of anti-cancer drugs if they are administered together, while targeting p21 can improve the efficacy of such combinations.
Keywords: DNA damage, Replication stress, Mitotic inhibitor, MDM2 antagonist, Melanoma, Polyploidy, p21, p53
Highlights
-
•
Mitotic inhibitors induce polyploidy which leads to replication stress and DNA damage.
-
•
MDM2 antagonists reduce mitotic inhibitor-induced DNA damage by blocking DNA re-replication.
-
•
Loss of p21 alleviates DNA protection by MDM2 antagonism and increases efficacy of combined MDM2 and mitotic inhibition.
Many anti-cancer therapies work by damaging DNA in cancer cells. Here we demonstrate that investigational cancer drugs that target MDM2 (MDM2 antagonists) can protect cancer cells from drug-induced DNA damage. They do so by increasing the levels of protein p21 which prevents cells from replicating DNA. Disabling of p21 increases drug-induced DNA damage and promotes killing of cancer cells. This study improves our understanding of how to combine different anti-cancer drugs to generate most effective treatment regimens and demonstrates the need for development of therapies that inactivate p21.
1. Introduction
A tremendous shift in the melanoma treatment landscape has recently unfolded. Clinically active targeted therapies (combined BRAF and MEK inhibitors) produce dramatic responses in melanoma patients harboring BRAF mutations. While initial responses are impressive, therapeutic resistance develops at a median of 9–11 months on treatment (Johnson and Sosman, 2015). Further, MEK inhibitor monotherapy has a much more modest efficacy in other subsets of melanoma, including NRAS-mutant melanoma (Vu and Aplin, 2016). In parallel, effective immune checkpoint inhibitors, including monoclonal antibodies to cytotoxic T lymphocyte antigen-4 (CTLA4; ipilimumab) and programmed death-1 (PD-1; nivolumab and pembrolizumab) produce durable responses in a subset of patients (Vilgelm et al., 2016). Although current therapeutics represent substantial advances compared to earlier treatment options, the majority of patients are either intrinsically resistant or acquire therapeutic resistance, and die of their disease. Clearly, additional treatment options remain a critical and unmet need.
Antagonists of MDM2-p53 interaction work by boosting the activity of wild type p53 and thus are best suited for treating malignancies where mutations in the TP53 gene are uncommon. This includes melanoma, which harbors a relatively low (~ 15%) rate of TP53 mutations (Hodis et al., 2012). However, the p53 pathway is suppressed in most melanomas via mutations, deletions or promoter methylation of the CDKN2A gene (Freedberg et al., 2008; Goldstein et al., 2007; Hodis et al., 2012). p14Arf, a product of this gene, negatively regulates MDM2, which is a main ubiquitin kinase that targets p53 for degradation (Kubbutat et al., 1998). In addition to CDKN2A inactivation, MDM2 gene is amplified in a subset of melanoma tumors (about 5% of cases) (Hodis et al., 2012). These alterations can diminish p53 activity in malignant cells. Therefore, targeting the MDM2-p53 interaction with specific small molecule antagonists may benefit melanoma patients with wild type TP53, and CDKN2A loss or MDM2 amplifications.
Early phase clinical trials of MDM2 antagonists showed evidence of anti-tumor activity in patients with leukemia and liposarcoma (Burgess et al., 2016). For instance, in the phase 1 study of RG7112 in liposarcoma, partial response was achieved in 1 out of 20 patients and 14 had stable disease (Vu et al., 2013). In a phase 1 leukemia trial, complete or partial response to RG7112 was seen in 5 out of 30 patients with AML (Andreeff et al., 2016). More promising results were obtained using the next generation MDM2 antagonist, RG7388 (idasanutlin), which induced complete responses in about a quarter of enrolled AML patients (Yee et al., 2014). However, clinical trials of MDM2 antagonists also reported serious on-target adverse events including GI toxicities and prolonged myelosuppression. These data suggest that using MDM2 antagonists at a lower dose and in combination with other therapies may be more effective than single agent therapy. Finding rational and effective combination partners for MDM2 inhibitors in melanoma which avoid excessive toxicity was a goal of the study discussed here.
We have recently reported that the combination of MDM2 antagonist with a senescence-inducing inhibitor of the mitotic kinase Aurora A (AURKA) has a potent anti-melanoma activity (Vilgelm and Richmond, 2015; Vilgelm et al., 2015). In mouse studies this drug combination induced senescence and immune clearance of cancer cells by antitumor leukocytes that were recruited into the tumor via NF-κB-dependent induction of CCL5, CCL1, and CXCL9. As a result, prominent responses were detected in vivo in several melanoma models. In addition, the AURKA and MDM2 combination therapy showed adequate bioavailability and low toxicity to the host (Vilgelm et al., 2015). Notably, we found that melanoma cells treated with AURKAi had high levels of DNA damage (Liu et al., 2013). The p53 protein is the master regulator of DNA damage responses. Therefore here we investigated whether activation of p53 using MDM2 antagonists can affect melanoma response to AURKAi-induced DNA damage.
2. Materials and Methods
2.1. Chemical Reagents, Cell Culture and Cell Transfection Protocols
Nutlin-3a was synthesized as described previously (Davis and Johnston, 2011; Davis et al., 2013). MLN8237 was kindly provided by Takeda Pharmaceuticals, Inc. Idasanutlin was provided by Roche Pharmaceuticals. Chemotherapeutic drugs were purchased from Selleck (Houston, TX). Stock solutions of drugs for in vitro studies were prepared in DMSO. Stock solutions of dNTPs were prepared as follows: adenosine (Sigma (St. Louis, MO), A4036; resuspended in sterile water to 10 mM), guanosine (Sigma, G6264; resuspended in sterile DMSO to 10 mM), thymidine (Sigma, T1895; resuspended in sterile water to 10 mM), cytosine (Sigma, C4654; resuspended in sterile water to 10 mM) in accordance with previously published literature (Aird et al., 2013). Cordycepin was purchased from Cayman Chemical (Ann Arbor, MI). All cell lines were obtained from ATCC, except for p21 isogenic HCT116 cells that were purchased from Horizon (Cambridge, United Kingdom). Cells were cultured in DMEM/F12 media supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. For knockdown experiments 1.5 × 105 cells per well were plated in 6 well plates. The next day SignalSilence p21 Waf1/Cip1 siRNA (#6456) or SignalSilence control siRNA were added to culture media (100 nM final concentration, all from Cell Signaling, Danvers, MA) along with 6 μL of Lipofectamine RNAi MAX (Invitrogen, part of the Thermo Fisher Scientific, Waltham, MA). siRNA transfection was repeated on the following day to increase knockdown efficiency. Cells were used for experiments 24 h after the second transfection.
2.2. Western Blotting and Immunofluorescent Staining
The western blotting procedure has been described previously (Su et al., n.d.). Primary antibodies included: p-Ser428 ATR (Cell Signaling, #2853), p-Ser345 Chk1 (Cell Signaling, #2348), p-Thr68 Chk2 (Cell Signaling, #2661), γH2AX (Cell Signaling, #2577), P-H3 (Cell Signaling, #3377), H3 (Cell Signaling, #4499), HSP90 (BD, #610418), MCM3 (Cell Signaling, #4012), MDM2 (SantaCruz, Dallas, TX, clone SMP14), p21 (Cell Signaling, #2947), p53 (Calbiochem, Billerica, MA, clone DO-1), P-S807/811 Rb (Cell Signaling, #8516), P-S4/S6 RPA (Bethyl, Montgomery, TX, A300-245), RPA (Abcam, Cambridge, MA, ab2175). All primary antibodies were used at 1:500 dilutions in 5% milk (total proteins) or in 5% BSA (phosphorylated proteins) prepared in TBST, except for β-actin-specific antibody that was used at 1:2000 dilution. Goat anti-mouse and goat anti-rabbit HRP-conjugated secondary antibodies (Jackson ImmunoResearch) were used at 1:7000 dilutions. For immunofluorescent analysis cells were grown on 8-well chamber slides and fixed with − 20 °C acetone for 10 min. Slides were blocked in 10% normal goat serum (Invitrogen, Carlsbad, CA) and primary antibodies were applied at 1:100 dilutions. Goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with Alexa488 or Alexa594 (Invitrogen) were used at 1:1000 dilutions. ImageJ was used to quantify IF images. Specifically, microphotographs of cells obtained in the DAPI channel were converted to 8 bit images and the threshold was set to 15. Next “analyze particles” function was applied to the images to generate masks of the cell nuclei. Partial nuclei on the edges of the image were excluded. Masks were imported in ROI manager and applied to original γH2AX and DAPI channel images to quantify integrated density (the sum of the values of the pixels in selected area) within the nuclei of individual cells.
2.3. Cell Viability and Comet Assay
Cell viability was measured by MTT assay performed using CellTiter 96 Non-Radioactive Cell Proliferation Assay kit (Promega, Madison, WI) as instructed by the manufacturer. IC50 values were determined using Prism software (Graphpad, La Jolla, CA). Comet assay was performed using OxiSelect Comet Assay Kit (Cell Biolabs, San Diego, CA) in accordance with the manufacturer's recommendations. After alkaline electrophoresis, cells were visualized and photographed at 10 × magnification. Olive tail moments of cells were calculated using Comet Score software (TriTek, Wilmington, DE).
2.4. Flow Cytometry Analysis of Cell Cycle, DNA Replication, γH2AX and PARP
BrdU incorporation combined with PI staining of cellular DNA content was used to analyze cell cycle and replication in melanoma cultures. Cells were grown in 6 well plates; 3 h prior to harvest 20 μM BrdU (BD Biosciences, Franklin Lakes, NJ) was added to the culture media. Cells were trypsinized, washed with ice-cold PBS, fixed with 70% ethanol and stored at 4 °C. The next day cell membranes were digested in 2 N HCl with 0.5% Triton X-100 solution for 30 min at room temperature, neutralized in 0.1 M Sodium Tetraborate for 2 min, and washed in PBS with 1% BSA. Pelleted cells were re-suspended in 50 μL 0.5% Tween-20 with 1% BSA in PBS and 20uL of FITC-labeled BrdU-specific antibody (FITC Mouse Anti-BrdU set, BD Pharmingen) and then incubated 1 h at room temperature. Next, cells were washed twice with PBS and 1% BSA and re-suspended in PBS containing 10 μg/mL RNase A and 20 μg/mL PI. After 30 min incubation samples were analyzed by flow cytometry analyzed on a custom 5-laser LSRII (BD Biosciences). For simultaneous analysis of BRDU incorporation, γH2AX and cleaved PARP BRDU-pre-treated cells were processed using Apoptosis, DNA Damage and Cell Proliferation Kit (BD Biosciences) according to manufacturer's recommendation. Imaging cytometry analysis of γH2AX foci was performed as follows. The Flowsight imaging flow cytometer (EMD Millipore) equipped with a 60 mw, 488 nm laser and a QI upgrade was used to analyze the single cell suspension of samples. 20 × Mag images were acquired at low speed and the highest resolution using the Flowsight software (EMD Milipore). Cells were gated away from debris using a plot of Channel 1 (Brightfield) area vs Channel 1 aspect ratio. The images were further refined by selecting for images that were RMS gradient high (channel 1), giving us focused images. Spot analysis in channel 2 was performed by the spot analysis application wizard in the IDEAS software (EMD Millipore). Roughly 1000 focused images were used in the analysis per each condition.
2.5. Mouse Experiments
All animal experiments were conducted in accordance with Vanderbilt University Animal Care and Use Committee (IACUC) guidelines and regulations (protocols M/10/034 and M1600023-00). To establish Hs294T xenografts, 2.5 × 106 cells were injected subcutaneously (SC) in both flanks of BALB/C nu/Foxn1 athymic nude mice. Our protocol for establishment of PDX has been described previously (Su et al., 2012; Vilgelm et al., 2015). The study was approved by the Vanderbilt University Institutional Review Board. For animal experiments, nutlin-3a was suspended in 2% Klucel, 0.5% Tween 80 and homogenized thoroughly prior to each use. Alisertib was dissolved in water. Mouse dosing were as follows: 200 mg/kg nutlin-3a, 30 mg/kg alisertib, and 150 mg/kg once a day idasanutlin. P21 siRNA (Cell Signaling, #6456) was mixed with in vivo JetPei (Polyplus-transfection SA, Illkirch, France) in accordance with manufacturer's recommendations and injected into the tumor. Mice were randomized for treatment when average tumor length reached 5–8 mm in diameter. Animal weight and tumor dimensions were measured 2 times a week. Tumor area was calculated as length x width. Experimenters were not blind to group assignment and outcome assessment.
2.6. Statistical Analysis
Mann-Whitney test and analysis of variances (ANOVA) were used for analysis comparing two samples and multiple samples, respectively. GraphPad Prizm software was used for the analysis of in vitro experiments. For in vivo experiments we compared the progression of tumor area (mm2) over time among groups of mice receiving different therapy with linear mixed effects regression analysis. To meet the normality assumptions for these parametric methods, a square root or the natural log transformations were used to ameliorate the heterogeneous variability in tumor area measurements over time. Mixed models estimate corrected variance estimates in the presence of correlated measurements taken in the same mouse (e.g., left and right flank) and for repeated measures on the same tumor over time. The Akaike information criterion was used to select among competing correlation structures. Standard residual analysis and goodness-of-fit statistics were evaluated.
At least 6 mice per treatment group were used. Guidance for sample size was in these experiments were supported under VA Merit Grant 5101BX000196-04. Sample size estimation was based on much simpler regression models requiring larger sample sizes than what would be expected compared to the mixed model approach used in our analyzes in this paper. For the comparisons of tumor size between treatments we chose all experiments to have at least 80% power to detect standardized effect sizes of 2 or greater; such effects sizes in our preliminary data ranges from 2.5 to 4.9. Especially considering the gain in information from longitudinal measurements, samples sizes for these experiments provide sufficient power, a priori, to detect biologically meaningful differences among treatments. All tests of statistical significance were two-sided. Findings were considered statistically significant if p < 0.05.
3. Results
3.1. MDM2i Inhibits Drug-Induced DNA Damage in Melanoma Cells
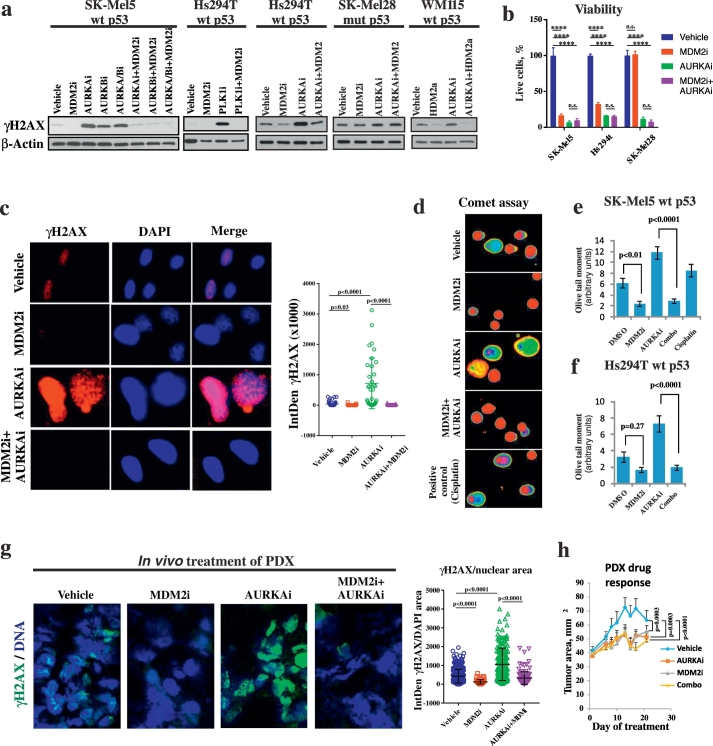
We have shown previously that the small molecule inhibitor of mitotic kinase AURKA, alisertib (AURKAi), induces DNA damage in melanoma cells (Liu et al., 2013). Here we treated cells with AURKAi in combination with the MDM2 antagonist nutlin-3a (MDM2i) and analyzed the induction of DNA damage marker γH2AX. Surprisingly, we found that addition of MDM2i protected melanoma cells from AURKAi-induced DNA damage (Fig.1a). Furthermore, treatment with MDM2 inhibitor alone also reduced the baseline DNA damage in melanoma cells as compared to vehicle treated cells. We also tested MDM2i in combination with other inhibitors of mitotic kinases such as small molecules targeting Aurora kinase A, B, C, Polo Like kinase (PLK1) and aurora kinase B only. Similar to finding with AURKAi, addition of MDM2i abrogated γH2AX induction by these mitotic inhibitors. A DNA-protective effect of MDM2i was observed only in cells with wt TP53 gene (SK-MEL5 and Hs294T), but not in cells with mutated TP53 (SK-Mel28), suggesting that it relies on functional p53. Similar findings were obtained using an investigational antagonist of HDM2, the human homolog of murine MDM2, idasanutlin (RG7388), which is currently being tested in several clinical trials for the treatment of various malignancies (Fig. 1a, right panel). Interestingly, despite inhibiting drug-induced DNA damage, MDM2i did not rescue cells from viability loss associated with drug treatment, indicating that inhibition of DNA damage does not interfere strongly with anti-tumor activity of tested mitotic inhibitors (Fig. 1b). These results are in agreement with clinical observations of melanomas responding poorly to the traditional genotoxic treatments such as radiation and chemotherapy.
Fig. 1.
MDM2i counteracts DNA damage induced by mitotic inhibitors in p53WT cells. (a) Western Blot analysis of DNA damage marker γH2AX in the indicated melanoma cells after 3 days of treatment with 10 μM nutlin-3a (MDM2i) or 1 μM idasanutlin (HDM2i) in combination with 1 μM alisertib (AURKAi), 1 μM borasertib (AURKBi), 5 μM danusertib (AURKA/Bi) or 5 nM volasertib (PLK1i). (b) Count of viable cells from indicated cell lines after 5 days treatment with vehicle, 1 μM AURKAi, 10 μM MDM2i, or the combination of both drugs. Mean ± SD from 2 biological replicates and results of ANOVA with Tukey's comparison are shown. (c) Immunofluorescent staining with γH2AX-specific antibody in SK-Mel5 cells treated as in (a). Images were quantified using ImageJ and DNA damage was measured as IntDen of γH2AX within the nucleus area. Five fields were quantified for each condition for the total of 56 cells in vehicle, 33 in MDM2i, 34 cells in AURKAi, and 23 in AURKAi + MDM2i-treated group. Raw values from individual cells, mean, SEM and Kruskal Wallis test results are shown. (d–f) Single cell electrophoresis (comet assay) in SK-Mel5 cells. Representative pseudo-colored images of fluorescent-labeled DNA after analysis using Comet Score software (d) and quantitative results (mean olive tail moment in individual cells ± SEM, e-f) are shown. About 200 cells were counted in each treatment group The Mann-Whitney test was used for statistical evaluation. Treatment with cisplatin (10 μM, 48 h) was used as a positive control. (g) Patient-derived human melanoma tissue was implanted SC into nude mice. When tumors reached the size of ~ 100 mm3 mice received daily treatments with nutlin-3a (200 mg/kg), MLN8237 (30 mg/kg), a combination of both, or vehicle control for 3 weeks. Representative images after IF staining for γH2AX. The amount of γH2AX per nuclear area was quantified using ImageJ across several fields. In total, 368 cells in vehicle, 59 in MDM2i, 176 cells in AURKAi, and 131 in AURKAi + MDM2i-treated group were quantified. Kruskal Wallis statistics are shown. (h) Tumor progression over time for tumors analyzed in (g). Tumor area (length x width) was measured every 3–4 days. Average tumor area ± SD is shown. N = 8 in combination groups and n = 10 in other treatment groups. Mixed-effects statistical model was used to assess group differences in tumor area over days. All in vitro experiments were repeated at least 3 times with consistent results and results from a representative experiment are shown. ****p < 0.0001, n.s. – not significant (p > 0.05).
Immunofluorescent staining revealed a classical punctate pattern of γH2AX expression in AURKAi-treated cells, known as γH2AX foci, which mark areas of DNA double stranded breaks (DSB). However, no foci induction was detected if cells were treated with a combination of AURKAi and MDM2i (Fig. 1c). Similar results were obtained when imaging flow cytometry was used to evaluate γH2AX foci. Data from 3 independent imaging cytometry experiments were analyzed using ANOVA after log transformation and blocking for inter-experiment variability, and the decrease in the numbers of DNA damage foci in AURKAi + MDM2i treatment groups compared to treatment with AURKAi alone was found to be statistically significant with p = 0.002 (Fig. S1). Furthermore, we also performed comet assay where single cells are subjected to gel electrophoresis which separates broken pieces of damaged DNA (comet tail) from intact DNA (comet head). The length and density of comet tails reflect the level of DSB in cells and are collectively quantified as olive tail moments. The appearance of comets (Fig. 1d) and the olive tail moments were significantly lower after combined MDM2 and AURKAi treatment, compared to the treatment with AURKAi alone in two tested melanoma cell lines SK-Mel5 and Hs294T (Mann-Whitney test p < 0.0001, Fig. 1e, f). These results show that AURKAi treatment increases the actual level of DNA breaks in cell nuclei and that addition of MDM2i blocks DNA breakage.
To confirm that DNA-protective effect of MDM2i is evident in vivo, and not an artifact of cell culture, we established melanoma patient-derived xenograft (PDX) tumors in nude mice. PDX tumors originated from a surgically resected patient tumor that had TP53 wt, BRAFV600E, and progressing on BRAF inhibitor therapy. PDX-bearing mice were treated with AURKAi and/or MDM2i or vehicle control. Similar to the results obtained in vitro, tumors from mice co-treated with AURKAi and MDM2i were protected from DNA damage, compared to single agent AURKAi treatment (Fig. 1g). These data suggest that p53 activation protects melanoma cells from DNA damage associated with mitotic inhibition.
MDM2i protects DNA from mitotic inhibitor-induced damage by preventing endoreduplication.
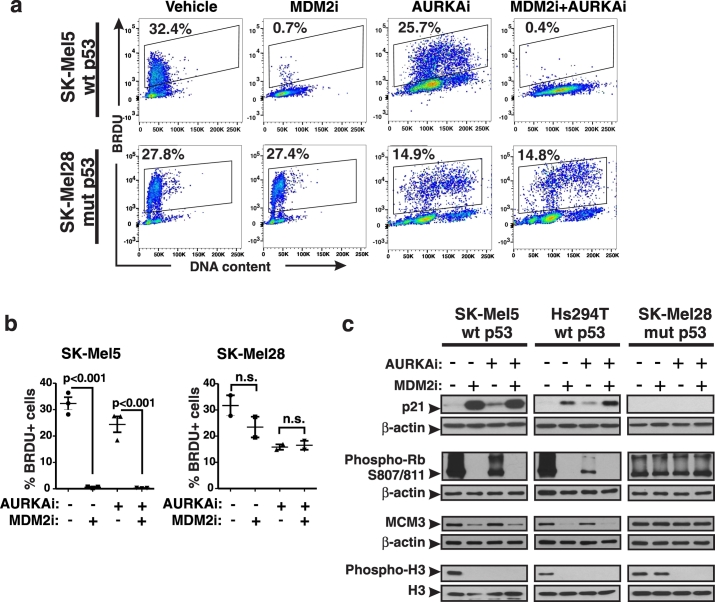
The p53 protein is a master regulator of cell fate in response to DNA damage. Depending on the circumstances of damage and cell context, p53 can either induce cell cycle arrest or apoptosis. Since no significant apoptosis was detected with the experimental settings applied in this study, we investigated whether MDM2i protects DNA by altering the cell cycle. Cell cycle distribution was evaluated with BRDU incorporation assay, which showed that DNA replication was nearly completely abrogated in MDM2i-treated cells with functional p53 (Fig. 2 a, b). We also studied expression of p21, which is a key effector of p53-induced cell cycle arrest. As expected, p21 levels were increased by MDM2i in cells with wt p53. p21 inhibits cyclin-dependent kinases (CDKs) that phosphorylate and inactivate RB to allow progression from G1 to S phase of cell cycle (Abbas and Dutta, 2009). We found that inhibitory phosphorylation of RB was prominently reduced by MDM2i (Fig. 2c). Furthermore, MDM2i hindered expression of an S phase-specific protein MCM3, which is one of the subunits of DNA helicase involved in DNA replication. These data suggest that MDM2i arrests cell cycle during the G1-S transition. We also studied M phase of cell cycle using phosphorylated histone H3 as a marker of cells undergoing mitosis. As expected from a mitotic inhibitor, AURKAi inhibited H3 phosphorylation (Fig. 2c). However, replication was not fully inhibited in AURKAi-treated cells based on their ability to incorporate BRDU and the presence of inactive phosphorylated RB. In addition, there was an increase in the number of polyploid cells with > 4n DNA content in AURKAi-treated samples. The degree of polyploidy after AURKAi treatment increased overtime (Fig. S2a). These data show that cells treated with AURKAi failed to complete mitosis but re-replicated their DNA resulting in polyploidy. Similarly, polyploidy was induced in cells treated with other inhibitors of mitotic kinases, such as Polo-like kinase inhibitor (PLK1i) and an inhibitor of AURKA and AURKB (AURKA/Bi) (Fig. S2b). Notably, addition of MDM2i blocked DNA re-replication and polyploidy in response to mitotic inhibition in cells expressing functional p53 (wt p53) (Fig. 2a, b).
Fig. 2.
MDM2i prevents induction of polyploidy by AURKAi. (a) Proliferation analysis using the BRDU incorporation assay. Cells were treated with 10 μM nutlin-3a (MDM2i) ± 1 μM alisertib (AURKAi) for 3 days. Gates show percent of BRDU-positive cells in relation to DNA content. Numbers next to gates indicate the percentage of cells. (b) Quantified results of the BRDU analysis shown in (a). Raw data, means, SEM, and the results of ANOVA with Tukey's comparison are indicated. (c) Western blot analysis of indicated cell cycle regulators in cells treated as in (a). Experiments were repeated at least 3 times with consistent results.
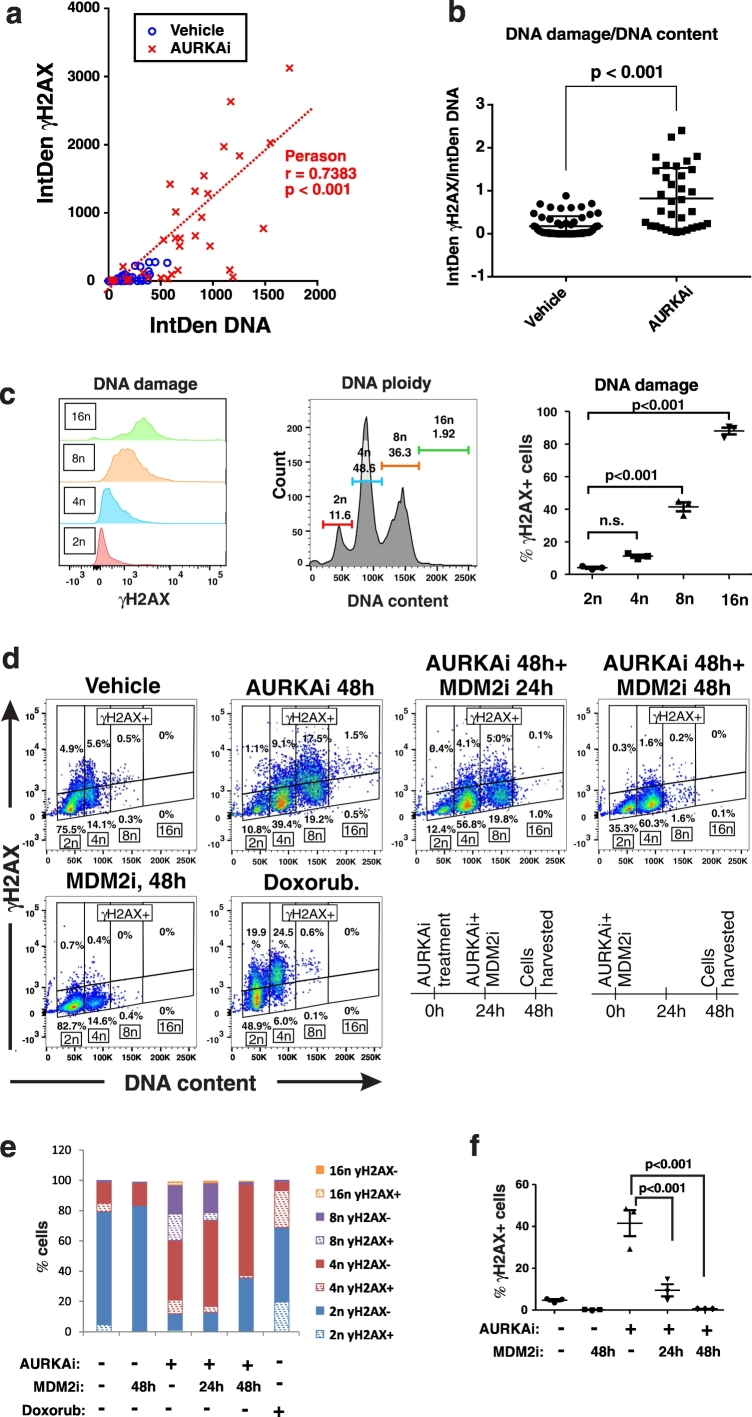
Based on these findings we hypothesized that induction of polyploidy is the mechanism whereby mitotic inhibition causes DNA damage in melanoma cells and that p53 activation abrogates this damage by preventing polyploidization. To test this hypothesis, we quantified γH2AX levels and DNA levels in individual cells after immunofluorescent staining. While vehicle treated cells had consistently low levels of DNA and γH2AX, many AURKAi-treated cells exhibited elevated DNA content (Fig.3a). Notably, cells with high DNA content showed high expression of γH2AX, and Pearson analysis showed significant correlation between DNA damage and DNA content in cells (r = 0.74, p < 0.001). We next tested whether increased DNA damage was simply proportional to the increasing DNA content in cells with cells containing large amount of DNA having higher probability of damage to occur compare to cells with small DNA content. However, after adjusting the levels of DNA damage by the total DNA content, AURKAi treated cells still exhibited over 4-fold higher DNA damage compare to vehicle-treated cells per same amount of DNA (Fig.3b). These data suggest that AURKAi-treatment leads to DNA damage in melanoma cells and this damage is associated with acquisition of polyploidy. We next performed flow cytometric analysis to accurately access cell ploidy (DNA content) while also quantifying DNA damage (γH2AX) in cell based assay. We found a positive association of cell ploidy and DNA damage in AURKAi-treated cells, where polyploid cells with DNA content of 8n and greater had higher levels of γH2AX expression and higher rates of γH2AX + cells compared to 2n and 4n cells (Fig. 2d). Gating of γH2AX positive and negative cells based on DNA content revealed that the majority of cells with DNA damage are polyploid (DNA content of 8n) (Fig. 3d, AURKAi 48 h). Furthermore, the percent of γH2AX cells increased with the increase of cell ploidy after AURKAi-treatment (Fig. 3e). As a positive control, cells were treated with chemotherapeutic drug doxorubicin which induced DNA damage without inducing of polyploidy. Neither polyploidy nor DNA damage were induced if AURKAi was administered in combination with MDM2i for 48 h (Fig. 3d, f, AURKAi 48 h + MDM2i 48 h). In contrast, only partial protection from polyploidy and DNA damage was detected if MDM2i was administered with a 24 h delay (Fig. 2d, f, AURKAI 48 h + MDM2 24 h). These data suggest that MDM2i prevents polyploidization induced by AURKAi, thus reducing DNA damage which is associated with polyploidy, however it has limited ability to reverse polyploidy and repair damage once it has occurred.
Fig. 3.
AURKAi-induced DNA damage is associated with polyploidy. (a) The scatter plot shows distribution of individual vehicle or AURKAi (1 μM alisertib, 3 days) treated SK-Mel5 cells based on their DNA content (x-axis) and DNA damage (y-axis). These parameters were derived from the ImageJ analysis of IF staining shown on Fig. 1b. DNA content is measured as integrated density of DAPI staining (IntDen DNA), and DNA damage is measured by IntDen of γH2AX staining within the same nucleus. Five fields were quantified for each condition for the total of 56 cells in vehicle and 34 cells in the AURKAi-treated group. The results of Pearson correlation test are shown. (b) DNA damage adjusted for DNA content in vehicle and AURKAi-treated SK-Mel5 cells shown in (a). Individual values, means, SEM, and Mann-Whitney test results are shown. (c) Expression of DNA damage marker γH2AX in SK-Mel5 cells with 2n, 4n, 8n, and > 8n ploidy after treatment with 1 μM alisertib. Over 10,000 cells were analyzed by flow cytometry. Values from 3 independent experiments are shown as a dot plot on the right. Statistical comparison was performed using ANOVA with Dunnett's post-test (d) Flow cytometry analysis of DNA damage and DNA ploidy in SK-Mel5 cells treated with vehicle or 1 μM alisertib (AURKAi) for 2 days. Groups of AURKAi-treated were also co-treated with 10 μM of nutlin-3a (MDM2i) for the full duration of 2 day treatment (AURKAi 48 h + MDM2 48 h) or only for the last 24 h of treatment (AURKAi 48 h + MDM2 24 h). Doxorubicin treatment (20 nM, 2 days) was used as positive control for DNA damage induction. A schematic depicting treatment schedule is shown at the bottom. (e) Distribution of SK-Mel-5 cells in the gates set based on cell ploidy (2n, 4n, 8n, 16n) and γH2AX expression (γH2AX- or γH2AX +) as shown in (d). (f)SK-Mel5 cells were treated as described in (d) and analyzed for γH2AX levels by flow cytometry. Dot plot shows data from 3 biological replicates, as well as mean ± SD and ANOVA with Tukey's post-test results.
3.2. Polyploid Cells Exhibit High Levels of Replication Stress which can Be Blocked by MDM2i
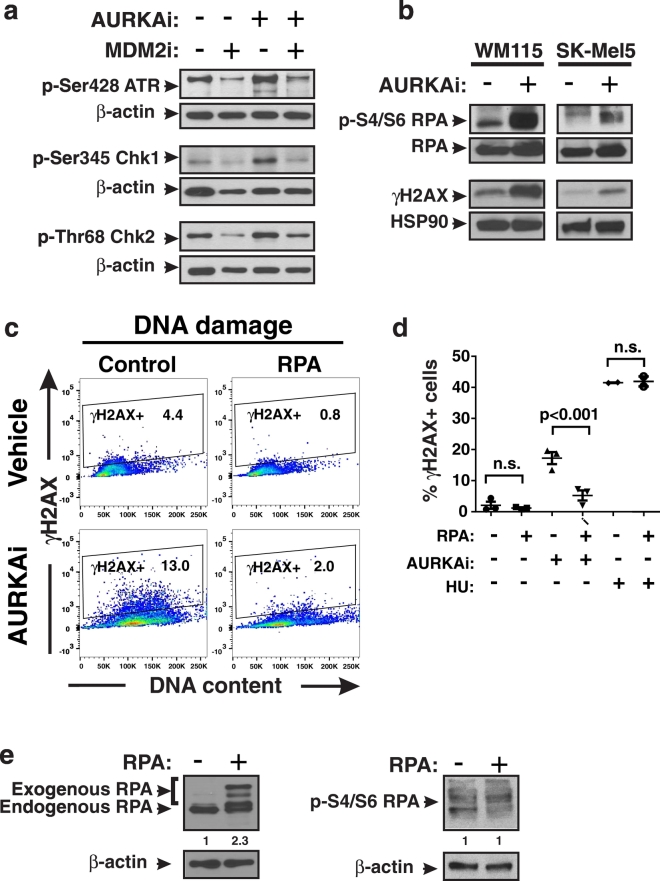
We next sought to examine the mechanism whereby DNA damage occurs in polyploid cells. Since DNA remained intact when DNA re-replication was blocked by MDM2i, we hypothesized that replication stress was the source of DNA damage in AURKAi-treated polyploid cells. We evaluated several kinases implicated in replication stress response cascade, including ATM, Chk1 and Chk2, and found that they were activated by phosphorylation in AURKAi-treated cells. Addition of MDM2i blocked this activation (Fig. 4a). Moreover, we detected increased phosphorylation of the replication protein A (RPA) which is a marker of replication stress (Fig. 4b). RPA directly binds and stabilizes single stranded DNA and is phosphorylated installed replication forks as a part of replication stress response. These findings indicate that polyploid cells generated by treatment with mitotic inhibitors are prone to replication stress. We investigated several potential mechanisms of replication stalling that have been previously reported to occur in cancer cells. These included 1) exhaustion of nucleotides necessary for DNA synthesis (Bester et al., 2011); 2) collision of replication machinery with over-active transcription (Kotsantis et al., 2016); and 3) exhaustion of RPA needed for stabilization of single stranded DNA in replication forks (Toledo et al., 2013). We ruled out mechanisms 1 and 2 since there was no abrogation of AURKAi-induced DNA damage detected after addition of extra dNTPs (Aird et al., 2013) or RNA transcription inhibitor cordycepin (Jones et al., 2013) to the cells' culture media (Fig. S3a, b). In contrast, introduction of additional copies of the RPA gene into cells via transfection rescued them from AURKAi-induced DNA damage (ANOVA: p < 0.001, Fig. 4c). No DNA protection was seen in RPA-expressing cells when hydroxyurea (HU) was used to induce replication stress (Fig. 4d). The level and phosphorylation of RPA after transfection is shown on Fig. 4e. These data suggest that the endogenous level of RPA is not sufficient to support replication of polyploid cells with large DNA content, likely due the generation of more replication forks in polypoid compared to diploid cells. In summary, our findings demonstrate that pharmacological p53 activation by MDM2i inhibits replication stress in mitosis-deficient cells by blocking DNA re-replication.
Fig. 4.
Polyploidy is associated with high levels of replication stress. (a) Phospho-protein analysis of indicated replication stress response kinases in SK-Mel5 cells treated with 10 μM nutlin-3a (MDM2i) ± 1 μM alisertib (AURKAi) for 3 days. (b) Western blot analysis of replication stress marker phospho-S4/S6 RPA after 3 days of treatment with 1 μM alisertib. Experiments were repeated at least 3 times with consistent results. (c) Comparison of γH2AX induction in control Hs294T cells and Hs294T cells overexpressing RPA after treatment with 1 μM alisertib (AURKAi) for 3 days. Gates show percent of γH2AX + cells. (d) Quantification of the percentages of γH2AX + cells shown in panel C from 3 biological replicates in vector control and RPA transfected Hs294T cells. Cells were treated with 1 μM alisertib (AURKAi), 3 mM hydroxyurea (HU) or vehicle for 3 days. Different conditions were compared using ANOVA with Tukey's post-test. (E) Analysis of RPA expression and phosphorylation after stable transection with RPA in Hs294T cells. Loading control-normalized relative densitometry values shown below the blots were obtained using ImageJ.
3.3. MDM2i Counteracts DNA Damage Induced by Chemotherapy
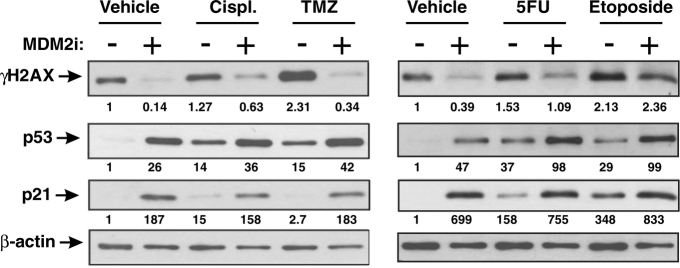
Our data demonstrate that mitotic inhibition causes DNA damage via replication stress and this can be blocked by MDM2i. We next investigated whether MDM2i can counteract DNA damage induced by other anti-cancer drugs that are known to induce replicative stress (Dobbelstein and Sorensen, 2015). Specifically we used chemotherapeutic drugs that hinder replicative forks by crosslinking nucleotide bases between two DNA strands (temozolomide and cisplatin), or by reducing pool of available dNTPs (5-FU) (Dobbelstein and Sorensen, 2015). We also used a topoisomerase inhibitor, etoposide, that inhibits the ability of topoisomerase to re-ligate DNA after unwinding during replication (Montecucco et al., 2015). It has been reported to form a physical obstacle to ongoing replication forks (Dobbelstein and Sorensen, 2015). Similar to our findings with mitotic inhibitors, MDM2i abrogated DNA damage induced by cisplatin, temozolomide and 5-FU in melanoma cells, but not by etoposide, suggesting drug-dependent activity (Fig. 5). Collectively our data suggest that MDM2i can counteract DNA damage induced by a variety of anti-cancer drugs that promote replicative stress.
Fig. 5.
MDM2i protects DNA from damage induced by traditional chemotherapeutics. Western blot analysis of lysates of Hs294T cells treated with 1.5 μM cisplatin (Cispl.), 100 μM temozolomide (TMZ), 10 μM 5-fluorouracil (5FU), or 1 μM etoposide ± 10 μM nutlin-3a (MDM2i) for 3 days. Densitometry analysis of the blot images was performed in ImageJ. The numerical data below the images represents the densitometry values for all tested proteins that were divided by the values in loading control to adjust for variation in sample loading and normalized to vehicle-treated baseline samples. The experiment was repeated at least 3 times with consistent results.
3.4. P21 is Essential for DNA Protective Effect of MDM2i
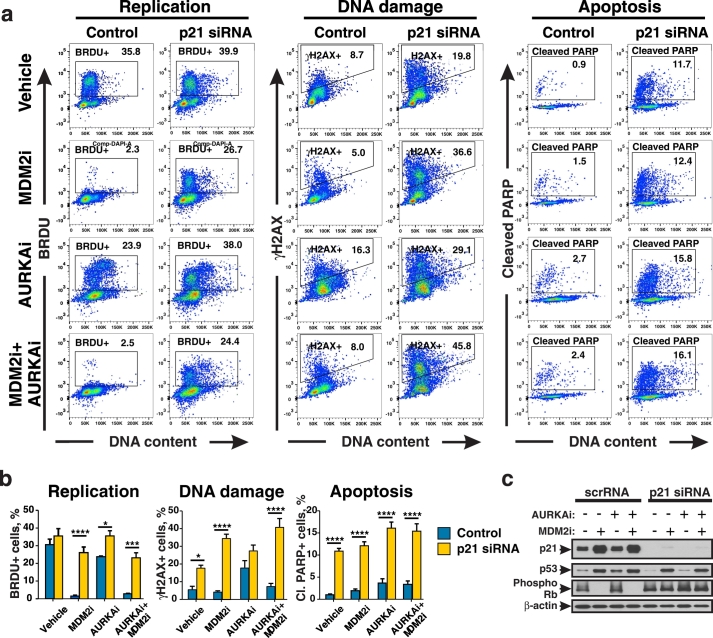
We next tested whether blocking p53-induced cell cycle arrest by targeting p21 can abrogate the DNA-protective effect of MDM2i. Expression of γH2AX (DNA damage marker), BRDU incorporation (replication marker), and cleavage of PARP (apoptosis marker), were compared in cells transfected with either p21-specific siRNA (p21KD) or non-targeting control siRNA (Fig. 6a, b). The percentages of BRDU + replicating cells after MDM2i treatment were significantly higher in cells expressing p21 siRNA compared to control non-targeting siRNA cells (ANOVA with Bonferroni, p < 0.001, n = 3). This demonstrates that p21-deficient cells failed to block replication in response to MDM2i. Consequently treatment with MDM2i did not protect p21KD cells from AURKAi-induced DNA damage based on a significantly higher percent of γH2AX + cells after MDM2i and AURKAi treatment in p21 siRNA samples compared to non-targeting siRNA (ANOVA with Bonferroni, p < 0.0001). Notably, p21-deficiency promoted PARP cleavage in melanoma cells indicative of apoptosis induction. Specifically, about 15% of p21 deficient cells showed PARP cleavage after combined MDM2 and AURKAi treatment compare to 3% of cells in control group (ANOVA with Bonferroni, p < 0.0001). While inactivation of p21 did not interfere with p53 activation by MDM2i, it abrogated activation of cell cycle arrest mediator RB (Fig. 6c). The efficiency of p21 knockdown was verified by Western blot (Fig. 6c). Similar data were obtained when isogenic HCT116 cells with and without CDKN1A (p21 gene) knockout were used (Fig. S4a). We found that p21 −/− cells had greater DNA damage after AURKAi treatment compared to parental wild type p21 cells. Furthermore, p21 −/− cells responded to MDM2i and AURKAi co-treatment with the induction of apoptosis rather than cell cycle arrest. Similarly to inhibition of AURKA, p21 knockout cells exhibited elevated γH2AX levels after treatment with an inhibitor of another mitotic kinase, PLK1, as well as with chemotherapeutic drugs cisplatin and temozolomide, as compared to p21 expressing cells (Fig. S4b).
Fig. 6.
P21 deficiency sensitizes cells to MDM2i and AURKAi therapy. (a) Hs294T cells were transfected with control siRNA or p21-specific siRNA and treated with 10 μM nutlin-3a (MDM2i) ± 1 μM alisertib (AURKAi) for 3 days and pulsed with BRDU for 2 h. Representative histograms of flow cytometric analysis of BRDU, γH2AX and cleaved PARP are shown. (b) Quantitation from 3 independent experiments performed as described in (a). Indicated groups were compared using ANOVA with Bonferroni test. (c) Western blot analysis of lysates from Hs294T cells treated as described in (a). *p < 0.05, ***p < 0.001, ****p ≤ 0.0001. All experiments were repeated at least 3 times with consistent results.
3.5. Targeting p21 Improves Anti-Tumor Activity of MDM2i and Mitotic Inhibition
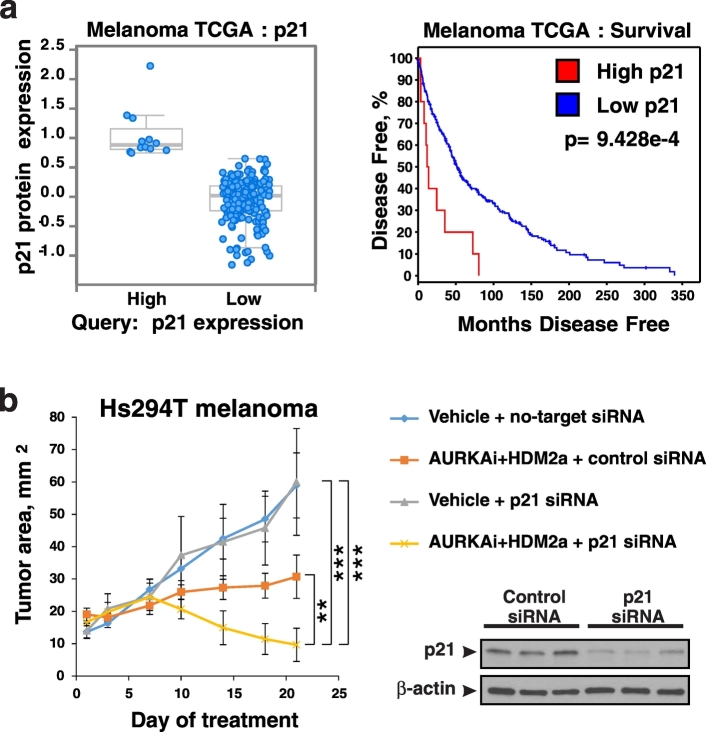
Based on finding described above we hypothesized that therapeutic targeting of p21 may enhance anti-tumor activity of MDM2 antagonists. In order to determine if p21 is a suitable therapeutic target in melanoma we analyzed the TCGA dataset of 479 melanoma cases which were stratified into 2 groups based on p21 protein expression as shown on Fig. 7a, right panel using cBio portal analysis tools (Cerami et al., 2012; Gao et al., 2013). Patients whose tumors expressed high levels of p21 protein had worse disease-free survival in comparison to patients with low p21 (cBio portal analysis: p = 9.428*10− 4, Fig. 7a, left panel). This suggests that targeting p21 is unlikely to worsen patient outcome. We next generated mice-bearing Hs294T human melanoma xenografts and treated them with AURKAi alisertib and MDM2 antagonist idasanutlin; both agents are in clinical development. Tumors were injected with in vivo transfection reagent pre-mixed with either p21 siRNA or non-targeting siRNA. We found that tumors co-treated with p21 RNAi were more sensitive to AURKAi + MDM2i therapy compared to the control RNAi group (linear mixed model, p < 0.01). In fact, tumor regression was seen in mice that received p21 siRNA combined with drug treatment in contrast to tumor stabilization in control mice co-treated with non-specific siRNA. These findings show that targeting p21 in tumor cells can improve therapeutic response of melanoma tumors to MDM2i combined with mitotic antagonists.
Fig. 7.
P21 as a potential therapeutic target in melanoma. (a) Analysis of the TCGA dataset of 479 melanoma specimens using cBio portal. Tumors were sorted into high and low expression of p21, based on p21 protein expression by RPPA analysis (left panel). Disease-free survival was compared between these groups (right panel). (b) Nude mice bearing Hs294T melanoma xenograft tumors were treated QD with 30 mg/kg alisertib (AURKAi) and 150 mg/kg idasanutlin (HDM2a). Animals also received injections of control siRNA (no-target siRNA) or p21 siRNA mixed with in vivo siRNA delivery reagent JetPei directly into the tumor twice a week. Tumor area (length x width) was measured every 3–4 days. Average tumor area ± SD is shown. N = 6 in vehicle groups and n = 7 in AURKAi and HDM2a treatment groups. Mixed-effects statistical model was used to assess group differences in tumor area over days. Expression of p21 in tumors from vehicle-treated mice injected with control or p21 siRNA was evaluated in whole tumor lysates by western blot with human p21-specific antibodies (right panel).
4. Discussion
The p53 protein is the key tumor suppressor in humans and is considered to be inactivated in virtually all cancer cases. Reactivation of p53 is a promising strategy that is actively pursued by pharmaceutical companies. A number of small molecules and peptide drugs that activate p53 by disrupting its interaction with ubiquitin ligase MDM2 are currently in clinical development, including idasanutlin, CGM097, HDM201, AMG232, DS3032b, and APG-115. Since DNA damaging chemotherapy remains standard of care for many malignancies, clinical trials often test MDM2 inhibitors in combination with DNA damaging drugs. This could be a concern since MDM2 antagonism can alleviate drug-induced DNA damage based on data above. Furthermore, we show that not only traditional chemotherapy, but also target-specific anti-cancer drugs, such as inhibitors of mitotic kinases, can induce DNA damage. Notably, MDM2 antagonists abrogate targeted therapy-induced damage as well. If DNA damage is important for anti-tumor activity of a given drug, than combining it with MDM2 antagonist may negatively impact overall treatment outcome. Therefore this study has important implications for clinical development of MDM2 antagonists.
Here we also provide an insight on mechanism of action of small molecule inhibitors of mitotic kinases, such as Aurora A and B (alisertib, danusertib, barasetrib) and Polo-like kinases (volasertib), that are currently in clinical development (see (Penna et al., 2017) for review). We and others have previously reported that specific targeting of these mitotic kinases causes DNA damage in cancer cells (Driscoll et al., 2014; Liu et al., 2013). In addition, induction of DNA damage markers was seen in cells treated with agents targeting mitotic microtubules, such as kinesin 5 inhibitors (Orth et al., 2012) and microtubule poisons (Colin et al., 2015). However, the mechanism of DNA damage induction by mitotic inhibitors has remained unclear.
It has been established that inhibition of mitosis can lead to polyploidy (Elhajouji et al., 1998; Tsuiki et al., 2001). When replicated cells with doubled DNA content (G2 phase of cell cycle) cannot progress through mitosis, they often can bypass it (mitotic slippage) and initiate another round of DNA replication resulting in polyploidy (Storchova and Pellman, 2004). We discovered here that re-replication of polyploid DNA causes replication stress which leads to DNA damage. One of the key mechanisms of replication stress is associated with slowing and stalling of replication forks which occurs when cells run out of essential replication resources such as nucleotides, replication machinery components, and histones that package the replicated DNA (Zeman and Cimprich, 2014). In contrast to normal diploid cells, polyploid cells must replicate a larger amount of DNA and thus may experience a deficit of replication resources. Indeed, we found that increasing levels of replication protein A (RPA), which stabilizes single-stranded DNA during replication, at least in part, rescues polyploid cells from DNA damage. It is plausible that replication of polyploid genome generates more single stranded DNA than could be stabilized by endogenous RPA, which leads to fork stalling and replicative stress. This conclusion is in agreement with prior reports that global exhaustion of RPA causes replication catastrophe and DNA damage (Glanzer et al., 2014; Toledo et al., 2014). Of note, both genomic instability and polyploidy are common characteristics of cancer cells (Coward and Harding, 2014; Hanahan and Weinberg, 2011; Storchova and Pellman, 2004). However, the mechanisms driving genomic instability, as well as the benefits of being in a polyploid state, are still not fully understood. Our results demonstrate a potential link between these two phenomena, showing that polyploidization of cancer cells can facilitate DNA damage and genomic instability, which may contribute to the extraordinary capacity of cancers to adapt to a changing microenvironment and acquire resistance to therapy.
We found that MDM2 antagonists can protect DNA in cancer cells treated with inhibitors of mitotic kinases AURKA, AURKB, all Aurora kinases, and PLK1. Investigation of the mechanism responsible for DNA-protective effect of MDM2i showed that p53 accumulation in MDM2i-treated melanoma cells blocked DNA replication via the induction of p21 and RB. The p21 protein induces G1 cell cycle arrest by binding and inactivating CDK/cyclin complexes. This results in hypophosphorylation and activation of RB, which, in turn, sequesters the transcription factor E2F1 that regulates expression of genes involved in replication (Sherr and Roberts, 1999). Activation of p21 in cells treated mitotic inhibitor stopped DNA re-replication (endoreduplication) after mitotic slippage thus preventing polyploidy. Hence, we concluded that MDM2i protects DNA in mitotic inhibitor-treated cells by blocking replication stress associated with polyploidy.
Despite its DNA protective properties, MDM2i did not limit anti-tumor activity of AURKAi in our melanoma model. In fact, melanoma cell viability was further reduced after treatment with combined AURKAi and MDM2i as we showed previously (Vilgelm et al., 2015). Furthermore, our previous study showed an improved anti-tumor effect of combined treatment with MDM2i and AURKAi in mice bearing melanoma patient-derived xenografts as compared to the activity of the respective single agents. This was due to the induction of delayed caspase-independent cell death and increase of anti-tumor immune response (Vilgelm et al., 2015). Similarly, a study using leukemia cells showed that the MDM2i, nutlin-3a, inhibited AURKAi-induced endoreduplication which was associated with improved cell killing (Kojima et al., 2008). However, other groups have reported that inhibition of endoreduplication by MDM2i made cells resistant to the drugs targeting mitotic kinases AURKA, AURK, and PLK1 (Cheok et al., 2010; Sur et al., 2009). Moreover, several previous publications indicated that MDM2i can protect cells with functional p53 from the toxicity of the microtubule poison taxol (Carvajal et al., 2005; Shen et al., 2012; Tokalov and Abolmaali, 2010). Consequently it has been suggested to use MDM2i in combination of mitotic inhibitors for treatment of tumors with mutated p53 in order to protect normal cells from drug-induced genotoxicity. However this approach is unlikely to have clinical utility since significant toxicities to normal hematopoetic and GI tissues were seen in clinical trials of MDM2i. Another critical point is that cancers with mutated or inactivated p53 tend to be resistant to genotoxic therapy. This is not surprising since p53 is a key inducer of apoptosis in response to DNA damage, while other p53 family members, p63 and p73, can only partially compensate for this activity (Vilgelm et al., 2008). Thus patients with p53-deficient tumors are not likely to benefit from combinations of anti-mitotic therapy and MDM2i.
Here we sought a strategy to improve outcomes of MDM2i therapy in patients with wild type p53 tumors. We found that p53-dependent cell cycle arrest was compromised in p21-deficient cells and they were unable to arrest after mitotic slippage. Furthermore, p21-deficient cells accumulated more DNA damage upon AURKAi-treatment. Notably, MDM2i co-treatment did not protect DNA in p21-deficient cells. Transient knockdown of p21 was sufficient to induce apoptosis even in the absence of any drugs. Moreover, cells with stable p21 knockout exhibited greater apoptosis induction after MDM2i and AURKAi treatment as compared to cells with WT p21. This suggests that p21 is important for cancer cell survival and resistance to DNA damaging therapy. Consistent with this, p21-RNAi tumors were more sensitive to the AURKAi and MDM2i combination, compared to p21-expressing tumors. These findings can explain cell line-dependent outcomes of MDMi treatment where cell fate is determined by the balance between cytostatic and cytotoxic activity of p53. Specifically, cell fate may depend on cell-type specific basal expression of pro-apoptotic and cell cycle arrest-inducing targets of p53, balance of pro-death and pro-survival mitochondrial proteins, the pre-existing level of replication stress and DNA damage, and other factors (Blagosklonny, 2007). We show that p21 promotes cell cycle arrest and survival of melanoma cell in response to DNA damage, while without p21, tumor cell fate is shifted towards cell death.
These findings suggest that caution should be used when combining MDM2i with DNA-damaging therapy. Perhaps selecting patients with tumors that have low basal p21 expression may enhance the efficacy of such combinations. Alternatively, co-targeting p21 in tumor cells combined with systemic inhibition of MDM2 may be a promising approach. TP53 is a key tumor suppressor gene, thus targeting its downstream mediator p21 to treat cancer may seem counterintuitive. However animal models show that p21 is not essential for p53-mediated tumor suppression. Only about 40% of p21 knockout mice develop tumors, and this tends to occur later in life, as compared to p53 knockout mice that develop tumors within first 6 months of age (Donehower et al., 1992; Martin-Caballero et al., 2001). Moreover, p21-deficient mice are protected from radiation-induced tumorigenesis compared to wild type mice (Martin-Caballero et al., 2001), suggesting that p21 may, perhaps, facilitate survival of cells with mutations induced by radiation. Analysis of the TCGA data base of human melanomas also indicated that downregulation of p21 is unlikely to be tumor-promoting. In fact, we show that high p21 protein expression is associated with poor survival. This suggests that p21 can be a reasonable target for cancer therapy. There are several drugs that have been reported to inhibit p21, including sorafenib and UC2288 (Inoue et al., 2011; Wettersten et al., 2013). However one potential pitfall of systemic p21 targeting is that it will likely sensitize not only tumors but also normal cells to p53-activating therapy. Considering severe GI and hematopoetic toxicity of MDM2 antagonists reported by clinical studies (Andreeff et al., 2016; Vu et al., 2013; Yee et al., 2014), further sensitization of non-malignant cells is a major concern. Here we tested local delivery of p21-targeted therapy in preclinical melanoma model. We found that polymer-based delivery of p21 siRNA directly into tumors sensitized them to treatment with MDM2 antagonist idasanutlin and mitotic inhibitor alisertib. With the rapid development of alternative cancer treatment strategies like targeted drug delivery and therapeutic RNAi (Barata et al., 2016; Harris and Chiu, 2017), clinical translation of tumor-specific p21 targeting to complement MDM2i therapy may be feasible.
In summary, our findings demonstrate that pharmacological p53 activation with MDM2i inhibits replication stress in mitosis-deficient cells by blocking DNA endoreduplication. This study has important implications for clinical development of emerging cancer targeted therapeutics like MDM2 antagonists and mitotic kinase inhibitors.
Acknowledgments
Acknowledgement
The authors would like to thank Alexia N. Melo, Christine M. Eischen, Huzefa M. Dungrawala, and David Cortez for their valuable advice. We also thank Rebecca Shattuck-Brandt for help with manuscript preparation.
Funding sources
This work was supported by grants from the Department of Veterans Affairs (5101BX000196-04), NIH (CA116021 (AR), CA116021-S1 (AR), K23 CA204726 (DBJ), and GM084333 (JNJ)), Senior Research Career Scientist Award to AR, Harry J. Lloyd Charitable Trust award (019720-001, AEV), and Vanderbilt Clinical Oncology Career Development program (AEV) funded through NIH K12 training grant (CA90625). Support for Core Facilities utilized in this study was provided by Vanderbilt Ingram Cancer Center (P30 CA68485). Funding agencies played no role in the design, execution, interpretation or writing of this manuscript.
Conflicts of Interest
Brian Higgins is an employee of Roche. Douglas Johnson serves on scientific advisory boards to BMS, Genoptix, Incyte, and Merck. Other authors have no financial and non-financial competing interests to report.
Author Contribution
Conceptualization, A.E.V. and A.R.; Methodology, A.E.V., D.F., and G.D.A.; Investigation, A.E.V., P.C., K.M., D.F., C.A.J., and D.R.; Formal Analysis, S.C.C., G.D.A., A.E.V., and D.F.; Writing – Original Draft, A.E.V and A.R.; Funding Acquisition, A.R. and A.E.V; Resources, B.H., B.A.V., J.N.J., D.B.J., and M.K.; Supervision, A.R.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.09.016.
Appendix A. Supplementary data
Supplementary figures
References
- Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird K.M., Zhang G., Li H., Tu Z., Bitler B.G., Garipov A., Wu H., Wei Z., Wagner S.N., Herlyn M., Zhang R. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeff M., Kelly K.R., Yee K., Assouline S., Strair R., Popplewell L., Bowen D., Martinelli G., Drummond M.W., Vyas P. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin. Cancer Res. 2016;22:868–876. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barata P., Sood A.K., Hong D.S. RNA-targeted therapeutics in cancer clinical trials: current status and future directions. Cancer Treat. Rev. 2016;50:35–47. doi: 10.1016/j.ctrv.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M.V. Mitotic arrest and cell - Fate why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle. 2007;6:70–74. doi: 10.4161/cc.6.1.3682. [DOI] [PubMed] [Google Scholar]
- Burgess A., Chia K.M., Haupt S., Thomas D., Haupt Y., Lim E. Clinical overview of MDM2/X-targeted therapies. Front. Oncol. 2016;6:7. doi: 10.3389/fonc.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal D., Tovar C., Yang H., Vu B.T., Heimbrook D.C., Vassilev L.T. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res. 2005;65:1918–1924. doi: 10.1158/0008-5472.CAN-04-3576. [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data (vol 2, pg 401, 2012) Cancer Discov. 2012;2:960. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok C.F., Kua N., Kaldis P., Lane D.P. Combination of nutlin-3 and VX-680 selectively targets p53 mutant cells with reversible effects on cells expressing wild-type p53. Cell Death Differ. 2010;17:1486–1500. doi: 10.1038/cdd.2010.18. [DOI] [PubMed] [Google Scholar]
- Colin D.J., Hain K.O., Allan L.A., Clarke P.R. Cellular responses to a prolonged delay in mitosis are determined by a DNA damage response controlled by Bcl-2 family proteins. Open Biol. 2015;5 doi: 10.1098/rsob.140156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward J., Harding A. Size does matter: why polyploid tumor cells are critical drug targets in the war on cancer. Front. Oncol. 2014;4:123. doi: 10.3389/fonc.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.A., Johnston J.N. Catalytic, enantioselective synthesis of stilbene cis-Diamines: a concise preparation of (−)-Nutlin-3, a potent p53/MDM2 inhibitor. Chem. Sci. 2011;2:1076–1079. doi: 10.1039/C1SC00061F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.A., Vilgelm A.E., Richmond A., Johnston J.N. Preparation of (−)-Nutlin-3 using enantioselective organocatalysis at decagram scale. J. Org. Chem. 2013;78:10605–10616. doi: 10.1021/jo401321a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M., Sorensen C.S. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 2015;14:405–423. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr., Butel J.S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Driscoll D.L., Chakravarty A., Bowman D., Shinde V., Lasky K., Shi J., Vos T., Stringer B., Amidon B., D'Amore N., Hyer M.L. Plk1 inhibition causes post-mitotic DNA damage and senescence in a range of human tumor cell lines. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhajouji A., Cunha M., Kirsch-Volders M. Spindle poisons can induce polyploidy by mitotic slippage and micronucleate mononucleates in the cytokinesis-block assay. Mutagenesis. 1998;13:193–198. doi: 10.1093/mutage/13.2.193. [DOI] [PubMed] [Google Scholar]
- Freedberg D.E., Rigas S.H., Russak J., Gai W., Kaplow M., Osman I., Turner F., Randerson-Moor J.A., Houghton A., Busam K. Frequent p16-independent inactivation of p14ARF in human melanoma. J. Natl. Cancer Inst. 2008;100:784–795. doi: 10.1093/jnci/djn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y.C., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer J.G., Liu S.Q., Wang L., Mosel A., Peng A.M., Oakley G.G. RPA inhibition increases replication stress and suppresses tumor growth. Cancer Res. 2014;74:5165–5172. doi: 10.1158/0008-5472.CAN-14-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.M., Chan M., Harland M., Hayward N.K., Demenais F., Bishop D.T., Azizi E., Bergman W., Bianchi-Scarra G., Bruno W. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J. Med. Genet. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harris J., Chiu B. Clinical considerations of focal drug delivery in cancer treatment. Curr. Drug Deliv. 2017 doi: 10.2174/1567201814666170224143706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P., Nickerson E., Auclair D., Li L., Place C. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Hwang S.H., Wecksler A.T., Hammock B.D., Weiss R.H. Sorafenib attenuates p21 in kidney cancer cells and augments cell death in combination with DNA-damaging chemotherapy. Cancer Biol. Ther. 2011;12:827–836. doi: 10.4161/cbt.12.9.17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.B., Sosman J.A. Therapeutic advances and treatment options in metastatic melanoma. JAMA Oncol. 2015;1:380–386. doi: 10.1001/jamaoncol.2015.0565. [DOI] [PubMed] [Google Scholar]
- Jones R.M., Mortusewicz O., Afzal I., Lorvellec M., Garcia P., Helleday T., Petermann E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–3753. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- Kojima K., Konopleva M., Tsao T., Nakakuma H., Andreeff M. Concomitant inhibition of Mdm2-p53 interaction and Aurora kinases activates the p53-dependent postmitotic checkpoints and synergistically induces p53-mediated mitochondrial apoptosis along with reduced endoreduplication in acute myelogenous leukemia. Blood. 2008;112:2886–2895. doi: 10.1182/blood-2008-01-128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsantis P., Silva L.M., Irmscher S., Jones R.M., Folkes L., Gromak N., Petermann E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016;7 doi: 10.1038/ncomms13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat M.H., Ludwig R.L., Ashcroft M., Vousden K.H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol. Cell. Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hawkins O.E., Su Y.J., Vilgelm A.E., Sobolik T., Thu Y.M., Kantrow S., Splittgerber R.C., Short S., Amiri K.I. Targeting aurora kinases limits tumour growth through DNA damage-mediated senescence and blockade of NF-?B impairs this drug-induced senescence. EMBO Mol. Med. 2013;5:149–166. doi: 10.1002/emmm.201201378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Caballero J., Flores J.M., Garcia-Palencia P., Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61:6234–6238. [PubMed] [Google Scholar]
- Montecucco A., Zanetta F., Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95–108. doi: 10.17179/excli2015-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Loewer A., Lahav G., Mitchison T.J. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell. 2012;23:567–576. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penna L.S., Henriques J.A., Bonatto D. Anti-mitotic agents: Are they emerging molecules for cancer treatment? Pharmacol. Ther. 2017 doi: 10.1016/j.pharmthera.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Shen H.C., Dong W., Gao D.W., Wang G.H., Ma G.Y., Liu Q., Du J.J. MDM2 antagonist Nutlin-3a protects wild-type p53 cancer cells from paclitaxel. Chinese Sci. Bull. 2012;57:1007–1012. [Google Scholar]
- Sherr C.J., Roberts J.M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Storchova Z., Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- Su Y.J., Vilgelm A.E., Kelley M.C., Hawkins O.E., Liu Y., Boyd K.L., Kantrow S., Splittgerber R.C., Short S.P., Sobolik T. RAF265 inhibits the growth of advanced human melanoma tumors. Clin. Cancer Res. 2012;18:2184–2198. doi: 10.1158/1078-0432.CCR-11-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., Vilgelm, A. E., Kelley, M. C., Hawkins, O. E., Liu, Y., Boyd, K. L., Kantrow, S., Splittgerber, R. C., Short, S. P., Sobolik, T., et al. (n.d.)RAF265 inhibits the growth of advanced human melanoma tumors. Clin. Cancer Res. 18, 2184–2198. [DOI] [PMC free article] [PubMed]
- Sur S., Pagliarini R., Bunz F., Rago C., Diaz L.A., Kinzler K.W., Vogelstein B., Papadopoulos N. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokalov S.V., Abolmaali N.D. Protection of p53 wild type cells from taxol by nutlin-3 in the combined lung cancer treatment. BMC Cancer. 2010;10:57. doi: 10.1186/1471-2407-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA (vol 155, pg 1088, 2013) Cell. 2014;156:374. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Tsuiki H., Nitta M., Tada M., Inagaki M., Ushio Y., Saya H. Mechanism of hyperploid cell formation induced by microtubule inhibiting drug in glioma cell lines. Oncogene. 2001;20:420–429. doi: 10.1038/sj.onc.1204126. [DOI] [PubMed] [Google Scholar]
- Vilgelm A., Richmond A. Combined therapies that induce senescence and stabilize p53 block melanoma growth and prompt antitumor immune responses. Oncoimmunology. 2015;4 doi: 10.1080/2162402X.2015.1009299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm A., El-Rifai W., Zaika A. Therapeutic prospects for p73 and p63: rising from the shadow of p53. Drug Resist. Updat. 2008;11:152–163. doi: 10.1016/j.drup.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm A.E., Pawlikowski J.S., Liu Y., Hawkins O.E., Davis T.A., Smith J., Weller K.P., Horton L.W., McClain C.M., Ayers G.D. Mdm2 and aurora kinase a inhibitors synergize to block melanoma growth by driving apoptosis and immune clearance of tumor cells. Cancer Res. 2015;75:181–193. doi: 10.1158/0008-5472.CAN-14-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm A.E., Johnson D.B., Richmond A. Combinatorial approach to cancer immunotherapy: strength in numbers. J. Leukoc. Biol. 2016;100:275–290. doi: 10.1189/jlb.5RI0116-013RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu H.L., Aplin A.E. Targeting mutant NRAS signaling pathways in melanoma. Pharmacol. Res. 2016;107:111–116. doi: 10.1016/j.phrs.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu B., Wovkulich P., Pizzolato G., Lovey A., Ding Q.J., Jiang N., Liu J.J., Zhao C.L., Glenn K., Wen Y. Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med. Chem. Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettersten H.I., Hwang S.H., Li C.W., Shiu E.Y., Wecksler A.T., Hammock B.D., Weiss R.H. A novel p21 attenuator which is structurally related to sorafenib. Cancer Biol. Ther. 2013;14:278–285. doi: 10.4161/cbt.23374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee K., Martinelli G., Vey N., Dickinson M.J., Seiter K., Assouline S., Drummond M., Yoon S.S., Kasner M., Lee J.H. Phase 1/1b study of RG7388, a potent MDM2 antagonist, in acute myelogenous leukemia (AML) patients (pts) Blood. 2014;124 [Google Scholar]
- Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures