Abstract
Since impaired mitochondrial ATP production in cardiomyocytes is thought to lead to heart failure, a drug that protects mitochondria and improves ATP production under disease conditions would be an attractive treatment option. In this study, we identified small-molecule drugs, including the anti-parasitic agent, ivermectin, that maintain mitochondrial ATP levels under hypoxia in cardiomyocytes. Mechanistically, transcriptomic analysis and gene silencing experiments revealed that ivermectin increased mitochondrial ATP production by inducing Cox6a2, a subunit of the mitochondrial respiratory chain. Furthermore, ivermectin inhibited the hypertrophic response of human induced pluripotent stem cell-derived cardiomyocytes. Pharmacological inhibition of importin β, one of the targets of ivermectin, exhibited protection against mitochondrial ATP decline and cardiomyocyte hypertrophy. These findings indicate that maintaining mitochondrial ATP under hypoxia may prevent hypertrophy and improve cardiac function, providing therapeutic options for mitochondrial dysfunction.
Keywords: Mitochondrial ATP, Cardiomyocytes, Hypertrophy, Hypoxia, Phenotypic screening
Abbreviations: ATP, adenosine triphosphate; BNP, brain natriuretic peptide; CsA, cyclosporin A; ET-1, endothelin 1; FCCP, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone; FPKM, fragments per kilobase of transcript per million mapped reads; FRET, fluorescence resonance energy transfer; HIF, hypoxia inducible factor; iPSC, induced pluripotent stem cell; mPTP, mitochondrial permeability transition pore; OXPHOS, oxidative phosphorylation; ROS, reactive oxygen species; UPR, unfolded protein response
Highlights
-
•
High-throughput image-based phenotypic screens based on mitochondrial ATP under hypoxia were developed.
-
•
Mitochondrial ATP protectors, including ivermectin, were identified in cardiomyocytes under hypoxia.
-
•
Ivermectin increased mitochondrial ATP production by inducing Cox6a2.
-
•
Mitochondrial ATP protectors inhibited hypertrophic responses in human iPSC-derived cardiomyocytes.
Impaired mitochondrial ATP production in cardiomyocytes is one of the major causes of heart failure. A drug that maintains ATP production would, therefore, be an attractive heart failure treatment option. We performed high-throughput image-based screens under hypoxia, and identified mitochondrial ATP protectors, including the anti-parasite drug, ivermectin. We show that ivermectin increased mitochondrial ATP production by inducing Cox6a2 expression and inhibited the hypertrophic response of human iPSC-derived cardiomyocytes. Pharmacological inhibition of importin β, a target of ivermectin, exhibited similar protective effects. Our studies indicate that compounds maintaining mitochondrial ATP under hypoxia may prevent pathological hypertrophy and improve cardiac function.
1. Introduction
Despite recent progress in medical therapies, heart failure remains one of the leading causes of death and healthcare burden worldwide (Ziaeian and Fonarow, 2016). Cardiomyocytes contain numerous mitochondria to support the heart's demand for adenosine triphosphate (ATP), and studies have implicated mitochondrial dysfunction in human patients and animal models of heart diseases, as demonstrated by reduced ATP levels (Beer et al., 2002), decreased enzymatic activity in the oxidative phosphorylation (OXPHOS) system (Ahuja et al., 2013), and enhanced reactive oxygen species (ROS) production (Borchi et al., 2010; Ide et al., 2000). Associations of mitochondrial dysfunction with heart diseases are also supported by previous studies, in which cardiomyopathy was observed in patients with mutations in critical mitochondrial proteins (Meyers et al., 2013). Since the heart becomes vulnerable to hypoxia in patients with heart disease and mitochondria require oxygen to synthesize ATP, hypoxia may be one of the key pathogenic factors causing impaired OXPHOS in heart diseases (Krishnan et al., 2009). Although targeting mitochondria has garnered increasing attention (Brown et al., 2017), there is no currently approved therapy that targets this organelle. Therefore, it would be valuable to identify novel therapeutic approaches for heart diseases based on mitochondrial ATP generation under pathological conditions, such as hypoxia. Although it had been challenging to directly measure mitochondrial ATP, a fluorescence resonance energy transfer (FRET)-based ATP biosensor, ATeam, has been recently reported, which made it possible to monitor spatiotemporal changes in ATP levels in a specific organelle, including mitochondria, in living cells (Imamura et al., 2009).
In this study, we identified small-molecule drugs, including the anti-parasitic drug, ivermectin, which maintain mitochondrial ATP levels under hypoxia in cardiomyocytes, through high-throughput phenotypic screens of a drug library using the aforementioned mitochondrial FRET-based ATP biosensor. We also assess the antihypertrophic effects of these mitochondrial modulators in human induced pluripotent stem cell (iPSC)-derived cardiomyocytes. Moreover, the transcriptomic analyses provide mechanistic insights into how these modulators exert their effects on mitochondrial ATP. This study provides previously unappreciated insights into the regulation of mitochondrial ATP, and has therapeutic implications for diseases involving mitochondrial dysfunction.
2. Materials and Methods
2.1. Chemicals and Compounds
The Prestwick Chemical Library was purchased from PerkinElmer. Dimethyl sulfoxide (DMSO), 2-deoxy-d-glucose (2-DG), diazoxide, cyclosporin A (CsA), N-acetyl cysteine (NAC), IPA-3, bifonazole, verapamil, tunicamycin, and thapsigargin were purchased from Wako. Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), deferoxamine mesylate, importazole, ivermectin, and oligomycin A were purchased from Sigma-Aldrich. Mito-TEMPO was purchased from Enzo Life Sciences. Compound 968 was purchased from Calbiochem. Dexrazoxane was purchased from TCI Chemicals. Cariporide was purchased from Cayman Chemical. Nifuroxazide was purchased from MP Biomedicals. 2-Methylthioadenosine triphosphate (2-meSATP) was purchased from Tocris. Azoramide was purchased from Ark Pharm, Inc.
2.2. Plasmids and siRNA
The plasmid vectors expressing ATeam (AT1.03 and mitAT1.03-YEMK in pcDNA3.1) (Imamura et al., 2009) were obtained under license to Osaka University. Control siRNA (ON-TARGETplus siRNA-SMARTpool, Non-targeting pool, D-001810-10-05) and Cox6a2 siRNA (ON-TARGETplus siRNA-SMARTpool, L-043169-01-0005) were purchased from Dharmacon/Thermo Fisher Scientific.
2.3. Cell Culture
HL-1 cardiomyocytes, obtained from William Claycomb (Louisiana State University, New Orleans, LA, USA), were cultured as previously described (Claycomb et al., 1998). In brief, Claycomb medium (Sigma-Aldrich), supplemented with 2 mM l-glutamine, 100 U/mL penicillin/streptomycin, 100 μM norepinephrine in 30 mM l-ascorbic acid, and 10% fetal bovine serum (FBS), was replaced every other day. The culture flasks and plates were precoated with a solution of 0.02% (weight/volume) gelatin containing 5 μg/mL fibronectin (Sigma-Aldrich). HeLa cells (ECACC, 93021013) were purchased from DS Pharma Biomedical, and cultured in DMEM (Gibco) containing 10% FBS and 100 U/mL penicillin/streptomycin. Human iPSC-derived cardiomyocytes (CMC-100-010-001) were purchased from Cellular Dynamics International (CDI) and cultured in maintenance medium (CDI, CMM-100-120-001) according to the manufacturer's instructions. Cells were maintained at 37 °C in a humidified 5% CO2 incubator.
2.4. Generation of HL-1 Cardiomyocytes Stably Expressing ATeam
HL-1 cardiomyocytes were reverse-transfected with plasmids expressing either mitAT1.03-YEMK (mito-ATeam) for mitochondrial ATP measurement or AT1.03 (cyto-ATeam) for cytosolic ATP measurement using Lipofectamine LTX (Invitrogen) in Opti-MEM (Gibco), according to the manufacturer's instructions. Four days after transfection, selection using 0.25 mg/mL geneticin (Gibco) was started. Populations of cells expressing ATeam at moderate levels were then selected using FACS Aria II cell sorter (Becton Dickinson) based on YFP fluorescence. The enriched ATeam stable cells were cultured as described above in the presence of geneticin.
2.5. Animals
The care and use of the animals and the experimental protocols used in this study were approved by the Experimental Animal Care and Use Committee of Takeda Pharmaceutical Company Limited. All experiments were performed in accordance with the guidelines and regulations of Takeda Pharmaceutical Company Limited (Shonan Research Center IACUC Guidelines). C57BL/6J mice were purchased from CLEA Japan, Inc. All mice used in this study were maintained in pathogen-free barrier animal facilities.
2.6. Generation of Mito-ATeam Knock-in Mice
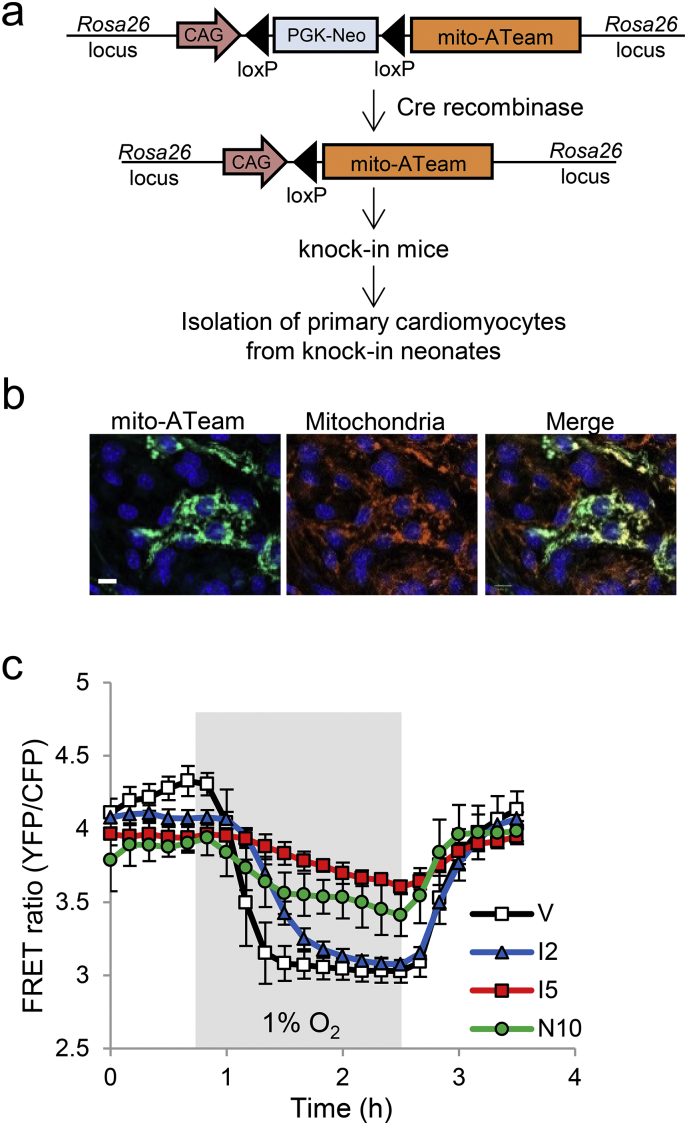
The targeting vector pROSA26-mitAT1.03-YEMK was constructed, in which a CAG-loxP-Neo cassette-loxP-mitAT1.03-YEMK unit was inserted into the ROSA26 locus by the Red/ET recombination system (Gene Bridges) to modify the bacterial artificial chromosome with the mouse ROSA26 gene (Advanced Geno Techs Co., RP23-184A7), as shown in Fig. 3a. pROSA26-mitAT1.03-YEMK was electroporated into embryonic stem cells (ESCs) derived from C57BL/6J mice. Clones with the inserted allele were screened by genomic PCR after G418 selection. The Neo cassette was removed by transient expression of pCAG-Cre plasmids in recombinant ESCs. Chimeric mice were generated by the tetraploid complementation method described previously (Yamamoto et al., 2013). The chimeric mice were crossed with C57BL/6J mice to obtain mice heterozygous for the mutant allele. Heterozygous mice were intercrossed to obtain animals homozygous for the mutant allele and wild-type littermates.
Fig. 3.
Effects of ivermectin and nifuroxazide on mitochondrial ATP levels under hypoxia in primary cardiomyocytes. (a) A schematic of mito-ATeam knock-in animals for the isolation of primary cardiomyocytes. (b) Representative images of mito-ATeam in primary cardiomyocytes isolated from mito-ATeam knock-in neonatal hearts stained for mitochondria (red) and nuclei (blue); mito-ATeam is in green. Scale bar, 10 μm. (c) Quantified FRET ratios of mito-ATeam knock-in primary cardiomyocytes pre-treated with vehicle (V), ivermectin (2 or 5 μM; I2 or I5), or nifuroxazide (10 μM, N10) for 24 h followed by hypoxia/reoxygenation. Data are presented as means ± SD (n = 3 biologically independent samples).
2.7. Isolation of Primary Neonatal Cardiomyocytes
Neonatal murine cardiomyocytes were isolated from mito-ATeam knock-in mice and wild-type littermates at 1 day of age and maintained as described previously (Sreejit et al., 2008). Isolated primary cardiomyocytes were cultured for up to 5 days for subsequent studies.
2.8. Imaging
All confocal images were acquired with a high-content screening system, Cell Voyager 7000 (CV7000; Yokogawa), which was equipped with a confocal scanner unit, live-cell incubator (maintained at 37 °C and 5% CO2) with a customized lid for hypoxia experiments, and built-in liquid handler. Hypoxic gas (1% O2, 5% CO2, and 94% air) was supplied using a gas mixer (Tokken, Inc.).
2.8.1. Live-Cell Imaging of ATP in HL-1 Cardiomyocytes
ATeam stable cells were seeded at a density of 10,000 cells per well into 96-well imaging plates and treated with compounds typically for 24 h. DMSO (0.1%) was used as a negative control. ATeam biosensors were excited at 445 nm, and emission was recorded at 480/17 nm for CFP and 543/22 nm for YFP at × 20 (0.75 NA UPLSAPO objective, 37 °C) or × 60 (1.2 NA UPLSAPO objective, 37 °C) magnification. Images were acquired before and after adding 10 μM oligomycin A, or under normoxia (21% O2) followed by hypoxia (1% O2) and reoxygenation (21% O2) for the indicated times at 5- to 10-min intervals. Image analysis was performed using CV7000 analysis software (Yokogawa); a region of interest (ROI) was defined based on CFP fluorescence intensity; fluorescence intensity of YFP (as well as that of CFP) in the ROI was quantified. The YFP/CFP emission ratio (FRET ratio) per ROI was then calculated by dividing YFP fluorescence intensity by CFP fluorescence intensity using TIBCO Spotfire (TIBCO). Ratiometric FRET images were obtained using ImageJ software (National Institutes of Health).
2.8.2. Live-Cell Imaging of Mitochondrial ATP in Primary Cardiomyocytes
Neonatal cardiomyocytes were seeded at a density of 200,000 cells per well into 96-well imaging plates and treated with compounds for 24 h in Dulbecco's modified Eagle's medium (DMEM)/F-12 (Gibco) supplemented with 0.2% bovine serum albumin (BSA) and 100 U/mL penicillin/streptomycin. Live-cell imaging experiments and image analysis were performed as described above.
2.9. Phenotypic Screening of the Prestwick Chemical Library
The mito-ATeam stable HL-1 cardiomyocytes were plated at a density of 8000 cells per well in 384-well imaging plates (Greiner Bio-One) using a Multidrop dispenser (Thermo Fisher Scientific). In the primary screening round, the cells were treated with 1256 Prestwick compounds at a final concentration of 10 μM (n = 1 well) by adding a 5 × concentration of compounds (8 μL/well) using a BioMek liquid handling automation workstation (Beckman Coulter). DMSO (0.1%) was used as a negative control and 30 μM Mito-TEMPO as a positive control. The next day, ATP imaging using a CV7000 system was performed as described above under normoxia (for initial ratio) followed by hypoxia (1% O2) for 1.5 h. The FRET ratio was calculated as described above and normalized by the initial value; positive hits were selected from wells with > 0.931 of normalized ratio (average + 3 standard deviations [SDs] of negative control) at the time point showing the best z-score per plate. To confirm primary hits, compounds were re-screened using 96-well plates, and dose-dependency was examined at lower concentrations. DMSO (0.1%) was used as a negative control and 30 μM deferoxamine as a positive control. Primary hits were also evaluated based on oligomycin A response (selected at < 0.9 of normalized FRET ratio), mitochondrial membrane potential (selected at > 80%), cell viability (selected at > 80%), and caspase-3/7 activity.
2.10. Human Cardiomyocyte Hypertrophy Model
Cardiac hypertrophy assays in human iPSC-derived cardiomyocytes were conducted as previously reported with some modifications (Carlson et al., 2013). In brief, cardiomyocytes were plated at a density of 20,000 viable cells per well on Cellmatrix Type I-C-coated 96-well plates or at 5000 viable cells per well on Cellmatrix Type I-C-coated 384-well imaging plates (Greiner Bio-One). On day 3 post-plating, the medium was replaced with Williams' Medium E (Gibco) supplemented with Cocktail B from the Hepatocyte Maintenance Supplement Pack (Gibco). On day 6 post-plating, the medium was replaced with supplemented Williams' Medium E containing 10 nM human endothelin-1 (ET-1; Peptide Institute) and test compounds for 18 h. For immunocytochemistry of brain natriuretic propeptide (proBNP), 10 μg/mL of Brefeldin A were added for the last 3 h. Eighteen hours after stimulation with ET-1 and test compounds, quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed as described in the Supplemental information.
2.11. Immunocytochemistry, Imaging, and Quantification
Eighteen hours after stimulation of iPSC cardiomyocytes with ET-1 and test compounds, an equal volume of 4% paraformaldehyde (PFA) was added to the medium (resulting in 2% PFA); the cells were incubated for 10 min at room temperature, followed by fixation with 4% PFA for 10 min at room temperature. After rinsing twice with phosphate-buffered saline (PBS), cells were incubated with a primary antibody against proBNP (Abcam; ab13115) at 1:500 in permeabilization buffer containing 0.1% Triton X-100 (Sigma-Aldrich, T8787-100ML) and 5% donkey serum (Abcam, ab7475) in PBS at 4 °C overnight. After rinsing twice with PBS, the samples were incubated with Alexa Fluor 568-labelled donkey anti-mouse IgG (Molecular Probes, A10037) at 1:1000 in permeabilization buffer at room temperature for 2 h. Hoechst 33342 was added to the samples and incubated at room temperature for 20 min. After rinsing twice with PBS, confocal images were acquired with a CV7000 system at × 20 magnification. Hoechst 33342 was excited at 405 nm and fluorescence recorded at 445/45 nm; Alexa Fluor 568 was excited at 561 nm and fluorescence recorded at 617/73 nm. Cells were defined using CV7000 analysis software based on Hoechst 33342, and fluorescence intensities of proBNP in the ROIs around nuclei (expand distance of 17 μm) were quantified.
2.12. RNA-Seq and Analyses
Library preparation, RNA-seq data processing, and pathway/upstream analyses using IPA (Ingenuity® Pathway Analysis, Qiagen) are described in the Supplemental information. The raw and processed data reported in this paper were deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE96989.
2.13. Statistical Analysis
Statistical analysis was carried out using EXSUS version 8.0 (CAC Croit Corp.). Statistical differences were analyzed using two-tailed unpaired Student's t-test or Alpin-Welch's t-test to compare two groups, or one-way ANOVA with Dunnett's multiple-comparison test to compare more than two groups. p < 0.05 was considered statistically significant. Data are presented as means ± SD. All experiments were performed with at least three biological replicates (unless noted otherwise). Panels in Figs. 1a, c; 2d, e; 3b; 6a, and Fig. S1c show a representative image of at least three independent experiments.
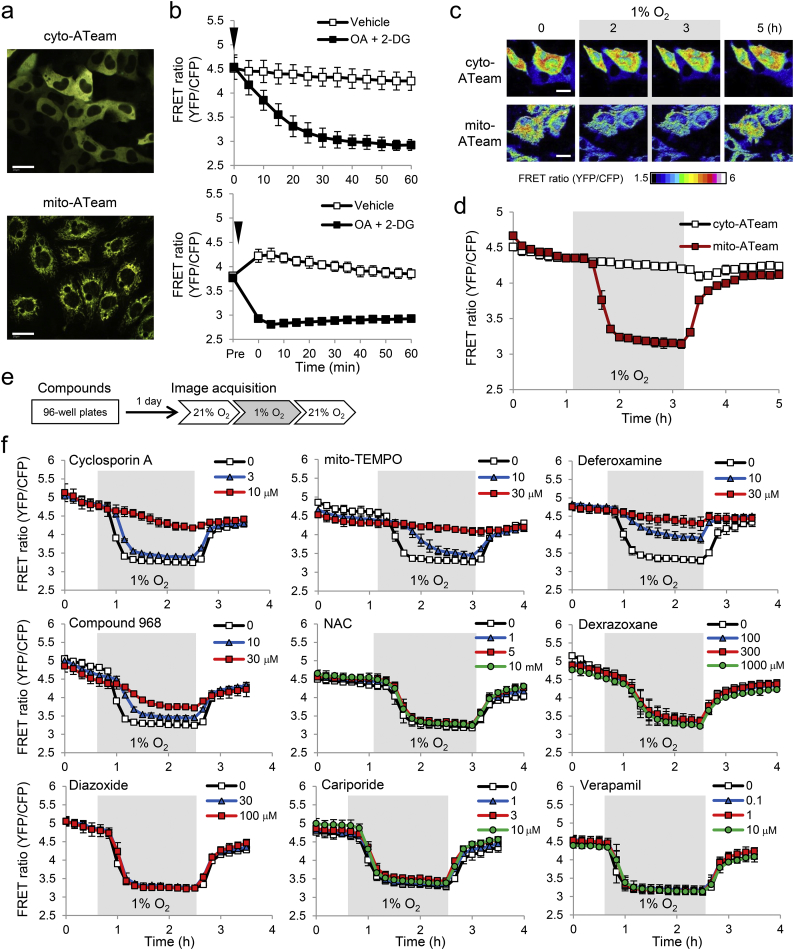
Fig. 1.
Live-cell imaging of mitochondrial ATP under hypoxia and effects of small-molecule compounds. (a) Representative images of fluorescence resonance energy transfer (FRET) signal in HL-1 cardiomyocytes stably expressing either cyto-ATeam (upper image) or mito-ATeam (lower image). Scale bars, 20 μm. (b) Quantified FRET ratio (YFP/CFP) in response to 10 μM oligomycin A (OA) and 10 mM 2-deoxyglucose (2-DG) treatment in HL-1 cardiomyocytes stably expressing either cyto-ATeam (upper panel) or mito-ATeam (lower panel). Data are presented as means ± SD (n = 3 biologically independent samples). The arrow heads indicate the time point when vehicle or OA + 2-DG was added. (c) Representative images of FRET signal in HL-1 cardiomyocytes stably expressing either cyto-ATeam (upper panel) or mito-ATeam (lower panel) under hypoxia (1% O2) followed by reoxygenation (21% O2). Scale bars, 20 μm. (d) Quantified FRET ratio in HL-1 cardiomyocytes stably expressing either cyto-ATeam (black line) or mito-ATeam (red line) under 2-h hypoxia (1% O2) followed by 2-h reoxygenation (21% O2). Data are presented as means ± SD (n = 3 biologically independent samples). (e) HL-1 cardiomyocytes stably expressing mito-ATeam were treated with compounds typically for 24 h, and subjected to FRET imaging under hypoxia followed by reoxygenation as shown in (d). (f) Effects of cyclosporin A, mito-TEMPO, deferoxamine, Compound 968, N-acetyl cysteine (NAC), dexrazoxane, diazoxide, cariporide, or verapamil on mitochondrial ATP under hypoxia are shown. Data are presented as means ± SD (n = 3 biologically independent samples).
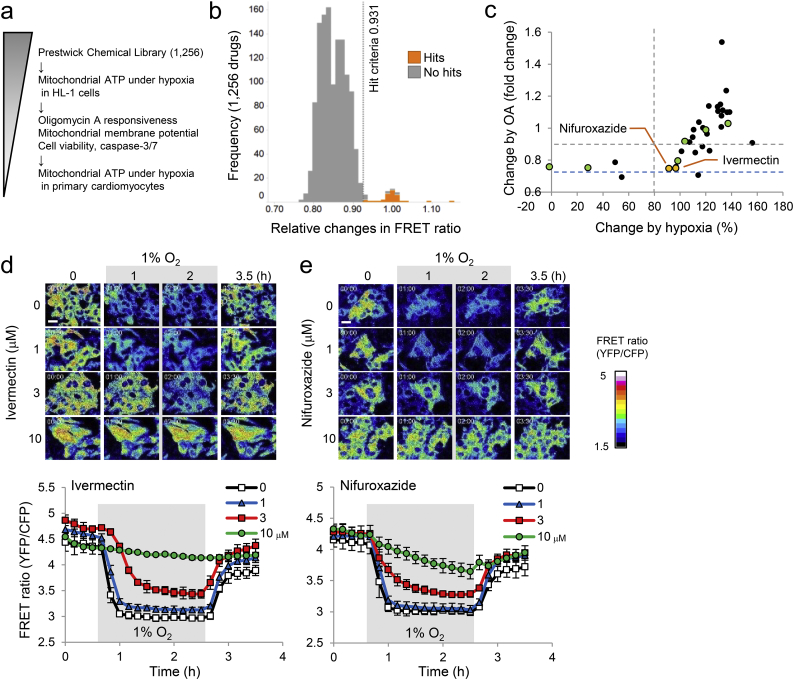
Fig. 2.
Identification of ivermectin and nifuroxazide as mitochondrial ATP modulators under hypoxia by phenotypic screens. (a) Assay workflow to identify small molecules that maintain mitochondrial ATP levels under hypoxia with cardiac anti-hypertrophic effects. (b) Stacked histogram of the primary screen. Thirty-four primary hits (orange) were identified by the hit criteria (vehicle average + 3 SD = 0.931) indicated by dashed line and other parameters. (c) Scatter plot of results of mito-ATeam assays (protection against hypoxia- or oligomycin A [OA]-mediated mitochondrial ATP decrease). Each data point represents the average of three independent experiments, normalized to initial values (OA) or negative and positive controls (hypoxia) (initial value was set to 1). Grey dashed lines indicate the thresholds (> 80% for x axis and < 0.9 for y axis). Blue dashed line indicates OA response in vehicle group. Twenty-six compounds shown in black were excluded based on the counter-assays (cell viability and mitochondrial membrane potential) (Fig. S2a). Out of eight selected compounds, ivermectin and nifuroxazide (shown in orange) exhibited protective effects on mitochondrial ATP under hypoxia, but not in response to OA, without activating caspase-3/7 (Fig. S2b). Source data are available online for this figure (Table S1). (d and e) Representative images of fluorescence resonance energy transfer (FRET) signal in mito-ATeam stable HL-1 cardiomyocytes treated with ivermectin (d, upper panel) or nifuroxazide (e, upper panel) for 24 h under hypoxia (1% O2), followed by reoxygenation (21% O2). Scale bars, 20 μm. Quantified FRET ratios of mito-ATeam in HL-1 cardiomyocytes treated with ivermectin (0, 1, 3, 10 μM, d, lower panel) or nifuroxazide (0, 1, 3, 10 μM, e, lower panel) under hypoxia/reoxygenation are shown. Data are presented as means ± SD (n = 3 biologically independent samples).
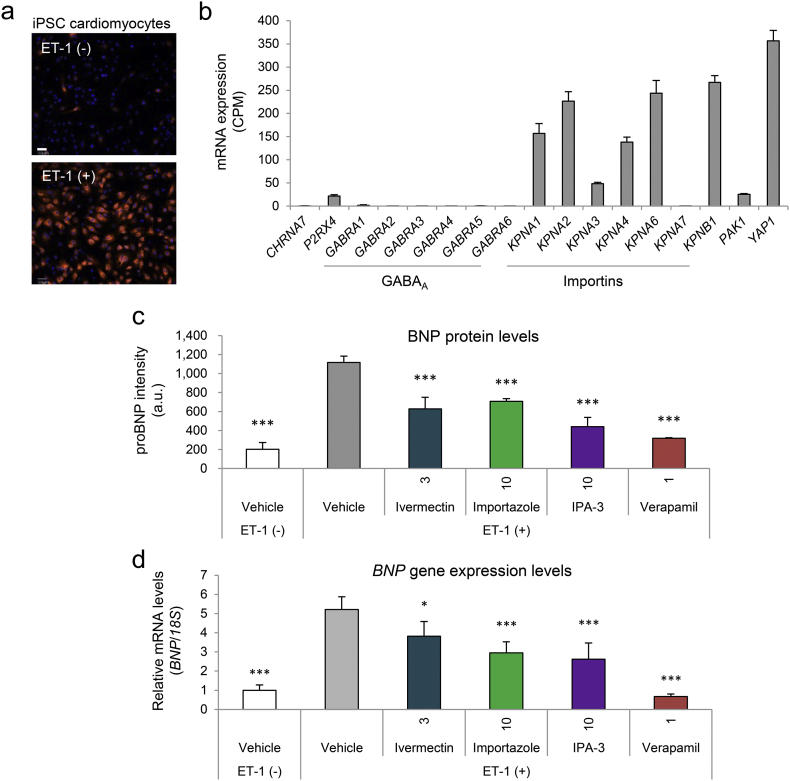
Fig. 6.
Antihypertrophic effects of mitochondrial ATP modulators in human induced pluripotent stem cell (iPSC)-derived cardiomyocytes. (a) Representative images of human iPSC-derived cardiomyocytes stimulated without (upper image) or with 10 nM human endothelin 1 (ET-1, lower image) for 18 h and stained with antibodies against proBNP (red); nuclei are in blue. Scale bar, 50 μm. (b) Gene expression levels of known ivermectin targets by AmpliSeq analysis in iPSC cardiomyocytes (untreated). Data are presented as means ± SD (n = 4 biologically independent samples). CPM, counts per million. (c and d) (c) Quantified proBNP intensities (arbitrary unit, a.u.) and (d) RT-qPCR of the canonical hypertrophy marker BNP normalized to 18S in iPSC cardiomyocytes untreated (white bar), or treated with vehicle, ivermectin (3 μM), importazole (10 μM), IPA-3 (10 μM), or verapamil (1 μM) in the presence of ET-1, is shown. Data are presented as means ± SD (n = 4 biologically independent samples). ⁎p < 0.05, ⁎⁎⁎p < 0.001 vs. vehicle (ET-1 alone) by one-way ANOVA with Dunnett's multiple-comparison test.
Additional methods, including Live-Cell Imaging of Mitochondrial ATP in HeLa Cells, Mitochondrial Membrane Potential Measurement, Calcium Imaging in Primary Cardiomyocytes, Cell Viability Assay, Caspase-3/7 Activity Assay, RNA-seq and Analyses, SDS-PAGE and Western Blotting, Crude Mitochondrial Isolation, Complex IV Activity, Mitochondrial ATP Production in Permeabilized Cells, and RT-qPCR are available in Supplemental Materials and Methods.
3. Results
3.1. Live-Cell Imaging of Mitochondrial ATP Under Hypoxia
To monitor ATP levels in the cytosol and mitochondria of living cells, HL-1 cardiomyocyte cell lines stably expressing either cyto-ATeam (Fig. 1a, upper image) or mito-ATeam (Fig. 1a, lower image) were established. The decrease in the FRET ratio was confirmed in the cytosol (Fig. 1b, upper panel) and the mitochondrial matrix (Fig. 1b, lower panel) upon ATP depletion by oligomycin A (OA), an inhibitor of complex V of the mitochondrial respiratory chain, and by 2-deoxyglucose (2-DG), a glycolysis inhibitor. Although ATeam FRET can be reduced as pH lowers in a cell-free system (Imamura et al., 2009), a mitochondrial uncoupler, which induces acidification of the mitochondrial matrix (Yang et al., 2016), did not decrease the FRET ratio in living cells (Fig. S1a), suggesting that intramitochondrial acidification has little impact on ATeam FRET. Collectively, these results indicate that ATeam serves as an ATP indicator in living cells. Of note, mitochondrial ATP decreased more rapidly than cytosolic ATP did in response to OA and 2-DG (Fig. 1b). We next compared changes in mitochondrial and cytosolic ATP in response to hypoxia (1% O2), using a customized lid for 96- and 384-well plates equipped with a high content analyzer CV7000. As previously reported (Kioka et al., 2014), mitochondrial ATP decreased robustly with hypoxia in HL-1 cardiomyocytes, whereas cytosolic ATP did not (Fig. 1c and d). These results suggest that mitochondrial ATP is more sensitive to hypoxia than cytosolic ATP is, and that cytosolic ATP may be compensated by anaerobic metabolism under stress conditions.
3.2. Effects of Small-Molecule Compounds on Mitochondrial ATP Decline in Hypoxia
Using mito-ATeam stable HL-1 cardiomyocytes, we assessed the effects of compounds reported to exhibit protective effects on mitochondria and/or cardioprotective effects on mitochondrial ATP levels during hypoxia (Fig. 1e). Mitochondrial ATP decline was substantially prevented by the mitochondrial permeability transition pore (mPTP) inhibitor CsA (Hausenloy et al., 2012), mitochondria-targeted antioxidant mito-TEMPO (Gelvan et al., 1991; Luo et al., 2013), iron chelator deferoxamine (Paraskevaidis et al., 2005), and mitochondrial glutaminase inhibitor Compound 968 (Gao et al., 2015) (Fig. 1f). These compounds had a minimal effect on mitochondrial ATP decline caused by OA and on mitochondrial membrane potential (Fig. S1b–d), suggesting that these compounds did not compromise mitochondrial function. Other compounds, including the topoisomerase II inhibitor dexrazoxane (Cvetković and Scott, 2005), K+ ATP channel opener diazoxide (Flagg et al., 2010), Na+/H+ exchange inhibitor cariporide (Avkiran and Marber, 2002; Ruiz-Meana et al., 2003), and Ca2 + channel blocker verapamil (Nayler et al., 1980), did not display protective effects (Fig. 1f), suggesting that these compounds exert their effects through different mechanisms. Given that NAC did not prevent mitochondrial ATP decline (Fig. 1f), mitochondria-localized superoxide and free radicals may modulate mitochondrial ATP levels. These results indicate that known mito-protective compounds may have unappreciated effects on mitochondrial ATP; our study could identify compounds that protect mitochondrial ATP selectively under hypoxia.
3.3. Identification of Ivermectin as a Mitochondrial ATP Modulator by Phenotypic Screening
To screen for drugs that may prevent mitochondrial ATP decline during hypoxia, we performed phenotypic screening of the Prestwick Chemical Library (Fig. 2a). Primary screens of 1256 compounds identified 34 primary hits (Fig. 2b and Table S1); eight compounds exhibited > 80% mitochondrial membrane potential retention and > 80% cell viability (Fig. S2a). Importantly, ivermectin and nifuroxazide did not affect OA-mediated mitochondrial ATP decrease (Fig. 2c and Table S1), indicating that their protective effects on mitochondrial ATP levels are specific to hypoxic conditions. In addition, ivermectin and nifuroxazide demonstrated concentration-dependent mitochondrial ATP protection (Fig. 2d and e), without affecting caspase-3/7 activity (Fig. S2b).
To determine the drug effects in primary cardiomyocytes, we established mito-ATeam knock-in mice and isolated primary cardiomyocytes from neonates (Fig. 3a). Mitochondrial localization of mito-ATeam in these cells was confirmed (Fig. 3b). Mitochondrial ATP levels decreased during hypoxia; this decline was prevented by ivermectin and nifuroxazide (Fig. 3c).
3.4. Identification of Importin β as a Potential Molecular Target of Ivermectin's Action
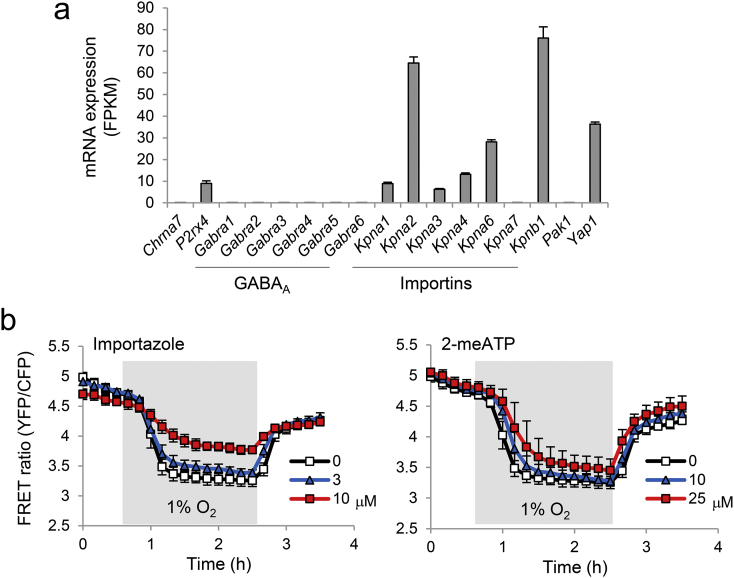
Nifuroxazide is an anti-diarrheal agent that inhibits signal transducer and activator of transcription 3 (STAT3) (Nelson et al., 2008), a transcription factor that plays well-described roles in cardiac hypertrophy and failure (Haghikia et al., 2011). Although ivermectin shows anti-tumor activity (Hashimoto et al., 2009; Liu et al., 2016; Nishio et al., 2016), enhances cardiomyocyte contractility (Yang et al., 2014), and is neuroprotective (Andries et al., 2007), few reports have addressed its effects on mitochondrial function in cardiomyocytes or cardiac hypertrophy. Therefore, we focused on ivermectin and its mechanism of action by assessing the effects of known ivermectin molecular targets on mitochondrial ATP under hypoxia. Among such targets, including α7 nicotinic acetylcholine receptor (Krause et al., 1998), P2X4 receptor (Khakh et al., 1999), GABAA receptor (Adelsberger et al., 2000), importin α/β (Kosyna et al., 2015; Wagstaff et al., 2012), PAK1 (Hashimoto et al., 2009), and YAP1 (Nishio et al., 2016), we selected targets that are expressed in HL-1 cardiomyocytes and can be pharmacologically modulated. From RNA-seq analysis, P2rx4 (P2X4), Kpna2 (importin α1), and Kpnb1 (importin β1) were expressed in HL-1 cardiomyocytes; Pak1, Chrna7 (α7 nAChR), and Gabra (GABAA receptor) expressions were low (Fig. 4a). We tested commercially available modulators for importin β (importazole as an inhibitor) and P2X4 (2-meSATP as a positive allosteric modulator) and found that importazole, but not 2-meSATP, prevented mitochondrial ATP decline, suggesting that importin β is involved (Fig. 4b).
Fig. 4.
Identification of importin β as a potential molecular target of ivermectin. (a) Expression levels of genes involved in ivermectin action were determined by RNA sequencing (RNA-seq) of HL-1 cardiomyocytes. Data are presented as means ± SD (n = 3 biologically independent samples). FPKM, fragments per kilobase of transcript per million mapped reads. (b) mito-ATeam stable HL-1 cardiomyocytes were treated with vehicle, the importin β inhibitor importazole (3 or 10 μM), or the P2X4 positive allosteric modulator 2-meSATP (10 or 25 μM) for 24 h. Quantified FRET ratios of mito-ATeam under hypoxia/reoxygenation are shown. Data are presented as means ± SD (n = 3 biologically independent samples).
3.5. Critical Role of Cox6a2 in Increased Mitochondrial ATP Production by Ivermectin
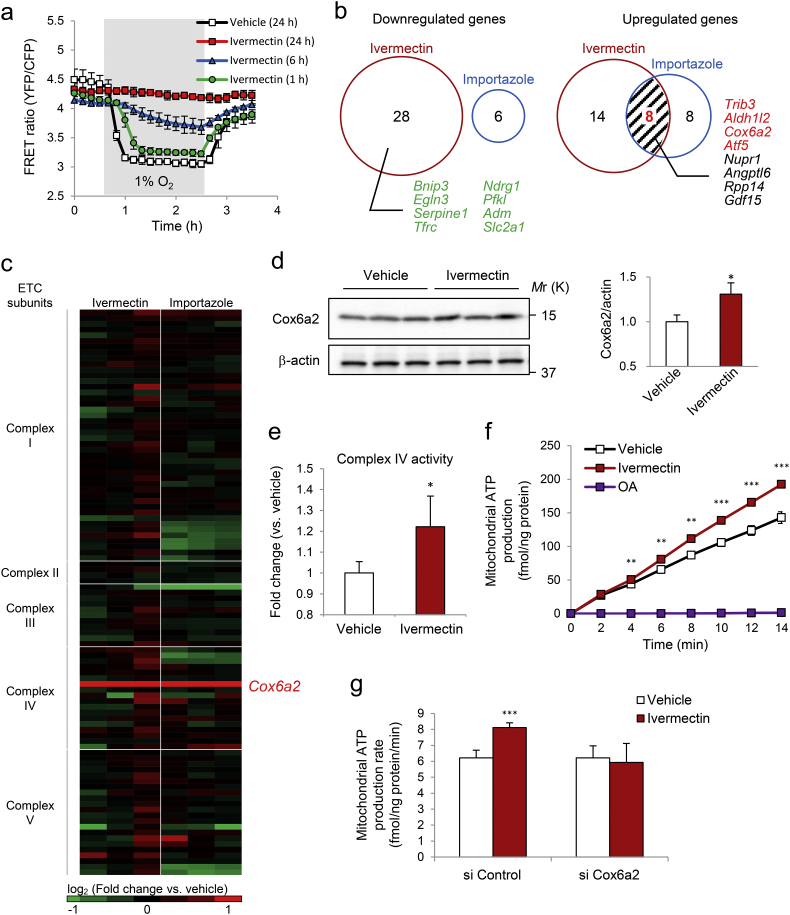
To understand how ivermectin affects HL-1 cardiomyocytes, we examined whether ivermectin displayed any acute treatment effect. A shorter treatment period resulted in weaker protection against mitochondrial ATP decline during hypoxia (Fig. 5a), indicating that ivermectin may affect transcription levels rather than acting directly on the target(s). We thus performed RNA-seq analysis in HL-1 cardiomyocytes treated with ivermectin or importazole to identify overlapping genes between the treatment groups. Surprisingly, there was no overlapping downregulated gene. Of note, hypoxia inducible factor (HIF)-1α target genes such as Bnip3 and Egln3 were downregulated in the ivermectin treatment group (Fig. 5b), consistent with a previous report (Kosyna et al., 2015). These results suggest that 1) ivermectin and importazole exert differential effects on HIF-1α pathways presumably because ivermectin inhibits importin α/β heterodimers (Wagstaff et al., 2012), whereas importazole specifically inhibits importin β (Soderholm et al., 2011), and 2) the action of ivermectin on mitochondrial ATP was not due to HIF-1α pathway activation, which plays a major role in preconditioning (Prabhakar and Semenza, 2012). However, eight upregulated genes overlapped, including Trib3, Aldh1l2, Cox6a2, and Atf5, with p values < 0.01 (Fig. 5b). Cox6a2, a heart/muscle-specific isoform of Cox6, is one of the subunits of Complex IV, and important for the activity of cytochrome c oxidase (COX) (Quintens et al., 2013; Radford et al., 2002). Intriguingly, the induction was specific to Cox6a2 among other mitochondrial respiratory chain complex subunits (Fig. 5c and Fig. S3a). Cox6a2 protein levels were also increased by ivermectin (Fig. 5d). Accordingly, COX activity significantly increased in the mitochondria isolated from ivermectin-treated HL-1 cardiomyocytes (Fig. 5e). To confirm whether ivermectin enhances ATP production via Complex IV, we directly measured ADP-to-ATP conversion driven by Complex IV substrates (ascorbate and N,N,N′,N′-tetramethyl-p-phenylenediamine [TMPD]) in the presence of the Complex III inhibitor, antimycin A, in permeabilized HL-1 cardiomyocytes; ivermectin significantly increased mitochondrial ATP production (Fig. 5f), and this increase was abolished by Cox6a2 knockdown (Fig. 5g and Fig. S3b). These results suggest that ivermectin enhances ATP production in the mitochondria by upregulating the Cox6a2 transcript.
Fig. 5.
Increased Cox6a2 levels, Complex IV activity, and mitochondrial ATP production following ivermectin treatment in HL-1 cardiomyocytes. (a) Fluorescence resonance energy transfer (FRET) ratio of mito-ATeam stable HL-1 cardiomyocytes treated with ivermectin for various time periods. Data are presented as means ± SD (n = 3 biologically independent samples). (b) Venn diagrams of the comparative analysis of differentially expressed genes identified from RNA-seq data on ivermectin (5 μM, 24 h) and importazole (10 μM, 24 h) treatment in HL-1 cardiomyocytes (n = 3 biologically independent samples for each condition, fold change [log2] ≥ 1, p < 0.05 vs. vehicle treatment). Numbers in red indicate the total number of genes common to two conditions. Genes in red signify p < 0.01. (c) Heatmap of RNA-seq data of genes encoding subunits of mitochondrial respiratory chain complexes following compound treatment in HL-1 cardiomyocytes as in b (n = 3 biologically independent samples for each condition). Red, upregulated genes; green, downregulated genes; fold change (log2) ≥ 1. (d) Western blot analysis of the indicated proteins in HL-1 cardiomyocytes treated with 5 μM ivermectin for 24 h. Data are presented as means ± SD (n = 3 biologically independent samples). ⁎p < 0.05 vs. vehicle by two-tailed unpaired Aspin-Welch's t-test. (e) Complex IV activity of isolated mitochondria isolated from HL-1 cardiomyocytes treated with 5 μM ivermectin for 24 h. Data are presented as means ± SD (n = 4 or 6 biologically independent samples). ⁎p < 0.05 vs. vehicle by two-tailed unpaired Aspin-Welch's t-test. (f) Mitochondrial ATP production in digitonin-permeabilized HL-1 cardiomyocytes treated with 5 μM ivermectin for 24 h. OA, oligomycin A (10 μM). Data are presented as means ± SD (n = 3 biologically independent samples). ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001 vs. vehicle by two-tailed unpaired Student's t-test. (g) Mitochondrial ATP production rate in digitonin-permeabilized HL-1 cardiomyocytes transfected with control siRNA or Cox6a2 siRNA followed by treatment with 5 μM ivermectin for 24 h. Data are presented as means ± SD (n = 6 biologically independent samples). ⁎⁎⁎p < 0.001 vs. vehicle by two-tailed unpaired Student's t-test.
3.6. Antihypertrophic Effects of Ivermectin in Human iPSC-derived Cardiomyocytes
To assess translatability to humans, we assessed the effects on a cardiac hypertrophy model using human iPSC-derived cardiomyocytes stimulated with ET-1 (Carlson et al., 2013). ET-1 enhanced proBNP levels, a pathological hypertrophy marker (Fig. 6a), which were significantly suppressed not only by ivermectin, but also by the importin β inhibitor, importazole, and the PAK1 inhibitor, IPA-3, (both ivermectin molecular targets were expressed in iPSC-derived cardiomyocytes) (Fig. 6b and c). ET-1-induced mRNA expression of BNP was also suppressed by these compounds (Fig. 6d), suggesting that ivermectin may act by inhibition of importin β and/or PAK1.
3.7. Induction of the Unfolded Protein Response (UPR) and Mitochondrial UPR (UPRmt) by Ivermectin in Human iPSC-derived Cardiomyocytes
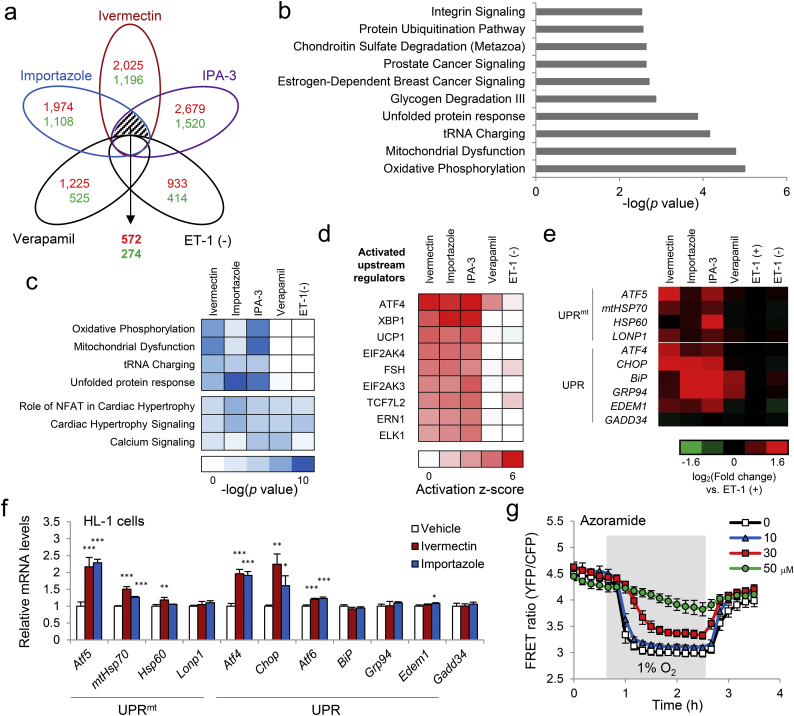
To examine how ivermectin affects iPSC cardiomyocytes, we analyzed transcriptomes in iPSC cardiomyocytes untreated or treated with ivermectin, importazole, IPA-3, or verapamil in the presence of ET-1. We identified 572 upregulated and 274 downregulated overlapping genes among the ivermectin, importazole, and IPA-3 treatment groups, but not in the verapamil or untreated groups (Fig. 7a). To identify specific pathways, we performed the IPA, which revealed that the genes commonly enriched in the ivermectin, importazole, and IPA-3 groups were involved in OXPHOS, tRNA charging, and UPR (Fig. 7b). Comparison analysis of differentially expressed genes also revealed that these pathways were enriched in the ivermectin, importazole, and IPA-3 groups (Fig. 7c). Upstream regulator analysis also identified the UPR regulators, activating transcription factor 4 (ATF4) and X-box binding protein 1 (XBP1) (Fig. 7d). UPRmt-related genes, such as ATF5 and mtHSP70 (Fiorese et al., 2016), were also upregulated by ivermectin, importazole, and IPA-3 (Fig. 7e). Expression levels of these genes were unchanged by ET-1 alone (Fig. 7e), suggesting that additional activation of UPR and/or UPRmt plays a protective role in ET-1-induced hypertrophic responses.
Fig. 7.
Induction of mitochondrial unfolded protein response (UPR) following mitochondrial ATP modulator treatment in human iPSC-derived cardiomyocytes. (a) Venn diagram of differentially expressed genes identified from AmpliSeq data in iPSC-derived cardiomyocytes untreated [ET-1 (−)], or treated with 3 μM ivermectin, 10 μM importazole, 10 μM IPA-3, or 1 μM verapamil in the presence of 10 nM ET-1 for 18 h (n = 4 biologically independent samples for each condition, fold change ≥ 1.5, p < 0.05 vs. ET-1 treatment). Red, the number of upregulated genes; green, the number of downregulated genes for each treatment. (b) Pathways enriched in commonly overlapped genes only among ivermectin, importazole, and IPA-3 were identified by Ingenuity Pathway Analysis (IPA). (c) The gene expression signature was subjected to the pathway association analysis (IPA pathways) and hierarchical cluster analysis. The heatmap shows representative IPA pathways enriched in two clusters (ivermectin, importazole, and IPA-3 groups, and all five groups). (d) Upstream analysis was performed using IPA and was subjected to hierarchical cluster analysis. The heatmap shows the top nine upstream regulators clustered into ivermectin, importazole, and IPA-3 groups. (e) Heatmap of AmpliSeq data of genes involved in mitochondrial UPR (UPRmt) and UPR in iPSC cardiomyocytes treated with the indicated compounds as in A. Red, upregulated genes; green, downregulated genes; fold change (log2) ≥ 1.6. (f) Relative mRNA levels of genes involved in UPRmt and UPR in HL-1 cardiomyocytes treated with the indicated compounds. Data are presented as means ± SD (n = 3 biologically independent samples). ⁎p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001 vs. vehicle by one-way ANOVA with Dunnett's multiple-comparison test. (g) FRET ratio of mito-ATeam stable HL-1 cardiomyocytes treated with azoramide. Data are presented as means ± SD (n = 3 biologically independent samples).
Some genes involved in UPR and UPRmt were also upregulated in HL-1 cardiomyocytes treated with ivermectin and importazole (Fig. 7f and Fig. S3c). It is noteworthy that the induction of C/EBP homologous protein (Chop) mRNA by ivermectin and importazole was less than that induced by known ER stressors, thapsigargin and tunicamycin (Fig. S3d), suggesting that ivermectin and importazole did not simply induce ER stress. These transcriptional profiles, therefore, strongly suggest that ivermectin and related compounds induce UPRmt and UPR. We then assessed the effect of azoramide, which increases chaperone capacity and improves ER function without inducing ER stress (Fu et al., 2015). Azoramide prevented mitochondrial ATP decline during hypoxia in a concentration-dependent manner (Fig. 7g), suggesting that enhanced ER function improves mitochondrial ATP homeostasis under hypoxic conditions. As ER-sarcoplasmic reticulum (SR) are important for calcium homeostasis and cardiomyocyte function, and ER-SR dysfunction is implicated in heart failure (Dorn and Scorrano, 2010), we tested whether ivermectin affects Ca2 + transients in beating primary cardiomyocytes. Indeed, ivermectin increased the peak of intracellular Ca2 + oscillations (Fig. S4). Together, these results suggest that ivermectin potentiates ER function via pro-survival UPR functions, preventing mitochondrial ATP decline under hypoxia and hypertrophic responses.
4. Discussion
Our unique phenotypic assays of mitochondrial ATP identified small molecules that maintain mitochondrial ATP levels under hypoxia with anti-hypertrophic effects in human iPSC cardiomyocytes, and that control ER homeostasis mediated by UPR. This study provides previously unappreciated insights into the regulation of mitochondrial ATP (Koopman et al., 2013; Schwarz et al., 2014) and mitochondrial-ER communication (Eisner et al., 2013), and has therapeutic implications for diseases involving mitochondrial dysfunction (Wang et al., 2016).
Mitochondrial dysfunction may be an underlying mechanism of cardiac remodeling and pathological cardiac hypertrophy in heart disease. Emerging evidence suggests that mitochondrial dysfunction causes cardiomyopathy and heart failure. During heart disease development and progression, oxygen supply to the heart decreases, leading to HIF-1α stabilization in patients with pathological hypertrophy (Krishnan et al., 2009). Hypoxia can reduce electron transport and proton pumping, leading to mitochondrial inner membrane depolarization and impaired ATP synthesis, causing generation of ROS (Giordano, 2005; Solaini et al., 2010). With the expectation that maintaining mitochondrial ATP production under hypoxia may improve cardiac function, we performed phenotypic screens in cardiomyocytes expressing a biosensor for mitochondrial ATP and identified small molecules, including ivermectin, with such effects. Although there have been reported drug screens based on mitochondrial function (Andreux et al., 2014) and protection against ischemia-reperfusion-induced cell death (Guo et al., 2012), none of these screens identified our hits, highlighting the uniqueness of our screening system that combined the mitochondrial ATP biosensor, mito-ATeam, and hypoxia in a high-throughput method.
Since mito-ATeam is expressed and detects ATP in the mitochondrial matrix, there are several possible mechanisms to maintain mitochondrial ATP levels under hypoxia: 1) Enhanced mitochondrial ATP synthesis, 2) increased ATP import and/or impaired ATP export, and 3) inhibition of ATP consumption. We found that small-molecule compounds, including approved drugs such as CsA (mPTP inhibitor) and deferoxamine (iron chelator), maintain mitochondrial ATP levels under hypoxia (Fig. 1f), suggesting that multiple mechanisms of action can modulate mitochondrial ATP levels. For instance, by binding to cyclophilin D and inhibiting the mPTP opening, CsA may prevent ATP export from the matrix to cytosol, resulting in the maintenance of mitochondrial matrix ATP levels during hypoxia. Since deferoxamine is known to stabilize HIF-1α, we tested FG-4592, which inhibits HIF-prolyl hydroxylases and thereby stabilizes HIF-1α. However, we found that FG-4592 did not prevent mitochondrial ATP decline (unpublished data), suggesting that deferoxamine's action does not result from HIF-1α stabilization. Given that a mitochondria-specific radical scavenger mito-TEMPO, but not NAC, protected mitochondrial ATP (Fig. 1f), we speculate that deferoxamine exerts its effect by chelating iron and blocking Fenton reactions, thereby inhibiting hydroxyl radical production (Xie et al., 2016). These results suggest that mitochondrial ROS is involved in mitochondrial ATP reduction under hypoxia.
Through the high-content phenotypic screening of the drug library, we identified ivermectin, which increases Cox6a2 expression levels and enhances Complex IV activity and mitochondrial ATP production (Fig. 5), thereby maintaining mitochondrial ATP levels under hypoxia (Fig. 2). Although ivermectin was shown to exert antitumor activities in various tumor types (Hashimoto et al., 2009; Liu et al., 2016; Nishio et al., 2016), ivermectin did not affect proliferation or activate caspase-3/7 in HL-1 cardiomyocytes (Fig. S2), indicating that the effects of ivermectin may be context-specific. Currently, the precise transcriptional regulation of Cox6a2 is not fully understood, and it remains to be determined how ivermectin and importazole induce the Cox6a2 transcript specifically among other subunits of the mitochondrial respiratory chain. Since Cox6a2 is a heart/muscle-specific isoform, heart/muscle-specific transcription factors, such as myocyte enhancer factor-2 (Mef2) (Wan and Moreadith, 1995), and/or transcriptional regulation may be involved. In addition, our transcriptomic analysis demonstrated upregulation of UPR-related transcription factors (Fig. 7). Indeed, gene promotor predictions (SABiosciences) indicate that, in addition to Mef2, Atf6 and Xbp1 may have binding sites in the mouse Cox6a2 promoter, but not in the Cox6a1 promotor, raising the possibility that these UPR-related transcription factors may be involved in the specific upregulation of Cox6a2 by ivermectin.
Among ivermectin's mammalian targets, associations of P2X4 receptor (Yang and Liang, 2012), PAK1 (Liu et al., 2011), and importin α/β (Cortés et al., 2010) with heart failure have been reported. We found that chemical inhibition of importin β by importazole also inhibited mitochondrial ATP reduction under hypoxia (Fig. 4b) and attenuated pathological cardiac hypertrophy (Fig. 6). It has been shown that importin α/β expression levels in the hearts of patients with ischemic and dilated cardiomyopathy were higher than those in healthy controls (Cortés et al., 2010). These findings indicate that importin β inhibition could be a promising target for the treatment of heart failure with hypertrophy. Due to the limited availability of specific compounds for other mammalian ivermectin targets, we could not conclude the degree to which those targets are involved in the effects of ivermectin on mitochondrial ATP modulation. Further investigations via knockdown or knockout of these genes are warranted.
Our results indicate that the anti-hypertrophic effects of ivermectin and its related compounds are associated with the pro-survival UPR that potentiates ER functions. UPRmt activation may also be involved; however, questions remain regarding the precise mechanisms by which these compounds induce UPRmt and by which UPRmt activation suppresses cardiomyocyte hypertrophy. For the former, a possibility includes importin α/β inhibition affecting nuclear-cytoplasmic shuttling regulators of UPRmt, such as CHOP and ATF5 (Chung et al., 2017; Fiorese et al., 2016). Although CHOP is known to mediate cardiomyocyte death and subsequent cardiac hypertrophy when highly expressed (Fu et al., 2010), a recent report sheds light on a critical role of CHOP in UPRmt and protection against muscle mitochondrial dysfunction (Chung et al., 2017). Moreover, it is not known whether ATF5 affects cardiac hypertrophy. Knockout or knockdown of these genes to assess their pathophysiological roles in cardiomyocytes is necessary. Given that azoramide prevents mitochondrial ATP decline under hypoxia, enhanced ER function, as demonstrated by elevated calcium transient amplitudes in cardiomyocytes (Fig. S4), may affect mitochondrial function.
In conclusion, we identified ivermectin, the approved anti-parasitic drug, using our unique phenotypic screen based on assessing mitochondrial ATP in cardiomyocytes under hypoxia. We found that ivermectin and its related compounds induce Cox6a2 expression, which may increase ATP production via OXPHOS, leading to anti-hypertrophic responses in human iPSC-derived cardiomyocytes. Our results provide therapeutic options for diseases with mitochondrial dysfunction, including heart failure.
Acknowledgments
Acknowledgments
We are grateful to Atsushi Miyawaki (RIKEN Brain Science Institute) for supervising the imaging studies. We thank Osaka University and Hiromi Imamura (Kyoto University) for providing ATeam plasmids and William Claycomb (Louisiana State University) for providing HL-1 cardiomyocytes. We also thank Tsuyoshi Nakamura, Masato Tsutsumi, and Takayoshi Matsubara (Yokogawa Electric Corporation) for customizing the imaging equipment for the hypoxia experiments and their excellent technical assistance with the imaging studies. We are grateful to Seigo Izumo for supporting and encouraging our research and a fruitful discussion. We thank Shuichi Takagahara for performing FACS; Mitsugu Nakata for the establishment of ES cells; Takashi Yano and Naoya Nishimura for the establishment of knock-in animals; Kunio Matsuoka for breeding mice; Tomohiro Itou for help with the isolation of primary cardiomyocytes and caspase-3/7 assay; Hirokazu Tozaki for help with RNA-seq; Koji Yamamoto for help with depositing the transcriptomic data into GEO; Kenichi Imahashi for critical reading of the manuscript; and Editage (www.editage.jp) for English language editing.
Funding Source
The study was supported by Takeda Pharmaceutical Company Limited.
Conflicts of Interest
The authors are/were employees of Takeda Pharmaceutical Company Limited.
Author Contributions
H.N. designed the research, performed the experiments, analyzed the data and wrote the paper. T.S., A.A., K.N. and M.M. performed experiments and edited/critiqued the manuscript. S.Y., K.K., K.M. and H.F. helped design and supervise specific experiments related to their areas of expertise and edited/critiqued the manuscript. M.N. supervised the research and edited/critiqued the manuscript. Y.T. directed the research, analyzed the data, and wrote the paper.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.09.022.
Contributor Information
Hiroaki Nagai, Email: hiroaki.nagai@takeda.com.
Yoshiyuki Tsujihata, Email: yoshiyuki.tsujihata@takeda.com.
Appendix A. Supplementary Data
References
- Adelsberger H., Lepier A., Dudel J. Activation of rat recombinant alpha(1)beta(2)gamma(2S) GABA(A) receptor by the insecticide ivermectin. Eur. J. Pharmacol. 2000;394:163–170. doi: 10.1016/s0014-2999(00)00164-3. [DOI] [PubMed] [Google Scholar]
- Ahuja P., Wanagat J., Wang Z., Wang Y., Liem D.A., Ping P., Antoshechkin I.A., Margulies K.B., MacLellan W.R. Divergent mitochondrial biogenesis responses in human cardiomyopathy. Circulation. 2013;127:1957–1967. doi: 10.1161/CIRCULATIONAHA.112.001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux P.A., Mouchiroud L., Wang X., Jovaisaite V., Mottis A., Bichet S., Moullan N., Houtkooper R.H., Auwerx J. A method to identify and validate mitochondrial modulators using mammalian cells and the worm C. elegans. Sci Rep. 2014;4:5285. doi: 10.1038/srep05285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries M., Van Damme P., Robberecht W., Van Den Bosch L. Ivermectin inhibits AMPA receptor-mediated excitotoxicity in cultured motor neurons and extends the life span of a transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis. 2007;25:8–16. doi: 10.1016/j.nbd.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Avkiran M., Marber M.S. Na +/H + exchange inhibitors for cardioprotective therapy: progress, problems and prospects. J. Am. Coll. Cardiol. 2002 doi: 10.1016/s0735-1097(02)01693-5. [DOI] [PubMed] [Google Scholar]
- Beer M., Seyfarth T., Sandstede J., Landschütz W., Lipke C., Köstler H., Von Kienlin M., Harre K., Hahn D., Neubauer S. Absolute concentrations of high-energy phosphate metabolites in normal, hypertrophied, and failing human myocardium measured noninvasively with 31P-SLOOP magnetic resonance spectroscopy. J. Am. Coll. Cardiol. 2002;40:1267–1274. doi: 10.1016/s0735-1097(02)02160-5. [DOI] [PubMed] [Google Scholar]
- Borchi E., Bargelli V., Stillitano F., Giordano C., Sebastiani M., Nassi P.A., d'Amati G., Cerbai E., Nediani C. Enhanced ROS production by NADPH oxidase is correlated to changes in antioxidant enzyme activity in human heart failure. Biochim. Biophys. Acta. 2010;1802:331–338. doi: 10.1016/j.bbadis.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Brown D.A., Perry J.B., Allen M.E., Sabbah H.N., Stauffer B.L., Shaikh S.R., Cleland J.G.F., Colucci W.S., Butler J., Voors A.A., Anker S.D., Pitt B., Pieske B., Filippatos G., Greene S.J., Gheorghiade M. Expert consensus document: mitochondrial function as a therapeutic target in heart failure. Nat. Rev. Cardiol. 2017;14:238–250. doi: 10.1038/nrcardio.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C., Koonce C., Aoyama N., Einhorn S., Fiene S., Thompson A., Swanson B., Anson B., Kattman S. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J. Biomol. Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- Chung H.K., Ryu D., Kim K.S., Chang J.Y., Kim Y.K., Yi H.S., Kang S.G., Choi M.J., Lee S.E., Jung S.B., Ryu M.J., Kim S.J., Kweon G.R., Kim H., Hwang J.H., Lee C.H., Lee S.J., Wall C.E., Downes M., Evans R.M., Auwerx J., Shong M. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 2017;216:149–165. doi: 10.1083/jcb.201607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb W.C., Lanson N.A., Stallworth B.S., Egeland D.B., Delcarpio J.B., Bahinski A., Izzo N.J. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés R., Roselló-Lletí E., Rivera M., Martínez-Dolz L., Salvador A., Azorín I., Portolés M. Influence of heart failure on nucleocytoplasmic transport in human cardiomyocytes. Cardiovasc. Res. 2010;85:464–472. doi: 10.1093/cvr/cvp336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetković R.S., Scott L.J. Dexrazoxane: a review of its use for cardioprotection during anthracycline chemotherapy. Drugs. 2005 doi: 10.2165/00003495-200565070-00008. [DOI] [PubMed] [Google Scholar]
- Dorn G.W., Scorrano L. Two close, too close: sarcoplasmic reticulum-mitochondrial crosstalk and cardiomyocyte fate. Circ. Res. 2010 doi: 10.1161/CIRCRESAHA.110.225714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner V., Csordas G., Hajnoczky G. Interactions between sarco-endoplasmic reticulum and mitochondria in cardiac and skeletal muscle - pivotal roles in Ca(2)(+) and reactive oxygen species signaling. J. Cell Sci. 2013;126:2965–2978. doi: 10.1242/jcs.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese C.J., Schulz A.M., Lin Y.F., Rosin N., Pellegrino M.W., Haynes C.M. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagg T.P., Enkvetchakul D., Koster J.C., Nichols C.G. Muscle KATP channels: recent insights to energy sensing and myoprotection. Physiol. Rev. 2010;90:799–829. doi: 10.1152/physrev.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H.Y., Okada K.I., Liao Y., Tsukamoto O., Isomura T., Asai M., Sawada T., Okuda K., Asano Y., Sanada S., Asanuma H., Asakura M., Takashima S., Komuro I., Kitakaze M., Minamino T. Ablation of C/EBP homologous protein attenuates endoplasmic reticulum-mediated apoptosis and cardiac dysfunction induced by pressure overload. Circulation. 2010;122:361–369. doi: 10.1161/CIRCULATIONAHA.109.917914. [DOI] [PubMed] [Google Scholar]
- Fu S., Yalcin A., Lee G.Y., Li P., Fan J., Arruda A.P., Pers B.M., Yilmaz M., Eguchi K., Hotamisligil G.S. Phenotypic assays identify azoramide as a small-molecule modulator of the unfolded protein response with antidiabetic activity. Sci. Transl. Med. 2015;7:292ra98. doi: 10.1126/scitranslmed.aaa9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvan D., Saltman P., Powell S.R. Cardiac reperfusion damage prevented by a nitroxide free radical. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4680–4684. doi: 10.1073/pnas.88.11.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F.J. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Invest. 2005 doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Olm-Shipman A., Walters A., Urciuoli W.R., Devito S., Nadtochiy S.M., Wojtovich A.P., Brookes P.S. A cell-based phenotypic assay to identify cardioprotective agents. Circ. Res. 2012;110:948–957. doi: 10.1161/CIRCRESAHA.111.263715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghikia A., Stapel B., Hoch M., Hilfiker-Kleiner D. STAT3 and cardiac remodeling. Heart Fail. Rev. 2011;16:35–47. doi: 10.1007/s10741-010-9170-x. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Messerli S.M., Sudo T., Maruta H. Ivermectin inactivates the kinase PAK1 and blocks the PAK1-dependent growth of human ovarian cancer and NF2 tumor cell lines. Drug Discov. Ther. 2009;3:243–246. [PubMed] [Google Scholar]
- Hausenloy D.J., Boston-Griffiths E.A., Yellon D.M. Cyclosporin A and cardioprotection: from investigative tool to therapeutic agent. Br. J. Pharmacol. 2012 doi: 10.1111/j.1476-5381.2011.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T., Tsutsui H., Kinugawa S., Suematsu N., Hayashidani S., Ichikawa K., Utsumi H., Machida Y., Egashira K., Takeshita A. Direct evidence for increased hydroxyl radicals originating from superoxide in the failing myocardium. Circ. Res. 2000;86:152–157. doi: 10.1161/01.res.86.2.152. [DOI] [PubMed] [Google Scholar]
- Imamura H., Nhat K.P., Togawa H., Saito K., Iino R., Kato-Yamada Y., Nagai T., Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Proctor W.R., Dunwiddie T.V., Labarca C., Lester H.A. Allosteric control of gating and kinetics at P2X(4) receptor channels. J. Neurosci. 1999;19:7289–7299. doi: 10.1523/JNEUROSCI.19-17-07289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioka H., Kato H., Fujikawa M., Tsukamoto O., Suzuki T., Imamura H., Nakano A., Higo S., Yamazaki S., Matsuzaki T., Takafuji K., Asanuma H., Asakura M., Minamino T., Shintani Y., Yoshida M., Noji H., Kitakaze M., Komuro I., Asano Y., Takashima S. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman W.J.H., Distelmaier F., Smeitink J.A., Willems P.H. OXPHOS mutations and neurodegeneration. EMBO J. 2013;32:9–29. doi: 10.1038/emboj.2012.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosyna F.K., Nagel M., Kluxen L., Kraushaar K., Depping R. The importin α/β-specific inhibitor Ivermectin affects HIF-dependent hypoxia response pathways. Biol. Chem. 2015;396:1357–1367. doi: 10.1515/hsz-2015-0171. [DOI] [PubMed] [Google Scholar]
- Krause R.M., Buisson B., Bertrand S., Corringer P.J., Galzi J.L., Changeux J.P., Bertrand D. Ivermectin: a positive allosteric effector of the alpha7 neuronal nicotinic acetylcholine receptor. Mol. Pharmacol. 1998;53:283–294. doi: 10.1124/mol.53.2.283. [DOI] [PubMed] [Google Scholar]
- Krishnan J., Suter M., Windak R., Krebs T., Felley A., Montessuit C., Tokarska-Schlattner M., Aasum E., Bogdanova A., Perriard E., Perriard J.C., Larsen T., Pedrazzini T., Krek W. Activation of a HIF1α-PPARγ Axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9:512–524. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Liu W., Zi M., Naumann R., Ulm S., Jin J., Taglieri D.M., Prehar S., Gui J., Tsui H., Xiao R.P., Neyses L., Solaro R.J., Ke Y., Cartwright E.J., Lei M., Wang X. Pak1 as a novel therapeutic target for antihypertrophic treatment in the heart. Circulation. 2011;124:2702–2715. doi: 10.1161/CIRCULATIONAHA.111.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Fang S., Sun Q., Liu B. Anthelmintic drug ivermectin inhibits angiogenesis, growth and survival of glioblastoma through inducing mitochondrial dysfunction and oxidative stress. Biochem. Biophys. Res. Commun. 2016;480:415–421. doi: 10.1016/j.bbrc.2016.10.064. [DOI] [PubMed] [Google Scholar]
- Luo M., Guan X., Luczak E.D., Lang D., Kutschke W., Gao Z., Yang J., Glynn P., Sossalla S., Swaminathan P.D., Weiss R.M., Yang B., Rokita A.G., Maier L.S., Efimov I.R., Hund T.J., Anderson M.E. Diabetes increases mortality after myocardial infarction by oxidizing CaMKII. J. Clin. Invest. 2013;123:1262–1274. doi: 10.1172/JCI65268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers D.E., Basha H.I., Koenig M.K. Mitochondrial cardiomyopathy: pathophysiology, diagnosis, and management. Tex. Heart Inst. J. 2013;40:385–394. [PMC free article] [PubMed] [Google Scholar]
- Nayler W.G., Ferrari R., Williams A. Protective effect of pretreatment with verapamil, nifedipine and propranolol on mitochondrial function in the ischemic and reperfused myocardium. Am. J. Cardiol. 1980;46:242–248. doi: 10.1016/0002-9149(80)90064-8. [DOI] [PubMed] [Google Scholar]
- Nelson E.A., Walker S.R., Kepich A., Gashin L.B., Hideshima T., Ikeda H., Chauhan D., Anderson K.C., Frank D.A. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood. 2008;112:5095–5102. doi: 10.1182/blood-2007-12-129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M., Sugimachi K., Goto H., Wang J., Morikawa T., Miyachi Y., Takano Y., Hikasa H., Itoh T., Suzuki S.O., Kurihara H., Aishima S., Leask A., Sasaki T., Nakano T., Nishina H., Nishikawa Y., Sekido Y., Nakao K., Shin-Ya K., Mimori K., Suzuki A. Dysregulated YAP1/TAZ and TGF-beta signaling mediate hepatocarcinogenesis in Mob1a/1b-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E71–80. doi: 10.1073/pnas.1517188113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevaidis I.A., Iliodromitis E.K., Vlahakos D., Tsiapras D.P., Nikolaidis A., Marathias A., Michalis A., Kremastinos D.T. Deferoxamine infusion during coronary artery bypass grafting ameliorates lipid peroxidation and protects the myocardium against reperfusion injury: immediate and long-term significance. Eur. Heart J. 2005;26:263–270. doi: 10.1093/eurheartj/ehi028. [DOI] [PubMed] [Google Scholar]
- Prabhakar N.R., Semenza G.L. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol. Rev. 2012;92:967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintens R., Singh S., Lemaire K., De Bock K., Granvik M., Schraenen A., Vroegrijk I.O., Costa V., Van Noten P., Lambrechts D., Lehnert S., Van Lommel L., Thorrez L., De Faudeur G., Romijn J.A., Shelton J.M., Scorrano L., Lijnen H.R., Voshol P.J., Carmeliet P., Mammen P.P., Schuit F. Mice deficient in the respiratory chain gene Cox6a2 are protected against high-fat diet-induced obesity and insulin resistance. PLoS One. 2013;8:e56719. doi: 10.1371/journal.pone.0056719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford N.B., Wan B., Richman A., Szczepaniak L.S., Li J.-L., Li K., Pfeiffer K., Schägger H., Garry D.J., Moreadith R.W. Cardiac dysfunction in mice lacking cytochrome-c oxidase subunit VIaH. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H726–H733. doi: 10.1152/ajpheart.00308.2001. [DOI] [PubMed] [Google Scholar]
- Ruiz-Meana M., Garcia-Dorado D., Pina P., Inserte J., Agulló L., Soler-Soler J. Cariporide preserves mitochondrial proton gradient and delays ATP depletion in cardiomyocytes during ischemic conditions. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H999–H1006. doi: 10.1152/ajpheart.00035.2003. [DOI] [PubMed] [Google Scholar]
- Schwarz K., Siddiqi N., Singh S., Neil C.J., Dawson D.K., Frenneaux M.P. The breathing heart - mitochondrial respiratory chain dysfunction in cardiac disease. Int. J. Cardiol. 2014 doi: 10.1016/j.ijcard.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Soderholm J.F., Bird S.L., Kalab P., Sampathkumar Y., Hasegawa K., Uehara-Bingen M., Weis K., Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaini G., Baracca A., Lenaz G., Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta Bioenerg. 2010;1797:1171–1177. doi: 10.1016/j.bbabio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Sreejit P., Kumar S., Verma R.S. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell. Dev. Biol. Anim. 2008;44:45–50. doi: 10.1007/s11626-007-9079-4. [DOI] [PubMed] [Google Scholar]
- Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D. a. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B., Moreadith R.W. Structural characterization and regulatory element analysis of the heart isoform of cytochrome c oxidase VIa. J. Biol. Chem. 1995;270:26433–26440. doi: 10.1074/jbc.270.44.26433. [DOI] [PubMed] [Google Scholar]
- Wang W., Karamanlidis G., Tian R. Novel targets for mitochondrial medicine. Sci. Transl. Med. 2016;8:326rv3. doi: 10.1126/scitranslmed.aac7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X., Kang R., Tang D. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Ooshima Y., Nakata M., Yano T., Matsuoka K., Watanabe S., Maeda R., Takahashi H., Takeyama M., Matsumoto Y., Hashimoto T. Generation of gene-targeted mice using embryonic stem cells derived from a transgenic mouse model of Alzheimer's disease. Transgenic Res. 2013;22:537–547. doi: 10.1007/s11248-012-9651-x. [DOI] [PubMed] [Google Scholar]
- Yang R., Liang B.T. Cardiac P2X4 receptors: targets in ischemia and heart failure? Circ. Res. 2012;111:397–401. doi: 10.1161/CIRCRESAHA.112.265959. [DOI] [PubMed] [Google Scholar]
- Yang T., Shen J.B., Yang R., Redden J., Dodge-Kafka K., Grady J., Jacobson K.A., Liang B.T. Novel protective role of endogenous cardiac myocyte P2X4 receptors in heart failure. Circ. Hear. Fail. 2014;7:510–518. doi: 10.1161/CIRCHEARTFAILURE.113.001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Nagasawa K., Munch C., Xu Y., Satterstrom K., Jeong S., Hayes S.D., Jedrychowski M.P., Vyas F.S., Zaganjor E., Guarani V., Ringel A.E., Gygi S.P., Harper J.W., Haigis M.C. Mitochondrial sirtuin network reveals dynamic SIRT3-dependent deacetylation in response to membrane depolarization. Cell. 2016;167:985–1000.e21. doi: 10.1016/j.cell.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziaeian B., Fonarow G.C. Epidemiology and aetiology of heart failure. Nat. Rev. Cardiol. 2016:1–11. doi: 10.1038/nrcardio.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.