Abstract
The 52-kDa FK506-binding protein (FKBP52), a regulator of steroid hormone receptor signaling, is potentially involved in a variety of hormone-dependent cancer types. The present study investigated the expression and clinical implications of FKBP52 in breast cancer. Immunohistochemistry was performed on samples from 145 breast cancer patients and on 66 unmatched breast non-cancerous tissues (as controls) to determine the expression level of FKBP52. Publicly available microarray and RNA-seq datasets used in the present study were downloaded from the European Bioinformatics Institute ArrayExpress. Kaplan-Meier survival analysis was also performed. FKBP52 expression was moderately higher in the tumors than that in the non-cancerous tissues, but this difference was not statistically significant (P=0.176). However, available microarray datasets exhibited a significant difference in FKBP52 mRNA levels between breast tumors and controls. In the 145 breast cancer patients, elevated FKBP52 expression was significantly associated with advanced Tumor-Node-Metastasis (TNM) stage (P=0.015), lymph node metastasis (P=0.015) and tumors with poor histological differentiation (P=0.047). FKBP52 expression was negatively associated with estrogen receptor expression (P=0.033), but positively associated with human epidermal growth factor receptor 2 expression (P=0.033). However, there was no association between FKBP52 and progesterone receptor expression. Survival analyses demonstrated that FKBP52 was indicative of a poor overall survival rate (P=0.026), which was consistent with the result of Kaplan-Meier analysis, exhibiting a negative association between the mRNA of FKBP52 and overall survival (OS) (P=0.044). Other than for FKBP52 [hazard ratio (HR), 2.315; 95% confidence interval (CI), 1.077–4.975; P=0.032], univariate analysis revealed that clinical stage exhibited a significant influence on the prognosis of the breast cancer patients (HR, 2.148; 95% CI, 1.011–4.566; P=0.047). However, multivariate analysis revealed that only clinical stage, not FKBP52, was an independent prognostic factor (HR, 2.721; 95% CI, 1.169–6.335; P=0.020). Patients were further classified according to their OS. Compared with the controls (3.94±2.992), FKBP52 expression in breast cancer patients with OS of ≤3 years (5.39±3.409; P=0.042) or OS of ≤5 years (5.88±3.473; P=0.005) was significantly increased, respectively. However, no significant difference in FKBP52 expression was observed between controls and individuals with an OS time of >3 years (4.84±3.769; P=0.109) or >5 years (5.32±3.372; P=0.090). Elevated FKBP52 expression may be involved in tumor progression and invasion, given its positive association with TNM stage and lymph node metastasis. Although it is not an independent predictor, FKBP52 has promise as a biological marker for estimating the progression of breast cancer.
Keywords: breast cancer, 52-kDa FK506-binding protein, immunohistochemistry
Introduction
FK506-binding proteins (FKBPs), known as such as they bind to the immunosuppressive drug FK506, were initially found to be intracellular receptors of immunosuppressive drugs (1). FKBPs are highly conserved proteins, owing to their peptidylprolyl isomerase (PPIase) domains, which catalyze the isomerization of peptidylprolyl imide bonds (from cis to trans) in protein substrates (2). In view of their isomerase activity and the capability to interact with other proteins, more attention has been focused on their modulatory function in several signal transduction pathways in the cell. A previous study supported the concept that certain FKBPs serve a role in cancer-associated pathways (3).
The FK506-binding protein 52 gene (FKBP52), an FKBP, is located at chromosome 12 (12p13.33) (4) and contains 10 exons and 9 introns spanning ~10 kb of genomic DNA. The FKBP52 protein contains a PPIase domain and a C-terminal tetratricopeptide repeat (TPR) domain. In addition to promoting binding to the immunosuppressive drug FK506, the PPIase domain is vital for correct protein folding. Through the TPR domain, FKBP52 binds the 90-kDa heat shock protein (Hsp90), and forms an Hsp90 co-chaperone. This complex regulates steroid receptor signaling, including regulation of receptor maturation, hormone binding and nuclear translocation, and is involved in a wide variety of endocrine-associated diseases (5,6). Through its functional domains, FKBP52 also interacts with other proteins (7). The most widely discussed role of FKBP52 is its regulation of steroid hormone receptor (SHR) activity in hormone-dependent cancer.
FKBP52 is proposed to inhibit the nuclear movement of the tumor suppressor protein p53 by forming p53-hsp90-immunophilin-dynein complexes, resulting in the inactivation of p53 (8). This indicates that FKBP52 may be able to promote cancer initiation and progression. In addition, an elevated level of FKBP52 was observed in several types of cancer, including prostate cancer, hepatocellular carcinoma and breast cancer (9–13); notably though, the majority of these studies focused on cancer cell lines. Furthermore, previous studies on the role of FKBP52 in breast cancer do not form a consensus of results. Certain studies reported that FKBP52 was expressed at a higher level in breast cancer and precancerous lesions (14,15), but another (16) reported that the expression of FKBP52 decreased in breast cancer cell line-formed mammospheres, suggesting that FKBP52 could elicit a tumor suppressor function. Therefore, the current study investigated the association of FKBP52 with clinical features and the outcome in patients with breast cancer.
Patients and methods
Patients and clinicopathological features
Archived paraffin-embedded pathological specimens, complete clinicopathological features and follow-up data were retrieved for 145 breast cancer patients (median age, 51 years; age range, 18–84 years) diagnosed in the Cancer Hospital of Shantou University Medical College (Shantou, China) between October 2001 and November 2011. All participating patients, initially diagnosed with invasive breast cancer, underwent surgery without radiation, chemo- or endocrine therapy. If patients were suffering from a different type of cancer, they were excluded from research group. The unmatched adjacent normal tissues of 66 patients were also obtained from surgical resections. Some patients were excluded from analysis due to lack of pathological data. Clinical Tumor-Node-Metastasis stage was grouped in accordance with the American Joint Committee on Cancer 6th Edition Cancer Staging Manual (2002) (17,18). In the present study, stage III and V disease were designated as being advanced stages, while stages I and II were designated as early stages (19). A previous study identified that the histological grade of breast cancer exhibited an effect on the prognosis (20). The most common method used to grade breast tumors is the Bloom Richardson grading system (also known as the Nottingham Grading System), which classifies the following groups: Grade 1 (G1), well-differentiated slow-growing; grade 2 (G2), moderately differentiated; and grade 3 (G3), poorly-differentiated highly proliferative (21–23). Well-differentiated G1 tumors are close to the cell of origin, and exhibit a low malignant grade. In comparison, poorly differentiated G3 tumors exhibit a poorer differentiation, increased degree of malignancy, and poor prognosis. G2 tumors are in between G2 and G3 (21–23). As certain samples were not stained for Ki-67, no further division of the luminal breast cancer specimens into luminal A or luminal B subtypes could be performed. Consequently, the individual breast subgroups were divided into three subgroups: Luminal, human epidermal growth factor receptor 2 (HER-2)-enriched and triple-negative breast cancer (TNBC). The observation period ranged between 7 and 151 months (median, 40 months). Informed consent for the use of their samples was obtained from all the patients. This study was approved by the Medical Ethics Committee of the Cancer Hospital of Shantou University Medical College.
Immunohistochemistry (IHC) assay
IHC for FKBP52 was performed using a standard Envision complex method (24). Briefly, sections (4 µm) were fixed in 10% buffered formalin at room temperature for 24 h and embedded in paraffin. The sections were deparaffinized by xylene for 1 h at room temperature, rehydrated using decreasing concentrations of ethanol (100, 95, 90, 80 and 70%, 5 min each) and washed in PBS. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide at room temperature for 30 min. Next, tissue sections were autoclaved at 121°C in citrate buffer (pH 6.0) for 10 min, and incubated with rabbit anti-FKBP52 monoclonal antibody (dilution, 1:100; catalog no. ab54991; Abcam, Cambridge, UK) at 4°C overnight. Slides were subsequently washed in PBS and incubated with biotinylated secondary antibody (GTVision™ I Detection System kit; Anti Mouse/Rabbit Detection System; Gene Tech Co., Ltd., Hong Kong, China; used as supplied) for 30 min at 37°C. Staining was performed using 3, 3-diaminobenzidine (DAB-0031/1031; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China; used as supplied) at room temperature for 2 or 3 min, and counterstained with hematoxylin (PT001; Shanghai Bogoo Biotechnology Co., Ltd., Shanghai, China; used as supplied) at room temperature for 2 min. A negative control was obtained by replacing the primary antibody with normal goat serum (used as provided, AR0009; Wuhan Boster Biological Technology, Ltd., Wuhan, China) at 4°C overnight.
IHC staining for FKBP52 was scored as previously described (25), by a combination of intensity (0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining) and proportion (0, <5% of tumor cells stained; 1, 5–25% cells stained; 2, 26–50% cells stained; 3, 51–75% cells stained; 4, >75% cells stained) scores. If the product of multiplication between staining intensity and the proportion of positive cells was >2 (the upper quartiles), expression was defined as FKBP52-positive (+), but if the score was ≤2, the sample was designated as FKBP52-negative (−). Two pathologists independently assessed the cellular location and intensity of immunostaining in each section, in a blinded manner.
Gene expression data
The microarray datasets employed in this study are publicly available from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) of the European Bioinformatics Institute, and include 4 independent cohorts of breast cancer [accession numbers: E-GEOD-42568 (26), E-GEOD-15852 (27), E-GEOD-21422 (28), E-GEOD-29044]. The CEL files containing the raw data from each experiment were directly downloaded from the ArrayExpress website. Details of these datasets are summarized in Table I.
Table I.
Independent datasets from ArrayExpress.
| Sample size, n | |||||
|---|---|---|---|---|---|
| Accession number | Array | Control | Breast cancer | Log-2 FC (cases/controls)a | P-value |
| E-GEOD-42568 | HG-U133_Plus_2 | 17 | 104 | 0.777 | <0.001 |
| E-GEOD-21422 | HG-U133A | 5 | 14 | 0.626 | 0.044 |
| E-GEOD-15852 | HG-U133A | 43 | 43 | 0.138 | <0.001 |
| E-GEOD-29044 | HG-U133_Plus_2 | 49 | 43 | 0.343 | 0.001 |
Fold-change of Log-2 mRNA signal intensity of 52-kDa FK506-binding protein between tumor tissue and healthy tissue was measured.
In the present study, KM Plotter (http://kmplot.com/analysis/), a tool for themeta-analysis-based biomarker assessment that includes gene expression and survival data of more than 4,000 breast cancer patients was used (29). The tool was used to perform Kaplan Meier survival analysis to further assess the association between FKBP52 mRNA expression and OS. Patients with breast cancer were split by the median expression of FKBP52 into two groups, namely patients with high or low expression of FKBP52.
Statistical analysis
Data are expressed as the mean ± standard deviation. Statistical analyses were performed using software SPSS (version 13.0) (SPSS, Inc., Chicago, IL, USA) and R (version 3.0.2; www.r-project.org). The difference in FKBP52 expression between tumors and non-cancerous tissues was detected by Mann-Whitney U test, and the difference in FKBP52 mRNA expression retrieved from online datasets between breast cancer cases and healthy controls included in this study was detected by an unpaired Student's t-test. Associations between FKBP52 expression and clinicopathological features were analyzed using the χ2 test. Survival curves were calculated using the Kaplan-Meier method with log-rank test. Cox regression analysis was used to study the effects of FKBP52 expression on overall survival (OS) time. OS was defined as the time from surgery to the date of last contact or mortality from any cause. The three-year survival period or five-year survival period is a professional term for evaluating the survival of a tumor patient (30,31). Breast cancer patients with an OS time of >3 or 5 years were defined as the better prognostic group, while those with an OS time of ≤3 or 5 years were classified into the poor prognostic group. Student's t-test was used for differential expression analysis of FKBP52 between control and better/poor prognosis-tumor samples. For gene expression microarray analyses, data were normalized using Robust Multi-array Analysis with R-package ‘affy’ (32). The normalized expression values (on a log-2 scale) of probes representing the same gene were averaged. A two-tailed P-value of <0.05 was considered to indicate a statistically significant difference.
Results
Difference in FKBP52 expression between breast tumors and normal tissues
FKBP52 expression was evaluated in 145 breast cancer patient samples and 66 unmatched breast non-cancerous tissues by IHC. As shown in Fig. 1, different staining intensities were observed; positive staining of FKBP52 was mainly observed in the cytoplasm of the majority of tumor cells. FKBP52 expression was slightly higher in the tumors than that in the non-cancerous tissues (Fig. 2), but this difference was not significant (Table II; P=0.176). Conversely, different independent datasets from public databases demonstrated that a significant upregulation of FKBP52 mRNA was found in breast tumors compared with that in the corresponding controls (Table I).
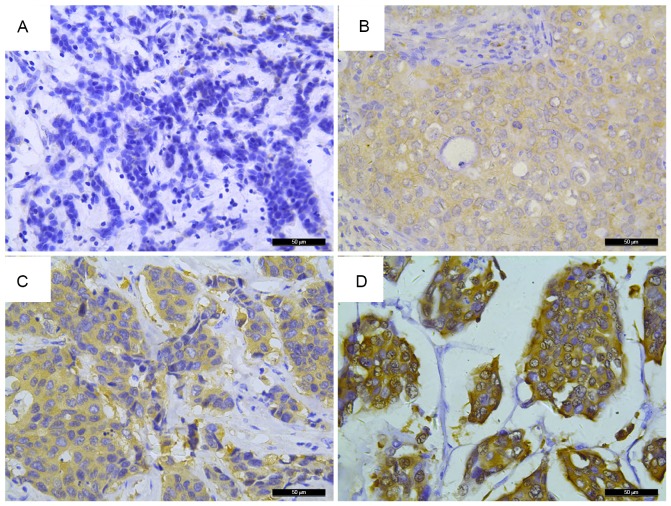
Figure 1.
FKBP52 expression in breast tumors. Immunohistochemical analysis detected (A) an absence of FKBP52 staining in breast tumors; (B) weak staining of FKBP52 in breast tumors; (C) moderate staining of FKBP52 in breast tumors; and (D) strong FKBP52 staining in breast tumors. Original magnification, ×400. FKBP52, 52-kDa FK506-binding protein.
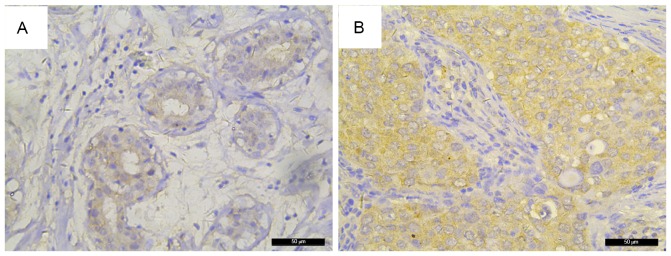
Figure 2.
FKBP52 expression in breast cancer and unmatched breast non-cancerous tissues. Immunohistochemical analysis found that FKBP52 expression was significantly higher in breast non-cancerous tissues. (A) Non-cancerous breast tissue; (B) breast cancer tissue. Original magnification, ×400. FKBP52, 52-kDa FK506-binding protein.
Table II.
Differential expression of 52-kDa FK506-binding protein in 66 unmatched breast non-cancerous samples and 145 breast tumor tissues.
| Term | Expression levela | Median (P25, P75) | Mann-Whitney U-value | P-value |
|---|---|---|---|---|
| Healthy tissue | 3.94±2.992 | 4.00 (1.75, 6.00) | 4234.5 | 0.176 |
| Tumor tissue | 4.75±3.605 | 4.00 (2.00, 8.00) |
Mean ± standard deviation. P25/P75, upper and lower quartiles.
Association of FKBP52 with clinicopathological features in a cohort of 145 breast cancer patients
To evaluate the association of FKBP52 expression with clinicopathological features, tumor sections from 145 primary breast cancer patients were subsequently divided into two groups according to their IHC scores: 50 (34.5%) tumors exhibited low expression of FKBP52 [FKBP52(−) group] and 95 (65.5%) tumors exhibited high expression of FKBP52 [FKBP52(+) group]. As shown in Table III, FKBP52(+) was detected in 36 patients with N1 stage disease (36/66, 54.5%) and 50 patients with N2-N3 stage disease (50/67, 74.6%), which demonstrated that the FKBP52 (+) rate was significantly higher in patients with more affected lymph nodes (P=0.015). Similarly, elevated FKBP52 expression was associated with advanced TNM stage (57/78, 73.1% vs. 26/50, 52.0%; P=0.015). Additionally, FKBP52(+) rate was positively associated with patients with a histological grade of G3 (P=0.047). Patients with G3 tumors had the highest FKBP52 (+) rate (54/74, 73.0%), successively followed by those with G2 (21/36, 58.3%) and G1 tumors (8/18, 44.4%). For molecular receptors, FKBP52 was negatively associated with estrogen receptor (ER) expression (47/82, 57.3% vs. 42/56, 75.0%; P=0.033), but positively associated with HER-2 expression (42/56, 75% vs. 47/82, 57.3%; P=0.033). Similarly, there was a statistically significant difference among the three breast cancer subgroups (P=0.036): FKBP52 expression was the highest in the HER-2-enriched subtype of breast cancer (23/27, 85.2%), second highest in the luminal-subtype breast cancer (17/27, 63.0%) and third highest in TNBC (48/83, 57.8%). By contrast, no significant differences were found between FKBP52 expression and other clinicopathological features assessed in this study, including age, tumor size and PR (Table III).
Table III.
Association between FKBP52 expression and clinicopathological features.
| FKBP52 expression, n (%) | |||||
|---|---|---|---|---|---|
| Clinicopathological feature | Patients, n (%) | Negative (n=50) | Positive (n=95) | χ2 | P-value |
| Age at diagnosis, years | |||||
| ≤50 | 69 (49.3) | 28 (40.6) | 41 (59.4) | 1.862 | 0.172 |
| >50 | 71 (50.7) | 21 (29.6) | 50 (70.4) | ||
| Primary tumor stage | |||||
| T1-T2 | 88 (66.2) | 32 (36.4) | 56 (63.6) | 0.120 | 0.729 |
| T3-T4 | 45 (33.8) | 15 (33.3) | 30 (66.7) | ||
| Regional lymph node stage | |||||
| N0-N1 | 66 (49.6) | 30 (45.5) | 36 (54.5) | 5.868 | 0.015 |
| N2-N3 | 67 (50.4) | 17 (25.4) | 50 (74.6) | ||
| TNM stage | |||||
| 1–2 | 50 (39.1) | 24 (48.0) | 26 (52.0) | 5.937 | 0.015 |
| 3–4 | 78 (60.9) | 21 (26.9) | 57 (73.1) | ||
| Histological grade | |||||
| G1 | 18 (14.1) | 10 (55.6) | 8 (44.4) | 6.1 | 0.047 |
| G2 | 36 (28.1) | 15 (41.7) | 21 (58.3) | ||
| G3 | 74 (57.8) | 20 (27.0) | 54 (73.0) | ||
| Estrogen receptor | |||||
| Negative | 56 (40.6) | 14 (25.0) | 42 (75.0) | 4.544 | 0.033 |
| Positive | 82 (59.4) | 35 (42.7) | 47 (57.3) | ||
| Progesterone receptor | |||||
| Negative | 71 (51.4) | 21 (29.6) | 50 (70.4) | 2.245 | 0.134 |
| Positive | 67 (48.6) | 28 (41.8) | 39 (58.2) | ||
| HER-2 | |||||
| Negative | 82 (59.4) | 35 (42.7) | 47 (57.3) | 4.544 | 0.033 |
| Positive | 56 (40.6) | 14 (25.0) | 42 (75.0) | ||
| Molecular subtypes | |||||
| Luminal | 83 (60.6) | 35 (42.2) | 48 (57.8) | ||
| HER-2 enriched | 27 (19.7) | 4 (14.8) | 23 (85.2) | ||
| TNBC | 27 (19.7) | 10 (37.0) | 17 (63.0) | 6.659 | 0.036 |
FKBP52, 52-kDa FK506-binding protein; TNM, Tumor-Node-Metastasis; HER-2, human epithelial growth factor receptor 2; TNBC, triple-negative breast cancer.
Effect of FKBP52 expression on the OS of breast cancer patients
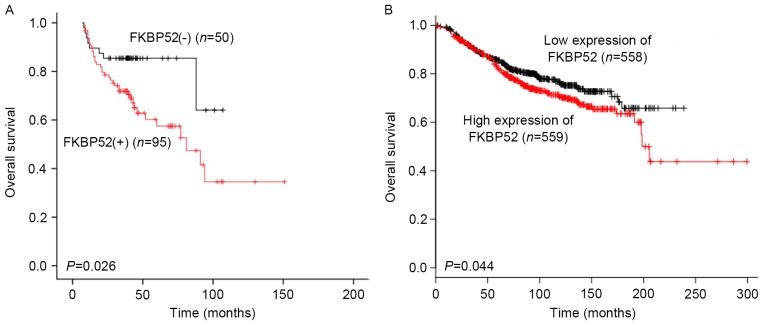
To examine whether the expression status of FKBP52 has any prognostic value for breast cancer, univariate and multivariate analyses using the Kaplan-Meier method and Cox regression analysis were performed. As shown in Fig. 3A, the Kaplan-Meier survival curve revealed that for the 145 breast cancer patients, the OS rate in the FKBP52(−) group was significantly higher than that in the FKBP52(+) group (P=0.026). The KM Plotter tool was used to further assess the association between the mRNA expression of FKBP52 and the OS of the breast cancer patients. As shown in Fig. 3B, low expression of FKBP52 predicted a significantly better OS rate in the breast cancer patients (P=0.044). In addition to FKBP52 (HR, 2.315; 95% CI, 1.077–4.975; P=0.032), TNM stage was another adverse predictor for breast cancer patients (HR: 2.148; 95% CI: 1.011–4.566, P=0.047) upon univariate analysis. However, multivariate analysis demonstrated that TNM stage, but not FKBP52, was an independent prognostic factor (HR, 2.721; 95% CI, 1.169–6.335; P=0.020) (Table IV).
Figure 3.
Kaplan-Meier survival analysis plot of the effect of FKBP52 expression on survival. (A) Effect of FKBP52 expression as assessed by immunohistochemical analysis on OS rate in all breast cancer patients. (B) Effect of FKBP52 mRNA expression on OS of breast cancer patients included in the Kaplan-Meier plot. FKBP52, 52-kDa FK506-binding protein; OS, overall survival.
Table IV.
Cox proportional hazard regression model analysis of overall survival in patients with breast cancer.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Terms | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (>50 years) | 1.011 (0.562–1.818) | 0.972 | ||
| Tumor size (T3-T4) | 1.630 (0.852–3.117) | 0.140 | ||
| Nodal status (N2-N3) | 1.594 (0.819–3.102) | 0.170 | ||
| TNM stage (3–4) | 2.148 (1.011–4.566) | 0.047 | 2.721 (1.169–6.335) | 0.020 |
| Histological grade | 0.427 | |||
| G2 | 0.627 (0.290–1.356) | 0.236 | ||
| G3 | 0.672 (0.254–1.778) | 0.423 | ||
| FKBP52 (positive) | 2.315 (1.077–4.975) | 0.032 | 2.343 (0.956–5.742) | 0.063 |
| Estrogen receptor (positive) | 0.926 (0.503–1.707) | 0.806 | ||
| Progesterone receptor (positive) | 0.664 (0.358–1.230) | 0.193 | ||
| HER-2 (positive) | 1.266 (0.689–2.327) | 0.447 | ||
HR, hazard ratio; CI, confidence interval; TNM, Tumor-Node-Metastasis; FKBP52, 52-kDa FK506-binding protein; HER-2, human epithelial growth factor receptor 2.
Association between controls and better/poor prognosis-tumor samples
Breast cancer patients were divided into two groups according to their OS: Better-prognosis group (OS >3 or 5 years) and poor-prognosis group (OS ≤3 or 5 years). As shown in Table V, FKBP52 expression in the patients with an OS ≤3-years (5.39±3.409; P=0.042) and an OS ≤5-years (5.88±3.473; P=0.005) was significantly higher than that in the controls (3.94±2.992). However, no statistical significance was determined for the comparison between the controls and the better-prognosis group (OS >3 years: 4.84±3.769, P=0.109; OS >5 years: 5.32±3.372, P=0.090).
Table V.
Difference between controls and poor/better prognosis tumor patients.
| Group | Patients, n | Survival timea | P-valueb |
|---|---|---|---|
| Controls | 66 | 3.94±2.992 | |
| Patients by OS time, yearsc | |||
| ≤3 | 33 | 5.39±3.409 | 0.042 |
| >3 | 93 | 4.84±3.769 | 0.109 |
| ≤5 | 40 | 5.88±3.473 | 0.005 |
| >5 | 28 | 5.32±3.372 | 0.090 |
Data presented as mean ± standard deviation.
Comparison of controls and better/poor prognosis-tumor groups.
OS >3 or 5 years represented the better-prognostic group; OS ≤3 or 5 years represented the poor-prognostic group. OS, overall survival.
Discussion
As a component of the Hsp90 co-chaperones, FKBP52 potentiates the gene activation by glucocorticoid (5), androgen (33,34) and progesterone (35) receptors through its involvement in nuclear receptor maturation. Our understanding of the mechanisms by which Hsp90 co-chaperones regulate SHR signaling and the role that they play in endocrine-associated physiological processes has progressed, although certain previous studies controversially suggested that FKBP52 had a potential role in breast cancer (14–16). In the present study, IHC was used to assess the difference in FKBP52 expression at the protein level between breast cancer and non-cancerous tissues. FKBP52 was slightly upregulated in breast tumor samples, but this did not reach statistical significance. However, by utilizing publicly available gene expression data, FKBP52 expression was found to be higher in breast tumors (Table I). The limited sample size in the current study may have led to the inconsistency in protein and mRNA expression levels observed between the samples analyzed and those obtained from previously generated datasets. Further study of FKBP52 expression with a larger sample size is therefore required to confirm the results of the present study.
In addition to the fact that FKBP52 was observed to be upregulated in breast cancer cell lines, previous reports further demonstrated that FKBP52 was predominantly expressed in ER-positive cells (14,36). However, IHC analysis in the present study revealed that the expression of FKBP52 was negatively associated with ER expression, but positively associated with HER-2 expression. These inconsistencies may reflect the complex association between FKBP52 and molecular receptors in breast cancer. The difference between cancer cells and organisms could also result from the cytoplasmic retention of ER by Hsp90-cochaperons; only nuclear staining of ER is assessed clinically. The difference in FKBP52 expression between luminal, HER-2-enriched and triple-negative-subtype breast cancer also indicated that higher FKBP52 expression was associated with negative ER or positive HER-2 expression. Considering its clinical significance FKBP52 may be an adverse prognostic factor, although further studies are required to confirm this. The current study also indicated that elevated FKBP52 expression tended to be observed in breast cancer patients with lymph node metastasis, poor cell differentiation and advanced TNM stage, suggesting that FKBP52 may be a useful biomarker for the evaluation of differentiation and metastasis in human breast carcinoma. FKBP52 could also serve as a novel therapeutic target for breast cancer patients. Through IHC analysis using tissue microarrays, Liu et al (11) found that FKBP52 regulation was relevant to hepatocellular carcinoma staging, with relatively high expression at stages I and II, but a marked decline at stage III, demonstrating that FKBP52 could be used for early HCC diagnosis. When considering that it was highly expressed in breast cancer patients with advanced TNM stage, FKBP52 was hypothesized to be expressed at different levels depending on the tissue types, as proposed by previous studies (10,37,38).
In the present study, FKBP52 protein expression was an adverse predictor for OS in breast cancer patients. However, multivariate survival analysis demonstrated that FKBP52 was not an independent factor, partially due to its association with TNM stage. Analysis using a publically available online tool, KM Plotter, revealed a valid association between FKBP52 mRNA expression and OS. This association may be caused by FKBP52-associated breast cancer resistance to chemotherapies (39). Thus, FKBP52 could be used as an objective biological marker to estimate the outcome of breast cancer. There was no statistically significant association between lymph node metastasis and OS, which may be a result of the insufficient sample size in the current study. A cohort with a larger sample size would be required for a further study.
The present study revealed that FKBP52 may serve a role in promoting breast cell growth, and could be one of the key factors affecting the prognosis of breast cancer patients. However, it cannot be excluded that FKBP52 is a biomarker for increased breast cancer risk. A comprehensive functional study should therefore be performed to elucidate this association.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81272931 and 81572588), the Guangdong Provincial Key Laboratory, Guangdong Provincial Natural Science Foundation (grant no. S2013010015969) and a Youth Research Grant from Shantou University Medical College Cancer Hospital (grant no. 2014/10).
References
- 1.Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature. 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 2.Theuerkorn M, Fischer G, Schiene-Fischer C. Prolyl cis/trans isomerase signalling pathways in cancer. Curr Opin Pharmacol. 2011;11:281–287. doi: 10.1016/j.coph.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Romano MF. FKBPs: Opportunistic modifiers or active players in cancer? Curr Opin Pharmacol. 2011;11:279–280. doi: 10.1016/j.coph.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Cioffi DL, Hubler TR, Scammell JG. Organization and function of the FKBP52 and FKBP51 genes. Curr Opin Pharmacol. 2011;11:308–313. doi: 10.1016/j.coph.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T, Gaber R, Picard D, Smith DF. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 2003;22:1158–1167. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirkl F, Buchner J. Functional analysis of the Hsp90-associated human peptidyl prolyl cis/trans isomerases FKBP51, FKBP52 and Cyp40. J Mol Biol. 2001;308:795–806. doi: 10.1006/jmbi.2001.4595. [DOI] [PubMed] [Google Scholar]
- 7.Davies TH, Sánchez ER. FKBP52. Int J Biochem Cell Biol. 2005;37:42–47. doi: 10.1016/j.biocel.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Galigniana MD, Harrell JM, O'Hagen HM, Ljungman M, Pratt WB. Hsp90-binding immunophilins link p53 to dynein during p53 transport to the nucleus. J Biol Chem. 2004;279:22483–22489. doi: 10.1074/jbc.M402223200. [DOI] [PubMed] [Google Scholar]
- 9.Teiten MH, Gaigneaux A, Chateauvieux S, Billing AM, Planchon S, Fack F, Renaut J, Mack F, Muller CP, Dicato M, Diederich M. Identification of differentially expressed proteins in curcumin-treated prostate cancer cell lines. OMICS. 2012;16:289–300. doi: 10.1089/omi.2011.0136. [DOI] [PubMed] [Google Scholar]
- 10.Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu Q, Xu GJ, Song JD, Zhao FK. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121:2596–2605. doi: 10.1002/ijc.23016. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Li C, Xing Z, Yuan X, Wu Y, Xu M, Tu K, Li Q, Wu C, Zhao M, Zeng R. Proteomic mining in the dysplastic liver of WHV/c-myc mice-insights and indicators for early hepatocarcinogenesis. FEBS J. 2010;277:4039–4053. doi: 10.1111/j.1742-4658.2010.07795.x. [DOI] [PubMed] [Google Scholar]
- 12.Guerrero-Preston R, Hadar T, Ostrow KL, Soudry E, Echenique M, Ili-Gangas C, Pérez G, Perez J, Brebi-Mieville P, Deschamps J, et al. Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncol Rep. 2014;32:505–512. doi: 10.3892/or.2014.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostrow KL, Park HL, Hoque MO, Kim MS, Liu J, Argani P, Westra W, Van Criekinge W, Sidransky D. Pharmacologic unmasking of epigenetically silenced genes in breast cancer. Clin Cancer Res. 2009;15:1184–1191. doi: 10.1158/1078-0432.CCR-08-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward BK, Mark PJ, Ingram DM, Minchin RF, Ratajczak T. Expression of the estrogen receptor-associated immunophilins, cyclophilin 40 and FKBP52, in breast cancer. Breast Cancer Res Treat. 1999;58:267–280. doi: 10.1023/A:1006390804515. [DOI] [PubMed] [Google Scholar]
- 15.Desmetz C, Bascoul-Mollevi C, Rochaix P, Lamy PJ, Kramar A, Rouanet P, Maudelonde T, Mangé A, Solassol J. Identification of a new panel of serum autoantibodies associated with the presence of in situ carcinoma of the breast in younger women. Clin Cancer Res. 2009;15:4733–4741. doi: 10.1158/1078-0432.CCR-08-3307. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Zhao F, Cui Y. Proteomics using mammospheres as a model system to identify proteins deregulated in breast cancer stem cells. Curr Mol Med. 2013;13:459–463. doi: 10.2174/1566524011313030015. [DOI] [PubMed] [Google Scholar]
- 17.Connolly JL. Changes and problematic areas in interpretation of the AJCC cancer staging manual, 6th edition, for breast cancer. Arch Pathol Lab Med. 2006;130:287–291. doi: 10.5858/2006-130-287-CAPAII. [DOI] [PubMed] [Google Scholar]
- 18.Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark G, Edge SB, Hayes DF, et al. Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol. 2002;20:3628–3636. doi: 10.1200/JCO.2002.02.026. [DOI] [PubMed] [Google Scholar]
- 19.Hwang SB, Bae JW, Lee HY, Kim HY. Circulating tumor cells detected by RT-PCR for CK-20 before surgery indicate worse prognostic impact in triple-negative and HER2 subtype breast cancer. J Breast Cancer. 2012;15:34–42. doi: 10.4048/jbc.2012.15.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller DV, Leontovich AA, Lingle WL, Suman VJ, Mertens ML, Lillie J, Ingalls KA, Perez EA, Ingle JN, Couch FJ, Visscher DW. Utilizing nottingham prognostic index in microarray gene expression profiling of breast carcinomas. Mod Pathol. 2004;17:756–764. doi: 10.1038/modpathol.3800114. [DOI] [PubMed] [Google Scholar]
- 21.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 22.Pereira H, Pinder SE, Sibbering DM, Galea MH, Elston CW, Blamey RW, Robertson JF, Ellis IO. Pathological prognostic factors in breast cancer. IV: Should you be a typer or a grader ? A comparative study of two histological prognostic features in operable breast carcinoma. Histopathology. 1995;27:219–226. doi: 10.1111/j.1365-2559.1995.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 23.Parham DM. Mitotic activity and histological grading of breast cancer. Pahtol Annu. 1995;30:189–207. [PubMed] [Google Scholar]
- 24.Patel RM, Folpe AL. Immunohistochemistry for human telomerase reverse transcriptase catalytic subunit (hTERT): A study of 143 benign and malignant soft tissue and bone tumours. Pathology. 2009;41:527–532. doi: 10.1080/00313020903121614. [DOI] [PubMed] [Google Scholar]
- 25.Han YP, Ma CK, Wang SQ, Enomoto A, Zhao Y, Takahashi M, Ma J. Evaluation of osteopontin as a potential biomarker for central nervous system embryonal tumors. J Neurooncol. 2014;119:343–351. doi: 10.1007/s11060-014-1484-4. [DOI] [PubMed] [Google Scholar]
- 26.Clarke C, Madden SF, Doolan P, Aherne ST, Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown J, et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 27.Ni Pau IB, Zakaria Z, Muhammad R, Abdullah N, Ibrahim N, Emran Aina N, Abdullah Hisham N, Hussain Syed SN. Gene expression patterns distinguish breast carcinomas from normal breast tissues: The Malaysian context. Pathol Res Pract. 2010;206:223–228. doi: 10.1016/j.prp.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li XP, Cao GW, Sun Q, Yang C, Yan B, Zhang MY, Fu YF, Yang LM. Cancer incidence and patient survival rates among the residents in the Pudong New Area of Shanghai between 2002 and 2006. Chin J Cancer. 2013;32:512–519. doi: 10.5732/cjc.012.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer suevival in China, 2003–2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 32.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 33.De Leon JT, Iwai A, Feau C, Garcia Y, Balsiger HA, Storer CL, Suro RM, Garza KM, Lee S, Kim YS, et al. Targeting the regulation of androgen receptor signaling by the heat shock protein 90 cochaperone FKBP52 in prostate cancer cells. Proc Natl Acad Sci USA. 2011;108:11878–11883. doi: 10.1073/pnas.1105160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung-Flynn J, Prapapanich V, Cox MB, Riggs DL, Suarez-Quian C, Smith DF. Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol Endocrinol. 2005;19:1654–1666. doi: 10.1210/me.2005-0071. [DOI] [PubMed] [Google Scholar]
- 35.Hirota Y, Tranguch S, Daikoku T, Hasegawa A, Osuga Y, Taketani Y, Dey SK. Deficiency of immunophilin FKBP52 promotes endometriosis. Am J Pathol. 2008;173:1747–1757. doi: 10.2353/ajpath.2008.080527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Mark PJ, Ward BK, Minchin RF, Ratajczak T. Estradiol-regulated expression of the immunophilins cyclophilin 40 and FKBP52 in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2001;284:219–225. doi: 10.1006/bbrc.2001.4952. [DOI] [PubMed] [Google Scholar]
- 37.Solassol J, Mange A, Maudelonde T. FKBP family proteins as promising new biomarkers for cancer. Curr Opin Pharmacol. 2011;11:320–325. doi: 10.1016/j.coph.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Ott M, Litzenburger UM, Rauschenbach KJ, Bunse L, Ochs K, Sahm F, Pusch S, Opitz CA, Blaes J, von Deimling A, et al. Suppression of TDO-mediated tryptophan catabolism in glioblastoma cells by a steroid-responsive FKBP52-dependent pathway. Glia. 2015;63:78–90. doi: 10.1002/glia.22734. [DOI] [PubMed] [Google Scholar]
- 39.Yang WS, Moon HG, Kim HS, Choi EJ, Yu MH, Noh DY, Lee C. Proteomic approach reveals FKBP4 and S100A9 as potential prediction markers of therapeutic response to neoadjuvant chemotherapy in patients with breast cancer. J Proteome Res. 2012;11:1078–1088. doi: 10.1021/pr2008187. [DOI] [PubMed] [Google Scholar]