Abstract
Dental caries, which affects billions of people, is a chronic infectious disease that involves Streptococcus mutans, which is nevertheless a poor predictor of individual caries development. We therefore investigated if adhesin types of S.mutans with sucrose-independent adhesion to host DMBT1 (i.e. SpaP A, B or C) and collagen (i.e. Cnm, Cbm) match and predict individual differences in caries development. The adhesin types were measured in whole saliva by qPCR in 452 12-year-old Swedish children and related to caries at baseline and prospectively at a 5-year follow-up. Strains isolated from the children were explored for genetic and phenotypic properties. The presence of SpaP B and Cnm subtypes coincided with increased 5-year caries increment, and their binding to DMBT1 and saliva correlated with individual caries scores. The SpaP B subtypes are enriched in amino acid substitutions that coincided with caries and binding and specify biotypes of S. mutans with increased acid tolerance. The findings reveal adhesin subtypes of S. mutans that match and predict individual differences in caries development and provide a rationale for individualized oral care.
Keywords: Streptococcus mutans, Dental caries, Chronic infections, Virulence, SpaP, Adhesion
Highlights
-
•
Adhesin subtypes of Streptococcus mutans match and predict individual caries development.
-
•
Adhesin binding to salivary DMBT1 correlates with individual caries scores.
-
•
The adhesin types coincide with distinct biotypes of S. mutans.
Dental caries, which affects billions of people, involves the bacterium Streptococcus mutans, which is nevertheless a poor predictor of caries development. The present findings provide the first evidence that S. mutans adhesin subtypes match and predict individual 5-year caries development in Swedish children. The binding strength of the adhesin subtypes correlates with individual caries scores, and the adhesin subtypes specify biotypes of S. mutans that also differ in acid tolerance. The present findings provide a rationale for individualized oral care and improved systemic health because chronic caries infection and carrying high-virulence strains pose a systemic disease risk.
1. Introduction
Dental caries, a persistent infectious disease, affects billions of people with large individual differences in numbers of caries lesions and activity (Selwitz et al., 2007; Nordlund et al., 2009; Kassebaum et al., 2015). At least half of school children and the vast majority of adults worldwide experience accordingly caries, and the economic burden of caries and dental diseases represents about 4.6% of global health expenditures (Listl et al., 2015). However, in Sweden and countries with a low caries prevalence, many children are either free of caries or have a low disease level while 15–20% have a high caries burden (Selwitz et al., 2007; Källestål, 2005). These high caries cases are poorly explained by life style-related variables, such as sugar consumption, oral hygiene, or fluoride use (i.e. relative risks 0.9–1.2) and seem to be largely unaffected by traditional prevention based on the same factors (Källestål, 2005). Accordingly, life style, saliva, and bacteria are poor predictors of caries development (Selwitz et al., 2007; Nordlund et al., 2009), and better etiological models and diagnostic and preventive tools are needed.
In spite of many advances in etiological and biochemical mechanisms related to caries disease during the last decades (Selwitz et al., 2007; Nobbs et al., 2009), dental caries is still generally considered a life style condition in which plaque acidification from sugar consumption shifts the oral ecology toward aciduric and acid-producing species; of these, Streptococcus mutans is the most well-known (Selwitz et al., 2007; de Soet et al., 2000; Cornejo et al., 2012; Palmer et al., 2013; Aas et al., 2008). The bacterium was early identified as the primary caries pathogen and vaccine candidate, but the inability of S. mutans, or any other species, to match or predict individual caries development has hampered its use in individualized oral care (Selwitz et al., 2007; Nordlund et al., 2009).
S. mutans (serotypes c > > e > f and k) infects the oral cavity of 40–80% of subjects depending on age, ethnicity and disease prevalence and colonize individuals with a dominant and largely unique genotype transmitted from parent to child through saliva (Lapirattanakul et al., 2008; Esberg et al., 2012). Cariogenic properties besides acid production that dissolves enamel are oxygen tolerance, bacteriocin production and adhesion and colonization at tooth surfaces (Cornejo et al., 2012; Palmer et al., 2013). Together, sucrose-independent adhesion of SpaP or Cnm adhesins to host salivary agglutinin/DMBT1 and collagen, respectively, and sucrose-dependent adhesion of glycosyltransferases to bacterial polysaccharides allow S. mutans to colonize naked and cavitated tooth surface and promote plaque growth (Nobbs et al., 2009).
Salivary agglutinin, originally identified by its ability to agglutinate S. mutans, is identical to DMBT1 or gp340 (Prakobphol et al., 2000). Salivary agglutinin or DMBT1 is a pattern-recognition receptor composed of multiple domains designed to bind S. mutans and a wide array of microbes along with many innate and adaptive immunity factors (Madsen et al., 2010; Loimaranta et al., 2005; Loimaranta et al., 2009). DMBT1 thus modulates innate and adaptive immunity, including complement activation, NF-κb signaling via Toll receptors and cellular proliferation (Madsen et al., 2010). A 6.2 kb dmbt1 deletion variant has been associated with cancer (Madsen et al., 2010) and inflammatory bowel disease by increased NF-κb mediated inflammation in human cases (Renner et al., 2007). The corresponding salivary size variant (designated gp340 or DMBT1 size variant I) coincides with increased levels of caries and S. mutans adhesion in children (Jonasson et al., 2007).
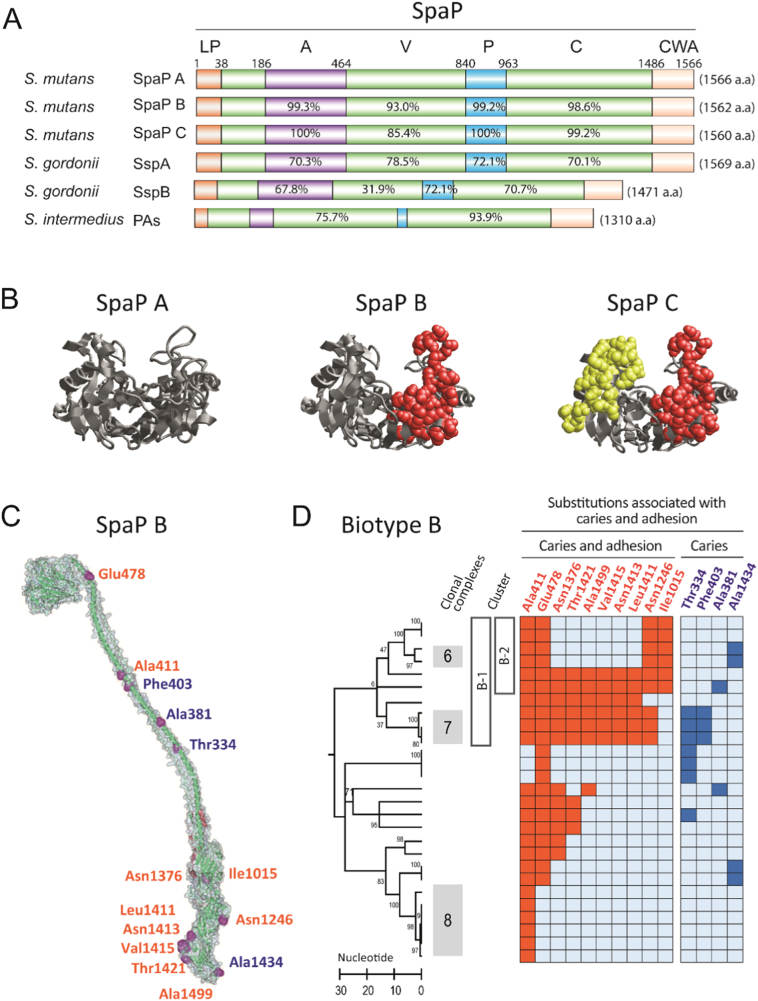
The AgI/II adhesin SpaP of S. mutans and AgI/II orthologs in various oral streptococci have been extensively explored for structural organization and interaction with its salivary receptor DMBT1 (Brady et al., 2010). AgI/II adhesins have an unusual tertiary structure where a central variable domain (V-domain) is presented at the tip of a stalk formed by intertwined, flanking alanine- and proline-rich regions (Larson et al., 2010). The carboxy-terminal domain (C-domain) to which a small N-terminal domain is bound is attached to the cell-wall via a cell-wall anchoring region (Heim et al., 2014). The SpaP binding sites for the DMBT1 agglutinin localize to the V-domain and the C-domain (Heim et al., 2013), and SpaP holds a supramolecular functional architecture at the cell surface (Heim et al., 2015). The SpaP adhesin harbors variants A, B and C (also referred to as A, B1, and B2) with clustered amino acid substitutions and different DMBT1 binding levels despite similar levels of SpaP expression (Esberg et al., 2012). The interaction of SpaP and AgI/II orthologs with DMBT1 depends on whether DMBT1 is in the fluid- or surface-bound form and also depending on the S. mutans strain (Loimaranta et al., 2005; Heim et al., 2013), suggesting that SpaP polymorphisms may modulate adhesion and aggregation by DMBT1 and consequently caries activity.
S. mutans also harbors collagen-binding Cnm and Cbm adhesins in 15% and 3% of clinical isolates, respectively, and more frequently in serotype e, f and k than in c isolates (Avilés-Reyes et al., 2017). Cnm/Cbm are highly homologous and consist, similar to collagen-binding proteins in Staphylococcus aureus and other bacteria (Kang et al., 2013; Xu et al., 2004), of an N-terminal collagen-binding domain presented on a stalk formed by several threonine-rich repeat domains and a cell wall anchoring region (Nomura et al., 2009, 2012). Whereas S. mutans (serotypes c, e, f and k) may cause infective endocarditis (Hoshino et al., 2005), serotypes f and k in which Cnm and Cbm are more frequent coincide with inflammatory bowel disease (Kojima et al., 2012) and Cnm phenotypes with hemorrhagic stroke (Nakano et al., 2011; Watanabe et al., 2016). The Cnm/Cbm phenotypes also increase strain virulence in endocarditis (Nomura et al., 2014). As potential virulence mechanisms in these so-called extra-oral infections, Cnm/Cbm mediate invasion of endothelial cells (Abranches et al., 2011, Review), formation of thrombus or heart valve vegetations or inhibition of platelet aggregation and wound healing (Nakano et al., 2011; Avilés-Reyes et al., 2017).
The aim of this study was to clarify the role of S. mutans as a caries pathogen by matching sucrose-independent adhesin types SpaP A, B, C and Cnm/Cbm with individual differences in caries development. We analyzed 452 Swedish children for the presence of S. mutans adhesin types and related them to baseline caries and 5-year increment and to cariogenic properties.
2. Methods
2.1. Study Participants and Registration of Caries
A total of 452 12-year-old children were collected as two independent samples (n = 218, n = 234) from 13 clinics in the northern county of Västerbotten, Sweden (Fig. S1). Included in the first sample were children born in 1996 and caries cases (≥ 1 Decayed and Filled Surfaces, DFS, in the permanent dentition) and caries-free controls in a 1:1 ratio; and for the second sample, they were born in 1998 and caries cases (≥ 2 DFS) and controls in a 2.1 ratio, and receiving ordinary dental care at Public Dental Service Clinics. The exclusion criterion was unwillingness to participate in the study. Both samples were re-examined after 5 years, for a total of 390 examined children with 14% drop-out rate (62/452) for having moved out of the area (20 children) or repeatedly missed the examination (42 children). The children received operative treatment and prevention of caries between 12 and 17 years of age, and 15% of the children orthodontic treatment with multibrackets after 12 years of age (as established from dental records), according to ordinary routines and policies at the clinics. The study was approved by the Ethics Committee for Human Experiments at Umeå University, Sweden, and informed consent was obtained from the children and their parents before participation. All parents signed consent to participate.
Caries was recorded by three dentists (intra- and inter-examiner kappa ≥ 0.979) by a mirror, probe and two bitewing radiographs and mean number of Decayed (enamel caries included), Filled Surfaces in the permanent dentition (DeFS) was the primary caries outcome measure. The 5-year caries increment (ΔDeFS) was calculated by subtracting latest from earliest DeFS, dividing that value by the number of observed years and multiplying the result by 5. The 1:1 ratio (first sample) and increased 2:1 ratio (second sample) of caries cases versus controls and DeFS index generated a continuous gradient of discriminatory caries DeFS scores in the entire sample at baseline (Fig. S1, Table S1).
2.2. Genotyping of S. Mutans Adhesin Types in Whole Saliva
Whole saliva, collected by chewing on paraffin for 5 min that was stored frozen at − 80 °C, was genotyped for cnm, cbm and spaP A, B and C status using bacterial DNA prepared with the GenElute™ bacterial genome DNA kit (Sigma-Aldrich, Sweden) and quantitative polymerase chain reaction (qPCR) using the KAPA SYBR Fast Universal qPCR kit (Tectum, Sweden). The primers for cnm and cbm were as described in Table S2 and did not cross-react in between or with other templates. The primers for spaP A, B, and C were from the spaP sequences (Table S2) and selected from prior testing and lack of cross-reactivity between A, B, and C or with DNA from oral streptococci with spaP analogs. The genotyping used internal standards and quantitative calibration curves based on dilutions of DNA purified from a reference genotype of each adhesin type, and cut-off values for A (3000 pg), B (3000 pg), C (6000 pg), cnm (3000 pg), and cbm (1000 pg).
2.3. Quantification of S. Mutans in Whole Saliva
S. mutans in whole saliva was quantified by culturing and qPCR (Yano et al., 2002). Serial dilutions of whole saliva were cultured on MSB agar plates and counted for colony-forming units of S. mutans (designated as ms). Plaque DNA purified from whole saliva samples was measured by qPCR using the KAPA SYBR FAST Universal qPCR kit (Sigma-Aldrich, Sweden) and Corbett Rotor-Gene 6000 and S. mutans specific primers (Table S2). Quantitative calibration curves from DNA prepared from serial dilutions of S. mutans strain Ingbritt (Esberg et al., 2012) were used to transform the qPCR responses into colony-forming units.
2.4. Isolation and Typing of S. Mutans Strains from Plaque
Plaque was collected from buccal surfaces of teeth 34–36 (premolars and the first molar of the left lower jaw) in caries-free children or from caries lesion in children with tooth decay (Esberg et al., 2012). Strains of S. mutans were isolated from the plaque samples by culturing on MSB agar plates and typed by Rapid ID 32 STREP Kit (Bio Merieux, La Balme les Grottes, France) metabolic tests (Hoshino et al., 2005). A total of 321 strains from 214 out of 217 infected children, one strain from each of the 214 children and additional isolates from 70 extreme or 26 cnm-positive children thereof, were isolated and phenotypically characterized (Fig. S1, Table S3). Of these isolates, one strain was mutated. The isolates were genotyped using qPCR and DNA purified by the GeneElute Bacterial Genomic DNA Kit (Sigma-Aldrich, Sweden). Typing of cnm and cbm status relied on specific primers and KAPA SYBR Fast Universal qPCR kit as described (Table S2) (Nomura et al., 2012), and typing of serotypes c, e, f and k was performed using specific primers and the PCR polymerase kit (GE Healthcare, Sweden) as described (Table S2, Shibata et al., 2003; Nakano et al., 2004).
2.5. Sequencing of SpaP and Cnm Genes
The full length of spaP and cnm genes were amplified by PCR using iProof High fidelity DNA polymerase (Bio-Rad, Sweden) and specific primers for spaP (Table S2) (Esberg et al., 2012) and cnm (Table S2) (Nomura et al., 2009). Amplified fragments were sequenced both forward and reverse by dideoxy chain termination/cycling sequencing on an ABI 3730 XL machine (Eurofins Genomics, Germany). Primers for spaP and cnm sequencing are listed in Table S2. Assembly and analyses of the full gene sequences were done with DNA Sequence Polymorphism DnaSP software (Librado and Rozas, 2009) and Molecular Evolutionary Genetics Analysis MEGA 5 software (Tamura et al., 2011). SpaP protein models were developing using the known crystal structures of S. mutans SpaP (Protein Data Bank IDs 3IOX and 3QE5) and CCP4MG software (Potterton et al., 2004).
2.6. Sequence Typing of S. Mutans Isolates
Sequence typing of S. mutans isolates (n = 144) was done by sequencing of housekeeping gene segments of map, sod, rpoB, ppaC, pfl, pyk, and tuf and inclusion of the spaP sequences (Esberg et al., 2012; Bishop et al., 2009; Edwards et al., 2008). DnaSP software (Librado and Rozas, 2009) was used to generate sequence characteristics (Table S4) and sequence types of the concatenated sequences, and MEGA 5 software (Tamura et al., 2011) to generate neighbor joining trees from concatenated sequences. The neighbor joining tree of all concatenated sequences was more stable (upon bootstrapping) than that of spaP or housekeeping genes alone and appeared to cluster isolates with host caries status better (Edwards et al., 2008). The eBURST software was used to group isolates into clonal complexes based on sharing of at least six out of eight alleles with at least one other member in the group (Feil et al., 2004).
2.7. Acid Tolerance
Isolates were screened for differences in acid tolerance by growth on Todd Hewitt medium agar plates at buffered pH values under aerobic conditions (Svensäter et al., 2003). The isolates (5 μL inoculum, OD495 2.0) were grown in 5% CO2 at 37 °C for 5 d, and acid tolerance was scored as the presence or absence of colony-forming units at different pHs from 5.0 to 7.0. All isolates yielded colony-forming units at pH 7.0, but these failed to grow at pH 6.0, 5.5, and 5.25 for isolates of low, moderate, and high acid tolerance, respectively. Isolates of S. mutans were also tested for acid tolerance by quantitative growth curves in BHI broth at pH 7.0 and pH 5.0 at 37 °C under aerobic and anaerobic conditions.
2.8. Hydroxyapatite Adhesion Assay
Adhesion of metabolically 35S-Met-labeled S. mutans isolates to saliva-coated hydroxyapatite beads (Bio-Rad Laboratories) was measured in 96 well microtiter plates as described (Esberg et al., 2012; Loimaranta et al., 2005; Jonasson et al., 2007). Briefly, after hydration of the hydroxyapatite beads in adhesion buffer at 4 °C overnight (5 mg beads in 125 μL per well), the beads were coated with 125 μL parotid saliva diluted in adhesion buffer 1:1 or with DMBT1 (2.5 μg/mL) or collagen type I (Sigma-Aldrich, Sweden) for 1 h and washed. The parotid saliva was collected by acid stimulation and Lashley cups and pooled from three donors before use (Esberg et al., 2012), and DMBT1 was purified biochemically from parotid saliva of several donors as described (Loimaranta et al., 2005; Eriksson et al., 2007). The coated beads were incubated with 35S-labeled bacteria (125 μL, 108 cells/ml; 1000 cells/cpm) for 1 h under agitation. After repeated washes (3 × 200 μL adhesion buffer), the beads were counted with a scintillation counter (MicroBeta2, PerkinElmer) and the proportion of bound bacteria out of the total amount of added bacteria (percent adhesion) was calculated.
2.9. Microtiter well Adhesion Assay
Adhesion of isolates to collagen was also measured in 96 well microtiter plates (Nakano et al., 2011). Briefly, wells (NUNC, Sigma-Aldrich, Sweden) were incubated overnight at 4 °C with 100 μL collagen type I (30 μg/mL, Sigma-Aldrich, Sweden, C224910) in 10 mM phosphate buffered saline, (PBS), pH 7.2. The wells were washed three times with PBS and incubated for 3 h at 37 °C with a suspension of S. mutans (100 μL, 1.0 OD600 in PBS) from overnight cultures in Jordan broth. Adherent cells after washes with PBS were stained by adding 200 μL 0.05% crystal violet in water for 5 min, washed and measured by absorbance at 595 nm after addition of 96% ethanol.
2.10. Construction of SpaP and Cnm Knockout Strains
To construct spaP and cnm knockout mutants in S. mutans wild type strain 49 (no. 23 in Fig. 2, Fig. S2), a PCR overlapping strategy was used involving insertion of a selective marker (ermAD or add9) within the target gene. The DNA was purified using GeneElute Bacterial Genomic DNA Kit (Sigma-Aldrich, Sweden), PCR amplifications were done with iProof High fidelity DNA polymerase (Bio-Rad, Sweden) and all primers (Table S2) were from Eurofins Genomics (Germany). Briefly, the 5′- and 3′-regions of the target gene were fused to the selective marker by end homology PCR and sub cloned into ZeroBlunt TOPO vector and transformed into a TOPO10 E. coli strain (Bio-Rad, Sweden). Correct generated disruption fragments were confirmed by sequencing (Eurofins Genomics, Germany). For construction of knockout mutants the cloned disruption fragments were amplified using T7 and Sp6 primers, transformed into isolates of S. mutans strain 49 by using a competence-stimulating peptide (SGSLSTFFRLFNRSFTQALGK) (Li et al., 2001). Correctly integrated fragments were confirmed by PCR and sequencing (Eurofins Genomics, Germany) (see also Fig. S5).
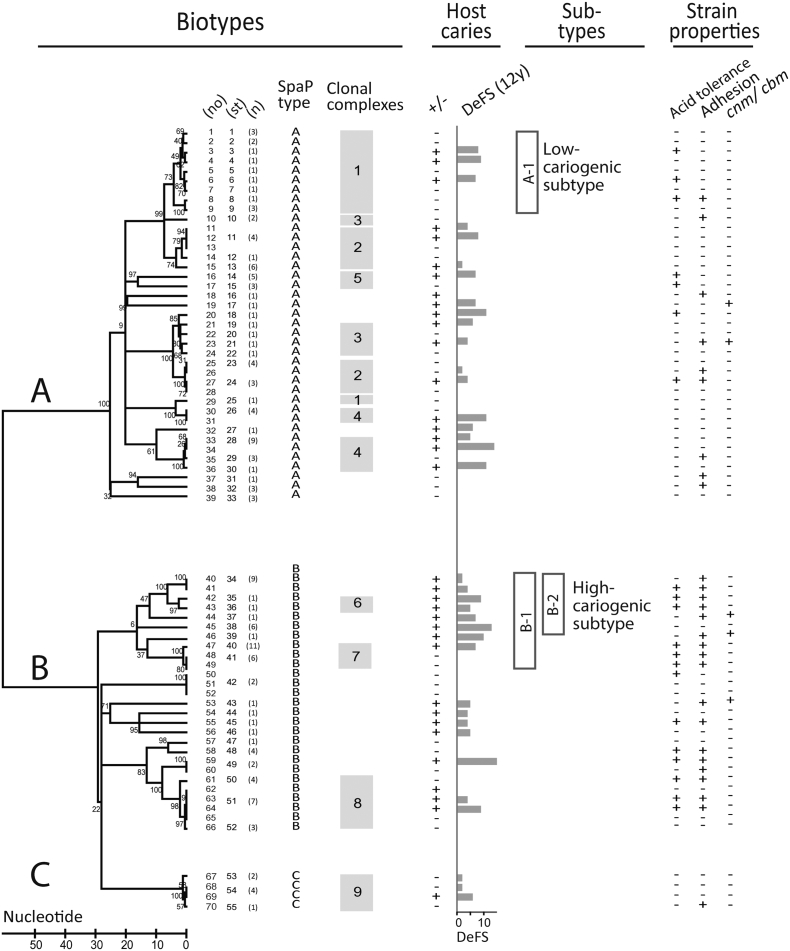
Fig. 2.
S. mutans biotypes A, B and C with corresponding SpaP adhesin subtypes of high (B-1) and low (A-1) cariogenicity. Clustering of S. mutans isolates from 35 caries (+) or 35 caries-free (−) extremes of infected children with caries status (+, − or DeFS at 12 years of age) based on neighbor joining and clonal complex analyses of spaP and housekeeping gene sequences. The isolates grouped both into biotypes A, B, and C with distinct SpaP A, B, and C adhesin types and clonal complexes C1–C9, and into genetically related subtypes that coincided with high caries (i.e. B-1, B-2 and C6, C7) or low caries (i.e. A-1 and C1) cases. The high- and low-cariogenic types also differed in acid tolerance and adhesion to DMBT1 and saliva. The clonal complexes share at least six identical alleles with at least one member in the group, all reflecting housekeeping alleles except for C8 which also shared the same spaP B allele. A total of 55 sequence types (ST) occurred among the 70 children. Numbers in parentheses mark STs shared in 15 children or identical STs in 25 children from which 74 additional strains were analyzed (mean 3, range 2 to 8 isolates/child).
2.11. Multivariate Partial Least Squares (PLS) Statistics
Multivariate PLS regression was performed with Simca+ P12.01 (Umetrics AB, Umeå, Sweden) (Eriksson et al., 2004). We used PLS, which relates two data matrices X and Y to each other by a linear multivariate prediction model, to identify amino acid substitutions of SpaP (the X-matrix) that modify adhesion and caries (the Y-matrix). The PLS model gives the X variable's ability to explain (R2) and – via cross-validation – predict (Q2, < 10% reflects a weak model) the variation in Y along with regression coefficients (CoeffCS) and the relative variable importance in projection (VIP) of each X variable (VIP-values > 1.0 indicate influential X variables).
2.12. Univariate Statistics
Data were expressed as means ± standard deviations (SD) and 95% confidence intervals (95% CI). The statistical analyses were performed with Chi-squared, Fisher's exact and Mann-Whitney U tests, or Spearman's rank correlation using SPSS or GraphPad softwares. All statistical analyses, univariate or multivariate, used independent isolates or subjects and two-tailed tests and p values < 0.05 were considered significant.
3. Results
3.1. Relationship Between Infection with S. Mutans and Caries Development
Infection by S. mutans was measured in 452 12-year-old children by qPCR and culturing of whole salivas and related to caries scores at 12 and 17 years of age and 5-year caries increment (Fig. S1, Table 1). About half of the children were infected with S. mutans at baseline (48%) and experienced a ~ 2-fold increased caries DeFS scores at 12 and 17 years of age and 5-year caries increment compared to non-infected children (Table 1). The S. mutans infection load, however, did not coincide with baseline or prospective caries (Table 1). Thus, children infected with S. mutans had 1.8-fold increased caries increment.
Table 1.
Stability of infection by S. mutans and adhesin types and their association with caries in 452 Swedish children.
| Group or adhesin typea, b | DeFS-12yc |
DeFS-17yc |
ΔDeFS-5yd |
Colonization stability, %g | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | Mean ± SD | pe | n | Mean ± SD | pe | n | Mean ± SD | pe | ||
| 452 children | |||||||||||
| S. mutans positivef | 217 | 48 | 3.6 ± 3.0 | 8 × 10− 13 | 185 | 9.2 ± 8.8 | 4 × 10− 8 | 185 | 5.5 ± 7.1 | 3 × 10− 5 | 78 |
| S. mutans negative | 234 | 52 | 1.7 ± 2.1 | Ref. | 204 | 4.9 ± 5.7 | Ref. | 204 | 3.1 ± 4.9 | Ref. | |
| S. mutans positive | |||||||||||
| High > 250,000 cfu | 49 | 23 | 3.9 ± 3.0 | 0.284 | 43 | 10.3 ± 9.8 | 0.544 | 43 | 6.4 ± 7.8 | 0.502 | |
| Low < 250,000 cfu | 168 | 77 | 3.5 ± 3.0 | Ref. | 142 | 8.8 ± 8.5 | Ref. | 142 | 5.3 ± 6.8 | Ref. | |
| SpaP A | 111 | 51 | 3.0 ± 2.8 | Ref. | 96 | 8.4 ± 8.2 | Ref. | 96 | 5.2 ± 7.1 | Ref. | 82 |
| SpaP B | 71 | 33 | 4.2 ± 3.5 | 0.034 | 55 | 11.4 ± 11.1 | 0.045 | 55 | 6.7 ± 8.8 | 0.110 | 88 |
| SpaP C | 19 | 9 | 3.3 ± 1.8 | 0.365 | 18 | 7.5 ± 4.8 | 0.746 | 18 | 4.3 ± 4.1 | 0.929 | 89 |
| Cnm | 26 | 12 | 4.4 ± 2.6 | 0.012 | 20 | 10.4 ± 8.7 | 0.182 | 20 | 6.0 ± 7.2 | 0.460 | 99 |
| Cbm | 7 | 3 | 2.9 ± 3.7 | 0.631 | 5 | 3.8 ± 2.8 | 0.201 | 5 | 2.4 ± 1.6 | 0.599 | 100 |
| SpaP B+ | 71 | 25 | 4.2 ± 3.5 | 3 × 10− 6 | 55 | 11.4 ± 11.1 | 5 × 10− 4 | 55 | 6.7 ± 8.8 | 8 × 10− 4 | |
| SpaP B− | 363 | 75 | 2.2 ± 2.4 | Ref. | 319 | 6.0 ± 6.5 | Ref. | 319 | 3.8 ± 5.4 | Ref. | |
Children infected with S. mutans and specific adhesin types or being negative as determined by qPCR of whole saliva; data missing from one child.
Infected children are either spaP A, B or C and Cnm distribute in equal portions in A-, B- or C-positive children.
DeFS, Decayed, enamel included, Filled Surfaces at 12 and 17 years of age.
ΔDeFS-5y = 5 year caries increment from 12 to 17 years of age.
Two-sided p-value from Mann-Whitney U test compared to a reference (Ref.).
Cut off value of 10,000 cfu.
Colonization stability of S. mutans and adhesin types marks the same type at both 12 and 17 years of age at an individual level.
3.2. Colonization of Specific S. Mutans Adhesin Types in Children in a 5-year Period
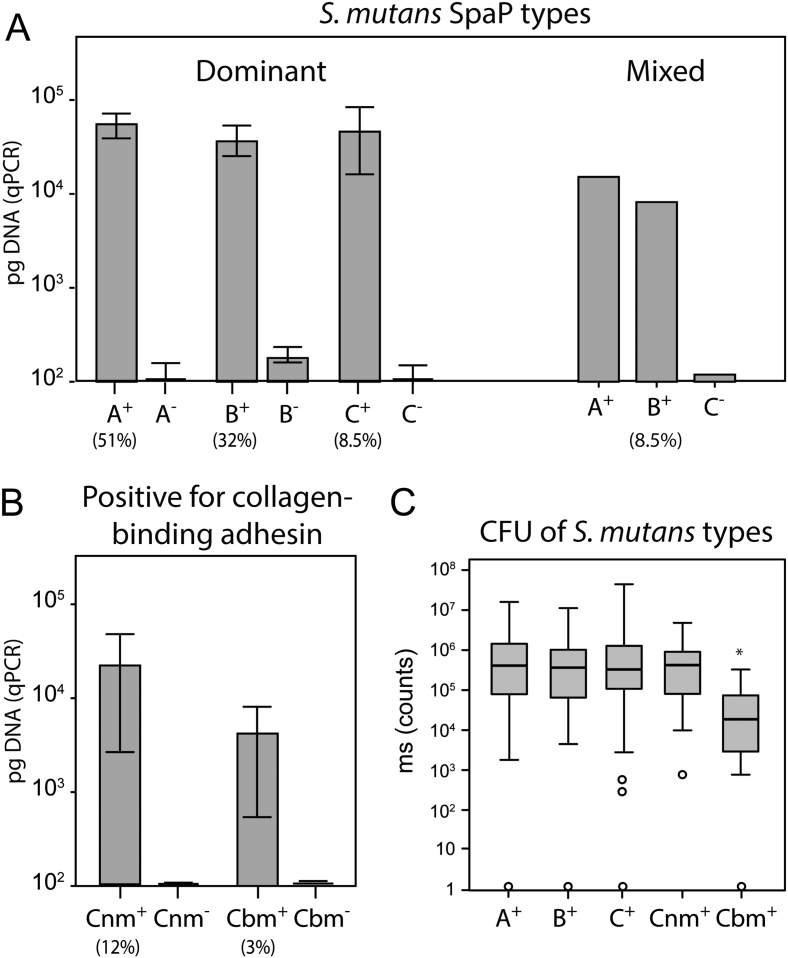
To establish the colonization pattern of specific S. mutans adhesin types in the 452 children, SpaP (A, B, or C) and collagen-binding (Cnm or Cbm) adhesin types were determined in the 452 children at 12 and 17 years of age using qPCR (Fig. 1A, B; Table 1). The S. mutans-positive children (n = 217) were either infected with a single dominant SpaP A (51%), B (32%), or C (8.5%) adhesin type or a mixture of two SpaP adhesin types (8.5%) at baseline. A portion of S. mutans-positive but none of the negative children were either Cnm-positive (26 children, 12%) or Cbm-positive (7 children, 3%) at baseline irrespective of SpaP A, B, or C status (Table 1). S. mutans and its adhesin types generally colonized the children at both 12 and 17 years with a colonization stability ranging from 78% to 100% (Table 1). Thus, children were stably colonized by specific SpaP A, B or C adhesin types that in some children also were Cnm or Cbm positive.
Fig. 1.
Determination of S. mutans adhesin types in whole saliva. Children infected (+) or not infected (−) with A) a dominant or mixed SpaP A, B or C adhesin types and B) a Cnm or Cbm adhesin type of S. mutans by quantitative PCR of whole salivas (pg DNA responses, 95% CI). C) Total colony-forming units, CFU, of S. mutans (ms counts) in whole saliva of infected children as shown by box plots (*p = 0.003, Mann-Whitney U test).
3.3. Relationship of Adhesin Types to Individual Caries Development
We next matched the S. mutans adhesin types with individual differences in caries at 12 and 17 years and in the 5-year caries increment (Table 1). The presence of S. mutans SpaP B and Cnm coincided with 1.4- and 1.5-fold increased caries scores at baseline compared to SpaP A (p = 0.034, p = 0.012), which was most prevalent and together with SpaP C and Cnm coincided with low caries scores at baseline. The increased association of SpaP B and Cnm with caries and comparably low caries scores of SpaP A, C, and Cbm remained at 17 years of age and for the 5-year caries increment (Table 1). In addition, the differential association of the adhesin types with caries did not coincide with differences in numbers of S. mutans in saliva except for a slight reduction in Cbm-positive children (Fig. 1C). Thus, the SpaP B and Cnm adhesin types coincided with increased baseline and prospective caries development although the children experienced adolescence and puberty and were treated for caries by operative and preventive means (Table S1) during the study period.
3.4. Multilocus Sequence Typing (MLST) of S. Mutans Using Housekeeping and SpaP Gene Sequences
To explore further the adhesin types for genetic relatedness and grouping of specific subtypes with caries cases, we isolated and typed S. mutans strains from all infected children as SpaP A, B, or C in good agreement with the saliva typing (Table S3) and subjected a subset of 70 isolates from 35 caries and 35 caries-free extremes to MLST using spaP and housekeeping genes (Fig. 2, Fig. S2; Table S4). The discriminatory ability of sequence typing related to host variables may increase with genes encoding bacterial proteins subject to host adaptation (Edwards et al., 2008), and inclusion of spaP improved the grouping of isolates with caries cases and controls (Fig. 2, Table 2).
Table 2.
Association of S. mutans SpaP subtypes with caries in 35 caries and 35 nearly caries-free extremes of children.
| Groupa | SpaP type | Sub-type | DMBT1 binding |
DeFS-12yb |
DeFS-17yb |
ΔDeFS-5yc |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ± SD | pd | n | Mean ± SD | pd | n | Mean ± SD | pd | n | Mean ± SD | pd | |||
| SpaP A | A | 39 | 13.3 ± 3.6 | 0.862 | 39 | 3.2 ± 4.2 | 0.518 | 34 | 9.0 ± 10.1 | 0.204 | 34 | 5.5 ± 6.5 | 0.282 | |
| A | A-1 | 9 | 13.2 ± 1.7 | 0.712 | 9 | 2.7 ± 4.0 | 0.882 | 8 | 5.3 ± 7.6 | 0.801 | 8 | 3.2 ± 5.2 | 0.803 | |
| A | C1 | 10 | 12.9 ± 1.8 | Ref. | 10 | 2.4 ± 3.9 | Ref. | 9 | 4.7 ± 7.5 | Ref. | 9 | 2.9 ± 5.0 | Ref. | |
| SpaP B | B | 27 | 17.8 ± 6.6 | 0.002 | 27 | 3.8 ± 4.4 | 0.320 | 22 | 11.3 ± 5.0 | 0.114 | 22 | 7.3 ± 10.4 | 0.104 | |
| B | B-1 | 10 | 21.8 ± 8.3 | < 0.001 | 10 | 5.7 ± 4.3 | 0.074 | 7 | 12.3 ± 10.6 | 0.033 | 7 | 8.4 ± 7.3 | 0.029 | |
| B | B-2 | 6 | 19.8 ± 5.3 | 0.002 | 6 | 6.7 ± 3.9 | 0.041 | 4 | 19.0 ± 10.9 | 0.023 | 4 | 12.4 ± 7.2 | 0.019 | |
| B | C6 | 2 | 17.7 ± 2.6 | 0.053 | 2 | 7.0 ± 2.8 | 0.119 | 2 | 28.0 ± 1.4 | 0.030 | 2 | 18.2 ± 0.1 | 0.029 | |
| SpaP C | C | C-1 | 4 | 12.8 ± 3.1 | 0.396 | 4 | 2.5 ± 2.5 | 0.480 | 3 | 4.3 ± 2.3 | 0.299 | 3 | 1.0 ± 0.1 | 0.925 |
| Adhesione | ||||||||||||||
| SpaP B | B | High | 17 | 20.7 ± 6.5 | < 0.001 | 17 | 4.8 ± 4.3 | 0.073 | 15 | 16.1 ± 16.0 | < 0.001 | 15 | 10.3 ± 11.5 | 0.003 |
| B | Low | 10 | 12.8 ± 2.1 | 10 | 2.2 ± 4.2 | 7 | 1.0 ± 1.2 | 7 | 0.9 ± 1.0 | |||||
| SpaP A | A | High | 16 | 16.6 ± 1.5 | < 0.001 | 16 | 1.9 ± 3.5 | 0.090 | 13 | 6.1 ± 8.0 | 0.353 | 13 | 3.1 ± 3.8 | 0.260 |
| A | Low | 23 | 10.9 ± 1.5 | 23 | 4.2 ± 4.4 | 21 | 10.8 ± 10.9 | 21 | 6.9 ± 7.4 | |||||
| Acid tolerancef | ||||||||||||||
| SpaP B | B | High | 13 | 4.4 ± 4.6 | 0.558 | 13 | 17.8 ± 16.7 | 0.001 | 13 | 11.6 ± 11.8 | 0.001 | |||
| SpaP A | B | Low | 14 | 3.3 ± 4.3 | 9 | 2.0 ± 2.2 | 9 | 1.1 ± 1.3 | ||||||
| A | High | 7 | 5.3 ± 4.2 | 0.115 | 6 | 12.8 ± 13.1 | 0.585 | 6 | 7.8 ± 9.0 | 0.820 | ||||
| A | Low | 32 | 2.8 ± 4.1 | 28 | 8.1 ± 9.4 | 28 | 4.9 ± 6.0 | |||||||
Children infected with S. mutans SpaP types and subtypes from sequencing and functional tests of isolates in 35 caries and 35 nearly caries-free extremes of children.
Caries DeFS (Decayed, enamel included, Filled Surfaces) at 12 and 17 years of age.
ΔDeFS (5 y) = 5-years prospective caries increment from 12 to 17 years of age.
2-sided p-value from Mann-Whitney U test.
Highest (50%) and lowest (50%) adhesion phenotypes among the 70 isolates.
Low (0) and high (1, 2) acid tolerance.
Quantitative neighbor joining and qualitative clonal complex methods were used to analyze the sequence variations and grouped the SpaP A, B, and C adhesin types into corresponding biotypes A, B, and C that each had a unique housekeeping allele profile (Fig. 2). Accordingly, each biotype A, B, or C had a unique panel of clonal complexes C1–C9 in which each clonal complex shared allelic profile: C1–C5 in biotype A; C6–C8 in biotype B; and C9 in biotype C. Thus, biotypes A, B, and C in S. mutans may differ in adhesion-related events because of corresponding SpaP A, B, and C adhesin types and in other properties because of different housekeeping gene profiles.
To identify SpaP A and B adhesin biotypes with markedly low and high cariogenicity, we further matched their specific subtypes with caries status of the strain donor (Fig. 2, Table 2). A subset of biotype B isolates that grouped together in the neighbor joining (i.e. B-1, B-2) and clonal complex (i.e. C6, C7) analyses also clustered with children of high caries experience and increment. Also, genetically related isolates of biotypes A (i.e. A-1, C1) and C (i.e. C9) grouped with low-caries children. Children infected with B-1 isolates had a 2.6-fold increased 5-year caries increment compared to children with A-1 (C1) isolates (p = 0.029) (Table 2), compared to the overall 1.3-fold difference between the biotype B and A isolates (p = 0.11) (Table 1). Thus, subtypes of SpaP adhesin biotype B display markedly high cariogenicity and subtypes of SpaP adhesin biotype A markedly low cariogenicity.
3.5. Association of SpaP Adhesin Biotype B Isolates with DMBT1-Binding and Acid Tolerance
Having found subtypes of the SpaP adhesin biotypes A, B and C with different cariogenicity and spaP and housekeeping allele profiles, including genes potentially involved in acid tolerance (Cotter and Hill, 2003), we next investigated the subtypes for binding to DMBT1 and saliva and acid tolerance (Fig. 2, Fig. S3).
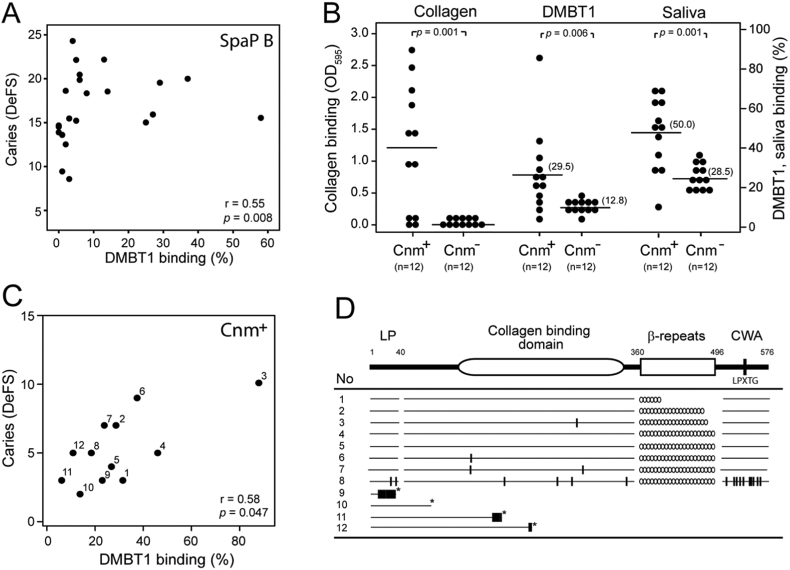
SpaP B isolates bound DMBT1 and saliva better than SpaP A and C isolates and high-cariogenicity SpaP B-1 isolates bound better than low-cariogenicity A-1 (C1) isolates (Fig. 2, Figs. S3, S4; Table 2). High versus low DMBT1-binding of SpaP B isolates coincided with a 11.4-fold increased 5-year caries increment in children (Table 2), and binding of SpaP B isolates to DMBT1 and saliva correlated with individual numbers of caries lesions (Fig. 3A). In contrast, the binding level in SpaP A isolates did not coincide with caries increment or individual caries scores (r = 0.06, p = 0.75) (Table 2).
Fig. 3.
SpaP B- and Cnm-mediated binding to DMBT1 correlates with individual caries scores. A) Correlation of binding of SpaP B to DMBT1 with individual numbers of caries DeFS lesions (binding to saliva generated virtually identical results, data not shown). B) Binding of Cnm-positive and Cnm-negative isolates to collagen, DMBT1, and saliva. Each isolate is marked by a dot and mean adhesion by a black line (Mann-Whitney U test). C) Correlation of binding of Cnm isolates (no. 1–12) to DMBT1 and collagen (r = 0.62, p = 0.031, respectively; data not shown). D) Deduced Cnm protein sequences of isolates no. 1–12. Substitutions are indicated with (|), translational stop codons with (*), and numbers of β-repeats with (o). LP = leading peptide. CWA = cell-wall anchoring motif.
Biotype B and subtype B-1 isolates had a high relative acid tolerance compared to biotype A and subtype A-1 (C1) isolates (Fig. 2, Fig. S3), which coincided with a 10.5-fold increased 5-year caries increment as compared to isolates with a low acid tolerance (Table 2).
3.6. Association of Substitutions in SpaP B Isolates with DMBT1-Binding and Caries
Having found an association between adhesion and caries in SpaP adhesin B subtypes and because they differ in sequence mainly outside the V-domain (Esberg et al., 2012, Fig. 4AB), we hypothesized that single amino acid substitutions in the C- and A-domains may specify the high-cariogenicity subtype B-1 isolates. We therefore used PLS analyses to screen single SpaP substitutions in SpaP B (and SpaP A) isolates for association with adhesion and caries (Fig. 4CD, Table S5). Substitutions that coincided with caries and adhesion were associated with high-cariogenicity SpaP adhesin B-1 (C6, C7) isolates, and 10 out of the 14 caries-associated substitutions in SpaP correlated positively with DMBT1-binding. SpaP A adhesin biotype A isolates behaved differently (Fig. S5, Table 2). In addition, the clustered substitutions that distinguish SpaP A, B, and C all localized on opposite sides of a potential binding pocket in the DMBT1-binding V-domain (Fig. 4B).
Fig. 4.
Adhesins SpaP A, B and C of different cariogenicity. A) Domain (A, V, P and C) structure and sequence identity of SpaP A, B and C and orthologs in various streptococci (reference SpaP A). The SpaP A, B and C adhesins differ by clustered substitutions in the V-domain and by single substitutions in the A-, P- and C- domains. LP = leading peptide, CWA = cell wall anchoring region. B) The clustered amino acid substitutions specific to SpaP A, B, and C localize on opposite sides of a pocket in the V-domain; A as reference, B and C share 29 substitutions (side chains in red), C holds 32 unique substitutions (side chains in yellow). C) SpaP structure with a tip-localized V-domain, intertwined P and A domains followed by C- and N-terminal complexes. Marked are all substitutions associated positively with caries and adhesion (red) or with caries alone (blue) in SpaP B isolates. D) Amino acid substitutions in SpaP B isolates (marked by a row) that coincide positively with caries and adhesion (to DMBT1 and saliva) or with caries alone upon PLS analyses. The substitutions are enriched in B-1 and B-2 isolates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. Association of Cnm Isolates with DMBT1-Binding and Acid Tolerance
Knowing that Cnm and Cbm isolates of serotypes f and k have been associated with systemic infection risk, we next explored all S. mutans isolates for adhesin (Cnm or Cbm) and serotype status and binding to collagen (Fig. S4, Table S3). Cnm and Cbm status, which coincided with collagen binding, was frequent with serotype k (100%) and f (50%) reported in systemic infections, but less common in serotype e (11%) and c (4%) isolates (Table S3).
We next explored whether the caries association of Cnm phenotypes similarly could be referred to binding to DMBT1 and saliva and to acid tolerance (Fig. 3B–D). Of the 26 cnm-positive children, 12 had S. mutans isolates with a dominant Cnm-positive phenotype (Table S3), and these isolates bound markedly better to DMBT1, saliva and collagen than Cnm-negative isolates (Fig. 3B). Binding of Cnm to DMBT1 was indicated by a cnm knockout mutant (Fig. S5), and by the more avid binding of Cnm than SpaP isolates to saliva and DMBT1 (Fig. 3B). The binding of the isolates to DMBT1 and collagen correlated positively with individual numbers of caries lesions (Fig. 3C), and the isolates had unique Cnm sequences or truncated Cnm proteins with weak or no binding (Fig. 3D). In addition, Cnm isolates showed low acid tolerance in aerobic but high acid tolerance under anaerobic conditions (Fig. S6).
4. Discussion
The present findings provide the first evidence for specific adhesin types of S. mutans that match and predict individual caries development in people and thus provide a rationale for individualized oral care. The presence of SpaP B and Cnm subtypes coincided with more cross-sectional caries but also predicted increased 5-year caries increment in 452 Swedish children. In addition, binding of those strains to salivary DMBT1 correlated with individual caries scores. A lack of such evidence for a specific S. mutans caries pathogen has hampered its clinical use and favored the idea of caries resulting from an ecological shift toward various acid-tolerant species by plaque acidification from sugar consumption. However, because children not infected with S. mutans also developed caries, although to lesser extent, both life style and non-mutans bacteria may also contribute to caries development. We assume a heterogeneous etiology of caries in which both life style and host susceptibility influence the role of the microbiota and its components in caries development. Moreover, both SpaP and glycosyltransferases, two long standing vaccine candidates involving host-microbe and bacteria-bacteria adhesion events, respectively, may play important roles in caries development depending on S. mutans biotype, disease prevalence, and life style profile in different populations and individuals.
Our present demonstration in S. mutans of specific biotypes A, B, and C with corresponding SpaP A, B, and C adhesin types, distinct housekeeping alleles and behaviors may in part explain previous difficulties in distinguishing S. mutans strains of low to high cariogenicity. The biotypes apparently differ in adhesion and in acid tolerance, and plausibly in as-yet-unknown cariogenic effects. However, similar to the H. pylori BABA and Streptococcus parasanguinis Fap1 adhesin structures (Bugaytsova et al., 2017; Ramboarina et al., 2010), the SpaP protein structure may adapt to pH and together with housekeeping genes, such as superoxide dismutase and elongation factor Tu, directly participate in acid tolerance (Cotter and Hill, 2003). Moreover, the different V-domains of SpaP A, B, and C may reflect an adaptation to host DMBT1 variants associated with different individuals and caries susceptibility (Jonasson et al., 2007) or microbial partner receptor configurations or epitopes, whereas differences in acid tolerance may match different biofilm conditions in various Actinomyces-Streptococcus communities. Of note, AgI/II adhesins participate in intra and inter generic interactions between Actinomyces-Streptococcus species (Nobbs et al., 2009). Biotypes A, B and C could accordingly represent members of distinct biofilm communities.
The present findings provide a rationale for individualized oral care in terms of risk assessment and prevention. Infection with SpaP B (16% prevalence), B-1 (3.5%) or Cnm (6%) subtypes and absence of infection with S. mutans (52%) would identify reasonable portions of high- versus low-risk children, respectively. The fact that S. mutans infection per se and type, but not infection load in terms of gross numbers of the organism, predicted 5-year caries increment highlights the importance of plaque quality. The distinct adhesin and biotype behaviors could also identify subjects and caries subtypes that benefit from different treatments. The children infected with SpaP B and Cnm types may benefit from anti-microbial treatment with chlorhexidine or antibiotics and novel replacement with low-cariogenic types, such as A-1 isolates, and Cnm phenotypes may be inhibited by high oxygen conditions, improved oral hygiene, or other means.
The high agreement between adhesin type and cross-sectional and prospective caries, although the children received caries prevention care and experienced puberty and adolescence as well as orthodontic treatment during the study, strengthens the adhesin-specific differences in cariogenicity. Moreover, the high colonization stability and persistence of the adhesin types in individual carriers over 5 years, and that children received multibrackets after 12 years of age, support this conclusion. However, it is difficult to estimate strain cariogenicity in absolute terms because of these experiences, along with the fact that treatment of caries with fillings reduces surfaces at future risk and that children differ in life style, socio-economic factors and genetic susceptibility. Moreover, the high S. mutans genetic diversity generated a repertoire of SpaP B-1 and B-2 subtypes of different cariogenic potentials and frequencies. The relative cariogenicity and natural history of the adhesin types should accordingly be replicated and further explored in longitudinal studies on initiation and progression of caries at different surfaces in the primary and permanent dentitions.
Binding strength of SpaP B- and Cnm-positive isolates for DMBT1 correlated with individual caries scores, and the high cariogenicity B-1 isolates were enriched in substitutions that correlated with both adhesion and caries. The localization of these substitutions in the C and A domains suggests that SpaP polymorphisms outside the V-domain play a more important role in strain binding and cariogenicity than previously anticipated. Of interest, the collagen-binding affinity of the S. aureus CNA adhesin specifies strain differences in virulence in experimental arthritis (Xu et al., 2004). The Cnm and SpaP B adhesin subtypes, which both display increased saliva binding, may evoke different adhesion, aggregation, and innate immunity events by DMBT1 that favor surface-close colonization and tooth demineralization rather than overall plaque colonization of S. mutans (Loimaranta et al., 2005; Heim et al., 2013). The mechanism underlying the correlation between binding affinity and individual caries scores may involve S. mutans adaptation to individual conditions or receptor modifications during life-long chronic caries infection and inflammation. We thus assume that variation in DMBT1-binding reflects a direct virulence mechanism and not a DMBT1-mediated pattern recognition of pathogenicity markers such as leucine-rich repeats (Butler et al., 2009). Accordingly, S. mutans, SpaP and Cnm/Cbm, as opposed to many other bacteria, bind to DMBT1 via non-leucine-rich repeats (Loimaranta et al., 2009).
The stable colonization of collagen binding Cnm or Cbm adhesin types of serotype f or k in ~ 1% children could lead to an increased risk for such diverse extra-oral infections as inflammatory bowel disease, hemorrhagic stroke and endocarditis. Of note, inflammatory bowel disease commonly debuts in adolescence and involves polymorphisms in the S. mutans receptor DMBT1. Moreover, although the prevalence of Cbm-positive children is too low for firm conclusions, the 7 children with Cbm-positive saliva had markedly low caries levels. This emphasizes that it remains to explore if extra-oral infections by S. mutans SpaP/Cnm/Cbm adhesins and serotype settings involve the same adhesin and biotype heterogeneity as observed in caries development. It also remains to be explored if expression of Cnm represses SpaP function and phenotype characteristics and if serotype c and e strains with Cnm expression are as virulent as their serotype f and k counterparts. Of note, Cnm isolates seemed to have both increased acid tolerance under anaerobic conditions and saliva/DMBT1 binding regardless of SpaP A, B or C background.
The present study emphasizes that careful sequence analysis and evaluation of S. mutans genotypes in concert with dental caries incidence serves as a useful example and model to better understand strain variation and disease association in relation to bacterial virulence in chronic infections. These results highlight the importance of developing novel approaches to diagnose high-risk patients and improve prevention and treatment of chronic infectious disease. Our findings may also have relevance beyond dental caries and translate to improved systemic health. For example, missing teeth as a consequence of caries and periodontitis is a reported risk factor for cardio-vascular disease (Nakano et al., 2011; Vedin et al., 2015). Therefore, chronic caries infections and associated inflammation from carriage of high virulence strains would likely contribute to poor outcomes related to systemic disease risk as well as to diminished oral health.
Acknowledgments
Acknowledgments
This work was supported by the Swedish Research Council (10906), the Faculty of Medicine of Umeå University, and Västerbotten County Council. We acknowledge financial support provided through a regional agreement between Umeå University and Västerbotten County Council in the field of Medicine, Odontology, and Health (377341) and through a Fund for Cutting-Edge Medical Research grant from the County Council of Västerbotten (135041). We also thank the Swedish dental Society and local foundations at Umeå University. The work was in part performed at Umeå Centre for Chronic Infectious Disease. Ingmarie Bernhardsson, Ewa Strömqvist-Engbo, Ulla Öhman and Prof. Lennart Hammarström, KI Stockholm, are acknowledged for assistance. We thank all families and public health care personnel who participated in the study.
The funders had no role in study design; data collection, analysis or interpretation; writing of the report.
A patent application has been filed; no. PCT/SE/2017/000028.
Conflicts of Interest
The authors have no competing interests to declare.
Author Contributions
N.S. designed research; A.E., N. Sheng performed most of the research; R.C., K.P. performed research; N.S., L.M., N. Sheng collected the clinical samples; A.E., N. Sheng, L.M., R.C., K.P.,T.B., N.S. analyzed data; N.S. wrote the paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.09.027.
Appendix A. Supplementary data
Supplementary material
References
- Aas J.A., Griffen A.L., Dardis S.R., Lee A.M., Olsen I., Dewhirst F.E., Leys E.J., Paster B.J. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J., Miller J.H., Martinez A.R., Simpson-Haidaris P.J., Burne R.A., Lemos J.A. The collagen-binding protein Cnm is required for Streptococcus mutans adherence to and intracellular invasion of human coronary artery endothelial cells. Infect. Immun. 2011;79:2277–2284. doi: 10.1128/IAI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilés-Reyes A., Miller J.H., Lemos J.A., Abranches J. Collagen-binding proteins of Streptococcus mutans and related streptococci. Mol Oral Microbiol. 2017;32:89–106. doi: 10.1111/omi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C.J., Aanensen D.M., Jordan G.E., Kilian M., Hanage W.P., Spratt B.G. Assigning strains to bacterial species via the internet. BMC Biol. 2009;7:3. doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J.L., Maddocks S.E., Larson M.R., Forsgren N., Persson K., Deivanayagam C.C., Jenkinson H.F. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaytsova J.A., Björnham O., Chernov Y.A., Gideonsson P., Henriksson S., Mendez M., Sjöström R., Mahdavi J., Shevtsova A., Ilver D., Moonens K., Quintana-Hayashi M.P., Moskalenko R., Aisenbrey C., Bylund G., Schmidt A., Åberg A., Brännström K., Königer V., Vikström S., Rakhimova L., Hofer A., Ögren J., Liu H., Goldman M.D., Whitmire J.M., Ådén J., Younson J., Kelly C.G., Gilman R.H., Chowdhury A., Mukhopadhyay A.K., Nair G.B., Papadakos K.S., Martinez-Gonzalez B., Sgouras D.N., Engstrand L., Unemo M., Danielsson D., Suerbaum S., Oscarson S., Morozova-Roche L.A., Olofsson A., Gröbner G., Holgersson J., Esberg A., Strömberg N., Landström M., Eldridge A.M., Chromy B.A., Hansen L.M., Solnick J.V., Lindén S.K., Haas R., Dubois A., Merrell D.S., Schedin S., Remaut H., Arnqvist A., Berg D.E., Borén T. Adaption of Helicobacter pylori to chronic infection and gastric disease by pH-responsive BabA mediated adherence. Cell Host Microbe. 2017;21:376–389. doi: 10.1016/j.chom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Rasmussen M.D., Lin M.F., Santos M.A., Sakthikumar S., Munro C.A., Rheinbay E., Grabherr M., Forche A., Reedy J.L., Agrafioti I., Arnaud M.B., Bates S., Brown A.J., Brunke S., Costanzo M.C., Fitzpatrick D.A., de Groot P.W., Harris D., Hoyer L.L., Hube B., Klis F.M., Kodira C., Lennard N., Logue M.E., Martin R., Neiman A.M., Nikolaou E., Quail M.A., Quinn J., Santos M.C., Schmitzberger F.F., Sherlock G., Shah P., Silverstein K.A., Skrzypek M.S., Soll D., Staggs R., Stansfield I., Stumpf M.P., Sudbery P.E., Srikantha T., Zeng Q., Berman J., Berriman M., Heitman J., Gow N.A., Lorenz M.C., Birren B.W., Kellis M., Cuomo C.A. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo O.E., Lefe'bure T., Pavinski Bitar P.D., Lang P., Richards V.P., Eilertson K., Do T., Beighton D., Zeng L., Ahn S.-J., Burne R.A., Siepel A., Bustamante C.D., Stanhope M.J. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol. Biol. Evol. 2012;30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Hill C. Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003;67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.T., Fry N.K., Harrison T.G. Clonal population structure of Legionella pneumophila inferred from allelic profiling. Microbiology. 2008;154:852–864. doi: 10.1099/mic.0.2007/012336-0. [DOI] [PubMed] [Google Scholar]
- Eriksson L., Antti H., Gottfries J., Holmes E., Johansson E., Lindgren F., Long I., Lundstedt T., Trygg J., Wold S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm) Anal. Bioanal. Chem. 2004;380:419–429. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- Eriksson C., Frängsmyr L., Danielsson-Niemi L., Loimaranta V., Holmskov U., Bergman T., Leffler H., Jenkinsson H., Strömberg N. Variant size- and glycoforms of the scavenger receptor cysteine-rich protein gp-340 with differential bacterial aggregation. Glycoconj. J. 2007;24:131–142. doi: 10.1007/s10719-006-9020-1. [DOI] [PubMed] [Google Scholar]
- Esberg A., Burström-Löfgren A., Öhman U., Strömberg N. Host and Bacterial phenotype variation in adhesion of Streptococcus mutans to matched human hosts. Infect. Immun. 2012;80:3869–3879. doi: 10.1128/IAI.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E.J., Li B., Aanensen D.M., Hanage W.P., Spratt B.G. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial phenotypes from multilocus sequence typing data. J. Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim K.P., Crowley P.J., Brady L.J. An intramolecular interaction involving the N terminus of a streptococcal adhesin affects its conformation and adhesive function. J. Biol. Chem. 2013;288(288):13762–13774. doi: 10.1074/jbc.M113.459974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim K.P., Crowley P.J., Long J.R., Kailasan S., McKenna R., Brady L.J. An intramolecular lock facilitates folding and stabilizes the tertiary structure of Streptococcus mutans adhesin P1. Proc. Natl. Acad. Sci. 2014;111:15746–15751. doi: 10.1073/pnas.1413018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim K.P., Sullan R.M.A., Crowley P.J., El-Kirat-Chatel S., Beaussart A., Tang W., Besingi R., Dufrene Y.F., Brady L.J. Identification of a supramolecular functional architecture of Streptococcus mutans adhesin P1 on the bacterial cell surface. J. Biol. Chem. 2015;290:9002–9019. doi: 10.1074/jbc.M114.626663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T., Fujiwara T., Kilian M. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 2005;43:6073–6085. doi: 10.1128/JCM.43.12.6073-6085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonasson A., Eriksson C., Jenkinson H.F., Källestål C., Johansson I., Strömberg N. Innate immunity glycoprotein gp-340 variants may modulate human susceptibility to dental caries. BMC Infect. Dis. 2007;7:57. doi: 10.1186/1471-2334-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källestål C. The effect of five years´ implementation of caries-preventive methods in Swedish high-risk adolescents. Caries Res. 2005;39:20–26. doi: 10.1159/000081652. [DOI] [PubMed] [Google Scholar]
- Kang M., Ko Y., Liang X., Ross C.L., Liu Q., Murray B.E., Höök M. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J. Biol. Chem. 2013;(28):20520–20531. doi: 10.1074/jbc.M113.454462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassebaum N.J., Bernabé E., Dahiya M., Bhandari B., Murray C.J., Marcenes W. Global burden of untreated caries: a systematic review and metaregression. J. Dent. Res. 2015;94:650–658. doi: 10.1177/0022034515573272. [DOI] [PubMed] [Google Scholar]
- Kojima A., Nakano K., Wada K., Takahashi H., Katayama K., Yoneda M., Higurashi T., Nomura R., Hokamura K., Muranaka Y., Matsuhashi N., Umemura K., Kamisaki Y., Nakajima A., Ooshima T. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci Rep. 2012;2:332. doi: 10.1038/srep00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapirattanakul J., Nakano K., Nomura R., Hamada S., Nakagawa I., Ooshima T. Demonstration of mother-to-child transmission of mutans streptococci using multilocus sequence typing. Caries Res. 2008;42:466–474. doi: 10.1159/000170588. [DOI] [PubMed] [Google Scholar]
- Larson M.R., Rajashankar K.R., Patel M.H., Robinette R.A., Crowley P.J., Michalek S., Brady L.J., Deivanayagam C. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of a- and PPII-helices. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Lau P.C., Lee J.H., Ellen R.P., Cvitkovitch D.G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 2001;83:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Listl S., Galloway J., Mossey P.A., Marcenes W. Global economic impact of dental diseases. J. Dent. Res. 2015;94:1355–1361. doi: 10.1177/0022034515602879. [DOI] [PubMed] [Google Scholar]
- Loimaranta V., Jakubovics N.S., Hytönen J., Finne J., Jenkinson H.F., Strömberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect. Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loimaranta V., Hytonen J., Pulliainen A.T., Sharma A., Tenovuo J., Strömberg N., Finne J. Leucine-rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J. Biol. Chem. 2009;284:18614–18623. doi: 10.1074/jbc.M900581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen J., Mollenhauer J., Holmskov U. Gp-340/DMBT1 in mucosal innate immunity. Innate Immun. 2010;16:160–167. doi: 10.1177/1753425910368447. [DOI] [PubMed] [Google Scholar]
- Nakano K., Nomura R., Shimizu N., Nakagawa I., Hamada S., Ooshima T. Development of a PCR method for rapid identification of new Streptococcus mutans serotype k strains. J. Clin. Microbiol. 2004;42:4925–4930. doi: 10.1128/JCM.42.11.4925-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Hokamura K., Taniguchi N., Wada K., Kudo C., Nomura R., Kojima A., Naka S., Muranaka Y., Thura M., Nakajima A., Masuda K., Nakagawa I., Speziale P., Shimada N., Amano A., Kamisaki K., Tanaka T., Umemura K., Ooshima T. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat. Commun. 2011;2:485. doi: 10.1038/ncomms1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs A.H., Lamont R.J., Jenkinsson H.F. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura R., Nakano K., Taniguchi N., Lapirattanakul J., Nemoto H., Grönroos L., Alaluusua S., Ooshima T. Molecular and clinical analyses of the gene encoding the collagen-binding adhesin of Streptococcus mutans. J. Med. Microbiol. 2009;58:469–475. doi: 10.1099/jmm.0.007559-0. [DOI] [PubMed] [Google Scholar]
- Nomura R., Nakano K., Naka S., Nemoto H., Masuda K., Lapirattanakul J., Alaluusua S., Matsumoto M., Kawabata S., Ooshima T. Identification and characterization of a collagen-bindning protein, Cbm, in Streptococcus mutans. Mol Oral Microbiol. 2012;27:308–323. doi: 10.1111/j.2041-1014.2012.00649.x. [DOI] [PubMed] [Google Scholar]
- Nomura R., Otsugo M., Naka S., Teramoto N., Kojima A., Muranaka Y., Matsumoto-Nakano M., Ooshima T., Nakano K. Contribution of the interaction of Streptococcus mutans serotype k strains with fibrinogen to the pathogenicity of infective endocarditis. Infect. Immun. 2014;82:5223–5234. doi: 10.1128/IAI.02164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund Å., Johansson I., Källestål C., Ericson T., Sjöström M., Strömberg N. Improved ability of biological and previous caries multimarkers to predict caries disease as revealed by multivariate PLS modelling. BMC Oral Health. 2009;9:28. doi: 10.1186/1472-6831-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S.R., Miller J.H., Abranches J., Zeng L., Lefebure T., Richards V.P., Lemos J.A., Stanhope M.J., Burne R.A. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton L., Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G.N., Cohen S., Perrakis A., Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Prakobphol A., Xu F., Hoang V.M., Larsson T., Bergstrom J., Johansson I., Frängsmyr L., Holmskov U., Leffler H., Nilsson C., Borén T., Wright J.R., Strömberg N., Fisher S.J. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- Ramboarina S., Garnett J.A., Zhou M., Li Y., Peng Z., Taylor J.D., Lee W., Bodey A., Murray J.W., Alguel Y., Bergeron J., Bardiaux B., Sawyer E., Isaacson R., Tagliaferri C., Cota E., Nilges M., Simpson P., Ruiz T., Wu H., Matthews S. Structural insights into serine-rich fimbriae from Gram-positive bacteria. J. Biol. Chem. 2010;285:32446–32457. doi: 10.1074/jbc.M110.128165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M., Bergmann G., Krebs I., End C., Lyer S., Hilberg F., Helmke B., Gassler N., Autschbach F., Bikker F., Strobel-Friedekind O., Gronert-Sum S., Benner A., Blaich S., Wittig R., Hudler M., Lightenberg A.J., Madsen J., Holmskov U., Annese V., Latiano A., Schirmacher P., Nieuw Amerongen A.V., D'amato M., Kioschis P., Hafner M., Poutska A., Mollenhauer J. DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn's disease. Gastroenterology. 2007;133:1499–1509. doi: 10.1053/j.gastro.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Selwitz R.H., Ismail A.I., Pitts N.B. Dental caries. Lancet. 2007;369:51–59. doi: 10.1016/S0140-6736(07)60031-2. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Ozaki K., Seki M., Kawato T., Tanaka H., Nakano Y., Yamashita Y. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J. Clin. Microbiol. 2003;41:4107–4112. doi: 10.1128/JCM.41.9.4107-4112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Soet J.J., Nyvad B., Kilian M. Strain-related acid production by oral streptococci. Caries Res. 2000;34:486–490. doi: 10.1159/000016628. [DOI] [PubMed] [Google Scholar]
- Svensäter G., Borgström M., Bowden G.H.W., Edwardsson S. The acid-tolerant microbiota associated with plaque from initial caries and healthy tooth surfaces. Caries Res. 2003;37:395–403. doi: 10.1159/000073390. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedin O., Hagström E., Gallup D., Neely M.L., Stewart R., Koenig W., Budaj A., Sritara P., Wallentin L., White H.D., Held C. Periodontal disease in patients with chronic coronary heart disease: prevalence and association with cardiovascular risk factors. Eur. J. Prev. Cardiol. 2015;22:771–778. doi: 10.1177/2047487314530660. [DOI] [PubMed] [Google Scholar]
- Watanabe I., Kuriyama N., Miyatani F., Nomura R., Naka S., Nakano K., Ihara M., Iwai K., Matsui D., Ozaki E., Koyama T., Nishigaki M., Yamamoto T., Tamura A., Mizuno T., Akazawa K., Takada A., Takeda K., Yamada K., Nakagawa M., Tanaka T., Kanamura N., Friedland R.P., Watanabe Y. Oral Cnm-positive Streptococcus mutans expressing collagen binding activity is a risk factor for cerebral microbleeds and cognitive impairment. Sci Rep. 2016;6 doi: 10.1038/srep38561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Rivas J.M., Brown E.L., Liang X., Höök M. Virulence potential of the staphylococcal adhesin CNA in experimental arthritis is determined by its affinity for collagen. J Infect Dis. 2004;189:2323–2333. doi: 10.1086/420851. [DOI] [PubMed] [Google Scholar]
- Yano A., Kaneko N., Ida H., Yamaguchi T., Hanada N. Real-time PCR for quantification of Streptococcus mutans. FEMS Microbiol. Lett. 2002;19:23–30. doi: 10.1111/j.1574-6968.2002.tb11451.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material