Abstract
Colorectal cancer (CRC) is a common digestive malignancy and emerging studies have closely linked its initiation and development with gut microbiota changes. Fusobacterium nucleatum (Fn) has been recently identified as a pathogenic bacteria for CRC; however, its prognostic significance for patients is poorly investigated and is less for patients within late stage. Therefore, in this study, we made efforts to analyze its level and prognostic significance in a retrospective cohort of 280 stage III/IV CRC patients. We found that the Fn level was abnormally high in tumor tissues and correlated with tumor invasion, lymph node metastasis status, and distant metastasis. We also identified it as an independent adverse prognostic factor for cancer-specific survival (CSS) and disease-free survival (DFS). The following subgroup analysis indicated that Fn level could stratify CSS and DFS in stage IIIB/C and IV patients but failed in stage IIIA patients. In addition, stage III/IV patients with low Fn level were found to benefit more from adjuvant chemotherapy than those with high Fn level, in terms of DFS. Finally, we analyzed the expression and clinical significance of epithelial-to-mesenchymal transition (EMT) markers (E-cadherin and N-cadherin) and cancer stem cell (CSC) markers (Nanog, Oct-4, and Sox-2) in CRC tissues. The results indicated that N-cadherin, Nanog, Oct-4, and Sox-2 were adverse prognostic factors in these patients, while the opposite was true for E-cadherin. More importantly, expression of E-cadherin, N-cadherin, and Nanog was significantly correlated with Fn level in tumor tissues, suggesting the potential involvement of Fn in EMT-CSC cross talk during CRC progression. Taken together, these findings indicate that Fn is a novel predictive biomarker for clinical management in stage III/IV patients, and targeting Fn may be an effective adjuvant approach for preventing CRC metastasis and chemotherapy resistance.
Keywords: colorectal cancer, Fn, EMT, cancer stem cell, prognosis
Introduction
Colorectal cancer (CRC) is a fatal digestive malignancy that is commonly diagnosed in both males and females worldwide.1 In USA, it is the third most common form of cancer and will account for an estimated 135,430 newly diagnosed cases and 50,260 CRC-specific deaths in 2017.2 In China, its incidence has reached ~37.63 per 100,000 in 2015 according to the latest report.3 The pathogenesis of CRC is a complicated multistep process involving various inherent and environmental factors such as genetic predisposition and unhealthy lifestyles.4 Although dramatic reduction has been achieved in CRC mortality because of the introduction of screening programs and multidisciplinary treatments, ~60% of CRC patients are still diagnosed with advanced stage with their 5-year survival rate ranging from 14% to 71%.5 In addition, there are few effective therapeutical approaches and prognostic biomarkers available for metastatic CRC currently, frequently leading to inappropriate decision making.6 Targeted therapy (such as epidermal growth factor receptor antagonists) represents an emerging clinical strategy for these patients; however, primary and acquired therapy resistance limit its actual efficiency.7 Molecular biomarker tests hold promise for personalized therapy, while a considerable proportion of them may be overestimated and fail to be recommended for prognosis prediction or therapy selection due to insufficient evidence.8,9 Therefore, it can be concluded that our existing achievements appear to be insufficient to improve the clinical outcome of CRC patients and therefore substantial efforts are still essential to identify other potential CRC-related driving factors.
Recently, increasing studies have suggested that gut microbiota dysbiosis is correlated with tumor initiation and development.10 Microbiota dysbiosis may contribute to the malignant progression of cancer cells through various mechanisms such as metabolism signals, inflammation induction, and immunosuppression.11 Furthermore, microbiota is also crucial for the therapeutical efficacy of some anticancer drugs such as cyclophosphamide, which may associate with its regulation of T-cell responses.12 In gastrointestinal malignancies, a close correlation between microbiota and carcinogenesis has been well established in gastric cancer, where Helicobacter pylori is most extensively studied and has been identified as a risk factor for screening.13 However, with regard to CRC, related studies are emerging although advanced metagenomic techniques are able to provide more potential pathogenic microbiota.14 For example, Tsoi et al proved that Peptostreptococcus anaerobius is increased in CRC tissues and promotes the growth of CRC cells through inducing intracellular cholesterol synthesis.15 Wang et al demonstrated that Enterococcus faecalis can drive the malignant transformation in normal colon epithelial cells via its bystander effect.16 Despite increasing evidences supporting the oncogenic role of some specific bacteria in CRC, their clinical significance is still poorly investigated and whether these bacteria can be further developed as clinical biomarkers for patient management remains unknown.
Previously, using pyrosequencing, we found that Fusobacterium nucleatum (Fn) is abnormally abundant in 1,2-dimethylhydrazine-induced CRC animal models as compared with healthy controls.17 Then, we used the same method to further confirm that it is also significantly more abundant in human CRC tissues than in adjacent normal tissues, suggesting its potential correlation with CRC development.18 Further investigation revealed that Fn promotes the proliferation and invasiveness of CRC cells through activating toll-like receptors/MyD88/NF-Kb/miR-21 signaling.19 Given these findings, we speculate that Fn may be a promising clinical biomarker for CRC patients. Therefore, in this study, we aimed to investigate the level and clinical significance of Fn in stage III/IV CRC patients, who are clinically characterized with positive regional/distant metastasis and have a dramatically worse outcome than those within stage I/II. Since epithelial-to-mesenchymal transition (EMT) and cancer stem cell (CSC) are both widely considered as major molecular factors driving cancer development, we also made efforts to detect the expression of representative EMT and CSC markers in these patients and identify their potential correlations with Fn.20,21 Taken together, our findings not only suggest Fn as a novel therapeutical target and prognostic biomarker for CRC patients within late stage, but also highlight the crucial link between dysregulated microbiota and oncogenic molecular events in CRC progression.
Materials and methods
Patient data and specimens
A total of 280 pairs of tumor and adjacent normal tissues were collected from stage III/IV CRC patients who underwent radical surgery at Department of General Surgery, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital and Shanghai Tenth People’s Hospital between October 1, 2007 and September 25, 2015. All the patients were pathologically confirmed as CRC with positive lymph node metastasis (LNM). Preoperative distant metastasis (including lung, liver, and ovary) was identified by enhanced computed tomography (CT) scanning. Tumor-node-metastasis (TNM) stage was determined according to the latest guidelines of the Union for International Cancer Control (8th edition). Neither preoperative chemotherapy nor radiotherapy was performed on patients. For postoperative chemotherapy, a standard FOLFOX scheme (5-fluorouracil [5-fu] [Shanghai Xudong Haipu Pharmaceutical Co., LTD, Shanghai, China] + oxaliplatin [Jiangsu HengRui Medicine Co., LTD, Lianyungang, Jiangsu, China] + leucovorin [Jiangsu HengRui Medicine Co., LTD, Lianyungang, Jiangsu, China]) was applied. Regular follow-up was conducted according to the Clinical Practice Guidelines in Oncology proposed by the National Comprehensive Cancer Network. In brief, patients were recommended to undergo physical examination, carcinoembryonic antigen (CEA) test, and enhanced CT scan every 3–6 months for the first 2 years, and then 6–12 months for the following 3 years. Patient prognosis was assessed by cancer-specific survival (CSS) and disease-free survival (DFS). CSS was calculated from the date of surgery to the date of death caused by CRC, while DFS was calculated from the date of surgery to the date of local recurrence or regional/distant metastasis. The basic clinical features of patients are summarized in Table 1. This study was approved by the ethics committees of both the hospitals mentioned above. Written informed consents were obtained from patients or their legal guardians for using their specimens in medical researches.
Table 1.
Correlations between Fn level and clinicopathological parameters in stage III/IV CRC patients
| Characteristics | Total |
Fn level
|
p-value | |
|---|---|---|---|---|
| Low | High | |||
| Gender | 0.705 | |||
| Female | 122 | 42 | 80 | |
| Male | 158 | 51 | 107 | |
| Age | 0.822 | |||
| ≤65 years | 111 | 36 | 75 | |
| >65 years | 169 | 57 | 112 | |
| Tumor location | 0.579 | |||
| Rectal | 130 | 41 | 89 | |
| Colon | 150 | 52 | 98 | |
| Tumor size | 0.357 | |||
| ≤5 cm | 214 | 68 | 146 | |
| >5 cm | 66 | 25 | 41 | |
| Tumor differentiation | 0.650 | |||
| Poor | 74 | 23 | 51 | |
| Well/moderate | 206 | 70 | 136 | |
| Tumor invasion | 0.015 | |||
| T1–T2 | 96 | 41 | 55 | |
| T3–T4 | 184 | 52 | 132 | |
| Lymph node metastasis | 0.008 | |||
| N1 | 81 | 17 | 64 | |
| N2a | 95 | 41 | 54 | |
| N2b | 104 | 35 | 69 | |
| Distant metastasis | 0.020 | |||
| Absent | 218 | 80 | 138 | |
| Present | 62 | 13 | 49 | |
| Ki-67 expression | 0.381 | |||
| <30% | 78 | 29 | 49 | |
| ≥30% | 202 | 64 | 138 | |
| Serum CEA level | 0.274 | |||
| ≤5 ng/mL | 99 | 37 | 62 | |
| >5 ng/mL | 181 | 56 | 125 | |
| BMI | 0.202 | |||
| <18.5 kg/m2 | 22 | 11 | 11 | |
| 18.5–24.99 kg/m2 | 178 | 58 | 120 | |
| ≥25.0 kg/m2 | 80 | 24 | 56 | |
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; CEA, carcinoembryonic antigen; BMI, body mass index.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
The levels of Fn in human CRC and adjacent normal tissues were detected by qRT-PCR. Briefly, paraffin-embedded tissues were deparaffinized in xylene and lysed in buffer ATL (Qiagen NV, Venlo, the Netherlands) and Proteinase K (Qiagen NV). Then, the genomic DNAs were extracted using QIAamp DNA FFPE Tissue Kit according to the manufacturer’s instructions (Qiagen NV). The quality of obtained DNAs was verified by an ultraviolet spectrophotometer and eligible DNA samples were preserved at −20°C. The PCR reaction was performed on a 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) using SYBR Premix Ex Taq (TaKaRa, Kusatsu, Shiga, Japan). The reaction conditions were applied as follows: initial denaturation at 95°C for 10 minutes, denaturation at 95°C for 1 minute, primer annealing at 60°C for 20 seconds, and primer extension at 56°C for 60 seconds. The sequences of primers were as follows: Fn, forward: 5′-CTTAGGAATGAGACAGAGATG-3′ and reverse: 5′-TGATGGTAACATACGAAAGG-3′; β-actin, forward: 5′-CCTCCATCGTCCACCGCAAATG-3′ and reverse: 5′-TGCTGTCACCTTCACCGTTCCA-3′. The 2−ΔΔCT method was utilized to calculate the relative level of Fn gene and β-actin served as an internal control gene. All the experiments were repeated in triplicate.
Immunohistochemistry (IHC) and staining evaluation
Experimental procedures of IHC were carried out according to our previous study.22 In brief, paraffin-embedded tissues were continuously cut into 4-μm-thick sections, dewaxed in xylene, and rehydrated in gradient concentrations of ethanol. Antigen retrieval was achieved by microwave heating and endogenous peroxidase activity was blocked by incubation with 3% H2O2 solution. Then, sections were incubated with the primary antibody against E-cadherin (1:250; Abcam, Cambridge, UK), N-cadherin (1:250; Abcam), Nanog (1:200; Abcam), Sox-2 (1:200; Abcam), and Oct-4 (1:200; Abcam) at 4°C overnight. Sections incubated with only antibody dilution buffer were utilized as negative controls. Following several washes with phosphate-buffered saline solution, sections were treated with the secondary antibody (1:250; Abcam) at 37°C for 30 minutes. Finally, protein staining was visualized by incubating sections with a diaminobenzidine kit (Thermo Fisher Scientific) for 5 minutes. The sections were counterstained with hematoxylin (Thermo Fisher Scientific) for 10 minutes, dehydrated, sealed, and transferred for microscopic examination.
Staining evaluation was independently carried out by two investigators who were blind to the clinical features and outcome of patients. Any controversial cases were determined by a well-skilled pathologist. The evaluation criteria were based on staining intensity (SI) and percentage of positive cells (PP). SI is scored as follows: 0, negative; 1, weak; 2, moderate; 3, strong. PP is scored as follows: 0, 0%–10%; 1, 11%–25%; 2, 26%–50%; 3, 51%–75%; 4, 76%–100%. A final staining score was calculated by multiplying the PP score with SI score. The cutoff value of the final score was determined by receiver operating characteristic (ROC) curve analysis. The sections that scored more or less than the cutoff value were regarded as high or low expression cases, respectively.
Statistical analysis
Data are presented as mean ± standard deviation and statistical analyses were performed on SPSS 20.0 statistical software (IBM Corporation, Armonk, NY, USA). The Fn level between CRC and adjacent normal tissues was compared by Mann–Whitney test. The cutoff value of the ROC curve was estimated by Youden index. The correlations between biomarkers and clinicopathological parameters were analyzed by chi-square test. The CSS and DFS curves based on Kaplan–Meier model were depicted using GraphPad Prism 5 (GraphPad Software, Inc, La Jolla, CA, USA) and intergroup difference was compared by log-rank test. Independent factors affecting CSS/DFS were identified by univariate and multivariate analysis based on Cox proportional hazards regression model. The impact of Fn level on chemotherapy benefits was evaluated using treatment-by-biomarker interaction analysis in a 2×2 factorial design.23 The correlations of Fn level with expression of EMT/CSC markers in CRC tissues were evaluated by Spearman’s rank correlation coefficient. A p-value <0.05 was considered statistically significant.
Results
Fn level in CRC and adjacent normal tissues of stage III/IV CRC patients
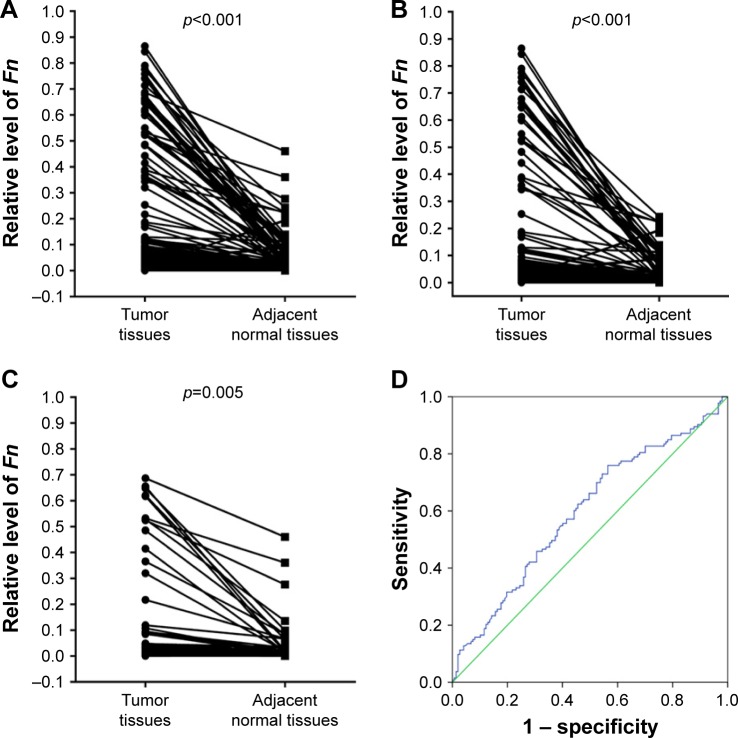
The relative level of Fn in CRC and adjacent normal tissues was detected by qRT-PCR. As shown in Figure 1A, for the whole study cohort, Fn level is significantly higher in CRC tissues than in adjacent normal tissues (CRC vs normal: 0.1092±0.2150 vs 0.0245±0.0553, n=280, p<0.001). In subgroups classified by tumor stage, this difference remains statistically significant in both stage III patients (CRC vs normal: 0.1043±0.2165 vs 0.0216±0.0450, n=218, p<0.001, Figure 1B) and stage IV patients (CRC vs normal: 0.1266±0.2106 vs 0.0348±0.0817, n=62, p=0.005, Figure 1C). Then, the ROC curve was used to calculate an optimal cutoff value for defining the Fn level (Figure 1D). The optimal cutoff value of Fn level in CRC tissues was 0.0282. Therefore, we classified the entire cohort into a high level group (n=187) and a low level group (n=93) according to this cutoff value.
Figure 1.
Fn level in tumor and adjacent normal tissues of stage III/IV CRC patients.
Notes: (A) Fn level is significantly higher in tumor tissues than in adjacent normal tissues of the whole cohort (tumor vs normal: 0.1092±0.2150 vs 0.0245±0.0553, n=280, p<0.001). (B) Fn level is significantly higher in tumor tissues than in adjacent normal tissues of stage III patients (tumor vs normal: 0.1043±0.2165 vs 0.0216±0.0450, n=218, p<0.001). (C) Fn level is significantly higher in tumor tissues than in adjacent normal tissues of stage IV patients (tumor vs normal: 0.1266±0.2106 vs 0.0348±0.0817, n=62, p=0.005). (D) ROC curve analysis determined the cutoff value of Fn level in tumor tissues to be 0.0282.
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; ROC, receiver operating characteristic.
Correlations between Fn level and clinicopathological parameters in stage III/IV CRC patients
As shown in Table 1, Fn level was found to significantly associate with tumor invasion (p=0.015), LNM status (p=0.008), and distant metastasis (p=0.020). No significant association was observed between Fn level and other clinicopathological parameters including age (p=0.822), gender (p=0.705), tumor location (p=0.579), tumor size (p=0.357), tumor differentiation (p=0.650), body mass index (p=0.202), preoperative serum CEA level (p=0.274), and Ki-67 positive rate (p=0.381).
Prognostic significance of Fn in stage III/IV CRC patients
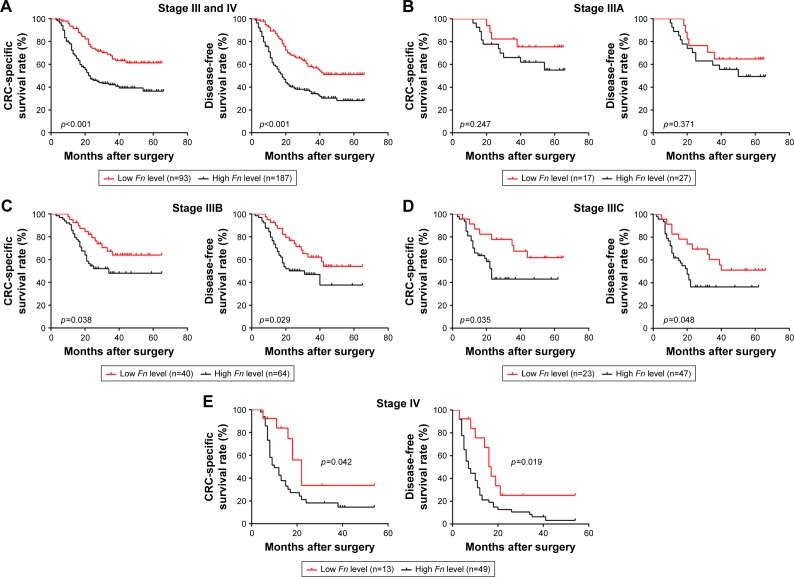
The impact of Fn on patient prognosis was illustrated using Kaplan–Meier survival curves. For the whole cohort, patients with low Fn level had a significantly better CSS and DFS than those with high Fn level (CSS, p<0.001; DFS, p<0.001, Figure 2A). As shown in Tables 2 and 3, the univariate analysis suggested that Fn level, tumor invasion, LNM status, distant metastasis, and serum CEA level were significant factors for CSS (p<0.001, p=0.015, p=0.002, p<0.001, p=0.046), while Fn level, tumor differentiation, tumor invasion, LNM status, distant metastasis, and serum CEA level were for DFS (p<0.001, p=0.009, p=0.005, p=0.015, p<0.001, p=0.018). The multivariate analysis suggested that Fn level, LNM status, distant metastasis, and serum CEA level were independent factors affecting CSS (p<0.001, p=0.001, p<0.001, p=0.031), while Fn level, tumor differentiation, tumor invasion, LNM status, distant metastasis, and serum CEA level were affecting DFS (p<0.001, p=0.003, p=0.022, p=0.008, p<0.001, p=0.027). To further identify whether Fn has the capacity to stratify patient prognosis within the same stage, subgroup analysis was performed according to LNM status and distant metastasis. Surprisingly, we found that stage IIIA patients with low Fn level had no better CSS and DFS than those with high Fn level (CSS: p=0.247; DFS: p=0.371, Figure 2B). But, high Fn level was still significantly associated with worse CSS and DFS in other stage III patients (stage IIIB: CSS: p=0.038, DFS: p=0.029, Figure 2C; stage IIIC: CSS: p=0.035, DFS: p=0.048, Figure 2D). With regard to its prognostic role in stage IV patients, a statistically significant association between high Fn level and worse clinical outcome is also obviously found (CSS: p=0.042; DFS: p=0.019, Figure 2E).
Figure 2.
Prognostic significance of Fn in stage III/IV CRC patients.
Note: CSS and DFS curves of stage III/IV CRC patients (A), stage IIIA patients (B), stage IIIB patients (C), stage IIIC patients (D), stage IV patients (E).
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; CSS, cancer-specific survival; DFS, disease-free survival.
Table 2.
Univariate and multivariate analysis for prognostic factors in cancer-specific survival of stage III/IV CRC patients
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Gender | 0.835 | 0.594–1.175 | 0.300 | |||
| Age | 1.351 | 0.943–1.935 | 0.101 | |||
| Tumor location | 1.188 | 0.843–1.676 | 0.326 | |||
| Tumor size | 1.199 | 0.817–1.757 | 0.354 | |||
| Tumor differentiation | 0.731 | 0.497–1.074 | 0.110 | |||
| Ki-67 positivity | 0.914 | 0.629–1.328 | 0.638 | |||
| Body mass index | 0.806 | 0.597–1.087 | 0.158 | |||
| Tumor invasion | 1.595 | 1.097–2.319 | 0.015 | 1.341 | 0.920–1.955 | 0.127 |
| Lymph node metastasis | 1.426 | 1.136–1.789 | 0.002 | 1.430 | 1.158–1.766 | 0.001 |
| Distant metastasis | 3.507 | 2.425–5.071 | <0.001 | 3.243 | 2.232–4.712 | <0.001 |
| Serum CEA level | 1.466 | 1.006–2.136 | 0.046 | 1.515 | 1.038–2.212 | 0.031 |
| Fn level | 2.302 | 1.541–3.437 | <0.001 | 2.222 | 1.483–3.329 | <0.001 |
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; CEA, carcinoembryonic antigen; HR, hazard ratio; CI, confidence interval.
Table 3.
Univariate and multivariate analysis for prognostic factors in disease-free survival of stage III/IV CRC patients
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Gender | 0.821 | 0.603–1.119 | 0.212 | |||
| Age | 1.092 | 0.796–1.498 | 0.585 | |||
| Tumor location | 1.149 | 0.842–1.568 | 0.381 | |||
| Tumor size | 1.132 | 0.796–1.609 | 0.490 | |||
| Ki-67 positivity | 0.998 | 0.708–1.408 | 0.993 | |||
| Body mass index | 0.871 | 0.665–1.143 | 0.319 | |||
| Tumor differentiation | 0.636 | 0.453–0.893 | 0.009 | 0.592 | 0.417–0.841 | 0.003 |
| Tumor invasion | 1.634 | 1.163–2.297 | 0.005 | 1.499 | 1.060–2.119 | 0.022 |
| Lymph node metastasis | 1.287 | 1.050–1.579 | 0.015 | 1.294 | 1.069–1.566 | 0.008 |
| Distant metastasis | 3.965 | 2.843–5.531 | <0.001 | 3.914 | 2.788–5.495 | <0.001 |
| Serum CEA level | 1.512 | 1.075–2.128 | 0.018 | 1.483 | 1.046–2.101 | 0.027 |
| Fn level | 2.133 | 1.496–3.041 | <0.001 | 2.000 | 1.396–2.865 | <0.001 |
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; CEA, carcinoembryonic antigen; HR, hazard ratio; CI, confidence interval.
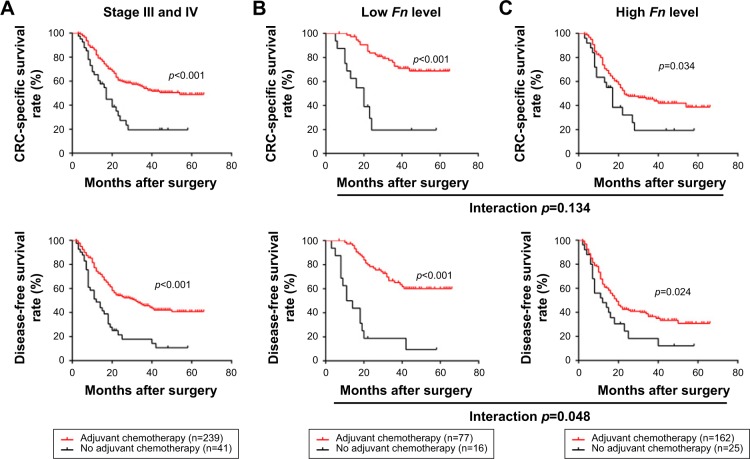
Adjuvant chemotherapy (AC) is the primary therapeutical modality for surgically treated CRC patients, especially for those within stage III/IV. Hence, we next made efforts to identify whether Fn level is associated with AC benefits in stage III/IV patients. In this study, majority of patients (n=239) received standard AC treatment postoperatively, while the rest (n=41) failed due to some factors such as poor physical condition and financial problems. As shown in Figure 3A, the survival analysis demonstrated that patients receiving AC had a dramatically better CSS and DFS than those receiving no AC (CSS: p<0.001; DFS, p<0.001). In the subgroups classified by Fn level, we found that AC treatment was associated with a significantly better clinical outcome in both patients with low Fn level (CSS: p<0.001, DFS: p<0.001, Figure 3B) and high Fn level (CSS: p=0.034, DFS: p=0.024, Figure 3C). However, the interaction analysis based on factorial design indicated that patients with low Fn level benefit more from AC than those with high Fn level, in terms of DFS (CSS: p=0.134; DFS: p=0.048).
Figure 3.
Correlations between Fn level and chemotherapy benefits in stage III/IV patients.
Notes: (A) CSS and DFS of the whole cohort, (B) CSS and DFS of low Fn level group, and (C) CSS and DFS of high Fn level group stratified by chemotherapy reception. An interaction analysis indicates that patients with low Fn level benefit more from chemotherapy than those with high Fn level, in terms of DFS (CSS: p=0.134; DFS: p=0.048).
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; CSS, cancer-specific survival; DFS, disease-free survival.
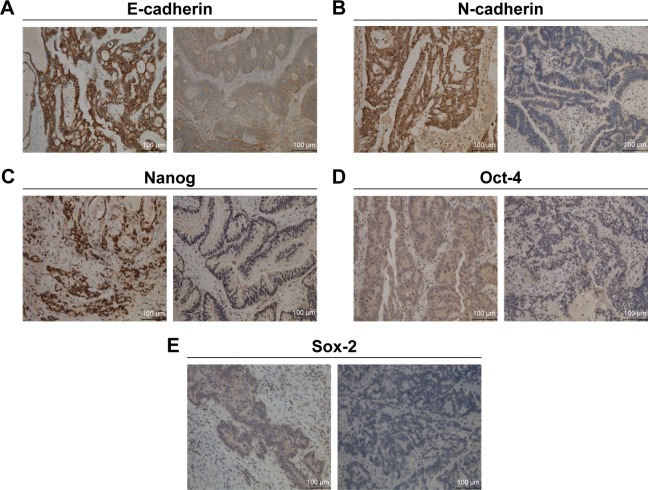
Expression and clinical significance of EMT and CSC markers in stage III/IV CRC patients
The representative images of IHC assay are shown in Figure 4. ROC curves were employed to estimate the cutoff values of staining scores for these markers and the results are shown in Figure S1. The cutoff value is 2.5 for E-cadherin and Sox-2, 3.5 for Oct-4, and 5 for N-cadherin and Nanog. Therefore, we used these cutoff values for the following statistical analysis and the correlations between their expression and clinicopathological features are summarized in Table S1. We noted that expression of these markers was significantly correlated with prognosis-related clinical features. For instance, both E-cadherin and N-cadherin expression was correlated with LNM status and distant metastasis (all p<0.05). Naong and Sox-2 expression was correlated with LNM status, while Oct-4 expression was correlated with distant metastasis (all p<0.05).
Figure 4.
Representative immunohistochemical staining images of EMT and CSC markers in CRC tissues.
Notes: High (left) and low (right) expression of E-cadherin (A), N-cadherin (B), Nanog (C), Oct-4 (D), Sox-2 (E). Magnification: ×200.
Abbreviations: CRC, colorectal cancer; EMT, epithelial-to-mesenchymal transition; CSC, cancer stem cell.
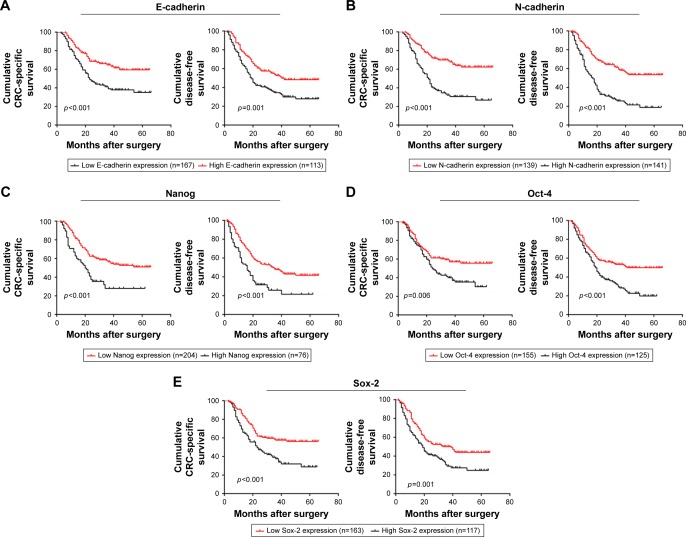
The prognostic significance of EMT and CSC markers was analyzed using Kaplan–Meier survival curves. Patients with high E-cadherin expression had a significantly better CSS and DFS than those with low E-cadherin expression (CSS: p<0.001, DFS: p=0.001, Figure 5A), while the opposite was true for N-cadherin (CSS: p<0.001, DFS: p<0.001, Figure 5B), Nanog (CSS: p<0.001, DFS: p<0.001, Figure 5C), Oct-4 (CSS: p=0.006, DFS: p<0.001, Figure 5D), and Sox-2 (CSS: p<0.001, DFS: p=0.001, Figure 5E).
Figure 5.
Prognostic significance of epithelial-to-mesenchymal transition and cancer stem cell markers in stage III/IV CRC patients.
Note: CSS and DFS curves stratified by E-cadherin expression (A), N-cadherin expression (B), Nanog expression (C), Oct-4 expression (D), Sox-2 expression (E).
Abbreviations: CRC, colorectal cancer; CSS, cancer-specific survival; DFS, disease-free survival.
Correlations of Fn with EMT and CSC markers in stage III/IV CRC patients
The correlations between Fn level and expression of EMT/CSC markers in CRC tissues are summarized in Table 4. Fn level was negatively correlated with E-cadherin expression (r=−0.301, p<0.001), but positively correlated with expression of N-cadherin (r=0.377, p<0.001) and Nanog (r=0.362, p<0.001). No significant association was observed between Fn level and Sox-2 expression (r=0.105, p=0.078) or Oct-4 expression (r=0.099, p=0.097).
Table 4.
Correlations of Fn with EMT/CSC markers in stage III/IV CRC patients
| Markers | N |
Fn level
|
r | p-value | |
|---|---|---|---|---|---|
| Low | High | ||||
| E-cadherin | <0.001 | ||||
| Low | 167 | 36 | 131 | −0.301 | |
| High | 113 | 57 | 56 | ||
| N-cadherin | <0.001 | ||||
| Low | 139 | 71 | 68 | 0.377 | |
| High | 141 | 22 | 119 | ||
| Sox-2 | 0.078 | ||||
| Low | 163 | 61 | 102 | 0.105 | |
| High | 117 | 32 | 85 | ||
| Oct-4 | 0.097 | ||||
| Low | 155 | 58 | 97 | 0.099 | |
| High | 125 | 35 | 90 | ||
| Nanog | <0.001 | ||||
| Low | 204 | 89 | 115 | 0.362 | |
| High | 76 | 4 | 72 | ||
Abbreviations: Fn, Fusobacterium nucleatum; CRC, colorectal cancer; EMT, epithelial-to-mesenchymal transition; CSC, cancer stem cell.
Discussion
Fn is a gram-negative anaerobe that is enriched in the oral cavity but hardly detected in other body organs under physiological conditions.24 However, under pathological conditions, it disseminates and colonizes into extraoral sites to function as pathogenic bacteria for various diseases such as inflammatory bowel disease, organ abscess, and adverse pregnancy outcome.25–27 In human malignancies, it is perhaps most relevant to CRC, although some emerging evidences have suggested its implication in esophageal and pancreatic cancer.28,29 Using RNA sequencing, Castellarin et al for the first time proposed that Fn infection might be prevalent in CRC patients.30 Then, increasing studies made efforts to investigate its potential oncogenic mechanisms in CRC where its regulatory role in tumor immunity is the most extensively discussed.31–33 In addition, Fn is found to be abundant in premalignant lesions with positive CpG island methylator phenotype, implying its involvement in epigenetic changes of early tumorigenesis.34 However, despite these novel findings about its oncogenic role, its prognostic significance in CRC patients remains unclear and whether it has the potential utility for improving the current TNM-based prognostic system still needs to be validated.
In this study, the level and clinical significance of Fn were analyzed in a cohort of 280 surgically treated stage III/IV patients. Firstly, we found that the Fn level is significantly higher in tumor tissues than that in adjacent normal tissues in both stage III and IV patients, supporting its promoting role in CRC initiation and development. A recent study proposed that this promoting role may be partly attributed to its participation in oncogenic biofilm formation.35 The following correlation analysis demonstrated that Fn level is significantly correlated with tumor invasion, LNM status, and distant metastasis. This further confirmed our previous finding that Fn enhances the malignant characteristics of CRC cells in vitro and in vivo.19 Li et al proved that Fn level is positively associated with the presence of LNM but not with tumor invasion in a relatively smaller cohort of CRC patients (n=101), partly consistent with our present result.36 Furthermore, Castellarin et al found that 74.4% (29/39) of CRC patients with high Fn level had positive LNM as compared with 44.8% (26/58) of those with low Fn level, also indicating a close correlation between Fn and LNM.30 Therefore, given these evidences, we concluded that Fn level might be a promising indicator for CRC metastasis in CRC patients, especially for those with positive LNM.
Although Fn level has been identified as an unfavorable prognostic factor in several studies, its specific prognostic significance for stage III/IV patients remains unknown.37,38 Using the Kaplan–Meier model, our survival analysis showed that stage III/IV patients with high Fn level had a significantly worse CSS and DFS than those with low Fn level. The following univariate and multivariate analysis not only further confirmed a significant correlation between Fn level and patient survival, but also revealed its independence in prognosis prediction. Given these results, we preliminarily proposed that Fn level might serve as a predictor for clinical outcome of stage III/IV patients. Several studies have recently suggested the limitation of traditional LNM status in prognosis stratification of stage III patients, strongly urging us to investigate whether the Fn level has the capacity to provide an accurate stratification for these patients.39,40 We therefore subsequently performed a subgroup analysis and found that Fn level could stratify the CSS and DFS of both stage IIIB and IIIC patients, but failed in stage IIIA patients. This result suggested that Fn level might be an effective prognostic indicator only for stage IIIB or IIIC patients. We also speculate that this result is partly attributed to the survival paradox that stage IIIA patients, clinically characterized as T1–2N1–2aM0, have a significantly better prognosis than other stage III and even most stage II patients, with a 5-year overall survival rate ranging from 81.6% to 85.6% as reported.41,42 This abnormally favorable prognosis may contribute to the failed prognostic stratification of Fn level in stage IIIA patients and we therefore suggest that detecting the Fn level in these patients may provide limited beneficial information for clinical management. Furthermore, we found that high Fn level is associated with worse outcome in stage IV patients despite the limited samples, implying its potential to be a prognostic predictor for surgically treated patients with distant metastasis. Finally, it should be noted that our study was unable to investigate the prognostic value of fecal Fn level in CRC patients, although its diagnostic potential has been highly advocated in several previous studies.43,44 Hence, whether its fecal level has any prognostic value or serves as a dynamic noninvasive marker like CEA in CRC surveillance still requires our extensive clinical validations in future.
Increasing evidences have supported that gut bacteria play a major role in modulating the anticancer efficacy of various CRC-related chemotherapeutic drugs such as 5-Fu, irinotecan, and oxaliplatin.45 To identify the correlation between Fn level and chemotherapy benefits in stage III/IV patients, a subgroup analysis was carried out based on Fn level and we found that patients receiving chemotherapy had a significantly better prognosis than those receiving no chemotherapy in both the high and low Fn level group. However, the following interaction analysis on DFS indicated that patients with low Fn level benefited more from chemotherapy than those with high Fn level, suggesting that Fn might be a predictive biomarker for chemotherapy response in stage III/IV patients. These results also implied its potential involvement in chemotherapy resistance of metastatic CRC cells. Yu et al have recently found that Fn can induce chemotherapy resistance of CRC cells through modulating autophagy via toll-like receptor/microRNAs signaling cascade, strongly supporting our results.46 Furthermore, it is reported that chemotherapy may in turn influence the gut bacteria of cancer patients.47 Therefore, whether the Fn level is changed during chemotherapy treatment and this change has any impact on therapy efficacy or even drug toxicity is also worthy of further investigation.
Finally, we analyzed the expression and clinical significance of EMT and CSC markers in stage III/IV patients, based on the consideration that both the molecular events play a major part in disease progression and therapy resistance of cancer patients.48 Our results showed that these markers are correlated with not only some clinicopathological features, but also CSS and DFS in stage III/IV patients. These findings are consistent with those of previous studies regarding their clinical significance in CRC patients.49–51 More importantly, through correlation analysis, we found that the Fn level was negatively correlated with E-cadherin expression, but positively correlated with N-cadherin expression in CRC tissues. Since loss of E-cadherin and gain of N-cadherin are defined as classical hallmarks of EMT, we speculated that Fn might contribute to CRC development partly by inducing this oncogenic molecular phenotype.52 This speculation is partly supported by a recent study that proved that Fn promotes CRC growth and invasion through regulating E-cadherin/β-catenin signaling.53 Our previous study also found that Fn upregulates miR-21 level to induce colitis-associated cancer by repressing E-cadherin, implying that Fn may induce EMT through upregulating miR-21.19,54 Then, we observed a positive correlation between Fn level and Nanog expression in CRC tissues, indicating that Fn might be involved in CSC phenotype. Nanog, as a well-established CSC marker, is also found to participate in the EMT program in cancer development, suggesting that Fn may partly induce EMT through regulating CSC phenotype.55,56 However, for further clarifying the correlation of Fn with EMT and CSC phenotype, extensive cellular assays are needed. In addition, it is reported that statins enhance the chemosensitivity of CRC cells through impairing CSC phenotype and whether Fn screening may be useful to discriminate between patients who most likely benefit from statins during chemotherapy still requires more clinical validations.57
In summary, our study indicates that Fn level is positively correlated with malignant progression and may serve as an independent prognostic indicator in stage III/IV patients. In addition, our findings also suggest that the Fn level is helpful for predicting chemotherapy benefits in these patients. Finally, we found that Fn level is correlated with several EMT and CSC markers in their tumor tissues, suggesting its potential involvement in EMT-CSC cross talk during CRC development. These findings not only suggest the immense potential of Fn as a clinically actionable biomarker for precise treatment in stage III/IV patients, but also provide a promising adjuvant therapeutic strategy for them that targeting Fn may be helpful for preventing CRC metastasis and improving chemotherapy efficacy.
Supplementary materials
The ROC curve analysis is used to determine the cutoff values of staining scores of epithelial–mesenchymal transition and cancer stem cell markers.
Notes: (A) E-cadherin; (B) N-cadherin; (C) Nanog; (D) Oct-4; (E) Sox-2.
Abbreviation: ROC, receiver operating characteristic.
Table S1.
Correlations between epithelial-to-mesenchymal transition/cancer stem cell markers and clinicopathological characteristics
| Characteristics | Total | E-cadherin
|
p-value | Total | N-cadherin
|
p-value | Total | Sox-2
|
p-value | Total | Oct-4
|
p-value | Total | Nanog
|
p-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | Low | High | Low | High | |||||||||||
| Gender | 0.761 | 0.892 | 0.621 | 0.279 | 0.434 | |||||||||||||||
| Female | 122 | 74 | 48 | 122 | 60 | 62 | 122 | 69 | 53 | 122 | 72 | 50 | 122 | 86 | 36 | |||||
| Male | 158 | 93 | 65 | 158 | 79 | 79 | 158 | 94 | 64 | 158 | 83 | 75 | 158 | 118 | 40 | |||||
| Age | 0.960 | 0.607 | 0.878 | 0.892 | 0.559 | |||||||||||||||
| ≤65 years | 111 | 66 | 45 | 111 | 53 | 58 | 111 | 64 | 47 | 111 | 62 | 49 | 111 | 83 | 28 | |||||
| >65 years | 169 | 101 | 68 | 169 | 86 | 83 | 169 | 99 | 70 | 169 | 93 | 76 | 169 | 121 | 48 | |||||
| Tumor location | 0.708 | 0.117 | 0.748 | 0.803 | 0.538 | |||||||||||||||
| Rectal | 130 | 76 | 54 | 130 | 58 | 72 | 130 | 77 | 53 | 130 | 73 | 57 | 130 | 97 | 33 | |||||
| Colon | 150 | 91 | 59 | 150 | 81 | 69 | 150 | 86 | 64 | 150 | 82 | 68 | 150 | 107 | 43 | |||||
| Tumor size | 0.297 | 0.233 | 0.904 | 0.664 | 0.731 | |||||||||||||||
| ≤5 cm | 214 | 124 | 90 | 214 | 102 | 112 | 214 | 125 | 89 | 214 | 120 | 94 | 214 | 157 | 57 | |||||
| >5 cm | 66 | 43 | 23 | 66 | 37 | 29 | 66 | 38 | 28 | 66 | 35 | 31 | 66 | 47 | 19 | |||||
| Tumor differentiation | 0.555 | 0.311 | 0.422 | 0.992 | 0.134 | |||||||||||||||
| Poor | 74 | 42 | 32 | 74 | 33 | 41 | 74 | 46 | 28 | 74 | 41 | 33 | 74 | 49 | 25 | |||||
| Well/moderate | 206 | 125 | 81 | 206 | 106 | 100 | 206 | 117 | 89 | 206 | 114 | 92 | 206 | 155 | 51 | |||||
| Tumor invasion | 0.275 | 0.555 | 0.630 | 0.120 | 0.010 | |||||||||||||||
| T1–T2 | 96 | 53 | 43 | 96 | 50 | 46 | 96 | 54 | 42 | 96 | 47 | 49 | 96 | 79 | 17 | |||||
| T3–T4 | 184 | 114 | 70 | 184 | 89 | 95 | 184 | 109 | 75 | 184 | 108 | 76 | 184 | 125 | 59 | |||||
| Lymph node metastasis | 0.029 | 0.024 | 0.046 | 0.609 | 0.003 | |||||||||||||||
| N1 | 81 | 39 | 42 | 81 | 31 | 50 | 81 | 43 | 38 | 81 | 45 | 36 | 81 | 59 | 22 | |||||
| N2a | 95 | 58 | 37 | 95 | 56 | 39 | 95 | 65 | 30 | 95 | 56 | 39 | 95 | 80 | 15 | |||||
| N2b | 104 | 70 | 34 | 104 | 52 | 52 | 104 | 55 | 49 | 104 | 54 | 50 | 104 | 65 | 39 | |||||
| Distant metastasis | 0.039 | 0.012 | 0.396 | <0.001 | 0.118 | |||||||||||||||
| Absent | 218 | 123 | 95 | 218 | 117 | 101 | 218 | 124 | 94 | 218 | 134 | 84 | 218 | 154 | 64 | |||||
| Present | 62 | 44 | 18 | 62 | 22 | 40 | 62 | 39 | 23 | 62 | 21 | 41 | 62 | 50 | 12 | |||||
| Ki-67 positivity | 0.042 | 0.382 | 0.483 | 0.394 | 0.041 | |||||||||||||||
| <30% | 78 | 54 | 24 | 78 | 42 | 36 | 78 | 48 | 30 | 78 | 40 | 38 | 78 | 50 | 28 | |||||
| ≥30% | 202 | 113 | 89 | 202 | 97 | 105 | 202 | 115 | 87 | 202 | 115 | 87 | 202 | 154 | 48 | |||||
| Serum CEA level | 0.199 | 0.971 | 0.174 | 0.764 | 0.276 | |||||||||||||||
| ≤5 ng/mL | 99 | 54 | 45 | 99 | 49 | 50 | 99 | 63 | 36 | 99 | 56 | 43 | 99 | 76 | 23 | |||||
| >5 ng/mL | 181 | 113 | 68 | 181 | 90 | 91 | 181 | 100 | 81 | 181 | 99 | 82 | 181 | 128 | 53 | |||||
| BMI | 0.472 | 0.887 | 0.367 | 0.394 | 0.166 | |||||||||||||||
| <18.5 kg/m2 | 22 | 12 | 10 | 22 | 12 | 10 | 22 | 14 | 8 | 22 | 11 | 11 | 22 | 13 | 9 | |||||
| 18.5–24.99 kg/m2 | 178 | 111 | 67 | 178 | 88 | 90 | 178 | 98 | 80 | 178 | 104 | 74 | 178 | 128 | 50 | |||||
| ≥25.0 kg/m2 | 80 | 44 | 36 | 80 | 39 | 41 | 80 | 51 | 29 | 80 | 40 | 40 | 80 | 63 | 17 | |||||
Abbreviations: CEA, carcinoembryonic antigen; BMI, body mass index.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (no 81230057, 81372615, 81472262, and 81200264); Emerging Cutting-Edge Technology Joint Research projects of Shanghai (SHDC12012106), Tongji University Subject Pilot Program (no 162385), and China Postdoctoral Science Foundation (no 2017M610278). We thank Dr Ruting Xie (Department of Pathology, Shanghai Tenth People’s Hospital, Tongji University School of Medicine) for her crucial assistance in sample collection and IHC assay.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Aran V, Victorino AP, Thuler LC, Ferreira CG. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer. 2016;15(3):195–203. doi: 10.1016/j.clcc.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 6.De Greef K, Rolfo C, Russo A, et al. Multidisciplinary management of patients with liver metastasis from colorectal cancer. World J Gastroenterol. 2016;22(32):7215–7225. doi: 10.3748/wjg.v22.i32.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troiani T, Napolitano S, Della Corte CM, et al. Therapeutic value of EGFR inhibition in CRC and NSCLC: 15 years of clinical evidence. ESMO Open. 2016;1(5):e000088. doi: 10.1136/esmoopen-2016-000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh RB, Prasad V. Blood-based screening for colon cancer: a disruptive innovation or simply a disruption? JAMA. 2016;315(23):2519–2520. doi: 10.1001/jama.2016.7914. [DOI] [PubMed] [Google Scholar]
- 9.Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35(13):1453–1486. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 10.Tsilimigras MC, Fodor A, Jobin C. Carcinogenesis and therapeutics: the microbiota perspective. Nat Microbiol. 2017;2:17008. doi: 10.1038/nmicrobiol.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15(2):109–128. doi: 10.1038/nrmicro.2016.171. [DOI] [PubMed] [Google Scholar]
- 12.Dart A. Tumour microenvironment: that gut feeling. Nat Rev Cancer. 2016;16(12):756–757. doi: 10.1038/nrc.2016.122. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor A, O’Morain CA, Ford AC. Population screening and treatment of Helicobacter pylori infection. Nat Rev Gastroenterol Hepatol. 2017;14(4):230–240. doi: 10.1038/nrgastro.2016.195. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol. 2016;14(8):508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 15.Tsoi H, Chu ES, Zhang X, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152(6):1419–1433. doi: 10.1053/j.gastro.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Yang Y, Huycke MM. Commensal bacteria drive endogenous transformation and tumour stem cell marker expression through a bystander effect. Gut. 2015;64(3):459–468. doi: 10.1136/gutjnl-2014-307213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q, Jin Z, Wu W, et al. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9(6):e90849. doi: 10.1371/journal.pone.0090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–866. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16(1):8. doi: 10.1186/s12943-016-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, You Y, Jiang H, Wang ZZ. Epithelial–mesenchymal transition (EMT): a biological process in the development, stem cell differentiation and tumorigenesis. J Cell Physiol. 2017;232(12):3261–3272. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, Liu L, Li H, et al. Dual specificity phosphatase 5 is a novel prognostic indicator for patients with advanced colorectal cancer. Am J Cancer Res. 2016;6(10):2323–2333. [PMC free article] [PubMed] [Google Scholar]
- 23.Dalerba P, Sahoo D, Paik S, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med. 2016;374(3):211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahara T, Shibata T, Kawamura T, et al. Fusobacterium detected in colonic biopsy and clinicopathological features of ulcerative colitis in Japan. Dig Dis Sci. 2015;60(1):205–210. doi: 10.1007/s10620-014-3316-y. [DOI] [PubMed] [Google Scholar]
- 26.Atanasova KR, Yilmaz O. Prelude to oral microbes and chronic diseases: past, present and future. Microbes Infect. 2015;17(7):473–483. doi: 10.1016/j.micinf.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockham S, Stamford JE, Roberts CT, et al. Abnormal pregnancy outcomes in mice using an induced periodontitis model and the haematogenous migration of Fusobacterium nucleatum sub-species to the murine placenta. PLoS One. 2015;10(3):e0120050. doi: 10.1371/journal.pone.0120050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22(22):5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuhashi K, Nosho K, Sukawa Y, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6(9):7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito T, Nishikawa H, Wada H, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22(6):679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 32.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1(5):653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito M, Kanno S, Nosho K, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137(6):1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol Med. 2017;23(1):18–30. doi: 10.1016/j.molmed.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Li YY, Ge QX, Cao J, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016;22(11):3227–3233. doi: 10.3748/wjg.v22.i11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 39.Huang B, Chen C, Ni M, Mo S, Cai G, Cai S. Log odds of positive lymph nodes is a superior prognostic indicator in stage III rectal cancer patients: a retrospective analysis of 17,632 patients in the SEER database. Int J Surg. 2016;32:24–30. doi: 10.1016/j.ijsu.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Li Q, Liang L, Jia H, et al. Negative to positive lymph node ratio is a superior predictor than traditional lymph node status in stage III colorectal cancer. Oncotarget. 2016;7(44):72290–72299. doi: 10.18632/oncotarget.10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu QD, Zhou M, Medeiros K, Peddi P. Positive surgical margins contribute to the survival paradox between patients with stage IIB/C (T4N0) and stage IIIA (T1-2N1, T1N2a) colon cancer. Surgery. 2016;160(5):1333–1343. doi: 10.1016/j.surg.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 42.Chu QD, Zhou M, Medeiros KL, Peddi P, Kavanaugh M, Wu XC. Poor survival in stage IIB/C (T4N0) compared to stage IIIA (T1-2 N1, T1N2a) colon cancer persists even after adjusting for adequate lymph nodes retrieved and receipt of adjuvant chemotherapy. BMC Cancer. 2016;16:460. doi: 10.1186/s12885-016-2446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong SH, Kwong TN, Chow TC, et al. Quantitation of faecal Fuso-bacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66(8):1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 45.Alexander JL, Wilson ID, Teare J, Marchesi JR, Nicholson JK, Kinross JM. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356–365. doi: 10.1038/nrgastro.2017.20. [DOI] [PubMed] [Google Scholar]
- 46.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopala SV, Yooseph S, Harkins DM, et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent acute lymphoblastic leukemia (ALL) at the time of disease diagnosis. BMC Genomics. 2016;17(1):635. doi: 10.1186/s12864-016-2965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishiwata T. Cancer stem cells and epithelial–mesenchymal transition: novel therapeutic targets for cancer. Pathol Int. 2016;66(11):601–608. doi: 10.1111/pin.12447. [DOI] [PubMed] [Google Scholar]
- 49.Yan X, Yan L, Liu S, Shan Z, Tian Y, Jin Z. N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer. Mol Med Rep. 2015;12(2):2999–3006. doi: 10.3892/mmr.2015.3687. [DOI] [PubMed] [Google Scholar]
- 50.Meng HM, Zheng P, Wang XY, et al. Over-expression of Nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9(4):295–302. doi: 10.4161/cbt.9.4.10666. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H, Hu YU, Wang W, et al. Expression of Oct-4 is significantly associated with the development and prognosis of colorectal cancer. Oncol Lett. 2015;10(2):691–696. doi: 10.3892/ol.2015.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial–mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi C, Yang Y, Xia Y, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65(9):1470–1481. doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 55.Pan Q, Meng L, Ye J, et al. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial–mesenchymal transition (EMT) Cancer Lett. 2017;392:26–38. doi: 10.1016/j.canlet.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 56.Migita T, Ueda A, Ohishi T, et al. Epithelial–mesenchymal transition promotes SOX2 and NANOG expression in bladder cancer. Lab Invest. 2017 Feb 27; doi: 10.1038/labinvest.2017.17. Epub. [DOI] [PubMed] [Google Scholar]
- 57.Kodach LL, Jacobs RJ, Voorneveld PW, et al. Statins augment the chemosensitivity of colorectal cancer cells inducing epigenetic reprogramming and reducing colorectal cancer cell ‘stemness’ via the bone morphogenetic protein pathway. Gut. 2011;60(11):1544–1553. doi: 10.1136/gut.2011.237495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ROC curve analysis is used to determine the cutoff values of staining scores of epithelial–mesenchymal transition and cancer stem cell markers.
Notes: (A) E-cadherin; (B) N-cadherin; (C) Nanog; (D) Oct-4; (E) Sox-2.
Abbreviation: ROC, receiver operating characteristic.
Table S1.
Correlations between epithelial-to-mesenchymal transition/cancer stem cell markers and clinicopathological characteristics
| Characteristics | Total | E-cadherin
|
p-value | Total | N-cadherin
|
p-value | Total | Sox-2
|
p-value | Total | Oct-4
|
p-value | Total | Nanog
|
p-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | Low | High | Low | High | |||||||||||
| Gender | 0.761 | 0.892 | 0.621 | 0.279 | 0.434 | |||||||||||||||
| Female | 122 | 74 | 48 | 122 | 60 | 62 | 122 | 69 | 53 | 122 | 72 | 50 | 122 | 86 | 36 | |||||
| Male | 158 | 93 | 65 | 158 | 79 | 79 | 158 | 94 | 64 | 158 | 83 | 75 | 158 | 118 | 40 | |||||
| Age | 0.960 | 0.607 | 0.878 | 0.892 | 0.559 | |||||||||||||||
| ≤65 years | 111 | 66 | 45 | 111 | 53 | 58 | 111 | 64 | 47 | 111 | 62 | 49 | 111 | 83 | 28 | |||||
| >65 years | 169 | 101 | 68 | 169 | 86 | 83 | 169 | 99 | 70 | 169 | 93 | 76 | 169 | 121 | 48 | |||||
| Tumor location | 0.708 | 0.117 | 0.748 | 0.803 | 0.538 | |||||||||||||||
| Rectal | 130 | 76 | 54 | 130 | 58 | 72 | 130 | 77 | 53 | 130 | 73 | 57 | 130 | 97 | 33 | |||||
| Colon | 150 | 91 | 59 | 150 | 81 | 69 | 150 | 86 | 64 | 150 | 82 | 68 | 150 | 107 | 43 | |||||
| Tumor size | 0.297 | 0.233 | 0.904 | 0.664 | 0.731 | |||||||||||||||
| ≤5 cm | 214 | 124 | 90 | 214 | 102 | 112 | 214 | 125 | 89 | 214 | 120 | 94 | 214 | 157 | 57 | |||||
| >5 cm | 66 | 43 | 23 | 66 | 37 | 29 | 66 | 38 | 28 | 66 | 35 | 31 | 66 | 47 | 19 | |||||
| Tumor differentiation | 0.555 | 0.311 | 0.422 | 0.992 | 0.134 | |||||||||||||||
| Poor | 74 | 42 | 32 | 74 | 33 | 41 | 74 | 46 | 28 | 74 | 41 | 33 | 74 | 49 | 25 | |||||
| Well/moderate | 206 | 125 | 81 | 206 | 106 | 100 | 206 | 117 | 89 | 206 | 114 | 92 | 206 | 155 | 51 | |||||
| Tumor invasion | 0.275 | 0.555 | 0.630 | 0.120 | 0.010 | |||||||||||||||
| T1–T2 | 96 | 53 | 43 | 96 | 50 | 46 | 96 | 54 | 42 | 96 | 47 | 49 | 96 | 79 | 17 | |||||
| T3–T4 | 184 | 114 | 70 | 184 | 89 | 95 | 184 | 109 | 75 | 184 | 108 | 76 | 184 | 125 | 59 | |||||
| Lymph node metastasis | 0.029 | 0.024 | 0.046 | 0.609 | 0.003 | |||||||||||||||
| N1 | 81 | 39 | 42 | 81 | 31 | 50 | 81 | 43 | 38 | 81 | 45 | 36 | 81 | 59 | 22 | |||||
| N2a | 95 | 58 | 37 | 95 | 56 | 39 | 95 | 65 | 30 | 95 | 56 | 39 | 95 | 80 | 15 | |||||
| N2b | 104 | 70 | 34 | 104 | 52 | 52 | 104 | 55 | 49 | 104 | 54 | 50 | 104 | 65 | 39 | |||||
| Distant metastasis | 0.039 | 0.012 | 0.396 | <0.001 | 0.118 | |||||||||||||||
| Absent | 218 | 123 | 95 | 218 | 117 | 101 | 218 | 124 | 94 | 218 | 134 | 84 | 218 | 154 | 64 | |||||
| Present | 62 | 44 | 18 | 62 | 22 | 40 | 62 | 39 | 23 | 62 | 21 | 41 | 62 | 50 | 12 | |||||
| Ki-67 positivity | 0.042 | 0.382 | 0.483 | 0.394 | 0.041 | |||||||||||||||
| <30% | 78 | 54 | 24 | 78 | 42 | 36 | 78 | 48 | 30 | 78 | 40 | 38 | 78 | 50 | 28 | |||||
| ≥30% | 202 | 113 | 89 | 202 | 97 | 105 | 202 | 115 | 87 | 202 | 115 | 87 | 202 | 154 | 48 | |||||
| Serum CEA level | 0.199 | 0.971 | 0.174 | 0.764 | 0.276 | |||||||||||||||
| ≤5 ng/mL | 99 | 54 | 45 | 99 | 49 | 50 | 99 | 63 | 36 | 99 | 56 | 43 | 99 | 76 | 23 | |||||
| >5 ng/mL | 181 | 113 | 68 | 181 | 90 | 91 | 181 | 100 | 81 | 181 | 99 | 82 | 181 | 128 | 53 | |||||
| BMI | 0.472 | 0.887 | 0.367 | 0.394 | 0.166 | |||||||||||||||
| <18.5 kg/m2 | 22 | 12 | 10 | 22 | 12 | 10 | 22 | 14 | 8 | 22 | 11 | 11 | 22 | 13 | 9 | |||||
| 18.5–24.99 kg/m2 | 178 | 111 | 67 | 178 | 88 | 90 | 178 | 98 | 80 | 178 | 104 | 74 | 178 | 128 | 50 | |||||
| ≥25.0 kg/m2 | 80 | 44 | 36 | 80 | 39 | 41 | 80 | 51 | 29 | 80 | 40 | 40 | 80 | 63 | 17 | |||||
Abbreviations: CEA, carcinoembryonic antigen; BMI, body mass index.