SUMMARY
K-Ras is targeted to the plasma membrane by a C-terminal membrane anchor that comprises a farnesyl-cysteine-methyl-ester and a polybasic domain. We used quantitative spatial imaging and atomistic molecular dynamics simulations to examine molecular details of K-Ras plasma membrane binding. We found that the K-Ras anchor binds selected plasma membrane anionic lipids with defined head groups and lipid side chains. The precise amino acid sequence and prenyl group define a combinatorial code for lipid binding that extends beyond simple electrostatics; within this code lysine and arginine residues are non-equivalent and prenyl chain length modifies nascent polybasic domain lipid preferences. The code is realized by distinct dynamic tertiary structures of the anchor on the plasma membrane that govern amino acid side-chain-lipid interactions. An important consequence of this specificity is the ability of such anchors when aggregated to sort subsets of phospholipids into nanoclusters with defined lipid compositions that determine K-Ras signaling output.
INTRODUCTION
Ras proteins oscillate between inactive GDP-bound and active GTP-bound states to regulate cell survival and proliferation. Three isoforms of Ras are ubiquitously expressed in human cells: K-Ras, H-Ras, and N-Ras, of which K-Ras and N-Ras are frequently mutated in human cancers (Prior et al., 2012). Ras proteins contain near identical G-domains that bind guanine nucleotides and interact with a common set of activators and effectors but have divergent C-terminal hypervariable regions (Hancock, 2003). For biological activity, Ras proteins must localize to the plasma membrane (PM). A Ras PM anchor consists of two components: a C-terminal S-farnesyl cysteine carboxylmethyl ester, common to all isoforms; and a second signal that comprises mono-palmitoylation of N-Ras, duo-palmitoylation of H-Ras and a polybasic domain (PBD) of six contiguous lysines in K-Ras4B, the predominantly expressed splice variant of K-Ras, hereafter referred to as K-Ras (Hancock et al., 1989, 1990).
On the PM, Ras proteins diffuse laterally as monomers and dimers and assemble into higher order oligomers and nanoclusters (Muratcioglu et al., 2015; Nan et al., 2015; Plowman et al., 2005; Zhou and Hancock, 2015). H-Ras, N-Ras, and K-Ras nanoclusters are spatially non-overlapping as are GTP- and GDP-loaded Ras nanoclusters (Abankwa et al., 2008, 2010; Gorfe et al., 2007a, 2007b; Güldenhaupt et al., 2012; Kapoor et al., 2012; Prior et al., 2003a; Weise et al., 2011; Zhou et al., 2014). A nanocluster contains approximately six Ras proteins, has a radius of ~9 nm (Plowman et al., 2005), and a lifetime of <1 s (Murakoshi et al., 2004; Plowman et al., 2005). Ras nanoclusters are platforms for effector binding and signal transmission (Hancock, 2003; Plowman et al., 2005; Tian et al., 2007). The recruitment and activation of RAF, MEK, and ERK within nanoclusters results in key emergent properties including high fidelity and low noise signal transmission within the Ras-mitogen-activated protein kinase (MAPK) signaling circuit (Kholodenko et al., 2010; Tian et al., 2007).
K-Ras localization to and spatial distribution on the PM are determined by the C-terminal farnesyl anchor and PBD (Hancock et al., 1989, 1990). Many other small GTPases contain PBDs, each with a specific sequence of arginines and lysines adjacent to a prenyl anchor; PBDs are also commonly found in other PM-interacting proteins. The presumption has been that all these PBDs are functionally identical and mediate non-specific electrostatic interactions with anionic phospholipids that are asymmetrically concentrated in the inner leaflet of the PM (Plowman et al., 2008). However, K-Ras nanoclusters contain a high level of monovalent phosphatidylserine (PtdSer) and phosphatidic acid but little multivalent phosphoinositol 4,5-bisphosphate (PIP2) (Cho et al., 2015; Zhou et al., 2014), suggesting that PBD interactions may not simply reflect electrostatics. Here, we now decode cryptic structural and dynamic features of the K-Ras membrane anchor that define exquisite specificity for highly selected PM anionic phospholipids.
RESULTS
K-Ras PM Localization and Nanoclustering Requires Specific PtdSer Acyl Chain Structures
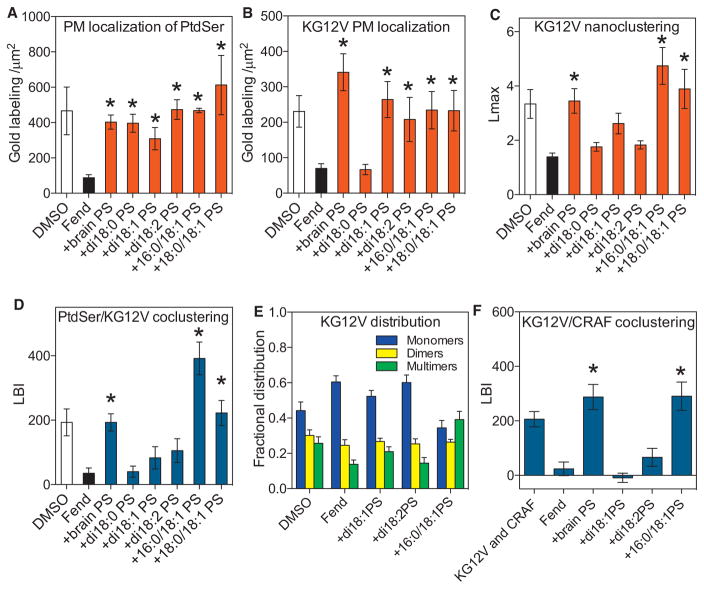
The K-Ras PBD interacts with anionic lipids for PM localization and lateral segregation. Electron microscopy (EM) spatial mapping experiments show that K-Ras nanoclusters are enriched in PtdSer but not PIP2 (Zhou et al., 2014). Depleting PM PtdSer causes loss of K-Ras from the PM and reduced nanoclustering (Cho et al., 2015; Zhou et al., 2014). Conversely enhancing PM PtdSer clustering by depolarizing the PM enhances K-Ras clustering (Zhou et al., 2015). These results suggest that the K-Ras anchor might selectively interact with specific anionic lipids by recognizing more than just charged lipid head groups. To test this, we examined whether K-Ras discriminates between PtdSer with different acyl chain structures. We reduced PM PtdSer content by treating baby hamster kidney (BHK) cells with R-Fendiline for 24 hr (Cho et al., 2015; van der Hoeven et al., 2013) and then added-back defined species of exogenous PtdSer including natural brain PtdSer, synthetic fully saturated di18:0 PtdSer, monounsaturated di18:1 PtdSer, di-unsaturated di18:2 PtdSer, and asymmetric 16:0/18:1 PtdSer or 18:0/18:1 PtdSer. To verify that each PtdSer species was delivered to the inner PM leaflet, we monitored the localization of GFP-LactC2, a specific PtdSer binding probe (Yeung et al., 2008). PM sheets were prepared from BHK cells expressing GFP-LactC2 and labeled with anti-GFP coupled to 4.5 nm gold particles and imaged by EM. The number of gold particles per 1 μm2 was used as a measure of PtdSer level. Fendiline treatment markedly reduced PtdSer PM content, which was restored to control levels by all PtdSer lipids (Figures 1A and S1). Because LactC2 interacts with the PtdSer head group, all exogenous PtdSer lipids were efficiently incorporated into the inner leaflet of the PM. Lipidomic analysis confirmed minimal acyl chain remodeling of the exogenous PtdSer lipids over the time course of the experiment (Figure S2H). Identical experiments were then carried out in fendiline-treated cells expressing GFP-K-RasG12V, the constitutively GTP-bound oncogenic mutant of K-Ras. Add-back of fully saturated di18:0 PtdSer did not recover GFP-K-RasG12V PM localization whereas add-back of other symmetric unsaturated and asymmetric PtdSer lipids successfully recovered GFP-K-RasG12V PM localization (Figure 1B). K-RasG12V nanoclustering was evaluated by spatial statistics to analyze the immunogold point patterns (Figures S1A–S1D), with the maximum value of the univariate K-function, Lmax used to quantify the extent of clustering. However, other summary parameters yield identical conclusions (Figure S2). Fendiline treatment markedly disrupted K-RasG12V nanoclustering on the PM as evidenced by a significant reduction in Lmax from control (Figure 1C). Strikingly, however, only PtdSer lipids with asymmetric acyl chains, such as natural brain PtdSer, a mixture but with 18:0/18:1 PtdSer as the main component, synthetic 16:0/18:1 PtdSer, or 18:0/18:1 PtdSer, fully recovered K-RasG12V nanoclustering (Figure 1C). PtdSer add-back experiments in PSA3 cells that had PM PtdSer levels reduced by growth in ethanolamine-depleted serum (Lee et al., 2012; Zhou et al., 2014) yielded identical results to those in fendiline-treated cells (Figures S2D–S2F).
Figure 1. K-Ras Interacts with Selected Species of PtdSer.
(A and B) BHK cells stably expressing the PtdSer probe GFP-LactC2 (A) or GFP-K-RasG12V (B) were treated with vehicle (DMSO) or 10 μM fendiline (Fend) for 24 hr before incubation with exogenous PtdSer lipids for 1 hr. Intact PM sheets were fixed, labeled with 4.5 nm gold-anti-GFP, and imaged by EM. PtdSer and K-RasG12V content, respectively, of the inner PM leaflet is quantified as the number of gold particles per 1 μm2 and shown as mean ± SEM (n ≥ 15 for each condition). Significant differences between fendiline-treated cells with and without PtdSer add-back were evaluated by one-way ANOVA (*p < 0.05).
(C) The gold point patterns from (B) were analyzed with univariate K-functions expressed as L(r)-r. The extent of GFP-K-RasG12V nanoclustering is quantified by the peak value Lmax of the L(r)-r curve (Figure S1). The results are mean ± SEM (n ≥ 15 for each condition). Statistical significance of differences between fendiline-treated cells with and without PtdSer add-back were evaluated in bootstrap tests (*p < 0.05).
(D) BHK cells co-expressing GFP-LactC2 and RFP-K-RasG12V were treated with vehicle (DMSO) or 10 μM fendiline for 24 hr and incubated with exogenous PtdSer for 1 hr. Intact PM sheets were labeled with 6 nm gold-anti-GFP and 2 nm gold-anti-RFP and imaged by EM. Co-localization of the two populations of gold particles and hence of LactC2 (PtdSer) and K-RasG12V was analyzed by integrated bivariate K-functions (= LBI) (Figure S1). Statistical significance of differences between fendiline-treated cells with and without PtdSer add-back were evaluated in bootstrap tests (*p < 0.05).
(E) A local L(r)-r analysis at r = 15 nm was used estimate to relative proportions of monomers, dimers, and higher order oligomers (nanoclusters) in the GFP-K-RasG12V gold PM point patterns in (B) and (C). Significant differences between fendiline-treated cells with and without PtdSer add-back were evaluated by one-way ANOVA (*p < 0.05).
(F) BHK cells co-expressing GFP-K-RasG12V and RFP-CRAF were treated with vehicle (DMSO) or 10 μM fendiline for 24 hr before 1 hr incubation with exogenous PtdSer. PM sheets from the cells were labeled with 6 nm gold-anti-GFP and 2 nm gold-anti-RFP and imaged by EM. The extent of co-clustering between K-RasG12V and CRAF was analyzed by integrated bivariate K-functions (= LBI). The results are mean ± SEM (n ≥15 for each condition). Statistical significance between fendiline-treated cells with and without PtdSer add-back were evaluated in bootstrap tests (*p < 0.05).
See also Figures S1 and S2.
We next analyzed co-localization between PtdSer and K-RasG12V after lipid add-back. PM sheets from BHK cells co-expressing GFP-LactC2 and RFP-K-RasG12V were co-labeled with 6 nm and 2 nm gold particles coupled to anti-GFP and anti-RFP, respectively, and the gold particle distributions quantified with bivariate K-functions (Lbiv(r)-r) (Figures S1E–S1H). We use a defined integral of the Lbiv(r)-r function termed Lbiv-integrated (LBI) as a summary parameter (Zhou et al., 2014, 2015). The larger the LBI value, the more extensive the co-localization of the 2 nm and 6 nm gold patterns. The extensive co-clustering of GFP-LactC2 with RFP-K-RasG12V was markedly reduced after fendiline treatment, as reflected by a significant reduction in the LBI value (Figure 1D). Add-back of brain PtdSer, 16:0/18:1 PtdSer, or 18:0/18:1 PtdSer significantly enhanced co-localization between PtdSer and K-RasG12V, while di18:0 PtdSer, di18:1 PtdSer, or di18:2 PtdSer did not (Figure 1D). Taken together, these data show that the K-Ras anchor is unable to interact with fully saturated PtdSer on the PM, can interact with symmetric mono- and di-unsaturated PtdSer to support PM binding, but can only assemble PtdSer with asymmetric acyl chains into nanoclusters. Further interrogation of the univariate point patterns also showed K-Ras anchored by symmetric di-unsaturated PtdSer present as monomers and dimers, with few higher order oligomers, whereas asymmetric PtdSer enhanced the formation of higher order oligomers (Figures 1E and S2C). This selective role of asymmetric PtdSer was also evident in the ability of K-RasG12V to recruit CRAF to the PM, a functional assay of nanocluster assembly (Plowman et al., 2008; Tian et al., 2007). In EM and fluorescence lifetime imaging microscopy (FLIM) experiments, GFP-K-RasG12V and RFP-CRAF co-clustering on the PM that was reduced by fendiline treatment could only be restored by add back of brain PtdSer or asymmetric 16:0/18:1 PtdSer (Figures 1F and S2G).
PBD Sequence and Prenyl Anchor Define Lipid Selectivity
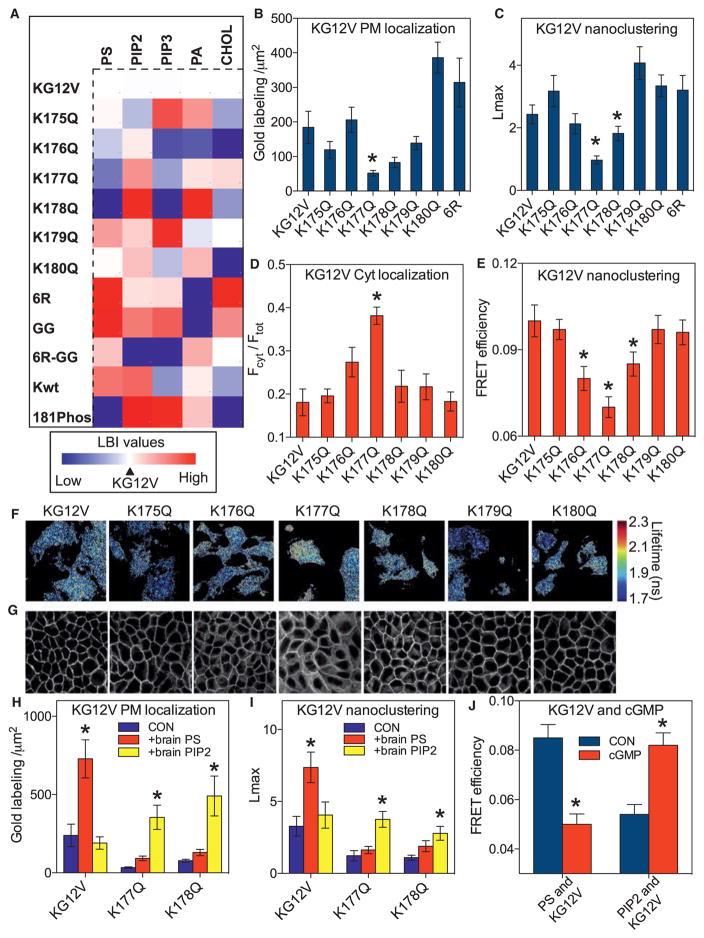
The selectivity of the K-Ras PM anchor for specific PtdSer lipids indicates that the PBD is more than a charge detector. To investigate this selectivity, we generated a set of single point mutations to reduce the net charge of the K-RasG12V PBD by 1: the mutations scanned through the PBD from K175 to K180 sequentially replacing each lysine with an uncharged glutamine. The lipid composition of nanoclusters formed by the PBD mutants was evaluated by bivariate EM spatial mapping. Each RFP-K-RasG12V PBD mutant was sequentially co-expressed with a set of lipid binding probes for PtdSer (GFP-LactC2), PIP2 (GFP-PH-PLCδ), phosphoinositol 3,4,5-triphosphate (PIP3) (GFP-PH-Akt), phosphatidic acid (GFP-Spo20), and cholesterol (GFP-D4H) (Maekawa and Fairn, 2015). Figure 2A shows the results as a heatmap matrix of LBI values centered on the K-RasG12V control. Each K-RasG12V PBD mutant generates nanoclusters with a different lipid composition, despite having the same net PBD charge. A similar profile was generated from fluorescence resonance energy transfer (FRET) efficiencies measured in FLIM experiments using the same probes (Figures S3F–S3I). We also used FLIM to directly measure the interaction of each RFP-K-RasG12V PBD mutant with fluorescently labeled Top-Fluor-PtdSer (PtdSer) that is effectively asymmetric having one saturated (C16 palmitoyl) chain and one acyl chain containing a BODIPY moiety. FRET efficiencies followed the same trend as the corresponding LactC2 LBI values (Figure S3J). Phosphorylation of Ser181 by protein kinase C (PKC) (Bivona et al., 2006) or protein kinase G (PKG) (Cho et al., 2016) also reduces the net charge on the K-Ras PBD. We therefore conducted lipid mapping of RFP-K-RasG12V nanoclusters after acute activation of PKG. EM-bivariate (Figure 2A) and FLIM-FRET (Figure 2J) analyses show that phosphorylation of Ser181 (181-phos) significantly changed nanocluster lipid content, reducing PtdSer and increasing PIP2.
Figure 2. The K-Ras Membrane Anchor Is a Combinatorial Code for Lipid Sorting.
(A) A heatmap was constructed with EM-derived mean LBI values (n ≥15 for each condition) to quantify co-clustering between GFP-lipid-binding domains and RFP-K-RasG12V mutants. The mutants include K → Q point mutations at each lysine in the PBD (K175-K180), replacement of all lysines with arginines (6R), replacement of farnesyl with geranylgeranyl (GG). K-RasG12V phosphorylated at Ser181 (S181-Phos) and wild-type K-Ras (Kwt) were also evaluated. For each lipid, the LBI value for K-RasG12V and the cognate lipid probe was assigned as midpoint (marked in white), with lower and higher values marked in blue and red, respectively. The heatmap thus shows positive or negative deviations from the lipid content of K-RasG12V nanoclusters. For raw LBI values see Figures S3A–S3E. The LactC2 domain did not compete for PtdSer because the PM binding and nanoclustering of GFP-K-RasG12V and each PBD mutant was unaffected by RFP-LactC2 expression (Figures S3K–S3N). Co-expression of the LactC2 domain also had no effect on K-RasG12V signaling (Figures S3L and S3M).
(B) PM sheets from BHK cells expressing each GFP-K-RasG12V PBD mutant were labeled with anti-GFP-4.5 nm gold and imaged by EM. Localization to the inner PM was quantified as the number of gold particles per 1 μm2 and is shown as mean ± SEM (n ≥15 for each condition). Significant differences from control were evaluated by one-way ANOVA (*p < 0.05).
(C) The gold point patterns from (B) were analyzed for the extent of GFP-K-RasG12V nanoclustering as in Figure 1C. The results show mean Lmax values ± SEM (n ≥ 15 for each condition). Significant differences between control and each mutant were evaluated in bootstrap tests (*p < 0.05).
(D) MDCK cells stably expressing each GFP-K-RasG12V PBD mutant were imaged by confocal microscopy. Fractional cytosolic mislocalization (mean ± SEM of >15 images) was estimated with a custom ImageJ algorithm. Statistical significance was evaluated by one-way ANOVA (*p < 0.05).
(E) BHK cells co-expressing GFP-K-RasG12V and RFP-K-RasG12V PBD mutants were imaged in a FLIM microscope to measure GFP fluorescence lifetime and mean FRET efficiency ± SEM (n ≥ 50 cells for each condition) calculated. Significant differences were evaluated by one-way ANOVA (*p < 0.05).
(F) Representative images of cells from (E) showing GFP fluorescence lifetime values.
(G) Representative confocal images from (D).
(H) Lipid add-back experiments using brain PtdSer or brain PIP2 were carried out as in Figure 1 in BHK cells expressing GFP-K-RasG12V, GFP-K-RasG12V,K177Q, or GFP-K-RasG12V,K178Q (not treated with fendiline). Inner PM leaflet localization was quantified as the number of gold particles per 1 μm2 and is shown as mean ± SEM (n ≥ 15 for each condition). Significant differences between cells with and without lipid add-back were evaluated by one-way ANOVA (*p < 0.05).
(I) Gold point patterns from (H) were analyzed for the extent of nanoclustering. The results show mean Lmax values ± SEM (n ≥ 15 for each condition). Bootstrap tests were used to evaluate statistical significance of differences induced by lipid supplementation (*p < 0.001).
(J) Co-localization between PtdSer (GFP-LactC2) or PIP2 (GFP-PH-PLCδ) and RFP-K-RasG12V was tested in a FLIM experiment as in (A) before and after treatment with 500 μM cGMP for 15 min to activate PKG and phosphorylate S181 in the PBD.
See also Figure S3.
To explore if the prenyl group contributes to lipid-sorting specificity, we mutated the K-Ras CAAX box from CVIM to CCIL, which directs geranylgeranylation (GG) rather than farnesylation (Hancock et al., 1991). The lipid composition of geranylgeranylated K-RasG12V (K-RasG12V-GG) nanoclusters was strikingly different from farnesylated K-RasG12V (Figure 2A). Next, we replaced all six lysines of the PBD with arginines to generate K-RasG12V-6R. Because lysine and arginine carry the same net charge, a PBD of 6R should not significantly alter pure electrostatic interactions. Nevertheless, K-RasG12V-6R nanoclusters had a distinct lipid composition with a significant enrichment of PtdSer and cholesterol compared to K-RasG12V with a wild-type 6K PBD (Figures 2A and S3A–S3E). Finally, we generated a K-RasG12V construct with both a 6R PBD and a GG prenyl group. The lipid composition of K-RasG12V-6R-GG nanoclusters was distinct from that of K-RasG12V-6R and K-RasG12V-GG (Figure 2A). Taken together, these results show that each distinct PBD sequence exhibits lipid-sorting specificity, and the complete anchor defines a combinatorial code where the prenyl group modulates the lipid selectivity of the precise PBD sequence. To explore the potential role of conformational changes in the G-domain on lipid interactions of the anchor, we examined wild-type RFP-K-Ras, which in serum-starved cells is >94% GDP loaded. The LBI heatmap (Figure 2A) shows that nanoclusters of GDP-loaded wild-type K-Ras have a different lipid composition from constitutively GTP-bound K-RasG12V.
Validation of Altered Lipid Sorting of Selected PBD Mutants
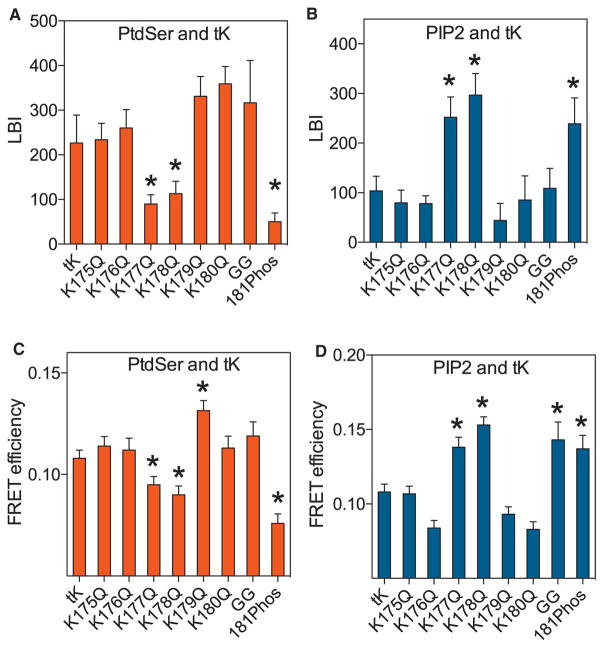
Nanoclusters of K-RasG12V-K177Q and K-RasG12V-K178Q were relatively depleted of PtdSer but enriched with PIP2 (Figure 2A). One interpretation of this change in lipid content is that the mutations reduce the affinity of the anchor for PtdSer; if so, then the overall PM interactions of these mutants should phenocopy the behavior of the wild-type anchor in cells under conditions of PtdSer depletion. Univariate EM-spatial mapping confirmed this prediction. The extent of inner PM gold labeling for GFP-K-RasG12V-K177Q or GFP-K-RasG12V-K178Q was significantly lower than GFP-K-RasG12V (Figure 2B), observations sustained by confocal imaging of MDCK cells stably expressing the same proteins (Figures 2D and 2G). GFP-K-RasG12V-K177Q or GFP-K-RasG12V-K178Q also had significantly lower Lmax values indicating reduced nanoclustering (Figure 2C). Conversely and concordant with an increased affinity for PtdSer, K-RasG12V-6R exhibited significantly enhanced nanoclustering compared to K-RasG12V (Figure 2C). FLIM-FRET assays in intact BHK cells also revealed changes in FRET efficiency that are entirely consistent with the EM Lmax values (Figures 2E and 2F). We evaluated the “switched” lipid specificity of K-RasG12V-K177Q or K178Q from PtdSer to PIP2 in lipid add-back experiments. PM localization and nanoclustering of K-RasG12V was enhanced by exogenous PtdSer but not PIP2 (Figures 2H and 2I), whereas PM localization and nanoclustering of K-RasG12V-K177Q and K-RasG12V-K178Q was enhanced by exogenous PIP2 but not PtdSer (Figures 2H and 2I). Efficient incorporation of PIP2 into the inner PM leaflet by this protocol was validated in previous studies (Cho et al., 2015; Zhou et al., 2015). Finally, we examined the isolated K-Ras anchor, RFP-tK, with and without PBD single mutations. Each RFP-tK PBD mutant matched the cognate full-length K-RasG12V PBD mutant in terms of relative affinity for PtdSer and PIP2 in EM (Figures 3A and 3B) and FLIM-FRET assays (Figures 3C and 3D). Moreover, replacing the farnesyl group to generate RFP-tK-GG, or phosphorylating the RFP-tK anchor by activating PKG, induced similar changes in PtdSer and PIP2 interactions as in full-length K-RasG12V (Figures 3A–3D).
Figure 3. K-Ras Minimal Membrane Anchor Has a Similar Lipid Binding Specificity as Full-Length GTP-Bound K-RasG12V.
(A and B) PM sheets of BHK cells co-expressing each RFP-tK PBD K → Q point mutant, RFP-tK-GG, or phosphorylated RFP-tK with GFP-LactC2 (A) or GFP-PH- PLCδ (B) were assayed for co-clustering using EM and bivariate analysis (n ≥ 15 for each condition) exactly as in Figure 2C to evaluate interaction with PtdSer (LactC2) and PIP2 (PH-PLCδ).
(C and D) BHK cells co-expressing the same set of RFP-tK mutants with GFP-LactC2 (C) or GFP-PH-PLCδ (D) were imaged in a FLIM microscope to measure GFP fluorescence lifetime and mean FRET efficiency ± SEM (n ≥ 60 cells for each condition) calculated. Statistical significance of differences from GFP-tK was evaluated by one-way ANOVA (*p < 0.05).
Structural Dynamics of the K-Ras Anchor on Membrane
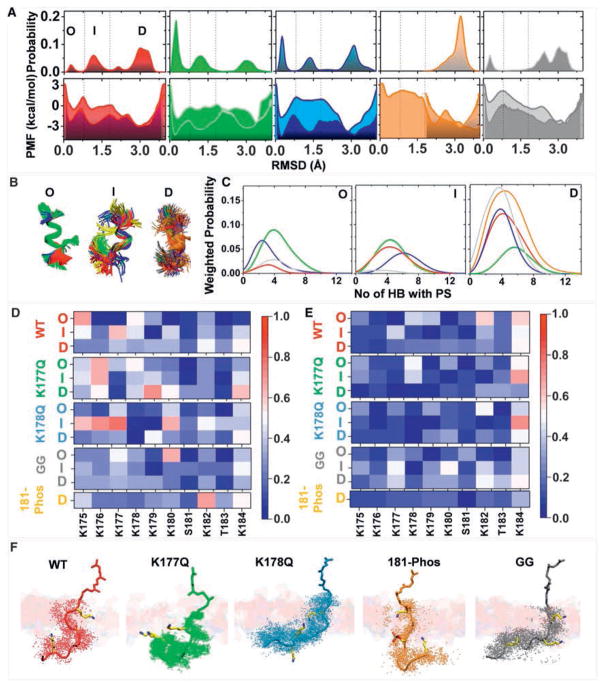
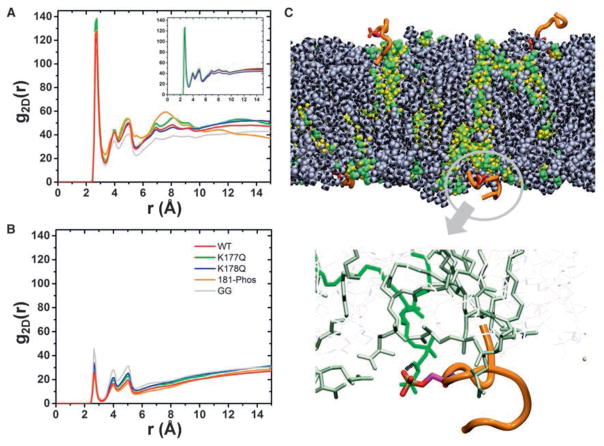
The lipid mapping data suggest a combinatorial lipid sorting code defined by the prenyl anchor and PBD sequence. The simplest realization of this code would be each PBD sequence and prenyl combination adopting a unique structure or dynamics that impacts interaction with lipids. To directly explore this hypothesis, we conducted microsecond-level all-atom molecular dynamics (MD) simulations of tK, the minimal membrane anchoring domain of K-Ras, on a bilayer composed of 80% POPC (16:0/18:1 PC) and 20% POPS (16:0/18:1 PS). Analysis of the Cα atom root-mean-square deviation (RMSD) of residues 177–182 (Figure 4A) suggested that the peptide samples at least three structural forms: disordered conformations with no secondary structure (D) and semi-ordered conformations with one or two helical turns referred to as intermediate (I) and ordered (O) (Figure 4B). Although 1 μs-long conventional MD (cMD) was not expected to sample multiple inter-state transitions, two of the four peptides visited more than one state. Moreover, we obtained a similar distribution of conformers when sampling was doubled to an aggregate length of 8 μs by including tK extracted from cMD of full-length K-Ras (Prakash et al., 2016) (Figure S4A). The existence of three conformational states is also supported by a 2D free energy surface derived from metadynamics (metaMD), an enhanced sampling method used to calculate free energy profile in a variety of systems including peptide-membrane complexes (Cui et al., 2011; Deighan and Pfaendtner, 2013). The challenge of metaMD convergence for peptide-bilayer systems has been discussed previously (Cui et al., 2011). After testing various reaction coordinates and sampling approaches (see the STAR Methods) we found that metaMD along two collective variables (CVs), end-to-end distance, and RMSD, converged fairly well (Figures S4B and S4C). The resulting free energy surface also compares very well with that derived from the 8 μs cMD data. We therefore used the metaMD PMF to estimate the free energy difference between states O and D and obtained a relatively small ΔG = −2.5 kcal/mol.
Figure 4. Different K-Ras Anchor Variants Sample Different Backbone Conformations that Interact Differentially with Membrane.
(A) Free energy profiles as a function of root-mean-square deviation (RMSD) of Cα atoms of residues 177–182 obtained after re-weighting the metaMD (lighter shade, bottom panel) and from the Boltzmann inversion of the probability distribution of the RMSD (top panel) from unbiased cMD simulations (darker shade, bottom panel). The metaMD PMF was extracted from the 2D PMFs shown in Figure S4. The three most common free energy basins demarcated by the vertical dashed lines represent distinct groups of conformations referred to as ordered O, intermediate I, and disordered D.
(B) Superposition of representative backbone conformations of O, I, and D.
(C) Weighted probability of the number of hydrogen bonds between protein (donor) and PtdSer (acceptor) in O, I, and D; the percentages in (A) were used for weighting as highlighted by the matching colors. Cutoffs for hydrogen bond calculation in this and the rest of the figures were donor-acceptor distance of 3.0 Å and donor-hydrogen-acceptor angle of 30°.
(D and E) Heatmaps of the probability of hydrogen bonding of individual residues with PtdSer (D) and PC (E) separately for each tK-variant and each group.
(F) Representative structures of bilayer-bound peptides (shown in stick model) highlighting similar membrane insertion but varied relative orientations of the backbone. For each peptide system, a snapshot was chosen based on the most probable location of K175, K180, and Far/GG185 Cα atoms relative to the bilayer center. For this analysis, we considered only the dominant conformations sampled during the simulations, which is D for all, except K177Q where it is I. The extent of the backbone conformational fluctuation is highlighted by the dots representing the instantaneous coordinates of the Cα atoms. A few selected residues with high hydrogen bonding potential are highlighted in licorice. A portion of one leaflet of the bilayer is shown in transparent slab with the membrane-penetrating farnesyl or GG moieties sticking out. Color code: tK-WT (red), tK-K177Q (green), tK-K178Q (blue), 181-Phos (orange), and tK-GG (gray); licorice presentation: carbon (yellow), nitrogen (blue), and oxygen (red).
See also Figure S4.
To check if the conformational space sampled by cMD represents a reasonable estimate of the underlying free energy surface, we Boltzmann-inverted the probability distribution shown in Figure 4A assuming that a single peptide simulated for a longer duration would sample the same phase space as the four peptides. The resulting energy versus RMSD plot overlaps surprisingly well with that from the metaMD-derived 1D PMF (Figure 4A). Combining this observation (namely: tK samples the three states in the unbiased simulation nearly as well as the biased one) with the relatively small apparent barriers between the states (Figure 4A), we conclude that the three conformations we observe represent inter-converting equilibrium states. We therefore hypothesized that small changes in the PBD sequence or membrane composition would be expected to alter their relative populations. We then conducted a detailed structural analysis of the cMD data to examine the potential link between conformation and lipid interaction. As shown in Figure 4A, ~64% of the total tK conformations sampled the D state whereas the remaining 35% are in the I (29%) or O (6%) state. These structure groups differed in their hydrogen bonding interactions with PtdSer (Figure 4C) and in the contribution of individual residues to membrane binding via hydrogen bonding with PtdSer and PC lipids (Figures 4D and 4E). For instance, K177 interacts with PtdSer in the I and D states but not in the O state, K178 interacts similarly in all three (Figure 4D). Therefore, contrary to common assumptions, the backbone of membrane-bound tK is not completely disordered; rather it adopts a few relatively well-defined dynamic structures including a combined 35% of structures with a helical content. The different structures in turn dictate the ability of individual Lys side chains to interact with bilayer lipids.
We next simulated, under identical conditions and using both cMD and metaMD, the K177Q (tK-K177Q) and K178Q (tK-K178Q) point mutants, tK phosphorylated at S181 (181-Phos), and tK with the farnesyl replaced by geranylgeranyl (tK-GG). As in wild-type tK, the PMFs of the mutants derived from cMD and metaMD are very similar (Figures 4A and S4C). Both show that the free energy profile is different among these mutants, as is the ΔG between the different states. For example, for K177Q and K178Q the free energy difference between O and D is small (−0.6 and −2 kcal/mol, respectively) whereas D is significantly more stable in 181-Phos and tK-GG (ΔG = −4.9 and −2.9 kcal/mol). The similarities of the 1D PMFs from cMD and metaD suggest that the two methods sampled the major conformational states in similar ratios, which justifies the use of the unbiased cMD data for a closer look at peptide-lipid atomic interactions.
From the probability distributions in Figure 4A, it is clear that the mutations K177Q and K178Q significantly increased the population of ordered conformers (from 6% in the WT to 42% and 25% in K177Q and K178Q, respectively). The individual side chains of tK-K177Q and tK-K178Q interacting with the bilayer were also altered (Figures 4D–4F). In contrast, 181-Phos and tK-GG shifted the population toward the D state (Figure 4A) and again modulated side chain interactions with the bilayer (Figures 4D–4F). In general, the N-terminal half of the PBD preferentially interacts with PtdSer in tK-K177Q and tK-K178Q whereas the C-terminal half of the PBD was favored in tK-181-Phos and tK-GG. Specifically, only residues K177, K180, and K182 interact with PtdSer in tK-GG and tK-181Phos whereas interaction of these peptides with PC is more pronounced than in the other Q mutants (Figures 4D and 4E).
The five systems were similar overall in terms of membrane insertion and orientation of the backbone with respect to the membrane plane (Figure 4F), but the predominantly D conformations of 181-Phos and especially tK-GG were more peripheral (Figure 4F). This is despite both anchors having additional interactions with membrane lipids: in tK-181-Phos the phosphate group accepts a hydrogen bond from the NH3 moiety of a PtdSer head group (Figures 5C and S5B), whereas in tK-GG the extra tail carbons of the longer prenyl group make additional vdW contacts with bilayer lipids (Figure S5C). We also observed that for all farnesylated anchors the number of hydrogen bonds with PtdSer progressively increases from the O state to the D state (Figure 4C). This suggests that the disordered or open conformation is favored for direct hydrogen bonding interaction with PtdSer over the organized helical conformation, again demonstrating a link between backbone conformation and lipid interaction. At a more global level, the 2D radial pair distribution functions (RDF) show substantial clustering of PtdSer around all the farnesylated anchors but intriguingly less so for tK-GG, as can be seen from the secondary and tertiary peaks in Figures 5A and 5B, and illustrated in Figure 5C. PtdSer represents only 20% of the lipids in the simulation box yet it accounts for 35%–70% of the total number of lipids that are near the farnesylated PBDs (Figure S5D), with an average enrichment of ~30% for WT tK, tK-K177Q, and tK-K178Q and ~40% for tK-181-Phos. In contrast the PtdSer preference of tK-GG is much smaller with an average enrichment of 10%. In sum, the computational analysis shows that the structure of K-Ras lipid anchor is not random and its binding to anionic membrane is not dictated solely by electrostatic attraction; the interaction of individual side chains with specific lipids varies with the backbone conformation and is modulated by mutation or prenylation. As a result, even mutations that a priori may appear structurally innocuous can have substantial effect on the dynamics of the PBD and its interaction with lipids. These data thus provide a mechanistic insight into the results of the K-Ras lipid mapping experiments in intact PM.
Figure 5. Lipid Clustering Profile of K-Ras Anchor Variants.
(A and B) Two-dimensional (2D) radial distribution function (RDF) of head group oxygen atoms of PtdSer (A) and PC (B) around nitrogen of Lys or oxygen atoms of Ser/Thr side chains of the peptides. The inset in (A) shows convergence of the RDF for WT-tK by comparing data from 500–600 ns (black), 500–700 ns (red), 500–800 ns (blue), 500–900 ns (pink), and 500–1,000 ns (green); similar results were obtained for the other peptides.
(C) A snapshot of 181-Phos showing the four peptides in the simulation box (orange) and the clusters of PtdSer around each peptide (green/yellow spheres). PC lipids are shown in gray/black spheres. A bi-dentate hydrogen bond between the phosphorylated Ser and PtdSer is highlighted in the bottom panel.
See also Figure S5.
K-Ras Polybasic Domain Mutants Have Distinct Biological Activities
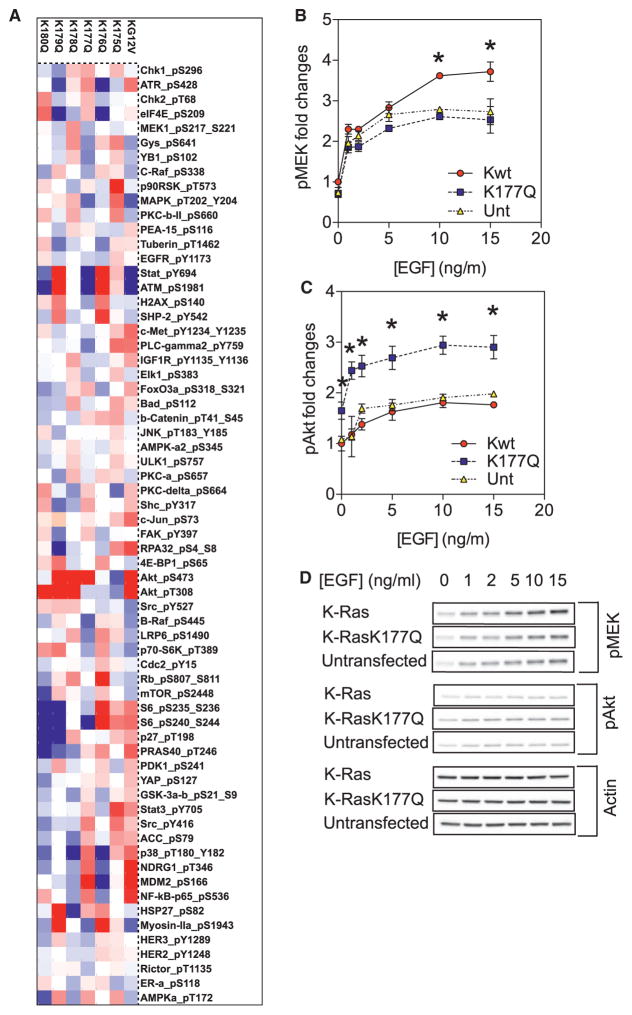
To systematically evaluate how PBD sequence potentially contributes to K-Ras function, we performed large-scale reverse phase protein array (RPPA). Each K-RasG12V PBD point mutant when transiently expressed at equivalent levels (Figure S6C) generated a significantly different global phosphorylation map (Figures 6A, S6A, and S6B). The results show that the actual sequence of the PBD determines signal output, concordant with distinct lipid-binding specificities determining PM localization and nanoclustering. To specifically examine MAPK and PI3K-AKT signaling, MDCK cells stably expressing wild-type K-Ras or K-Ras-K177Q were serum-starved and stimulated with various doses of EGF for 5 min. Cells expressing GFP-K-Ras-K177Q exhibited reduced MEK activation, but markedly elevated Akt phosphorylation, in response to EGFR activation compared to cells expressing GFP-K-Ras (Figures 6B–6D). These data are consistent with the spatial and imaging data showing that nanoclusters of GFP-K-Ras-K177Q are enriched in PIP2, the lipid substrate for PI3K, and together illustrate that changes in K-Ras nanocluster lipid composition also have important attendant consequences for signal output.
Figure 6. K-Ras PBD Mutations Change Signal Output.
(A) Lysates of BHK cells expressing equal amounts of GFP-tagged K-RasG12V (KG12V) or each K-RasG12V PBD mutant were collected after overnight serum-starvation. Reverse phase protein array (RPPA) was conducted. Relative protein levels for each sample were determined by interpolation from the standard curve of each antibody and normalized for protein loading. Raw values were converted to Log2 values. Results of phosphorylated proteins are shown as a heatmap of Log2 values centered on the 50th centile (white) and with a range of −0.3 (blue) and 0.3 (red).
(B and C) MDCK cells stably expressing GFP-K-Ras or GFP-K-Ras-K177Q were serum-starved and stimulated with various doses of EGF (0–15 ng/mL). Whole cell lysates were blotted for phosphorylated MEK (B) or phosphorylated AKT (C) and quantified, the results are mean ± SEM (n = 5). In each plot, all values have been normalized against untransfected control without EGF stimulation. Significant differences between control and each mutant were evaluated by one-way ANOVA (*p < 0.05).
(D) Representative blots are shown.
See also Figure S6.
DISCUSSION
We used quantitative spatial imaging analyses and atomistic molecular dynamics to systematically examine the mechanism of association of the K-Ras bi-partite PBD-prenyl membrane anchor with the PM. Traditionally, PBDs have been thought to interact with PM exclusively via electrostatics where the total number of basic residues determines the strength of electrostatic association with anionic lipids. However, we show here that the molecular mechanism of K-Ras PM binding is considerably more complex. Rather, the precise PBD amino acid sequence acting with the prenyl group comprises a combinatorial code for lipid sorting. This code translates into dynamic tertiary structures on the PM that exhibit exquisite binding specificity for defined anionic phospholipids. An important consequence is the ability of such anchors to sort or retain specific subsets of phospholipids into nanoclusters with a defined lipid composition that in turn determine K-Ras signaling output.
Several lines of evidence show that lipid interactions of the PBD are not simply governed by charge. PBDs of six arginines or lysines have the same net positive charge, yet K-Ras-6R is more extensively PM localized and generates nanoclusters with a different lipid composition than K-Ras with a wild-type (6K) PBD. C20-anchored K-Ras-6R also has a different lipid composition than C15-anchored K-Ras-6R. K-RasG12V with six different single point mutations of K → Q assemble into nanoclusters with markedly different lipid compositions even though each PBD mutant has the same net charge. MD simulations and free energy calculations show that the 6K K-Ras anchor adopts a combination of disordered, intermediate, and ordered helical structures that present different lysines for interaction with the bilayer. Not all lysines of the PBD therefore participate equally in hydrogen bonding with the bilayer with side chains of K177, K178 and K180 being especially important. The 6K K-Ras anchor favors a disordered extended state and associates with PtdSer extensively. Single point mutations K177Q or K178Q shift the conformational equilibrium to a more ordered state and compromise hydrogen bonding with PtdSer, consistent with the reduced nanoclustering and PM localization of K-Ras-K177Q and K178Q in intact cells. Compromised interactions with PtdSer may in turn allow the enhanced interaction with multivalent PIP2 observed with these mutants in cells. We speculate that the low PIP2 content of the PM (<1 mol%) may not be effective for PM anchoring of PBD proteins, especially in competition with proteins that have more specific PH/PX or MARCKS-related PIP2-binding domains (McLaughlin et al., 2002). In support of this concept, PM binding of K-Ras-K177Q and K178Q was restored when the PIP2 content of the inner PM was increased by exogenous PIP2.
The ability of the K-Ras lipid anchor and its mutants to sample ordered and semi-ordered conformations in addition to an extended conformation is reminiscent of intrinsically disordered proteins (IDPs) (Granata et al., 2015). Just as in IDPs (Granata et al., 2015), the global free energy minimum of the K-Ras anchor is populated by the extended conformation but it also adopts partial order. The latter is shared by most IDPs, which either harbor ordered segments or undergo disorder to order transition upon binding to a reaction partner (Granata et al., 2015). Indeed, the ability to easily transition between order and disorder may be functionally relevant. For example, the myristoylated polybasic peptide MARCKS, which closely resembles the K-Ras anchor, is largely extended on membrane (Ellena et al., 2003) but adopts an α-helical conformation upon binding to calmodulin (Yamauchi et al., 2003).
Quantification of the different anchor variants further showed that PtdSer clustering around the anchor was not merely a function of electrostatic interactions; thus replacing C15-farnesyl by the longer C20-geranylgeranyl, a substitution that does not affect charge distribution, nearly abolished the specificity for PtdSer clustering. The C15 to C20 switch induced markedly different conformational dynamics of the anchor peptide because the longer prenyl-chain length increased vdW interactions with bilayer lipids but concurrently decreased hydrogen bonding with PtdSer head groups at the interface. The EM data also showed that K-RasG12V-GG exhibited less selectivity for different anionic membrane lipids than farnesylated K-RasG12V perhaps reflecting reduced head group interactions. In sum, K-Ras interactions with membrane lipids are not simply governed by electrostatics. The precise sequence of the PBD and prenyl group together determine the conformational dynamics of the anchor, the residues participating in membrane binding, and hence which anionic lipid interactions are favored. Subtle changes in PBD sequence, even single K → Q or K → R substitutions, as well as phosphorylation therefore have substantial effects on structural dynamics and hence lipid interactions. The Ras G-domain also participates in PM binding, engaging differently in GTP-bound and GDP-bound conformations and concomitantly modulating interactions of the anchor with the PM (Abankwa et al., 2010; Weise et al., 2011). Concordantly, whereas the lipid preferences of GTP-bound K-RasG12V and the minimal membrane anchor anchoring tK are similar, those of GDP-bound K-Ras are quite different, consistent also with the lateral segregation of K-Ras.GTP and K-Ras.GDP into different nanoclusters.
In the complex environment of the PM, the K-RasG12V anchor exhibits remarkable specificity for distinct subclasses of PtdSer, preferentially assembling asymmetric PtdSer into nanoclusters. K-RasG12V does not interact with fully saturated PtdSer at all, whereas mono- and di-unsaturated PtdSer can support K-RasG12V PM binding but cannot be assembled into nanoclusters. We propose that these differences reflect the lateral availability of PtdSer in the PM. Fully saturated PtdSer is likely retained in cholesterol-rich complexes more extensively than asymmetric PtdSer (Bach et al., 1992; Maekawa and Fairn, 2015) concordant with the demonstration of a cholesterol-sensitive pool of PtdSer that does not interact with K-RasG12V (Cho et al., 2015). Moreover, membranes composed of asymmetric PtdSer sustain less perturbation when undergoing electrostatic interactions with external components than fully saturated PtdSer (Blois et al., 2006; Broniec et al., 2007). These biophysical properties may account for the preferential sorting of asymmetric PtdSer into K-RasG12V nanoclusters. These results also illustrate the functional importance of nanoclusters, because only asymmetric PtdSer was able to fully restore K-RasG12V interactions with CRAF in PtdSer-depleted cells.
Most importantly, the distinct lipid binding specificity of each K-RasG12V PBD mutant translated into different extents of PM localization and nanoclustering and resulted in nanoclusters with different lipid compositions. The integrated effects of these differences was a unique signaling profile for each mutant, indicating a critical role for anchor lipid binding specificity in governing effector recruitment, activation, and hence K-RasG12V signal output (Zhou et al., 2014, 2015). Our findings have broader implications in understanding the function of the lipid-binding domains of other PM-binding proteins. Certain lipid-binding domains (e.g., C2 and PH/PX domains) have long been known to possess defined structures and bind to specific lipids with high affinity. By contrast, many small GTPases have membrane anchors analogous to K-Ras comprising a PBD of lysine and arginine residues operating in conjunction with a prenyl lipid. Our results here suggest that each of these anchors, by virtue of their different primary sequences, will exhibit a distinct secondary structure on the PM and hence have the capacity to selectively interact with and sort different cohorts of anionic lipids with attendant consequences for function. We thus need to re-evaluate the types of protein structure that can encode lipid-binding specificity.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Culture Conditions for In Vitro Systems
-
METHOD DETAILS
EM-Spatial Mapping to Quantify the Extent of Nanoclustering and Co-localization of Proteins and Lipids on the Plasma Membrane
Western Blotting to Determine the Activities of the MAPK and PI3K Pathways in Cells
FLIM-FRET Imaging to Visualize and Quantify the Extent of Aggregation / Co-localization of Proteins and Lipids on the Plasma Membrane
Confocal Imaging to Visualize the Intracellular Localization of K-Ras PBD Mutants in Cells
Classical Molecular Dynamics Simulation to Solve the Structures of K-Ras Anchors
Metadynamics Simulations to Calculate the Conformational Transitions of K-Ras Anchors
Reverse Phase Protein Array (RPPA) to Quantify Effects of Polybasic Domain Mutations on Signal Output
Lipidomics to Monitor the Extent of Acyl Chain Modification of Exogenous PtdSer Species after Uptake by BHK Cells
QUANTIFICATION AND STATISTICAL ANALYSIS
DATA AND SOFTWARE AVAILABILITY
Supplementary Material
Acknowledgments
This work was supported by the Cancer Research and Prevention Institute of Texas (CPRIT) (RP130059 and RP170233) to J.F.H., the NIH (R01GM100078) to A.A.G., and an NIH Pathway to Independence Award (K99-CA188593) to K.-J.C. We thank the Texas Advanced Computing Center (TACC) and the Extreme Science and Engineering Discovery Environment (XSEDE) (Project: MCB150054) for computational resources and Dr. Abdallah-Sayyed Ahmad for helpful discussions.
Footnotes
Supplemental Information includes six figures and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.11.059.
AUTHOR CONTRIBUTIONS
Y.Z., A.A.G., and J.F.H. designed the experiments. Y.Z., P.P., H.L., K.-J.C., A.A.G., and J.F.H. performed experiments and conducted data analysis. Y.Z., P.P., A.A.G., and J.F.H. wrote the manuscript.
References
- Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, Hancock JF. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci USA. 2010;107:1130–1135. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach D, Wachtel E, Borochov N, Senisterra G, Epand RM. Phase behaviour of heteroacid phosphatidylserines and cholesterol. Chem Phys Lipids. 1992;63:105–113. [Google Scholar]
- Barducci A, Bonomi M, Parrinello M. Metadynamics. Wiley Interdiscip Rev Comput Mol Sci. 2011;1:14. [Google Scholar]
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Blois A, Holmsen H, Martino G, Corti A, Metz-Boutigue MH, Helle KB. Interactions of chromogranin A-derived vasostatins and monolayers of phosphatidylserine, phosphatidylcholine and phosphatidylethanolamine. Regul Pept. 2006;134:30–37. doi: 10.1016/j.regpep.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Broniec A, Gjerde AU, Ølmheim AB, Holmsen H. Trifluoperazine causes a disturbance in glycerophospholipid monolayers containing phosphatidylserine (PS): effects of pH, acyl unsaturation, and proportion of PS. Langmuir. 2007;23:694–699. doi: 10.1021/la061628b. [DOI] [PubMed] [Google Scholar]
- Cho KJ, van der Hoeven D, Zhou Y, Maekawa M, Ma X, Chen W, Fairn GD, Hancock JF. Inhibition of acid sphingomyelinase depletes cellular phosphatidylserine and mislocalizes K-Ras from the plasma membrane. Mol Cell Biol. 2015;36:363–374. doi: 10.1128/MCB.00719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KJ, Casteel DE, Prakash P, Tan L, van der Hoeven D, Salim AA, Kim C, Capon RJ, Lacey E, Cunha SR, et al. AMPK and eNOS signaling regulates K-Ras plasma membrane interactions via cGMP-dependent protein kinase 2. Mol Cell Biol. 2016;36:3086–3099. doi: 10.1128/MCB.00365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Lyman E, Voth GA. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighan M, Pfaendtner J. Exhaustively sampling peptide adsorption with metadynamics. Langmuir. 2013;29:7999–8009. doi: 10.1021/la4010664. [DOI] [PubMed] [Google Scholar]
- Diggle PJ, Mateu J, Clough HE. A comparison between parametric and non-parametric approaches to the analysis of replicated spatial point patterns. Adv Appl Probab. 2000;32:331–343. [Google Scholar]
- Ellena JF, Burnitz MC, Cafiso DS. Location of the myristoylated alanine-rich C-kinase substrate (MARCKS) effector domain in negatively charged phospholipid bicelles. Biophys J. 2003;85:2442–2448. doi: 10.1016/s0006-3495(03)74667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorin G, Klein ML, Hénin J. Using collective variables to drive molecular dynamics simulations. Mol Physiol. 2013;111:3345–3362. [Google Scholar]
- Gorfe AA, Babakhani A, McCammon JA. H-ras protein in a bilayer: interaction and structure perturbation. J Am Chem Soc. 2007a;129:12280–12286. doi: 10.1021/ja073949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007b;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- Granata D, Baftizadeh F, Habchi J, Galvagnion C, De Simone A, Camilloni C, Laio A, Vendruscolo M. The inverted free energy landscape of an intrinsically disordered peptide by simulations and experiments. Sci Rep. 2015;5:15449. doi: 10.1038/srep15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kötting C, Gerwert K. N-Ras forms dimers at POPC membranes. Biophys J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Janosi L, Gorfe AA. Segregation of negatively charged phospholipids by the polycationic and farnesylated membrane anchor of Kras. Biophys J. 2010;99:3666–3674. doi: 10.1016/j.bpj.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Triola G, Vetter IR, Erlkamp M, Waldmann H, Winter R. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc Natl Acad Sci USA. 2012;109:460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Uchida Y, Emoto K, Umeda M, Kuge O, Taguchi T, Arai H. Impaired retrograde membrane traffic through endosomes in a mutant CHO cell defective in phosphatidylserine synthesis. Genes Cells. 2012;17:728–736. doi: 10.1111/j.1365-2443.2012.01622.x. [DOI] [PubMed] [Google Scholar]
- Limongelli V, Bonomi M, Parrinello M. Funnel metadynamics as accurate binding free-energy method. Proc Natl Acad Sci USA. 2013;110:6358–6363. doi: 10.1073/pnas.1303186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Fairn GD. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J Cell Sci. 2015;128:1422–1433. doi: 10.1242/jcs.164715. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci USA. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, Dyba MA, Stefanisko K, Tarasov SG, Gursoy A, et al. GTP-dependent K-Ras dimerization. Structure. 2015;23:1325–1335. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Tamgüney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc Natl Acad Sci USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol Cell Biol. 2008;28:4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Zhou Y, Liang H, Hancock JF, Gorfe AA. Oncogenic K-Ras binds to an anionic membrane in two distinct orientations: a molecular dynamics analysis. Biophys J. 2016;110:1125–1138. doi: 10.1016/j.bpj.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003a;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Parton RG, Hancock JF. Observing cell surface signaling domains using electron microscopy. Sci STKE. 2003b;2003:PL9. doi: 10.1126/stke.2003.177.pl9. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- van der Hoeven D, Cho KJ, Ma X, Chigurupati S, Parton RG, Hancock JF. Fendiline inhibits K-Ras plasma membrane localization and blocks K-Ras signal transmission. Mol Cell Biol. 2013;33:237–251. doi: 10.1128/MCB.00884-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise K, Kapoor S, Denter C, Nikolaus J, Opitz N, Koch S, Triola G, Herrmann A, Waldmann H, Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J Am Chem Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- Yamauchi E, Nakatsu T, Matsubara M, Kato H, Taniguchi H. Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca2+-calmodulin. Nat Struct Biol. 2003;10:226–231. doi: 10.1038/nsb900. [DOI] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang H, Rodkey T, Ariotti N, Parton RG, Hancock JF. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol Cell Biol. 2014;34:862–876. doi: 10.1128/MCB.01227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wong CO, Cho KJ, van der Hoeven D, Liang H, Thakur DP, Luo J, Babic M, Zinsmaier KE, Zhu MX, et al. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 2015;349:873–876. doi: 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.