Abstract
Whether a transplanted allograft is stably accepted, rejected or achieves immunological tolerance is dependent on the frequency and function of alloreactive lymphocytes, making the identification and analysis of alloreactive T and B cells in transplant recipients critical for understanding mechanisms, and the prediction of allograft outcome. In animal models, tracking the fate of graft-reactive T and B cells allows investigators to uncover their biology and develop new therapeutic strategies to protect the graft. In the clinic, identification and quantification of graft-reactive T and B cells allows for the early diagnosis of immune reactivity and therapeutic intervention to prevent graft loss. In addition to rejection, probing of T and B cell fate in vivo provides insights into the underlying mechanisms of alloimmunity or tolerance that may lead to biomarkers predicting graft fate. In this review, we discuss existing and developing approaches to track and analyze alloreactive T and B cells in mice and humans and provide examples of discoveries made utilizing these techniques. These approaches include mixed lymphocyte reactions (MLRs), trans-vivo delayed-type hypersensitivity (DTH), enzyme-linked immunospot (ELISpot) assays, the use of antigen receptor transgenic lymphocytes, and utilization of peptide-MHC complex (pMHC) multimers, along with imaging techniques for static multiparameter analysis or dynamic in vivo tracking. Such approaches have already refined our understanding of the alloimmune response and are pointing to new ways to improve allograft outcomes in the clinic.
Introduction
In the absence of immunosuppression, allografts in inbred mice succumb primarily to acute T cell-mediated rejection, whereas in outbred mice, allografts can also be rejected in a T cell-independent but complement/neutrophil-dependent manner, underscoring the heterogeneity of rejection processes in the absence of immunosuppression1. In the clinic, conventional pharmacological immunosuppression is largely effective at preventing and treating T cell-mediated rejection2,3, so most allografts are lost from antibody-mediated rejection (ABMR). Both preexisting and de novo donor-specific antibodies (DSA) predict poor graft outcomes compared to DSA-negative recipients, with de novo DSA-mediated ABMR being associated with IFNγ-inducible, natural killer cell and T cell transcripts and inferior graft survival compared to preexisting DSA4. These observations suggest that the accurate quantification of donor-specific T, B and plasma cell responses may allow for an earlier diagnosis and the development of therapeutic interventions that result in improved long-term outcomes. To this end, traditional methods of identifying alloreactive T cells and DSA are being improved upon and new techniques have become available. In this review, we will discuss evolving methods for identifying, isolating and tracking alloreactive T and B cells in mouse models and in the clinical setting.
Detecting alloreactive T cells following alloantigen stimulation
T cell-mediated allograft rejection is thought to depend on cytokine production, cytotoxicity and provision of help to other lymphocytes. Thus, tracking the phenotype and function of alloreactive T cells in animal models and clinical studies of transplantation may lead to better diagnosis of transplantation rejection and tolerance. We note that in addition to alloreactive T cells, autoreactive T cells from preexisting autoimmune conditions, or activated when cryptic epitopes become exposed, can also participate in damaging the graft5,6, but this review will focus on tracking alloreactive T cells. Historically, alloreactive T cells have been defined by their ability to respond to stimulation with alloantigen. By coculturing peripheral blood mononuclear cells (PBMCs) from the donor with PBMCs from the recipient in vitro, a technique known as an MLR, recipient alloreactive T cells can be tracked based on their activation, proliferation, or production of cytokines. MLRs can theoretically measure responses to antigen presented by both direct and indirect pathways, although accumulation of T cells that recognize alloantigen directly is thought to overshadow the response of indirect T cells that may start at a lower frequency. To identify T cells specific for indirectly presented antigen, donor PBMCs lysed prior to coculture with live host PBMCs have been used as a source of donor antigen to be presented to host T cells by host antigen-presenting cells7. The MLR has also been adapted to quantify the frequency of T cells with cytotoxicity against donor cells by sequentially diluting responder cells in limiting dilution assays and measuring cytotoxic activity against donor targets following stimulation8. Both naïve and memory CD4+ and CD8+ T cells from human peripheral blood have been shown to proliferate in an MLR, while granzyme B and perforin are preferentially expressed by memory CD8+ T cells9.
In association with the MLR, flow cytometry or ELISpot have been used to determine the number of cytokine-producing cells following stimulation, with flow cytometry identifying the cells containing the intracellular cytokine and ELISpot detecting the frequency of cells secreting a given cytokine. Similar to the MLR, T cells activated in an allospecific ELISpot may predominantly respond to directly presented alloantigen and the indirect response can be probed by coculturing intact recipient PBMCs with lysed donor PBMCs10. ELISpot has a lower limit of detection, which makes it advantageous for detecting rare cytokine-secreting cells but has the risk of underreporting, particularly in samples where there are many cytokine-producing cells11. Because effector and memory, but not naïve, T cells can produce IFNγ after ≤24 hours of in vitro stimulation, 24h ELISpot assays have been used to determine the number of donor-reactive memory IFNγ-producing T cells12. Indeed, the presence of donor-reactive IFNγ-producing cells prior to transplantation as detected by ELISpot has been associated with worse clinical outcomes following renal transplantation13.
Flow cytometry analysis following an MLR has also been used to identify T cells that upregulate activation markers or proliferate in response to donor stimulation. Activated T cells increase their expression of many surface molecules, including those used for costimulation or coinhibition, antigen presentation, cytokine responsiveness, chemotaxis and other functions14,15. Of these, CD2516,17, CD6917,18, CD7116,17, CD13419, CD13719,20, CD15419 and LFA1α21 have been used in transplantation to identify alloreactive T cells. The glycoproteins CD4421–23 or CD45 isoforms24, markers of antigen experience, are also used to distinguish naive from effector and memory cells. Markers of cell-cycle entry such as Ki67 can also identify recently activated T cells23. Additionally, labeling responder PBMCs with intracellular dyes such as carboxyfluorescein succinimidyl ester (CFSE), whose fluorescence intensity is halved with each cell division, has been used to identify proliferating donor-reactive T cells following culture in an MLR25. While upregulation of many surface markers of activation can be detected within 24–48 hours of stimulation, in vitro assays detecting cell division such as CFSE dilution, require >48 hours. Stimulated cells expressing activation markers or diluted CFSE can be isolated by fluorescence-activated cell sorting (FACS) and subjected to additional analysis. For example, next generation sequencing of T cell receptors (TCRs) has recently been used to identify alloreactive T cell clones that expanded following ex vivo stimulation with donor PBMCs26. Identifying these alloreactive clones in pretransplant patients allows for their prospective tracking in circulating and graft-infiltrating T cells posttransplantation. Such an analysis of circulating T cells revealed clonal T cell deletion as 1 of the mechanisms of clinical tolerance in recipients of combined donor bone marrow and renal allografts27. Allospecific regulatory T cells (Tregs) also express markers of activation and proliferation in response to their cognate alloantigen in an MLR, and can be identified by staining with Foxp3-specific antibodies17,24,28.
While in vitro stimulation assays are simple and valuable, they may not reliably reflect in vivo activation with regard to kinetics, extent of proliferation, or expression of surface markers and cytokines29,30. Thus, in vivo methods of stimulating allospecific cells may more faithfully reflect an immune response to an allograft in terms of functionality and phenotype. In experimental models, one can study the frequency of alloreactive T cells within a polyclonal repertoire in vivo by observing cell proliferation after adoptive cell transfer into a recipient mouse expressing alloantigen. For example, this has been done by transferring CFSE-labeled C57BL/6 (H-2b) cells into a C57BL/6xDBA F1 mouse (H-2b/H-2d)25. This strategy that models graft-versus-host disease ensures that the recipient mouse does not reject the transferred cells because of their shared MHC haplotype, while the transferred cells can react to the host H-2d haplotype. The frequency of transferred cells with diluted CFSE can then be determined as a measure of alloreactivity, though this approach can obviously not be used in humans.
The trans-vivo DTH model has adapted the concept of the MLR to an in vivo setting that allows for the study of alloimmunity in patient samples31. In this assay, graft recipient PBMCs and lysed donor cells as a source of donor antigen are coinjected into the footpad of a severe combined immunodeficiency (SCID) mouse lacking its own functional adaptive immune system. If the patient has immune memory to donor antigens presented through the indirect pathway, DTH can be observed in the mouse footpad within 24 hours, in the form of increased swelling when compared with injection of PBMCs alone. Because the lysate lacks intact donor antigen-presenting cells, this method is presumed to not probe the direct pathway of allorecognition. Cytokine and receptor blocking antibodies have been injected into the SCID host to determine which factors are necessary for the stimulation or suppression of alloreactive cells in the trans-vivo DTH assay. Through the observation of linked-suppression to a third party antigen, this model has revealed that in human allograft recipients tolerant to their grafts following cessation of immunosuppression, alloreactive immune responses are actively suppressed in an IL-10- or TGFβ-dependent manner32. While this assay is useful for mechanistic studies, its utility as a routine clinical assay is limited because it is labor intensive, challenging to standardize, and requires stored donor cell lysates to be available.
A disadvantage of studying alloreactive T cells by stimulation with alloantigen ex vivo or by the trans-vivo assay is that stimulation by these methods may not reflect the nuances associated with processing of donor antigens derived from engrafted tissue, and thus T cells stimulated in these assays may not behave similarly to those stimulated in the context of a transplant. Responder cells used in these assays also do not reflect the effects that circulating immunosuppressive drugs may have on the alloresponse in vivo. Nevertheless, ex vivo stimulation and trans-vivo DTH assays have allowed for the analysis of a polyclonal alloreactive repertoire in a wide range of mouse models and experimental human studies.
Using model alloantigens and T cells with known specificity
In mice, TCR-transgenic T cells specific for intact or processed donor MHC, or for model antigens expressed transgenically under ubiquitous, tissue-specific, or inducible promoters in transplanted organs, have permitted the study of monoclonal alloimmune responses in both naïve and sensitized mice. TCR-Tg T cells can be adoptively transferred into recipients prior to transplantation, allowing control of graft-specific T cell precursor frequency. Furthermore, these cells can be reisolated based on staining with antibodies specific to congenic markers or to their clonotypic TCR. Such experiments have revealed, for example, that the frequency of donor-reactive T cells in the host prior to transplantation determines the efficacy of costimulation blockade therapy33. Mice bearing Tg TCRs specific to antigens restricted to presentation on either donor or host MHC have been used to study direct or indirect allorecognition, respectively. Comparison of direct and indirect T cells in a mouse model of chronic heart allograft rejection has shown that direct recognition of alloantigen occurs for a brief time following transplantation, whereas indirect allorecognition can be more long-lived provided that donor antigen remains available for processing and presentation23.
Adoptive transfer of fluorescently labeled TCR-Tg T cells combined with intravital imaging by 2-photon microscopy has been used in mouse models of kidney34 and pancreatic islet35 transplantation to visualize TCR-Tg alloreactive T cells traversing graft-associated endothelium while in contact with dendritic cells. This approach has revealed a mechanism for antigen-dependent but chemokine-independent migration of effector alloreactive T cells, and the chemokine-dependent migration of antigen-nonspecific effector T cells, into vascularized grafts36. Intravital imaging has also allowed the observation of the spatial relationship between allospecific cytotoxic T lymphocytes (CTLs) and their targets in a mouse model of skin transplantation37.
Generation of TCR-Tg mice is time consuming, explaining why most preclinical studies using these cells rely on a small number of commonly available TCR-Tg mice (see Table 1). In contrast, the endogenous response to a given allopeptide/MHC complex involves multiple TCRs with varied avidity, such that a single TCR transgene reactive to 1 pMHC specificity cannot accurately represent the function and phenotypes of an endogenous polyclonal TCR repertoire recognizing the same pMHC. Retrogenic (Rg) mice bearing retrovirus-transduced TCRs as a source of TCR-Rg T cells are a much faster alternative to the development of TCR-Tg mice, and this approach also eliminates the need to backcross the mice. The TCR-Rg technology provides the opportunity to produce mice expressing more TCRs of interest, for example to compare the behavior of diverse TCRs specific to a single model antigen. This technique has been utilized to study autoreactive38–40 T cells and may become useful in studying alloreactive T cells as well.
Table 1.
Transgenic T Cell Receptor lines used in mouse models of transplantation.
| Transgenic TCR | Peptide origin | MHC restriction | Direct vs Indirect | References | |
|---|---|---|---|---|---|
|
|
|||||
| CD4 | OT-II | Ovalbumin | I-Ab | Indirect* | 105,106 |
| TCR75 | Kd | I-Ab | Indirect | 23,41,107 | |
| TEa | I-E alpha | I-Ab | Indirect | 23,105,107 | |
| Marilyn | H-Y | I-Ab | Indirect* | 23,107 | |
| 4C | ? | I-Ad | Direct | 108 | |
| DO11.10 | Ovalbumin | I-Ad | Indirect* | 106,109 | |
| ABM | ? | I-Abm12 | Direct | 23,110,111 | |
| CD8 | OT-I | Ovalbumin | Kb | Indirect* | 35,112 |
| MataHari | H-Y | Db | Indirect* | 113 | |
| 2C | LSPFPFDL | Ld | Direct | 114,115 | |
Use of a graft donor sharing the host MHC haplotype will ensure direct, in addition to indirect, allorecognition.
It is important to note that some TCR-Tg T cells may become induced Tregs following adoptive transfer and antigen experience, whereas others don’t seem to convert to Tregs in vivo41. Another key caveat of adoptive transfer of monoclonal allospecific T cells is that they may not behave as innocuous tracers of the endogenous alloimmune response, but in some cases, may alter the intensity and quality of endogenous alloimmunity. Indeed, our group showed that adoptive transfer of ≥ 103 CD4+ indirect alloreactive TCR-Tg T cells/mouse potentiated alloantibody production and expansion of endogenous alloreactive CD8+ T cells, and altered the quality of the intra-graft innate infiltrate42.
Studying the polyclonal allospecific T repertoire with pMHC multimers
In addition to using monoclonal allospecific Tg or Rg T cells as a tracer population of alloreactive T cells, one can track the polyclonal alloreactive endogenous response to a single donor antigen by staining these cells using pMHC multimers. pMHC multimers consist of 4 or more pMHC complexes linked together and associated with a fluorophore. Because a single pMHC-TCR association is not strong enough to reliably persist throughout staining and flow cytometry, combining multiple pMHCs together into a multimer increases the likelihood that at least 1 pMHC remains bound to its target T cell so that it may be detected by flow cytometry. As many as 12 pMHC complexes have been linked into a single multimer, with increased numbers of pMHC providing a greater ability to identify low avidity T cells43. There is controversy over the contribution of TCR-peptide versus TCR-MHC interactions in direct allorecognition, and analogously over the nature of binding of alloreactive TCR to donor pMHC multimers. In support of a key role for direct TCR-MHC interactions, crystal structures have shown alloreactive TCRs binding donor MHC in a geometry that would provide little contact with allopeptide44. Conversely, experimental models have shown an attenuated alloresponse when stimulator cells or grafts have impaired peptide loading onto donor class II MHC45. Crystal structures have also displayed variation in the conformation of a dEV8 peptide when presented on Kb versus Kbm3 , leading to positive selection of 2C cells with dEV8:Kb and negative selection of 2C cells with dEV8:Kbm3. These findings suggest that MHC and peptide cooperate to shape the epitope recognized by an alloreactive TCR46.
The design of pMHC multimers requires knowledge of the peptide that alloreactive T cells recognize. Because the endogenous peptide that is recognized by T cells in concert with a given donor MHC molecule is often not known, tetramers are currently used in mouse models expressing model antigens with well-established peptide sequences. Although a technique frequently used for tracking tumor-specific T cells47,48, relatively few studies in transplantation have made use of pMHC multimers for tracking alloreactive T cells. Theoretically, pMHC multimers can be used to study host T cells that recognize alloantigens both directly and indirectly, depending on whether the MHC in the tetramer matches the haplotype of the graft donor or recipient, respectively. Indirect T cell recognition in mice has been modeled via expression of a model antigen by the donor graft, such as ovalbumin, and tracking of SIINFEKL:Kb tetramer-binding CD8+ T cells after transplantation42,49. Class II MHC multimers can be used to track CD4+ Tconv and Treg cells but the use of class II MHC multimers has been rarer as they have been technically challenging to generate. Nevertheless, this technique has recently been applied to transplantation in a study comparing the effects on donor-reactive T cells of CTLA4-Ig when administered at the time of immunization with model antigen-expressing donor splenocytes, versus 1 week later50. Human studies using tetramer staining to identify donor-specific T cells have been even more difficult because > 1000 classical HLA alleles have been discovered in the human population51. One potential solution is to select study participants who share at least 1 HLA allele and study the response to peptides presented on this MHC molecule. This technique has been used to identify CTLs and CD8+ regulatory T cells specific to a fetus-derived minor antigen in mothers, as well as to a noninherited maternal antigen (NIMA) in offspring52. Indeed, exposure to NIMA may impact the outcome of transplanted organs from family members53.
A single sample can be stained with multiple unique pMHC multimers to identify and compare cell populations with different pMHC specificities. Typically, these populations are resolved by conjugating each pMHC complex to a unique fluorophore. However, this strategy limits the number of multimers used to the number of channels available on the flow cytometer. Mass cytometry has expanded the number of channels that can be utilized for a single cell but that number remains in the double digits. Interestingly, a recent study has described a method capable of costaining 1 peripheral blood sample with 1000 different pMHC multimers by conjugating each type of multimer to a unique DNA barcode54. The proportion of T cells with each pMHC specificity is then determined by sorting all multimer-bound cells (identified by conjugation of all pMHC multimers to 1 common fluorophore), sequencing the barcodes in the sorted population to enable functional T cell analysis with large-scale epitope recognition profiling for various diseases. While this method has not yet been used to study alloimmunity55, it may be a useful strategy for comparing T cells spanning many allospecificities, particularly in clinical samples for which there is not enough material to subject to multiple staining conditions.
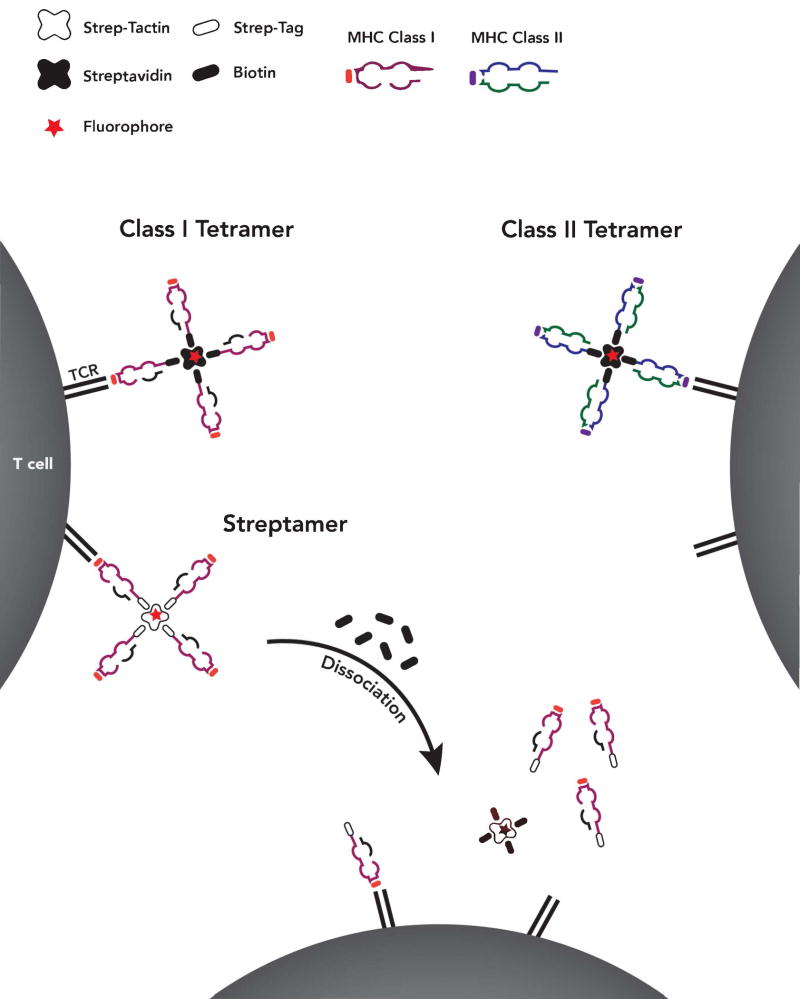
One drawback to the use of pMHC multimers for functional studies is that the stable binding of the TCR to a pMHC may alter the potential of that T cell to proliferate or exhibit effector function. To circumvent this problem, an adapted multimer structure called the Streptamer was designed that allows for the induced dissociation of the pMHC complex56 and subsequent culture or adoptive transfer of isolated epitope-specific T cells57,58. Instead of the biotinylated pMHC used in traditional multimers, Streptamers consist of pMHC complexes conjugated to Strep-tag and are assembled on a Strep-Tactin backbone in place of streptavidin. Biotin successfully competes with Strep-tag for Strep-Tactin binding and therefore, adding free biotin to Streptamers will lead to multimer disassembly and dissociation of individual pMHC complexes from the targeted T cell (Figure 1). Two additional reversible pMHC multimer forms, the DTB-pMHC multimer59 and NTAmer60, have been developed to improve upon the shelf life and ease of preparation of Streptamers though they are not commercially available. These forms of reversible pMHC multimers have only been described for Class I MHC but could theoretically be made for Class II MHC as well. While a promising solution to a potential limitation of traditional pMHC multimers, the use of reversible pMHC multimers to analyze the alloresponse should our knowledge not yet been reported.
Figure 1.
Alloreactive T cells present in the polyclonal repertoire can be identified using pMHC multimers bearing allopeptide presented on donor or recipient MHC. Traditional pMHC multimers consist of multiple pMHC monomers in association with a multivalent streptavidin backbone. The association of pMHC and streptavidin occurs via engineered biotin tails on the pMHC monomers. The streptavidin backbone is covalently linked to a fluorophore, allowing detection of multimer-binding cells by flow cytometry. While individual pMHC monomers quickly dissociate from their target T cells, the higher avidity of pMHC multimers allows for long-term binding and observation of allospecific T cells using flow cytometry. In contrast, streptamers are adapted pMHC multimers whose disassembly into pMHC monomers can be induced by the addition of free biotin. In the Streptamer, the biotinylated tails on the pMHC monomer are replaced with a similar moiety called Strep-Tag. Instead of the biotin-Streptavidin association used in traditional pMHC multimers, Strep-Tag associates with a Strep-Tactin backbone. When free biotin is added to a suspension of Streptamer-bound cells, the biotin competes with Strep-Tag for association with Strep-Tactin, thus causing the individual pMHC monomers to dissociate from Strep-Tactin. Streptamers are an alternative to traditional multimers used when the multimer-bound cells are to be isolated by FACS for further functional analysis, as traditional multimers may interfere with T cell function.
Quantifying allograft-reactive antibodies
In contrast to the lack of routine assays for quantification of donor-specific T cell responses in the clinic, a state-of-the-art approach for quantifying human B cell responses in the clinic is well established and involves the detection of graft-specific antibodies. The specificities of these antibodies can be divided into 3 broad categories: HLA-specific, non-HLA-reactive and polyreactive antibodies. The contribution of HLA-specific antibodies (HSA), especially donor HLA-specific antibodies (DSA) to antibody-mediated rejection (ABMR) and graft loss is well documented 61–65 and aided by the availability of clinical grade reagents and assays to detect HSA and DSA in the serum of transplant recipients and of patients on the transplant wait list 66–68. In addition, investigations into the outcomes of HLA-identical sibling transplantations suggest that minor histocompatibility antigens may impact graft outcome.
Other than detection of antibodies to blood group antigens, the assessment of non-HLA antibodies has been challenging for clinical practice although recent availability of high-throughput protein arrays as well as solid phase assays have increased the ability to detect auto/non-HLA antibodies in the clinic 69,70. Specifically, there is emerging evidence that antibodies directed at several endothelial antigenic targets on macrovascular and microvascular endothelium, including the angiotensin type I receptor (AT1R), endothelin type A receptor (ETAR) and the bioactive C-terminal fragment of perlecan (LG3) contribute to ABMR. Finally, Zorn and colleagues 71 reported on the high incidence of polyreactive Abs that cross-react with multiple self-antigens, HLA and apoptotic cells, in pre and posttransplantation sera from kidney transplant recipients.
Despite significant assay refinement, there remains several limitations to DSA quantification, including the sequestration of antibodies by the graft that limits or delays their detection, a so called “prozone effect” that results in false negatives when there is high titer of DSA, and the lack of standardized DSA quantification that predicts ABMR and graft loss 68,72. In contrast, the ability to detect and quantify the B cells responding to the allograft would provide complementary advantages to HSA/DSA quantification, including a potentially earlier detection of emerging humoral responses, and insights into the evolving cellular and molecular bases for de novo DSA production73. These insights may allow for immunosuppression to be tailored more effectively to control antibody responses and ABMR. New assays for the quantification of HLA-specific B cells, non-HLA-specific and polyreactive B cells in the context of allograft transplantation in mouse models and humans are discussed.
Tracking allospecific B cells
The early studies tracking the fate of donor-specific B cells in experimental mouse models used BCR-Tg mice bearing BCRs specific for known MHC molecules. The 3–83 Igi BCR-knock-in mice that express a BCR with dual specificity for H-2Kk and H-2Kb were used to show the deletional fate of alloreactive B cells in an anti-CD154-induced model of peripheral transplantation tolerance74, and that the presence of memory alloreactive B cells prevented the induction of anti-CD154-mediated allograft acceptance75. Parsons et al.76 used bone marrow cells from a related 3–83 BCR-Tg mouse to generate mixed bone marrow chimeras, and to show that emerging donor-specific B cells were deleted in the presence of allogeneic bone-marrow cells but acquired an anergic phenotype in the presence of heart allografts. However, limited availability of BCR-Tg mice with allo-MHC specificity and notable caveats of using mice bearing nonphysiological frequencies of monoclonal populations of B cells have hampered the widespread application of this approach for studying the fate of donor-specific B cells in mouse models.
Like T cells, pMHC tetramers have been used to study donor MHC-specific B cells in mice. In contrast to the pMHC for T cells, the peptide presented by pMHC plays minimal role in the ability of B cells to recognize allogeneic MHC, thus any appropriate MHC allele incorporated into a tetramer can be used to detect donor MHC-specific B cells. However, 1 major caveat with this approach is that the MHC tetramer not only identifies B cells reactive to pMHC epitopes, but also B cells reactive to non-MHC components of the tetramer, including streptavidin and fluorochromes. Thus, considerable efforts have been made to ensure the identification of B cells that are MHC-specific77,78 (Figure 2). Kwun et al.79 used allogeneic MHC Class I tetramers labeled with APC-Cy7 fluorochrome, in combination with syngeneic MHC Class I tetramers labeled with APC, to identify donor-MHC Class I specific B cells as the APC-Cy7+APC− population (Figure 2, Left Panel). The reasoning was that B cells recognizing the APC fluorochrome, biotin and streptavidin would be excluded, as would B cells that are cross-reactive to self and allogeneic MHC. Chen et al.80 used a different approach by staining with 2 identical allogeneic tetramers labeled with 2 different fluorophores, FITC and PE (Figure 2, Middle Panel). By blocking with syngeneic tetramers and soluble streptavidin, and gating on the FITC+PE+APC− events, they could identify an enriched population of donor-MHC I specific B cells. Subsequently the same group extended their studies by identifying B cells that were specific for donor MHC Class II using pMHC tetramers81.
Figure 2.
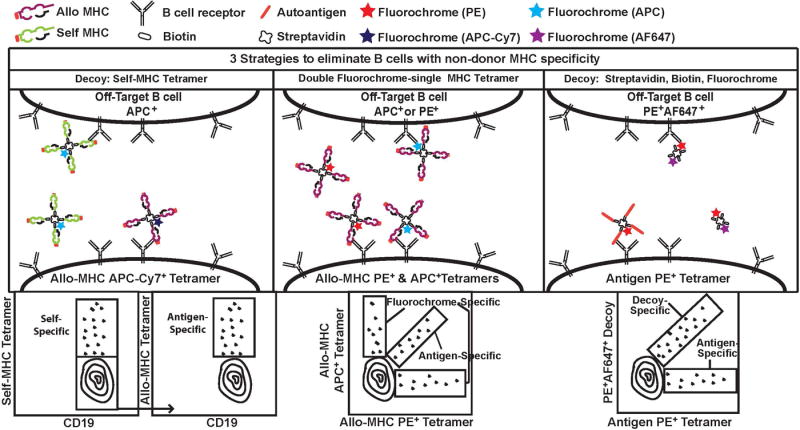
Strategies to identify donor-MHC-specific and autoreactive B cells. Because pMHC tetramers comprise MHC and non-MHC components, including streptavidin, biotin and fluorophores that are also recognized by B cells, 3 approaches have been utilized to enrich for B cells that are MHC-specific or autoreactive, and to exclude B cells that are specific for non-MHC components of the pMHC tetramer. Left Panel (Top), Allogeneic MHC tetramer is labeled with APC-Cy7 and the decoy self-MHC tetramer is labeled with APC. B cells specific for APC, biotin, streptavidin and self-MHC are APC+ and can be distinguished from donor MHC-specific B cells (APC-Cy7+). Left Panel (Bottom), Representative sequential gating strategy. Center Panel (Top), A second strategy uses donor-MHC tetramers conjugated to PE or APC. Gating only on double-positive (PE+APC+) cells eliminates the single-positive, fluorochrome-specific B cells. Right Panel (Top), A third strategy to detect autoantigen-specific B cells uses APC-tetramers presenting autoantigen, and a decoy comprising a streptavidin-biotin core conjugated to APC and a second fluorochrome, AF647. Autoantigen-specific B cells are APC+AF647− while the off-targets cells are APC+AF647+. Middle and Left Panel (Bottom), Representative gating strategy.
A related tetramer-based strategy was used to identify low-affinity autoantigen-specific B cells78,82 (Figure 2, Right Panel). Taylor et al, used tetramers comprising of a biotinylated model self-antigen (ovalbumin) complexed to a PE-labeled streptavidin core, and with decoy tetramers comprising biotin and streptavidin labeled with both Alexa Fluor (AF)647 and PE78. Hamilton et al, showed that B cells specific for 2 linear peptides from RNA-binding proteins could be similarly identified with tetramers comprising of biotinylated linear peptides and streptavidin conjugated to PE, and used a similar decoy consisting of biotin-streptavidin core conjugated to PE and AF64782. These studies demonstrated that a tetramer-based strategy can, in principle, be used to identify donor-specific B cells that are specific for non-MHC and autoreactive molecules.
By using pMHC tetramer-based systems, it became possible to measure the expansion and differentiation of antigen-specific B cells after sensitization or allograft transplantation. Following antigen encounter, B cells undergo a process of class switching, resulting in the down-regulation of IgD and upregulation of several costimulatory molecules, such as MHC Class II, CD80, and CD86. These activated B cells then enter a germinal center reaction and acquire Fas(CD95)/GL7 expression, which can be observed in ≤80% of allospecific B cells posttransplantation80. The germinal center B cells differentiate into short-lived plasmablasts, which can be defined by their expression of CD138 and the transcription factor IRF4, as well as into memory B cells. Memory B cells have been challenging to detect in mice because of their low numbers and the absence of unique and definitive markers. Isotype-switched IgG postgerminal center B cells have been historically defined as class-switched memory B cells, but the IgM+ memory B cell population cannot be detected by this method83. Thus, genetically-modified mice in which B cells become irreversibly ‘marked’ by the expression of a fluorescent protein as they enter germinal centers during a T cell-dependent response, have been used to identify memory B cells. By crossing Rosa26-loxP-EYPF reporter mice with transgenic mice expressing Cre recombinase driven by the promoter of the gene encoding activation-induced cytidine deaminase (Aicda), Yang et al.81 quantified the generation of memory B cells with donor-MHC Class II specificity, and showed that the frequency correlates with the strength of the recall antibody response. These studies therefore show how tracking alloreactive B cells can inform on their in vivo fate after transplantation.
There are considerable efforts devoted to enable the tracking of memory donor-specific B cells in humans, and substantial frequencies of memory B cells have been reported as circulating in human peripheral blood 84–87. These cells have been detected indirectly, following their activation and differentiation into antibody-secreting cells ex vivo, and then quantified by IgG ELISpot assays or by assessment of secreted IgG in the supernatant88–95. The ELISpot assay provides information on the frequency of HLA-specific B cell, but this approach is handicapped by the low frequencies of HLA-specific antibody-secreting B cells, by the challenge of developing ELISpot assays for the array of HLA alleles encountered in the clinic, and by the assay duration that requires 6–10 days of ex vivo culture. Quantifying IgG in the supernatant and assessing their specificity by ELISA or using clinical grade HLA-coated beads overcomes some of the limitations imposed by the ELISpot assay, but this approach only provides an indirect assessment of the frequency of HLA-specific B cells that assumes that plasma cells secrete comparable amounts of IgG. Furthermore, there is no direct way to assess the phenotype and functional characteristics of graft-specific B cells, and the 6–10-day culture is still required.
The approach to directly identify HLA-specific B cells with fluorescently labeled HLA Class I tetramers was reported by Zachary and colleagues96–98, but its clinical translation is hampered by the limited availability of validated HLA Class I and II tetramers that prevents the assessment of the full donor MHC-reactive B cell repertoire. More recently, a related approach to identify HLA-specific B cells using microspheres coated with HLA molecules was described 99. In this assay, HLA-specific B cells bind to, and form HLA bead-B cell rosettes (BBR) that are identified by flow cytometry. Importantly these single HLA antigen-coated beads are being used in the clinic, where their multiplexing power enables the simultaneous analysis of Abs (and B cells) directed to a large panel of clinically relevant HLA class I or II antigens 66,68. With this approach, the identity and frequency of B cells specific for HLA alleles, as well as their phenotypes, can be rapidly determined in time-frames that are comparable to HSA/DSA assays. For instance, the frequency of CD19+ B cells that bind to single HLA-A*0201 allele-coated beads were shown to be increased in sensitized transplant recipients with biopsy-proven ABMR compared to healthy volunteers or nonsensitized stable transplant recipients. Nevertheless, the expanded numbers of B cells that bound the HLA beads were only modestly enriched for B cells with a memory phenotype, suggesting a need for further validation of the specificity of the B cell-bead interaction. Notably, the authors also used this bead assay to quantify B cells with specificity for human albumin, tetanus toxin, EBNA and MOG, suggesting that this platform may be expanded to detect B cells specific for non-HLA antigens.
It is currently possible, but technically challenging and labor intensive to detect B cells expressing polyreactive B cell receptors. Zorn and colleagues have generated EBV-immortalized B cell clones, and tested their secreted antibodies for binding to a panel of HLA antigens and apoptotic cells74. Serum samples that had higher reactivity to apoptotic cells also displayed reactivity to a broader array of MHC Class I alleles compared to samples with low reactivity to apoptotic cells. Moreover, the observation of 4 distinct B cell clones cross-reactive to self and numerous HLA class I alleles that did not have “shared” epitopes provided proof of principle evidence that human polyreactive antibodies can cross-react to HLA, multiple self-antigens and apoptotic cells. The extent to which polyreactivity contributes to high panel-reactive antibodies (PRA), and the provenance of these polyreactive B cells, either as “natural” low affinity B cells or postgerminal center B cells, require further investigation.
Tracking in vivo generated plasma cells
Tetramer-based detection systems have allowed for the identification of antigen-specific naïve, activated, and memory B cells. However, it has been challenging to assess fully-differentiated allospecific plasma cells with this approach. Plasma cells express high levels of CD138, which is also expressed on other cell types including pro B cells and select subsets of myeloid, T and NK cells100,101 (http://www.immgen.org). Furthermore, plasma cells express little to no CD19 or B cell receptor, making the identification of antigen-specific plasma cells by cell-surface expression and flow cytometry extremely challenging. Thus, an alternative approach has been developed to detect the frequency of plasma cells secreting MHC-specific antibodies using an ELISpot assay that utilizes immobilized anti-IgG and biotinylated MHC monomers81,102,103. Sicard et al.103 reported that the frequency of plasma cells posttransplantation was increased ~104-fold in the spleen and ~103-fold in the bone-marrow, in contrast to the plasma cells generated following viral infections that were predominantly enriched in the bone marrow. Furthermore, spleen-resident plasma cells differentiated into cells that secreted more donor-specific IgG compared to their bone marrow-resident counterparts, providing unexpected insights into the biology of donor MHC-specific plasma cells.
Long-lived plasma cells are responsible for serological memory, and there is considerable interest in identifying desensitization protocols that successfully eliminate these cells in patients with high PRA. Perry et al, reported anti-HLA antibody production in vitro by B cells isolated from the bone marrow of sensitized kidney transplant recipients, and the impact of bortezomib-mediated proteasome inhibition for treating humoral rejection104. However, this approach has limited clinical applicability because of the need for bone marrow or lymphoid organ sampling to access the long-lived plasma cells that are responsible for serological memory in sensitized pretransplant and posttransplant individuals, as the blood only harbors short-lived plasma blasts. The determination of successful desensitization will therefore most likely utilize DSA detection as a proxy for the presence or absence of long-lived plasma cells.
Conclusion
In summary, tracking of alloreactive T cells has allowed the study of the phenotype, genotype and function of cells mediating the alloimmune response (Table 2). Identification of polyclonal alloreactive T cells through restimulation has been useful in both mouse and human studies whereas mouse models of transplantation using grafts expressing known antigens are allowing precise prospective tracking of an alloimmune T cell response. Some tools such as TCR-Tg mice have been used widely in the field of transplantation and have been key to our current understanding of alloimmunity, while the use of pMHC multimers to track endogenous alloreactive T cells is gaining traction. Other techniques such as Streptamer-based cell isolation and intravital imaging have not yet been extensively used in transplantation and may be powerful tools for future studies. Despite progress in experimental models, it remains extremely challenging to identify alloreactive T cells in the clinic. In contrast, available MHC tetramers have allowed for the quantification of donor HLA-specific B cells in both mouse and humans. While access to both the secondary lymphoid organs and bone marrow has allowed for more detailed analysis of the evolution of the B cell response following allotransplantation in mice, widely accessible immune monitoring for pre and posttransplant patients will likely be restricted to the analysis of naïve and memory B cells in the peripheral blood.
Table 2.
Strategies for tracking alloreactive T cells.
| Tracking Strategy |
Which T cells are alloreactive |
Animal Models |
Human Studies |
Allorecognition pathways detected |
Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|---|
|
| |||||||
| In vitro restimulation: MLR | Cells expressing surface markers of activation and proliferation by flow cytometry | ✓ | ✓ | Direct and indirect* | Simple preparation and readout | In vitro stimulation differs from in vivo stimulation conditions | 11,14,20,25 |
| Cells with diluted CFSE following CFSE labeling | Allows study of polyclonal response to alloantigen | Does not reflect effects of immunosuppression | |||||
| IFNy-producing cells by cytokine flow cytometry | Surface markers can be detected within 24 hours of stimulation | CFSE dilution requires ≥48 hours for detection | |||||
|
| |||||||
| In vitro restimulation: IFNy ELISpot | IFNy-secreting cells | ✓ | ✓ | Direct and indirect* | Simple preparation and readout | In vitro stimulation differs from in vivo stimulation conditions | 10–12 |
| Allows study of polyclonal response to alloantigen | Does not reflect effects of immunosuppression | ||||||
| More sensitive than intracellular IFNy detection by flow cytometry | Underreporting in samples with many IFNy-producing cells | ||||||
|
| |||||||
| Trans-vivo DTH | Cells expressing surface markers of activation and proliferation by flow cytometry | ✓ | ✓ | Indirect | Allows study of polyclonal response to alloantigen | Does not fully recapitulate allograft rejection processes | 31,32 |
| Cells with diluted CFSE following CFSE labeling | Includes influence of factors produced by local cells and other nuances of in vivo stimulation | Does not reflect effects of immunosuppression | |||||
| Labor intensive and requires careful assessment of footpad swelling | |||||||
| IFNy producing cells by cytokine flow cytometry | Surface markers can be detected within 24 hours of stimulation | CFSE dilution requires ≥48 hours for detection | |||||
|
| |||||||
| Transgenic TCR | Tg or Rg mouse-derived T cells labeled by CFSE, transgenic fluorescent marker or congenic marker | ✓ | Direct or indirect | Allows analysis of naïve or antigen-experienced allospecific T cells | Single TCR is not reflective of a polyclonal response | 23,33 | |
| Allows control of precursor frequency by adoptive transfer | Generation of new TCR-Tg mice is slow | ||||||
|
|
|
||||||
| Retrogenic TCR** | ✓ | Direct or indirect | Faster than generation of TCR-Tg mice | Single TCR is not reflective of a polyclonal response | 116 | ||
|
| |||||||
| pMHC Multimer | Multimer-bound T cells | ✓ | ✓ | Direct or indirect | Allows study of a polyclonal response to a single alloantigen specificity | Requires knowledge of allopeptide identity | 42,43,50,52 |
| Multimer binding to TCR impacts T cell function | |||||||
|
|
|
||||||
| pMHC Streptamer** | ✓ | ✓ | Direct or indirect | Lower impact of TCR binding on T cell function than traditional pMHC multimers; allows functional analysis after sorting | Requires knowledge of allopeptide identify | 56 | |
| More costly and difficult to prepare than traditional pMHC multimers | |||||||
In cultures containing host and donor PBMCs, donor or recipient PBMCs can theoretically present alloantigen, although direct responses may predominate. If donor PBMCs are lysed prior to coculture, only PBMCs from the recipient will contain intact antigen-presenting cells and all allopresentation will occur through the indirect pathway.
Application of the technique for this purpose has not been reported. However, pMHC Streptamers are structurally and functionally like pMHC multimers and TCR-Rgs are like TCR-Tgs such that many of the applications of TCR-Tgs and pMHC multimers should be possible using TCR-Rgs and pMHC Streptamers, respectively.
Acknowledgments
Funding:
This work was supported in part by grants (R01 AI072630; P01AI097113) from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health. JSY received an American Heart Association postdoctoral fellowship award (15POST25700452) and was funded by a Respiratory Biology Training Grant (T32 HL07605). CM was funded by the Growth, Development and Disabilities Training Program (T32 HD 7009-41).
Abbreviations
- ABMR
antibody-mediated rejection
- AF
Alexa Fluor
- APC
allophycocyanin
- AT1R
angiotensin type I receptor
- BCR-Tg
B cell receptor-transgenic
- CFSE
carboxyfluorescein succinimidyl ester
- DSA
donor specific antibodies
- DTH
delayed type hypersensitivity
- ETAR
endothelin type A receptor
- ELISPOT
enzyme-linked immunosorbent assay
- FACS
fluorescence-activated cell sorting
- HAS
HLA-specific antibodies
- MLR
mixed lymphocyte reaction
- PBMC
peripheral blood mononuclear cells
- PE
phycoerythrin
- SCID
severe combined immunodeficiency
- TCR-Rg
T cell receptor-retrogenic
Footnotes
Author Contributions:
All authors contributed to the writing of the article.
Conflict of Interest Statement:
The authors have declared that no conflict of interest exists.
References
- 1.Reichenbach DK, Li Q, Hoffman RA, et al. Allograft outcomes in outbred mice. Am J Transplant. 2013;13(3):580–588. doi: 10.1111/ajt.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halloran PF, Chang J, Famulski K, et al. Disappearance of T cell-mediated rejection despite continued antibody-mediated rejection in late kidney transplant recipients. J Am Soc Nephrol. 2015;26(7):1711–1720. doi: 10.1681/ASN.2014060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sellares J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant. 2012;12(2):388–399. doi: 10.1111/j.1600-6143.2011.03840.x. [DOI] [PubMed] [Google Scholar]
- 4.Aubert O, Loupy A, Hidalgo L, et al. Antibody-mediated rejection due to preexisting versus de novo donor-specific antibodies in kidney allograft recipients. J Am Soc Nephrol. 2017;28(6):1912–1923. doi: 10.1681/ASN.2016070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haque MA, Mizobuchi T, Yasufuku K, et al. Evidence for immune responses to a self-antigen in lung transplantation: role of type V collagen-specific T cells in the pathogenesis of lung allograft rejection. J Immunol. 2002;169(3) doi: 10.4049/jimmunol.169.3.1542. 1542 LP-1549. [DOI] [PubMed] [Google Scholar]
- 6.Rolls HK, Kishimoto K, Dong VM, et al. T-cell response to cardiac myosin persists in the absence of an alloimmune response in recipients with chronic cardiac allograft rejection. Transplantation. 2002;74(7):1053–1057. doi: 10.1097/00007890-200210150-00028. [DOI] [PubMed] [Google Scholar]
- 7.Haller GW, Lima B, Kunisaki SM, et al. MHC alloantigens elicit secondary, but not primary, indirect in vitro proliferative responses. J Immunol. 2002;169(7):3613. doi: 10.4049/jimmunol.169.7.3613. [DOI] [PubMed] [Google Scholar]
- 8.Langhorne JLKF, Lindahl KF. Role of non-H-2 antigens in the cytotoxic T cell response to allogeneic H-2. Eur J Immunol. 1982;12(2):101–106. doi: 10.1002/eji.1830120202. [DOI] [PubMed] [Google Scholar]
- 9.Macedo C, Orkis EA, Popescu I, et al. Contribution of naïve and memory T-cell populations to the human alloimmune response. Am J Transplant. 2009;9(9):2057–2066. doi: 10.1111/j.1600-6143.2009.02742.x. [DOI] [PubMed] [Google Scholar]
- 10.Benichou G, Valujskikh A, Heeger PS. Contributions of direct and indirect T cell alloreactivity during allograft rejection in mice. J Immunol. 1999;162(1):352–8. [PubMed] [Google Scholar]
- 11.Karlsson AC, Martin JN, Younger SR, et al. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J Immunol Methods. 2003;283(1):141–153. doi: 10.1016/j.jim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Heeger PS, Greenspan NS, Kuhlenschmidt S, et al. Pretransplant frequency of donor-specific, IFN-γ-producing lymphocytes is a manifestation of immunologic memory and correlates with the risk of posttransplant rejection episodes. J Immunol. 1999;163(4) 2267 LP-2275. [PubMed] [Google Scholar]
- 13.Hricik DE, Augustine J, Nickerson P, et al. Interferon gamma ELISPOT testing as a risk-stratifying biomarker for kidney transplant injury: results from the CTOT-01 multicenter study. Am J Transplant. 2015;15(12):3166–3173. doi: 10.1111/ajt.13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shipkova M, Wieland E. Surface markers of lymphocyte activation and markers of cell proliferation. Clinica Chimica Acta. 2012;413(17–18):1338–1349. doi: 10.1016/j.cca.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Pilat N, Sayegh MH, Wekerle T. Costimulatory pathways in transplantation. Semin Immunol. 2011;23(4):293–303. doi: 10.1016/j.smim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohler T, Canivet C, Galvani S, et al. Pharmacodynamic monitoring of the conversion from mycophenolate mofetil to enteric-coated mycophenolate sodium in stable kidney-allograft recipients. Int Immunopharmacol. 2008;8(5):769–773. doi: 10.1016/j.intimp.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 17.Ekong UD, Luo X, Yu M, Wang D, Miller SD, O’Gorman MR. Lymphocyte activation markers may predict the presence of donor specific alloreactivity in pediatric living related liver transplant recipients. Hum Immunol. 2011;72(5):392–397. doi: 10.1016/j.humimm.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Haan A, van der Gun I, van der Bij W, de Leij LFMH, Prop J. Detection of alloreactive T cells by flow cytometry: a new test compared with limiting dilution assay1. Transplantation. 2002;74(4):562–570. doi: 10.1097/00007890-200208270-00023. [DOI] [PubMed] [Google Scholar]
- 19.Van Rijen MML, Metselaar HJ, Hommes M, Ijzermans JNM, Tilanus HW, Kwekkeboom J. Mycophenolic acid is a potent inhibitor of the expression of tumour necrosis factor- and tumour necrosis factor-receptor superfamily costimulatory molecules. Immunology. 2003;109(1):109–116. doi: 10.1046/j.1365-2567.2003.01635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litjens NH, de Wit EA, Baan CC, Betjes MG. Activation-induced CD137 is a fast assay for identification and multi-parameter flow cytometric analysis of alloreactive T cells. Clin Exp Immunol. 2013;174(1):179–191. doi: 10.1111/cei.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong YY, Kishihara K, Yoshida H, Mak TW, Nomoto K. Generation of T cells with differential responses to alloantigens in CD45 exon 6-deficient mice. J Immunol. 1995;154(11):5725. [PubMed] [Google Scholar]
- 22.Scifo C, Mekaelian L, Munyazesa E, Schmitt-Verhulst A-M, Guimezanes A. Selection of T-cell receptors with a recurrent CDR3β peptide-contact motif within the repertoire of alloreactive CD8+ T cells. Eur J Immunol. 2011;41(8):2414–2423. doi: 10.1002/eji.201141494. [DOI] [PubMed] [Google Scholar]
- 23.Ali JM, Negus MC, Conlon TM, et al. Diversity of the CD4 T cell alloresponse: the short and the long of It. Cell Rep. 2016;14(5):1232–1245. doi: 10.1016/j.celrep.2015.12.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallon L, Traitanon O, Sustento-Reodica N, et al. Cellular and molecular immune profiles in renal transplant recipients after conversion from tacrolimus to sirolimus. Kidney Int. 2015;87(4):828–838. doi: 10.1038/ki.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suchin EJ, Langmuir PB, Palmer E, Sayegh MH, Wells AD, Turka LA. Quantifying the frequency of alloreactive T cells in vivo: new answers to an old question. J Immunol. 2001;166(2):973–81. doi: 10.4049/jimmunol.166.2.973. [DOI] [PubMed] [Google Scholar]
- 26.Emerson RO, Mathew JM, Konieczna IM, Robins HS, Leventhal JR. Defining the alloreactive T cell repertoire using high-throughput sequencing of mixed lymphocyte reaction culture. PLoS One. 2014;9(11):1–7. doi: 10.1371/journal.pone.0111943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris H, DeWolf S, Robins H, et al. Tracking donor-reactive T cells: Evidence for clonal deletion in tolerant kidney transplant patients. Sci Transl Med. 2015;7(272):272ra210–272ra210. doi: 10.1126/scitranslmed.3010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto R, You S, Zaitsu M, Chatenoud L, Wood KJ. Delayed anti-CD3 therapy results in depletion of alloreactive T cells and the dominance of Foxp3+CD4+ graft infiltrating cells. Am J Transplant. 2013;13(7):1655–1664. doi: 10.1111/ajt.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazekas De St Groth B, Smith AL, Higgins CA. T cell activation: in vivo veritas. Immunol Cell Biol. 2004;82(3):260–268. doi: 10.1111/j.0818-9641.2004.01243.x. [DOI] [PubMed] [Google Scholar]
- 30.Storni T, Bachmann MF. On the role of APC-activation for in vitro versus in vivo T cell priming. Cell Immunol. 2003;225(1):1–11. doi: 10.1016/j.cellimm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Carrodeguas L, Orosz CG, Waldman WJ, Sedmak DD, Adams PW, Vanbuskirk AM. Trans vivo analysis of human delayed-type hypersensitivity reactivity. Hum Immunol. 1999;60(8):640–651. doi: 10.1016/s0198-8859(99)00002-6. [DOI] [PubMed] [Google Scholar]
- 32.VanBuskirk AM, Burlingham WJ, Jankowska-Gan E, et al. Human allograft acceptance is associated with immune regulation. J Clin Invest. 2000;106(1):145–155. doi: 10.1172/JCI9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford ML, Koehn BH, Wagener ME, et al. Antigen-specific precursor frequency impacts T cell proliferation, differentiation, and requirement for costimulation. J Exp Med. 2007;204(2):299–309. doi: 10.1084/jem.20062319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walch JM, Zeng Q, Li Q, et al. Cognate antigen directs CD8+ T cell migration to vascularized transplants. J Clin Invest. 2013;123(6):2663–2671. doi: 10.1172/JCI66722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Dai H, Yatim KM, et al. CD8+ Effector T cell migration to pancreatic islet grafts Is dependent on cognate antigen presentation by donor graft cells. J Immunol. 2016;197(4) doi: 10.4049/jimmunol.1600832. 1471 LP-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberbarnscheidt MH, Walch JM, Li Q, et al. Memory T cells migrate to and reject vascularized cardiac allografts independent of the chemokine receptor CXCR3. Transplantation. 2011;91(8):827–832. doi: 10.1097/TP.0b013e31820f0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celli S, Albert ML, Bousso P. Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat Med. 2011;17(6):744–749. doi: 10.1038/nm.2376. [DOI] [PubMed] [Google Scholar]
- 38.Bettini M, Blanchfield L, Castellaw A, et al. TCR affinity and tolerance mechanisms converge to shape T cell diabetogenic potential. J Immunol. 2014;193(2) doi: 10.4049/jimmunol.1400043. 571 LP-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alli R, Nguyen P, Geiger TL. Retrogenic modeling of experimental allergic encephalomyelitis associates T cell frequency but not TCR functional affinity with pathogenicity. J Immunol. 2008;181(1) doi: 10.4049/jimmunol.181.1.136. 136 LP-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold PY, Burton AR, Vignali DAA. Diabetes incidence is unaltered in glutamate decarboxylase 65-specific TCR retrogenic nonobese diabetic mice: generation by retroviral-mediated stem cell gene transfer. J Immunol. 2004;173(5):3103–3111. doi: 10.4049/jimmunol.173.5.3103. [DOI] [PubMed] [Google Scholar]
- 41.Chai J-G, Ratnasothy K, Bucy RP, Noelle RJ, Lechler R, Lombardi G. Allospecific CD4+ T cells retain effector function and are actively regulated by Treg cells in the context of transplantation tolerance. Eur J Immunol. 2015;45(7):2017–2027. doi: 10.1002/eji.201545455. [DOI] [PubMed] [Google Scholar]
- 42.Miller ML, Chen J, Daniels MD, et al. Adoptive transfer of tracer-alloreactive CD4+ T cell receptor transgenic T cells alters the endogenous immune response to an allograft. Am J Transplant. 2016;16(10):2842–2853. doi: 10.1111/ajt.13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Zeng X, Sigal N, et al. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc Natl Acad Sci U S A. 2016;113(13):E1890–E1897. doi: 10.1073/pnas.1602488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiser JB, Darnault CGA, Gregoire C, et al. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1(4):291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 45.Felix NJ, Brickey WJ, Griffiths R, et al. H2-DMα(−/−) mice show the importance of major histocompatibility complex-bound peptide in cardiac allograft rejection. J Exp Med. 2000;192(1):31–40. doi: 10.1084/jem.192.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luz JG, Huang M, Garcia KC, et al. Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility complex complexes. J Exp Med. 2002;195(9):1175. doi: 10.1084/jem.20011644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manlove LS, Berquam-Vrieze KE, Pauken KE, Williams RT, Jenkins MK, Farrar MA. Adaptive immunity to Leukemia is inhibited by cross-reactive induced regulatory T cells. J Immunol. 2015;195(8) doi: 10.4049/jimmunol.1501291. 4028 LP-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson AM, Mylin LM, Thompson MM, Schell TD. Modification of a tumor antigen determinant to improve peptide/MHC stability is associated with increased immunogenicity and cross-priming a larger fraction of CD8+ T cells. J Immunol. 2012;189(12) doi: 10.4049/jimmunol.1102221. 5549 LP-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krummey SM, Chen C-W, Guasch SA, et al. Enhanced requirement for TNFR2 in graft rejection mediated by low-affinity memory CD8+ T cells during heterologous immunity. J Immunol. 2016;197(5) doi: 10.4049/jimmunol.1502680. 2009 LP-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young JS, Chen J, Miller ML, et al. Delayed cytotoxic T lymphocyte-associated protein 4-immunoglobulin treatment reverses ongoing alloantibody responses and rescues allografts from acute rejection. Am J Transplant. 2016;16(8):2312–2323. doi: 10.1111/ajt.13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams TM. Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J Mol Diagn. 2001;3(3):98–104. doi: 10.1016/S1525-1578(10)60658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halteren AGSV, Jankowska-gan E, Joosten A, et al. Naturally acquired tolerance and sensitization to minor histocompatibility antigens in healthy family members. Blood. 2009;114(11):2263–2272. doi: 10.1182/blood-2009-01-200410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai J, Lee J, Jankowska-Gan E, et al. Minor H Antigen HA-1-specific regulator and effector CD8+ T cells, and HA-1 microchimerism, in allograft tolerance. J Exp Med. 2004;199(7) doi: 10.1084/jem.20031012. 1017 LP-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bentzen AK, Marquard AMA-Ohoo, Lyngaa R, et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol. 2016;34(10):1037–1045. doi: 10.1038/nbt.3662. [DOI] [PubMed] [Google Scholar]
- 55.Guillonneau C, David L, Anegon I. Improved analyses of CD8+ T cell specificities using multimers of peptide MHC complexes coupled to DNA barcodes. Transplantation. 2017;101(2):219–221. doi: 10.1097/TP.0000000000001601. [DOI] [PubMed] [Google Scholar]
- 56.Knabel M, Franz TJ, Schiemann M, et al. Reversible MHC multimer staining for functional isolation of T-cell populations and effective adoptive transfer. Nat Med. 2002;8(6):631–637. doi: 10.1038/nm0602-631. [DOI] [PubMed] [Google Scholar]
- 57.Wang XC, Pang H, Xu X, et al. Streptamer versus tetramer-based selection of functional cytomegalovirus-specific T cells. J Formos Med Assoc. 2013;112(6):338–345. doi: 10.1016/j.jfma.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 58.Xu XN, Purbhoo MA, Chen N, et al. A novel approach to antigen-specific deletion of CTL with minimal cellular activation using alpha3 domain mutants of MHC class I/peptide complex. Immunity. 2001;14(5):591–602. doi: 10.1016/s1074-7613(01)00133-9. [DOI] [PubMed] [Google Scholar]
- 59.Guillaume P, Baumgaertner P, Angelov GS, Speiser D, Luescher IF. Fluorescence-activated cell sorting and cloning of bona fide CD8+ CTL with reversible MHC-peptide and antibody fab′ conjugates. J Immunol. 2006;177(6):3903–12. doi: 10.4049/jimmunol.177.6.3903. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt J, Guillaume P, Irving M, Baumgaertner P, Speiser D, Luescher IF. Reversible major histocompatibility complex I-peptide multimers containing Ni2+-nitrilotriacetic acid peptides and histidine tags improve analysis and sorting of CD8+ T cells. J Biol Chem. 2011;286(48):41723–41735. doi: 10.1074/jbc.M111.283127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Halloran PF, Reeve JP, Pereira AB, Hidalgo LG, Famulski KS. Antibody-mediated rejection, T cell-mediated rejection, and the injury-repair response: new insights from the Genome Canada studies of kidney transplant biopsies. Kidney Int. 2014;85(2):258–264. doi: 10.1038/ki.2013.300. [DOI] [PubMed] [Google Scholar]
- 62.Wiebe C, Gibson IW, Blydt-Hansen TD, et al. Rates and determinants of progression to graft failure in kidney allograft recipients with de novo donor-specific antibody. Am J Transplant. 2015;15(11):2921–2930. doi: 10.1111/ajt.13347. [DOI] [PubMed] [Google Scholar]
- 63.Wiebe C, Nevins TE, Robiner WN, Thomas W, Matas AJ, Nickerson PW. The synergistic effect of class II HLA epitope-mismatch and nonadherence on acute rejection and graft survival. Am J Transplant. 2015;15(8):2197–2202. doi: 10.1111/ajt.13341. [DOI] [PubMed] [Google Scholar]
- 64.Lefaucheur C, Viglietti D, Bentlejewski C, et al. IgG donor-specific anti-human HLA antibody subclasses and kidney allograft antibody-mediated injury. J Am Soc Nephrol. 2016;27(1):293–304. doi: 10.1681/ASN.2014111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viglietti D, Loupy A, Vernerey D, et al. Value of donor-specific anti-HLA antibody monitoring and characterization for risk stratification of kidney allograft loss. J Am Soc Nephrol. 2017;28(2):702–715. doi: 10.1681/ASN.2016030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.El-Awar N, Lee J, Terasaki PI. HLA antibody identification with single antigen beads compared to conventional methods. Hum Immunol. 2005;66(9):989–997. doi: 10.1016/j.humimm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Bray RA, Tarsitani C, Gebel HM, Lee JH. Clinical cytometry and progress in HLA antibody detection. Methods Cell Biol. 2011;103:285–310. doi: 10.1016/B978-0-12-385493-3.00012-7. [DOI] [PubMed] [Google Scholar]
- 68.Gebel HM, Bray RA. HLA antibody detection with solid phase assays: great expectations or expectations too great? Am J Transplant. 2014;14(9):1964–1975. doi: 10.1111/ajt.12807. [DOI] [PubMed] [Google Scholar]
- 69.Delville M, Charreau B, Rabant M, Legendre C, Anglicheau D. Pathogenesis of non-HLA antibodies in solid organ transplantation: Where do we stand? Hum Immunol. 2016;77(11):1055–1062. doi: 10.1016/j.humimm.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 70.Dragun D, Catar R, Philippe A. Non-HLA antibodies against endothelial targets bridging allo- and autoimmunity. Kidney Int. 2016;90(2):280–288. doi: 10.1016/j.kint.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 71.Zorn E, See SB. Polyreactive natural antibodies in transplantation. Curr Opin Organ Transplant. 2017;22(1):8–13. doi: 10.1097/MOT.0000000000000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tambur AR, Herrera ND, Haarberg KM, et al. Assessing antibody strength: comparison of MFI, C1q, and titer information. Am J Transplant. 2015;15(9):2421–2430. doi: 10.1111/ajt.13295. [DOI] [PubMed] [Google Scholar]
- 73.Chong AS, Sciammas R. Memory B cells in transplantation. Transplantation. 2015;99(1):21–28. doi: 10.1097/TP.0000000000000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Ma L, Shen J, Chong AS. Peripheral deletion of mature alloreactive B cells induced by costimulation blockade. Proc Natl Acad Sci U S A. 2007;104(29):12093–12098. doi: 10.1073/pnas.0705240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burns AM, Ma L, Li Y, et al. Memory alloreactive B cells and alloantibodies prevent anti-CD154-mediated allograft acceptance. J Immunol. 2009;182(3):1314–1324. doi: 10.4049/jimmunol.182.3.1314. [DOI] [PubMed] [Google Scholar]
- 76.Parsons RF, Vivek K, Rostami SY, et al. Acquisition of humoral transplantation tolerance upon de novo emergence of B lymphocytes. J Immunol. 2011;186(1):614–620. doi: 10.4049/jimmunol.1002873. [DOI] [PubMed] [Google Scholar]
- 77.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331(6021):1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. 2012;209(3):597–606. doi: 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwun J, Oh BC, Gibby AC, et al. Patterns of de novo allo B cells and antibody formation in chronic cardiac allograft rejection after alemtuzumab treatment. Am J Transplant. 2012;12(10):2641–2651. doi: 10.1111/j.1600-6143.2012.04181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J, Yin H, Xu J, et al. Reversing endogenous alloreactive B cell GC responses with anti-CD154 or CTLA-4Ig. Am J Transplant. 2013;13(9):2280–2292. doi: 10.1111/ajt.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Chen J, Young JS, et al. Tracing donor-MHC class II reactive B cells in mouse cardiac transplantation: delayed CTLA4-Ig treatment prevents memory alloreactive B-cell generation. Transplantation. 2016;100(8):1683–1691. doi: 10.1097/TP.0000000000001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamilton JA, Li J, Wu Q, et al. General approach for tetramer-based identification of autoantigen-reactive B cells: characterization of La- and snRNP-reactive B cells in autoimmune BXD2 mice. J Immunol. 2015;194(10):5022–5034. doi: 10.4049/jimmunol.1402335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dogan I, Bertocci B, Vilmont V, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10(12):1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 84.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298(5601):2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 85.Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171(10):4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 86.Lanzavecchia A, Bernasconi N, Traggiai E, Ruprecht CR, Corti D, Sallusto F. Understanding and making use of human memory B cells. Immunol Rev. 2006;211:303–309. doi: 10.1111/j.0105-2896.2006.00403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol. 2009;39(5):1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karahan GE, de Vaal YJ, Roelen DL, Buchli R, Claas FH, Heidt S. Quantification of HLA class II-specific memory B cells in HLA-sensitized individuals. Hum Immunol. 2015;76(2–3):129–136. doi: 10.1016/j.humimm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 89.Bromage E, Stephens R, Hassoun L. The third dimension of ELISPOTs: quantifying antibody secretion from individual plasma cells. J Immunol Methods. 2009;346(1–2):75–79. doi: 10.1016/j.jim.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 90.Lycke NY, Coico R. Measurement of immunoglobulin synthesis using the ELISPOT assay. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im0714s17. Chapter 7:Unit 7.14. [DOI] [PubMed] [Google Scholar]
- 91.Han M, Rogers JA, Lavingia B, Stastny P. Peripheral blood B cells producing donor-specific HLA antibodies in vitro. Hum Immunol. 2009;70(1):29–34. doi: 10.1016/j.humimm.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 92.Perry DK, Pollinger HS, Burns JM, et al. Two novel assays of alloantibody-secreting cells demonstrating resistance to desensitization with IVIG and rATG. Am J Transplant. 2008;8(1):133–143. doi: 10.1111/j.1600-6143.2007.02039.x. [DOI] [PubMed] [Google Scholar]
- 93.Heidt S, Roelen DL, de Vaal YJH, et al. A novel ELISPOT assay to quantify HLA-specific B cells in HLA-immunized individuals. Am J Transplant. 2012;12(6):1469–1478. doi: 10.1111/j.1600-6143.2011.03982.x. [DOI] [PubMed] [Google Scholar]
- 94.Lynch RJ, Silva IA, Chen BJ, Punch JD, Cascalho M, Platt JL. Cryptic B cell response to renal transplantation. Am J Transplant. 2013;13(7):1713–1723. doi: 10.1111/ajt.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lucia M, Luque S, Crespo E, et al. Preformed circulating HLA-specific memory B cells predict high risk of humoral rejection in kidney transplantation. Kidney Int. 2015;88(4):874–887. doi: 10.1038/ki.2015.205. [DOI] [PubMed] [Google Scholar]
- 96.Lucas DP, Leffell MS, Zachary AA. Tetramer staining for the detection of HLA-specific B cells. Methods Mol Biol. 2013;1034:313–318. doi: 10.1007/978-1-62703-493-7_17. [DOI] [PubMed] [Google Scholar]
- 97.Zachary AA, Kopchaliiska D, Montgomery RA, Leffell MS. HLA-specific B cells: I. A method for their detection, quantification, and isolation using HLA tetramers. Transplantation. 2007;83(7):982–988. doi: 10.1097/01.tp.0000259017.32857.99. [DOI] [PubMed] [Google Scholar]
- 98.Zachary AA, Kopchaliiska D, Montgomery RA, Melancon JK, Leffell MS. HLA-specific B cells: II. Application to transplantation. Transplantation. 2007;83(7):989–994. doi: 10.1097/01.tp.0000259019.68244.d7. [DOI] [PubMed] [Google Scholar]
- 99.Degauque N, Elong Ngono A, Akl A, et al. Characterization of antigen-specific B cells using nominal antigen-coated flow-beads. PLoS One. 2013;8(12):e84273. doi: 10.1371/journal.pone.0084273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Connell FP, Pinkus JL, Pinkus GS. CD138 (syndecan-1), a plasma cell marker immunohistochemical profile in hematopoietic and nonhematopoietic neoplasms. Am J Clin Pathol. 2004;121(2):254–263. doi: 10.1309/617D-WB5G-NFWX-HW4L. [DOI] [PubMed] [Google Scholar]
- 101.Heng TS, Painter MW. The immunological genome project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 102.Chen J, Wang Q, Yin D, Vu V, Sciammas R, Chong AS. Cutting edge: CTLA-4Ig inhibits memory B cell responses and promotes allograft survival in sensitized recipients. J Immunol. 2015;195(9):4069–4073. doi: 10.4049/jimmunol.1500940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sicard A, Phares TW, Yu H, et al. The spleen is the major source of antidonor antibody-secreting cells in murine heart allograft recipients. Am J Transplant. 2012;12(7):1708–1719. doi: 10.1111/j.1600-6143.2012.04009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Perry DK, Burns JM, Pollinger HS, et al. Proteasome inhibition causes apoptosis of normal human plasma cells preventing alloantibody production. Am J Transplant. 2009;9(1):201–209. doi: 10.1111/j.1600-6143.2008.02461.x. [DOI] [PubMed] [Google Scholar]
- 105.Richards DM, Zhang N, Dalheimer SL, Mueller DL. Allopeptide-specific CD4+ T cells facilitate the differentiation of directly alloreactive graft-infiltrating CD8+ T cells. Am J Transplant. 2007;7(10):2269–2278. doi: 10.1111/j.1600-6143.2007.01934.x. [DOI] [PubMed] [Google Scholar]
- 106.Richards DM, Dalheimer SL, Ehst BD, et al. Indirect minor histocompatibility antigen presentation by allograft recipient cells in the draining lymph node leads to the activation and clonal expansion of CD4+ T cells that cause obliterative airways disease. J Immunol. 2004;172(6):3469–79. doi: 10.4049/jimmunol.172.6.3469. [DOI] [PubMed] [Google Scholar]
- 107.Ali J, Bolton E, Saeb-Parsy K, Bradley JA, Pettigrew G. Targeting indirect pathway CD4 T-cell alloresponses in the prevention of chronic transplant rejection. Lancet. 2015;385(Suppl 1):S17. doi: 10.1016/S0140-6736(15)60332-4. [DOI] [PubMed] [Google Scholar]
- 108.Brennan TV, Hoang V, Garrod KR, et al. A new T-cell receptor transgenic model of the CD4+ direct pathway: level of priming determines acute versus chronic rejection. Transplantation. 2008;85(2):247–255. doi: 10.1097/TP.0b013e31815e883e. [DOI] [PubMed] [Google Scholar]
- 109.Zhang Z, Kaptanoglu L, Haddad W, et al. Donor T cell activation initiates small bowel allograft rejection through an IFN-gamma-inducible protein-10-dependent mechanism. J Immunol. 2002;168(7):3205–3212. doi: 10.4049/jimmunol.168.7.3205. [DOI] [PubMed] [Google Scholar]
- 110.Sayegh MH, Wu Z, Hancock WW, et al. Allograft rejection in a new allospecific CD4+ TCR transgenic mouse. Am J Transplant. 2003;3(4):381–389. doi: 10.1034/j.1600-6143.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 111.Schmaler M, Broggi MAS, Lagarde N, et al. IL-7R signaling in regulatory T cells maintains peripheral and allograft tolerance in mice. Proc Natl Acad Sci U S A. 2015;112(43):13330–13335. doi: 10.1073/pnas.1510045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cai YH, Alvarez A, Alcaide P, et al. Abrogation of functional selectin-ligand expression reduces migration of pathogenic CD8+ T cells into heart. J Immunol. 2006;176(11):6568–75. doi: 10.4049/jimmunol.176.11.6568. [DOI] [PubMed] [Google Scholar]
- 113.Valujskikh A, Lantz O, Celli S, Matzinger P, Heeger PS. Cross-primed CD8(+) T cells mediate graft rejection via a distinct effector pathway. Nat Protoc. 2002;3(9):844–851. doi: 10.1038/ni831. [DOI] [PubMed] [Google Scholar]
- 114.Diamond AS, Gill RG. An essential contribution by IFN-γ to CD8+ T cell-mediated rejection of pancreatic islet allografts. J Immunol. 2000;165(1):247–55. doi: 10.4049/jimmunol.165.1.247. [DOI] [PubMed] [Google Scholar]
- 115.Chau LA, Rohekar S, Wang JJ, et al. Thymic re-entry of mature activated T cells and increased negative selection in vascularized allograft recipients. Clin Exp Immunol. 2002;127(1):43–52. doi: 10.1046/j.1365-2249.2002.01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, Vignali DA. Generation of T-cell receptor retrogenic mice. Nat Protoc. 2006;1(1):406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]