Abstract
The aim of the study was to determine the phylogenetic groups of E. coli strains isolated from seemingly healthy broiler and broiler condemned suspected of colibacillosis in a Brazilian slaughterhouse. Samples from respiratory tract and edible giblets (liver and heart) of broilers with and without macroscopic lesions of colibacillosis were collected at slaughter. There were 84 strains isolated from broilers condemned of which 11 were obtained from swabs of the heart, 7 from the liver, and 66 from the respiratory tract. Of the 53 E. coli strains isolated from broilers not condemned, 5 were isolated from the heart, 4 from the liver, and 44 from the respiratory tract. E coli strains were tested via PCR for phylogenetic groups A, B1, B2, C, D, E, and F. Phylogroups A and B1 were the most common phylogroups of E. coli obtained from healthy and sick-appearing broiler carcasses. The results of the study showed that phylogroups B2 and E were associated with the heart samples and phylogroup A was associated with respiratory tract samples, phylogroup B1 with not condemned carcass, and phylogroup D with liver samples.
1. Introduction
Brazil is the second largest producer of chicken meat and the largest exporter of this product [1]. Moreover, the Brazilian poultry industry is expanding to other regions beside southern Brazil (the traditional hub of poultry production) to include the Central/Western region of the country [2]. Currently, one of the greatest challenges for Brazilian poultry producers is to reduce the economic losses caused by the condemnation of carcasses in slaughterhouses, including the air sac disease, due the E. coli infection [3].
E. coli is a member of the normal microbiota in poultry intestine but the colonization of the respiratory tract by pathogenic E. coli strains is associated with extraintestinal disease, resulting in morbidity and mortality of poultry by septicemia [4, 5].
The pathogenesis of colibacillosis is not completely understood, but some E. coli have many virulence properties associated with host tissue colonization, iron uptake systems, serum resistance, production of toxins, and defensins [5]. These strains were classified as Avian Pathogenic E. coli (APEC), a subpathotype of the Extraintestinal Pathogenic E. coli (ExPEC).
The molecular detection of virulence genes by PCR is the most common approach for the study of ExPEC. However, there is no consensus about the minimum predictive factors of APEC, since this subset group of strains is quite heterogenous [6–8]. In addition, while APEC acts as a primary pathogen, some commensal strains of E. coli, devoid of virulence factors, can also cause colisepticemia, acting as opportunistic secondary pathogens in stressed and immunocompromised birds, mainly in the presence of respiratory diseases as infectious bronchitis or mycoplasmosis [3, 5].
E. coli consists of commensal and pathogenic strains of great diversity, and the classification of these microorganisms in phylogenetic groups through the combination of gene clusters is important to understand E. coli pathogenesis and interaction with its hosts [9]. An understanding of the E. coli genomic structure showed that the strains belonging to the different phylogroups may be associated with the source of isolation [10]. Since its introduction in 2000 [11], phylogenetic typing using PCR has become widely used due to its simplicity and speed, allowing the differentiation of virulent strains (B2 or D) and commensal lineages (A and B1). This method was improved in 2013 [10] and a quadruplex PCR can identify seven phylogroups (A, B1, B2, C, D, E, and F).
Phylogenetic characterization is an important tool to improve the understanding of E. coli population and the relationship between strains and disease [12]. Due to the diversity and importance of E. coli infections in poultry as well as the economic impact of these infections, this study determined the phylogenetic groups of E. coli isolated from healthy-appearing broiler carcasses and carcasses condemned as suspected of colibacillosis at slaughter.
2. Materials and Methods
2.1. Sample Collection and Bacterial Examination
Samples from respiratory tract and edible giblets were collected from one slaughterhouse under Federal Sanitary Inspection localized in Tocantins State, Brazil, from May 2011 to April 2012. This facility slaughters broiler chickens from São Paulo, Tocantins, Goiás States, and the Federal District. Lesions were classified during slaughter by the Federal Inspectors of Brazilian Ministry of Agriculture, and slaughter followed the Brazilian Ministry of Agriculture guidelines [13].
Samples from the respiratory tract (trachea and air sacs) and the edible giblets liver and heart of broilers without macroscopic lesions (which were not condemned and passed the inspection) were collected after chilling process. Swabs from the trachea, air sacs, liver, and heart from carcasses condemned as suspected of colibacillosis were also collected during inspection of the carcass and the corresponding viscera at the evisceration line. The swabs of each organ were placed in a sterile test tube containing 0.9 mL of 0.85% saline and refrigerated until processing. The samples were appropriately packed in an isothermal box with ice packs and transported to the School of Veterinary and Animal Science at the Federal University of Tocantins for bacteriological culture of E. coli.
The swabs were processed individually according to [14] for isolation and identification of E. coli. Briefly, swabs were streaked onto MacConkey Agar and incubated at 37°C for 24 h. Then, after checking the growth of colonies on MacConkey Agar, up to three lactose positive and negative colonies were characterized biochemically according to [14] using agar triple sugar iron (TSI), citrate, indole, methyl red, and urea tests. Tubes were then incubated at 37°C for 24 h. Only one confirmed lactose positive strain was chosen from each carcass sample for further analysis. E. coli strains were maintained in Nutrient Agar at room temperature until molecular analysis.
E. coli strains were obtained from 59 batches in the abattoir. The distribution of batches per state is shown in Table 1. In each batch of animals to be slaughtered, 10 samples were collected, 5 from healthy-appearing broiler carcasses and 5 from carcasses condemned by colibacillosis at slaughter.
Table 1.
Distribution of batches of animals to be slaughtered according to origin of sample to be collected and the state of the batches.
| Samples | GO | TO | DF | SP | Total |
|---|---|---|---|---|---|
| Respiratory tract | 17 | 6 | 11 | 1 | 35 |
| Heart | 4 | 4 | 2 | 3 | 13 |
| Liver | 5 | 1 | 3 | 2 | 1 |
| Total | 26 | 11 | 16 | 6 | 59 |
2.2. DNA Extraction and Phylogenetic Group Determination
E. coli strains were sent to the Laboratory of Applied Bacteriology at the Veterinary School of The Federal University of Minas Gerais for phylogenetic group determination. Bacterial DNA was obtained by plating previously isolated colonies of E. coli onto MacConkey Agar. After plating, the samples were incubated for 18–24 h at 37°C. The bacterial DNA was then obtained by resuspending the subcultured cells in 1.5 mL microtubes containing 100 µL of TE (10 mM Tris-HCl; EDTA 1 mM, pH 8.0); DNA was extracted according to [15]. E. coli strains were assigned to one of the seven phylogenetic groups A, B1, B2, C, D, E, and F based on the presence and absence of the gene or DNA fragments chuA, yjaA, arpA, and trpA genes and TSPE4.C2 via PCR assay according to [10]. The typing scheme to classify E. coli in the phylogroups is well described in the article of [10] and was performed in the present study accordingly. The EDL933 (phylogroup E) and E2348/69 (phylogroup B2) were used as positive controls.
2.3. Statistical Analysis
The chi-squared test was used at 95% significance to estimate the differences among the proportion of phylogenetic groups identified in the organs of broilers and to assess the proportion of phylogenetic groups in colisepticemic and healthy carcasses.
Correspondence analysis (CA) [16] was used to study the phylogroups, origin of the samples (liver, heart, and respiratory tract), and the characteristic of the carcass/viscera at inspection (condemned or not condemned) using the Stata®/SE 12.0 [17]. In CA analysis, the relationship between the categories was represented in a two-dimensional graph with the value of the third dimension shown in parentheses. The degree of relation between the categories was demonstrated by evaluating which variables were plotted closely together. The CA examines the relationship between categorical and nominal data using a contingency table of the categorical variables and transforms nonmetric data to a metric data, allowing data to be mapped and visualized [18].
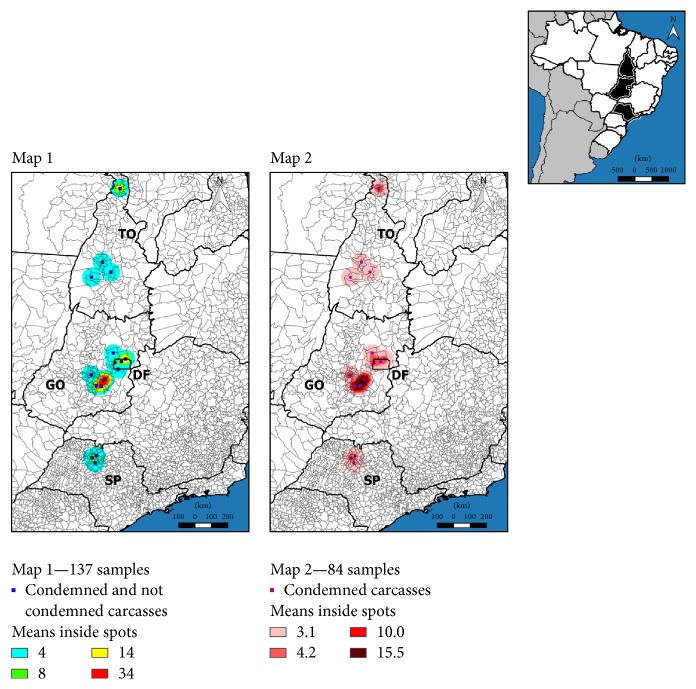
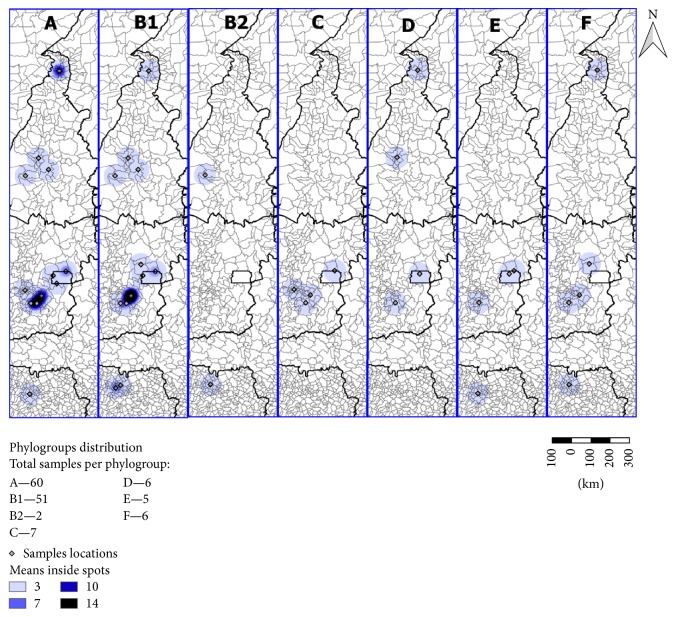
All spatial analytical procedures were performed with program Quantum GIS [19]. Two maps were created to get the georeference of the E. coli strains isolated from the samples collected (Figure 1). One was for all strains of E. coli isolated from the respiratory tract and the edible giblets (Figure 1/Map 1) and the other was for E. coli strains isolated from samples collected from condemned broilers (Figure 1/Map 2). Figure 2 was created to show the distribution of the phylogenetic groups of E. coli strains isolated from broilers at slaughter (condemned and not condemned) according to the origin of the batch of animals to be slaughtered. The coordinates of the samples were obtained by taking the centroid point of the municipality to which they belonged—this created a layer of points. These points were submitted to an adaptive radius with a quartic density kernel to illustrate the concentration of the samples as many of them belonged to the same location.
Figure 1.
Spatial kernel density estimator of the total E. coli strains isolated in the respiratory tract, liver, and heart of broilers at a slaughterhouse according to the city of origin of the batch of broilers (Map 1) and the kernel density estimator of the E. coli strains isolated from the respiratory tract, liver, and heart of broilers condemned suspect of colibacillosis according to the origin of the batch of broilers (Map 2).
Figure 2.
Spatial kernel density estimator of the distribution of the phylogenetic groups of E. coli strains isolated from the respiratory tract, liver, and heart of broilers at a slaughterhouse according to the city of origin of the batch of broilers to be slaughtered.
3. Results
In total, 59 batches of animals were analyzed, a total of 590 broiler carcasses. One hundred and thirty-seven strains of E. coli were isolated and used in the study. E. coli isolates were obtained from respiratory tract (trachea and air sacs) and edible giblets (liver and heart) of broiler carcasses. There were 84 strains isolated from broilers condemned suspected of colibacillosis of which 11 were obtained from swabs of the heart, 7 from the liver, and 66 from the respiratory tract. Of the 53 E. coli isolated from samples of inspection passed broilers, 5 were isolated from the heart, 4 from the liver, and 44 from the respiratory tract. Table 2 shows the phylogenetic group distribution of E. coli obtained from samples of condemned and not condemned broilers slaughtered in a Brazilian slaughterhouse. The phylogenetic groups of E. coli according to the origin of isolation in the carcasses are shown in Table 3. There was no association between phylogroups and the condition of the carcasses (P = 0.20), which means condemned or not condemned. Phylogroups B2 (P < 0.001) and E (P = 0.044) were associated with the heart samples.
Table 2.
Distribution of phylogroups of E. coli strains isolated from respiratory tract, liver, and heart of broilers condemned and not condemned during inspection.
| Condition of the carcasses | Phylogroup | |||||||
|---|---|---|---|---|---|---|---|---|
| A | B1 | B2 | C | D | E | F | Total | |
| Condemned | 30 | 35 | 1 | 6 | 3 | 4 | 5 | 84 |
| Not condemned | 30 | 16 | 1 | 1 | 3 | 1 | 1 | 53 |
| Total | 60 | 51 | 2 | 7 | 6 | 5 | 6 | 137 |
Table 3.
Distribution of phylogroups of E. coli strains isolated from the heart, liver, and respiratory tract of broiler during inspection.
| Organ of origin | Phylogroup | |||||||
|---|---|---|---|---|---|---|---|---|
| A | B1 | B2 | C | D | E | F | Total | |
| Heart | 5 | 4 | 2∗ | 1 | 1 | 2∗ | 1 | 16 |
| Liver | 3 | 7 | 0 | 0 | 1 | 0 | 0 | 11 |
| Respiratory tract | 52 | 40 | 0 | 6 | 4 | 3 | 5 | 110 |
| Total | 60 | 51 | 2 | 7 | 6 | 5 | 6 | 137 |
∗Identification of phylogroups B2 (P < 0,001) and E (P = 0,044) was associated with heart samples.
We used CA to analyze the degree of relationship between the categories phylogroups, origin of the sample (liver, heart, and respiratory tract), and characteristic of the carcass at the inspection (condemned or not condemned) (Figure 3). The two-dimensional representation explains 42.52% of the total variation with 15.59% explained by the 1st dimension, 13.86% by the 2nd dimension, and 13.07% by the 3rd. According to this analysis, phylogroup A was plotted near the respiratory tract samples, while phylogroup B1 was near the not condemned respiratory samples. Samples obtained from condemned broilers were plotted close to both phylogroups A and B1; the liver samples were close to phylogroup D. Although phylogroups B2 and E were plotted away from the other variables, both phylogroups were closer to the heart samples than the other variables.
Figure 3.
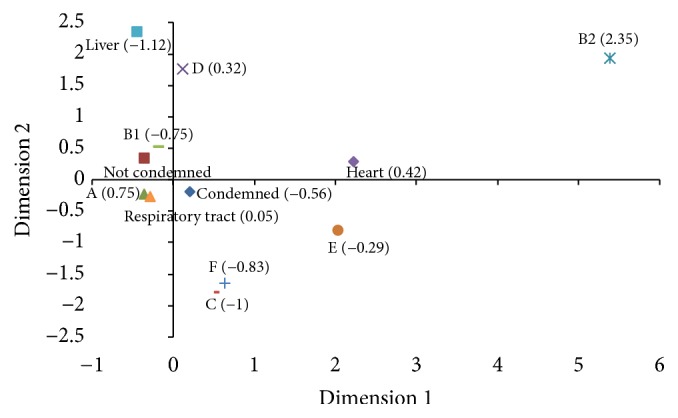
Correspondence analysis for the categories. This two-dimensional representation explains 42.52% of the total variation with 15.59% explained by 1st dimension, 13.86% by the 2nd dimension, and 13.07% by the 3rd. The value of the third dimension is shown in parenthesis.
Figure 1 shows the kernel function of the total E. coli strains isolated in the slaughterhouse according to the city of origin of the broiler chickens' batches to be slaughtered (Figure 1/Map 1) and the kernel function of the E. coli strains isolated from broiler chickens condemned suspect of colibacillosis according to the origin of the batch of the broilers (Figure 1/Map 2). The map showed that E. coli strains isolated from healthy and sick-appearing broiler carcasses originated mainly from batches of animals from the Goiás State and the Federal District. Figure 2 shows the distribution of the phylogenetic groups of E. coli strains isolated from the respiratory tract and the edible giblets of broiler chickens at slaughter according to the origin of the batch of broilers to be slaughtered and showed that phylogroups A and B1 were detected in São Paulo, Tocantins, Goiás States and the Federal District, but mostly in Goiás State; phylogroup B2 in São Paulo and Tocantins States; phylogroup C in Goiás State; phylogroup D in Tocantins and Goiás States and the Federal District; phylogroup E in São Paulo and Goiás States and the Federal District; phylogroup F São Paulo, Tocantins and Goiás States.
4. Discussion
A PCR technique to identify the seven phylogenetic groups was recently developed by Clermont et al. [10], and its use is rare in the literature [20–22]. The cited articles in the discussion section of the present study used the triplex PCR-based method, which identifies the phylogroups A, B1, B2, and D. Previously, our group showed the relation of the seven phylogroups according to the host (cattle calves, buffalo calves, and poultry) [20]. Therefore, to the best of our knowledge, this is the first report that analyzed the association of the seven phylogroups of E. coli, the origin of the samples (liver, heart, and respiratory tract), and the characteristic of the carcass/viscera at inspection (condemned or not condemned) using the quadruplex PCR-based method in Brazil.
Our study showed that phylogroups A followed by B1 are the most common phylogroups of E. coli obtained from not condemned broiler carcasses. This suggests that systemic contamination occurs via the respiratory tract because E. coli strains from the respiratory tract are primarily phylogroup A and to a lesser extent B1. The main form of infection with E. coli is via inhalation of feces-contaminated dust [23]. Koga et al. [24] suggest that poultry carcass contamination occurs by commensal E. coli. This observation is reinforced by the fact that E. coli isolated from the healthy group of our study was mainly phylogroup A. Moreover, the CA showed a relationship between phylogroup A strains and the respiratory tract—an organ considered to be the first step of systemic infection by E. coli [5]. In addition, commensal strains of E. coli may act as opportunistic agents in secondary infections in lots of birds affected by infectious bronchitis virus and other endemic respiratory pathogens in Brazil.
On the contrary, phylogroups B1 followed by A are the most common in strains obtained from condemned broilers. Similar results were found in Brazil [25, 26], Japan [27], Iran [28], and Australia [29]. A study in South Brazil showed that the percentage of phylogroup B1 isolated in refrigerated chicken carcasses increased over time while phylogroups B2 and D decreased [24]. Moreover, phylogroups A and B1 strains predominate in birds [20, 30], similar to our findings. However, phylogroups A and D were the most common phylogroups of E. coli isolated from poultry in Italy [31], China [32], Canada [33], and Iran [34], indicating that the frequency of phylogroups might vary between different geographic regions.
Overall, our results show that phylogroups B2, C, D, E, and F are not common E. coli phylogroups isolated from broilers. These results agree with other studies [20, 24, 31, 32]. On the contrary, phylogroup B2 was detected at high frequencies in other studies [25, 26, 35]. Phylogroup D was also detected at a low frequency (4.37%). In the CA, phylogroup D fell close to the liver samples in the CA map, while phylogroups B2 and E fell close to and associated with heart samples. In a Brazilian study, E. coli isolated from cellulitis lesions in broiler chickens were predominated by phylogroup D [36]. Phylogroups B2 and D strains are considered extraintestinal pathogenic E. coli [37] while phylogroup E includes human and animal intestinal strains and human EHEC O157:H7 strains [38]. In sum, our results suggest that phylogroups B2 and D are also extraintestinal pathogenic E. coli for broilers and indicate that phylogroup E may be associated with extraintestinal pathogenic E. coli for broilers, but further studies are needed to confirm this association, since the number of phylogroup E strains identified in the study was low.
The kernel function showed that the E. coli strains isolated originated mainly from batches from the Goiás State and the Federal District. The number of broilers slaughtered under Federal Inspection in the Middle-West region of Brazil has increased over the years, and the main cause of condemnation is colibacillosis and airsacculitis. In addition, E. coli strains were the main bacterial agent isolated in slaughterhouses with Federal Inspection in the State of Goiás [39]. Over 90% of Brazilian poultry production is based on integrated systems with good sanitary status, quality, and sustainability. The slaughterhouses are under the Federal Inspection Service to minimize risk. Colibacillosis in poultry is not well understood and is highly associated with the presence of risk factors such as stress, presence of other pathogens, and immunity status of birds [5].
Studies suggest that APEC is a reservoir of human ExPEC and should be considered a potential zoonotic risk [35, 40]. Our study did not evaluate the presence of virulence genes, but ExPEC phylogroups B2 and D were detected. Studies on the phylogenetic background of E. coli are important because there are data showing a link between phylogeny and virulence. Here, the background is important for the acquisition and expression of different virulence factors and epidemiology studies [41]. Over 95% of E. coli strains can be correctly assigned using the extended quadruplex method [10].
5. Conclusions
Our results indicate that all phylogroups are detected in E. coli isolated from healthy and sick-appearing broiler carcasses slaughtered in Brazil, but phylogroups A and B1 are the most common phylogroups. Phylogroup A was isolated mainly from the respiratory tract, while B1 was mainly detected in carcasses condemned. Phylogroups B2 and E were associated with E. coli isolated from heart samples, while phylogroup D was associated with liver samples.
Acknowledgments
This work was supported by the Conselho Nacional de Desenvolvimento Técnico e Científico (CNPq) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (Fapemig). Andrey P. Lage and Marcos B. Heinemann are indebted to CNPq for the fellowships received. Fernanda M. Coura thanks CNPq and Capes (13827/13-8) for her fellowship.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.ABPA. Annual Report. Brazilian Association of Animal Protein. 2016.
- 2.Prudêncio da Silva V., van der Werf H. M. G., Soares S. R., Corson M. S. Environmental impacts of French and Brazilian broiler chicken production scenarios: An LCA approach. Journal of Environmental Management. 2014;133:222–231. doi: 10.1016/j.jenvman.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Kabir S. M. L. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. International Journal of Environmental Research and Public Health. 2010;7(1):89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakazato G., de Campos T. A., Stehling E. G., Brocchi M., da Silveira W. D. Virulence factors of avian pathogenic Escherichia coli (APEC) Pesquisa Veterinaria Brasileira. 2009;29(7):479–486. doi: 10.1590/s0100-736x2009000700001. [DOI] [Google Scholar]
- 5.Guabiraba R., Schouler C. Avian colibacillosis: Still many black holes. FEMS Microbiology Letters. 2015;362(15) doi: 10.1093/femsle/fnv118.fnv118 [DOI] [PubMed] [Google Scholar]
- 6.Ewers C., Janßen T., Kießling S., Philipp H., Wieler L. H. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Veterinary Microbiology. 2004;104(1-2):91–101. doi: 10.1016/j.vetmic.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Johnson T. J., Wannemuehler Y., Doetkott C., Johnson S. J., Rosenberger S. C., Nolan L. K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. Journal of Clinical Microbiology. 2008;46(12):3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunha M. P. V., de Oliveira M. G. X., de Oliveira M. C. V. Virulence profiles, phylogenetic background, and antibiotic resistance of Escherichia coli isolated from Turkeys with Airsacculitis. The Scientific World Journal. 2014;2014:8. doi: 10.1155/2014/289024.289024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croxen M. A., Law R. J., Scholz R., Keeney K. M., Wlodarska M., Finlay B. B. Recent advances in understanding enteric pathogenic Escherichia coli. Clinical Microbiology Reviews. 2013;26(4):822–880. doi: 10.1128/cmr.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont O., Christenson J. K., Denamur E., Gordon D. M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environmental Microbiology Reports. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 11.Clermont O., Bonacorsi S., Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Applied and Environmental Microbiology. 2000;66(10):4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenaillon O., Skurnik D., Picard B., Denamur E. The population genetics of commensal Escherichia coli. Nature Reviews Microbiology. 2010;8(3):207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 13.Brasil. Ministério da Agricultura, Pecuária e do Abastecimento. Secretaria de Defesa Agropecuária. 1998.
- 14.Brenner D. J. Facultatively anaerobic gram-negative rods: Family I. Enterobacteriaceae Rahn. In: Krieg., R N., Holt., G J., Holt J. G., editors. Bergeys Manual of Systematic Bacteriology. Baltimore, Md, USA: Williams & Wilkins; 1984. pp. 408–420. [Google Scholar]
- 15.Pitcher D. G., Saunders N. A., Owen R. J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Letters in Applied Microbiology. 1989;8(4):151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 16.Greenacre M., Blasius J. Multiple correspondence analysis and related methods. Boca Raton, Fla, USA: Chapman & Hall/CRC; 2006. (Statistics in the Social and Behavioral Sciences Series). [DOI] [Google Scholar]
- 17.StataCorp. Stata Statistical Software: Release 12. College Station, Tex, USA, 2011.
- 18.Hair J. F., Black W. C., Babin B. J., Anderson R. E. Multivariate Data Analysis. Pearson Prentice Hall; 2010. [Google Scholar]
- 19.Sherman G. E., Sutton T., Blazek R., et al. Quantum GIS User Guide - Version 1.7. Wroclaw. 2011 [Google Scholar]
- 20.Coura F. M., Diniz S. D. A., Silva M. X., et al. Phylogenetic Group Determination of Escherichia coli Isolated from Animals Samples. The Scientific World Journal. 2015;2015:4. doi: 10.1155/2015/258424.258424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iranpour D., Hassanpour M., Ansari H., Tajbakhsh S., Khamisipour G., Najafi A. Phylogenetic groups of escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. BioMed Research International. 2015;2015:12. doi: 10.1155/2015/846219.846219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Um M. M., Barraud O., Kérourédan M., et al. Comparison of the incidence of pathogenic and antibiotic-resistant Escherichia coli strains in adult cattle and veal calf slaughterhouse effluents highlighted different risks for public health. Water Research. 2016;88:30–38. doi: 10.1016/j.watres.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Dho-Moulin M., Fairbrother J. M. Avian pathogenic Escherichia coli (APEC) Veterinary Research. 1999;30(2-3):299–316. [PubMed] [Google Scholar]
- 24.Koga V. L., Rodrigues G. R., Scandorieiro S., et al. Evaluation of the antibiotic resistance and virulence of Escherichia coli strains isolated from chicken carcasses in 2007 and 2013 from Paraná, Brazil. Foodborne Pathogens and Disease. 2015;12(6):479–485. doi: 10.1089/fpd.2014.1888. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi R. K. T., Aquino I., Ferreira A. L. D. S., Vidotto M. C. EcoR phylogenetic analysis and virulence genotyping of avian pathogenic Escherichia coli strains and Escherichia coli isolates from commercial chicken carcasses in southern Brazil. Foodborne Pathogens and Disease. 2011;8(5):631–634. doi: 10.1089/fpd.2010.0726. [DOI] [PubMed] [Google Scholar]
- 26.Maluta R. P., Logue C. M., Casas M. R. T., et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0105016.e105016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiki M., Usui M., Akiyama T., et al. Phylogenetic grouping, epidemiological typing, analysis of virulence genes, and antimicrobial susceptibility of Escherichia coli isolated from healthy broilers in Japan. Irish Veterinary Journal. 2014;67(1, article 14) doi: 10.1186/2046-0481-67-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagheri M., Ghanbarpour R., Alizade H. Shiga toxin and beta-lactamases genes in Escherichia coli phylotypes isolated from carcasses of broiler chickens slaughtered in Iran. International Journal of Food Microbiology. 2014;177:16–20. doi: 10.1016/j.ijfoodmicro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Obeng A. S., Rickard H., Ndi O., Sexton M., Barton M. Antibiotic resistance, phylogenetic grouping and virulence potential of Escherichia coli isolated from the faeces of intensively farmed and free range poultry. Veterinary Microbiology. 2012;154(3-4):305–315. doi: 10.1016/j.vetmic.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Gordon D. M., Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. 2003;149(12):3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]
- 31.Pasquali F., Lucchi A., Braggio S., et al. Genetic diversity of Escherichia coli isolates of animal and environmental origins from an integrated poultry production chain. Veterinary Microbiology. 2015;178(3-4):230–237. doi: 10.1016/j.vetmic.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Wang X.-M., Liao X.-P., Zhang W.-J., et al. Prevalence of serogroups, virulence genotypes, antimicrobial resistance, and phylogenetic background of avian pathogenic escherichia coli in south of China. Foodborne Pathogens and Disease. 2010;7(9):1099–1106. doi: 10.1089/fpd.2010.0542. [DOI] [PubMed] [Google Scholar]
- 33.Aslam M., Toufeer M., Narvaez Bravo C., et al. Characterization of Extraintestinal Pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. International Journal of Food Microbiology. 2014;177:49–56. doi: 10.1016/j.ijfoodmicro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Ghanbarpour R., Sami M., Salehi M., Ouromiei M. Phylogenetic background and virulence genes of Escherichia coli isolates from colisepticemic and healthy broiler chickens in Iran. Tropical Animal Health and Production. 2011;43(1):153–157. doi: 10.1007/s11250-010-9667-2. [DOI] [PubMed] [Google Scholar]
- 35.Ewers C., Li G., Wilking H., et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? International Journal of Medical Microbiology. 2007;297(3):163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Barbieri N. L., de Oliveira A. L., Tejkowski T. M., et al. Genotypes and pathogenicity of cellulitis isolates reveal traits that modulate APEC virulence. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0072322.e72322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhuri R. R., Henderson I. R. The evolution of the Escherichia coli phylogeny. Infection, Genetics and Evolution. 2012;12(2):214–226. doi: 10.1016/j.meegid.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Clermont O., Olier M., Hoede C., et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infection, Genetics and Evolution. 2011;11(3):654–662. doi: 10.1016/j.meegid.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Minharro S., Linhares G. F. C., Andrade M. A., Rocha P. T., Santana P., Santana Â. P. Envolvimento de Escherichia coli, de Mycoplasma gallisepticume de Mycoplasma synoviae em lesões de sacos aéreos em frangos abatidos no estado de Goiás. Ciência Animal Brasileira. 2001;2:111–117. [Google Scholar]
- 40.Bauchart P., Germon P., Brée A., Oswald E., Hacker J., Dobrindt U. Pathogenomic comparison of human extraintestinal and avian pathogenic Escherichia coli—search for factors involved in host specificity or zoonotic potential. Microbial Pathogenesis. 2010;49(3):105–115. doi: 10.1016/j.micpath.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Escobar-Páramo P., Clermont O., Blanc-Potard A.-B., Bui H., Le Bouguénec C., Denamur E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Molecular Biology and Evolution. 2004;21(6):1085–1094. doi: 10.1093/molbev/msh118. [DOI] [PubMed] [Google Scholar]