Abstract
Most soybean cultivars possess broad leaflets; however, a recessive allele on the Ln locus is known to cause the alteration of broad to narrow leaflets. The recessive allele ln has also been considered to increase the number of seeds per pod (NSP) and has the potential to improve yield. Recently, Gm-JAG1 (Glyma20g25000), a gene controlling Ln, has been shown to complement leaf shape and silique length in Arabidopsis mutants. However, whether Gm-JAG1 is responsible for those traits in soybean is not yet known. In this study, we investigated the pleiotropic effect of soybean Ln gene on leaflet shape and NSP by using two independent soybean Gm-jag1 mutants and four ln near isogenic lines (NILs). The leaflet shape was evaluated using a leaf image analysis software, SmartLeaf, which was customized from SmartGrain. The leaflets of both the Gm-jag1 mutants were longer and narrower than those of the wild-type plants. Interestingly, the image analysis results clarified that the perimeter of the mutant leaflets did not change, although their leaflet area decreased. Furthermore, one mutant line with narrow leaflets showed significantly higher NSP than that in the wild (or Ln) genotype, indicating that soybean Ln gene pleiotropically controls leaflet shape and NSP.
Keywords: Ln gene, Glycine max (L.) Merrill, pleiotropic effect, leaflet shape, seed number per pod
Introduction
The genetic factor controlling broad and narrow shape of soybean leaflets was first reported by Takahashi and Fukuyama (1919). In heterozygous plants, the broad leaflet trait is inherited in an incompletely dominant manner, leading to the development of an intermediate leaf shape in heterozygous plants (Takahashi and Fukuyama 1919, Takahashi 1934). The gene responsible for this trait was named Ln and ln for broad and narrow leaflet shapes, respectively, on the Ln locus (Bernard and Weiss 1973). Interestingly, Takahashi (1934) first reported that the narrow leaflet trait (ln) is closely associated with the increase in the number of seeds per pod (NSP). Weiss (1970) suggested that it has a pleiotropic effect on increasing the NSP, which is also obviously influenced by the vigor and genetic background of plants. In this case, the genotype at Ln is important to control not only leaflet shape but also NSP, one of the seed yield components, in soybean.
Recently, the Ln locus was positioned into a soybean linkage map (Jeong et al. 2011) and Gm-JAG1, Glyma20g25000 in the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Gmax; Schmutz et al. 2010), was reported to be a responsible gene on the Ln locus (Jeong et al. 2012). The JAGGED (JAG) gene encodes a zinc finger protein family of plant transcription factors in Arabidopsis thaliana. The jag mutant line, which possesses a nonsense mutation on the JAG gene, leads to the development of jagged shape along the lateral margins of leaves and the narrowing of petals owing to the defects in both cell division and cell expansion (Ohno et al. 2004). Although narrow petals are known to contain elongated and round cells, the relationship between leaf and cell shapes is not yet completely known. Furthermore, Jeong et al. (2012) did not investigate the effect of Gm-JAG1 on leaf shape and NSP in soybean. Thus, whether Gm-JAG1 pleiotropically controls leaf shape and NSP as Ln in soybean is not yet known.
The leaflet size and shape are known to change with developing stages and environments; therefore, reliably evaluating these traits is difficult. Sawada (1992) reported that the central leaflet of the fifth main leaf from the base on the main stem has a distinctive ratio of length to width among soybean cultivars. Automatic image analysis allows the sequential measurement of many samples and could be used to measure the length and width of leaflets as well as to describe the features of leaflet shape, such as leaf area and perimeter. SmartGrain software, which allows easy manipulations, has been developed for high-throughput analysis of seed shape (Tanabata et al. 2012). SmartGrain might enable the analysis of leaflet shape, although this software is not suitable for the analysis of entire leaves.
Induced mutant libraries are a powerful tool for conducting genetic and functional analyses in soybean (Anai 2012, Cooper et al. 2008, Xia et al. 2012). The Gm-jag1 mutant lines are attractive materials to determine the pleiotropic effect of ln. In this study, we evaluated the pleiotropic effects of Ln on leaflet shape and NSP by using the Gm-jag1 mutant lines and an updated image analysis software of SmartGrain for leaf shape analysis. The leaflets of the two independent mutant lines were longer and narrower than those of the wild-type plants. Furthermore, one mutant line with narrow leaflets showed significantly increased NSP compared to that of the wild-type plants. To our knowledge, this is the first study to indicate that soybean Ln gene pleiotropically controls leaflet shape and NSP.
Materials and Methods
Characterization of ‘Enrei’ mutants for Gm-JAG1 and development of backcrossed lines
Tsuda et al. (2015) constructed a mutant library (M2′ generation) of broad leaflet cultivar ‘Enrei’ (EN) with a high mutation density by repeating chemical mutagen (ethyl methanesulfonate) treatment and obtained a total of eleven mutant types consisting of one null and ten missense mutations (named ‘EnT lines’) for Gm-JAG1 (Glyma20g25000), although their phenotypes were not described. No M3′ seeds were obtained for two missense mutant lines. Fewer than nine M3′ plants per line were grown in the greenhouse controlled at 25°C under natural light condition with supplemental lighting of 79 W/m2 by using metal-halide lamps (M360FCELSP-W/BUD; Iwasaki Electric, Tokyo, Japan) from 5:00 to 19:00. The leaflet shape and genotype of Gm-JAG1 of respective plants were evaluated per the methods hereinafter described. The individuals showing narrow leaflets and mutant genotype were used for backcrossing to EN for evaluating the detailed leaflet shape and the relationship between genotypes and phenotypes to eliminate other mutation sites and to develop segregating progenies at the Ln locus.
For assessing the genotype of the mutants, DNA was extracted from a young leaf by using an automated purification system (BioSprint 96 DNA Plant Kit; Qiagen, Hilden, Germany), and the genotype for each mutation site was determined using high resolution melting (HRM) analysis according to Tsuda et al. (2015). The primer pairs for the HRM analysis are shown in Table 1.
Table 1.
Primers used for genotyping the nine EnT mutant lines by using high-resolution melting analysis
| Target mutant lines | Forward primer | Reverse primer |
|---|---|---|
| EnT-0112, EnT-0160, EnT-0541 EnT-0621, EnT-0685, EnT-0987 |
CTACACCTTCACACCCTTCTTTTCT | CAAGGAATATACCACAGGGATGG |
| EnT-0634, EnT-1084 | GGTAATCGAGAATGGGGAGTT | CTTGTGAGAAGGAGTGGTAAAAGAA |
| EnT-1619 | CATGACACTGTTTGATACTGCGA | CTCTCATCTCTTTAGAAACCATCT |
NIL pairs carrying natural mutant and wild alleles at the Ln locus
In order to confirm that Gm-JAG1 was the gene responsible for leaflet shape and had pleiotropic effects on NSP, we used several NIL lines, along with the abovementioned mutant lines. A narrow leaflet phenotype derived from ‘Tachinagaha’ (ln cultivar) was introduced to ‘Sachiyutaka’ (Norin No. 116; SC) and ‘Fukuyutaka’ (Norin No. 73; FK) to develop two NILs, ‘Zenkei 101’ (SC_ln) and ‘Zenkei 102’ (FK_ln) by backcrossing six times based on the leaflet shape in the Western Region Agricultural Research Center. These cultivars and NILs were registered and conserved in NARO Genebank. FK (JP29668) and EN (JP28862) in addition to ‘Tachinagaha’ (JP67666) is now available, and the others will be available soon. In addition, ‘Clark’ (CL) and ‘Harosoy’ (HR) and their NILs with the ln allele, CL_ln and HR_ln were registered as PI 548533, PI 548573, PI 547413, and PI 547690, respectively, in the Germplasm Resources Information Network (GRIN), USA. According to the pedigree of CL_ln and HR_ln, a narrow leaflet phenotype derived from ‘T204’ (ln cultivar) was introduced to CL and HR by backcrossing five times (Bernard et al. 1991).
DNA sequencing of the Glyma20g25000 region
The DNA sequence of the Glyma20g25000 region for each mutant and near isogenic line has not been previously identified. Each sequence was determined by the following procedure.
The DNA was extracted from 10 mg seed powder by using an automated purification system (BioSprint 96 DNA Plant Kit). Next, the sequence of the Glyma20g25000 region was amplified using the following primer pair: forward, 5′-AGCTTTAGTTTTATCCCTACCCACAC-3′ and reverse, 5′-CCACATAACATAACAGAAACATACCA-3′; the sequence was then determined using the primer-walking method by using BigDye Terminator v3.1 Cycle Sequencing Kit and 3500xl Genetic Analyzer (Thermo Fisher Scientific, Massachusetts, USA).
Planting design
Seeds were sown on June 26, 2015 in a field in Tsukuba (Institute of Crop Science, NARO; 36°01′N, 140°06′E), the soil type of which was Andosol, and on July 14, 2015 in a field in Tsukubamirai (Institute of Crop Science, NARO; 36°00′N, 140°01′E), which has gray lowland soil. Chemical fertilizer (N:P:K) was applied at the rate of 20:280:60 (kg/ha) and 30:100:100 (kg/ha) in Tsukuba and Tsukubamirai, respectively. Each line or cultivar was planted in single-row plots following a randomized block design with 3–4 replications. Each plot consisted of a row of 4–7 plants that were spaced 0.2 m apart, and each row was 0.8 or 0.7 m apart (Table 2).
Table 2.
Plant materials and planting design
| Plant materials | Phenotype of leaflet shape (narrow leaflet origin) | Genotype | Cultivar or Generation | Planting design | |
|---|---|---|---|---|---|
|
| |||||
| in Tsukuba | in Tsukubamirai | ||||
|
|
|
||||
| EN | Broad | Ln/Ln | Enrei | 7 plants, 4 reps. | 4 plants, 3 reps. |
|
|
|
||||
| 685w | Broad | Ln/Ln | BC2F3 | 7 plants, 4 reps. | 4 plants, 3 reps. |
| 685h | Intermediate | Ln/L10F | BC2F3 | 7 plants, 4 reps. | 4 plants, 3 reps. |
| 685m | Narrow (EnT-0685) | L10F/L10F | BC2F3 | 7 plants, 4 reps. | 4 plants, 3 reps. |
|
|
|
||||
| 541w | Broad | Ln/Ln | M4′ | (Not planted) | 4 plants, 3 reps. |
| 541h | Intermediate | Ln/M1null | M4′ | (Not planted) | 4 plants, 3 reps. |
| 541m | Narrow (EnT-0541) | M1null/M1null | M4′ | (Not planted) | 4 plants, 3 reps. |
|
|
|
||||
| SC | Broad | Ln/Ln | Sachiyutaka | 7 plants, 4 reps. | (Not planted) |
| SC_ln | Narrow (Tachinagaha) | ln/ln | BC6 | 7 plants, 4 reps. | (Not planted) |
|
|
|
||||
| FK | Broad | Ln/Ln | Fukuyutaka | 7 plants, 4 reps. | (Not planted) |
| FK_ln | Narrow (Tachinagaha) | ln/ln | BC6 | 7 plants, 4 reps. | (Not planted) |
|
|
|
||||
| CL | Broad | Ln/Ln | Clark | 4 plants, 3 reps. | (Not planted) |
| CL_ln | Narrow (T204) | ln/ln | BC5 | 4 plants, 3 reps. | (Not planted) |
|
|
|
||||
| HR | Broad | Ln/Ln | Harosoy | 4 plants, 3 reps. | (Not planted) |
| HR_ln | Narrow (T204) | ln/ln | BC5 | 4 plants, 3 reps. | (Not planted) |
Reps. in planting design means replications; EnT lines are derived from M1′ plants (the generation after two mutagen treatments), and their genotypes indicate deduced amino acid mutations in the Gm-JAG1 (Glyma20g25000) gene; EnT-0685 (the tenth leucine changed to phenylalanine: L10F) and EnT-0541 (the start codon was altered: M1null).
Evaluation of leaflet shape-related traits by using image analysis
A fully expanded central leaflet of the fifth leaf from the base on the main stem was collected according to Sawada (1992). The image of the leaflet was captured using a scanner GT-X970 (EPSON, Tokyo, Japan) and saved as a jpeg image file. The leaflet image was analyzed using SmartLeaf software (http://phenotyping.image.coocan.jp/smartleaf/), which was an updated image analysis software of SmartGrain (Tanabata et al. 2012) adapted for leaf shape analysis. SmartLeaf can automatically measure the length, width, perimeter, and area of a leaf and calculate the length/width ratio (L/W ratio), centroid position, circularity, and L*a*b* colorimetric values.
Evaluation of NSP and other seed yield components
Single seed weight (SSW), seed weight per plant (SWP), number of pods per plant (NP), and NSP were measured for individual plants. First, pods were separated from the stem. They were classified based on the number of seed cavity per pod by visual inspection based on the method described by Jeong et al. (2011), and then the pod number was recorded for each plant. After the pods were threshed, the seed number was counted. Sufficiently air-dried seed samples were weighed, and the water content of the seeds was immediately measured using a grain moisture tester (PM-830-2; Kett, Tokyo, Japan). SSW was determined by weighing all seeds except pest-damaged ones from each plant. SWP was calculated as the potential ability of plants to produce seeds by multiplying SSW and the total number of seed cavities per plant. The SSW and SWP were adjusted to reflect a moisture content of 15%. NSP was calculated by dividing the total number of seed cavities per plant by NP.
Statistical analysis
Statistical analysis was performed using the statistical package R version 3.1.2 (http://www.r-project.org). The statistical significance was evaluated using analysis of variance followed by Student’s t-test for comparisons between two groups or by using the Tukey–Kramer HSD test for comparisons among more than two groups. Significance was determined using an alpha level of 5%.
Results
Identification of ln mutants by using the reverse genetic approach
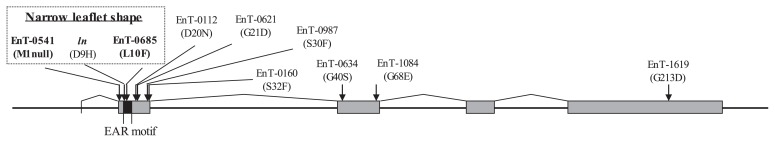
Of the nine M3′ mutant lines with one null (altered initiation codon) and eight missense (amino acid substitution) mutations for Gm-JAG1 ( Glyma20g25000; Fig. 1), only two mutant lines, ‘EnT-0541’ (the start codon was altered: M1null) and ‘EnT-0685’ (tenth leucine changed to phenylalanine: L10F), exhibited narrow leaflet characteristics. Whether the narrow leaflet phenotypes of these mutants were controlled by Gm-JAG1 was confirmed by genetically analyzing the relationship between the mutations and leaflet shape in the segregating progenies of ‘EnT-0541’ (M4′) and ‘EnT-0685’ (BC2F3; Fig. 2). The two sets, including the wild, heterozygous, and mutant genotypes derived from ‘EnT-0541’ and ‘EnT-0685’, were named as follows: 541w, 541h, 541m, 685w, 685h, and 685m, respectively (Table 2). According to genomic sequences in the Glyma20g25000 regions of 541m and 685m (respective DDBJ accession numbers were LC203763 and LC203764), no mutations were confirmed except for the respective base substitutions detected by HRM analysis (Supplemental Fig. 1). The mutation of 685m (L10F) existed in the ethylene response factor-associated amphiphilic repression (EAR) motif (Ohta et al. 2001) like the mutation of ln cultivars (ninth aspartic acid changed to histidine: D9H) reported by Jeong et al. (2012). Conversely, the genomic sequences for the four ln NILs, SC_ln, FK_ln, CL_ln, and HR_ln, showing narrow leaflet phenotype (Table 2), contained the same base substitution (D9H) in the Glyma20g25000 region (Supplemental Fig. 1). The five Ln cultivars, EN, SC, FK, CL, and HR, yielded sequences identical to ‘Williams 82’ in the Phytozome. All the cultivars showed broad leaflet phenotype.
Fig. 1.
Schematic diagram of mutations of Gm-JAG1 (Glyma20g25000) in the high-density mutant library of ‘Enrei’. Glyma20g25000 consists of 2,436 bp (black line), including 771 bp coding sequences connected by 5 exons. The first exon consists of a sequence of 2 bases, and the remaining exons are included in 4 boxes. The mutant line ‘EnT-0685’ and a natural variation of ln (ninth aspartic acid changed to histidine: D9H) have a respective unique mutation on the ERF-associated amphiphilic repression (EAR) motif (8aa-LDLNNLP-14aa; black box). Only the progenies of ‘EnT-0541’ and ‘EnT-0685’ with the mutant genotype showed narrow leaflet shape like the ln genotype. The detailed information of 1 null and 8 missense mutant lines and screening method has been described in Tsuda et al. (2015).
Fig. 2.
Representative leaflet shapes of EN (a), 541m (b), and 685m (c) planted in Tsukubamirai. These plant materials are shown in Table 2.
High-throughput analysis of leaflet shape by using the enhanced SmartGrain software
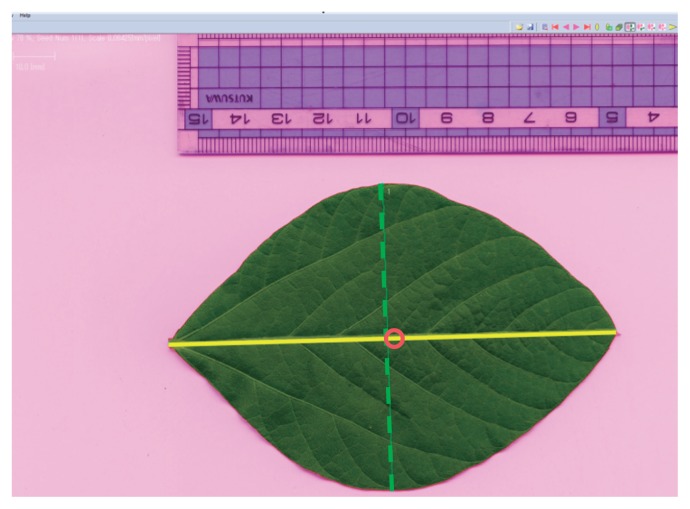
The leaflet shape was resolved into length, width, L/W ratio, perimeter, and leaf area by using SmartLeaf, an updated image analysis software of SmartGrain (Table 3) adaptable for leaf shape analysis. Unlike manual measurement, SmartLeaf automatically recognizes the leaflet area only from images and successively reports various parameters characterizing the leaflet (Fig. 3). This automation reduced the measuring time, and personal equation could be eliminated at any time if leaf images were obtained.
Table 3.
Traits related to leaflet shape
| a) Experiment conducted in Tsukuba | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Plant materials | Genotype | Length (L) [cm] | Width (W) [cm] | L/W ratio | Perimeter [cm] | Area [cm2] |
| EN | Ln/Ln | 9.53 | 6.09 | 1.57 | 25.16 | 39.82 |
|
| ||||||
| 685w | Ln/Ln | 8.74c | 5.46a | 1.61c | 22.80 | 32.22a |
| 685h | Ln/L10F | 9.60b | 4.81b | 2.00b | 23.96 | 31.67a |
| 685m | L10F/L10F | 10.25a | 3.65c | 2.82a | 24.00 | 26.12b |
|
| ||||||
| SC | Ln/Ln | 9.26b | 6.32a | 1.47b | 25.21b | 38.94a |
| SC_ln | ln/ln | 11.41a | 4.55b | 2.51a | 27.32a | 34.19b |
|
| ||||||
| FK | Ln/Ln | 9.10b | 6.54a | 1.39b | 24.89b | 39.26a |
| FK_ln | ln/ln | 11.28a | 4.53b | 2.50a | 27.12a | 33.85b |
|
| ||||||
| CL | Ln/Ln | 8.87b | 5.65a | 1.56b | 23.22 | 32.82a |
| CL_ln | ln/ln | 10.44a | 3.57b | 2.93a | 24.83 | 25.18b |
|
| ||||||
| HR | Ln/Ln | 8.52b | 4.93a | 1.73b | 21.86 | 27.69a |
| HR_ln | ln/ln | 9.82a | 3.55b | 2.78a | 23.23 | 23.23b |
| b) Experiment conducted in Tsukubamirai | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Plant materials | Genotype | Length (L) [cm] | Width (W) [cm] | L/W ratio | Perimeter [cm] | Area [cm2] |
| EN | Ln/Ln | 12.31 | 8.75 | 1.41 | 33.88 | 75.09 |
|
| ||||||
| 685w | Ln/Ln | 10.77 | 7.83a | 1.38c | 30.23 | 59.74 |
| 685h | Ln/L10F | 11.37 | 6.81b | 1.68b | 30.51 | 56.38 |
| 685m | L10F/L10F | 11.74 | 6.11b | 1.93a | 29.98 | 48.80 |
|
| ||||||
| 541w | Ln/Ln | 11.61b | 7.79a | 1.50c | 31.19 | 62.54a |
| 541h | Ln/M1null | 12.20b | 7.33a | 1.67b | 31.73 | 62.36a |
| 541m | M1null/M1null | 13.38a | 5.53b | 2.43a | 32.34 | 49.37b |
Different letters indicate significant difference at the 5% level between respective NILs.
Fig. 3.
Picture of an ‘Enrei’ leaflet analyzed using the SmartLeaf software. SmartLeaf accurately recognized the leaf and background indicated by pink region (light grey region in black and white; green region recognized in the SmartLeaf software). It also measured the length indicated by yellow line (solid line in black and white); width indicated by green line (broken line in black and white); their ratio; the centroid indicated by circle with a hole in the middle; circularity; perimeter; size; and the L*a*b* colorimetric values, which was the additional function to SmartLeaf.
The 685m and 541m leaflets were longer, narrower, and had larger L/W ratio and smaller area than those of the wild-type and EN leaflets (Table 3). These values for the two heterozygous mutant lines, 685h and 541h, were intermediate between the narrow and broad leaflet types. The L/W ratio characterizing the leaflet shape stably reflected the genotypes independently of growth conditions (Table 3a, 3b). Similarly, four ln NILs had similar leaf shape parameter values as those of the mutants. The two mutant lines, 685m and 541m, and two NILs, CL_ln and HR_ln, did not show significant differences in leaf perimeters from those of the wild (or Ln) and heterozygous genotypes, whereas SC and FK ln NILs had slightly longer leaf perimeter than that of the Ln genotypes.
Effects on NSP and other seed yield components
The mutant line 685m showed 10%–20% greater NSP than that of 685w and 685h, but had 7%–9% smaller SSW under both the growing conditions. No significant difference in NP and SWP was found among the three genotypes (Table 4a, 4b). The NSP, SSW, NP, and SWP of 541m was not significantly different from those of 541w and 541h (Table 4b). All the four NILs showed significantly higher NSP values in the ln genotype than in Ln. Like 685m, CL_ln and HR_ln showed smaller SSW than that of the respective recurrent parents. In contrast, SC_ln and FK_ln showed almost the same SSW as that of the recurrent parents. With regard to NP and SWP, the differences between Ln and ln varied depending on the genetic background, that is, CL_ln and HR_ln had significantly lower NP and SWP than those of CL and HR, whereas SC_ln and FK_ln had higher values than those of SC and FK.
Table 4.
Traits related to seed yield components
| a) Experiment conducted in Tsukuba | |||||
|---|---|---|---|---|---|
|
| |||||
| Plant materials | Genotype | NSP | SSW [mg] | NP | SWP [g] |
| EN | Ln/Ln | 2.12 | 342 | 69.7 | 50.5 |
|
| |||||
| 685w | Ln/Ln | 2.20b | 305a | 70.5 | 47.1 |
| 685h | Ln/L10F | 2.24b | 295a | 69.5 | 46.0 |
| 685m | L10F/L10F | 2.43a | 284b | 65.3 | 45.1 |
|
| |||||
| SC | Ln/Ln | 2.16b | 357 | 87.6b | 67.9b |
| SC_ln | ln/ln | 2.32a | 361 | 96.0a | 80.8a |
|
| |||||
| FK | Ln/Ln | 2.07b | 342a | 96.7 | 68.9b |
| FK_ln | ln/ln | 2.41a | 333b | 99.7 | 80.2a |
|
| |||||
| CL | Ln/Ln | 2.84b | 208a | 111.7a | 65.9a |
| CL_ln | ln/ln | 3.22a | 185b | 81.9b | 49.4b |
|
| |||||
| HR | Ln/Ln | 2.76b | 179a | 104.2a | 51.3a |
| HR_ln | ln/ln | 2.98a | 160b | 91.4b | 43.6b |
| b) Experiment conducted in Tsukubamirai | |||||
|---|---|---|---|---|---|
|
| |||||
| Plant materials | Genotype | NSP | SSW [mg] | NP | SWP [g] |
| EN | Ln/Ln | 2.25 | 292 | 46.7 | 30.6 |
|
| |||||
| 685w | Ln/Ln | 2.23b | 291 | 37.7 | 24.2 |
| 685h | Ln/L10F | 2.22b | 263 | 33.8 | 20.5 |
| 685m | L10F/L10F | 2.68a | 266 | 29.1 | 20.7 |
|
| |||||
| 541w | Ln/Ln | 2.23 | 250 | 26.2 | 14.4 |
| 541h | Ln/M1null | 2.11 | 230 | 24.4 | 11.5 |
| 541m | M1null/M1null | 2.19 | 240 | 22.6 | 12.0 |
Different letters indicate significant difference at the 5% level between respective NILs.
Discussion
Assessing genetically pleiotropic effects of a single locus is difficult, because the locus cannot be separated from the effects of closely linked genes. Mutation is an attractive choice to verify the pleiotropic gene function. The present study first showed that two lines (‘EnT-0541’ and ‘EnT-0685’) of the mutant lines screened from the high-density soybean mutant library (Tsuda et al. 2015) having independent missense and null mutations exhibited narrow leaflet phenotype. Therefore, this mutant library was confirmed to be useful to determine gene functions not only for seed components (Yano et al. 2017) but also for plant morphology.
The SmartLeaf software revealed the leaflet characteristics of four pairs of ln NILs (Table 3); the leaflet of ln NILs was the longer and narrower, and thus the L/W ratio was notably higher than that in the original Ln varieties. This suggests the importance of the L/W ratio in determining the narrow leaflet shape as previously reported (Chen and Nelson 2004, Dinkins et al. 2002, Jeong et al. 2011, Sawada 1992). The image analysis software revealed identical features of the two Gm-jag1 mutant lines, which showed higher L/W ratio than that of the original cultivar ‘Enrei’ and sib lines having a wild Gm-JAG1 allele. Interestingly, in this study, image analysis indicated no apparent changes in the perimeter of mutant leaflets despite the decrease in leaf area. The leaf morphology is controlled genetically in either the length or width (Tsukaya 2005). In this case, the perimeter consequentially changes. Although Gm-JAG1 coincidentally controls the length and width in the soybean leaflet, no apparent changes were noted in the perimeter. This observation might contribute to the elucidation of the function of JAGGED-like gene in cell division and/or cell expansion. SmartLeaf can allow the use of new evaluation methods for leaf morphology, which is known to vary across environments, and leaf positions, because of its high-throughput image processing.
Jeong et al. (2012) introduced the soybean Gm-JAG1 gene into Arabidopsis jag mutant line and showed that soybean Gm-JAG1 complemented the leaf shape and silique length in the Arabidopsis mutant. However, the function of Gm-JAG1 in soybean was not determined. In this study, we found that Gm-JAG1 is responsible for the function of Ln in soybean based on the narrow leaflet shape of the two different jag soybean mutant lines, ‘EnT-0541’ (M1null) and ‘EnT-0685’ (L10F; Fig. 2). Considering that the other seven mutants did not form narrow leaflets (Fig. 1), mutations on the EAR motif (Ohta et al. 2001) in Gm-JAG1 (Supplemental Fig. 2) were thought to affect the leaflet shape. Deduced amino acid substitutions occurred in the conserved EAR motif in both the 685m mutant line (L10F) and soybean ln NILs (D9H; Jeong et al. 2012). In addition, a mutation in the 541m mutant line (M1null) possibly resulted in the loss-of-function allele of Gm-JAG1. These mutations likely weaken the transcriptional property of Gm-JAG1 and change cell shape and/or proliferation rate, thereby leading to the formation of narrow leaflets in soybean. The jag mutant lines of Arabidopsis and tomato have irregular cell size and develop malformed leaves (David-Schwartz et al. 2009, Ohno et al. 2004); further, a mutant in rice showed abnormal flowers, especially stamens (Xiao et al. 2009). However, the narrow leaflet shape is retained in the elite cultivars of soybean. The reason for this difference can be attributed to the existence of a paralog in soybean, Gm-JAG2 (Gm10g42020 in Phytozome), which, like Gm-JAG1, is also expressed in young leaves, meristems, flowers, and young pods (Jeong et al. 2012). Determining the cause of such mild phenotype in soybean mutants requires the elucidation of the function of Gm-JAG2.
The NSP of the mutant line 685m with narrow leaflets significantly increased like that in the four ln NILs that were grown under the same conditions, indicating that soybean Ln gene pleiotropically controls NSP as well as leaflet shape (Table 4). This is consistent with the findings of Takahashi (1934) who showed that narrow leaflet phenotype is closely associated with NSP. The pleiotropy is conceivable in part because the leaf shape and silique length were also affected in the Arabidopsis jag mutant (Jeong et al. 2012). In contrast, no significant differences were found in the NSP, SSW, NP, and SWP among 541w, 541h, and 541m. Since the parental (M2′) line (‘EnT-0541’) was estimated to contain more than 15,000 base changes in the genome (Tsuda et al. 2015; no data for ‘EnT-0685’), such a high density of mutations in these lines might affect other traits such as SWP that was almost the half of that in EN (Table 4b). The gynoecium form (Ohno et al. 2004) and fruit length (Jeong et al. 2012) in Arabidopsis are known to be controlled by the JAG gene. However, the molecular genetic basis of the relationship between leaf and pod/fruit development, which are derived from flowers and their associated tissues, and EAR-motif of the JAG gene are poorly understood. The findings that a close relationship exists between the EAR motif and leaflet shape or NSP might allow the elucidation of the genetic network controlled by the JAG gene and the improvement of seed productivity in soybean and other crops.
Mutations in the EAR motif (Supplemental Fig. 2) certainly increased the NSP, whereas the SSW of the mutant line 685m and the two NILs, CL_ln and HR_ln, was significantly smaller than that of 685w or the recurrent parents of the NILs (Table 4a, 4b). Generally, a negative correlation exists between the total number of seeds and seed size per plant in various plant species. Our results might suggest that competition of resource allocation among seeds occurs inside a pod. Mandl and Buss (1981) proposed that the narrow leaflet trait offers neither yield advantage nor disadvantage compared with the broad leaflet trait. Similarly, in this study, 685m did not show considerable changes in SWP, because the increase in NSP led to a concomitant decrease in SSW. Therefore, we suggest that ln alone could not improve the seed yield. However, the SWP of SC_ln and FK_ ln was higher than that of their recurrent parents, suggesting that ln might improve seed yield under some particular genetic background(s) in soybean. Further studies are warranted to identify the genetic factors associated with these phenotypes and determine the synergistic effect of ln on NSP in such a genetic background.
Supplementary Information
Acknowledgments
We thank Yumiko Zaibu, Tokuko Kawaguchi, and Atsuko Narushima for technical assistance. This study was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement: NGB-3001, SFC-1001, and IVG-3005).
Literature Cited
- Anai, T. (2012) Potential of a mutant-based reverse genetic approach for functional genomics and molecular breeding in soybean. Breed. Sci. 61: 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, R.L. and Weiss, M.G. (1973) Qualitative genetics. In: Caldwell, B.E. (ed.) “Soybeans: improvement, production and uses,” American Society of Agronomy, USA, pp. 117–154. [Google Scholar]
- Bernard, R.L., Nelson, R.L. and Cremeens, C.R. (1991) USDA soybean genetic collection: Isoline collection. Soybean Genet. Newsl. 18: 27–57. [Google Scholar]
- Chen, Y. and Nelson, R.L. (2004) Evaluation and classification of leaflet shape and size in wild soybean. Crop Sci. 44: 671–677. [Google Scholar]
- Cooper, J.L., Till, B.J., Laport, R.G., Darlow, M.C., Kleffner, J.M., Jamai, A., El-Mellouki, T., Liu, S., Ritchie, R., Nielsen, N.et al. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol. 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Schwartz, R., Koenig, D. and Sinha, N.R. (2009) LYRATE is a key regulator of leaflet initiation and lamina outgrowth in tomato. Plant Cell 21: 3093–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins, R.D., Keim, K.R., Farno, L. and Edwards, L.H. (2002) Expression of the narrow leaflet gene for yield and agronomic traits in soybean. J. Hered. 93: 346–351. [DOI] [PubMed] [Google Scholar]
- Jeong, N., Moon, J.K., Kim, H.S., Kim, C.G. and Jeong, S.C. (2011) Fine genetic mapping of the genomic region controlling leaflet shape and number of seeds per pod in the soybean. Theor. Appl. Genet. 122: 865–874. [DOI] [PubMed] [Google Scholar]
- Jeong, N., Suh, S.J., Kim, M.H., Lee, S., Moon, J.K., Kim, H.S. and Jeong, S.C. (2012) Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell 24: 4807–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl, F.A. and Buss, G.R. (1981) Comparison of narrow and broad leaflet isolines of soybean. Crop Sci. 21: 25–27. [Google Scholar]
- Ohno, C.K., Reddy, G.V., Heisler, M.G. and Meyerowitz, E.M. (2004) The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H. and Ohme-Takagi, M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, S. (1992) Time of determination and variations within and between plants in leaf shape of soybean. Jpn. J. Crop Sci. 61: 96–100. [Google Scholar]
- Schmutz, J., Cannon, S.B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., Hyten, D.L., Song, Q., Thelen, J.J., Cheng, J.et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Takahashi, N. (1934) Linkage relation between the genes for the form of leaves and the number of seeds per pod of soybeans. Jpn. J. Genet. 9: 208–225. [Google Scholar]
- Takahashi, Y. and Fukuyama, J. (1919) Morphological and genetic studies on soybean. Hokkaido Agr. Exp. Stn. Rep. 10: 1–100. [Google Scholar]
- Tanabata, T., Shibaya, T., Hori, K., Ebana, K. and Yano, M. (2012) SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, M., Kaga, A., Anai, T., Shimizu, T., Sayama, T., Takagi, K., Machita, K., Watanabe, S., Nishimura, M., Yamada, N.et al. (2015) Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genomics 16: 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2005) Leaf shape: genetic controls and environmental factors. Int. J. Dev. Biol. 49: 547–555. [DOI] [PubMed] [Google Scholar]
- Weiss, M.G. (1970) Genetic linkage in soybeans. Linkage group IV. Crop Sci. 10: 368–370. [Google Scholar]
- Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., Anai, T., Sato, S., Yamazaki, T., Lü, S.et al. (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 109: 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H., Tang, J., Li, Y., Wang, W., Li, X., Jin, L., Xie, R., Luo, H., Zhao, X., Meng, Z.et al. (2009) STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J. 59: 789–801. [DOI] [PubMed] [Google Scholar]
- Yano, R., Takagi, K., Takada, Y., Mukaiyama, K., Tsukamoto, C., Sayama, T., Kaga, A., Anai, T., Sawai, S., Ohyama, K.et al. (2017) Metabolic switching of astringent and beneficial triterpenoid saponins in soybean is achieved by a loss-of-function mutation in cytochrome P450 72A69. Plant J. 89: 527–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.