Abstract
The ribosome is a complex macromolecular machine responsible for protein synthesis in the cell. It consists of two subunits, each of which contains both RNA and protein components. Ribosome assembly is subject to intricate regulatory control and is aided by a multitude of assembly factors in vivo, but can also be carried out in vitro. The details of the assembly process remain unknown even in the face of atomic structures of the entire ribosome and after more than three decades of research. Some of the earliest research on ribosome assembly produced the Nomura assembly map of the small subunit, revealing a hierarchy of protein binding dependencies for the 20 proteins involved and suggesting the possibility of a single intermediate. Recent work using a combination of RNA footprinting and pulse-chase quantitative mass spectrometry paints a picture of small subunit assembly as a dynamic and varied landscape, with sequential and hierarchical RNA folding and protein binding events finally converging on complete subunits. Proteins generally lock tightly into place in a 5′ to 3′ direction along the ribosomal RNA, stabilizing transient RNA conformations, while RNA folding and the early stages of protein binding are initiated from multiple locations along the length of the RNA.
Keywords: 30S subunit, ribosome assembly, RNA folding, protein binding, cooperativity
INTRODUCTION
The ribosome is an intricate molecular machine that is responsible for the synthesis of new proteins in the cell. The bacterial 70S ribosome consists of two parts, a small (30S) subunit composed of a single strand of RNA and 21 proteins, and a large (50S) subunit composed of two strands of RNA and 34 proteins. Assembling these components into intact subunits is a carefully controlled and choreographed process that is both demanding on the cell and efficient by necessity. The high-resolution atomic structures of both the 30S (63) and 50S (6) subunits were solved in 2000, followed by atomic resolution structures of the 70S ribosome (40, 49, 50), offering a detailed look at the end product of ribosome assembly. While the structures are useful for interpreting the extensive biochemical data concerning ribosome assembly, little information about the assembly process itself can be gained from their inspection. Ribosome assembly has been the subject of scrutiny for several decades by a variety of methods including RNA chemical footprinting, equilibrium binding assays, mass spectrometry, and simulation, primarily in vitro. Although the story of ribosome assembly is far from complete, great strides have been made in advancing the knowledge of how these individual protein and RNA molecules come together to form complete and functional ribosomes.
RIBOSOME BIOGENESIS IN BACTERIA
Synthesis and assembly of an intact ribosome in bacterial cells occurs in a series of steps, including rRNA transcription, ribosomal protein synthesis, RNA processing, RNA folding, protein binding, and protein and RNA modification. The nature and regulation of many of these steps have been extensively reviewed elsewhere and are presented only briefly here with appropriate references. Ribosomal RNA (rRNA) is transcribed as a single transcript including both the 16S rRNA for the small subunit and the 23S and 5S rRNAs for the large subunit in what is the rate-limiting step for ribosome synthesis in vivo (39). The transcription of the rRNA operon is tightly coupled to growth rate and subject to regulation by multiple factors (27, 39). At the same time, ribosomal proteins are synthesized, some of which regulate not only their own translation but that of proteins sharing the same operon (64). Both ribosomal proteins (5, 27, 41) and rRNA (11, 27, 62) are chemically modified, and although not all such modifications appear to be essential, several modifications of the 23S rRNA in particular are necessary to avoid assembly defects (27). The single rRNA primary transcript is processed into mature 5S, 16S, and 23S rRNAs by a series of RNAses (27, 52). Finally, the rRNAs and proteins fold and assemble in a hierarchical manner into complete subunits, in a process governed by a set of over 20 assembly factors (27, 62). These essential steps are not strictly sequential, and rRNA folding, protein binding, and rRNA processing occur both in parallel and cotranscriptionally (12, 30).
RECONSTITUTION OF RIBOSOMAL SUBUNITS
With the array of assembly factors present in vivo it is incredible that an in vitro reconstitution of intact ribosomal subunits using only the rRNA and proteins is even possible. Nevertheless, in vitro reconstitution of 30S subunits occurs with both reasonable speed and efficiency in the total absence of assembly factors, albeit significantly more slowly than in vivo. Reconstitution of the larger 50S subunits is comparatively slow and inefficient (17, 33) and as a result has not been studied as extensively. This review primarily focuses on the extensive body of work on 30S ribosome assembly in vitro.
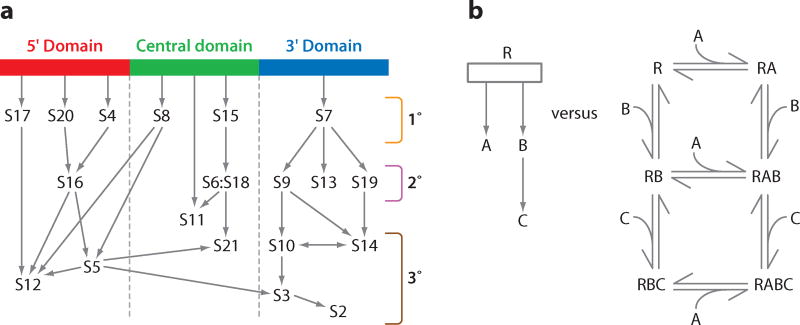
In the 1970s, seminal work by Nomura and colleagues on 30S subunit assembly resulted in a complete assembly map for the 30S subunit (21, 31). Reconstitution of 30S subunits using different subsets of the total complement of proteins allowed the hierarchy of binding dependencies to be determined, as represented in what is now known as the Nomura assembly map. Protein S1 binds relatively weakly to the 30S subunit and exchanges rapidly, so it is typically excluded (61). The 30S assembly map shown in Figure 1a is modified from the original map by Nomura to reflect the domain structure of the 30S subunit and to indicate the updated location of S13 in the S7 assembly branch, recently determined by RNA footprinting (18). From the assembly map the 30S ribosomal proteins can be classified according to their physical location and order in the binding hierarchy. Proteins bind principally to one of the 5′, central, or 3′ domains of the rRNA and are designated as primary (binding directly to the rRNA), secondary (binding is dependent on primary binding proteins), or tertiary, (binding is dependent on secondary binding proteins). To this day, the Nomura assembly map offers one of the most useful overviews of 30S subunit assembly and is widely reproduced. Herold & Nierhaus developed a similar assembly map for the 50S subunit (22), but assembly of the large ribosomal subunit is not so clearly organized by structural domains and has many more proteins that interact in a more complex binding hierarchy.
Figure 1.
The Nomura assembly map for the 30S subunit. (a) A modified version of the traditional view of the Nomura assembly map that has been reorganized according to domains, with arrows indicating the facilitating effect of binding between proteins. Proteins can be categorized as 5′, central, or 3′ domain proteins, and either primary binding proteins (1°), secondary binding proteins (2°), or tertiary binding proteins (3°), the last two of which depend upon proteins from the previous category for binding to the 16S rRNA. S11 is categorized in a species-dependent manner, acting as a tertiary binding protein in Escherichia coli, but as a primary binding protein in Aquifex aeolicus (45). (b) A comparison between a hypothetical assembly map (left) and an equivalent assembly mechanism (right) of a quaternary RABC complex between RNA (R) and three proteins (A, B, and C). Each reaction step needs to be characterized by thermodynamic equilibrium constants and both forward and reverse rate constants.
While the Nomura map has constituted the overview of the 30S subunit assembly process for over 30 years, there are a number of shortcomings with this representation. First, the Nomura map is not a mechanism, but rather a diagram containing logical information about dependencies during assembly. An example of a true mechanism compared with an equivalent assembly map is shown in Figure 1b. The assembly map does not contain information about the specific complexes that are present or the equilibria present. Second, all of the information in the Nomura map is thermodynamic, and it offers no information about the kinetics of protein binding or the specific energy changes that occur. Third there is no information about the folding pathway of the 16S rRNA that constitutes two-thirds of the mass of the 30S subunit. Subsequent work has attempted to shed light on these and other questions using a variety of methods to probe 30S assembly in vitro. The balance of this review focuses on these experiments.
A NEW REPRESENTATION FOR 16S rRNA
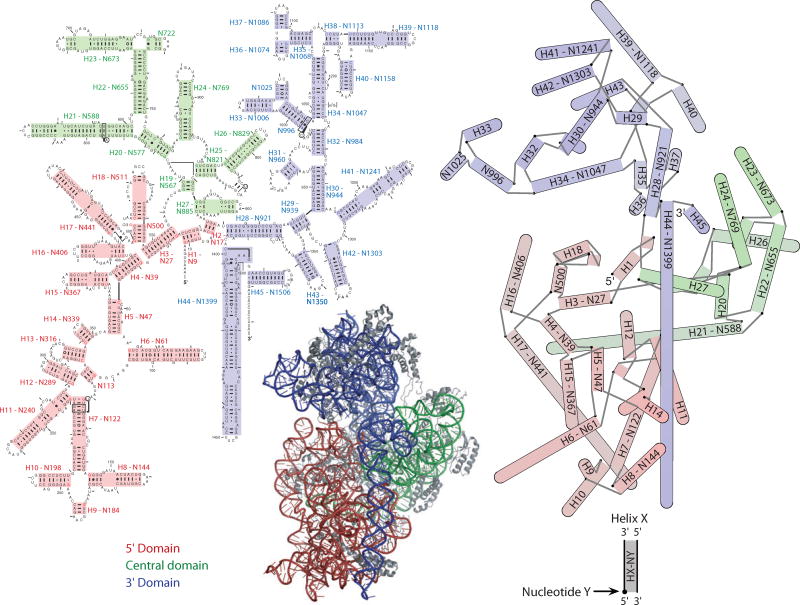
The 16S rRNA is usually represented as the phylogenetically derived secondary structure (10), or as a schematic ribbon and stick representation of the 3D structure, but neither is ideal. The former lacks information about the relative positions of the helices in space and the latter does not convey detailed nucleotide-level information well when rendered in 2D. The secondary structure can be annotated with interaction networks that show base-base connections through space (29), but the result is unwieldy for RNAs as large as the 16S. A hybrid representation has been developed, which displays the RNA helices in 2D but arranged according to their positions in the 3D structure. Axes for 51 helices in the 16S rRNA were defined from their 3D coordinates, all-versus-all distances were calculated between the ends of these axes, and the program NEATO (16) was used to generate an energy-minimized 2D projection. The resulting schematic is similar to the canonical view used to display the 30S subunit in 3D. A comparison of a phylogenetic secondary structure, the hybrid representation, and a cartoon rendering of the 30S subunit is provided in Figure 2. Each illustrates the domain structure of the 30S subunit. The hybrid representation contains less detailed information than the secondary structure but manages to capture the overall structure and shape of the 30S subunit in a much simpler way than the 3D image. Depth information is preserved by shading the helices in accordance with their depth in the 3D structure, with darker helices in back and lighter helices in front. Templates for this representation both with and without helix numbering are available as supplementary material (Supplemental Figure 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org).
Figure 2.
A comparison of different representations of the 30S subunit. (Left) A traditional secondary structure diagram of the 16S rRNA. (Bottom, center) A 3D image of the 30S subunit including proteins. (Right) A 2D projection of the 16S rRNA helices. Interhelical distances are calculated from the coordinates of the helix axes in the 3D structure, and these distances are optimized for projection into 2D by the program NEATO (16), such that their layout is faithful to the 3D structure. Helices are shown as cylinders, capped by a semicircle when the two helical strands are contiguous. A black dot indicates the first nucleotide of the helix, and gray lines indicate connecting strands between helices. Helices are shaded in the hybrid representation according to their position along the axis normal to the page. Darker colors are farther away, while lighter colors are closer. These positions correspond to the actual positions in the 3D structure and are also used to order the rendering of helices in the cases in which overlap occurs. Proteins in the 3D image are shown in gray. Both the secondary structure and the hybrid representation have helices numbered according to Brimacombe numbers as well as their first nucleotide. The secondary structure is the simplest representation but fails to capture any information about the actual shape of the 30S subunit, and annotation with the tertiary RNA contacts results in a congested diagram. The hybrid representation blends the simplicity of the secondary structure with 3D information and captures the overall shape of the subunit. The secondary structure is based on one at the Comparative RNA Web Site and Project (10), and the 3D image was generated with PyMol (13).
BINDING OF PRIMARY PROTEINS
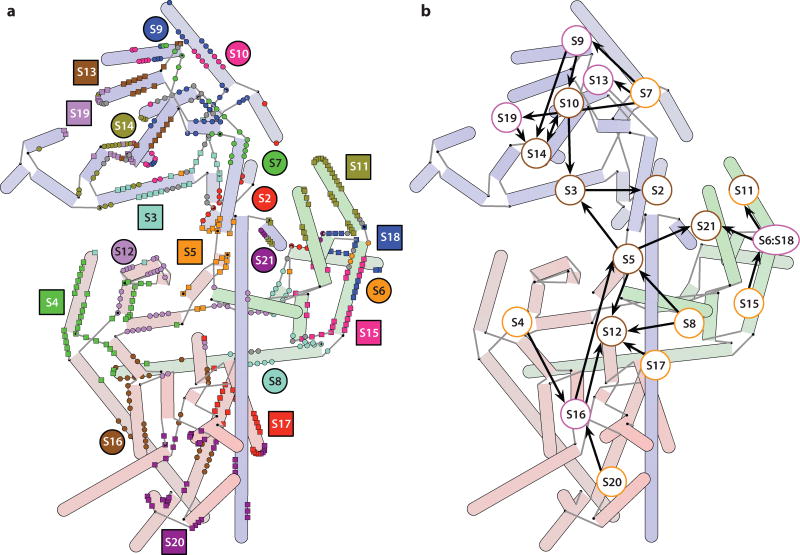
Six of the ribosomal proteins of the 30S subunit (S4, S7, S8, S15, S17, and S20) are primary binding proteins, capable of binding directly to the 16S rRNA. The RNA contacts made by these and the rest of the ribosomal proteins are shown in Figure 3a, overlaid on a hybrid schematic representation of the 16S rRNA. A new representation of the assembly map, where the proteins are overlaid on a hybrid schematic representation of the 16S rRNA according to their approximate positions in the 30S subunit, is shown in Figure 3b.
Figure 3.
Ribosomal protein-RNA contacts. (a) These contacts are mapped onto a hybrid 2D representation of the 16S rRNA. A contact is annotated on a residue basis whenever any non-hydrogen atoms from a nucleotide and an amino acid residue are within 4 Å of each other. The cases in which a single nucleotide contacts multiple proteins are indicated in gray. Protein labels are placed near the primary sites of contact. (b) The Nomura map overlaid on a hybrid schematic representation of the 16S rRNA. Labels for the proteins are located according to their approximate position in the 3D structure of the 30S subunit. The primary binding proteins appear to bind in the periphery of the 30S subunit, and the tertiary proteins cluster around the cleft containing the decoding site.
The low-temperature binary complex of the 5′ domain protein S4 and the 16S rRNA undergoes a temperature-dependent conformational change upon heating, as shown by chemical footprinting of the RNA (43). After the conformational change additional nucleotides are protected by S4, indicating a possible strengthening of the association by broadening the scope of the protein-RNA contacts. These additional contacts are implicated in the binding of tertiary protein S12 and secondary protein S16 to the RNA, both of which depend on S4 according to the Nomura map (Figures 1a and 3b). A binary complex of S4 and the 16S rRNA in which the two components have been heated separately does not display the same pattern of protection, suggesting that the protein confers the conformational change directly to the RNA, allowing subsequent proteins to bind and ensuring the hierarchy of assembly. Subsequent studies of the remaining five primary binding proteins indicated a range of behavior (15). The other two 5′ domain primary proteins S17 and S20 bound to the 16S rRNA in a temperature-independent manner, while the central domain primary proteins S8 and S15 conferred only small differences to the chemical footprint of the binary complex upon heating. The 3′ domain primary protein S7 behaved much in the same manner as S4, with significant changes to the chemical footprint of the binary complex with RNA upon heating. Taken together, these results suggest that one of the roles of the primary proteins is to promote certain conformations of the RNA and allow the binding of subsequent proteins.
Site-directed hydroxyl radical footprinting studies on the environment surrounding S15 indicated that although S8 may not mediate secondary protein binding, it does influence the organization of the RNA surrounding S15 (25), and that even proteins located a significant distance away from S15 were able to affect its environment (26). Similar studies on the environment of S20 revealed that its contacts with the 5′ domain RNA were formed early in the assembly process, whereas its contacts with the 3′ minor domain (helix H44–N1399) were not formed until later in the process, consistent with a 5′ to 3′ directionality for assembly.
INDEPENDENTLY FOLDING DOMAINS
In addition to reconstitutions of the entire 30S subunit, individual domains can be assembled independently using domain fragments of 16S rRNA. Three different studies have demonstrated the assembly of the 5′ domain rRNA with proteins S4, S16, S17, and S20 (60); the central domain rRNA of Thermus thermophilus with proteins S6, S8, S11, S15, and S18 (4); and the 3′ domain rRNA with proteins S2, S3, S7, S9, S10, S13, S14, and S19 (48). In each case, the assembled domains were similar in conformation to their counterparts in intact 30S subunits. However, the tertiary binding proteins in the 5′ and central domain reconstitutions predicted by the Nomura map (Figure 1) did not bind stably to the isolated domains, indicating dependencies on contacts to RNA or proteins bound in other domains. The dependence of the 3′ protein S3 on the 5′ protein S5 in the Nomura map is not absolute, because S3 is incorporated into the 3′ domain reconstitution. The 5′ domain rRNA is capable of folding into a native-like conformation in the presence of magnesium ions, but in the absence of ribosomal proteins (2). Tertiary contacts virtually identical to those found in intact 30S subunits were observed, suggesting that RNA folding forms the basis for protein binding during 5′ domain assembly.
CENTRAL DOMAIN ASSEMBLY
Perhaps the best understood region of the 30S subunit, in terms of assembly kinetics and thermodynamics, is the central domain. In the Nomura map proteins S15 and S8 are the primary binding proteins, and although S8 influences the RNA structure of the central domain (25), it is protein S15 that initiates a cascade of central domain protein binding by S6, S18, S11, and S21. An extensive series of biophysical studies has been carried out using fragments of the central domain RNA and individually purified proteins to better understand the molecular basis for the observed hierarchy.
The minimal binding site for the protein S15 was localized to a three-helix junction region in the central domain, helices H20, H21, and H22 (N577, N588, and N655 based on their starting nucleotide). The minimal site contains all the nucleotides implicated in S15 binding as determined by chemical probes and hydroxyl radical footprinting (44, 53). S15 binds to the minimal three-helix junction with a Kd value of ~35 nM using components from Bacillus stearothermophilus (7, 8). Binding of S15 induces a conformational change in the three-helix junction, suggested by an accelerated mobility of the RNA-protein complex in a nondenaturing polyacrylamide gel (7). Quantitative hydrodynamic experiments using transient electric birefringence demonstrated coaxial stacking of helices H21 and H22, while helices H20 and H22 formed an acute angle upon S15 binding (34). This conformational change implied that one of the functions of S15 binding was to organize a critical region in the central domain to initiate assembly.
A series of deletion RNA constructs for the central domain was prepared to identify the minimal binding region for the remaining central domain proteins. Surprisingly, almost half of the central domain can be deleted while still retaining binding of proteins S15, S6, S18, and S11. A minimal fragment composed of two adjacent three-helix junctions binds to S15, S6, and S18. The X-ray crystal structure of the quaternary complex was solved, revealing a structure essentially identical to the corresponding region in the complete 30S subunit (3, 63).
In the structure of the S15:S6:S18 RNA complex, a number of important conclusions can be drawn that provide a molecular picture of specific features of the Nomura assembly map. Proteins S6 and S18 form an intimate heterodimer, which explains their completely cooperative association to 16S rRNA. There are no protein-protein interactions observed between S15 and either of the downstream binding proteins, indicating that the thermodynamic effect on binding observed in the Nomura map must be mediated by stabilization of the RNA tertiary structure by binding to protein S15. A surprising interhelix base pair was revealed in the structure between nucleotides A665 and G724. This tertiary base pair is apparently unstable or not formed in the absence of S15 binding, and it is required for stable binding of the S6:S18 heterodimer.
From these studies, a generalized model for ribosome assembly can be proposed. The RNA tertiary structure in 16S rRNA is not stable in the absence of supporting protein contacts. Transient RNA folding can create the binding site for a primary binding protein, and the local RNA tertiary structure is stabilized by protein binding, consolidating the RNA folding gains. Binding of the primary binding protein sets up the next RNA conformational change, creating the binding site for a secondary binding protein. Thus, the assembly proceeds by an alternating series of RNA conformational changes and protein binding events, sequentially stabilizing the final RNA tertiary structure.
CHEMICAL PROBING OF RIBOSOMAL INTERMEDIATES
When 30S particles are reconstituted at low temperatures (~0°C), a 21S reconstitution intermediate (RI) is obtained, rather than complete 30S subunits (56). RI lacks the tertiary binding proteins in the central and 5′ domains (S2, S3, S10, S14, and S21) and is not competent to form a complete small subunit. Incubation of RI at high temperatures (~40°C) produces a second intermediate, RI*, that is characterized by a different sedimentation coefficient (26S). Conversion of RI to RI* is believed to occur by a large conformational rearrangement of the rRNA, because of the observed difference in sedimentation between the two particles, which have the same rRNA and protein composition. Once RI* has been formed, it is competent to bind the remaining five ribosomal proteins to make complete 30S subunits. From these intermediates a simplified assembly framework has been suggested that is composed of three steps: (a) 16S rRNA and 15 ribosomal proteins form RI, (b) conformational change where RI becomes RI*, and (c) RI* and five ribosomal proteins form the 30S subunit.
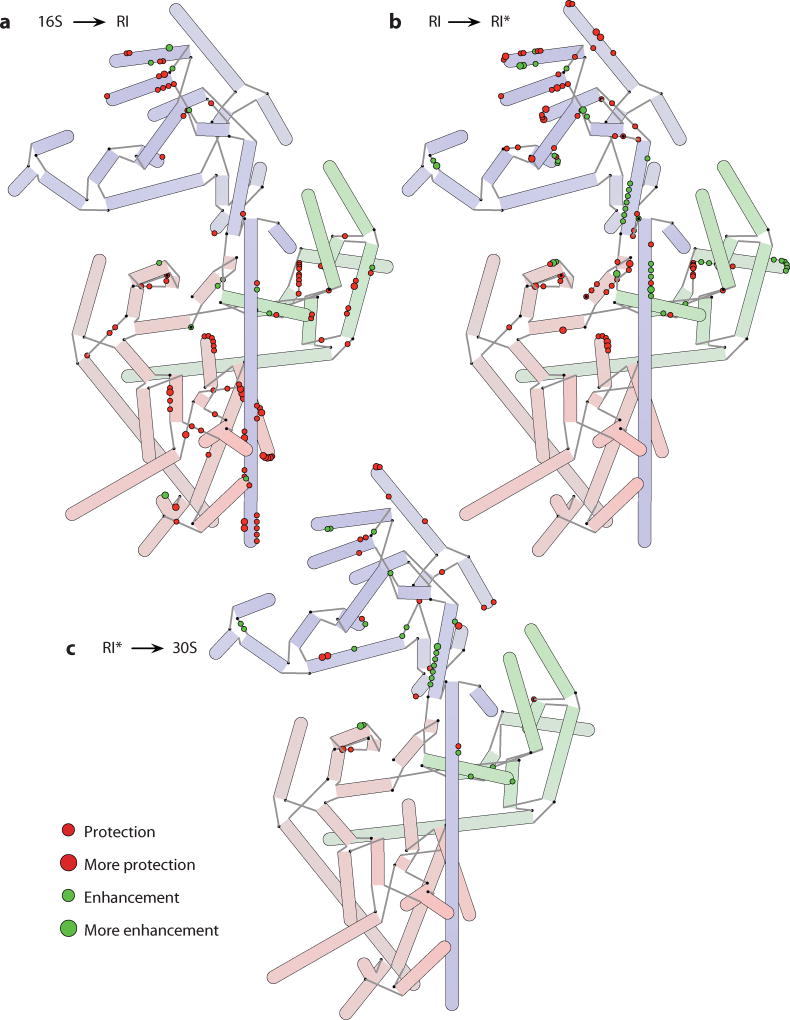
RNA footprinting can be used to quantify the accessibility of nucleotides to hydroxyl radical cleavage or chemical modification. An early series of studies described the results of both untargeted chemical footprinting (32) and site-directed hydroxyl radical footprinting from modified ribosomal proteins (19, 20, 44). More recent work has focused on assembly intermediates, including two studies that explore the changes that occur in 16S rRNA over the course of the transitions to RI, RI*, and 30S (23, 24). Comparing the accessibility to chemical modification of individual nucleotides between consecutive species, the changes to the 16S that occur during an assembly step are revealed in the differences. The data for each of the three transitions are summarized in Figure 4. Nucleotides exhibit either a protection from or an enhancement to chemical modification, resulting from either RNA folding or unfolding or protein binding. The primary differences observed for the 16S to RI transition are protections for nucleotides that are uniformly spread across the rRNA, suggestive of widespread RNA folding and protein binding and consistent with the addition of 15 ribosomal proteins (44). For the RI to RI* transition changes are primarily localized in the central and 3′ domains, and for the RI* to 30S transition most of the changes occur in the 3′ domain, suggesting a general 5′ to 3′ directionality for assembly. Of particular note is a series of enhancements in the core of the subunit that occurs in the RI to RI* transition. Because proteins do not dissociate during this step, it is clear that a major conformational rearrangement of RNA takes place at this time, resulting in the exposure of those nucleotides to chemical modification, consistent with the changes to sedimentation coefficient that have been observed.
Figure 4.
Hybrid representations of the 16S rRNA annotated with information about changes in accessibility to chemical modification during different steps in 30S subunit assembly (23, 24). Both decreased accessibility (protection) and increased accessibility (enhancement) are shown. The extent of protection or enhancement is indicated by the size of the circle used to annotate the nucleotides. (a) Changes to the 16S rRNA during the formation of a reconstitution intermediate (RI). (b) Changes to the 16S rRNA during the RI to RI* transition. (c) Changes to the 16S rRNA while a complete 30S subunit is formed from RI*. The RI to RI* transition in particular has many enhancements to modification suggestive of a large refolding of the RNA that exposes several nucleotides.
The RI to RI* transition exhibits the hallmarks of a kinetic folding trap. RNA secondary structures are extremely stable thermodynamically, and consequently, misfolded structures with incorrect base pairing can be stable. Slow folding and kinetic traps are characteristic of the folding pathways of large RNAs, such as the Tetrahymena Group I intron and RNase P (38, 57, 58). Typically, lower magnesium ion concentrations, addition of denaturants, and higher temperatures accelerate the refolding reactions that must occur to escape from stable misfolded kinetic traps (14, 28, 36, 47, 51, 54). The RI to RI* transition is consistent with the observed folding of these other large RNAs, and heating is required to complete the transition. Even though kinetic traps are commonly observed in RNA folding reactions, they do not appear to be essential features of the folding mechanism, as rapidly folding sequences or mutants can usually be identified (35, 37, 46, 59). The possible significance of the RI to RI* transition is discussed in the context of these and other data below.
KINETIC STUDIES OF PROTEIN BINDING
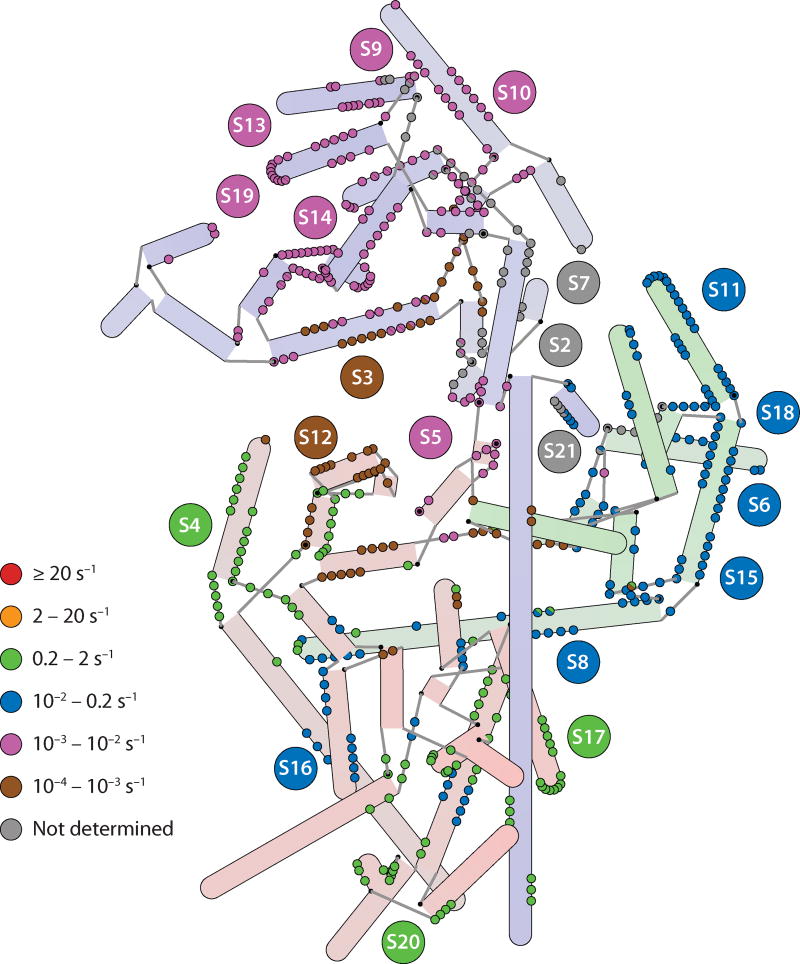
An isotope pulse-chase experiment was developed to monitor the assembly kinetics of all 30S proteins simultaneously. The 16S rRNA is first incubated with a pulse of isotopically labeled ribosomal proteins for varying lengths of time and then chased with an excess of unlabeled proteins, allowing the reaction to reach completion. The relative amount of unlabeled and labeled protein in the resulting 30S ribosomal subunits can be determined by quantitative mass spectrometry. The time course of the pulse-chase experiment can be analyzed to determine the binding rate of each protein. This technique was recently used to determine binding rates for 17 of the 20 ribosomal proteins in the small subunit (55). The binding rates of the ribosomal proteins are mapped onto the individual protein binding sites, as shown in Figure 5. In general, primary binding proteins bind more quickly than secondary or tertiary binding proteins, consistent with the hierarchical nature of assembly. Proteins in the 5′ domain generally bind more quickly overall than those in the central or 3′ domain, consistent with the idea of a 5′ to 3′ directionality for assembly (42). The slowly binding 5′ domain tertiary proteins S5 and S12 do not strictly follow the 5′ to 3′ directionality of assembly, but these two proteins bind at the intersection of all the domains near the decoding site of the 30S subunit.
Figure 5.
A hybrid representation of the 16S rRNA annotated with rate constants for protein binding as determined by pulse-chase quantitative mass spectrometry (55). Nucleotides that make protein contacts are color-coded according to the binding rate of the protein that they contact. In the case in which two proteins are contacted by a single nucleotide, semicircles are drawn. Nucleotides that contact proteins S2, S7, and S21 are marked in gray, as no rate constant was obtained for these proteins. Rates generally cluster by domain, with the fastest binding rates observed in the 5′ domain and the slowest in the 3′ domain, the exception being the 5′ domain protein S12, which is among the slowest binders (indicated by the brown circles in the 5′ domain).
TIME-RESOLVED HYDROXYL RADICAL FOOTPRINTING OF ASSEMBLY
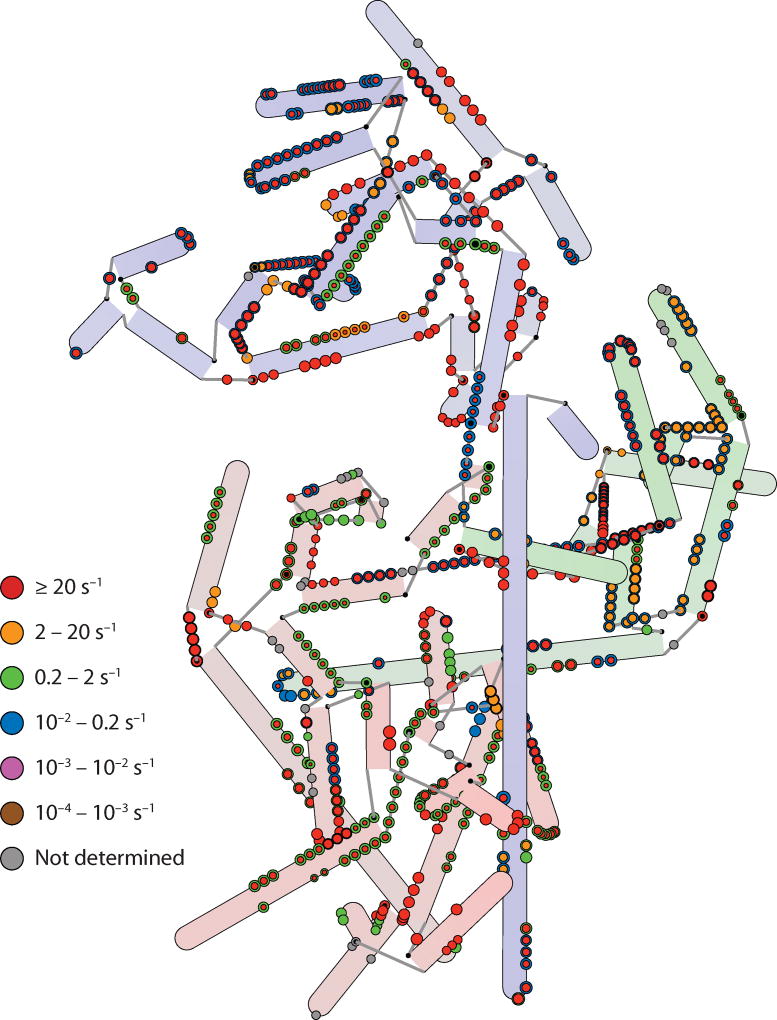
The most recent work on 30S assembly uses time-resolved hydroxyl radical footprinting to obtain information on 16S rRNA folding in the presence of proteins and 30S subunit assembly (1). Synchrotron X-ray radiation can generate a sufficiently concentrated burst of hydroxyl radicals in a short pulse to give a footprint, allowing kinetic footprinting information to be gathered about the protection from hydroxyl radical cleavage of individual nucleotides, and is summarized in Figure 6. Most nucleotides exhibited protection in two phases, suggesting an initial fast RNA folding or protein binding event followed by a second, slower event. In these experiments, the rRNA was prefolded in the presence of magnesium ions, and much of the 16S secondary structure is likely preformed at the start of the experiment. However, initial protections may still be due to secondary structure formation as it reorganizes during the early stages of protein binding while the initial tertiary contacts form. The second, slower protections are likely to be due to the formation of RNA tertiary contacts from large conformational changes such as the RI to RI* transition, or the final stages of protein binding. Within the set of nucleotide protections due to interactions with a particular protein, two different rates of protection were often observed. This general observation suggests that protein binding itself may be a two-phase process, with an initial loose association with some fraction of the binding site followed by a second set of interactions formed upon conformational rearrangement of the protein and/or RNA. Perhaps the most striking observation in the context of previous work was the seeming lack of any 5′ to 3′ directionality in the fast folding events. Initial burst phases of protection were observed for nucleotides across the entire 16S rRNA, suggesting that RNA folding nucleates from many different sites spread across the entire molecule.
Figure 6.
A hybrid representation of the 16S rRNA annotated with rate constants for protection from hydroxyl radical cleavage (1). In the cases in which two rate constants are calculated for two phases of protection, concentric circles are displayed, with the inner circle colored according to the rate constant of the initial burst of protection and the outer circle colored according to the second, slower protection. In all cases the area displayed is proportional to the amplitude of the protection, so smaller amplitudes are reflected by smaller circles. Fast rates are evident across the entire 16S, suggesting multiple nucleation sites for assembly. Whereas the initial burst rates are in almost all cases faster than the rates observed for protein binding (Figure 5), the second slower rates are on par with those observed for protein binding in many cases.
DISCUSSION
The view of assembly that emerges from consideration of the sum of the body of experimental data is rich with detail, but not without apparent discrepancies. Chemical footprinting (23, 24, 44) and pulse-chase experiments measuring the kinetics of protein binding using pulse-chase quantitative mass spectrometry (PC/QMS) (55) indicate a 5′ to 3′ directionality of assembly, but this was not observed in the time-resolved hydroxyl radical footprinting data (1). The key to reconciling these data sets is the observation that nucleotides that contact a particular protein do not always experience the same rate of protection. In the PC/QMS experiments, only tightly bound proteins are observed, as proteins that are loosely bound during the pulse can exchange during the chase. In contrast, the footprinting data can capture occupancy by both a weakly binding protein and a stably associated protein. It is likely that the faster phase of RNA protections correspond to thermodynamically favorable, but kinetically labile interactions of an initial encounter complex. The agreement of the measured rates for the slow phase in the RNA footprinting data and the rates from pulse-chase experiments is quite good. The general picture that emerges is consistent with the idea of alternating protein binding and RNA conformational changes that emerged from the studies of central domain fragments.
Most ribosomal protein binding sites have some fast fractional protection for at least part of the binding site. This indicates that some portion of each protein binding site on the 16S rRNA is preorganized in such a way to permit rapid protein binding, and that most of the RNA structure is capable of forming at least transiently in the absence of proteins. The hierarchy observed in the original Nomura map, as well as the kinetics observed by PC/QMS, indicates that protein binding to many of these sites is kinetically labile. Weakly bound proteins can dissociate during the ultracentrifugation step under nonequilibrium conditions after reconstitution or during the chase step in PC/QMS. The observed order of stable protein binding provides clues to the order of RNA conformational changes that lock down initial encounter complexes.
Furthermore, while the chemical footprinting data on assembly intermediates RI and RI* also suggest a 5′ to 3′ directionality, these data report on a specific set of steps during assembly. Indeed, the conversion of RI* to a complete 30S subunit gives rise to changes in nucleotide protections heavily concentrated in the 3′ domain, but this is a preordained result because four of the five proteins missing in the reconstituted RI* particle are 3′ domain proteins. The data on the transition between naked 16SrRNA and the RI particle show nucleotide protections across the entire subunit, owing to the addition of proteins from all three domains, and the RI to RI* transition describes a single, albeit significant step in the assembly process rather than a global rearrangement of the entire RNA, so its localization does not directly imply a 5′ to 3′ directionality.
There is also the question of whether RI represents a true assembly intermediate. Clearly RI is a prominent feature of in vitro reconstitution reactions at low temperature. It is possible to form and isolate an RI particle from reconstitutions, but this does not demand that all assembly trajectories necessarily converge on RI. The temperature dependence as shown by Arrhenius plots of the protein binding rates measured by pulse-chase mass spectrometry was linear over the measured range (15–40°C) (55). This indicates that the rate-determining step for each protein is the same at high and low temperatures, which is not consistent with a single temperature-dependent rate-limiting step such as the RI to RI* transition. The data indicate a series of multiple parallel folding pathways whose point of convergence is the final 30S subunit and not a single intermediate. The 30S subunit assembles over a complicated assembly landscape, and not a unique trajectory (55). The time-resolved hydroxyl radical footprinting data also suggest that many portions of the RNA are folding in parallel. The intermediate RI is most likely a misfolded RNA structure in which portions of the 3′ domain, and possibly the central domain, are mispaired, and the RI to RI* transition is the rate-limiting refolding of this stable kinetic trap.
The striking observation remains that 5′ domain proteins generally bind more quickly than 3′ domain proteins do, even if only in their final tight association. This is a curious result especially in the face of data showing that each domain can assemble independently. Reconciliation of all the in vitro data may ultimately be achieved by understanding what happens in vivo, where assembly occurs cotranscriptionally. The 5′ domain of the RNA is transcribed first, thereby having the opportunity to fold first into appropriate binding domains for the proteins. Because the 5′ domain proteins are present first in practice, binding of the central domain proteins and 3′ domain proteins may have evolved to be maximally efficient in their presence. Each domain is capable of assembling independently but may assemble most efficiently in the context of the entire subunit. In addition, cotranscriptional folding and assembly of the 5′ and central domains is likely to avoid formation of the RI kinetic trap.
The structure of the 16S rRNA is fairly monolithic, and it is possible to imagine that much of the RNA secondary and tertiary structure can form in the absence of the proteins dotting its periphery. The 5′ domain is highly folded in the absence of proteins (2), but it is not stable without the complement of ribosomal proteins. The body of work on central domain assembly suggests that protein binding does not so much induce a conformational change as it stabilizes an unstable RNA conformation. Transient RNA folding and unstable conformations may be populated enough to influence the footprinting data, but this does not imply a single stable conformation.
The ribosomal proteins serve to consolidate RNA folding gains as assembly proceeds. The three-helix junction binding site of S15 folds significantly in the presence of magnesium ions, and subsequent binding of S6:S18 is possible but greatly reduced in the absence of S15. S15 is nonessential for growth (9), which is consistent with a primary role for S15 in ribosome assembly rather than ribosome function. The hierarchy evident in the Nomura map is apparently not absolute in vivo. The binding sites for S6:S18 must be able to fold sufficiently well for transient protein binding. There is reciprocity of the thermodynamics of cooperativity, meaning that if S6:S18 binding stabilizes subsequent RNA tertiary structure, then that same tertiary structure can stabilize S6:S18 binding if it can form in the absence of S15. The parallel nature of assembly makes it possible and likely that such defects can be overcome.
The overall impression of 30S subunit assembly is that of a dynamic and fluid process. Proteins do not simply lock into place in a strict processive manner; they work their way into the network of RNA helices in a multiphasic manner. The 16S rRNA begins folding on its own but reacts and adapts to the influx of ribosomal proteins, adopting new local conformations around them and stabilizing others. There are many parallel folding and binding pathways, each of them proceeding simultaneously toward the final product of a complete 30S subunit.
The assembly process for ribosomes has been highly evolved for efficiency and accuracy, and it appears that some combination of parallel and sequential folding and binding events has been selected. Although the efficient in vitro assembly of 30S subunits has been instrumental in allowing biophysical characterization of specific steps in assembly, many features of the 30S assembly mechanism remain to be understood in vivo. The assembly of the 50S subunit is much more complex, and although it also forms a monolithic RNA tertiary structure, it is much more topologically intricate. The cotranscriptional nature of assembly and the participation of tens of exogenous cofactors and chaperones are critical for rapid and efficient assembly of the 50S subunit. The next frontier in ribosome assembly is tackling this formidable problem, which will no doubt require experimental innovation for both in vitro and in vivo methodologies. Ten years ago, it was difficult to imagine the “movie” of the process of translation, but extraordinary efforts in structural biology have made this possible. Similar efforts will hopefully generate the ribosome assembly movie in the coming decade.
Supplementary Material
SUMMARY POINTS.
Assembly of the 30S subunit occurs via multiple parallel folding pathways
The incorporation of proteins into the 30S subunit occurs in a hierarchical manner.
The 16S rRNA determines the overall fold of the 30S subunit.
Whereas the final tight binding of proteins occurs in a 5′ to 3′ manner, folding and assembly nucleate from several points in the ribosome independent of the domain structure. The directionality observed in vitro may be a side effect of optimization for in vivo assembly.
The three domains are capable of assembling independently.
Some ribosomal proteins help drive large conformational rearrangements of the RNA.
FUTURE ISSUES.
An unambiguous description of the precise events that confer protection against hydroxyl radical cleavage of the 16S rRNA is needed.
A statistical view of the simultaneous folding pathways that nucleate from different sites along the 16S rRNA is important to understanding the parallel nature of ribosome assembly.
The 3D structures of important assembly intermediates need to be determined.
A complete kinetic and energetic description of each step of the assembly trajectory are necessary for the construction of a true mechanism for assembly.
The specific roles of assembly cofactors need to be determined.
The assembly process in vivo needs to be fully explored and compared to the process in vitro.
Acknowledgments
This work was supported by grants from the NIH (F32-GM083510 to M.T.S. and R37-GM053757 to J.R.W.). The authors thank Drs. Gloria Culver and Sarah Woodson for critical comments on the manuscript.
Glossary
- rRNA
ribosomal RNA
- 16S rRNA
the RNA component of the 30S subunit
- 30S subunit
the small subunit of the ribosome, consisting of a single RNA and 21 proteins
- RNA footprinting
a method used to probe the folding of an RNA by determining the accessibility of a nucleotide to hydroxyl radical cleavage or chemical modification
- S2
ribosomal protein S2
- Reconstitution intermediate (RI)
an intermediate observed during in vitro 30S subunit assembly at low temperature (~0°C)
- RI*
a conformational variant of RI obtained by heating RI to high temperature (~40°C)
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–72. doi: 10.1038/nature07298. A high-resolution kinetic study of RNA footprinting during assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 2005;351:508–19. doi: 10.1016/j.jmb.2005.06.020. A study of the 5′ domain reveals native structure in the absence of proteins. [DOI] [PubMed] [Google Scholar]
- 3.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. Structure of the S15, S6, S18-rRNA complex: assembly of the 30S ribosome central domain. Science. 2000;288:107–13. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 4.Agalarov SC, Zheleznyakova EN, Selivanova OM, Zheleznaya LA, Matvienko NI, et al. In vitro assembly of a ribonucleoprotein particle corresponding to the platform domain of the 30S ribosomal subunit. Proc. Natl. Acad. Sci. USA. 1998;95:999–1003. doi: 10.1073/pnas.95.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold RJ, Reilly JP. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 1999;269:105–12. doi: 10.1006/abio.1998.3077. [DOI] [PubMed] [Google Scholar]
- 6.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 7.Batey RT, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16S rRNA: I. Defining the minimal RNA site. J. Mol. Biol. 1996;261:536–49. doi: 10.1006/jmbi.1996.0481. [DOI] [PubMed] [Google Scholar]
- 8.Batey RT, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16S rRNA: II. Specificity determinants of RNA-protein recognition. J. Mol. Biol. 1996;261:550–67. doi: 10.1006/jmbi.1996.0482. [DOI] [PubMed] [Google Scholar]
- 9.Bubunenko M, Korepanov A, Court DL, Jagannathan I, Dickinson D, et al. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA. 2006;12:1229–39. doi: 10.1261/rna.2262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–19. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J. Mol. Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 13.DeLano WL. The PyMOL molecular graphics system. 2002 http://www.pymol.org/
- 14.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the Tetrahymena ribozyme P4–P6 domain. Biochemistry. 2000;39:10975–85. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 15.Dutcă LM, Jagannathan I, Grondek JF, Culver GM. Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16S rRNA. J. Mol. Biol. 2007;368:853–69. doi: 10.1016/j.jmb.2007.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graphviz Graph Visualization Software. http://www.graphviz.org/
- 17.Green R, Noller HF. Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry. 1999;38:1772–79. doi: 10.1021/bi982246a. [DOI] [PubMed] [Google Scholar]
- 18.Grondek JF, Culver GM. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–66. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilek GM, Marusak R, Meares CF, Noller HF. Directed hydroxyl radical probing of 16S rRNA using Fe(II) tethered to ribosomal protein S4. Proc. Natl. Acad. Sci. USA. 1995;92:1113–16. doi: 10.1073/pnas.92.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilek GM, Noller HF. Site-directed hydroxyl radical probing of the rRNA neighborhood of ribosomal protein S5. Science. 1996;272:1659–62. doi: 10.1126/science.272.5268.1659. [DOI] [PubMed] [Google Scholar]
- 21.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974;249:3103–11. [PubMed] [Google Scholar]
- 22.Herold M, Nierhaus KH. Incorporation of six additional proteins to complete the assembly map of the 50S subunit from Escherichia coli ribosomes. J. Biol. Chem. 1987;262:8826–33. [PubMed] [Google Scholar]
- 23.Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat. Struct. Mol. Biol. 2004;11:179–86. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- 24.Holmes KL, Culver GM. Analysis of conformational changes in 16S rRNA during the course of 30S subunit assembly. J. Mol. Biol. 2005;354:340–57. doi: 10.1016/j.jmb.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 25.Jagannathan I, Culver GM. Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J. Mol. Biol. 2003;330:373–83. doi: 10.1016/s0022-2836(03)00586-2. [DOI] [PubMed] [Google Scholar]
- 26.Jagannathan I, Culver GM. Ribosomal protein-dependent orientation of the 16S rRNA environment of S15. J. Mol. Biol. 2004;335:1173–85. doi: 10.1016/j.jmb.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 27.Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–94. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc. Natl. Acad. Sci. USA. 2007;104:7045–50. doi: 10.1073/pnas.0608765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1993;231:581–93. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214–18. doi: 10.1038/2261214a0. The first appearance of an assembly map for the 30S ribosomal subunit. [DOI] [PubMed] [Google Scholar]
- 32.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 33.Nomura M. Assembly of bacterial ribosomes. Science. 1973;179:864–73. doi: 10.1126/science.179.4076.864. [DOI] [PubMed] [Google Scholar]
- 34.Orr JW, Hagerman PJ, Williamson JR. Protein and Mg(2+)-induced conformational changes in the S15 binding site of 16S ribosomal RNA. J. Mol. Biol. 1998;275:453–64. doi: 10.1006/jmbi.1997.1489. [DOI] [PubMed] [Google Scholar]
- 35.Pan J, Deras ML, Woodson SA. Fast folding of a ribozyme by stabilizing core interactions: evidence for multiple folding pathways in RNA. J. Mol. Biol. 2000;296:133–44. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 36.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J. Mol. Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 38.Pan T, Sosnick T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–75. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 39.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004;38:749–70. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 40.Petry S, Brodersen DE, Murphy FV, Dunham CM, Selmer M, et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–66. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 41.Polevoda B, Sherman F. Methylation of proteins involved in translation. Mol. Microbiol. 2007;65:590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 42.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16S rRNA into 30S ribosomal subunits. J. Mol. Biol. 1993;232:362–74. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 43.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16S rRNA complex. J. Biol. Chem. 1995;270:1238–42. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 44.Powers T, Noller HF. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 45.Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleo-protein complex. J. Mol. Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J. Mol. Biol. 1998;281:609–20. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 47.Rook MS, Treiber DK, Williamson JR. An optimal Mg(2+) concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl. Acad. Sci. USA. 1999;96:12471–76. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samaha RR, O’Brien B, O’Brien TW, Noller HF. Independent in vitro assembly of a ribonucleo-protein particle containing the 3′ domain of 16S rRNA. Proc. Natl. Acad. Sci. USA. 1994;91:7884–88. doi: 10.1073/pnas.91.17.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 50.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 51.Silverman SK, Deras ML, Woodson SA, Scaringe SA, Cech TR. Multiple folding pathways for the P4–P6 RNA domain. Biochemistry. 2000;39:12465–75. doi: 10.1021/bi000828y. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 1990;44:105–29. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 53.Svensson P, Changchien LM, Craven GR, Noller HF. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16S ribosomal RNA. J. Mol. Biol. 1988;200:301–8. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- 54.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, et al. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4–P6 RNA domain in monovalent cations. J. Mol. Biol. 2004;343:1195–206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 55.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. A nearly complete set of kinetic data on protein binding during assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30S ribosomes studied in vitro. J. Mol. Biol. 1969;40:391–413. doi: 10.1016/0022-2836(69)90161-2. An early description of the assembly of 30S subunits via RI. [DOI] [PubMed] [Google Scholar]
- 57.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–46. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 58.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–14. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 59.Treiber DK, Williamson JR. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J. Mol. Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 60.Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–80. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 61.Wilson DN, Nierhaus KH. Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 2005;40:243–67. doi: 10.1080/10409230500256523. [DOI] [PubMed] [Google Scholar]
- 62.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 63.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 64.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–70. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.