Abstract
Understanding the dynamics of Zika virus transmission and formulating rational strategies for its control require precise diagnostic tools that are also appropriate for resource-poor environments. We have developed a rapid and sensitive loop-mediated isothermal amplification (LAMP) assay that distinguishes Zika viruses of Asian and African lineages. The assay does not detect chikungunya virus or flaviviruses such as dengue, yellow fever, or West Nile viruses. The assay conditions allowed direct detection of Zika virus RNA in cultured infected cells; in mosquitoes; in virus-spiked samples of human blood, plasma, saliva, urine, and semen; and in infected patient serum, plasma, and semen samples without the need for RNA isolation or reverse transcription. The assay offers rapid, specific, sensitive, and inexpensive detection of the Asian-lineage Zika virus strain that is currently circulating in the Western hemisphere, and can also detect the African-lineage Zika virus strain using separate, specific primers.
INTRODUCTION
Asian-lineage Zika viruses have established mosquito-borne transmission in North, South, and Central America and in the Cape Verde Islands and are poised to enter Africa (1–5). The spread of Zika virus underscores the need for rapid and inexpensive Zika-specific diagnostic assays for surveillance and health care. Zika virus infection may be asymptomatic or cause fever, rash, arthralgia, and conjunctivitis lasting several days. Guillain-Barré syndrome in adults and microcephaly in newborns have been associated with Asian-lineage Zika virus infection (6–9). Zika viruses are primarily transmitted by Aedes spp. mosquitoes but can be directly transmitted through sexual contact and from mother to fetus (2–4, 10–14). The impending overlap of Asian- and African-lineage viruses requires their precise identification. Zika viruses circulate with other arboviruses in the Americas, specifically dengue viruses (DENV), yellow fever virus (YFV), and chikungunya virus (CHIKV), and patients infected with Zika virus, DENV, or CHIKV exhibit similar symptoms. Current diagnostic tests for these viruses include viral RNA detection by reverse transcription polymerase chain reaction (RT-PCR), post-exposure serology, and virus isolation (1, 3).
Loop-mediated isothermal amplification (LAMP) is a rapid, simple, and sensitive nucleic acid amplification assay (15). It uses Bst DNA polymerase, which has reverse transcriptase activity and amplifies specific DNAs without temperature cycling (16). Results can be determined by a variety of methods including turbidity, color change, or fluorescence detection and can be confirmed by amplicon restriction digestion, PCR, or sequencing. RT-LAMP is used to detect viral RNAs after RNA purification and inclusion of reverse transcriptase in the LAMP reaction. Specific RT-LAMP protocols for arboviruses have been deployed successfully in endemic countries (17–24).
We have developed a Zika virus LAMP assay that uses the reverse transcriptase activity of Bst DNA polymerase for direct detection of viral RNA in mosquito homogenates and human biofluids without previous RNA isolation. The assay provides clade-specific detection of Zika virus RNAs. Visual readouts were assessed immediately, and samples were retained for further analysis. The assay distinguished Asian- and African-lineage Zika viruses from other arthropod-borne viruses and from each other using specific primer sets. Sensitivity was comparable to quantitative RT-PCR (qRT-PCR) but without the need for RNA isolation or reverse transcription.
RESULTS
The LAMP assay specifically amplifies RNA from Asian- or African-lineage Zika viruses
We designed Zika virus LAMP primers to a unique genome region of the Puerto Rican Zika strain, PRVABC59 [3500 to 5300 nucleotides (nt)] via the LAMP software PrimerExplorer V4 (15). Of the five primer sets, a single set of four primers between positions 3694 and 3978 in the NS2A coding region was chosen for further work (Fig. 1A and table S1). We infected Vero cells with the PRVABC59 Zika virus strain and amplified the target sequence from purified cellular RNA without previous reverse transcription or inclusion of reverse transcriptase in the reaction (Fig. 1B and Table 1). We confirmed the target sequence by restriction fragment analysis and PCR of the LAMP product with the LAMP primers (fig. S1, A and B) and by direct sequence analysis of both the LAMP and PCR amplicons, which matched the target PRVABC59 sequence (Fig. 1A). Ribonuclease (RNase) A treatment abrogated signal from PRVABC59-infected Vero cell RNA (fig. S1C).
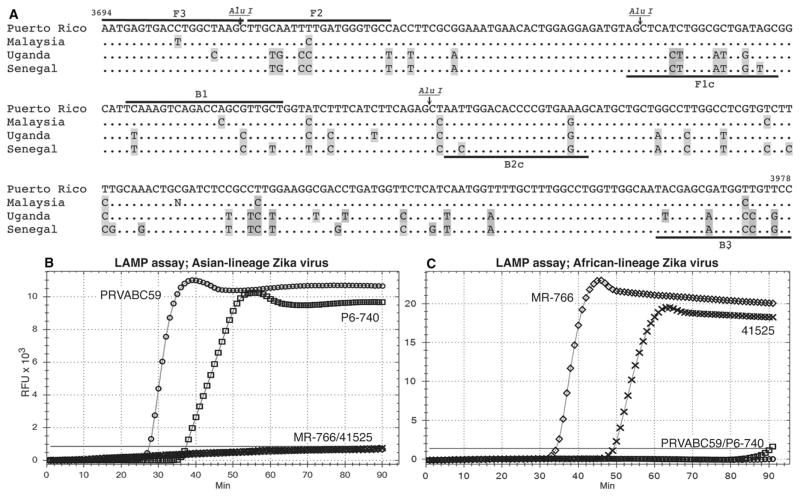
Fig. 1. Design and implementation of LAMP primers.
(A) LAMP target sequences in the Puerto Rican strain (PRVABC59) of Zika virus aligned with sequences from the Malaysia (P6-740), Uganda MR-766NIID (MR-766), and Senegal (41525) strains. Primer sequences are indicated by black bars. BIP and FIP primers are fusions of B1 and B2c and F1c and F2, respectively (B1, B2c, F1c, and F2 labels are shown). Alu I restriction sites are indicated. Sequence variations are in gray. (B) Asian-lineage–specific primers detect Zika virus RNAs exclusively from Vero cells infected with Asian strains, PRVABC59 and P6-740. (C) African-lineage–specific primers detect Zika virus RNAs exclusively from Vero cells infected with African strains, MR-766 and 41525. (B and C) PRVABC59 (circles), P6-740 (boxes), MR-766 (diamonds), and 41525 (crosses). The threshold of detection is defined by a horizontal gray line. X axis, minutes to LAMP amplicon incorporation of fluorescent dye; y axis, relative fluorescence units (RFUs).
Table 1. Detection of Zika virus RNA in cell and supernatant RNA preparations.
ND, not determined.
| Virus RNA | Asian Zika LAMP | African Zika LAMP | RT-PCR (primer specificity) | LAMP (primer specificity) |
|---|---|---|---|---|
| PRVABC59* | + | − | + (Zika) | − (WNV, DENV, YFV) |
| P6-740 | + | − | + (Zika) | − (WNV, DENV, YFV) |
| 41525 | − | + | + (Zika) | − (WNV, DENV, YFV) |
| MR-766 | − | + | + (Zika) | − (WNV, DENV, YFV) |
| WNV | − | ND | + (Pan-flavi) | + (WNV) |
| DENV-2 | − | ND | + (Pan-flavi) | + (DENV) |
| YFV | − | ND | + (Pan-flavi) | + (YFV) |
| Bussuquara | − | ND | + (Pan-flavi) | ND |
| St. Louis encephalitis | − | ND | + (Pan-flavi) | ND |
| Langat | − | ND | + (Pan-flavi) | ND |
| Deer tick | − | ND | + (Pan-flavi) | ND |
| Ilheus | − | ND | + (Pan-flavi) | ND |
| CHIKV | − | ND | + (CHIKV) | ND |
PRVABC59-infected Vero cell total RNA and C6/36 cell total RNA.
Total RNA from Vero cells infected under identical conditions with the P6-740 Malaysian, MR-766NIID Ugandan, or 41525 Senegalese strains of Zika virus were also tested for amplification. P6-740 RNA was amplified with the PRVABC59 primer set, but there was no amplification from total RNA from cells infected with the Senegalese or Ugandan viruses (Fig. 1B and Table 1). All four Zika virus RNA preparations could be amplified by conventional RT-PCR with Zika-specific primers (fig. S2A and Table 1). The PRVABC59 LAMP primers closely match sequences from 64 Zika virus strains within the Asian clade, but the 19 sequences in the African clade have a minimum of 17 base changes across the primer target sequences (fig. S3, A and B). Most base changes within the Asian clade are in strains from Southeast Asia and Micronesia. The 1966 Malaysian sequence P6-740 is most divergent, with five base changes, yet P6-740 was readily amplified with the PRVABC59 Zika virus LAMP primers (Fig. 1B). Among the American isolates, a sequence from Panama has two base changes, and nine others have single base changes across all primer target sequences (a total of six distinct base changes). Four of these do not alter the amino acid sequence. Primer target sequences in 47 of the Asian-lineage Zika viruses are identical to the PRVABC59 strain sequence.
To detect the African-lineage Zika viruses in an identical LAMP assay, we modified the primer sequences to match the MR-766NIID Ugandan prototype strain (fig. S3, A and B) and retested the MR-766NIID, 41525, P6-740, and PRVABC59 RNAs for amplification. LAMP amplification with these primers was limited to the two African strains, MR-766NIID and 41525 (Fig. 1C and Table 1).
We tested the PRVABC59 Zika virus LAMP primer set against RNAs from Bussuquara virus, St. Louis encephalitis virus, Langat virus, Powassan virus, Ilheus virus, DENV-2, West Nile virus (WNV), YFV, and CHIKV. All were negative for amplification with the Asian-lineage Zika virus primers (Table 1). All tested flavivirus RNAs were positive by RT-PCR with pan-flavivirus primers (25), and CHIKV RNA was positive with CHIKV-specific primers (fig. S2, B and C, and Table 1). We were able to amplify DENV-2, WNV, and YFV RNAs with previously described RT-LAMP primers (17, 20, 24) using our amplification protocol without reverse transcription (fig. S2D and Table 1). The DENV-2, WNV, and YFV RT-LAMP primers did not amplify PRVABC59 Zika virus RNA.
Zika virus RNA can be detected in infected mosquitoes with the LAMP assay
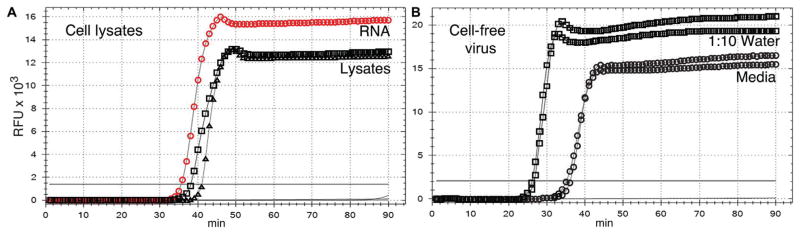
We detected the PRVABC59 Zika virus strain in infected C6/36 mosquito cells using the PRVABC59 LAMP primers (Table 2). To test direct amplification of virus RNA, we lysed infected and mock-infected Vero cells and C6/36 cells in water and used 2 μl of lysate (equivalent to 1000 cells) for amplification. Lysis in water allowed amplification of Zika virus RNA without further processing (Fig. 2A and Table 2). Viral RNA in infected Vero cell supernatant was also directly amplified (Fig. 2B). Tenfold dilution of supernatant in water decreased the time to detection from 36 to 26 min, indicating an increase in the efficiency of the reaction with dilution of the cell culture medium.
Table 2. Direct detection of the PRVABC59 Zika virus strain in cell lysates and mosquito tissue homogenates.
dpi, days post-infection; MD, mosquito diluent.
| Sample | Zika LAMP (number of positive/total) | PFU |
|---|---|---|
| Mock-infected cultured cells* | − | 0 |
| Infected cultured cells* | + | 0 |
| Infected Vero cell supernatant | + | 6.4 × 106 PFU/ml |
| Infected C6/36 cell supernatant | + | 7.4 × 106 PFU/ml |
| Mock-infected mosquitoes | − (0/3) | − (3/3) |
| Infected mosquitoes (7 dpi) | + (15/15) | + (14/15)† |
| Infected mosquitoes (14 dpi) | + (MD, 9/10; H2O, 10/10) | + (10/10)† |
| Infected mosquito midgut | + (4/5) | + (2/5) |
| Infected mosquito salivary gland | + (4/5) | + (2/5) |
| Infected mosquito carcass | + (1/5) | + (1/5) |
Vero cells or C6/36 cells, lysed in water.
Positive for plaque formation.
Fig. 2. Direct detection of Zika virus RNA in cells and supernatants.
(A) Lysates of 1000 infected Vero or C6/36 cells in water. Vero cell lysate (black triangles), C6/36 cell lysate (black squares), mock-infected Vero cells (no symbols; line below threshold), mock-infected C6/36 cells (no symbols; line below threshold), infected Vero cell RNA (positive control; red circles), buffer only (no symbols; line below threshold). (B) Duplicate aliquots of infected Vero cell supernatant amplified directly (circles) or diluted 10-fold in water (squares). X axis, minutes to LAMP amplicon incorporation of fluorescent dye; y axis, RFUs.
We carried out oral infections of Aedes aegypti mosquitoes with cell-free PRVABC59 Zika virus. At 7 days after infection, whole mosquitoes were homogenized in mosquito diluent for plaque assays and 2.5 μl (1/100th) of homogenates from 15 infected and 3 mock-infected mosquitoes were directly amplified. All 15 infected mosquito homogenates were positive, and the 3 mock-infected mosquito homogenates were negative (fig. S4 and Table 2). Plaque assays detected infectious virus in 14 of the 15 homogenates (Table 2). A representative mosquito-LAMP product was subjected to restriction digestion, PCR, and sequencing; the predicted PCR product and restriction fragments were observed (fig. S5, A and B), and the sequence was confirmed. RNase A–treated homogenates were negative for amplification (fig. S6A), and deoxyribonuclease (DNase) I digestion only reduced the time to amplification from 40 min (±2.1 min SE) to 34 min (±1.4 min SE) (fig. S6B).
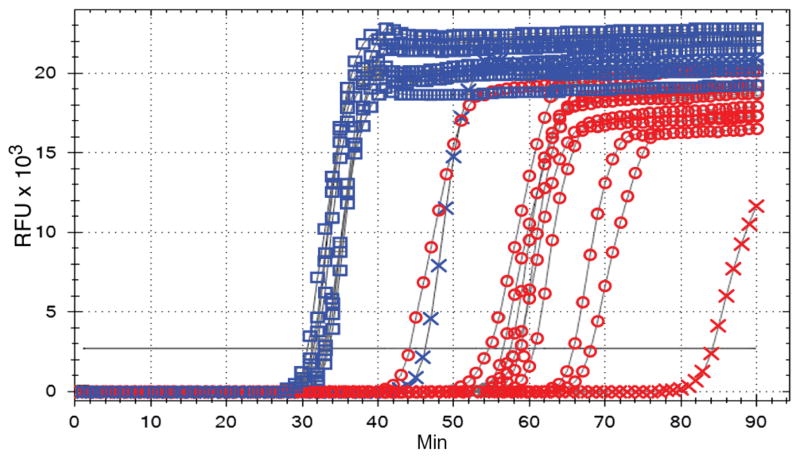
Ten additional infected mosquito homogenates and homogenates of dissected salivary glands, midguts, and carcasses from five infected mosquitoes were tested at 14 days after infection. We made sagittal sections of the mosquitoes and homogenized half in mosquito diluent with serum for plaque and LAMP assays and half in water for the LAMP assay alone. We detected viral RNA in all 10 virus-infected homogenates, but the average time to detect positive LAMP signal with mosquito diluent was 60.5 min (±9.4 min SE) versus 33.9 min in water (±4.3 min SE) (Fig. 3). A sample from a single homogenate with low virus titer corresponded to delayed time to detection (Table 2). Dissected tissues were all collected in mosquito diluent. Viral RNA was detected in four of five midguts, four of five salivary glands, and one of five mosquito carcasses (Table 2). Plaque assays matched results for one of four positive midguts and two of four positive salivary glands, but not the single carcass (table S3).
Fig. 3. Direct detection of Zika virus in mosquitoes.
A LAMP assay was performed on 2.5 μl (1/100th of total) from PRVABC59 strain–infected mosquitoes split sagittally; one-half of the mosquito carcass was homogenized in mosquito diluent (red circles) and the other half in water (blue squares). Blue and red crosses, single mosquito with low virus load (table S2, mosquito #3); x axis, minutes to LAMP amplicon incorporation of fluorescent dye; y axis, RFUs.
Other investigators have established methods for LAMP product detection by color change or turbidity (26, 27), and these methods have been implemented for the detection of arbovirus RNAs (17–24). We amplified mock and PRVABC59 strain–infected mosquito homogenates without RNA isolation or reverse transcription in a simple heat block and distinguished positive samples by visible turbidity after 1 hour (fig. S7).
We have determined that the basic requirement for LAMP amplification of Zika virus RNA is the lysis of infected cells. Homogenization of mosquitoes in water instead of mosquito diluent with serum reduced the time to detection. Reduced viral loads also correlated with extended time to detection. With or without serum or host DNA, the detection of Zika virus RNA in infected mosquitoes using the LAMP assay does not require RNA isolation or reverse transcription.
The limit of detection of RNA from the PRVABC59 Zika virus strain by the LAMP assay is comparable to qRT-PCR
We assayed 10-fold dilutions of the total RNA from PRVABC59-infected Vero cells by the LAMP assay and by conventional RT-PCR. Gel analyses of LAMP and RT-PCR products are shown in fig. S8. Both methods detected PRVABC59 RNA equivalent to 100 fg of total infected cell RNA.
We compared direct detection of virus in cell supernatants between the LAMP assay and a standard qRT-PCR assay for Zika virus detection (28). Tenfold dilutions of PRVABC59-infected Vero cell supernatants [6.4 × 106 plaque-forming units (PFU)/ml)] were prepared in water, and 2 μl from each dilution was amplified by both assays (n = 6 replicates). Additional twofold dilutions around the end point were prepared and assayed to better delineate the limit of detection (LOD) for each assay. The Zika virus LAMP assay LOD was 0.43 PFU, and the LOD for qRT-PCR was 0.17 PFU. Confidence intervals (CIs) and PFU/ml equivalents are reported in Table 3. Representative results of LAMP assays of serial dilutions of Vero cell supernatants are presented in Fig. 4A.
Table 3. LOD (30) of Zika virus RNA with the LAMP assay (n ≥ 6 replicates).
NA, not applicable.
| Sample | Assay | LOD | 95% CI | PFU/ml* |
|---|---|---|---|---|
| Vero cell supernatant | LAMP | 0.43 PFU | 0.24–4.89 | 214 |
| Vero cell supernatant | qRT-PCR | 0.17 PFU | 0.08–6.55 | 87 |
| Genome equivalents | LAMP | 111 copies | 58–1148 | NA |
| Genome equivalents | qRT-PCR | 53 copies | 38–98 | NA |
| Plasma (1:100)† | LAMP | 0.28 PFU | 0.19–0.87 | 31 |
| Plasma (1:1000)† | LAMP | 0.05 PFU | 0.03–0.42 | 21 |
| Plasma (1:1000)† | LAMP (turbidity) | 0.17 PFU | 0.08–2.52 | 86 |
| Spiked blood | LAMP | 0.05 PFU | 0.03–0.25 | 24 |
| Spiked urine | LAMP | 0.32 PFU | 0.22–3.26 | 159 |
| Spiked saliva | LAMP | 0.13 PFU | 0.09–0.86 | 63 |
| Spiked semen | LAMP | 5.57 PFU | 2.18–608.14 | 2785 |
Based on a 2-μl sample volume and rounded to nearest whole number.
Final dilution in water.
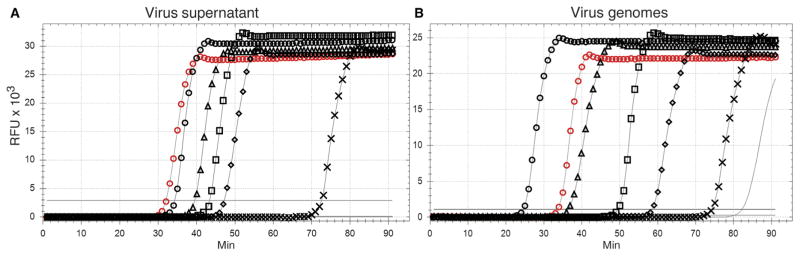
Fig. 4. Limit of detection of Zika virus RNA.
(A) A LAMP assay was performed on 2 μl of 10-fold dilutions of Vero cell supernatants containing 6.4 × 105 PFU/ml. Virus input: 1280 PFU (black circles), 128 PFU (black triangles), 12.8 PFU (black squares), 1.28 PFU (black diamonds), 0.128 PFU (black crosses), and 0.0128 PFU (no symbols; line below threshold). (B) A LAMP assay was performed on 10-fold dilutions of Zika virus genome copies: 105 copies (black circles), 104 copies (black triangles), 103 copies (black squares), 102 copies (black diamonds), 10 copies (black crosses), and 1 copy (line, no symbols). (A and B) Control infected Vero cell RNA (positive control; red circles) and no RNA (no symbols; line below threshold). Results are representative of a minimum of six replicates of each dilution series. X axis, minutes to LAMP amplicon incorporation of fluorescent dye; y axis, RFUs.
For direct, quantitative comparison of the LAMP and qRT-PCR assays, we constructed a complementary DNA (cDNA) clone of the full-length PRVABC59 Zika virus genome and transcribed genomic RNA in vitro. Genome equivalents were determined by molecular weight, and 10-fold dilutions were measured with both assays. Twofold dilutions at the end point were performed to determine the LOD for each assay (n ≥ 6 replicates) (Table 3). The Zika virus LAMP assay LOD was 111 genome copies (table S4A), and the qRT-PCR LOD was 53 genome copies (table S4B). Representative results of LAMP assays of serial dilutions of in vitro transcribed virus RNA are presented in Fig. 4B.
Detection limits for RNA from infected cells were comparable between LAMP and conventional RT-PCR assays. We also demonstrated the direct detection of virus RNA, without RNA isolation, from virus supernatants using both the LAMP assay and qRT-PCR at comparable levels. The 95% CI for the LOD with the LAMP assay lies entirely within the 95% CI for qRT-PCR. Similarly, there was a large overlap between the higher bounds of the 95% CI for detection of genome equivalents by qRT-PCR and the lower bounds of the 95% CI by the LAMP assay (Table 3). We did not conduct a formal statistical test of differences in mean LOD.
The LAMP assay detects PRVABC59 Zika virus strain RNA in virus-spiked samples of human blood, plasma, saliva, urine, and semen
We tested direct detection of Zika virus in human samples of blood, plasma, saliva, urine, and semen. Aliquots of each fluid were inoculated with PRVABC59 Zika virus from infected Vero cell supernatants at a final concentration of 106 PFU/ml. Tenfold dilutions of each biofluid were prepared in water, and 2 μl (undiluted, 2000 PFU) was amplified. Fluorescent dye incorporation was very low in undiluted biofluids (fig. S9, blue circles). A 1:100 dilution (20 PFUs) in water was necessary for adequate amplification. A 1:1000 dilution yielded full fluorescent dye incorporation. Biofluids without added virus were negative.
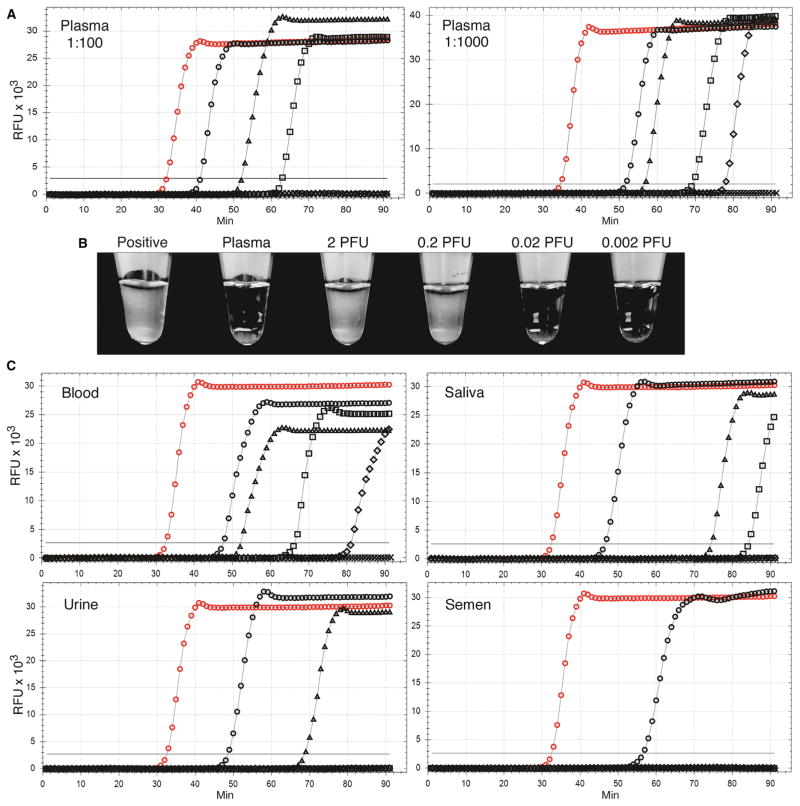
To quantify the limits of detection for each spiked biofluid, we first prepared 10-fold dilutions of the biofluid, then further diluted each dilution in water, and amplified 2 μl (n ≥ 6). Twofold serial dilutions of biofluid followed by dilution in water were assayed at the LOD. Dilutions of plasma with virus were assayed at final 1:100 and 1:1000 dilutions in water, and the LOD was 0.28 and 0.05 PFU, respectively (Table 3 and table S5A). Representative results of LAMP assays of serial dilutions are presented in Fig. 5. Matched amplifications of the 1:1000 plasma dilutions were also incubated in a heat block followed by visual assessment of turbidity, and the LOD was 0.17 PFU (table S5B). Representative results of the turbidity assays of plasma dilutions are presented in Fig. 5B. Limits of detection of other biofluids were determined at 1:1000 final dilution in water. The LOD was 0.05 PFU in whole blood, 0.32 PFU in urine, 0.13 PFU in saliva, and 5.57 PFU in semen (Table 3 and tables S6 to S9). Representative results of LAMP assays of the serial dilutions of these virus-spiked biofluids are presented in Fig. 5C.
Fig. 5. Detection of Zika virus spiked into human blood, plasma, saliva, urine, and semen samples.
Healthy human biofluids were spiked with the Zika virus PRVABC59 strain from infected Vero cell supernatants at a final concentration of 106 PFU/ml. (A) Amplification of 2 μl of serial 10-fold dilutions of plasma containing 106 PFU/ml of the PRVABC59 strain followed by dilution to 1:100 or 1:1000 in water. (B) Matching samples of the 1:1000 plasma dilutions were incubated in tubes in a heat block for 70 min and examined for turbidity. (C) Tenfold dilutions of blood, saliva, urine, and semen each spiked with 106 PFU/ml of PRVABC59 Zika virus strain followed by dilution to 1:1000 in water. Virus input: (A and C) 2000 PFU (black circles), 200 PFU (black triangles), 20 PFU (black squares), 2 PFU (black diamonds), 0.2 PFU (black crosses; below threshold), 0.02 PFU (no symbols; gray line below threshold), infected Vero cell RNA (positive control; red circles), and diluted biofluids without virus (no symbols; black line below threshold). Results are representative of a minimum of six replicates of each dilution series. X axis, minutes to LAMP amplicon incorporation of fluorescent dye; y axis, RFUs.
We detected Zika virus directly in biofluids with varying degrees of sensitivity. On the basis of the values at upper CIs, detection of Zika virus in plasma and blood was <2.1 × 102 and 1.26 × 102 PFU/ml, respectively. Detection was less than 1.63 × 103 PFU/ml in urine, 4.28 × 103 PFU/ml in saliva, and 3.04 × 105 PFU/ml in semen. Quantitative detection required 1000-fold dilution in water for efficient LAMP amplification.
Zika virus RNA can be detected in plasma, serum, and semen samples from infected patients
We next tested Zika virus–positive and Zika virus–negative human plasma and serum samples from cohorts of infected patients in Brazil and Nicaragua, respectively, as well as Zika virus RNA–positive human semen samples. All samples were previously tested by qRT-PCR for the presence or absence of Zika virus RNA at the respective points of care. The plasma and serum samples were coded before receipt so that the LAMP assay was conducted in a blinded manner. We assayed 32 serum samples, collected from patients in Managua, Nicaragua, which were thawed and diluted 1:100 in water for direct LAMP amplification of 2.5 μl in quadruplicate. We called the samples positive based on one or more positive replicates with a defined product melting point; of the 32 samples, we identified 22 that were positive (Table 4 and table S10). Upon decoding, the original qRT-PCR results had identified 20 Zika virus–positive samples of the 32 serum samples. Of the 22 positive samples identified by the LAMP assay, 18 matched the original qRT-PCR results (90%) and 4 were false positives. Of the 10 LAMP assay–negative samples, 8 matched known negatives (67%) and 2 were false negatives. Assays of samples diluted to 1:10 returned 28 positives, and dilution to 1:500 returned 14 positive samples.
Table 4.
Serum samples from Zika virus–infected patients in Nicaragua (20 positive and 12 negative by qRT-PCR).
| Sera (n = 32) | Serum RNA preparation (n = 32) | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Direct LAMP | qRT-PCR | LAMP | |||||
|
|
|
|
|||||
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| qRT-PCR | Positive | 18 (90%) | 2 (10%) | 19 (95%) | 1 (5%) | 19 (95%) | 1 (5%) |
|
| |||||||
| Negative | 4 (33%) | 8 (67%) | 0 | 12 (100%) | 1 (8%) | 11 (92%) | |
To compare qRT-PCR and LAMP amplification of the samples directly, we isolated RNA from 20 μl of each serum sample and used equal volumes of these RNA preparations in both assays, before decoding the original results (Table 4 and table S10). Of the 20 known positives, qRT-PCR and the LAMP assay both identified 19 of the positive samples (95%). RT-PCR yielded one false negative, whereas the LAMP assay yielded identification of one false negative and one false positive. We detected one positive sample with the LAMP assay that was missed by qRT-PCR (table S10), and qRT-PCR missed one positive sample that was missed by the LAMP assay.
We detected Zika virus RNA directly, without RNA isolation, in serum samples from infected individuals; the success and accuracy of direct detection were dependent on the dilution factor in water. LAMP amplification of Zika virus RNA from purified RNA preparations was comparable in error rate to qRT-PCR using the same RNAs but without the need for reverse transcription.
We also assayed 49 plasma samples collected from patients in Re-cife, Brazil. It should be noted that these samples had been previously thawed and refrozen. These samples were thawed and diluted 1:100 in water for direct LAMP amplification of 2.5 μl in quadruplicate. We called the samples based on one or more positive replicates with a defined product melting point and then decoded them to compare with previous results using qRT-PCR and serology (Table 5 and table S11). Of the 49 plasma samples in the cohort, 25 were previously identified as Zika virus–positive and 24 as Zika virus–negative by qRT-PCR. Six of the RT-PCR–negative samples were subsequently identified as positive by serology [using anti–Zika virus immunoglobulin M (IgM)] for a total of 31 antibody-positive and RT-PCR–positive plasma samples and 18 negative samples. Direct LAMP assay identified 33 positive and 16 negative samples. Twenty-one of the positive samples matched known positives (68%) and 12 were false positives. Of the 16 LAMP-negative samples, 6 matched known negative samples (33%) and 10 were false negatives.
Table 5.
Plasma samples from Zika virus–infected patients in Brazil (31 positive and 18 negative by qRT-PCR and serology).
| Plasma (n = 49) | RNA (n = 26) | RNA and plasma combined | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| qRT-PCR | LAMP | LAMP | LAMP | ||||||
|
|
|
|
|
||||||
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | ||
| qRT-PCR and serology | Positive | 25 (81%) | 6 (19%) | 20 (65%) | 10 (32%) | 16 (70%) | 7 (30%) | 26 (84%) | 5 (16%) |
|
| |||||||||
| Negative | 0 | 18 (100%) | 12 (67%) | 6 (33%) | 2 (9%) | 1 (33%) | 12 (67%) | 6 (33%) | |
In addition to the plasma samples, a subset of 26 stored RNA samples, originally assayed by qRT-PCR, was available for LAMP assay. Three of the 26 had been negative by qRT-PCR (Table 5 and table S11). Eighteen of the 26 RNAs were positive by the LAMP assay. Sixteen of the positive samples matched known positives (70%) and 2 were false positives. Of the eight LAMP-negative RNAs, one matched a known negative sample and seven were false negatives. Combined results from direct LAMP amplification of plasma samples and amplification of matching, stored RNAs yielded positive results for 26 of the 31 positive samples (84%) with 12 false positives and 6 false negatives.
An overview of the data indicates the presence of false-positive and false-negative results in the direct amplification of human plasma and serum samples when compared to previous data from qRT-PCR. The available RNA preparations from these samples were predominantly Zika virus–positive samples, which limited the assessment of false-positive results from RNA samples. We were unable to produce new RNA preparations from these samples for direct comparison of qRT-PCR and LAMP amplification.
In addition to the serum and plasma samples, we tested five known Zika virus RNA–positive semen samples from male U.S. travelers who were confirmed to be infected with Zika virus either serologically or by qRT-PCR from a sample of body fluid (serum, whole blood, or urine). The results of direct LAMP amplification of 1:100 dilutions of the semen samples yielded three of five positive samples (Table 6). The remaining two samples, which had comparable amounts of Zika virus RNA based on qRT-PCR of isolated RNAs, exhibited an unusual linear fluorescent dye incorporation that suggested excess DNA contamination. We digested all five samples with DNase I and were then able to directly detect Zika virus RNA in all five samples. These results suggest the need for DNase I digestion to directly detect viral RNA in semen samples.
Table 6.
Zika virus RNA–positive semen samples from male U.S. travelers confirmed to be infected with Zika virus.
| Semen (five samples) | |||
|---|---|---|---|
| Sample number | RNA copies/ml (log10) | LAMP | DNase + LAMP |
| 1 | 5.4 | + | + |
| 2 | 4.3 | − | + |
| 3 | 3.2 | − | + |
| 4 | 4.1 | + | + |
| 5 | 3.4 | + | + |
DISCUSSION
The Zika virus outbreak in the Americas resulted from the recent introduction of Asian-lineage strains into the Western hemisphere via Southeast Asia and Micronesia (29, 30). Their spread in a large and naïve population produced a full spectrum of disease in adults and birth defects in developing fetuses. Whether the Asian strains have acquired greater virulence or more efficient transmission is not known, but significant sequence variation from African strains is apparent. Such variation has made the specific detection of Asian versus African Zika viruses possible. Asian-lineage Zika viruses have now spread to the Cape Verde Islands (31) and are anticipated to enter Africa with uncertain outcomes in terms of human health. The ability to distinguish Asian and African Zika viruses will make it possible to better understand these outcomes. The need for rapid and inexpensive detection of the Asian-lineage Zika viruses in the Americas is already clear. The distinction of Asian and African Zika viruses may also become critical.
The PRVABC59 Zika virus strain was isolated in the United States in 2015 from a patient who had traveled to Puerto Rico. Phylogenetic analyses place it in the Asian clade with the prototype Malaysian strain P6-740 (30), which we detected by LAMP assay with the PRVABC59 primers. The sequence of P6-740 is the most distant from other members of the clade, and sequences of American isolates are very similar in comparison, so, while not certain, it is highly likely that the PRVABC59 primers would hybridize to all Zika virus RNA sequences in this clade. The African strains we tested, the Ugandan and Senegalese isolates, were not amplified, but we modified the primer sequences to match the prototype Ugandan strain and established an African clade–specific assay. The stringency of the assay precludes detection of other flavi-viruses and CHIKV, which circulate in the Western hemisphere, and allows specific detection of Zika virus isolates of the Asian or African lineage.
The LAMP assay we describe will be valuable for vector surveillance. We were able to detect Zika virus RNA in single mosquitoes without RNA extraction. The lowest virus load detected, 8.4 × 103 PFU/ml, is comparable to the lowest virus load observed in Zika virus–infected A. aegypti (32), which ranges from 104 to 106 PFU/ml through the course of infection. This sensitivity is sufficient to detect a single infected mosquito in a collection pool of 50 noninfected mosquitoes. Equipment requirements have been reduced to fluid handling and single-temperature heat blocks that can be run on batteries. Rapid field tests of individual mosquitoes in addition to pools of mosquitoes will provide an assessment of virus load within the vector population. Additionally, on-site identification of infected mosquitoes will permit highly focused control efforts. The field assay will increase efficiency and reduce overall cost, allowing an increase in the extent of surveillance in resource-limited regions.
Under controlled conditions with known quantities of cultured virus inoculated into human samples of blood, plasma, urine, saliva, and semen, dilution in water permitted efficient detection of Zika virus RNA (fig. S9 and Table 4). Detection of less than 210 PFU/ml in plasma should be sensitive enough to detect virus during acute infection, when titers in blood range from 102 to 106 PFU/ml (28, 33–35). Direct detection was less sensitive in control preparations of whole blood, urine saliva, and semen. These types of samples will require additional approaches to reach adequate sensitivity in the direct assay. Mucolytic treatment of semen with dithiothreitol and DNase is an example of a simple modification that yielded consistent amplification of viral RNAs.
Assays of clinical samples present challenges to the direct detection of Zika virus RNA by the LAMP assay. No assay can account for virus instability in biofluids, especially in late acute or convalescent patients with rising antibody titers and declining viremia, but the requirement for 100-fold sample dilution limits the sensitivity of the direct LAMP assay. This dilution factor is not necessary for LAMP amplification from purified RNA. Successful detection by the LAMP assay or by RT-PCR is also subject to sample storage conditions, especially the numbers of freeze-thaw cycles of plasma or serum. This was a particular issue with the tested plasma samples, and yet, the direct LAMP assay identified five seropositive samples that had gone undetected by qRT-PCR. Realistically, the detection of viruses in human clinical samples will continue to require RNA isolation to achieve maximum sensitivity and accuracy. When matched RNA preparations were used, the LAMP assay without reverse transcription matched RT-PCR with respect to specificity and levels of detection near a theoretical limit of single virus genome copies. In the near term, the LAMP assay will offer a rapid and inexpensive confirmatory assay for Zika virus RNA from human clinical samples.
The LAMP assay is limited as a quantitative assay: End point dilutions with appropriate standards are required for full quantitation because the relationship between time to positivity and target concentration is not linear. As with RT-PCR, the extreme sensitivity of the LAMP assay requires careful control of sample and assay preparation facilities and the complete physical separation of amplified product to avoid contamination of input samples or reagents. Also, prolonged incubation times, as with extended RT-PCR cycle numbers, can yield false-positive signals. In a real-time machine, such products can be distinguished by melting point, but they are indistinguishable by turbidity or colorimetric assays. Overall, the LAMP assay offers an inexpensive alternative to the RT-PCR assays currently in use for Zika virus detection, and this direct assay is fully functional for sensitive detection of Zika virus RNA in infected mosquitoes.
Public health efforts to control arbovirus infection rely on mosquito control and focused vaccination programs. There is as yet no approved vaccine for Zika virus, and this virus presents the additional challenge of sexual transmission (11, 14). Fast and simple virus RNA detection will support interdiction efforts in mosquitoes and quickly inform medical decisions for acutely infected patients. The direct LAMP assay, without RNA isolation or reverse transcription, can be used to detect a variety of flaviviruses with the use of appropriate primers. The ease and cost of any assay influence the degree of its implementation and, in turn, the extent of vector and reservoir host surveillance and of transmission control in low-resource settings. In the laboratory, increased speed of the direct LAMP assay will increase productivity in the development of animal models and the performance of vector competence studies, and it is adaptable to high-throughput platforms to further accelerate Zika virus research.
MATERIALS AND METHODS
Study design
The overall objective of the study was the establishment of a LAMP assay for the specific detection of Zika virus genomic RNA. The study required LAMP primer design and testing in cell culture systems, followed by implementation of the assay in infected mosquitoes for application to vector competence studies and surveillance. Further evaluation in specific human biofluids (blood, plasma, saliva, urine, and semen) was performed by inoculating these samples with tissue culture virus preparations to test assay function and to determine limits of detection. Comparisons with established RT-PCR assays were performed with tissue culture virus and recombinant virus RNA. Finally, initial validations were performed in Zika virus–positive and Zika virus–negative human clinical samples from Nicaragua and Brazil.
Sample sizes for serum, plasma, and semen samples were defined by their availability. All available samples were included in the assays, and results of all assays were reported. All clinical human sera and plasma samples were provided as blinded samples for assay, evaluation, and tabulation before decoding and revealing of their previously determined Zika virus status. Individual co-authors were responsible for sample provision and coding and decoding of samples. Other coauthors were responsible for assay and tabulation of results before decoding. Five semen samples were available for assay and were presented as Zika virus–positive samples before assay. A minimum of four experimental replicates were performed for each single clinical sample. Control sample and experimental sample replicates are indicated in the text and figure legends.
Viruses
Zika virus strain PRVABC59 (GenBank accession no. KU501215) was isolated at the Centers for Disease Control and Prevention (Fort Collins, CO) from serum of an infected patient who traveled to Puerto Rico in 2015. Zika virus strain P6-740 (GenBank accession no. HQ234499) was isolated from infected A. aegypti in Malaysia in 1966 and provided by the University of Texas Medical Branch (Galveston, TX) (36, 37). Strain 41525 (GenBank accession no. KU955591) was isolated from infected Aedes africanus in Dakar, Senegal, in 1984, and strain MR-766NIID (GenBank accession no. LC002520) was isolated from blood of a febrile sentinel rhesus monkey in the Zika forest of Uganda in 1947 (38). Additional flaviviruses for assessment included Bussuquara (accession no. AY632536), St. Louis encephalitis (accession no. EF158055), Langat (accession no. NC_003690), Powassan (accession no. KU886216), Ilheus (accession no. KC481679), DENV-2 (accession no. U87411), WNV (accession no. KR868734), YFV (accession no. NC_002031), and the alphavirus CHIKV (accession no. KT449801). Viruses were propagated in Vero cells (African green monkey kidney epithelial cells).
Virus infection in cell cultures
Vero and C6/36 (Aedes albopictus) cells were inoculated with Zika virus at a multiplicity of infection of 0.01 to 0.1. Virus was absorbed at room temperature for 1 hour. Vero cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), and C6/36 cells were cultured in minimum essential medium supplemented with 2% fetal bovine serum (FBS), L-glutamine, and nonessential amino acids. Virus supernatant was harvested when peak cytopathic effect (CPE) was observed. Supernatant titers were as follows: PRVABC59, 6.4 × 106 PFU/ml; 41525, 9.8 × 106 PFU/ml; P6-740, 8.5 × 105 PFU/ml; and MR-766NIID, 1.5 × 108 PFU/ml.
RNA isolation
Total cell RNA was isolated with TRIzol reagent (Ambion) when peak CPE was observed. RNA from viral stocks was purified using the Mag-Bind Viral DNA/RNA kit (Omega Bio-Tek) according to the manufacturer’s instructions.
Mosquito rearing and oral infection with Zika virus
The F12 generation of A. aegypti Poza Rica strain, collected from Poza Rica, Veracruz, Mexico, in 2012, was used (39). At days 5 to 7 after hatching, pupae were transferred to plastic cups and placed in half-gallon paperboard cartons for eclosion. Adult mosquitoes were supplied with raisins as a sugar source and water and maintained at 28°C, 80% relative humidity, and a 14-hour light/10-hour dark photocycle. At 5 to 7 days after eclosion, mosquitoes were deprived of sugar for 24 hours and water for 5 hours before an artificial blood meal containing PRVABC59 Zika virus (40). The infectious blood meal was prepared by mixing supernatant from Zika virus–infected Vero cells 1:1 with de-fibrinated calf blood and 1 mM adenosine triphosphate for a blood meal titer of 1.55 × 107 PFU/ml. The infectious blood meal was placed in a glass membrane feeder covered with hog gut and warmed with circulating water at 37°C. Mosquitoes were allowed to feed for 1 hour. Blood-engorged mosquitoes were sorted and supplied with sugar and water for 7 or 14 days after infection.
Mosquito sample preparation
Mosquito samples were collected on days 7 or 14 after blood meal. Whole bodies (wings and legs removed), half bodies (split sagittally), or mosquito tissues (midguts, salivary glands, and carcasses) were placed in tubes containing a single stainless steel BB and 250 μl of mosquito diluent [phosphate-buffered saline (PBS) supplemented with 20% FBS, penicillin/streptomycin (50 μg/ml), gentamicin (50 μg/ml), Fungizone (2.5 μg/ml)] or water (half bodies). Mosquitoes and tissues were homogenized using a Retsch MM400 Mixer Mill homogenizer at a frequency of 25 cycles per second for 1 min. Samples were subsequently centrifuged at 15,000g for 5 min, and supernatant was stored at −80°C.
Quantification of Zika virus infectious titer by plaque assay
Plaque assays were performed on Vero cells. Briefly, 25 μl of virus supernatant from infected cells or mosquito homogenates was diluted to 250 μl in PBS with 20% FBS, and 10-fold dilutions were absorbed on confluent Vero cells for 1 hour at room temperature. The cells were then overlaid with 3 ml of 1% agarose in DMEM supplemented with 2.5% FBS. After incubation for 4 days, 4% neutral red solution in PBS was added to the agar overlay, and plaques were counted at 6 to 12 hours after staining.
LAMP primer selection
A full alignment of the PRVABC59 Puerto Rican strain sequence (GenBank accession no. KU501215.1) with the sequence of dengue virus serotype 2 (GenBank accession no. M29095.1) was scanned for regions of greatest divergence. An 1800-nt region between Zika virus RNA positions 3500 and 5300 was selected for submission to Eiken’s LAMP software Primer Explorer V4 (http://primerexplorer.jp/e/; Eiken Chemical Company) (15). Five primer sets were identified and tested for amplification of RNA from PRVABC59-infected Vero cells. African-lineage–specific primers were selected directly from the primer target sequences in the sequence of the MR-766NIID Ugandan strain (GenBank accession no. LC002520).
LAMP target sequence alignment
Geneious version 8.1.6 (41) was used to prepare an alignment of the full-length LAMP amplified sequence (from F3 to B3; 284 bases) of PRVABC59 using National Center for Biotechnology Information BLAST. Duplicated, identical sequences were deleted, and aligned sequences were then edited to include only the individual LAMP primer sequences presented in fig. S3.
LAMP assay
Reactions were prepared as previously described (16). Briefly, 25-μl reactions were prepared with 8 U of Bst DNA polymerase [version 2.0 WarmStart; New England Biolabs (NEB)] in 20 mM tris (pH 8.8), 10 mM (NH4)2, 8 mM MgSO4, 50 mM KCl, 0.1% Tween 20, 1.4 mM deoxynucleotide triphosphates (dNTPs) (Thermo Scientific), and 2 μM SYTO-9 (Life Technologies). Final primer concentrations were 0.2 μM for F3 and B3 and 1.6 μM for FIP and BIP. After addition of template, reactions were incubated at 63°C for 90 min in a CFX96 Real-Time PCR System (Bio-Rad) with fluorescence readings at 1-min intervals. Lid temperature was 100°C. Reactions were followed with melt curve analysis; positive reaction product melting points varied from 86.5° to 87.5°C. Background subtraction, thresholds, and trace color and symbol were performed with Bio-Rad CFX Manager Software. Images of charts were generated by screen capture and assembled in Photoshop (Adobe). Human plasma samples were assayed in Applied Biosystems ABI 7500 Real-Time PCR System and analyzed with ABI software at the Laboratory of Virology and Experimental Therapeutics, Centro de Pesquisas Aggeu Magalhaes, Fundacao Oswaldo Cruz, in Recife-PE, Brazil.
Additional flavivirus LAMP primers for DENV-2, WNV, and YFV (17, 20, 24) were used under identical assay conditions, without reverse transcription, but with the addition of published loop-back primers. LAMP products were purified with the QIAquick PCR Clean-up Kit (Qiagen) according to the manufacturer’s instructions before submission of 500 ng for sequencing with B3 and F3 primers (Genewiz).
Turbidity was assessed with samples prepared in an identical reaction mix. Reactions were run in TempAssure PCR tubes (USA Scientific) and incubated at 63°C in a MyBlock HL Mini DryBath, Heated Lid (Benchmark Scientific) with lid temperature set at 100°C for 60 and 70 min before observation and photography with an Apple iPhone.
For PCR amplification, LAMP products were diluted 1:500 and 1 μl was amplified in a 25-μl reaction with 2.5 mM B3 and F3 LAMP primers in 75 mM tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 0.01% Tween 20, 1 mM dNTPs, 2 mM MgCl2, and 2 U of Taq polymerase (Thermo Scientific). Reactions were heated to 94°C for 3 min before 35 cycles of 10 s at 94°C, 10 s at 63°C, and 10 s at 72°C. Fifteen micro-liters from the reactions was separated on a 4% NuSieve agarose gel, and image was captured with ultraviolet (UV) transillumination and a Canon digital camera (model SD780 IS). Images were processed uniformly in Photoshop (Adobe). The 284–base pair (bp) amplicon was purified with the QIAquick PCR Clean-up Kit (Qiagen) according to the manufacturer’s instructions before submission of 500 ng for sequencing with B3 and F3 primers (Genewiz).
For restriction digestion analysis, 5 μg of LAMP product DNA was cut with Alu I in a 25-μl reaction with 20 U of enzyme for 1 hour at 37°C. Fifteen microliters of the digest was separated on 4 or 5% NuSieve gels along with 5 μg of unrestricted LAMP product.
DNase and RNase digestions
Ten-microliter aliquots of positive mosquito homogenate were incubated for 30 min at 37°C with or without 0.2 U of DNase I (Thermo Scientific) before heat inactivation at 70°C for 5 min. Ten-microliter aliquots of mock-infected and infected mosquito homogenates and infected Vero cell supernatant and total cellular RNA were incubated for 30 min at 37°C with or without 100 μg of RNase A (Qiagen) before heat inactivation at 70°C for 5 min. Two-microliter samples of each preparation were subjected to LAMP amplification.
Reverse transcription polymerase chain reaction
Ten nanograms of Zika virus or CHIKV RNAs was amplified in a onestep RT-PCR with iTaq reverse transcriptase and polymerase (Bio-Rad) according to the manufacturer’s instructions. PCR conditions were 3 min at 94°C and 35 cycles of 15 s at 94°, 15 s at 60°C, and 15 s at 72°C. Zika virus primers were as follows: nt9014, 5′-AGTGCCAGAGCT-GTGTGTAC-3′; nt9104, 5′-TCTAGCCCCTAGCCACATGT-3′. CHIKV primers were as follows: forward, 5′-GTACGGAAGGTAAACT-GGTATGG-3′; reverse, 5′-TCCACCTCCCACTCCTTAAT-3′ (sequence based on La Reunion strain, LR2006_OPY1; GenBank accession no. DQ443544.2).
Flavivirus RNAs were amplified from cDNAs with a pan-flavivirus primer set, FU1PM and cFD3 (25). These primers yield a different size product of about 1000 bp from each virus. Ten nanograms of total RNA from infected cells was first reverse-transcribed with SuperScript IV and random hexamers (Invitrogen) according to the manufacturer’s instructions. Two microliters of the reverse transcription reaction was amplified with Phusion DNA polymerase (NEB) according to the manufacturer’s instructions. PCR conditions were 30 s at 98°C, 4 cycles of 20 s at 98°C, 20 s at 55°C, 30 s at 72°C, and 34 cycles of 20 s at 98°C, 20 s at 62°C, and 30 s at 72°C. All RT-PCRs were run in a Bio-Rad T100 thermal cycler, amplicons were separated on 4% agarose gels, and image was captured with UV transillumination and a Canon digital camera (model SD780 IS). Images were processed uniformly in Photoshop (Adobe).
Limit of detection
A full-length Zika virus cDNA clone of RNA from strain PRVABC59 was inserted in the pACYC177 backbone (42). Briefly, PCR products with overlapping fragments were amplified using q5 high-fidelity polymerase (NEB) and gel-purified before assembly via circular poly-merase extension cloning (43). Cloning reactions were transformed into NEB Stable competent cells, plated on LB agar, and grown overnight at 37°C followed by additional growth (~8 hours) at room temperature. Small colonies were selected for growth in terrific broth overnight at 30°C. After plasmid purification (Zymo Research), restriction digestion and sequencing confirmed correct insertion of the Zika virus ge-nome. A full-length clone, pJW223, was transformed into MAX Efficiency Stbl2 Competent E. coli Cells (Thermo Fisher) under ampi-cillin selection (100 mg/ml). Plasmid purified from these cells was linearized with Eco RI–HF (NEB) and RNA-transcribed and template-digested with a MEGAscript T7 Transcription kit (Ambion/Life Technologies) according to the manufacturer’s instructions. Unit-length RNAs were confirmed by gel electrophoresis, concentration was determined by optical density, and copy number was calculated according to the molecular weight of the genome sequence (2.184 × 108 copies/ng RNA; Endmemo.com). Transcribed RNA was diluted to fixed copy numbers in 2 μl of aliquots for determination of the LOD in the LAMP assay on a CFX96 real-time PCR system (Bio-Rad) and in the standardized qRT-PCR Zika assay as previously described (28) using TaqMan probe and primers (Zika1087, Zika1108FAM, and Zika1163c) in a LightCycler 96 real-time PCR machine (Roche). A minimum of six technical replicates were assayed at each dilution. Probit analysis was performed using the IBM SPSS Statistics version 24 software to calculate the LOD and 95% CIs.
Preparation of biofluids inoculated with the PRVABC59 Zika virus strain
Whole blood and plasma were collected from an uninfected donor under Institutional Review Board protocol no. 09-1148H. Aliquots of each sample were inoculated with Zika virus–infected Vero cell supernatant (6.4 × 106 PFU/ml; strain PRVABC59) at a final concentration of 1 × 106 PFU/ml. Additional biofluids were collected from an uninfected donor and inoculated with the same virus supernatant at a final concentration of 1 × 106 PFU/ml. Inoculated biofluids were diluted 10-fold in water and 2 μl of each dilution subject to LAMP assay. For the determination of the LOD, inoculated biofluids were first diluted 10-fold in the specific biofluid, and then each dilution was diluted further in water for LAMP assay with six replicates at each dilution. Further twofold dilution series in biofluid were assayed in a 2-log range across the LOD with six replicates at each dilution. A minimum of six technical replicates were assayed at each dilution. Probit analysis was performed using the IBM SPSS Statistics version 24 software to calculate the LOD and 95% CIs.
Collection and assessment of human clinical samples
Serum samples
Serum samples were obtained from the Nicaraguan Pediatric Dengue Cohort Study, a prospective cohort study of ~3500 active participants 2 to 14 years of age in Managua, Nicaragua, that has been ongoing continuously since 2004. A detailed description of the study design, methods, and study population has been published previously (44). Zika virus was included in the cohort as of February 2016. Acute-phase (days 1 to 6 after onset of illness) and convalescent-phase (days 14 to 28) blood samples were collected from children presenting to the study health center, Centro de Salud Sócrates Flores Vivas in District II, with (i) fever or feverishness with two or more of the following symptoms: headache, muscle ache, joint pain, retro-orbital pain, rash, hemorrhagic manifestations, or leukopenia; (ii) undifferentiated fever; or (iii) rash with one or more of the following: conjunctivitis, arthralgia, myalgia, and/or periarticular edema regardless of fever (since February 2016). Acute samples were tested by RT-PCR for DENV RNA (45, 46) before 2016 and after February 2016 for both Zika virus and DENV RNA separately (28, 47) in triplex real-time RT-PCR assays testing for Zika virus, DENV, and CHIKV RNA (48).
Plasma samples
The Recife cohort samples used for the assays described here were collected from individuals presenting with acute febrile illnesses in an urgent health care clinic in the Recife Metropolitan Region from May 2015 to April 2016, as part of the International Research Consortium on Dengue Risk Assessment, Management and Surveillance (IDAMS) study (49). The ages of patients from whom sera were used for the assays varied from 9 to 62 years (mean age, 32), where 27 were females and 23 were males. Sample collection was performed on the first day of recruitment (day 1—acute sample), which, following the IDAMS protocol, corresponds to the period within the first 72 hours of the febrile period, and on the convalescent phase (days 10 to 30 after recruitment—convalescent sample). For molecular viral diagnosis, viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s instructions, and qRT-PCR for Zika virus was performed following the protocol of Lanciotti et al. (28, 50). Samples were also assayed for anti-DENV IgM and IgG and anti–Zika virus IgM by enzyme-linked immunosorbent assay (ELISA). The Panbio Dengue Capture ELISA was used for the anti-DENV IgM and IgG assays following the manufacturer’s protocol. Anti-Zika IgM was determined with the Centers for Diseases Control and Prevention protocol (28, 51). Samples were classified as Zika virus–positive if they were positive for Zika virus RNA by qRT-PCR in the acute phase and positive for anti–Zika virus IgM with titers more than two times those for anti-DENV IgM in the convalescent phase when the convalescent sample was available (37 of 49 patients).
Semen samples
Semen samples were obtained from men with confirmed Zika virus infection. Semen samples were treated with an equal volume of freshly prepared 10 mM dithiothreitol (Pierce Biotechnology), and RNA was extracted from samples using the MagMAX Viral RNA Isolation Kit (Ambion). Zika virus RNA was detected using the Qiagen QuantiTect Probe RT-PCR Kit using primers Zika4481/Zika4552c and probe Zika4507cFAM and PCR settings as described by Lanciotti et al. (28). A standard curve was generated by in vitro transcription of pCDNA3.1 containing a fragment of Zika virus sequence spanning genomic nucleotides 3576 to 4631.
Statistics
A minimum of six technical replicates were assayed at each dilution for determination of level of detection. Probit analysis was performed using the IBM SPSS Statistics version 24 software to calculate the limits of detection and 95% CIs.
Supplementary Material
Fig. S1. Confirmation of Zika-specific, RNA-dependent LAMP amplification.
Fig. S2. Control samples containing each virus RNA.
Fig. S3. Alignment of Zika LAMP primer sequences with target sequences in all known Zika virus sequences.
Fig. S4. Direct detection of Zika virus in mosquitoes.
Fig. S5. Confirmation of Zika-specific LAMP amplification from mosquito homogenate.
Fig. S6. Zika LAMP assay is dependent upon RNA.
Fig. S7. Visual LAMP assay of mosquito homogenates.
Fig. S8. Limits of PRVABC59-LAMP detection match RT-PCR.
Fig. S9. LAMP assay detects PRVABC59 virus in human biofluids.
Table S1. Zika LAMP primers.
Table S2. Plaque titration of split, infected mosquitoes.
Table S3. Plaque titration of midguts, salivary glands, and carcass.
Table S4A. LAMP assay LOD for genome copies.
Table S4B. TaqMan LOD for genome copies.
Table S5A. Plasma 1:1000 LOD.
Table S5B. Plasma 1:1000 turbidity LOD.
Table S6. Blood 1:1000 LOD.
Table S7. Urine 1:1000 LOD.
Table S8. Saliva 1:1000 LOD.
Table S9. Semen 1:1000 LOD.
Table S10. Nicaraguan serum and RNA samples.
Table S11. Brazilian plasma and RNA samples.
Acknowledgments
We thank C. D. Blair, J. S. Parker, J. W. Casey, R. J. Cohrs, G. N. Callahan, K. E. Olson, and T. Tellinghuisen for critical review of the manuscript.
Funding: The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (award nos. RO1AI067380 to J.W.-L., C.R., J.R.F., and G.D.E.; R01AI0833680 to W.C.B.; R21AI129464 to B.D.F., C.N., M.G., T.M., and E.T.A.M.; R01AI099631 to A.B.; and R21AI129488-01 to R.P. and J.R.) and the NIH/NIAID International Collaborations in Infectious Disease Research Program (U01AI088647). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional support was provided by the Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (APQ-0302-4.01/13 to E.T.A.M.) and the European Union (FP7-281803 IDAMS) (41). S.M.G.L. is supported by Fogarty Training Grant 2D43TW001130-08 “Training in dengue prevention and control.” N.K.D. and A.C.B. were supported by the Centers for Disease Control and Prevention. This submission represents the views solely of the authors and does not constitute those of the Centers for Disease Control and Prevention or the U.S. government. B.A., C.D.B., R.P., R.C.K., S.L.Q., and J.R. were supported by the Department of Microbiology, Immunology, and Pathology and the College of Veterinary Medicine and Biomedical Sciences at Colorado State University, and B.D.F., R.P., T.M., and N.C. were supported by the Office of the Vice President for Research, Colorado State University.
Footnotes
Author contributions: J.W.-L., C.R., C.N., S.M.G.L., G.D.E., W.C.B., B.D.F., and R.C.K. designed and implemented mosquito infections and assays. J.W.-L. prepared the Zika virus cDNA clone. M.G. designed CHIKV primers. J.R.F. designed Zika virus primers and produced flavivirus RNAs. R.P., G.D.E., and B.D.F. contributed to experimental design and manuscript preparation. N.K.D. and A.C.B. provided semen samples. T.M., T.J., and E.T.A.M. ran the studies in Brazil and contributed samples. E.H., G.K., and A.B. ran the studies in Nicaragua and contributed samples. E.H., E.T.A.M., and A.C.B. contributed to manuscript review. B.A. reviewed and revised statistical analyses. S.L.Q. contributed to experimental design and manuscript preparation and performed statistical analysis. N.C. produced virus stocks; dissected mosquitoes; ran all plaque, qRT-PCR, and LAMP assays in Brazil; and contributed to experimental design and manuscript preparation. C.D.B. selected LAMP primers, designed and implemented the assay, prepared sequence alignments, and contributed to experimental design and manuscript preparation. J.R. designed and performed LAMP experiments and prepared the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: No materials or clinical samples were transferred from Brazil to Colorado State University for the purposes of this study. Assays using clinical samples from Brazil were performed in Recife, Pernambuco, at the Department of Virology, FIOCRUZ-PE. To the extent that enough samples are available, they may be shared via a material transfer agreement for use in scientifically sound research in accordance with Brazilian laws, ethical principles, and governance of the IDAMS project that generated those samples. Inquiries should be addressed to E.T.A.M. (marques@pitt.edu). Assays of Nicaraguan samples were performed at the Colorado State University. If sufficient, Nicaraguan samples may be shared for use in scientifically sound and collaborative research. Inquiries should be addressed to E.H. of the University of California, Berkeley (eharris@berkeley.edu).
REFERENCES AND NOTES
- 1.Musso D, Gubler DJ. Zika virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika virus. N Engl J Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 3.Waggoner JJ, Pinsky BA. Zika virus: Diagnostics for an emerging pandemic threat. J Clin Microbiol. 2016;54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. Zika virus: History, emergence, biology, and prospects for control. Antivir Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.W. Report. www.who.int/csr/don/21-december-2015-zika-cape-verde/en/
- 6.Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, Araujo ESM, de Sequeira PC, de Mendonça MCL, de Oliveira L, Tschoeke DA, Schrago CG, Thompson FL, Brasil P, dos Santos FB, Nogueira RMR, Tanuri A, de Filippis AMB. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: A case study. Lancet Infect Dis. 2016;16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 7.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Rus KR, Vipotnik TV, Vodušek VF, Vizjak A, Pižem J, Petrovec M, Županc TA. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 8.Broutet N, Krauer F, Riesen M, Khalakdina A, Almiron M, Aldighieri S, Espinal M, Low N, Dye C. Zika virus as a cause of neurologic disorders. N Engl J Med. 2016;374:1506–1509. doi: 10.1056/NEJMp1602708. [DOI] [PubMed] [Google Scholar]
- 9.Roze B, Najioullah F, Fergé JL, Apetse K, Brouste Y, Cesaire R, Fagour C, Fagour L, Hochedez P, Jeannin S, Joux J, Mehdaoui H, Valentino R, Signate A Cabié; GBS Zika Working Group A. Zika virus detection in urine from patients with Guillain-Barre syndrome on Martinique, January 2016. Euro Surveill. 2016;21:30154. doi: 10.2807/1560-7917.ES.2016.21.9.30154. [DOI] [PubMed] [Google Scholar]
- 10.Besnard M, Lastere S, Teissier A, Cao-Lormeau VM, Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill. 2014;19:20751. [PubMed] [Google Scholar]
- 11.Foy BD, Kobylinski KC, Foy JLC, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, Tesh RB. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17:880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marano G, Pupella S, Vaglio S, Liumbruno GM, Grazzini G. Zika virus and the never-ending story of emerging pathogens and transfusion medicine. Blood Transfus. 2016;14:95–100. doi: 10.2450/2015.0066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musso D. Zika virus transmission from French Polynesia to Brazil. Emerg Infect Dis. 2015;21:1887. doi: 10.3201/eid2110.151125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau VM. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanner NA, Evans TC., Jr Loop-mediated isothermal amplification for detection of nucleic acids. Curr Protoc Mol Biol. 2014;105:15141–151414. doi: 10.1002/0471142727.mb1514s105. [DOI] [PubMed] [Google Scholar]
- 17.Kwallah A-o, Inoue S, Muigai AWT, Kubo T, Sang R, Morita K, Mwau M. A real-time reverse transcription loop-mediated isothermal amplification assay for the rapid detection of yellow fever virus. J Virol Methods. 2013;193:23–27. doi: 10.1016/j.jviromet.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Le Roux CA, Kubo T, Grobbelaar AA, van Vuren PJ, Weyer J, Nel LH, Swanepoel R, Morita K, Paweska JT. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid detection of Rift Valley fever virus in clinical specimens. J Clin Microbiol. 2009;47:645–651. doi: 10.1128/JCM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X, Li X, Mo Z, Jin F, Wang B, Zhao H, Shan X, Shi L. Rapid identification of Chikungunya and Dengue virus by a real-time reverse transcription-loop-mediated isothermal amplification method. Am J Trop Med Hyg. 2012;87:947–953. doi: 10.4269/ajtmh.2012.11-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parida M, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, Islam MA, Inoue S, Hosaka N, Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parida MM, Santhosh SR, Dash PK, Tripathi NK, Lakshmi V, Mamidi N, Shrivastva A, Gupta N, Saxena P, Babu JP, Rao PVL, Morita K. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2007;45:351–357. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parida MM, Santhosh SR, Dash PK, Tripathi NK, Saxena P, Ambuj S, Sahni AK, Rao PVL, Morita K. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J Clin Microbiol. 2006;44:4172–4178. doi: 10.1128/JCM.01487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teoh B-T, Sam S-S, Tan K-K, Johari J, Danlami MB, Hooi P-S, Md-Esa R, AbuBakar S. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect Dis. 2013;13:387. doi: 10.1186/1471-2334-13-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto M, Honda E, Ogura A, Nomoto A, Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 27.Mori Y, Kitao M, Tomita N, Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faria NR, Azevedo RSS, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, da Silva Vasami FG, de Lima Macedo FL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, de Almeida Medeiros DB, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, de Brito Simith D, Messina JP, Abade L, Lourenço J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, da Silva Lemos P, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez JLSG, Júnior, Mir D, Bello G, Delatorre E, Khan K, Creatore M, Coelho GE, de Oliveira WK, Tesh R, Pybus OG, Nunes MRT, Vasconcelos PFC. Zika virus in the Americas: Early epidemiological and genetic findings. Science. 2016;352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, del Carmen Castillo Signor L. Phylogeny of Zika virus in Western Hemisphere, 2015. Emerg Infect Dis. 2016;22:933–935. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peyrefitte CN, Boubis L, Coudrier D, Bouloy M, Grandadam M, Tolou HJ, Plumet S. Real-time reverse-transcription loop-mediated isothermal amplification for rapid detection of rift valley Fever virus. J Clin Microbiol. 2008;46:3653–3659. doi: 10.1128/JCM.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li MI, Wong PSJ, Ng LC, Tan CH. Oral susceptibility of Singapore Aedes (Stegomyia) aegypti (Linnaeus) to Zika virus. PLOS Negl Trop Dis. 2012;6:e1792. doi: 10.1371/journal.pntd.0001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bearcroft WG. Zika virus infection experimentally induced in a human volunteer. Trans R Soc Trop Med Hyg. 1956;50:442–448. [PubMed] [Google Scholar]
- 34.Simpson DI. Zika virus infection in man. Trans R Soc Trop Med Hyg. 1964;58:335–338. [PubMed] [Google Scholar]
- 35.Weinbren MP, Williams MC. Zika virus: Further isolations in the Zika area, and some studies on the strains isolated. Trans R Soc Trop Med Hyg. 1958;52:263–268. doi: 10.1016/0035-9203(58)90085-3. [DOI] [PubMed] [Google Scholar]
- 36.Marchette NJ, Garcia R, Rudnick A. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg. 1969;18:411–415. doi: 10.4269/ajtmh.1969.18.411. [DOI] [PubMed] [Google Scholar]
- 37.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. Genetic characterization of Zika virus strains: Geographic expansion of the Asian lineage. PLOS Negl Trop Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 39.Vera-Maloof FZ, Saavedra-Rodriguez K, Elizondo-Quiroga AE, Lozano-Fuentes S, Black WC., IV Coevolution of the Ile1,016 and Cys1,534 mutations in the voltage gated sodium channel gene of Aedes aegypti in Mexico. PLOS Negl Trop Dis. 2015;9:e0004263. doi: 10.1371/journal.pntd.0004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. Dengue virus type 2: Replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007;7:9. doi: 10.1186/1471-2180-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grubaugh ND, Weger-Lucarelli J, Murrieta RA, Fauver JR, Garcia-Luna SM, Prasad AN, Black WC, IV, Ebel GD. Genetic drift during systemic arbovirus infection of mosquito vectors leads to decreased relative fitness during host switching. Cell Host Microbe. 2016;19:481–492. doi: 10.1016/j.chom.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- 44.Kuan G, Gordon A, Avilés W, Ortega O, Hammond SN, Elizondo D, Nuñez A, Coloma J, Balmaseda A, Harris E. The Nicaraguan pediatric dengue cohort study: Study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol. 2009;170:120–129. doi: 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balmaseda A, Sandoval E, Pérez L, Gutiérrez CM, Harris E. Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am J Trop Med Hyg. 1999;61:893–897. doi: 10.4269/ajtmh.1999.61.893. [DOI] [PubMed] [Google Scholar]
- 46.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waggoner JJ, Ballesteros G, Gresh L, Mohamed-Hadley A, Tellez Y, Sahoo MK, Abeynayake J, Balmaseda A, Harris E, Pinsky BA. Clinical evaluation of a single-reaction real-time RT-PCR for pan-dengue and chikungunya virus detection. J Clin Virol. 2016;78:57–61. doi: 10.1016/j.jcv.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJV, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis. 2016;22:1295–1297. doi: 10.3201/eid2207.160326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaenisch T, Tam DTH, Kieu NTT, Van Ngoc T, Tran Nam N, Van Kinh N, Yacoub S, Chanpheaktra N, Kumar V, See LLC, Sathar J, Sandoval EP, Alfaro GMM, Laksono IS, Mahendradhata Y, Sarker M, Ahmed F, Caprara A, Benevides BS, Marques ETA, Magalhaes T, Brasil P, Netto M, Tami A, Bethencourt SE, Guzman M, Simmons C, Quyen NTH, Merson L, Dung NTP, Beck D, Wirths M, Wolbers M, Lam PK, Rosenberger K, Wills B. Clinical evaluation of dengue and identification of risk factors for severe disease: Protocol for a multicentre study in 8 countries. BMC Infect Dis. 2016;16:120. doi: 10.1186/s12879-016-1440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanciotti RS, Kosoy OL, Laven JJ, Panella AJ, Velez JO, Lambert AJ, Campbell GL. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordeiro MT, Brito CAA, Pena LJ, Castanha PMS, Gil LHVG, Lopes KGS, Dhalia R, Meneses JA, Ishigami AC, Mello LM, Alencar LXE, Guarines KM, Rodrigues LC, Marques ETA. Results of a Zika Virus (ZIKV) immunoglobulin M-specific diagnostic assay are highly correlated with detection of neutralizing anti-ZIKV antibodies in neonates with congenital disease. J Infect Dis. 2016;214:1897–1904. doi: 10.1093/infdis/jiw477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Confirmation of Zika-specific, RNA-dependent LAMP amplification.
Fig. S2. Control samples containing each virus RNA.
Fig. S3. Alignment of Zika LAMP primer sequences with target sequences in all known Zika virus sequences.
Fig. S4. Direct detection of Zika virus in mosquitoes.
Fig. S5. Confirmation of Zika-specific LAMP amplification from mosquito homogenate.
Fig. S6. Zika LAMP assay is dependent upon RNA.
Fig. S7. Visual LAMP assay of mosquito homogenates.
Fig. S8. Limits of PRVABC59-LAMP detection match RT-PCR.
Fig. S9. LAMP assay detects PRVABC59 virus in human biofluids.
Table S1. Zika LAMP primers.
Table S2. Plaque titration of split, infected mosquitoes.
Table S3. Plaque titration of midguts, salivary glands, and carcass.
Table S4A. LAMP assay LOD for genome copies.
Table S4B. TaqMan LOD for genome copies.
Table S5A. Plasma 1:1000 LOD.
Table S5B. Plasma 1:1000 turbidity LOD.
Table S6. Blood 1:1000 LOD.
Table S7. Urine 1:1000 LOD.
Table S8. Saliva 1:1000 LOD.
Table S9. Semen 1:1000 LOD.
Table S10. Nicaraguan serum and RNA samples.
Table S11. Brazilian plasma and RNA samples.