Abstract
Many chronic neurodegenerative disorders share a common pathogenic mechanism involving the aggregation and deposition of misfolded proteins. Recently, it was shown that these aggregated proteins could be transferred from one cell to another via extracellular nanovesicles called exosomes. Initially thought to be a means of cellular waste removal, exosomes have since been discovered to actively participate in cell-to-cell communication. Importantly, various inflammatory and signaling molecules, as well as small RNAs are selectively packaged in these vesicles. Considering the important role of environmental manganese (Mn) in Parkinson’s disease (PD)-like neurological disorders, we characterized the effect of Mn on exosome content and release using an MN9D dopaminergic cell model of PD, which was generated to stably express wild-type human α-synuclein (αSyn). Mn exposure (300 μM MnCl2) for 24 h induced the release of exosomes into the extracellular media prior to cytotoxicity, as determined by NanoSight particle analysis and electron microscopy. Strikingly, Western blot analysis revealed that Mn treatment in αSyn-expressing cells increases the protein Rab27a, which regulates the release of exosomes from cells. Moreover, next-generation sequencing showed more small RNAs in exosomes isolated from Mn-exposed cells than the control exosomes. Our miRNA profiling analysis led to the discovery of increased expression of certain miRNAs previously shown to regulate key biological pathways, including protein aggregation, autophagy, inflammation and hypoxia. Collectively, our results provide a glimpse of Mn’s role in modulating extracellular miRNA content through exosomal release from dopaminergic neuronal cells and thus potentially contributing to progressive neurodegeneration. Further characterization of extracellular miRNAs and their targets will have major impacts on biomarker discovery and translational strategies for environmentally linked neurodegenerative diseases including PD.
Keywords: exosome, manganese, α-synuclein, extracellular RNAs, neurotoxicity, Parkinson diseases, protein aggregation, miRNA biomarkers, translational research
Graphical Abstract

1. Introduction
Chronic exposure to high concentrations of the metal manganese (Mn) can cause neurotoxicity and manganism, which is a neurological syndrome consisting of movement abnormalities that shares many Parkinsonian features (Aschner et al., 2009; Dobson et al., 2004; Kwakye et al., 2015). Manganism does not present as clinical Parkinson’s disease (PD) because it lacks nigrostriatal dopaminergic neuron damage and the classic response to levodopa. In addition to high dose exposures, low dose exposures through contaminated air, food and water have been shown to cause neurobehavioral abnormalities (Fulk et al., 2017; Mergler et al., 1999; Racette et al., 2017; Sen et al., 2011). Unlike PD, Mn accumulates in and damages the globus pallidus rather than the substantia nigra (SN). Neurochemically, Mn exposure affects the GABAergic neurotransmitter system more than dopaminergic neurotransmission (Guilarte et al., 2006). Given the Mn deposition in the globus pallidus, the correspondingly increased T1-weighted MRI signal in the extra-pallidal basal ganglia (caudate and putamen) has been studied as a potential marker of neurotoxicity associated with Mn exposure (Criswell et al., 2015; Sen et al., 2011). In clinical settings, differential diagnosis of Mn-induced Parkinsonism is primarily based on the Unified Parkinson Disease Rating Scale, Motor Subsection 3 (UPDRS3). However, the UPDRS3 scale can only be applied when motor symptoms are present and MRI is too expensive to be widely accessible throughout the world as an effective early biomarker for Parkinsonism.
To bridge the gap in affordable, reliable quantitative diagnostic tests for these neurological disorders, molecular biomarker discovery becomes an increasingly important goal if we are to diagnose Parkinsonism in early, presumably asymptomatic stages. Current candidate biomarkers are heavily based on individual proteins related to the pathogenesis of PD in CSF and brain tissues, which if done antemortem, involves invasive techniques and surgeries. Hence, we urgently need to develop and validate noninvasive screening tests for the early detection of neurodegenerative diseases. In this context, blood-based assays for proteins, antibodies and circulating extracellular RNAs could prove particularly fruitful as biomarkers in diagnosing neurodegenerative diseases.
Micro-RNA (miRNA) is a class of non-coding RNA (ncRNA), whose final product is an approximately 18- to 22-nucleotide long functional RNA molecule that represses translation and regulates degradation of their target mRNAs by binding complementary regions of messenger transcripts. Recent findings in various disease processes ranging from cancer to cardiovascular disease have found that miRNAs play a crucial role in disease pathogenesis and have potential as biomarkers and therapeutic agents (Eacker et al., 2009; Lin and Gregory, 2015). Exploring miRNA involvement in PD has focused on miRNA expression in the midbrain and its role in the functioning of dopaminergic neurons. In late 2007, a study determined that miR-133b, which is specifically expressed in healthy midbrain dopaminergic neurons, was deficient in midbrain tissues isolated from PD patients (Kim et al., 2007). Since then, several miRNAs controlling autophagy regulation (miR-124, miR-320a and miR-325) and α-synuclein synthesis (miR-7 and miR153) in experimental models of PD have been identified (Alvarez-Erviti et al., 2013; Martins et al., 2011; Saugstad, 2010). More recently, Khoo et al. (Khoo et al., 2012) identified PD-predictive biomarkers through an analysis of global miRNA expression patterns of circulating miRNAs in PD patients and age-matched healthy individuals.
Importantly, miRNAs are also found in blood (Chen et al., 2008; Lawrie et al., 2008; Mitchell et al., 2008). Such circulating miRNAs are remarkably stable even under harsh conditions (Chen et al., 2008; Mitchell et al., 2008), thus making peripheral blood a potential non-invasive source of circulating miRNAs to facilitate biomarker discovery in humans. Exosomes are nano-sized vesicles (50–150 nm) that are released from many cell types into the extracellular space (EL Andaloussi et al., 2013; Thery et al., 2002; Zondler et al., 2017). These vesicles are widely distributed in various body fluids and can easily cross the blood–brain and other barriers. Tremendous interest has emerged over the years in exosome research because of its potential role in disease pathogenesis and biomarker discovery (Alderton, 2015; Anastasiadou and Slack, 2014; Couzin, 2005; Minton, 2015; Thery et al., 2002). Numerous examples exist of miRNAs as putative biomarkers of occupational and environmental exposures to neurotoxicants, including polycyclic aromatic hydrocarbons (Deng et al., 2014), dioxin (Feitelson and Lee, 2007), metal-rich particulate matter (Volinia et al., 2006), arsenic (Dai et al., 2009), and many others (Vrijens et al., 2015). Growing evidence suggests that circulating miRNAs play a significant role in the pathogenesis of many chronic diseases, including PD. For example, differences have been found in miRNA expression in circulating plasma miRNAs of PD patients compared to control subjects (Cardo et al., 2013; Khoo et al., 2012; Kim et al., 2007).
Recently, several miRNAs regulating the synthesis of α-synuclein (αSyn), the hallmark protein of PD, as well as autophagy and apoptosis, were identified in experimental models of PD (Alvarez-Erviti et al., 2013; Heman-Ackah et al., 2013; Kabaria et al., 2015; Wang et al., 2015). Yet, despite its prevalence, the effect of metals on extracellular miRNAs, specifically the mechanisms by which Mn exerts its neurotoxic effects and its relationship with αSyn secretion and transmission in the context of human health are not well studied thus far. Hence, in this study, we use Mn as an environmental neurotoxicant to examine its effects on αSyn aggregation, secretion and the cell-to-cell transmission of extracellular miRNAs via exosomes to manipulate recipient cell gene expression. To further elucidate the molecular mechanisms of Mn-induced neurotoxicity, we used miRNA deep-sequencing and custom miRNA PCR array technology to investigate the role of exosomes in Mn neurotoxicity.
2. Materials and Methods
2.1 Chemicals and reagents
Fetal bovine serum (FBS), L-glutamine, Lipofectamine 2000, Alexa fluorophore-tagged secondary antibodies, Hoechst nuclear stain, penicillin, streptomycin, and other cell culture reagents were purchased from Life Technologies (Gaithersburg, MD). Mouse β-actin antibody, DMEM media, Mn chloride (MnCl2.4H20) and all other chemicals were obtained from Sigma (St. Louis, MO). Antibodies for Rab27a, Caspase-3 and αSyn (Syn 211) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The antibody against αSyn (Syn1) was purchased from BD Bioscience and anti-GFP was obtained from Abcam (Boston, MA). Western blot supplies and the Bradford protein assay kit were purchased from Bio-Rad Laboratories (Hercules, CA). Exosome-depleted FBS was purchased from SBI Biosciences (Mountain view, CA).
2.2 Cell culture and stable expression of human wild-type αSyn
For αSyn release and exosome isolation experiments, we created a GFP-tagged αSyn stably expressing MN9D dopaminergic cell line. Expression plasmids for human full-length αSyn-pMAXGFP and control pMAXGFP vectors (Lonza) were transfected into MN9D cells using Lipofectamine 2000 reagent (Invitrogen) and grown in DMEM (D5648; Sigma) supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% FBS. For stable transfection, MN9D cells were selected after culturing in 400 μg/ml of geneticin for one week post-transfection, and then selected cells were cultured in media supplemented with 200 μg/ml of geneticin to maintain the stable transfection. GFP-positive αSyn-expressing (MN9D_SynGFP) and vector control (MN9D_EVGFP) cells were further selected by FACSAria III (BD Bioscience) high-speed sorting flow cytometer to obtain homogeneously transgene-expressing cell populations.
2.3 Western blotting
Whole-cell lysates or exosome lysates were prepared using modified RIPA buffer containing protease and phosphatase inhibitor cocktail (Thermo Scientific, Waltham, MA), as described previously (Brenza et al., 2016; Harischandra et al., 2014). Cell lysates containing equal amounts of protein were separated on a 12–15% SDS-polyacrylamide gel. After separation, proteins were electro-blotted onto a nitrocellulose membrane, and nonspecific-binding sites were blocked by treating with LI-COR blocking buffer. Syn-1 (BD Bioscience), GFP (Abcam), Syn 211 (Santa Cruz), Rab27a (Santa Cruz), Caspase-3 (Cell Signaling), and β-actin (Sigma) primary antibodies were used to blot the membranes. Membranes were then developed with IR800-conjugated anti-rabbit or Alexa Fluor 680-conjugated anti-mouse secondary antibody for 1 h at RT. Western and slot blot images were captured with the Odyssey IR Imaging system (LI-COR) and data were analyzed using Odyssey 2.0 software.
2.4 Immunocytochemistry (ICC)
For ICC, cells were plated on 50 μg/mL PDL-coated 12-mm glass coverslips. Cells were washed with PBS and incubated in 4% paraformaldehyde for 30 min at RT. After fixing, the cells were washed with PBS and incubated in blocking agent (2% BSA, 0.05% Tween-20, and 0.5% Triton X-100 in PBS) for 45 min. Cells were then incubated with antibodies against human αSyn (Syn 211; Santa Cruz, 1:500) and GFP (Abcam 1:2000) overnight at 4°C or the cytoskeleton marker Phalloidin (Alexa Fluor 647 phalloidin, Invitrogen) for 30 min at RT. After primary incubation, the cells were washed and incubated in the dark for 90 min with Alexa-488 and -555 dye-conjugated secondary antibodies (Invitrogen, 1:1000). Hoechst 44432 was used as a nuclear stain and the coverslips were then mounted on glass slides and viewed with 63× and 43× oil objectives using a Leica DMIRE2 confocal microscope.
2.5 Exosome isolation
Various methods for isolating exosomes from biological fluids have been developed by exploiting specific traits of exosomes, such as their density, shape, size and surface proteins (Sunkara et al., 2016). However, each of these isolation methods has its own unique set of advantages and limitations (Kowal et al., 2016). In this regard, coupling differential centrifugation with filtration isolates an appreciable quantity and purity of exosomes. Briefly, MN9D_SynGFP and MN9D_EVGFP cells at 70–80% confluency were treated with or without 300 μM Mn in medium containing 10% exosome-depleted FBS for 24 h. For each exosome sample, conditioned media from two T175 flasks were pooled with an original seeding density of 10 million cells per flask. After treatment, the cell culture supernatant was collected and spun at 300 x g for 10 min to remove cells and then again at 10,000 x g for 15 min to exclude cell debris from the supernatant. The resulting media was then passed through a 0.22-μm syringe filter (Millipore) to remove any remaining particles or cell debris, and the filtrate was centrifuged at 100,000 x g for 90 min using a Beckman Optima L-100 XP ultracentrifuge. The pellet containing exosomes was washed once with cold PBS and centrifuged again at 100,000 x g for 90 min using a Beckman optima MAX ultracentrifuge.
2.6 Nanoparticle tracking analysis (NTA)
Ultracentrifuged exosome samples were used for NTA, as previously described (Zamanian et al., 2015). Briefly, isolated exosomes were resuspended in 500–1000 μl PBS, from which approximately 300 μl was loaded into the sample chamber of an LM10 unit (NanoSight, Amesbury, UK) using a disposable syringe. Sample durations of 30–60 sec per sample were analyzed with NTA 2.3 software (NanoSight). Samples containing higher numbers of exosomes were diluted before the analysis and their relative concentrations were then calculated according to the dilution factor.
2.7 Transmission electron microscopy (TEM)
Purified exosomes were resuspended in 200 μl PBS. We mixed 20 μl of each sample with uranyl acetate 2% (w/v), incubated them for 5 min, and then 5 μl were applied to carbon-coated copper grids. Images were taken using a JEOL 2100 200-kV scanning and transmission electron microscope (STEM) with a Thermo Fisher Noran System 6 elemental analysis system. TEM was operated at 80 kV and images were obtained at 18000–20000× magnification.
2.8 Exosomal RNA extraction and characterization
For detection of RNA species in exosome samples isolated from MN9D_SynGFP and control MN9D_EVGFP cells that were either Mn-stimulated or vehicle-treated, we used the miRCURY RNA isolation kit (Exiqon; 30010) per manufacturer’s protocols. After extraction, a Nanodrop spectrophotometer was used initially to determine the concentration and quality of the RNA preparation. Then RNA quality, yield, and size of exosomal small RNA were analyzed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA) with the Agilent RNA 6000 Nano Kit as described previously (Zamanian et al., 2015).
2.9 Next-generation sequencing of exosomal RNA
After Mn stimulation, whole conditioned media was sent to SBI Bioscience for exosome isolation and preparation of exoRNA libraries compatible with sequencing on the Illumina NGS platform. The amplified indexed libraries were then resolved in a polyacrylamide gel from which the desired bands are excised in a streamlined gel purification method. The recovered amplified libraries were then analyzed on the Illumina sequencing platforms (HiSeq, MiSeq, and Genome Analyzer II) with about 300M reads per sample to generate substantial sequencing depth. After sequencing, reads were de-multiplexed based upon their unique index sequence and then assigned to their appropriate input samples identities. We used the Maverix exoRNA analysis pipeline (Maverix Biomics; San Mateo, CA) along with open-source tools, including bowtie for alignment and DESeq for differential expression analysis. Raw reads were trimmed and filtered based on quality to remove adaptor sequence and spike-in controls. The scores used for trimming and filtering were specific to the sequencing platform. Sequences were then mapped to a reference genome of choice to determine sequence identities and relative abundances of various RNA types such as ncRNA, antisense transcripts, and miRNA.
2.10 miRNA profiling and data validation
Total RNA that includes small non-coding miRNA was purified from exosomes isolated from untreated and Mn-stimulated MN9D_SynGFP cells using miRCURY RNA isolation kit (Exiqon; 30010) following manufacturer’s instructions. RNA quality was determined post-isolation using a Nanodrop 2000 instrument. For miRNA validation studies, SYBR green-based custom miScript miRNA PCR Array (Qiagen, MD) was used. Our miRNA PCR array consisted of 48 miRNAs comprising 43 previously identified miRNAs and five internal controls including miRTC controls (miRTC: Reverse transcriptase qPCR; Qiagen) at 45 thermal cycles while PPC (Positive qPCR Controls, Qiagen) Ct values ranged from 18 to 20. Our assay also consisted of C. elegans microRNA cel-miR-39 as synthetic microRNA spike-in control, and SNORD61 and SNORD68 were used as endogenous reference RNA for relative quantification of target miRNAs. However, in the final data analysis, SNORD61 was used as a housekeeping miRNA. RNA was reverse transcribed using the miScript II RT Kit (Qiagen) in HiSpec buffer, and diluted cDNA was mixed with universal primer and SYBR Green dye before being added to the wells of 96-well plates containing lyophilized primer. The plates were run on a Stratagene MX3005P instrument (Agilent technologies) and the expression of individual miRNAs was analyzed using the obtained Ct values. The plate assay was performed per manufacturer’s protocol and fold changes in miRNA expression were calculated using the Ct value of the normalizing control.
2.11 Statistical analysis
Prism 4.0 software was used to analyze data from two or more independent experiments, each with n ≥ 6. Student’s t-test was used to find significant differences between treatment and control groups. Differences with p ≤ 0.05 were considered significantly different.
3. Results
3.1 Manganese induces exosome release from an αSyn transgenic cell model of PD
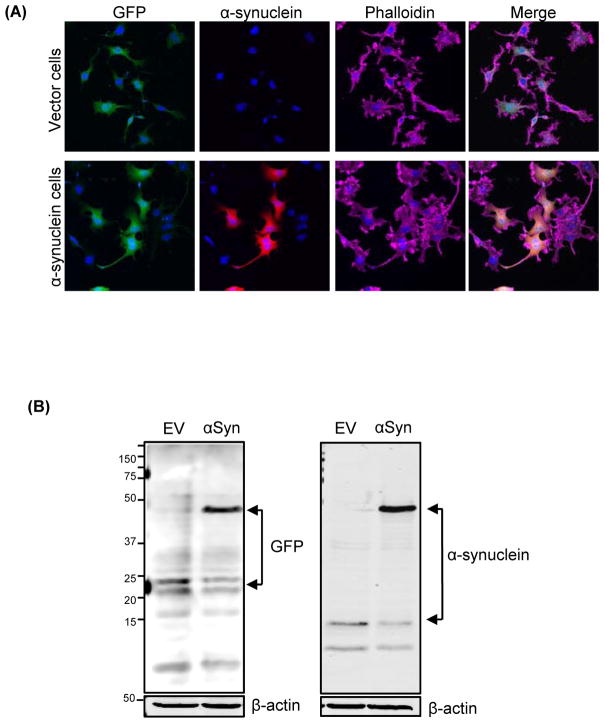
Transgenic dopaminergic neuronal cell lines constitutively expressing GFP-tagged human wild-type αSyn or vector control were established by stably transfecting MN9D mouse dopaminergic cells with either plasmid pMAXGFP_αSyn (MN9D_SynGFP or αSyn cells) or pMAXGFP_EV (MN9D_EVGFP or Vector cells). The MN9D cell model is widely used in PD research (Ay et al., 2015; Jin et al., 2014). ICC analyses with human-specific αSyn antibody (Syn 211) indicate that >90% of the αSyn cells were positive for the transgene human αSyn, and that all Vector cells were positive for GFP but not for human αSyn (Fig. 1A). Western blot analyses indicate a low-level expression of endogenous αSyn in both cell types and strong expression of the higher molecular weight (~45 kDa), GFP-tagged αSyn in αSyn cells (Fig. 1B).
Figure 1. Generation of GFP-tagged stable human α-synuclein-expressing MN9D cells.
(A) Immunocytochemical analysis depicting stably expressed GFP-tagged wild-type human αSyn protein in MN9D dopaminergic cells. The αSyn-expressing cells exhibited strong ubiquitous expression of αSyn, whereas Vector cells showed no detectable αSyn immunoreactivity. (B) Stable expression of αSyn was determined by Western blot analysis. A 45-kDa band corresponding to the molecular mass of GFP-fused human αSyn was detected in αSyn-expressing cells, whereas no human-specific αSyn expression appeared in Vector cells. However, both human αSyn-expressing and control cells showed low levels of endogenous mouse αSyn expression.
The miRNAs carried from cell-to-cell can silence corresponding mRNAs in target cells (Stoorvogel, 2012). Exosome biogenesis and release are mainly regulated by Rab GTPases, while Rab27a and Rab27b function in multivesicular bodies (MVB) docking at the plasma membrane (Ostrowski et al., 2010). Therefore, we exposed both αSyn cells and Vector cells to 300 μM Mn for 24 h and analyzed the expression of Rab27a, a GTPase responsible for MVB fusion with the plasma membrane and exosome release. Mn exposure markedly increased Rab27a expression in αSyn cells when compared via Western blot to Vector cells exposed to Mn under same conditions (Fig. 2A). This result suggests that Mn can modulate the cellular endosomal sorting mechanism to induce exosome vesicles. To determine whether Mn exposure induces exosome release, we collected the extracellular media following Mn exposure and isolated exosomes by ultracentrifugation method. To characterize the exosomes, NanoSight particle analyses were performed. The diameter of our isolated vesicles, 129 ± 7.05 nm (Fig. 2B), was comparable and consistent with previously published reports on cell culture-derived exosomes (Danzer et al., 2012; Emmanouilidou et al., 2010). To further confirm that these extracellular vesicles manifest the characteristic morphology of exosomes, we analyzed these vesicles with TEM. Our TEM micrographs readily detect the distinct exosome morphology and size (Fig. 2C) often reported in the literature (Alvarez-Erviti et al., 2011; Chen et al., 2013; Danzer et al., 2012), indicating that Mn exposure indeed induces cells to release exosome vesicles into their micro-environment.
Figure 2. Manganese induces exosome release from neuronal cells.
(A) Western blot analysis shows increased Rab27a expression upon Mn exposure in αSyn-expressing cells (upper panel) and GFP-tagged αSyn in both vehicle- and Mn-treated αSyn cells (lower panel). (B) NanoSight-generated histogram of particle size and abundance of isolated exosomes. Brownian movement of exosome particles (white arrows) captured by NanoSight LM10. (insert). (C) Electron micrographs of isolated exosomes revealing that transmission electron microscopy readily detected exosomes in ultracentrifuged conditioned medium.
3.2 Manganese-induced exosomes contain miRNAs
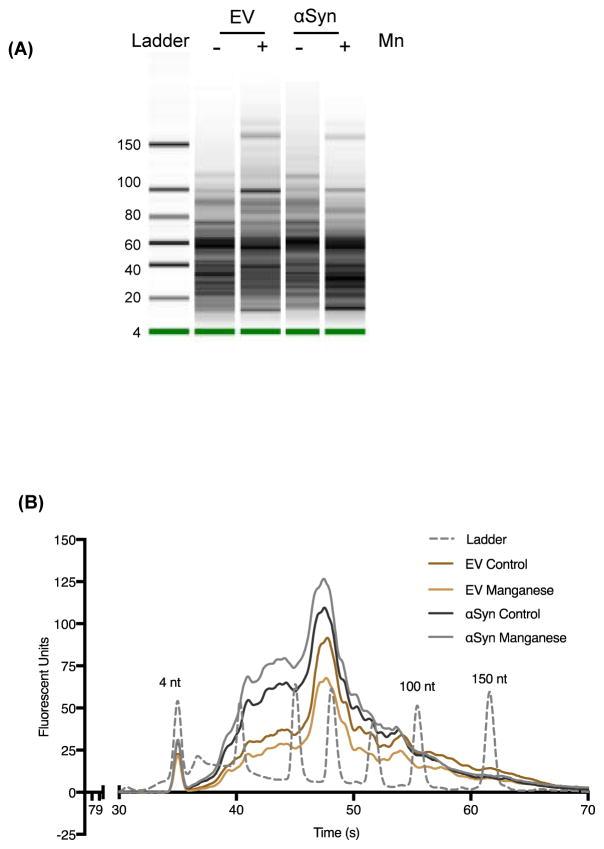
Since exosomes reportedly contain a unique RNA profile distinct from that of host cells (Valadi et al., 2007), we further analyzed the exosomes for small noncoding RNAs, such as miRNAs. Exosomal RNA was isolated using the miRCURY RNA isolation kit and small RNAs were analyzed with the Agilent 2100 Bioanalyzer Lab-on-a-Chip instrument system (Agilent Technologies). The exosomes we isolated do indeed contain small RNAs (Fig. 3A), of which about 86% were positive for miRNAs (Fig. 3B), indicating that these exosomes serve as cargo vessels for the transport and release of RNAs into extracellular spaces during Mn exposure. Interestingly, the total RNA load was higher in αSyn cell exosomes as compared to exosomes isolated from control vector cells.
Figure 3. Exosomal small RNA quantification through bioanalyzer.
(A) Exosomal RNAs determined using the Agilent Small RNA Chip. The small RNAs were predominant in the exosomal RNAs. (B) Small RNA analysis and quantification via bioanalyzer showing data for RNAs isolated from Vector cells and αSyn cells treated with Mn or vehicle. Exosomes isolated from αSyn cells contain significant amounts of small RNAs in the 25–150 nt range (4, 100 and 150 nt reference peaks labeled). Data represented here as average trace of four separate experiments for each group.
3.3 Manganese exposure leads to differential expression of small RNAs
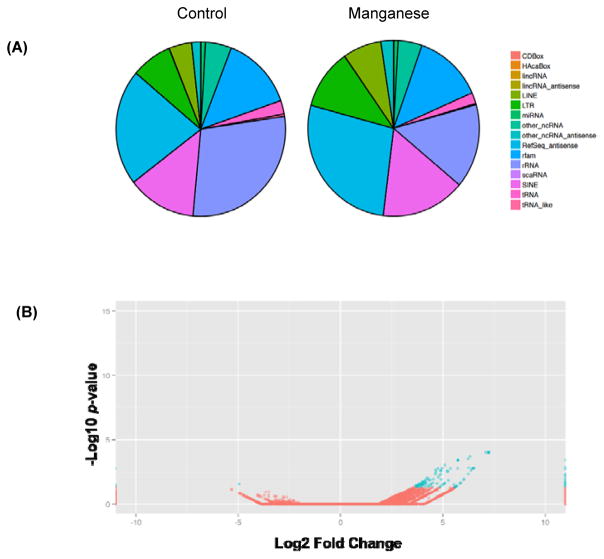
To examine the identity and abundance of small RNA changes upon Mn exposure, we performed next-generation exosome RNA sequencing in exosomes. Given the role of αSyn in Parkinsonism and other types of synucleinopathies, we focused on αSyn cells and the role of Mn in modulating small RNA cargo in exosomes. We isolated exosomes from αSyn cells treated with Mn or vehicle, purified the exoRNA and built Illumina NGS libraries. These libraries were then sequenced using 1x 50-bp single-end Illumina HiSeq NGS runs to provide enough depth for RNA sequence identification. Our small RNA subtype profiles (Fig. 4A) revealed gross changes in miRNA, ribosomal RNA (rRNA), transfer RNA (tRNA) and ncRNA expression, indicating that Mn exposure altered small RNA profiles in exosomes. We also compared the miRNA changes in Mn-stimulated exosomes to those in vehicle-treated exosomes. After filtering out those with low miRNA expression in either sample, our analysis indicates (Fig. 4B) substantially greater changes in miRNA expression in Mn-stimulated exosomes. Our volcano plot analysis of log2 fold changes in normalized expression with control group indicates that 43 miRNAs (blue dots) were differentially expressed among all miRNAs (red and blue dots) in the Mn-stimulated exosome sample. Together, these results indicate that Mn exposure significantly alters extracellular miRNAs released via exosomes.
Figure 4. Manganese exposure leads to differential expression of small RNAs.
(A) Percentage of reads mapping to a variety of small RNA types. Comparison of RNA profiles between vehicle- (left) and Mn-stimulated (right) samples reveals different abundances of small RNA subtypes. (B) A volcano plot shows fold change and significance for every miRNA tested. Plotted points identify both the magnitude and significance of differential expression for all miRNA for vehicle- and Mn-stimulated samples. P-values are corrected for multiple comparisons with p ≤ 0.05 (blue dots) considered significant.
3.4 Validation of miRNA targets through custom miScript PCR array technology
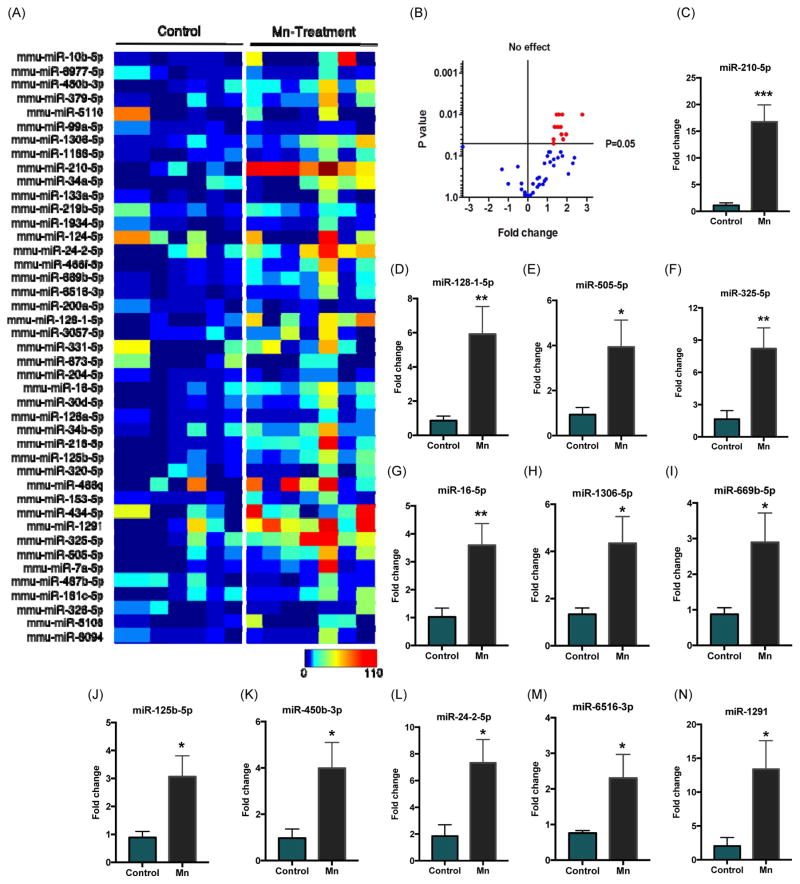
After identifying differentially expressed miRNAs through RNA-sequencing, we further performed miRNA qPCR to validate our RNA-Seq results. Our custom miRNA PCR array consisted of 48 miRNAs, including 43 previously identified miRNAs and five internal controls for expression normalization. Normalized data were analyzed with MATLAB to generate a heat map (Fig. 5A). Averaged fold changes were displayed as a volcano plot to show the fold change and significance of all differentially expressed miRNAs (Fig. 5B). P-values are corrected for multiple comparisons and changes with p≤0.05 were marked significant (Red dots). Among several miRNAs, twelve miRNAs including miR-210-5p, miR128-1-5p, miR-505-5p, miR-325-5p, miR-16-5p, miR-1306-5p, miR-669b-5p, miR-125b-5p, miR-450b-3p, miR-24-2-5p, miR-6516-3p and miR-1291 significantly increased to 2.5- to 15-fold (Fig. 5C–N) in Mn-stimulated exosomes in comparison to vehicle-stimulated exosomes.
Figure 5. Validation of miRNA targets through custom miScript PCR array technology.
(A) Heatmap visualizations of exosomal RNA expression through custom PCR array for all 43 differentially expressed miRNAs identified during RNA-Seq analysis. Each box represents fold change per biological replicate (n = 7 per group) for both vehicle- or Mn-stimulated exosome samples. (B) Volcano plot shows fold change and significance for all 43 miRNAs tested (blue and red dots). P-values are corrected for multiple comparisons with p ≤ 0.05 (red dots) considered significant. (C–N) Fold-change histograms for significantly upregulated miRNAs. Each group represented as mean ± SEM from seven separate experiments (* p < 0.05, ** p < 0.01 and *** p < 0.001).
4. Discussion
In the present study, we show that Mn exposure induces the release of extracellular miRNAs by altering the exosomal pathway in a model of PD. The miRNAs are small non-coding RNAs that post-transcriptionally regulate many metazoan genes by binding to partially complementary sites in target messenger RNAs. It is predicted that miRNAs account for 1–5% of the human genome and regulate at least 30% of protein-coding genes (Macfarlane and Murphy, 2010; Rajewsky, 2006). Although little is currently known about the specific targets and biological functions of miRNAs, they have emerged as key regulators of gene expression involved in several neurodegenerative disorders, including PD and Alzheimer’s disease. For instance, miRNAs post-transcriptionally regulate αSyn, the major component of Lewy bodies in PD (Kordower et al., 2008). Previously, miR-7 and miR-153 were shown to target and downregulate αSyn expression (Doxakis, 2010; Junn et al., 2009), while inhibition of miR-34b and miR-34c elevated αSyn levels leading to the formation of αSyn-containing aggregates (Kabaria et al., 2015).
Evidence also suggests that exposure to environmental neurotoxicants such as pesticides and metals can increase αSyn expression and protein aggregation, thereby contributing to the pathogenesis of PD (Betarbet et al., 2000; Giasson and Lee, 2000; Rokad et al., 2016). Thus, it is crucial to understand the molecular mechanisms underlying αSyn aggregation driven by environmental factors. Additionally, exposure to environmental toxicants is thought to account for most sporadic PD cases, however, the effect of neurotoxic chemicals on extracellular miRNA in regulating gene expression and disease pathogenesis is poorly studied. Chronic occupational exposure to high levels of Mn by welding, mining, dry cell battery manufacturing and Mn-rich agro-chemicals is known to cause PD-like manganism, which is similarly characterized by tremors, rigidity and psychosis (Olanow, 2004; Racette et al., 2012; Racette et al., 2016). Furthermore, Mn neurotoxicity leads to neuronal apoptosis, αSyn upregulation and aggregation in cell culture as well as in animal models (Cai et al., 2010; Harischandra et al., 2015; Hirata, 2002), providing direct experimental evidence for a close relationship between Mn and the αSyn proteins implicated in PD. To further understand the role of Mn in miRNA dysregulation, we created wild-type human αSyn constitutively expressing dopaminergic cells, which we exposed to inorganic Mn. We subsequently collected exosomes from conditioned culture media, and for the first time, analyzed environmental neurotoxicant-driven miRNA changes in exosomes obtained from an in vitro model of Mn overexposure.
Our results from the wild-type human αSyn-expressing cell culture model clearly demonstrates that Mn exposure significantly upregulated Rab27a (Fig. 2A), the small GTPase responsible for membrane fusion of multivesicular bodies (Ostrowski et al., 2010; Pfeffer, 2010), releasing exosomes to the extracellular environment. TEM analysis of enriched exosomes clearly shows characteristic ‘saucer-like’ morphology (Fig. 2C), or flattened sphere, bounded by a lipid bi-layer (Thery et al., 2002) that collapses during sample preparation for TEM (Raposo et al., 1996). Further analysis also indicates exosome particle diameter ranging between 50 – 150 nm, generally the accepted size range for exosomes (Greening et al., 2015; Lane et al., 2015). Complementary to TEM, our NTA of exosomes determined the size distribution well within that for exosomes with the mean particle size of 129 nm (Fig. 2B). These characteristics are consistent with the observed size and morphology of internal vesicles in multivesicular endosomes. However, given that the size range of exosomes often overlaps with a class of extracellular vesicles that result from outward budding of the cell membrane, such as microvesicles, it is likely that some of these shedding vesicles are also present in our exosome preparations.
Recently, miRNAs were identified in exosomes, suggesting that miRNAs can be transferred between cells through exosomes, thereby mediating target gene repression in neighboring or distant cells and subsequently modulating recipient cells. Exosomal miRNAs also play an important role in disease progression. For example, they can stimulate angiogenesis and facilitate metastasis in cancers (Anastasiadou and Slack, 2014; Melo et al., 2014), promote neurodegeneration (Thompson et al., 2016; Vella et al., 2016), and regulate host-parasitic interactions (Buck et al., 2014; Zamanian et al., 2015). Furthermore, it appears that the loading of miRNAs into exosomes is a selective process since exosomal miRNA signatures do not simply reflect the miRNA composition of the parent cell, but rather a distinct set of miRNAs assembled to carry out specific biological functions (Gibbings et al., 2009; Koppers-Lalic et al., 2014). In our study, we isolated and enriched small RNAs from exosomes and analyzed the small RNA content through the Agilent 2100 bioanalyzer (Fig. 3A). While exosomes isolated from both Vector and αSyn cells contained significant amounts of RNAs in the range of 20–150 nt, the RNA load present in exosomes isolated from Vector cells (both untreated and Mn-treated) was smaller compared to that in αSyn cell exosomes (both untreated and Mn treatment). This is particularly interesting, considering gene triplication and mutations are directly involved with familial PD (Singleton et al., 2003; Stefanis, 2012), while αSyn gene-environment interactions are speculated to govern the sporadic form of PD (Rokad et al., 2016; Ross and Smith, 2007). To further investigate possible Mn-induced miRNA changes in αSyn-expressing cells, we carried out genome-wide miRNA expression profiling for all known miRNAs using Illumina sequencing. Our global small RNA expression data profiles showed significant changes in miRNA, rRNA, antisense long non-coding RNAs (lncRNA) and short interspersed elements (SINE) in exosomes isolated from αSyn cells treated with Mn when compared to vehicle-treated cells (Fig. 4A). To determine potential changes in the miRNAs, we used DESeq to calculate fold change and statistical significance for all differentially expressed genes between vehicle- and Mn-treated αSyn cells. Volcano plots revealed 43 miRNAs differentially expressed in Mn-stimulated αSyn exosomes relative to control exosomes.
Following the identification of differentially expressed miRNAs through RNA-Seq, we performed miRNA qPCR to validate our RNA-Seq results using multiple experimental replicates. Our normalized data identified 12 miRNAs significantly upregulated (Fig. 5C–N) in Mn-stimulated αSyn exosomes when compared to exosomes isolated from vehicle-stimulated αSyn cells. Our data indicate that Mn exposure significantly increased expression of miR-16 (Fig. 5G), a member of the evolutionarily-conserved miR-15/107 family consisting of miR-15a, miR-15b, miR-16, miR-103 and miR-107, miR-195, miR-424, miR-497, miR-503 and miR-646 (Wang et al., 2014). This group of miRNAs shares a sequence (AGCAGC) near the 5′ end that complements with an overlapping list of mRNA targets (Nelson et al., 2011). Members of the miR-15/107 family play key roles in gene regulation involved in cell division, metabolism, stress response and angiogenesis and have been implicated in pathological processes including cancer, cardiovascular and neurodegenerative diseases (Finnerty et al., 2010; Muller et al., 2014). Recently, proteomic analysis of mouse brain tissues identified a number of additional putative miR-16 targets in vivo, including αSyn and transferrin receptor 1 (TfR1) (Parsi et al., 2015). Importantly, the Tf/TfR system facilitates the uptake of Mn (Aschner et al., 2007), which is a known binding partner of αSyn resulting in its aggregation (Uversky et al., 2001). Furthermore, gain-of-function studies in mouse hippocampal HT22 cells with miR-16 mimics show that both αSyn and TfR1 are significantly downregulated in vitro (Parsi et al., 2015). Our analysis also indicated that Mn exposure significantly increased miRNA-128 (Fig. 5D), which is widely and abundantly expressed in the postnatal developing brain, but its functions are not well understood (Lewis, 2014). A recent study revealed that miR-128 plays a crucial role in regulating the excitability of dopamine D1 receptor-expressing neurons in the striatum by targeting ERK2 expression (Tan et al., 2013), and increasing its expression protects against chemically-induced Parkinsonian motor deficits in mice (Tan et al., 2013). However, in the AAV-αSyn rat model of PD, miR-128 represses transcription factor EB (TFEB), a major transcriptional regulator of the autophagy-lysosome pathway, increasing the vulnerability of nigral and ventral tegmental area dopamine neurons to αSyn toxicity (Decressac et al., 2013).
Our analysis also showed a significant increase in miR-210, which is known to promote a hypoxic phenotype (Grosso et al., 2013). Others reported elevated levels of miR-210 in circulating miRNAs in plasma samples of patients suffering from cerebral ischemia and stroke (Ouyang et al., 2013). Importantly, miR-210 decreases mitochondrial function by directly targeting COX10 (cytochrome c oxidase assembly protein), an important factor of the mitochondrial electron transport chain, leading to the generation of reactive oxygen species (ROS) (Chen et al., 2010). Given that Mn neurotoxicity is mainly associated with mitochondrial dysfunction, leading to decreased oxidative phosphorylation and oxidative stress (Rao and Norenberg, 2004), this may reveal a miRNA-mediated link between cellular stress, oxidative phosphorylation, ROS and metal homeostasis. Our previous studies show that Mn-induced mitochondrial dysfunction activates PKCδ-dependent pro-apoptotic signaling pathways in dopaminergic neurons (Afeseh Ngwa et al., 2011; Kitazawa et al., 2005). Our results also showed that Mn exposure significantly increased miR-325, which previously was shown to be upregulated during TNF-α-mediated inflammation (Chakraborty et al., 2015) and hypoxia-reoxygenation injury (Bo et al., 2014), leading to oxidative stress. Importantly, miR-325 is known to suppress the apoptosis repressor with caspase recruit domain (ARC), an anti-autophagic protein, thus promoting the autophagic cascade (Bo et al., 2014).
Based on function classification, some miRNAs are classified as pro-inflammatory molecules, and some are anti-inflammatory (Ponomarev et al., 2013). Our miRNA data show a marked Mn-induced increase of miR-125b (Fig. 5J), a known pro-inflammatory miRNA, providing evidence for a novel mechanism whereby environmental toxicants modulate miRNA expression and exert inflammatory responses. The pro-inflammatory miRNA-125 increases M1 activation of macrophages by targeting IRF4, increasing responsiveness to IFN-γ, and most importantly for antigen presentation, by upregulating MHC class II, CD40, CD80 and CD86 (Chaudhuri et al., 2011). We also identified multiple miRNAs, including miR-450b and miR-669b (Fig. 5I), previously shown to target known aging-associated pathways (Dimmeler and Nicotera, 2013; Inukai and Slack, 2013). Interestingly, a recent plasma-based circulating miRNA biomarker study also identified miR-450b and miR-505 (Fig. 5K and 5E) as reliable biomarkers with highest predictive power of 91% sensitivity and 100% specificity in diagnosing PD (Khoo et al., 2012). In addition, our Mn treatment also markedly increased miR-24 (Fig. 5L), which has been shown to be upregulated in PD, MSA (Vallelunga et al., 2014), and other prominent synucleinopathies in humans. We also detected other miRNAs, including miR-1306, miR-6516 and miR-1291, which have not been directly linked to a disease mechanism, and thus would warrant further study. Although some miRNAs, such as miR-128, seem to have pleiotropic effects in biological systems, the role of this miRNA in Mn neurotoxicity has yet to be defined.
In conclusion, we report that the environmental neurotoxicant Mn can induce the release of miRNA-containing exosomes into the extracellular milieu, potentially modulating host cell gene expression. Our results identified a pool of miRNAs previously shown to regulate multiple biological pathways, including mitochondrial function, inflammation, autophagy and protein aggregation. Future studies should investigate how to utilize these miRNAs as biomarkers of Mn toxicity in humans as well as to identify new mechanistic pathways contributing to degenerative processes in environmentally linked Parkinsonisms.
Highlights.
Exposure to an environmental neurotoxin, Mn, can actively modulate exosome RNA cargo.
Mn induces Rab27a to increase extracellular RNA via exosomes.
Mn treatment alters exosomal small RNA profiling.
Mn upregulates several miRNAs related to key biological pathways to neurodegeneration.
Exosomal miRNAs could represent potential biomarker of Mn-induced toxicity.
Acknowledgments
This work was supported by National Institutes of Health R01 grants [ES026892, ES019267 and NS074443] to A.G.K. The W. Eugene and Linda Lloyd Endowed Chair to A.G.K is also acknowledged. Authors also like to thank Dr. Michael Cho (Iowa State University) for use of ultracentrifuge and NanoSight instruments and Mr. Chi-Fu Yen (Iowa State University) for assistance in MATLAB data analysis. We also thank Gary Zenitsky for assistance in preparing this manuscript.
Footnotes
Conflict of interest
A.G.K. and V.A. are shareholders of PK Biosciences Corporation (Ames, IA), which is interested in identifying novel biomarkers and potential therapeutic targets for PD. A.G.K and V.A. do not have any competing interests impacted by the present work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afeseh Ngwa H, Kanthasamy A, Gu Y, Fang N, Anantharam V, Kanthasamy AG. Manganese nanoparticle activates mitochondrial dependent apoptotic signaling and autophagy in dopaminergic neuronal cells. Toxicol Appl Pharmacol. 2011;256(3):227–240. doi: 10.1016/j.taap.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderton GK. Diagnosis: Fishing for exosomes. Nature reviews Cancer. 2015;15(8):453. doi: 10.1038/nrc3990. [DOI] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, Sargent IL, Wood MJ, Cooper JM. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiology of disease. 2011;42(3):360–367. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson’s disease. Cell death & disease. 2013;4:e545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadou E, Slack FJ. Cancer. Malicious exosomes. Science. 2014;346(6216):1459–1460. doi: 10.1126/science.aaa4024. [DOI] [PubMed] [Google Scholar]
- Aschner M, Erikson KM, Herrero Hernandez E, Tjalkens R. Manganese and its role in Parkinson’s disease: from transport to neuropathology. Neuromolecular Med. 2009;11(4):252–266. doi: 10.1007/s12017-009-8083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschner M, Guilarte TR, Schneider JS, Zheng W. Manganese: recent advances in understanding its transport and neurotoxicity. Toxicol Appl Pharmacol. 2007;221(2):131–147. doi: 10.1016/j.taap.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay M, Jin H, Harischandra DS, Asaithambi A, Kanthasamy A, Anantharam V, Kanthasamy AG. Molecular cloning, epigenetic regulation, and functional characterization of Prkd1 gene promoter in dopaminergic cell culture models of Parkinson’s disease. J Neurochem. 2015;135(2):402–415. doi: 10.1111/jnc.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3(12):1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Bo L, Su-Ling D, Fang L, Lu-Yu Z, Tao A, Stefan D, Kun W, Pei-Feng L. Autophagic program is regulated by miR-325. Cell Death Differ. 2014;21(6):967–977. doi: 10.1038/cdd.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenza TM, Ghaisas S, Ramirez JE, Harischandra D, Anantharam V, Kalyanaraman B, Kanthasamy AG, Narasimhan B. Neuronal protection against oxidative insult by polyanhydride nanoparticle-based mitochondria-targeted antioxidant therapy. Nanomedicine. 2016 doi: 10.1016/j.nano.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, Ceroni A, Babayan SA, Blaxter M, Ivens A, Maizels RM. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nature communications. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Yao T, Zheng G, Chen Y, Du K, Cao Y, Shen X, Chen J, Luo W. Manganese induces the overexpression of alpha-synuclein in PC12 cells via ERK activation. Brain Res. 2010;1359:201–207. doi: 10.1016/j.brainres.2010.08.055. [DOI] [PubMed] [Google Scholar]
- Cardo LF, Coto E, de Mena L, Ribacoba R, Moris G, Menendez M, Alvarez V. Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. Journal of neurology. 2013;260(5):1420–1422. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Zawieja DC, Davis MJ, Muthuchamy M. MicroRNA signature of inflamed lymphatic endothelium and role of miR-9 in lymphangiogenesis and inflammation. Am J Physiol Cell Physiol. 2015;309(10):C680–692. doi: 10.1152/ajpcell.00122.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O’Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. Journal of immunology. 2011;187(10):5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Hogan MC, Ward CJ. Purification of exosome-like vesicles from urine. Methods Enzymol. 2013;524:225–241. doi: 10.1016/B978-0-12-397945-2.00013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29(30):4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- Couzin J. Cell biology: The ins and outs of exosomes. Science. 2005;308(5730):1862–1863. doi: 10.1126/science.308.5730.1862. [DOI] [PubMed] [Google Scholar]
- Criswell SR, Nelson G, Gonzalez-Cuyar LF, Huang J, Shimony JS, Checkoway H, Simpson CD, Dills R, Seixas NS, Racette BA. Ex vivo magnetic resonance imaging in South African manganese mine workers. Neurotoxicology. 2015;49:8–14. doi: 10.1016/j.neuro.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatology international. 2009;29(7):749–754. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, Vanderburg CR, McLean PJ. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Molecular neurodegeneration. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110(19):E1817–1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Huang S, Zhang X, Zhang W, Feng J, Wang T, Hu D, Guan L, Li J, Dai X, Deng H, Zhang X, Wu T. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environmental health perspectives. 2014;122(7):719–725. doi: 10.1289/ehp.1307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmeler S, Nicotera P. MicroRNAs in age-related diseases. EMBO Mol Med. 2013;5(2):180–190. doi: 10.1002/emmm.201201986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson AW, Erikson KM, Aschner M. Manganese neurotoxicity. Ann N Y Acad Sci. 2004;1012:115–128. doi: 10.1196/annals.1306.009. [DOI] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285(17):12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10(12):837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30(20):6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer letters. 2007;252(2):157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulk F, Succop P, Hilbert TJ, Beidler C, Brown D, Reponen T, Haynes EN. Pathways of inhalation exposure to manganese in children living near a ferromanganese refinery: A structural equation modeling approach. Sci Total Environ. 2017;579:768–775. doi: 10.1016/j.scitotenv.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Lee VM. A new link between pesticides and Parkinson’s disease. Nat Neurosci. 2000;3(12):1227–1228. doi: 10.1038/81737. [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11(9):1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noel A, Pouyssegur J, Barbry P, Mazure NM, Mari B. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell death & disease. 2013;4:e544. doi: 10.1038/cddis.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilarte TR, Chen MK, McGlothan JL, Verina T, Wong DF, Zhou Y, Alexander M, Rohde CA, Syversen T, Decamp E, Koser AJ, Fritz S, Gonczi H, Anderson DW, Schneider JS. Nigrostriatal dopamine system dysfunction and subtle motor deficits in manganese-exposed non-human primates. Exp Neurol. 2006;202(2):381–390. doi: 10.1016/j.expneurol.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Harischandra DS, Jin H, Anantharam V, Kanthasamy A, Kanthasamy AG. alpha-Synuclein protects against manganese neurotoxic insult during the early stages of exposure in a dopaminergic cell model of Parkinson’s disease. Toxicol Sci. 2015;143(2):454–468. doi: 10.1093/toxsci/kfu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harischandra DS, Kondru N, Martin DP, Kanthasamy A, Jin H, Anantharam V, Kanthasamy AG. Role of proteolytic activation of protein kinase Cdelta in the pathogenesis of prion disease. Prion. 2014;8(1):143–153. doi: 10.4161/pri.28369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heman-Ackah SM, Hallegger M, Rao MS, Wood MJ. RISC in PD: the impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Frontiers in molecular neuroscience. 2013;6:40. doi: 10.3389/fnmol.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y. Manganese-induced apoptosis in PC12 cells. Neurotoxicol Teratol. 2002;24(5):639–653. doi: 10.1016/s0892-0362(02)00215-5. [DOI] [PubMed] [Google Scholar]
- Inukai S, Slack F. MicroRNAs and the genetic network in aging. J Mol Biol. 2013;425(19):3601–3608. doi: 10.1016/j.jmb.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kanthasamy A, Harischandra DS, Kondru N, Ghosh A, Panicker N, Anantharam V, Rana A, Kanthasamy AG. Histone hyperacetylation up-regulates protein kinase Cdelta in dopaminergic neurons to induce cell death: relevance to epigenetic mechanisms of neurodegeneration in Parkinson disease. J Biol Chem. 2014;289(50):34743–34767. doi: 10.1074/jbc.M114.576702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106(31):13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS letters. 2015;589(3):319–325. doi: 10.1016/j.febslet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo SK, Petillo D, Kang UJ, Resau JH, Berryhill B, Linder J, Forsgren L, Neuman LA, Tan AC. Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. J Parkinsons Dis. 2012;2(4):321–331. doi: 10.3233/JPD-012144. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Yang Y, Hirata Y, Kanthasamy A, Kanthasamy AG. Activation of protein kinase C delta by proteolytic cleavage contributes to manganese-induced apoptosis in dopaminergic cells: protective role of Bcl-2. Biochem Pharmacol. 2005;69(1):133–146. doi: 10.1016/j.bcp.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, Wurdinger T, Meijer GA, Pegtel DM. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8(6):1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14(5):504–506. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113(8):E968–977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakye GF, Paoliello MM, Mukhopadhyay S, Bowman AB, Aschner M. Manganese-Induced Parkinsonism and Parkinson’s Disease: Shared and Distinguishable Features. Int J Environ Res Public Health. 2015;12(7):7519–7540. doi: 10.3390/ijerph120707519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RE, Korbie D, Anderson W, Vaidyanathan R, Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci Rep. 2015;5:7639. doi: 10.1038/srep07639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. British journal of haematology. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Lewis S. Neurological disorders: microRNA gets motoring. Nat Rev Neurosci. 2014;15(2):67. doi: 10.1038/nrn3672. [DOI] [PubMed] [Google Scholar]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nature reviews Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Rosa A, Guedes LC, Fonseca BV, Gotovac K, Violante S, Mestre T, Coelho M, Rosa MM, Martin ER, Vance JM, Outeiro TF, Wang L, Borovecki F, Ferreira JJ, Oliveira SA. Convergence of miRNA expression profiling, alpha-synuclein interacton and GWAS in Parkinson’s disease. PLoS One. 2011;6(10):e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergler D, Baldwin M, Belanger S, Larribe F, Beuter A, Bowler R, Panisset M, Edwards R, de Geoffroy A, Sassine MP, Hudnell K. Manganese neurotoxicity, a continuum of dysfunction: results from a community based study. Neurotoxicology. 1999;20(2–3):327–342. [PubMed] [Google Scholar]
- Minton K. Exosomes: Apoptotic beads on a string. Nature reviews Molecular cell biology. 2015;16(8):453. [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Kuiperij HB, Claassen JA, Kusters B, Verbeek MM. MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiology of aging. 2014;35(1):152–158. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Mao G, Wilfred BR, Xie K, Jennings MH, Gao Z, Wang X. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res. 2011;39(18):8163–8172. doi: 10.1093/nar/gkr532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanow CW. Manganese-induced parkinsonism and Parkinson’s disease. Ann N Y Acad Sci. 2004;1012:209–223. doi: 10.1196/annals.1306.018. [DOI] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Stary CM, Yang GY, Giffard R. microRNAs: innovative targets for cerebral ischemia and stroke. Curr Drug Targets. 2013;14(1):90–101. doi: 10.2174/138945013804806424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsi S, Smith PY, Goupil C, Dorval V, Hebert SS. Preclinical Evaluation of miR-15/107 Family Members as Multifactorial Drug Targets for Alzheimer’s Disease. Mol Ther Nucleic Acids. 2015;4:e256. doi: 10.1038/mtna.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Two Rabs for exosome release. Nat Cell Biol. 2010;12(1):3–4. doi: 10.1038/ncb0110-3. [DOI] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Weiner HL. MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia. 2013;61(1):91–103. doi: 10.1002/glia.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racette BA, Criswell SR, Lundin JI, Hobson A, Seixas N, Kotzbauer PT, Evanoff BA, Perlmutter JS, Zhang J, Sheppard L, Checkoway H. Increased risk of parkinsonism associated with welding exposure. Neurotoxicology. 2012;33(5):1356–1361. doi: 10.1016/j.neuro.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racette BA, Searles Nielsen S, Criswell SR, Sheppard L, Seixas N, Warden MN, Checkoway H. Dose-dependent progression of parkinsonism in manganese-exposed welders. Neurology. 2016 doi: 10.1212/WNL.0000000000003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racette BA, Searles Nielsen S, Criswell SR, Sheppard L, Seixas N, Warden MN, Checkoway H. Dose-dependent progression of parkinsonism in manganese-exposed welders. Neurology. 2017;88(4):344–351. doi: 10.1212/WNL.0000000000003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. L(ou)sy miRNA targets? Nat Struct Mol Biol. 2006;13(9):754–755. doi: 10.1038/nsmb0906-754. [DOI] [PubMed] [Google Scholar]
- Rao KV, Norenberg MD. Manganese induces the mitochondrial permeability transition in cultured astrocytes. J Biol Chem. 2004;279(31):32333–32338. doi: 10.1074/jbc.M402096200. [DOI] [PubMed] [Google Scholar]
- Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokad D, Ghaisas S, Harischandra DS, Jin H, Anantharam V, Kanthasamy A, Kanthasamy AG. Role of neurotoxicants and traumatic brain injury in alpha-synuclein protein misfolding and aggregation. Brain Res Bull. 2016 doi: 10.1016/j.brainresbull.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Smith WW. Gene-environment interactions in Parkinson’s disease. Parkinsonism Relat Disord. 2007;13(Suppl 3):S309–315. doi: 10.1016/S1353-8020(08)70022-1. [DOI] [PubMed] [Google Scholar]
- Saugstad JA. MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab. 2010;30(9):1564–1576. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Flynn MR, Du G, Troster AI, An H, Huang X. Manganese accumulation in the olfactory bulbs and other brain regions of “asymptomatic” welders. Toxicol Sci. 2011;121(1):160–167. doi: 10.1093/toxsci/kfr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Stefanis L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2(2):a009399. doi: 10.1101/cshperspect.a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119(3):646–648. doi: 10.1182/blood-2011-11-389478. [DOI] [PubMed] [Google Scholar]
- Sunkara V, Woo HK, Cho YK. Emerging techniques in the isolation and characterization of extracellular vesicles and their roles in cancer diagnostics and prognostics. Analyst. 2016;141(2):371–381. doi: 10.1039/c5an01775k. [DOI] [PubMed] [Google Scholar]
- Tan CL, Plotkin JL, Veno MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O’Carroll D, Greengard P, Schaefer A. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science. 2013;342(6163):1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature reviews Immunology. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Thompson AG, Gray E, Heman-Ackah SM, Mager I, Talbot K, Andaloussi SE, Wood MJ, Turner MR. Extracellular vesicles in neurodegenerative disease - pathogenesis to biomarkers. Nat Rev Neurol. 2016;12(6):346–357. doi: 10.1038/nrneurol.2016.68. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J Biol Chem. 2001;276(47):44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Vallelunga A, Ragusa M, Di Mauro S, Iannitti T, Pilleri M, Biundo R, Weis L, Di Pietro C, De Iuliis A, Nicoletti A, Zappia M, Purrello M, Antonini A. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella LJ, Hill AF, Cheng L. Focus on Extracellular Vesicles: Exosomes and Their Role in Protein Trafficking and Biomarker Potential in Alzheimer’s and Parkinson’s Disease. Int J Mol Sci. 2016;17(2):173. doi: 10.3390/ijms17020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environmental health perspectives. 2015;123(5):399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, Wang H, Gong X, He X, Lu G, Lu F, Zhang S. MiR-124 Regulates Apoptosis and Autophagy Process in MPTP Model of Parkinson’s Disease by Targeting to Bim. Brain pathology. 2015 doi: 10.1111/bpa.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Danaher RJ, Miller CS, Berger JR, Nubia VG, Wilfred BS, Neltner JH, Norris CM, Nelson PT. Expression of miR-15/107 family microRNAs in human tissues and cultured rat brain cells. Genomics Proteomics Bioinformatics. 2014;12(1):19–30. doi: 10.1016/j.gpb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M, Fraser LM, Agbedanu PN, Harischandra H, Moorhead AR, Day TA, Bartholomay LC, Kimber MJ. Release of Small RNA-containing Exosome-like Vesicles from the Human Filarial Parasite Brugia malayi. PLoS Negl Trop Dis. 2015;9(9):e0004069. doi: 10.1371/journal.pntd.0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondler L, Feiler MS, Freischmidt A, Ruf WP, Ludolph AC, Danzer KM, Weishaupt JH. Impaired activation of ALS monocytes by exosomes. Immunol Cell Biol. 2017;95(2):207–214. doi: 10.1038/icb.2016.89. [DOI] [PubMed] [Google Scholar]