ABSTRACT
The human fungal pathogen Candida albicans develops drug resistance after long-term exposure to azole drugs in the treatment of chronic candidiasis. Gain-of-function (GOF) mutations in the transcription factor Tac1 and the consequent expression of its targets, drug efflux pumps Cdr1 and Cdr2, are a common mechanism by which C. albicans acquires fluconazole resistance. The mechanism by which GOF mutations hyperactivate Tac1 is currently unknown. Here, we define a transcriptional activation domain (TAD) at the C terminus of Tac1. GOF mutations within the Tac1 TAD, outside the context of full-length Tac1, generally do not enhance its absolute potential as a transcriptional activator. Negative regulation of the Tac1 TAD by the Tac1 middle region is necessary for the activating effect of GOF mutations or fluphenazine to be realized. We have found that full-length Tac1, when hyperactivated by xenobiotics or GOF mutations, facilitates the recruitment of the Mediator coactivator complex to the CDR1 promoter. Azole resistance and the activation of Tac1 target genes, such as CDR1, are dependent on the Tac1 TAD and subunits of the Mediator tail module. The dependence of different Tac1 target promoters on the Mediator tail module, however, varies widely. Lastly, we show that hyperactivation of Tac1 is correlated with its Mediator-dependent phosphorylation, a potentially useful biomarker for Tac1 hyperactivation. The role of Mediator in events downstream of Tac1 hyperactivation in fluconazole-resistant clinical isolates is complex and provides opportunities and challenges for therapeutic intervention.
KEYWORDS: Candida albicans, fluconazole, Mediator, Tac1
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen that usually exists as a benign human commensal but can cause life-threatening systemic infections in immunocompromised individuals (1–3). Azole drugs are well tolerated and widely used agents to treat fungal infections (4). Long-term exposure to azoles in chronic infections, however, can lead to drug resistance (5, 6). Increased transcription of drug efflux pumps is often observed in clinical isolates of azole-resistant C. albicans and other pathogenic fungi (7, 8). Gain-of-function (GOF) mutations in a zinc cluster transcription factor, Tac1, are commonly responsible for this increased drug efflux in drug-resistant C. albicans (7–10).
Tac1 regulates the expression of ATP-binding cassette transporters Cdr1 and Cdr2 by directly binding to drug-responsive elements (DREs) in their gene promoters (9–12). In azole-susceptible wild-type C. albicans, Tac1 activates CDR1 and CDR2 expression in response to certain xenobiotics, such as fluphenazine (FNZ) and estradiol (EST) (9, 11, 13). GOF mutations, identified in azole-resistant C. albicans isolates, hyperactivate Tac1 and lead to constitutively high expression of CDR1, CDR2, and a number of other genes (9, 10, 14). Overexpression of Cdr1 protein is the major contributor to resistance in TAC1GOF mutant strains (15). Hyperactivation of Tac1 does not seem to be required for its sequence-specific DNA binding, as Tac1 occupancy is observed at the CDR1 promoter in the absence and presence of GOF mutations (9, 11, 12). Another hyperactive Tac1 target, RTA3, encodes a putative lipid translocase that also has recently been found to contribute to azole resistance in TAC1GOF mutant strains (16).
Tac1 is a classical zinc cluster transcription factor (17). Zinc cluster transcription factors typically possess three domains: an N-terminal Zn(II)Cys6 DNA binding domain, a C-terminal transcriptional activation domain (TAD), and an ∼700-amino-acid middle domain that contains the middle homology region (MHR) (7, 9, 17, 18). Deletion of the MHR in several Zn(II)Cys6 transcription factors leads to their hyperactivation (17), but the structure and mechanism of the MHR is not well defined. Previous work clearly described a Tac1 N-terminal Zn(II)Cys6 DNA binding domain, while the MHR and C-terminal TAD are less well defined (7, 9, 17, 18). GOF mutations are usually located within Tac1 middle and putative activation domains (7). Tac1, akin to other zinc cluster transcription factors that regulate drug resistance through activation of efflux pumps (19–21), can also be hyperactivated by xenobiotics such as fluphenazine and estradiol (9, 11, 13). It is unknown whether this mechanism of Tac1 hyperactivation is used physiologically. Moreover, the mechanism(s) by which GOF mutations hyperactivate Tac1, whether all GOF mutations and xenobiotic activators use the same mechanism of hyperactivation, and whether hyperactive Tac1 activates all of its target promoters by the same mechanism are open questions. Specifically, the hyperactive Tac1 transcription coactivator target(s) has yet to be investigated.
In Saccharomyces cerevisiae and the fungal pathogen Candida glabrata, activation of Pdr5 efflux pump expression and the resulting multidrug resistance are driven by the transcription factor Pdr1 (pleiotropic drug resistance regulator), which requires the Mediator complex as a coactivator (22, 23). We hypothesized that Mediator is also a required transcription coactivator for hyperactivated Tac1 in C. albicans. Mediator is a multisubunit complex coactivator in eukaryotes that functions as an intermediary between DNA-bound transcription factors and the RNA polymerase II machinery (24, 25). The S. cerevisiae Mediator core complex is composed of 21 subunits that are structurally categorized as belonging to the head, middle, or tail module (26, 27). The tail module of Mediator is directly targeted by multiple transcription activation domains and plays a critical role in the activation of many highly inducible promoters (24, 25). A submodule composed of Med15, Med3, and Med2 (encoded by the TLO genes in C. albicans) forms a trimeric complex within the fungal Mediator tail module and is key to this module's function in S. cerevisiae and C. albicans (28–31). The Mediator complex also contains a variably associated Cdk8 kinase module, which can regulate transcription through mechanisms that are dependent on, and independent of, its kinase activity (24, 25, 32, 33). Direct phosphorylation of transcription factors by Cdk8 (Ssn3) can result in positive and negative regulation of their activity (32).
The goal of this study was to determine the coactivator requirements for hyperactive Tac1 regulation of the CDR1 and CDR2 promoters and to identify the sequences within Tac1 responsible for conferring these requirements. Our study has found that the Mediator tail module plays an important role in the activation of CDR1 expression and the subsequent fluconazole resistance in TAC1GOF mutant strains. Interestingly, this dependence is not as pronounced at the CDR2 promoter. Defining the Tac1 C-terminal TAD has allowed us to posit a model in which all mechanisms of Tac1 hyperactivation involve the release of a potent activation domain from autoinhibition, which enables the recruitment of Mediator. In light of a recent report of the reduction in C. glabrata azole MIC by a small-molecule inhibitor that blocks interactions between the Pdr1 activation domain and the tail module of Mediator (34), understanding the efflux pump activation mechanisms in other fungal pathogens is of critical importance.
RESULTS
Fluconazole resistance driven by TAC1GOF mutations is decreased in strains lacking an intact Mediator tail module.
We deleted MED3 and MED15 from several TAC1GOF mutant strains and measured the fluconazole MIC (Table 1) to determine whether hyperactive TAC1GOF mutants work through the tail module of Mediator to facilitate azole resistance. The absence of Med3 or Med15 leads to the dissociation of the Med15 trimeric subcomplex from both the S. cerevisiae and C. albicans Mediator tail modules (28–30, 35). The fluconazole MIC (Table 1) was reduced in a med3 null background for all tested TAC1GOF mutant strains by approximately one-half to two-thirds. Fluconazole resistance was decreased in a med15 null background to the same degree as a med3 deletion (Table 1). Deletion of either med3 or med15 only slightly sensitizes a wild-type TAC1 strain to fluconazole (Table 1).
TABLE 1.
Fluconazole MIC in TAC1GOF mutant strains in med3, med15, and cdr1 null backgrounds
| TAC1 allele | Fluconazole MICa (μg/ml) |
||||
|---|---|---|---|---|---|
|
MED3+/+
MED15+/+ |
med3Δ/Δ MED15+/+ |
MED3+/+ med15Δ/Δ CDR1+/+ | |||
| CDR1+/+ | cdr1Δ/Δ | CDR1+/+ | cdr1Δ/Δ | ||
| WT | 0.75 (yLM167) | 0.094 (yLM611) | 0.5–0.75 (yLM232) | —b | 0.5 (yLM240) |
| T225A | 16 (yLM168) | — | 4–6 (yLM233) | — | 6 (yLM241) |
| A736V | 16–24 (yLM169) | 1–1.5 (yLM612) | 6–8 (yLM234) | 1 (yLM615) | 6 (yLM242) |
| G980E | 16–24 (yLM170) | — | 8–12 (yLM235) | — | 8 (yLM243) |
| E461K | 8–12 (yLM490) | — | 4–6 (yLM492) | — | — |
| ΔM677 | 8–12 (yLM502) | — | 4 (yLM504) | — | — |
| N972D | 12 (yLM493) | 0.75–1 (yLM613) | 4 (yLM495) | 1 (yLM616) | — |
| Δ962–969 | 8–12 (yLM499) | — | 3 (yLM501) | — | — |
| N977D | 12–16 (yLM496) | 1 (yLM614) | 4–6 (yLM498) | 1 (yLM617) | — |
Fluconazole MICs were measured by Etest at 30°C on YPD plates. Plates were incubated for 36 h before readout. Intermediate values, between scale marks, are presented as intervals. The exact strain used for each MIC measurement is listed in parentheses.
—, not determined.
TAC1GOF-induced CDR1 activation is severely compromised by med3 or med15 deletion.
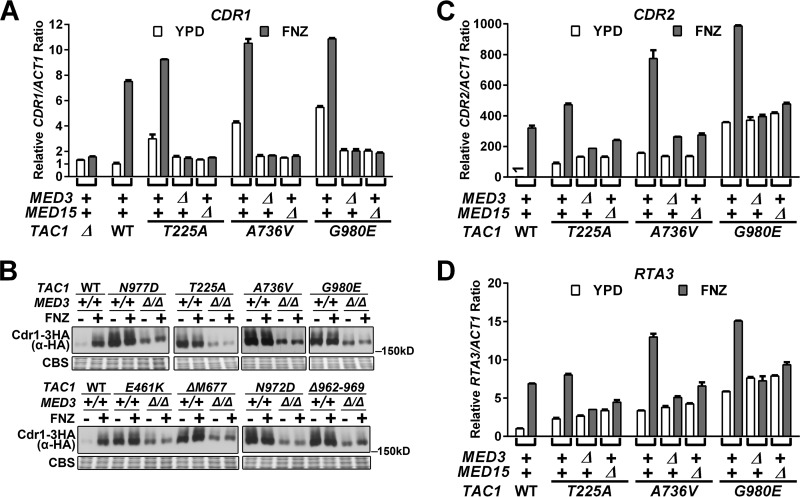
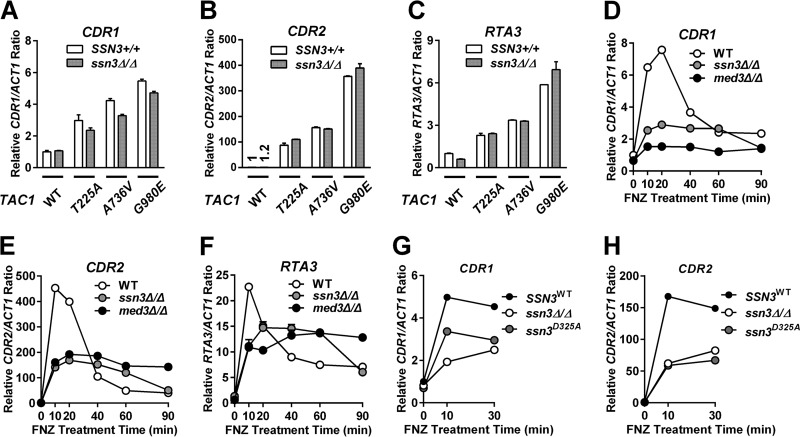
To test whether there was a decrease in the transcription of Tac1 target genes in the TAC1GOF med3Δ/Δ and TAC1GOF med15Δ/Δ strains that corresponded to the decreased fluconazole MIC, we measured CDR1, CDR2, and RTA3 mRNA abundance using reverse transcription-quantitative PCR (RT-qPCR). As shown in Fig. 1A, both TAC1GOF- and fluphenazine-activated levels of CDR1, the major contributor to fluconazole resistance in hyperactive TAC1 strains (15), are reduced to levels seen in the noninduced TAC1WT or tac1Δ/Δ strain (Fig. 1A; see also Table S1 in the supplemental material). We monitored Cdr1 protein by immunoblotting in strains with one copy of CDR1 tagged with a C-terminal 3× hemagglutinin (HA) tag (Fig. 1B). TAC1GOF- and fluphenazine-activated Cdr1 protein levels were reduced in the med3Δ/Δ strains (Fig. 1B). Notably, the TAC1GOF med3Δ/Δ strains expressed a significantly greater amount of Cdr1 protein than the wild-type TAC1/MED3 reference strain (Fig. 1B). This is in contrast to the almost equal amounts of CDR1 mRNA levels found in these strains (Fig. 1A and Table S1). Deletion of cdr1 in the TAC1GOF med3Δ/Δ strains resulted in fluconazole sensitivity comparable to that of TAC1 and MED3 wild-type reference strains (Table 1). This result suggests the difference in MIC between TAC1GOF med3Δ/Δ strains and their nonhyperactivated TAC1 counterparts can be attributed to higher levels of Cdr1 protein. More broadly, there appears to be a nonlinearity in the increases in CDR1 mRNA and protein in which protein levels show a large increase with initial changes in mRNA but eventually level out at higher mRNA levels (Fig. 1A and B). Unlike CDR1, TAC1GOF-driven expression of CDR2 (Fig. 1C and Table S2) and RTA3 (Fig. 1D) is unchanged in a med3 null strain. Deletion of cdr2 in three TAC1GOF med3Δ/Δ strains did not further sensitize them to fluconazole (Table S3), confirming that CDR2 is unlikely to contribute to the resistance observed in the med3Δ/Δ strains. CDR2 and RTA3 might, however, be responsible for the increased fluconazole MIC resulting from TAC1GOF mutations in a cdr1 null background (Table 1). The finding that TAC1GOF-driven CDR2 and RTA3 levels showed little MED3 dependence was supported by the absence of further reductions in MIC when med3 was deleted from TAC1GOF cdr1Δ/Δ strains (Table 1).
FIG 1.
Tac1GOF-mediated gene expression in med3 and med15 deletion strains. (A) RT-qPCR analysis of CDR1 mRNA levels in strains carrying TAC1T225A (ACY67), TAC1A736V (ACY13), or TAC1G980E (ACY71), their med3 deletion derivatives (yLM233, yLM234, and yLM235, respectively), and their med15 deletion derivatives (yLM241, yLM242, and yLM243, respectively) treated with 20 μg/ml fluphenazine (FNZ) or vehicle for 30 min before collection for mRNA extraction. TAC1 wild-type (DSY2937-35) and deletion (DSY2906) strains, in a wild-type Mediator background, were tested in parallel as a reference. The expression level in the uninduced TAC1 MED3 MED15 wild-type strain was set to 1. (B) Immunoblot analysis of lysates from TAC1GOF strains in a CDR1 HA-tagged background (yLM518 [N977D], yLM507 [T225A], yLM509 [A736V], yLM511 [G980E], yLM514 [E461K], yLM513 [ΔM677], yLM516 [N972D], and yLM520 [Δ962–969]) and their med3 deletion derivatives (yLM519, yLM508, yLM510, yLM512, yLM515, yLM557, yLM517, and yLM521, respectively). Cells were treated with 20 μg/ml fluphenazine or vehicle for 30 min. A MED3+/+ strain with wild-type TAC1 and one copy of HA-tagged CDR1 (yLM505) was treated and analyzed in parallel for comparison. Samples were resolved on a 6% SDS-PAGE gel and probed by an anti-HA antibody. Coomassie blue staining (CBS) was used as the loading control. (C and D) RT-qPCR measurements of the indicated samples analyzed from panel A for CDR2 (C) and RTA3 (D) expression. The expression level for each gene in the uninduced TAC1 MED3 MED15 wild-type strain was set to 1.
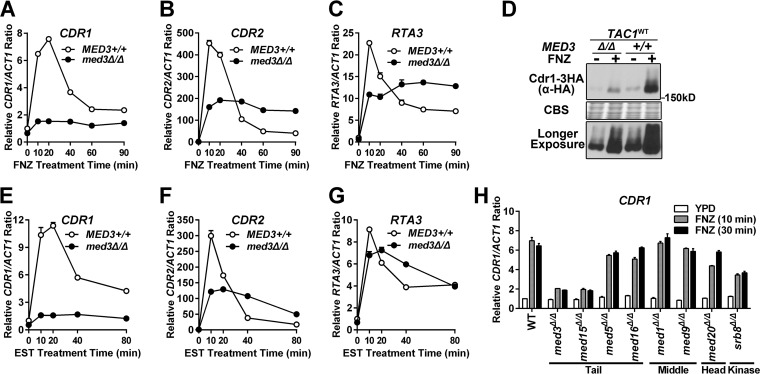
Xenobiotic-activated TAC1 induction of CDR1 is dependent on MED3.
To determine whether induction of Tac1-activated transcription by exposure to xenobiotics had coactivator requirements similar to those of Tac1GOF mutants, we tested the MED3 dependence of FNZ- and EST-hyperactivated Tac1 function. Both fluphenazine (Fig. 2A to C) and estradiol (Fig. 2E to G) treatment of a wild-type TAC1 strain results in a rapid peak of CDR1, CDR2, and RTA3 expression followed by a steady-state increase over the noninduced level. Both the rapid induction and steady-state expression of CDR1, in a wild-type TAC1 background, is strongly decreased by deletion of med3 (Fig. 2A and E). Fluphenazine induction of Cdr1 protein expression is also strongly decreased in the med3 null mutant (Fig. 2D). The rapid peak of fluphenazine (Fig. 2B and C)- and estradiol (Fig. 2F and G)-induced CDR2 and RTA3 expression is compromised in the med3Δ/Δ strain. The steady-state levels (>40 min after induction) of CDR2 and RTA3 expression, however, are somewhat higher in the med3Δ/Δ strain (Fig. 2B, C, F, and G). The impact of Mediator on CDR1 activation by hyperactive Tac1 appears to be specific to subunits of the trimeric tail submodule, as other Mediator subunits encoded by nonessential genes had a much smaller effect on CDR1 expression (Fig. 2H).
FIG 2.
Xenobiotic induction of Tac1 target genes in a med3Δ/Δ strain and other Mediator subunit deletion strains. (A to C) RT-qPCR analysis of fluphenazine (10 μg/ml)-induced transcriptional activation of CDR1 (A), CDR2 (B), and RTA3 (C) in MED3+/+ and med3Δ/Δ strains with wild-type TAC1 (DSY2937-35 and yLM232, respectively). The expression level for each gene in the untreated MED3 wild-type strain was set to 1. (D) Immunoblot analysis of Cdr1 protein levels in response to treatment with 20 μg/ml fluphenazine for 30 min in MED3+/+ and med3Δ/Δ strains carrying one copy of C-terminal 3×HA-tagged CDR1 (yLM505 and yLM506, respectively). The bottom panel shows a longer exposure of the immunoblot shown in the top panel. The samples were resolved on 6% SDS-PAGE gel and probed by an anti-HA antibody. Coomassie blue staining (CBS) was used as the loading control. (E to G) RT-qPCR analysis of 10 μg/ml estradiol (EST)-induced transcriptional activation of CDR1 (E), CDR2 (F), and RTA3 (G) in MED3+/+ and med3Δ/Δ strains with wild-type TAC1 (DSY2937-35 and yLM232, respectively). The expression level for each gene in the uninduced MED3 wild-type strain was set to 1. (H) RT-qPCR analysis of fluphenazine-induced CDR1 activation in Mediator subunit deletion strains (med3Δ/Δ [yLM95], med15Δ/Δ [cTTR01], med5Δ/Δ [AZC32], med16Δ/Δ [AZC34], med1Δ/Δ [AZC46], med9Δ/Δ [AZC42], med20Δ/Δ [AZC44], and srb8Δ/Δ [AZC52]) and a wild-type parental strain (SN152) treated with 10 μg/ml FNZ for the indicated periods of time. Mutants within the same Mediator module were grouped as labeled. The expression level in the untreated Mediator wild-type strain was set to 1.
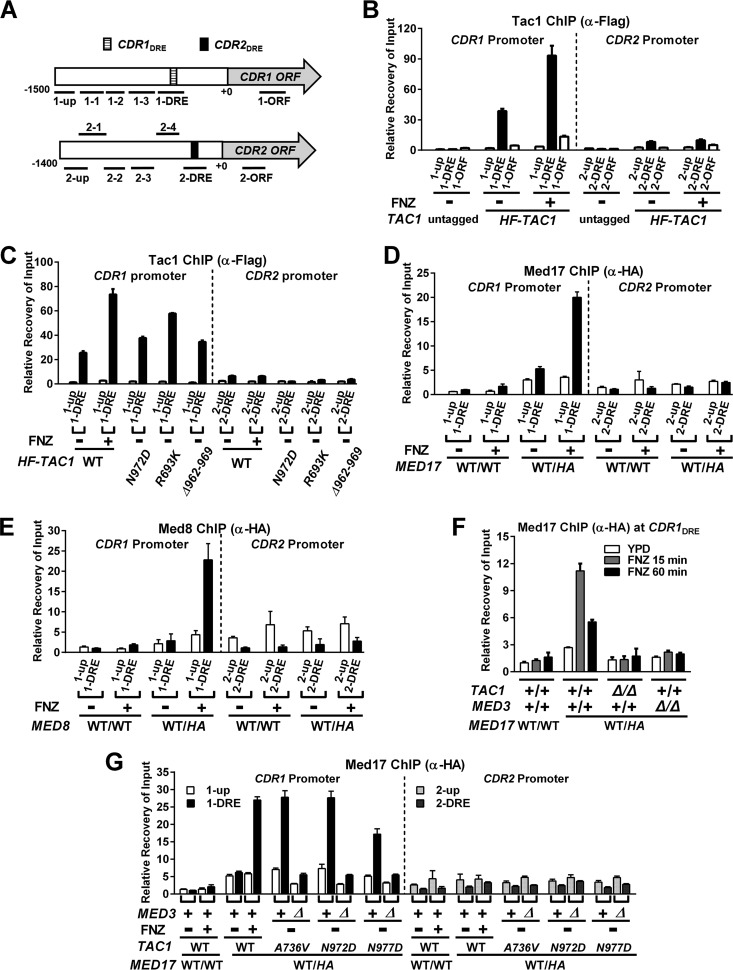
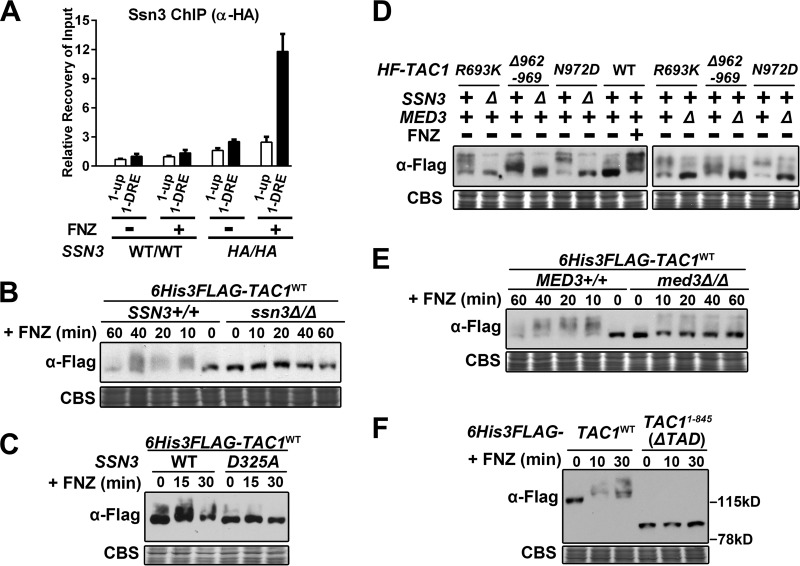
Hyperactivation of Tac1 results in an increased Mediator occupancy at the CDR1 promoter that is dependent on the Mediator tail module.
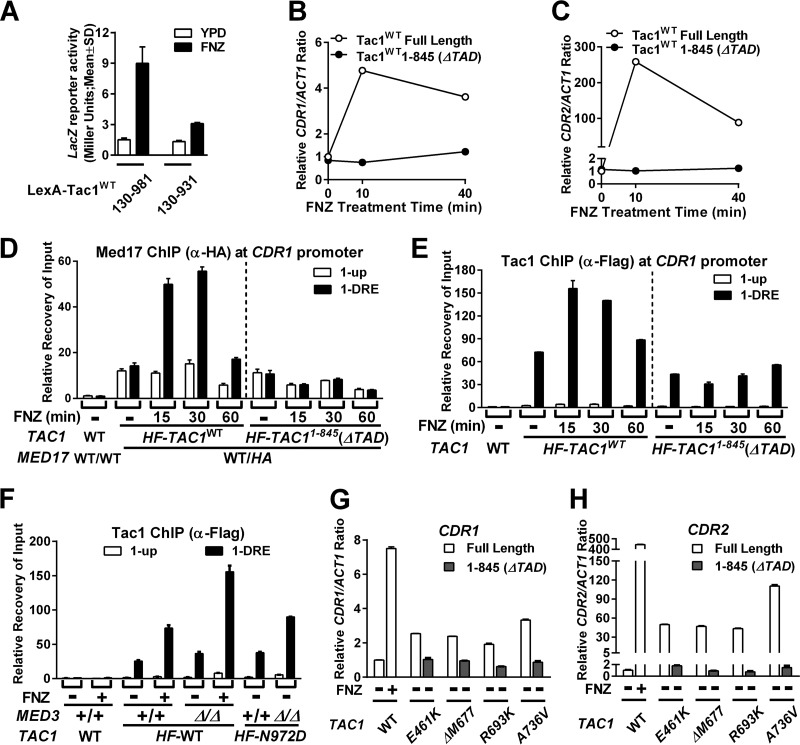
Chromatin immunoprecipitation (ChIP) assays were used to determine whether hyperactivation of Tac1 led to an increase in Mediator occupancy at Tac1-induced promoters, such as CDR1. Tac1 constitutively binds to the CDR1 drug-responsive element (CDR1DRE) (11–13) (Fig. 3A to C), and we found that its occupancy increases when the promoter is activated by FNZ (Fig. 3B) or Tac1GOF (Fig. 3C) mutants. The occupancy of Tac1 at the highly induced CDR2 promoter, however, is very low compared to that of the CDR1 promoter in both the absence and presence of hyperactivation (Fig. 3B and C). The lower enrichment in Tac1 occupancy at the CDR2 DRE (versus the untagged control), despite the high inducibility of CDR2, is similar to what has been observed previously (12). Med17, a highly conserved essential Mediator head module component, was 3×HA tagged on its C terminus to assess Mediator occupancy by ChIP in several strain backgrounds. In the absence of hyperactivation, a small increase (versus an untagged control) in Mediator occupancy is observed at the CDR1 promoter (Fig. 3D). There is no specific enrichment, however, in Mediator occupancy at the CDR1DRE compared with that of a region ∼1.4 kb upstream from the CDR1 open reading frame under these conditions (Fig. 3A, 1-up). Mediator occupancy increases specifically at the CDR1DRE when CDR1 is activated by FNZ treatment (Fig. 3D). Tagging another Mediator subunit, Med8, and performing ChIP showed similar results (Fig. 3E). The increased Mediator occupancy at the CDR1DRE observed after 15 min of induction with FNZ was found to decrease after 60 min of exposure to FNZ (Fig. 3F). This pattern is similar to the temporal induction of CDR1 transcription by fluphenazine (Fig. 2A). This increase in Mediator occupancy, like the FNZ induction of CDR1 expression (Fig. 1), is dependent on TAC1 and MED3 (Fig. 3F). Mediator occupancy at the CDR1 promoter is also increased, in a MED3-dependent manner, when Tac1 is hyperactivated by a variety of GOF mutations (Fig. 3G).
FIG 3.
Tac1 and Mediator occupancy at the CDR1 and CDR2 promoters under conditions of Tac1 hyperactivation and Mediator tail module null mutation. (A) Schematic view of the CDR1 and CDR2 loci showing the relative positions of the previously identified drug-responsive element (DRE) (11) at each promoter and the regions probed for Tac1 or Mediator occupancy in this study. (B) ChIP analysis of Tac1 occupancy at the CDR1 and CDR2 promoters in the presence and absence of 15 min of incubation with 10 μg/ml fluphenazine in strains expressing untagged (DSY2937-35) or N-terminal 6His3Flag-tagged Tac1 (HF-TAC1; yLM485) using an anti-Flag antibody. ChIP experiments were done at least two times and gave consistent results. Results from one representative experiment are shown as the means and standard deviations (SD) calculated from two sets of qPCR measurements. Unless otherwise specified, the ChIP data in this study were analyzed in a similar fashion. (C) ChIP analysis of Tac1 occupancy at the CDR1 and CDR2 promoters in strains expressing 6His3Flag-tagged Tac1GOF mutants (Tac1N972D [yLM534], Tac1R693K [yLM532] or Tac1Δ962–969 [yLM535]) using an anti-Flag antibody. Wild-type Tac1 occupancy in the absence and presence of fluphenazine treatment (10 μg/ml for 15 min) was analyzed in yLM485 for comparison. (D) ChIP analysis of Mediator subunit Med17 occupancy at the CDR1 and CDR2 promoters in the absence and presence of 10 μg/ml fluphenazine using Tac1WT strains without and with one copy of 3×HA-tagged MED17 (DSY2937-35 [WT/WT] and yLM482 [WT/HA], respectively) and an anti-HA antibody. (E) ChIP analysis of Mediator subunit Med8 occupancy at the CDR1 and CDR2 promoters in the absence and presence of 10 μg/ml fluphenazine using Tac1WT strains without and with one copy of 3×HA-tagged MED8 (DSY2937-35 [WT/WT] and yLM560 [WT/HA], respectively) and an anti-HA antibody. (F) ChIP analysis of Mediator subunit Med17 occupancy at the fluphenazine-activated CDR1 promoter in MED17 3×HA-tagged strains lacking either TAC1 (LM559) or MED3 (yLM483) using an anti-HA antibody. (G) ChIP analysis, using an anti-HA antibody, of Mediator subunit Med17 occupancy at the CDR1 and CDR2 promoters in TAC1GOF mutant strains in the MED3 wild-type and null backgrounds. yLM561, yLM563, and yLM565, and their med3 deletion derivatives (yLM562, yLM564, and yLM566, respectively), were constructed by tagging one copy of MED17 with a 3×HA tag in A736V, N972D, and N977D TAC1GOF mutant strains and their med3 deletion derivatives, respectively.
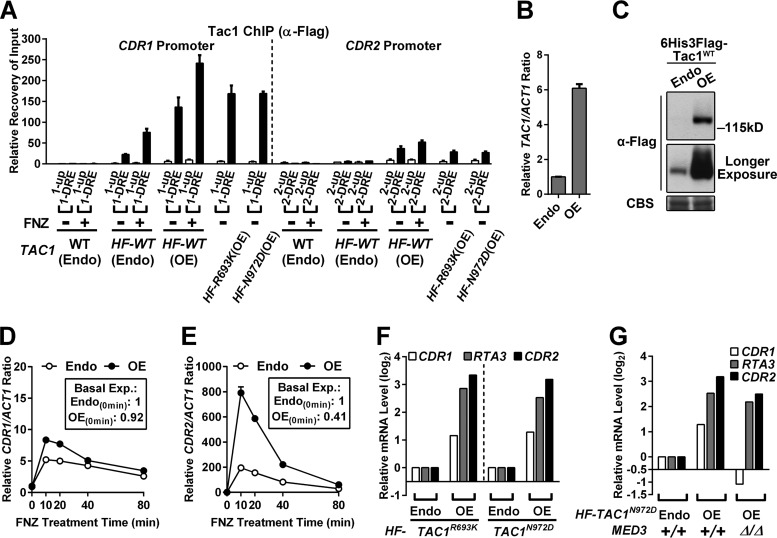
The modest effects of med3 deletion on CDR2 induction compared to CDR1 induction (Fig. 1 and 2) are consistent with ChIP assays that showed little to no effect of Tac1 hyperactivation on Mediator occupancy at the CDR2DRE (Fig. 3). Induction of CDR2 in TAC1GOF mutant strains also failed to result in increased Tac1 occupancy at the CDR2 promoter (Fig. 3C). Scanning the CDR2 (and CDR1) promoter with several additional ChIP primer pairs did not identify alternative Tac1 binding peaks in these promoters (Fig. S1). We were, however, able to detect increased Tac1 occupancy at the CDR2DRE (Fig. 4A) when Tac1 was overexpressed (Fig. 4B and C). The occupancy of overexpressed Tac1 also increased at the CDR1DRE (Fig. 4A). Overexpression of Tac1 does not activate CDR1 and CDR2 expression (Fig. 4D and E, boxes) or increase fluconazole MICs (Table 2) unless Tac1 is hyperactivated by FNZ or a GOF mutation (Fig. 4D to F and Table 2). Overexpression of Tac1, under hyperactivation conditions, results in a much greater increase in CDR2 and RTA3 activation than in that of CDR1 (Fig. 4D to F). The increased Tac1 expression did not change the previously observed differential dependence of the CDR2/RTA3 and CDR1 promoters on Mediator tail module subunits (Fig. 4G). Endogenous levels of Tac1 are more limiting for expression and occupancy of CDR2 and RTA3 promoters than for CDR1. The single-nucleotide variation, which differs from the CDR1DRE and CDR2DRE DNA sequences, does not affect Tac1 binding in vitro (9) or estradiol induction of a DRE-driven reporter in vivo (11). The possible reasons that the CDR2 (and RTA3) promoters exhibit lower Tac1 occupancy include other transcription factors differentially bound at these promoters that influence Tac1 binding or an intrinsically less accessible chromatin structure at these promoters. To determine whether chromatin remodeling played a differential role in the regulation of CDR1, CDR2, and RTA3, we checked their dependence on the Swi/Snf ATP-dependent chromatin remodeling complex (36–38). The absence of Swi/Snf chromatin remodeling activity, by deletion of the snf2 catalytic subunit (39, 40), abolishes FNZ induction of CDR2 and RTA3, as well as their overexpression driven by Tac1GOF mutants (Fig. S2A and S2B). Activity of the CDR1 promoter, induced by Tac1 hyperactivation, is not affected by deletion of snf2 (Fig. S2C). These results are consistent with a more restrictive chromatin structure at the CDR2 and RTA3 promoters compared to CDR1.
FIG 4.
Effect of Tac1 overexpression on Tac1 occupancy at and induction of the CDR1 and CDR2 promoters. (A) ChIP analysis of Tac1 occupancy at the CDR1 and CDR2 promoters in strains expressing N-terminal 6His3Flag-tagged wild-type Tac1 from its endogenous promoter [HF-WT (Endo); yLM485] or a promoter with high constitutive activity [HF-WT (OE); yLM530] and a control strain with native TAC1 [WT (Endo); DSY2937-35] treated with 10 μg/ml fluphenazine or vehicle for 15 min before fixation for ChIP assays using an anti-Flag antibody. Strains overexpressing 6His3Flag-tagged Tac1R693K [HF-R693K(OE); yLM540] or Tac1N972D [HF-N972D(OE); yLM541] were also tested in parallel. Columns representing the recovery rate at the two DREs were marked black. (B and C) RT-qPCR (B) and immunoblot (C) analyses of the HF-Tac1 overexpression strain (OE; yLM530) and a strain expressing an endogenous level of HF-Tac1WT (Endo; yLM485). For panel B, the endogenous expression level was set to 1. For panel C, samples were resolved on a 6% SDS-PAGE gel and probed by an anti-Flag antibody. Coomassie blue staining (CBS) was used as the loading control. (D and E) RT-qPCR time course analysis of fluphenazine induction of CDR1 (D) and CDR2 (E) in strains expressing N-terminal 6His3Flag-tagged wild-type Tac1 at endogenous (Endo; yLM485) or increased (OE; yLM530) levels after treatment with 10 μg/ml fluphenazine. The expression level for each gene in the uninduced endogenous TAC1 strain was set to 1. Numbers in the boxes denote the basal expression levels of CDR1 and CDR2 in the two strains. (F) RT-qPCR analysis of CDR1, CDR2, and RTA3 expression in strains expressing N-terminal 6His3Flag-tagged Tac1R693K or Tac1N972D at either endogenous (Endo; yLM532 and yLM534, respectively) or increased (OE; yLM540 and yLM541, respectively) levels. The expression levels of each gene in yLM532 and yLM534 were individually set to 1. Fold changes in CDR1, CDR2, and RTA3 expression in the Tac1 overexpression strains are presented in logarithmic form (log2). (G) RT-qPCR analysis of CDR1, CDR2, and RTA3 expression in a 6His3Flag-tagged Tac1N972D (HF-TAC1N972D) overexpression strain (yLM541) and its med3 deletion derivative (yLM595). A strain expressing an endogenous level of the same Tac1GOF mutant protein (Endo; yLM534) was tested in parallel for comparison. The abundance of each of the three tested genes in yLM534 was individually set to 1. Fold changes in CDR1, CDR2, and RTA3 expression caused by increased Tac1N972D levels were compared between the MED3+/+ and med3Δ/Δ backgrounds in their logarithmic forms (log2).
TABLE 2.
Fluconazole MIC of strains overexpressing Tac1 variants
| TAC1 allele | FNZb | Fluconazole MICa (μg/ml) |
|
|---|---|---|---|
| Endogenous expression | Overexpression | ||
| WT | − | 0.75 (yLM485) | 0.75 (yLM530) |
| WT | + | 2–3 (yLM485) | 6–8 (yLM530) |
| R693K | − | 8 (yLM532) | 64 (yLM540) |
| N972D | − | 8–12 (yLM534) | 64 (yLM541) |
Fluconazole MICs were measured by Etest at 30°C on YPD plates. Plates were incubated for 36 h before readout. Intermediate values, between scale marks, are presented as intervals. The exact strain used for each MIC measurement is listed in parentheses.
A plus sign indicates the fluconazole MIC was measured on a YPD plate supplemented with 10 μg/ml fluphenazine.
The C terminus of Tac1 encodes a MED3-dependent transcriptional activation domain.
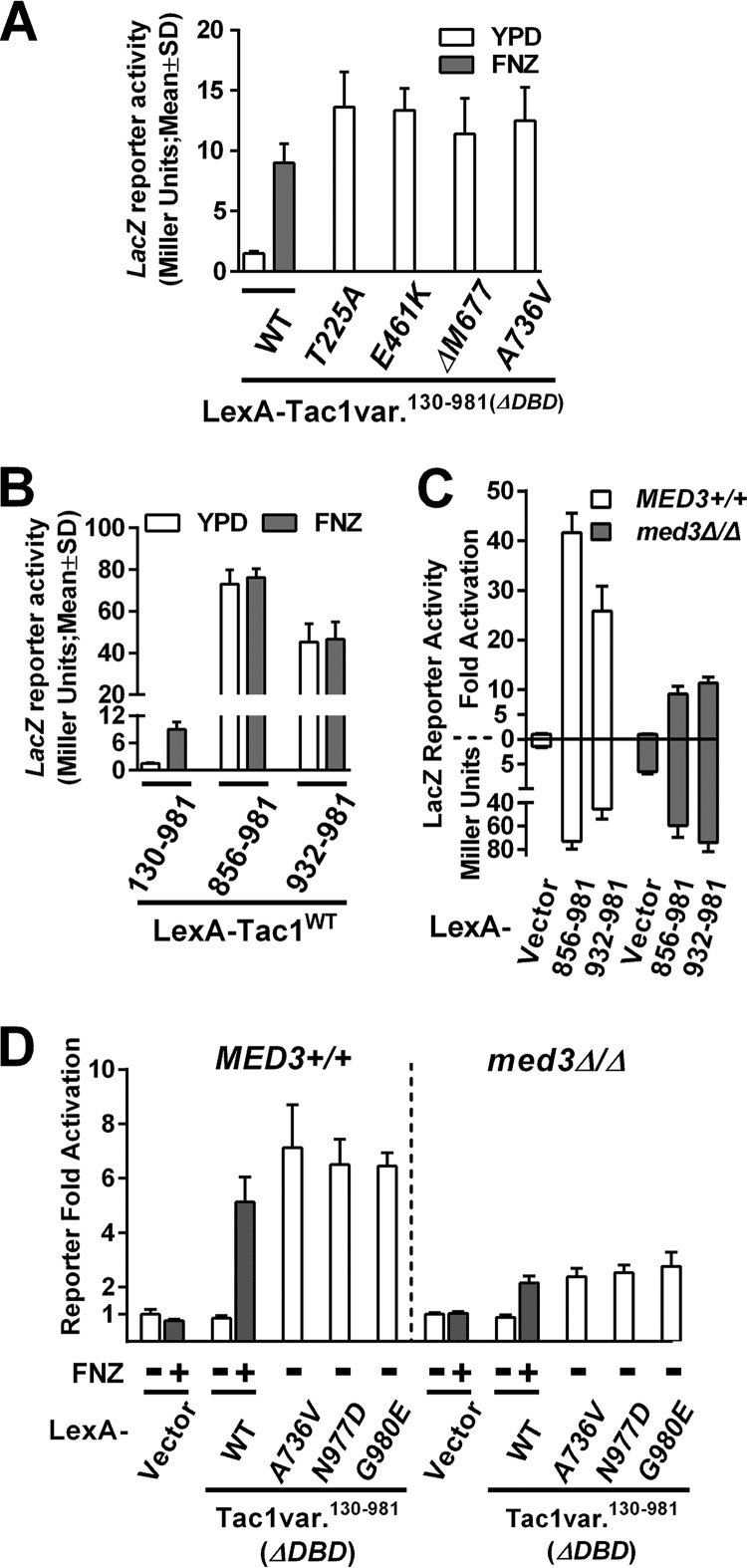
To determine the mechanism by which Tac1 is hyperactivated by GOF mutations, we dissected the posited transcriptional activation domain in the C terminus of Tac1 (7). A C. albicans one-hybrid reporter system (41) was adapted to measure the transcription activation potential of Tac1 fragments and their variants from GOF mutants. Fusion of a Tac1 fragment (amino acids [aa] 130 to 981), lacking only its DNA binding domain (DBD), to the LexA DNA binding domain activated expression of a lacZ reporter, with a LexA binding site upstream, in response to FNZ exposure (Fig. 5A). Fusion of Tac1 (aa 130 to 981) fragments containing individual GOF mutations resulted in activation of the lacZ reporter to a level comparable to that of FNZ induction of the wild-type Tac1 fragment (Fig. 5A). These results showed that the simplified one-hybrid system was suitable for further dissection of hyperactive Tac1 function. One-hybrid assays of two Tac1 C-terminal fragments, a longer (aa 856 to 981) and a shorter (aa 932 to 981) form, revealed highly activated reporter gene expression that was independent of fluphenazine treatment (Fig. 5B). The longer Tac1 C-terminal fragment (aa 856 to 981) is referred to here as the Tac1 TAD. The overall level of reporter activity is markedly higher for the C-terminal fragments than for the induced Tac1 (aa 130 to 981) fragment (Fig. 5B). This suggests that the Tac1 region between aa 130 and 856 negatively regulates the function of the Tac1 TAD. We next tested the MED3 dependence of activation by the LexA-Tac1 fragments in reporter assays. The fold activation of the reporter by the Tac1 TAD fragments relative to a vector control (the LexA DBD alone) is decreased in a med3 null mutant (Fig. 5C). The strains expressing either wild-type LexA-Tac1130–981 with FNZ exposure or a LexA-Tac1130–981 GOF mutant all showed decreased fold activation of the reporter in the absence of med3 (Fig. 5D). Interestingly, the fold activation of the reporter in the med3 null mutant is influenced by an observed derepression of reporter expression in the absence of activation (Fig. 5C, LexA vector control). This finding differentiates the behavior of the reporter from the regulation of endogenous Tac1 targets. When comparing levels of fold activation, however, the LexA-Tac1 constructs exhibit a dependence on MED3 in the reporter assay that is more similar to that of the CDR1 promoter than that of the CDR2/RTA3 promoters. The changes in the reporter assay are unlikely to be a result of changes in the expression of the LexA fusion constructs in the Mediator mutants. LexA fusion expression is controlled by the C. albicans ACT1 promoter. ACT1 is a gene that we have previously shown to be unaffected by the Mediator mutations tested in this paper. Since epitope tagging of the LexA-Tac1 fusions was incompatible with their functionality, it is difficult to rule out the possibility that differences in protein amount between the constructs in different strains influenced these results. However, Western blots of the epitope-tagged strains did not reveal any difference between the constructs (data not shown).
FIG 5.
Dissection of C-terminal Tac1 TAD using a β-galactosidase reporter assay. (A) β-Galactosidase reporter assays in a strain with a fusion of the LexA DBD to a Tac1 fragment containing aa 130 to 981 and a reporter with a LexA binding site upstream of lacZ [LexA-Tac1130–981(ΔDBD); yLM568] in the presence and absence of fluphenazine. Reporter activity was also measured in strains expressing LexA-Tac1130–981 variants, each carrying one TAC1GOF mutation (T225A [yLM569], E461K [yLM570], ΔM677 [yLM571], or A736V [yLM572]) without FNZ treatment. At least three independent transformants were tested for each construct, and the results are presented as means (column height) and standard deviations (error bar). (B) β-Galactosidase reporter assay on strains expressing LexA-Tac1130–981 (yLM568), LexA-Tac1856–981 (yLM579), or LexA-Tac1932–981 (yLM580) in the absence and presence of fluphenazine. (C) β-Galactosidase reporter assays of Tac1 C-terminal fragments in MED3+/+ one-hybrid strains expressing LexA-vector (vector; yLM567), LexA-Tac1856–981 (856–981; yLM579), or LexA-Tac1932–981 (932–981; yLM580) and in med3Δ/Δ one-hybrid strains individually expressing the same constructs (yLM588, yLM593, and yLM594, respectively). Results, in Miller units, are reported in the plot below the x axis. Data, after normalization to the corresponding basal reporter activity, are presented as the fold activation in the plot above the x axis. (D) β-Galactosidase reporter assays in MED3+/+ one-hybrid strains expressing LexA vector (yLM567) or individual LexA-Tac1130–981 variants (wild-type [yLM568], A736V [yLM572], N977D [yLM575] or G980E [yLM576]) and in a med3Δ/Δ one-hybrid strain expressing the same constructs (yLM588 and yLM589 to yLM592) with or without FNZ treatment. Data are presented as the fold activation after normalization to the basal reporter activity (not shown in this panel) in the corresponding strain background.
Tac1-mediated gene activation and Mediator occupancy at the CDR1 promoter requires the Tac1 TAD.
Deletion of a C-terminal TAD-containing fragment from the LexA-Tac1 fusion protein (LexA-Tac1130–931) reduces LacZ reporter expression in response to FNZ treatment compared to the full-length construct (LexA-Tac1130–981) (Fig. 6A). In the native context, expression of a C-terminally truncated Tac1 mutant (aa 1 to 845) fails to restore FNZ induction of CDR1 (Fig. 6B) and CDR2 (Fig. 6C) in a tac1 deletion mutant compared to the wild-type full-length protein. Coordinate with the inability to activate CDR1, FNZ-induced Mediator occupancy at the CDR1 promoter is lost in the C-terminal truncation (Tac1aa1–845) mutant strain (Fig. 6D). In addition, the absence of the Tac1 TAD also abrogates the FNZ-induced increase in Tac1 occupancy observed at the CDR1 promoter (Fig. 6E). Although the Tac1 TAD is required for the increase in Tac1 occupancy, increased Mediator occupancy does not appear to be necessary for the fluphenazine-induced Tac1 occupancy at the CDR1 promoter (Fig. 6F). Deletion of med3 does not reduce Tac1 occupancy under FNZ induction at the CDR1 promoter (Fig. 6F) despite the lack of Mediator recruitment in this strain. The CDR1DRE may become more accessible via cooperative interaction between the Tac1 TAD and chromatin remodeling complexes. The TAD is also required for Tac1 hyperactivation by GOF mutations located within the middle region. Activation of CDR1 (Fig. 6G) and CDR2 (Fig. 6H) by Tac1E461K, Tac1ΔM677, Tac1R693K, and Tac1A736V is dependent on the Tac1 TAD, as is the associated fluconazole resistance (Table 3). This result effectively eliminates the possibility that these middle-region GOF mutations directly confer an activation potential to the domain. The model consistent with these data is that middle-region GOF mutations weaken an inhibitory interaction between this domain and the C-terminal TAD.
FIG 6.
Tac1-mediated gene activation and Mediator and Tac1 occupancy at the CDR1 promoter in strains lacking the Tac1 TAD. (A) β-Galactosidase assays in one-hybrid strains expressing LexA-Tac1130–981 (yLM568) or LexA-Tac1130–931 (yLM578) in the absence and presence of fluphenazine. At least three independent transformants were tested for each construct, and results are presented as means (column height) and standard deviations (error bar). RT-qPCR analysis of fluphenazine-induced CDR1 (B) and CDR2 (C) expression in strains expressing 6His3Flag-tagged wild-type full-length Tac1 (Tac1WT Full Length; yLM485) or TAD-truncated Tac1 [Tac1WT1–845(ΔTAD); yLM539] in a tac1 null mutant background. (D) ChIP analysis, using an anti-HA antibody, of Mediator occupancy at the CDR1 promoter in strains expressing 6His3Flag full-length wild-type Tac1 (yLM597) or TAD truncated Tac1 (yLM598) and one copy of MED17 tagged by fusion of a 3×HA tag (MED17 WT/HA) after treatment with fluphenazine. A strain expressing untagged Med17 (WT/WT) and Tac1 (DSY2937-35) without treatment was analyzed as the reference. (E) ChIP analysis of Tac1 occupancy, using an anti-Flag antibody, at the CDR1 promoter in the strain lysates used for the experiments shown in panel D and using the same normalization strategy. (F) ChIP analysis of Tac1 occupancy, using an anti-Flag antibody, at the CDR1 promoter in strains expressing untagged native Tac1 (WT; yLM485), 6His3Flag-tagged wild-type Tac1 (HF-TAC1WT; yLM485), or 6His3Flag Tac1N972D (HF-TAC1N972D; yLM534) in a MED3+/+ background and 6His3Flag-tagged wild-type Tac1 (HF-TAC1WT; yLM547) or 6His3Flag Tac1N972D (HF-TAC1N972D; yLM549) in a med3Δ/Δ background in the absence or presence of fluphenazine. (G and H) RT-qPCR analysis of CDR1 (G) and CDR2 (H) levels in strains expressing full-length or TAD-truncated (aa 1 to 845; ΔTAD) Tac1E461K (yLM490 and yLM607), Tac1ΔM677 (yLM502 and yLM608), Tac1R693K (yLM602 and yLM609), or Tac1A736V (yLM169 and yLM610). Basal and FNZ-induced CDR1 and CDR2 expression levels in a strain with wild-type Tac1 (DSY2937-35) were analyzed as reference, and the basal expression level for each gene was set to 1.
TABLE 3.
Fluconazole MIC of strains expressing TAD-truncated Tac1GOF mutants
| TAC1 allele | Fluconazole MICa (μg/ml) |
|
|---|---|---|
| Full length | ΔTAD | |
| WT | 1 (yLM485) | 1 (yLM539) |
| E461K | 8 (yLM490) | 1 (yLM607) |
| ΔM677 | 8–12 (yLM502) | 0.75 (yLM608) |
| R693K | 8–12 (yLM602) | 0.75 (yLM609) |
| A736V | 16–24 (yLM169) | 1 (yLM610) |
Fluconazole MICs were measured by Etest at 30°C on YPD plates. Plates were incubated for 36 h before readout. Intermediate values, between scale marks, are presented as intervals. The exact strain used for each MIC measurement is listed in parentheses.
GOF mutations have modest and variable effects on Tac1 TAD activation potential.
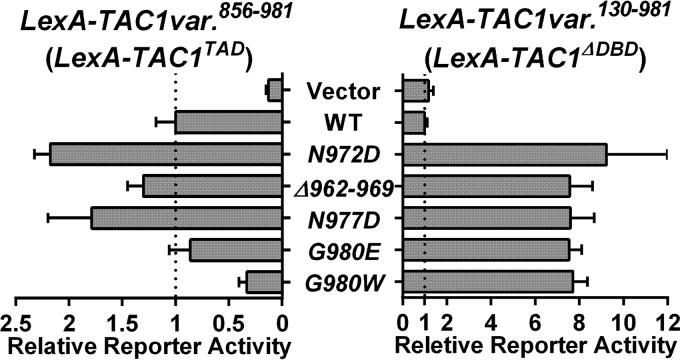
At least 7 Tac1GOF mutations (Δ962–969, N972D/I/S, N977D, and G980E/W) (7) are located within the Tac1 TAD. These GOF mutations could hyperactivate Tac1 by directly conferring greater activation potential to the Tac1 TAD or by weakening an interaction with an inhibitory domain. We used the lacZ reporter assay with five LexA-TAD variants containing GOF mutations that naturally occur in the Tac1 C terminus to test this hypothesis. All of the Tac1GOF mutations tested had only a modest effect on TAD activity in the reporter assay and, depending on the particular GOF mutation, could increase or decrease the activation potential (Fig. 7). These small changes in activation potential showed no clear correlation with the impact of the GOF mutations in the context of the entire Tac1 protein (aa 130 to 981) (Fig. 7). These data suggest that enhancement of the intrinsic Tac1 TAD activity plays a small or nonexistent role in Tac1 hyperactivation by GOF mutations.
FIG 7.
β-Galactosidase reporter assays on Tac1 TAD GOF mutants. The C. albicans one-hybrid strains expressing individual LexA-Tac1TAD(aa856–981) variants (wild type [yLM579], N972D [yLM581], Δ962–969 [yLM582], N977D [yLM583], G980E [yLM584], or G980W [yLM585]) and one-hybrid strains expressing individual LexA-Tac1ΔDBD(aa130–981) variants (wild-type [yLM568], N972D [yLM573], Δ962–969 [yLM574], N977D [yLM575], G980E [yLM576], or G980W [yLM577]) were tested using β-galactosidase assays. At least three transformants for each construct were measured. Data are reported after normalization to the activation activity of the wild-type variant in each group.
Tac1 proteins show a phosphorylation mobility shift on SDS-PAGE upon hyperactivation by xenobiotic or GOF mutations.
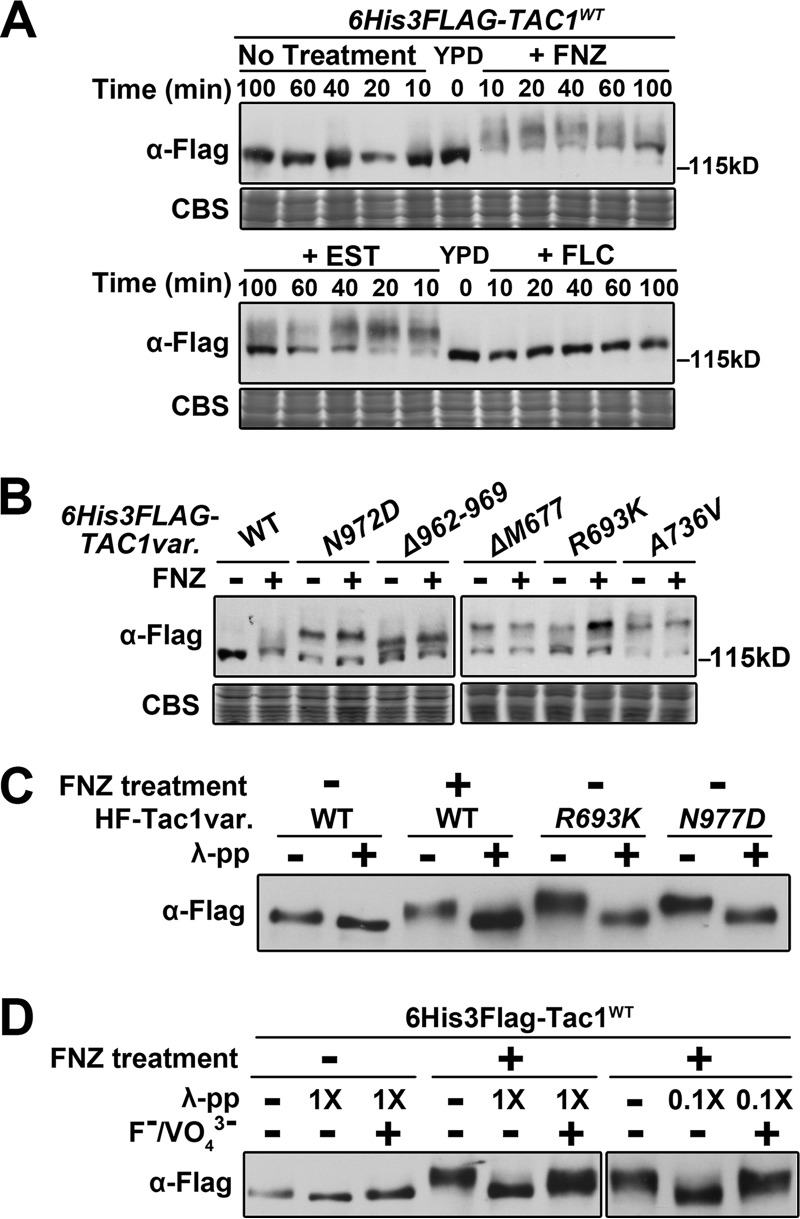
In the course of these studies, we noted that Tac1 hyperactivation was accompanied by an SDS-PAGE mobility shift. Zinc cluster transcription factors are often phosphorylated (17, 42), and phosphorylation of active S. cerevisiae Pdr1/3 has been demonstrated previously (43). It has been speculated that Tac1 activity is regulated by posttranslational modification, such as phosphorylation (7, 9, 10, 43), but modification has not been carefully documented. Immunoblotting showed that fluphenazine and estradiol treatment reduces the mobility of a 6His3Flag-Tac1 N-terminal fusion protein on SDS-PAGE from a single band into a slowly migrating smear (Fig. 8A). Treatment with fluconazole, which does not hyperactivate Tac1 (Fig. S3A and S3B), does not cause the mobility shift (Fig. 8A). The Tac1 signal showed a maximal mobility shift after ∼10 min of treatment that was gradually reversed over 60 to 100 min. This pattern mirrors the time course of CDR1 and CDR2 expression after treatment with FNZ or estradiol (Fig. S3A and S3B). Furthermore, titrations showed that the minimum concentration of FNZ (2 μg/ml) for significant upregulation of CDR1 (Fig. S3C) and CDR2 (Fig. S3D) expression is identical to that required for induction of the Tac1 mobility shift, and that these two phenomena track together over the range of concentrations used (Fig. S3E). Akin to the fluphenazine-induced Tac1 SDS-PAGE mobility shift, hyperactive Tac1GOF mutant proteins showed a constitutive SDS-PAGE mobility shift compared to the untreated wild-type Tac1, which was not further impacted by addition of fluphenazine (Fig. 8B). The origin of the unshifted band in each Tac1GOF mutant is unclear but may represent specific C-terminally truncated breakdown products of Tac1 or a subpopulation of Tac1GOF mutant protein that is not functionally hyperactivated. We purified FNZ-treated wild-type Tac1 protein and two Tac1GOF mutant proteins to determine whether the hyperactive Tac1 SDS-PAGE mobility shift results from phosphorylation. These purified proteins showed an SDS-PAGE mobility shift similar to that observed in extracts, which was largely eliminated by incubation with λ protein phosphatase (Fig. 8C). Inclusion of phosphatase inhibitors during the λ protein phosphatase incubation generated bands with intermediate SDS-PAGE mobility (Fig. 8D).
FIG 8.
SDS-PAGE and phosphatase analysis of the mobility shift of hyperactive Tac1. (A) Immunoblot of Tac1 SDS-PAGE mobility in lysates from a 6His3Flag-tagged wild-type Tac1 strain (yLM485) treated with 10 μg/ml FNZ, 10 μg/ml EST, or 40 μg/ml fluconazole (FLC) for the indicated periods of time. Samples were resolved on 6% SDS-PAGE gel and the blot was probed with an anti-Flag antibody. Coomassie blue staining (CBS) was used as the loading control. (B) Immunoblot analysis of SDS-PAGE mobility in lysates from strains expressing an endogenous level of individual 6His3Flag-tagged Tac1 variants (wild-type [yLM485], N972D [yLM534], Δ962–969 [yLM535], ΔM677 [yLM533], R693K [yLM532], and A736V [yLM531]) in the absence and presence of 10 μg/ml fluphenazine. Samples were resolved on 6% SDS-PAGE gel and the blot was probed with an anti-Flag antibody. CBS was used as the loading control. (C) Immunoblot analysis demonstrating the effect of λ protein phosphatase (λ-pp) treatment on the SDS-PAGE mobility shift of hyperactive Tac1 purified from strains that overexpressed 6His3Flag (HF)-tagged wild-type Tac1 (yLM530; grown in the presence and absence of fluphenazine), Tac1R693K (yLM540), or Tac1N977D (yLM536). Samples were resolved by 6% SDS-PAGE and the blot was probed by an anti-Flag antibody. (D) Immunoblot analysis of the effect of λ protein phosphatase treatment (in the presence and absence of 50 mM NaF and 20 mM Na3VO4 [F−/VO43−] phosphatase inhibitors) on the SDS-PAGE mobility shift of Tac1 purified from 6His3Flag-tagged wild-type Tac1 cells (yLM530) grown in the absence and presence of fluphenazine. Samples were resolved by 6% SDS-PAGE and the blot was probed by an anti-Flag antibody.
The Mediator Ssn3 kinase subunit is required for the Tac1 phosphoshift but has only a modest effect on the expression of Tac1 target genes.
The kinase activity of the Mediator complex, which is provided by its Ssn3 subunit, has been shown to directly regulate several sequence-specific DNA binding transcription factors in S. cerevisiae via phosphorylation (44–47). Akin to Mediator subunits Med17 and Med8 (Fig. 3), ChIP analysis showed an increase in HA-tagged Ssn3 occupancy at the CDR1 promoter after FNZ treatment (Fig. 9A). Tac1, in a strain incubated with fluphenazine (Fig. 9B and C) or possessing a GOF mutation (Fig. 9D), did not exhibit an SDS-PAGE phosphorylation shift in ssn3 null or kinase-dead mutant (SSN3D325A; adapted from reference 48) strains. Consistent with the idea that the Ssn3 kinase was targeted to Tac1 through its association with Mediator, we found that the hyperactive Tac1 SDS-PAGE phosphorylation shift does not occur in a med3 deletion strain (Fig. 9D and E) or in a strain where Tac1 lacks its TAD (Fig. 9F). This last piece of data could also be interpreted as the Tac1 TAD containing the primary Tac1 phosphorylation sites. A potential model that emerges from these data is that in the process of hyperactive Tac1 inducing its target genes, an interaction with Mediator facilitates the phosphorylation of Tac1. Consistent with this model, the binding of Tac1 to DNA is also required for the phosphorylation shift. A Tac1 point mutant (C43Y; adapted from reference 49), which is unable to bind DNA (Fig. S4A), does not activate transcription (Fig. S4B, C, E, and F) or undergo a mobility shift in response to hyperactivation (Fig. S4D and G).
FIG 9.
Effect of SSN3 and MED3 on the hyperactive Tac1 SDS-PAGE mobility shift. (A) ChIP analysis of Ssn3 occupancy at the CDR1DRE in strains with either two copies of native SSN3 (WT/WT; BWP17) or C-terminal 3×HA-tagged SSN3 (HA/HA; yLM481), grown in the presence and absence of fluphenazine, using an anti-HA antibody. Recovery rate (% Input) of DNA fragments containing the 1-DRE amplicon in uninduced BWP17 was set to 1 to calculate the relative recovery at additional regions. (B) Immunoblot analysis of the Tac1 SDS-PAGE phosphorylation shift in lysates from a 6His3Flag-tagged Tac1 SSN3+/+ strain (yLM485) and its ssn3Δ/Δ derivative (yLM552) grown in the absence and presence of fluphenazine. For all immunoblots in the figure, samples were resolved by 6% SDS-PAGE and probed by an anti-Flag antibody, and Coomassie blue staining (CBS) was used as the loading control. (C) Immunoblot analysis of the fluphenazine-induced Tac1 SDS-PAGE phosphorylation shift in lysates from strains (yLM596 and yLM660) that overexpressed 6His3Flag-tagged wild-type Tac1 in an ssn3Δ/Δ strain complemented by either wild-type SSN3 (yLM279) or a kinase-dead allele, SSN3D325A (yLM276). (D) Immunoblot analysis of the SDS-PAGE phosphorylation shift of Tac1GOF mutant proteins in lysates from strains expressing individual 6His3Flag (HF)-tagged Tac1GOF mutant variant (Tac1R693K [yLM532], Tac1Δ962–969 [yLM535], or Tac1N972D [yLM534]), their ssn3Δ/Δ derivatives (yLM553, yLM554, and yLM556, respectively), and their med3Δ/Δ derivatives (yLM550, yLM551, and yLM549, respectively). Cell lysates generated from a 6His3Flag-Tac1WT-expressing strain (yLM485) before and after fluphenazine treatment are included for comparison. (E) Immunoblot analysis of the Tac1 SDS-PAGE phosphorylation shift in lysates from a 6His3Flag-tagged Tac1 MED3+/+ strain (yLM485) and its med3Δ/Δ derivative (yLM547) grown in the presence of fluphenazine for the indicated period of time. (F) Immunoblot analysis of the Tac1 SDS-PAGE phosphorylation shift in lysates from strains expressing 6His3Flag-tagged wild-type Tac1 (yLM485) or TAD truncated Tac11–845 (yLM539) treated with fluphenazine for the indicated periods of time.
Despite the SSN3 requirement for the phosphorylation shift, TAC1GOF ssn3Δ/Δ strains were found to have only modestly decreased MICs when tested for fluconazole resistance (Table 4), while RT-qPCR revealed that CDR1, CDR2, and RTA3 gene expression levels were comparable between the TAC1GOF mutant strains and their ssn3 null derivatives (Fig. 10A to C). However, fluphenazine induction of CDR1, CDR2, and RTA3 was compromised in a TAC1WT strain lacking ssn3 over a range of concentrations tested (Fig. 10D to F and Fig. S5A and B) and also in an ssn3 kinase-dead mutant strain (Fig. 10G and H). A similar effect of the ssn3 null and kinase-dead mutants on Tac1 target gene expression is seen upon treatment with estradiol (Fig. S5C to G). Efforts to identify and mutate the phosphorylation sites on Tac1 have been hindered by the multisite nature of the phosphorylation. Although there is no clear evidence that Tac1 phosphorylation impacts its functional role in azole resistance, it does appear to be a faithful biomarker for Tac1 hyperactivation.
TABLE 4.
Fluconazole MIC of TAC1GOF mutants in an ssn3Δ/Δ background
| TAC1 allele | Fluconazole MICa (μg/ml) |
|
|---|---|---|
| SSN3+/+ | ssn3Δ/Δ | |
| WT | 0.75 (yLM167) | 0.75–1 (yLM236) |
| T225A | 12–16 (yLM168) | 12 (yLM237) |
| A736V | 16–24 (yLM169) | 12–16 (yLM238) |
| G980E | 16–24 (yLM170) | 12–16 (yLM239) |
Fluconazole MICs were measured by Etest at 30°C on YPD plates. Plates were incubated for 36 h before readout. Intermediate values, between scale marks, are presented as intervals. The exact strain used for each MIC measurement is listed in parentheses.
FIG 10.
Expression of TAC1 target genes in ssn3 deletion and kinase-dead mutant strains. (A to C) RT-qPCR analysis of CDR1 (A), CDR2 (B), or RTA3 (C) expression in SSN3+/+ strains with individual TAC1 variants (wild type [DSY2937-35], T225A [ACY67], A736V [ACY13], or G980E [ACY71]) and their ssn3Δ/Δ derivatives (yLM236, yLM237, yLM238, and yLM239, respectively). The expression level of each gene measured in the SSN3+/+ strain with wild-type TAC1 (DSY2937-35) was set to 1. The CDR2 expression levels in DSY2937-35 and yLM236 are denoted above each corresponding column. (D to F) RT-qPCR analysis of fluphenazine-induced expression of CDR1 (D), CDR2 (E), and RTA3 (F) in an ssn3Δ/Δ strain with wild-type TAC1 (yLM236). Parallel analyses of fluphenazine induction in a wild-type (WT; DSY2937-35) and a med3Δ/Δ mutant (yLM232) strain are presented for comparison. The basal expression level of each gene in DSY2937-35 was individually set to 1. (G and H) RT-qPCR analysis of fluphenazine-induced CDR1 (G) and CDR2 (H) expression in an ssn3Δ/Δ null strain (yLM265) complemented by either wild-type SSN3WT (yLM279) or a kinase-dead allele SSN3D325A (yLM276). The CDR1 and CDR2 expression levels in the SSN3WT strain (yLM279) collected before treatment were individually set to 1.
DISCUSSION
This study has helped elucidate the mechanism(s) by which GOF mutations hyperactivate Tac1. It has shown that TAC1GOF mutations and xenobiotic activators largely use the same mechanism of hyperactivation at a particular promoter but that the mechanism can differ according to the specific target promoter. The Mediator tail module plays an important role in the azole resistance driven by TAC1GOF mutations and serves an essential transcription coactivator role for Tac1-dependent activation of the CDR1 promoter. Our data suggest a model where CDR1DRE-bound Tac1 protein, when hyperactivated, recruits the Mediator complex through the tail module to facilitate transcription activation. This model is consistent with the classical working mechanism for Mediator coactivator function at highly inducible promoters (24, 25). Unlike the CDR1 promoter, activation of the CDR2 and RTA3 promoters shows a greater chromatin remodeling activity (Swi/Snf) dependence than Mediator tail module dependence. Therefore, it is likely that Tac1 regulon promoters work through at least two different mechanisms. At the CDR1 promoter, Tac1 occupancy is high at the DRE and gene induction is highly dependent on Mediator and its tail module. The CDR2 (and RTA3) promoters feature lower levels of Tac1 occupancy and low dependence on the Mediator tail module but high dependence on Swi/Snf function.
Given the strong dependence of CDR1 mRNA expression on MED3 and MED15 and the strong dependence of azole resistance on CDR1 (15), it was somewhat surprising that the MIC decreased by only 2- to 3-fold in the TAC1GOF med3 null strains. The residual levels of Cdr1 protein in the TAC1GOF med3 null strains make an important contribution to the remaining azole resistance. This finding suggests that even though the med3 deletion results in near-baseline levels of CDR1 transcription in TAC1GOF mutant strains, Cdr1 overexpression is under translational control by a mechanism dependent on TAC1GOF mutations and/or med3 deletion. The implication of residual azole resistance in Mediator tail module mutants is that an inhibitor of Tac1-TAD–Mediator tail module interactions, such as has been devised for C. glabrata Pdr1-Med15 interactions (34), would not be that potent in C. albicans TAC1GOF azole-resistant strains.
Although the Tac1 coactivator requirements appear to be promoter specific, we found that the coactivator mechanism used by Tac1 to induce a given promoter is the same regardless of the means of Tac1 hyperactivation. Disruption of the Mediator tail module equally affected CDR1 activation in TAC1GOF mutant strains and wild-type TAC1 strains induced by xenobiotic exposure. The same conclusion was reached for the CDR2 and RTA3 promoters as well, where hyperactive Tac1 relied on the Swi/Snf chromatin remodeling complex to activate transcription. Therefore, Tac1GOF mutants most likely function through the same activation mechanisms as xenobiotic-induced wild-type Tac1. The significance of this finding is that it may be possible to find a general mechanism to block Tac1 hyperactivation through a small-molecule screening approach. The actual mechanism of Tac1 hyperactivation appears to be antagonizing an inhibitory interaction between the Tac1 TAD and its middle region. Our finding that GOF mutations located in the middle domain are dependent on the Tac1 TAD does not support a model in which the GOF mutations lead to the acquisition of an activation potential in this region. Our finding that GOF mutations within a LexA-Tac1 TAD fusion fail to have a large impact on reporter gene expression helps eliminate a mechanism in which certain GOF mutations directly increase the Tac1 TAD potential. Our finding that the Tac1 TAD, in the absence of the middle region (LexA-Tac1856–981), shows much higher activation potential than the LexA-Tac1130–981 construct suggests that a region between amino acids 130 and 856 negatively regulates Tac1 TAD activity. Our current model posits that a direct interaction between the Tac1 TAD and the middle region can be disrupted by GOF mutations in either of these domains. It is an open question whether the xenobiotic inducers antagonize this interaction through direct interactions with Tac1 (50) or an indirect mechanism. The lack of structural information on zinc cluster middle homology regions makes it difficult to identify a particular region that might interact with the Tac1 TAD, although the locations of the GOF mutations in this region (which occur at aa 461 and within regions aa 225 to 255 and aa 673 to 841) could provide a starting point.
In this study, we also reported the phosphorylation of hyperactive Tac1 protein. Our data revealed the phosphorylation event requires Tac1-DNA interaction, hyperactivation of Tac1, Tac1-Mediator interaction, and Ssn3 kinase activity. Loss of ssn3 or its kinase activity modestly compromises xenobiotic induction of Tac1 targets, while steady-state overexpression of CDR1, CDR2, and RTA3 in TAC1GOF mutant strains is not affected. Therefore, it is possible that phosphorylation of Tac1 is required for its full activity under acute activation conditions. However, considering that the Cdk8 module regulates other steps during transcription initiation through its kinase activity, compromised xenobiotic induction in ssn3 mutants may not directly result from loss of Tac1 phosphorylation. Precisely testing the effect of phosphorylation requires the identification of phosphorylation sites on Tac1 protein, a task that has been made difficult by the multisite nature of the phosphorylation event. Phosphorylation with ambiguous functional relevance has been reported for multiple Zn(II)Cys6 transcription factors (42). Generation of significant changes in gel mobility and its tight correlation with hyperactivity makes Tac1 phosphorylation a good potential biomarker for its hyperactivity independent of its ultimate impact on Tac1 activity.
MATERIALS AND METHODS
Strain construction.
Plasmid construction is described in detail in the supplemental materials. C. albicans strains used in this study (see Table S4 in the supplemental material) were generated by transforming the parental strains (listed in the parental strains column of Table S4) with the DNA constructs specified in the corresponding constructs integrated column. Plasmids used in this study are listed in Table S5. The restriction enzyme(s) used for digestion of plasmids before transformation and primers used for confirmation of correct integration is also described in Table S5. Primers used in this study are listed in Table S6. C-terminal 3×HA tagging of MED8, MED17, and CDR1 was performed using the same method and tool construct described previously (29). Each tagging cassette was amplified from pFA-3HA-SAT1 by primer pair LM023/LM014 (MED8), ZL648/ZL649 (MED17), or ZL552/ZL553 (CDR1). Correct integration was tested by Kpp017/LM021 (MED8), ZL650/LM021 (MED17), or ZL554/LM021 (CDR1) at each 5′ junction and by Kpp063/Kpp018 (MED8), Kpp063/ZL651 (MED17), or Kpp063/ZL555 (CDR1) at each 3′ junction. Successful tagging was also confirmed by immunoblot analysis using an anti-HA antibody (3F10; Roche). Deletion of cdr1 in this study was achieved by a transient CRISPR-Cas9 system (51, 52), with modifications described in the supplemental material. All transformations were performed by electroporation and selected on YPD plus Clonat (1% yeast extract, 2% peptone, 2% glucose, 2% agar, 0.1 mM uridine, 100 μg/ml Clonat) or Sc-Ura (6.7 g/liter Difco yeast nitrogen base without amino acids [BD], 2 g/liter dropout mix synthetic without uracil [US Biological], 2% glucose, 2% agar). Activation of flippase expression and pop-out of the SAT1 marker were conducted by growing cells in YPMal (1% yeast extract, 2% peptone, 2% maltose, 0.1 mM uridine) liquid medium for 24 h. Successful recombinants were selected by sensitivity to 100 μg/ml Clonat.
Cell growth and drug treatment.
Cells were grown in liquid YPD medium (1% yeast extract, 2% peptone, 2% glucose, 0.1 mM uridine) at 30°C in this study. Fluphenazine treatment was performed by adding 6 mg/ml fluphenazine (Alfa Aesar) to mid-log-phase C. albicans cultures to a final concentration of 10 μg/ml if not otherwise specified. Estradiol treatment was performed by adding 2 mg/ml estradiol (Spectrum) to mid-log-phase C. albicans cultures to a final concentration of 10 μg/ml. Fluconazole treatment was performed by adding 4 mg/ml fluconazole (Tokyo Chemical Industry Co.) in dimethyl sulfoxide (DMSO) stock to a mid-log-phase yLM485 culture to a final concentration of 40 μg/ml. Cultures were collected after a certain period of exposure, as specified in the figure legends, for ChIP, RT-qPCR, or immunoblot analysis.
Fluconazole MIC measurement by Etest.
Overnight YPD cultures of strains to be tested were diluted in 0.85% NaCl solution to an optical density (OD) sufficient to form an even lawn on a YPD plate (supplemented with 0.1 mM uridine). After placement of fluconazole Etest strips (MIC range, 0.016 to 256 μg/ml; bioMérieux), plates were incubated for 36 h at 30°C before reading the MIC.
Immunoblotting.
Immunoblotting was used to compare Cdr1-3HA expression, 6His3Flag-Tac1 variant protein levels, and gel mobility between strains and conditions. Approximately 10 OD600 units of cells (108 cells) were collected, briefly washed with cold water, and frozen in liquid nitrogen. Whole-cell lysate was prepared by following the method described in reference 9, except that the trichloroacetic acid (TCA)-precipitated pellets were resuspended in 150 μl loading buffer (40 mM Tris-HCl, pH 6.8, 8 M urea, 5% SDS, 0.1 M EDTA, 1% β-mercaptoethanol, and 0.1 mg/ml bromophenol blue) instead of 50 μl. Samples were resolved by 6% SDS-PAGE, transferred to nitrocellulose membranes (120 mA; overnight), and probed by an α-HA (3F10; Roche) or α-Flag (F7425; Sigma) antibody. The lower-molecular-weight region of the gel, which typically did not contain the immunoblotting signals of interest in this study, was stained by Coomassie blue as the loading reference. Sensitive ECL substrate Clarity (Bio-Rad) was used to develop Tac1 (α-Flag) blots.
Protein purification.
6His3Flag-tagged Tac1WT was purified from fluphenazine-treated (20 μg/ml; ∼20 min) and nontreated cultures of yLM530. We also purified 6His3Flag-tagged Tac1R693K and Tac1N977D from non-FNZ-treated cultures of yLM540 and yLM536, respectively. Mid-log-phase cells were collected by centrifugation directly from mid-log-phase YPD cultures or after 10 min of exposure to 20 μg/ml fluphenazine. During the harvest step, cells spent an additional 10 min in contact with the growth media before being resuspended and washed with prechilled 1× lysis buffer (180 mM Tris-acetate [Tris-OAc] [pH 7.8], 400 mM potassium acetate [KOAc], 0.24 mM EDTA, 12% glycerol) with 1 mM dithiothreitol (DTT), 1× protease inhibitor cocktail (PIC) (53), and 1× phosphatase inhibitor cocktail (2 mM sodium fluoride, 2 mM imidazole, 1.2 mM sodium molybdate, 2 mM sodium orthovanadate, 4 mM sodium tartrate, and 1.5 mM sodium pyrophosphate) and then flash frozen in liquid nitrogen. DTT (1 mM), PIC (1×), and phosphatase inhibitor (1×) were added to all of the buffers used during purification unless otherwise specified. Cells were lysed by a mortar-and-pestle method, and lysate was sonicated, treated with Benzonase, and clarified by ultracentrifugation as described previously (29). Supernatant was loaded on DEAE Sepharose after dilution by addition of F-0 buffer (25 mM HEPES-KOH [pH 7.6], 0.01% NP-40, 10% glycerol, and the concentration of KOAc in millimolar after the dash [e.g., F-0 indicates 0 mM KOAc]) to a final salt concentration around 150 mM KOAc. After loading, the column was washed with 2 volumes of F-200 buffer. Tac1 protein, eluted from DEAE Sepharose by F-650 buffer containing 0.5 mM DTT and 1× protease/phosphatase inhibitor, was affinity captured by M2 Flag agarose (Sigma) and eluted in F-300 buffer supplemented with 2.5 mM β-mercaptoethanol, 1× PIC, 0.2× phosphatase inhibitor, and 100 μg/ml 3×Flag peptide. Phosphatase treatment analyses were performed by incubating the purified Tac1 protein samples with 32 U/μl or 3.2 U/μl λ protein phosphatase (NEB) in the presence and absence of 50 mM sodium fluoride and 20 mM sodium orthovanadate at 37°C.
RT-qPCR.
RNA samples were prepared from collected frozen cell pellets and reverse transcribed as described previously (54). qPCR was performed using the relative standard curve method (StepOne; Life Technologies). ACT1 relative abundance measured by ZL712/ZL713 or TEF1 relative abundance measured by ZL386/ZL387 was used as an internal reference to compare CDR1 (ZL540/ZL541), CDR2 (ZL542/ZL543), RTA3 (ZL544/ZL545), and TAC1 (ZL578/ZL579) expression among strains and conditions.
ChIP.
ChIP experiments were performed as described previously, with modifications (55). Fifty milliliters of the culture to be tested was fixed for 15 min by adding formaldehyde to a final concentration of 1%. Cross-linking was stopped by adding 2.5 ml of 2.5 M glycine. After washing 3 times with 20 ml cold 1× phosphate-buffered saline (PBS), cells were resuspended in 500 μl lysis buffer (50 mM HEPES-KOH, pH 7.5; 1% Triton X-100; 0.1% sodium deoxycholate; 1 mM EDTA; 150 mM NaCl) supplemented with protease inhibitor cocktail (Roche) and lysed by five 35-s bead beatings (Biospec). Cell lysates were brought to 1 ml with lysis buffer and probe sonicated (Fisher) for three 8-s intervals at 30% amplitude. Chromatin was further sheared by a Bio-Distruptor (high settings; 5 min four times; 30 s on/30 s off). Two hundred microliters of sonicated cell lysate was incubated with 1.5 μl α-HA antibody (F7; Santa Cruz) or 1.5 μl α-Flag antibody (M2, Sigma) at 4°C overnight. Chromatin was captured by an additional 2-h incubation with 15 μl lysis buffer equilibrated with protein G Dynabeads (Life Technologies). Beads were washed twice for 10 min in 500 μl wash buffer (50 mM HEPES-KOH, pH 7.5; 1% Triton X-100; 0.1% sodium deoxycholate; 1 mM EDTA; 500 mM NaCl), twice for 10 min in 500 μl deoxycholate buffer (10 mM Tris-HCl, pH 8.0; 0.5% sodium deoxycholate; 1 mM EDTA; 0.5% NP-40; 0.25 M LiCl), and briefly in 500 μl TE buffer (1 mM EDTA, 10 mM Tris-HCl, pH 8.0) at room temperature. Precipitated chromatin was eluted by incubating beads twice in 125 μl freshly made TES buffer (50 mM Tris-HCl, pH 8.0; 10 mM EDTA; 1.5% SDS) for 2 h at 65°C. Two eluates (∼250 μl total volume) were pooled and reverse cross-linked at 65°C overnight. Forty microliters of each input cell lysate was reverse cross-linked in 200 μl TES buffer. The next day, after a brief RNase treatment (1 μl DNase-free RNase [Roche]; 30 min) at room temperature, samples were mixed with 1.5 ml PB buffer (Qiagen) and loaded onto a PCR purification column (Qiagen). The final ChIP products and input reference samples were eluted in 150 μl EB buffer (Qiagen).
Analysis of ChIP data.
Results of ChIP experiments are presented in the form of relative recovery of input. Specifically, the abundance of DNA fragments containing a tested region first was compared with its abundance in the corresponding input reference sample by qPCR to calculate the absolute recovery rate. If not specified, absolute recovery rate at the 1-DRE region in uninduced DSY2937-35, a reference strain with wild-type TAC1 that is free of HA or Flag tagging, or Mediator subunit mutations, was set to 1 to calculate the relative recovery rate at 1-DRE and other regions tested among strains or conditions. Primers used for ChIP assay are listed in Table S6 with their testing regions specified in the corresponding note column.
Liquid β-galactosidase activity assays.
Twenty-five microliters of an overnight YPD culture was diluted in 4 ml of fresh YPD medium, grown for 5 to 6 h, and collected for β-galactosidase activity measurement using the SDS-chloroform method (56). Cultures used for testing fluphenazine induction of lacZ first were grown in YPD for 3 h after dilution from overnight culture and then grown for ∼200 min (treatment time was extended for LacZ protein maturation) in the presence of fluphenazine (18 μg/ml) before collection for measurement. Cell pellets were resuspended, further diluted in 800 μl Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7), and treated with 30 μl 0.1% SDS and 60 μl chloroform by vortex. After 10 min of equilibration at 30°C, reactions were started by addition of 160 μl 4 mg/ml o-nitrophenyl-β-d-galactopyranoside (RPI) Z buffer stock and incubated in a 30°C water bath shaker. Reactions were stopped by addition of 400 μl 1 M Na2CO3. β-Galactosidase activity, in Miller units, was calculated by the following simplified formula: 1,000 × A420/(T × C), where A420 is the absorbance of the reaction product at 420 nm, T is the reaction time in minutes, and C is the total amounts of cells at the total OD600 value used in the reaction. A420 and OD600 values were measured with a Beckman Coulter DU-7300 spectrophotometer.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH 5R21AI113390 to L.C.M.
We thank Dominique Sanglard and Joachim Morschhäuser for providing strains. We also thank Gerry Fink for providing the C. albicans CRISPR system.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/AAC.01344-17.
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01342-17.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol 36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 2.Martins N, Ferreira IC, Barros L, Silva S, Henriques M. 2014. Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment. Mycopathologia 177:223–240. doi: 10.1007/s11046-014-9749-1. [DOI] [PubMed] [Google Scholar]
- 3.Antinori S, Milazzo L, Sollima S, Galli M, Corbellino M. 2016. Candidemia and invasive candidiasis in adults: a narrative review. Eur J Intern Med 34:21–28. doi: 10.1016/j.ejim.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 4.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Messer SA, Boyken L, Hollis RJ, Rice C, Tendolkar S, Diekema DJ. 2004. In vitro activities of voriconazole, posaconazole, and fluconazole against 4,169 clinical isolates of Candida spp. and Cryptococcus neoformans collected during 2001 and 2002 in the ARTEMIS global antifungal surveillance program. Diagn Microbiol Infect Dis 48:201–205. doi: 10.1016/j.diagmicrobio.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med 125:S3–S13. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Sanglard D, Coste A, Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res 9:1029–1050. doi: 10.1111/j.1567-1364.2009.00578.x. [DOI] [PubMed] [Google Scholar]
- 8.Morschhauser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet Biol 47:94–106. doi: 10.1016/j.fgb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coste A, Turner V, Ischer F, Morschhauser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coste AT, Crittin J, Bauser C, Rohde B, Sanglard D. 2009. Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot Cell 8:1250–1267. doi: 10.1128/EC.00069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhauser J, Nantel A, Raymond M, Rogers PD. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell 6:2122–2138. doi: 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Micheli M, Bille J, Schueller C, Sanglard D. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol Microbiol 43:1197–1214. doi: 10.1046/j.1365-2958.2002.02814.x. [DOI] [PubMed] [Google Scholar]
- 14.Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d'Enfert C, Berman J, Sanglard D. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6:1889–1904. doi: 10.1128/EC.00151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes AR, Lin YH, Niimi K, Lamping E, Keniya M, Niimi M, Tanabe K, Monk BC, Cannon RD. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother 52:3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whaley SG, Tsao S, Weber S, Zhang Q, Barker KS, Raymond M, Rogers PD. 2016. The RTA3 gene, encoding a putative lipid translocase, influences the susceptibility of Candida albicans to fluconazole. Antimicrob Agents Chemother 60:6060–6066. doi: 10.1128/AAC.00732-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacPherson S, Larochelle M, Turcotte B. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schjerling P, Holmberg S. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res 24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother 48:3064–3079. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rognon B, Kozovska Z, Coste AT, Pardini G, Sanglard D. 2006. Identification of promoter elements responsible for the regulation of MDR1 from Candida albicans, a major facilitator transporter involved in azole resistance. Microbiology 152:3701–3722. doi: 10.1099/mic.0.29277-0. [DOI] [PubMed] [Google Scholar]
- 21.Harry JB, Oliver BG, Song JL, Silver PM, Little JT, Choiniere J, White TC. 2005. Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob Agents Chemother 49:2785–2792. doi: 10.1128/AAC.49.7.2785-2792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Naar AM. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 23.Shahi P, Gulshan K, Naar AM, Moye-Rowley WS. 2010. Differential roles of transcriptional mediator subunits in regulation of multidrug resistance gene expression in Saccharomyces cerevisiae. Mol Biol Cell 21:2469–2482. doi: 10.1091/mbc.E09-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansari SA, Morse RH. 2013. Mechanisms of Mediator complex action in transcriptional activation. Cell Mol Life Sci 70:2743–2756. doi: 10.1007/s00018-013-1265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poss ZC, Ebmeier CC, Taatjes DJ. 2013. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol 48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. 2014. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157:1430–1444. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. 2015. Molecular architecture of the yeast Mediator complex. eLife 4:e08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. 1999. Mediator protein mutations that selectively abolish activated transcription. Proc Natl Acad Sci U S A 96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang A, Petrov KO, Hyun ER, Liu Z, Gerber SA, Myers LC. 2012. The Tlo proteins are stoichiometric components of Candida albicans mediator anchored via the Med3 subunit. Eukaryot Cell 11:874–884. doi: 10.1128/EC.00095-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YC, Park JM, Min S, Han SJ, Kim YJ. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol Cell Biol 19:2967–2976. doi: 10.1128/MCB.19.4.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Myers LC. 2015. Fungal mediator tail subunits contain classical transcriptional activation domains. Mol Cell Biol 35:1363–1375. doi: 10.1128/MCB.01508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemet J, Jelicic B, Rubelj I, Sopta M. 2014. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie 97:22–27. doi: 10.1016/j.biochi.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez D, Hamidi N, Del Sol R, Benschop JJ, Nancy T, Li C, Francis L, Tzouros M, Krijgsveld J, Holstege FC, Conlan RS. 2014. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc Natl Acad Sci U S A 111:2500–2505. doi: 10.1073/pnas.1307525111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishikawa JL, Boeszoermenyi A, Vale-Silva LA, Torelli R, Posteraro B, Sohn YJ, Ji F, Gelev V, Sanglard D, Sanguinetti M, Sadreyev RI, Mukherjee G, Bhyravabhotla J, Buhrlage SJ, Gray NS, Wagner G, Naar AM, Arthanari H. 2016. Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 530:485–489. doi: 10.1038/nature16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haran J, Boyle H, Hokamp K, Yeomans T, Liu Z, Church M, Fleming AB, Anderson MZ, Berman J, Myers LC, Sullivan DJ, Moran GP. 2014. Telomeric ORFs (TLOs) in Candida spp. Encode mediator subunits that regulate distinct virulence traits. PLoS Genet 10:e1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hargreaves DC, Crabtree GR. 2011. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rando OJ, Winston F. 2012. Chromatin and transcription in yeast. Genetics 190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5:a017905. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cote J, Quinn J, Workman JL, Peterson CL. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 40.Richmond E, Peterson CL. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res 24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell CL, Brown AJ. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet Biol 42:676–683. doi: 10.1016/j.fgb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Leverentz MK, Reece RJ. 2006. Phosphorylation of Zn(II)2Cys6 proteins: a cause or effect of transcriptional activation? Biochem Soc Trans 34:794–797. doi: 10.1042/BST0340794. [DOI] [PubMed] [Google Scholar]
- 43.Mamnun YM, Pandjaitan R, Mahe Y, Delahodde A, Kuchler K. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol Microbiol 46:1429–1440. doi: 10.1046/j.1365-2958.2002.03262.x. [DOI] [PubMed] [Google Scholar]
- 44.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. 1999. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell 3:673–678. doi: 10.1016/S1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 45.Nelson C, Goto S, Lund K, Hung W, Sadowski I. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 46.Raithatha S, Su TC, Lourenco P, Goto S, Sadowski I. 2012. Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol Cell Biol 32:664–674. doi: 10.1128/MCB.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent O, Kuchin S, Hong SP, Townley R, Vyas VK, Carlson M. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol Cell Biol 21:5790–5796. doi: 10.1128/MCB.21.17.5790-5796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Noble SM. 2012. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS Pathog 8:e1002956. doi: 10.1371/journal.ppat.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston M, Dover J. 1987. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A 84:2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naar AM, Thakur JK. 2009. Nuclear receptor-like transcription factors in fungi. Genes Dev 23:419–432. doi: 10.1101/gad.1743009. [DOI] [PubMed] [Google Scholar]
- 51.Min K, Ichikawa Y, Woolford CA, Mitchell AP. 2016. Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1:e00130-16. doi: 10.1128/mSphere.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv 1:e1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers LC, Leuther K, Bushnell DA, Gustafsson CM, Kornberg RD. 1997. Yeast RNA polymerase II transcription reconstituted with purified proteins. Methods 12:212–216. doi: 10.1006/meth.1997.0473. [DOI] [PubMed] [Google Scholar]
- 54.Zhang A, Liu Z, Myers LC. 2013. Differential regulation of white-opaque switching by individual subunits of Candida albicans mediator. Eukaryot Cell 12:1293–1304. doi: 10.1128/EC.00137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. 2006. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell 22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 56.Guarente L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol 101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.