ABSTRACT
Two adjacent colistin resistance gene variants, termed mcr-3.3 and mcr-3-like, were identified in the chromosome of an Aeromonas veronii isolate obtained from retail chicken meat. The variants showed 95.20% and 84.19% nucleotide sequence identity, respectively, to mcr-3 from porcine Escherichia coli. Functional cloning indicated that only mcr-3.3 conferred polymyxin resistance in both E. coli and Aeromonas salmonicida. The mcr-3.3-mcr-3-like segment was also observed in other Aeromonas species, including A. media, A. caviae, and A. hydrophila.
KEYWORDS: colistin resistance, mcr-3, Aeromonas veronii
TEXT
Colistin, the last line of defense against multidrug resistant Gram-negative bacteria, especially carbapenem-resistant Enterobacteriaceae, is nowadays increasingly used in the treatment of serious clinical infections. However, its clinical efficacy has been jeopardized since the emergence of the plasmid-mediated colistin resistance gene mcr-1, first identified in 2016 (1), followed by seven other mcr-1 variants and a second resistance gene, mcr-2 (2). Recently, a third polymyxin resistance gene, mcr-3, was characterized from Escherichia coli, further affecting the efficacy of colistin (3).
Compared with mcr-1, the global spread of which was diligently documented (4–11), there are few data on the prevalence of mcr-3-positive Gram-negative bacteria. It is particularly important to identify the presence of these strains in food products as they can have a direct impact on public health. Other than three Enterobacteriaceae species, including Escherichia coli, Klebsiella pneumoniae, and Salmonella enterica serovar Typhimurium, which have been identified as mcr-3 positive, all mcr-3-like genes, whose deduced amino acids presented 75.6–94.8% amino acid identity to MCR-3, have been identified in 10 Aeromonas species of various origins across four continents (3). However, whether these mcr-3-like genes confer colistin resistance in Aeromonas and Enterobacteriaceae remains unknown.
In the current study, 16 chicken meat samples were collected from six geographically distant supermarkets and three farmers' markets in Qingdao, Shandong Province, China, in 2015. The chicken meat samples were then macerated in 10 ml of brain heart infusion broth for 5 min in aseptic sampling bags (Hope Bio-Technology Co., Beijing, China), and the resulting infiltrated fluids were stored in ESwab tubes (Copan Diagnostics, Murrieta, CA) for further analysis. Direct screening was then performed on all 16 samples to detect the mcr-3 gene, and only one sample was identified as mcr-3 positive. The mcr-3-positive strain 172 was isolated using brain heart infusion agar (Beijing Land Bridge, Beijing, China) with 1 mg/liter colistin and then identified as Aeromonas veronii by 16S rRNA sequencing and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis.
Antimicrobial susceptibility testing was performed on A. veronii 172 using the agar dilution method described by the Clinical and Laboratory Standards Institute (document VET01-A4) (12) and was interpreted according to Clinical and Laboratory Standards Institute document M45 (13). E. coli strain ATCC 25922 served as the quality control strain. A. veronii 172 was resistant to amoxicillin-clavulanic acid (64/32 mg/liter) and chloramphenicol (32 mg/liter) and had intermediate resistance to tetracycline (8 mg/liter) but was susceptible to ceftazidime (1 mg/liter), imipenem (1 mg/liter), meropenem (1 mg/liter), gentamicin (1 mg/liter), aztreonam (0.03 mg/liter), ciprofloxacin (0.5 mg/liter), and trimethoprim-sulfamethoxazole (0.5/9.5 mg/liter). Interestingly, this mcr-3-carrying isolate had an MIC for polymyxin B of 2 mg/liter and was borderline susceptible to colistin (2 mg/liter), according to the European Committee on Antimicrobial Susceptibility Testing clinical breakpoints (http://www.eucast.org/clinical_breakpoints/).
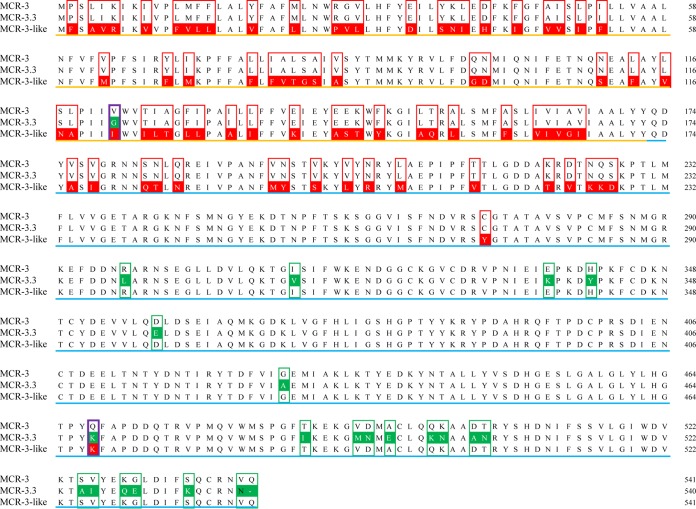
A. veronii 172 was then subjected to 300-bp paired-end whole-genome sequencing using the Illumina HiSeq 2500 system (Annoroad, Beijing, China). A total of 116 contigs were obtained by draft assembly using the CLC Genomics Workbench 9 (CLC Bio, Aarhus, Denmark). Among these, contig 13 (268.7 kb) contained two mcr-3 variants that were located adjacently and separated by only 66 bp. The two variants showed 95.20% and 84.19% nucleotide sequence identity to the original mcr-3 gene from pigs and were therefore termed mcr-3.3 and mcr-3-like, respectively (Fig. 1). Both variants carried several missense mutations, resulting in 95.75% (mcr-3.3) and 84.84% (mcr-3-like) amino acid sequence similarity to mcr-3 (Fig. 2). To identify the function of mcr-3 variant genes, the 1853-bp and 3558-bp DNA fragments, including mcr-3.3 and mcr-3.3-mcr-3-like fragments as well as their respective upstream sequences, were amplified and cloned into pUC19 before being electroporated into E. coli DH5α (TaKaRa). Transformants containing pUC19-mcr-3.3 and pUC19-mcr-3.3-mcr-3-like had colistin MICs of 2 mg/liter and 1 mg/liter, respectively, which were 4- to 8-fold higher than the MIC of DH5α containing pUC19 alone (0.25 mg/liter), while the MICs of transformants with pUC19-mcr-3-like were no different than that of pUC19-DH5α. However, the gap between mcr-3-like and mcr-3.3 is very small (only 66 bp), which means that the promoter region of mcr-3-like may be absent, leading to an insufficient expression of mcr-3-like. Therefore, to further confirm the invalidation of mcr-3-like in mediating colistin resistance, only the 1623-bp and 1626-bp coding sequences of mcr-3.3 and mcr-3-like were ligated into expression vector pET-28a (Novagen, Beijing, China) and transformed into E. coli BL21(DE3)plys (Transgen Biotech, Beijing, China). When induced by isopropyl-β-D-thiogalactopyranoside (IPTG) with a concentration of 0.1 mM, the transformants containing pET-mcr-3-like did not present any change in MIC compared with BL21(DE3)plys containing pET28a alone (8 mg/liter), while the MICs of transformants carrying pET-mcr-3.3 increased to 16 mg/liter, suggesting that only mcr-3.3 confers colistin resistance in E. coli. In addition, to investigate the function of two variants in the species of Aeromonas, the plasmids pUC19-mcr-3.3 and pUC19-mcr-3.3-mcr-3-like were separately electroporated into a competent Aeromonas salmonicida strain that originated from a chicken cloaca swab. To our surprise, MICs of both transformants soared to 64 mg/liter, which was 64-fold higher than the MIC of a transformant with pUC19 alone (1 mg/liter). Stability testing revealed that pUC19-mcr-3.3 existed stably in the host of A. salmonicida after 20 generations of subculture both without or with colistin (1 mg/liter). These results suggested that mcr-3.3, but not mcr-3-like, could confer colistin resistance in E. coli and Aeromonas species, the latter of which may not only serve as a reservoir for mcr-3 variants but also have the potential to develop a highly colistin-resistant phenotype.
FIG 1.
The genetic environment of the mcr-3.3-mcr-3-like segment in different Aeromonas isolates. Arrows indicate the directions of the genes. The regions in gray represent the two linked areas with high similarity.
FIG 2.
Alignment of the predicted amino acid sequences of MCR-3, MCR-3.3, and MCR-3-like. Orange lines denote putative transmembrane regions and blue lines indicate catalytic domains. Differences in amino acid residues between MCR-3 and MCR-3.3 are shown in green boxes, while red boxes indicate differences between MCR-3 and MCR-3-like.
Comparative genomic analysis between contig 13 and the whole complete genomic sequence of A. veronii AVNIH1 (GenBank accession no. CP014774.1) was performed using Mauve (http://darlinglab.org/mauve/mauve.html) and showed that almost all of the block outlines in contig 13 matched their counterparts in the chromosome of AVNIH1 (see Fig. S1 in the supplemental material). S1-nuclease pulsed-field gel electrophoresis and Southern blotting also confirmed that the two mcr-3 variants were located on the chromosome of A. veronii 172 (data not shown).
The mcr-3.3-mcr-3-like segment in A. veronii 172 was surrounded by both hypothetical and functional protein-coding genes. Unlike the mcr-3 in E. coli plasmid pWJ1, the fragment containing the mcr-3.3-mcr-3-like segment in A. veronii 172 lacked transfer elements (Fig. 1). However, dgkA, which was located downstream of mcr-3 in pWJ1, was also identified in the downstream regions of the mcr-3.3-mcr-3-like segment in A. veronii 172, Aeromonas caviae (GenBank accession no. JWJP01000016.1), and Aeromonas hydrophila (GenBank accession no. AOBN01000008.1). The upstream region of the mcr-3.3-mcr-3-like segment in A. hydrophila contained a complete insertion sequence, ISAs17, the open reading frame of which exhibited 99% nucleotide sequence identity to that originated from Aeromonas salmonicida. This ISAs17 element had imperfect inverted repeats of 50 bp right (IRR, TGGATTTGACCCCATAGATCCAGACACTTTTTCCCGCTTAGGGTTGTGAT) and 50 bp left (IRL, TAGATTGGCCCCTTGAAGATCCAGACACTTCGCCCCTCTTAACTTAAGAG) at its ends. In addition, direct target site duplications (5′-TGAG-3′) were also observed immediately upstream and downstream of ISAs17 (Fig. 1).
Apart from the mcr-3.3-mcr-3-like segment, BLAST analysis revealed the presence of other resistance genes in the genome of A. veronii 172, including β-lactam resistance genes cphA1 and ampS, phenicol resistance gene catA2, and tetracycline resistance gene tet(E). This was consistent with the multidrug-resistance phenotype observed during antibiotic susceptibility testing. In addition, the multilocus sequence type of this Aeromonas strain was submitted and assigned as a novel sequence type ST512 (https://cge.cbs.dtu.dk/services/MLST/).
In conclusion, an A. veronii strain containing both mcr-3.3 and mcr-3-like genes on its chromosome was isolated from chicken meat in China. This strain showed resistance to both β-lactams and phenicols and had borderline susceptibility to colistin, which was mediated by mcr-3.3 alone. The presence of these mcr-3 chromosomal variants is worrisome because acquisition of a plasmid carrying an mcr-3 variant may lead to high levels of colistin resistance in these Aeromonas strains. Since Aeromonas species are prevalent in aquatic environments, where they interact with bacteria from different origins, they may act as a reservoir for mcr-3 and contribute to its potential dissemination.
Accession number(s).
The 268746-bp nucleotide sequence of contig 13 carrying mcr-3.3-mcr-3-like segment has been deposited as GenBank accession no. MF495680.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Key Basic Research Program of China (grant 2013CB127200) and the National Natural Science Foundation of China (grants 31422055, 31602107, and 81661138002). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01272-17.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):pii=30280 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=22525. [DOI] [PubMed] [Google Scholar]
- 3.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernasconi OJ, Kuenzli E, Pires J, Tinguely R, Carattoli A, Hatz C, Perreten V, Endimiani A. 2016. Travelers can import colistin-resistant Enterobacteriaceae, including those possessing the plasmid-mediated mcr-1 gene. Antimicrob Agents Chemother 60:5080–5084. doi: 10.1128/AAC.00731-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 6.Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RN, Flamm RK. 2016. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother 60:5623–5624. doi: 10.1128/AAC.01267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen Z, Wang Y, Shen Y, Shen J, Wu C. 2016. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 16:293. doi: 10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 8.Gu DX, Huang YL, Ma JH, Zhou HW, Fang Y, Cai JC, Hu YY, Zhang R. 2016. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrob Agents Chemother 60:5099–5100. doi: 10.1128/AAC.00476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perreten V, Strauss C, Collaud A, Gerber D. 2016. Colistin resistance gene mcr-1 in avian-pathogenic Escherichia coli in South Africa. Antimicrob Agents Chemother 60:4414–4415. doi: 10.1128/AAC.00548-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, MCR Group, Corso A. 2016. First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob Agents Chemother 60:4412–4413. doi: 10.1128/AAC.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Tian G-B, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li H-Y, Doi Y, Fang Y, Ren H, Zhong L-L, Shen Z, Zeng K-J, Wang S, Liu J-H, Wu C, Walsh TR, Shen J. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 4th ed. CLSI VET01-A4 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed. CLSI M45 Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.