ABSTRACT
Vaborbactam (formerly RPX7009) is a new beta-lactamase inhibitor based on a cyclic boronic acid pharmacophore. The spectrum of beta-lactamase inhibition by vaborbactam and the impact of bacterial efflux and permeability on its activity were determined using a panel of strains with beta-lactamases cloned from various classes and a panel of Klebsiella pneumoniae carbapenemase 3 (KPC-3)-producing isogenic strains with various combinations of efflux and porin mutations. Vaborbactam is a potent inhibitor of class A carbapenemases, such as KPC, as well as an inhibitor of other class A (CTX-M, SHV, TEM) and class C (P99, MIR, FOX) beta-lactamases. Vaborbactam does not inhibit class D or class B carbapenemases. When combined with meropenem, vaborbactam had the highest potency compared to the potencies of vaborbactam in combination with other antibiotics against strains producing the KPC beta-lactamase. Consistent with broad-spectrum beta-lactamase inhibition, vaborbactam reduced the meropenem MICs for engineered isogenic strains of K. pneumoniae with increased meropenem MICs due to a combination of extended-spectrum beta-lactamase production, class C beta-lactamase production, and reduced permeability due to porin mutations. Vaborbactam crosses the outer membrane of K. pneumoniae using both OmpK35 and OmpK36, but OmpK36 is the preferred porin. Efflux by the multidrug resistance efflux pump AcrAB-TolC had a minimal impact on vaborbactam activity. Investigation of the vaborbactam concentration necessary for restoration of meropenem potency showed that vaborbactam at 8 μg/ml results in meropenem MICs of ≤2 μg/ml in the most resistant engineered strains containing multiple mutations. Vaborbactam is a highly active beta-lactamase inhibitor that restores the activity of meropenem and other beta-lactam antibiotics in beta-lactamase-producing bacteria, particularly KPC-producing carbapenem-resistant Enterobacteriaceae.
KEYWORDS: vaborbactam, meropenem-vaborbactam, KPC, major porins OmpK35 and OmpK36
INTRODUCTION
Beta-lactam antibiotics are among the most useful classes of antibiotics to treat bacterial infections due to their spectra of activity, mode of action, and excellent safety profile. Among them, carbapenems have a particular clinical utility due to their resistance to hydrolysis by numerous beta-lactamases; however, the recent dissemination of carbapenemases that inactivate nearly all beta-lactams (including carbapenem antibiotics) threatens the loss of this class. Carbapenemases include serine beta-lactamases from Ambler classes A and D (1) and all metallo-beta-lactamases from class B (2, 3). In many locations around the world, including the United States, class A Klebsiella pneumoniae carbapenemases (KPC) represent the most prevalent carbapenemases (4–6). Infections caused by KPC-producing bacteria have been associated with increased health care costs and increased lengths of stay, as well as frequent treatment failures, with mortality rates varying from 22% to 72% (7–9). The in vitro characterization of KPC-producing strains has established that the bacteria evolved a multifactorial path to resistance, often combining the production of a carbapenemase with mutations that alter the expression or function of porins or efflux proteins (10–14).
The use of beta-lactam–beta-lactamase inhibitor (BLI) combinations is an effective strategy to overcome beta-lactamase-mediated resistance to beta-lactam antibiotics. However, older BLIs, such as clavulanic acid, tazobactam, and sulbactam (15), do not inhibit class A carbapenemases. Avibactam, the first member of the diazabicyclo-octane class, has a broader spectrum of beta-lactamase inhibition, including inhibition of many class A, class C, and some class D enzymes, than older BLIs (16, 17). Although avibactam was originally developed to address the dissemination of class A extended-spectrum beta-lactamases (ESBLs) and class C enzymes and was specifically optimized for activity against these beta-lactamases (18), it was subsequently found to inhibit the KPC carbapenemase (19). The potent activity of ceftazidime-avibactam against KPC-producing Enterobacteriaceae has been well documented in numerous studies (20, 21), and data from uncontrolled clinical series have shown its clinical efficacy (22) but also failures and relapses with ceftazidime-avibactam treatment (23), including the development of resistance during or following therapy (24, 25). This highlights the importance of the continued development of novel beta-lactam–beta-lactamase inhibitor combinations.
Vaborbactam (formerly RPX7009; Fig. 1), a new BLI based on a cyclic boronic acid pharmacophore, was discovered during a program specifically focused on targeting KPC carbapenemases. The biochemical, microbiological, and pharmacological properties of candidate BLIs were optimized for use in combination with a carbapenem antibiotic. Vaborbactam emerged from this effort and is being developed in combination with meropenem for the treatment of serious Gram-negative bacterial infections, including those due to KPC-producing carbapenem-resistant Enterobacteriaceae (CRE) (26).
FIG 1.
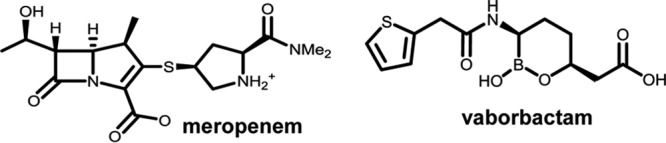
Chemical structures of meropenem and vaborbactam. Me2, dimethyl.
This communication provides a detailed characterization of the spectrum of beta-lactamase inhibition by vaborbactam and the impact of efflux and uptake on restoration of the potency of meropenem. A panel of engineered strains of Escherichia coli and K. pneumoniae producing various beta-lactamases and an isogenic set of KPC-producing strains of K. pneumoniae with various combinations of efflux and porin mutations were used in these studies.
RESULTS
Vaborbactam is a broad-spectrum inhibitor of diverse class A and class C beta-lactamases with potent inhibitory activity against KPC and other class A carbapenemases.
The profile of beta-lactamase inhibition by vaborbactam was evaluated using a panel of engineered strains of E. coli expressing diverse beta-lactamase genes coding for four molecular classes of beta-lactamases. Meropenem MICs against the strains producing various carbapenemases (class A carbapenemases KPC-2/KPC-3, SME, and NMC-A; class D carbapenemase OXA-48; class B carbapenemases VIM and NDM) ranged from ≤0.125 μg/ml to 16 μg/ml, whereas the meropenem MIC against the vector-only control strain, ECM6704, was ≤0.03 μg/ml (Table 1). As expected, the strains producing ESBLs (SHV, TEM, CTX-M) or class C enzymes (CMY, chromosomal AmpC from Enterobacter cloacae) had sensitivity to meropenem similar to that of parent strain ECM6704. Vaborbactam at 4 μg/ml decreased the meropenem MICs against the strains producing class A carbapenemases to the level for the vector-only control strain. Meropenem MICs against the strains producing OXA-48 or class B carbapenemases were not decreased.
TABLE 1.
MICs of ceftazidime and aztreonam alone or in combination with BLIs against the panel of engineered E. coli strains producing various cloned beta-lactamasesa
| Strain | Beta-lactamase | Class | Antibiotic MIC (μg/ml) in the absence or presence of BLIs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CAZ + VAB | CAZ + TZB | CAZ + CLA | ATM | ATM + VAB | ATM + TZB | ATM + CLA | MEM | MEM + VAB | |||
| ECM6704 | None | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 | |
| ECM6701 | KPC-2 | A-CARB | 4 | ≤0.125 | 4 | 2 | 32 | ≤0.125 | 16 | 16 | 2 | ≤0.03 |
| ECM6702 | KPC-3 | A-CARB | 16 | ≤0.125 | 16 | 8 | 32 | ≤0.125 | 16 | 16 | 2 | ≤0.03 |
| ECM6706 | SME-2 | A-CARB | 1 | ≤0.125 | ≤0.125 | 0.25 | >128 | 0.25 | 4 | 16 | 16 | ≤0.03 |
| ECM6696 | NMC-A | A-CARB | 0.5 | ≤0.125 | 0.25 | 0.25 | 64 | ≤0.125 | 8 | 8 | 1 | ≤0.03 |
| ECM6718 | SHV-5 | A-ESBL | 8 | 0.5 | ≤0.125 | ≤0.125 | 16 | 1 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6698 | SHV-12 | A-ESBL | 32 | 2 | ≤0.125 | ≤0.125 | 32 | 4 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6699 | SHV-18 | A-ESBL | 8 | 0.5 | ≤0.125 | ≤0.125 | 16 | 1 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6713 | TEM-10 | A-ESBL | 128 | 16 | 0.25 | 0.25 | 16 | 4 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6714 | TEM-26 | A-ESBL | 128 | 2 | ≤0.125 | 0.25 | 8 | 2 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6695 | CTX-M-3 | A-ESBL | 1 | ≤0.125 | ≤0.125 | ≤0.125 | 4 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6693 | CTX-M-14 | A-ESBL | 1 | ≤0.125 | ≤0.125 | ≤0.125 | 4 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6694 | CTX-M-15 | A-ESBL | 4 | ≤0.125 | ≤0.125 | ≤0.125 | 8 | 0.25 | ≤0.125 | ≤0.125 | ≤0.03 | ≤0.03 |
| ECM6692 | DHA-1 | C | 8 | 0.25 | ≤0.125 | 8 | 2 | 0.25 | ≤0.125 | 2 | ≤0.03 | ≤0.03 |
| ECM6691 | MIR-1 | C | 32 | 0.5 | 8 | 32 | 32 | 1 | 16 | 32 | ≤0.03 | ≤0.03 |
| ECM6705 | FOX-5 | C | 32 | 8 | 32 | 32 | 2 | 0.5 | 2 | 2 | ≤0.03 | ≤0.03 |
| ECM6715 | AmpC-ECL (P99-like) | C | 16 | 0.25 | 1 | 16 | 16 | 0.5 | 2 | 16 | ≤0.03 | ≤0.03 |
| ECM6700 | CMY-2 | C | 16 | 0.25 | 0.5 | 16 | 8 | 0.25 | 1 | 8 | ≤0.03 | ≤0.03 |
| ECM6697 | OXA-2 | D | 1 | 1 | 0.25 | ≤0.125 | ≤0.125 | ND | ND | ND | ≤0.03 | ≤0.03 |
| ECM6712 | OXA-10 | D | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ND | ND | ND | ≤0.03 | ≤0.03 |
| ECM6716 | OXA-48 | D-CARB | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ND | ND | ND | 0.125 | 0.125 |
| ECM6703 | NDM-1 | B | >128 | >128 | >128 | >128 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | 16 | 16 |
| ECM6711 | VIM-1 | B | 128 | 128 | 128 | 128 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | 1 | 1 |
All beta-lactamase inhibitors were tested at a fixed concentration of 4 μg/ml. BLIs, beta-lactamase inhibitors; CAZ, ceftazidime; ATM, aztreonam; MEM, meropenem; VAB, vaborbactam; TZB, tazobactam; CLA, clavulanic acid; ND, not done; A-CARB, class A carbapenemase; D-CARB, class D carbapenemase.
In order to fully characterize the spectrum of inhibition by vaborbactam, the activities of ESBL-labile agents ceftazidime and aztreonam were tested against a panel of beta-lactamase-producing strains alone and in combination with 4 μg/ml of vaborbactam, tazobactam, or clavulanic acid. In the absence of vaborbactam, the ceftazidime and aztreonam MICs against strains producing class A carbapenemases ranged from 0.5 to 16 μg/ml and from 32 to >128 μg/ml, respectively. As was seen with meropenem, vaborbactam strongly enhanced the activity of both antibiotics against these strains. In the presence of vaborbactam at 4 μg/ml, the MICs for both antibiotics were ≤0.125 μg/ml, the same as that seen for the strain containing the vector alone, indicating complete inhibition of beta-lactamase activity (Table 1).
Vaborbactam also restored the activity of ceftazidime and aztreonam against various ESBL-producing strains. It had potency comparable to that of tazobactam and clavulanic acid against CTX-M (CTX-M-3, CTX-M-14, CTX-M-15)-producing strains, but it was generally less potent against SHV (SHV-5, SHV-12, SHV-18) and TEM (TEM-10, TEM-26) producers (Table 1). In strains producing class C cephalosporinases, ceftazidime and aztreonam MICs decreased from 8 to 32 μg/ml and from 2 to 32 μg/ml, respectively, for the antibiotics alone to 0.25 to 8 μg/ml and to 0.25 to 1 μg/ml, respectively, for the antibiotics with vaborbactam at 4 μg/ml. Checkerboard experiments using aztreonam confirmed the higher potency (8- to 32-fold) of vaborbactam at reducing the MICs of antibiotics against strains producing KPC (and other class A carbapenemases) than against strains producing ESBLs (in particular, SHV/TEM) and class C beta-lactamases (see Table S3 in the supplemental material). Among the tested class D enzymes (OXA-2, OXA-10, OXA-48), only OXA-2 production resulted in a measurable increase in the ceftazidime MIC (from ≤0.125 to 1 μg/ml). The MIC for ECM6607 expressing OXA-2 remained 1 μg/ml in the presence of vaborbactam 4 μg/ml, indicating that OXA-2, similar to OXA-48, is not inhibited by this BLI. Vaborbactam did not have any inhibitory activity against the metallo-beta-lactamases NDM-1 and VIM-1 (Table 1). Evaluation of strains expressing additional metallo-beta-lactamases (multiple IMPs, SPM, CcrA, L1) confirmed this result (Table S4).
These studies demonstrate that vaborbactam is a broad-spectrum inhibitor of class A and class C beta-lactamases with particularly potent inhibitory activity against class A carbapenemases, such as KPC.
Vaborbactam restores the potency of meropenem against engineered strains of K. pneumoniae with increased meropenem MICs due to the combination of ESBL or class C beta-lactamase production and reduced permeability due to porin mutations.
While hydrolysis by carbapenemases is the major mechanism of carbapenem resistance in clinical isolates, low-level resistance to carbapenems in Enterobacteriaceae can occur when some class A or class C noncarbapenemases are produced by strains with mutations resulting in reduced outer membrane permeability (27, 28). The activity of meropenem alone and in combination with vaborbactam against a panel of engineered strains of K. pneumoniae with or without permeability defects that produced selected class A or class C beta-lactamases was evaluated. KPC- and OXA-48-producing strains were used as positive and negative controls, respectively, for restoration of the potency of meropenem (Table 2). Only plasmids that carried KPC-2 and KPC-3 were able to confer a modest meropenem MIC increase (from ≤0.06 to 0.5 μg/ml) compared to the MIC for wild-type strain K. pneumoniae KPM1001 (ATCC 43816) (Table S2). When the same plasmids with these beta-lactamase genes were transformed into isogenic strain KPM1176 (Table S2), which overexpressed the acrAB-tolC operon, had ompK35 downregulated due to a mutation in the ramR gene, and contained a loss-of-function mutation in ompK36, the meropenem MICs increased compared to those for the corresponding KPM1001 transformants; KPC- and OXA-48-producing strains each had MICs of 64 μg/ml, which corresponds to a 128-fold MIC increase compared to that for the vector-only control strain. As expected, vaborbactam enhanced the activity of meropenem against KPC-producing strains but not OXA-48-producing strains. An increase in the vaborbactam concentration was associated with a further reduction of the meropenem MIC for KPC-producing derivatives of KPM1176 (data not shown). CTX-M-15, CMY-2, and AmpC-ECL (P99-like) production but not SHV-12 production in KPM1176 was associated with a 4- to 8-fold increase in the meropenem MIC, but vaborbactam at 4 μg/ml reduced the MICs back to the level for the vector-only control strain (Table 2).
TABLE 2.
MICs of meropenem alone or in combination with vaborbactam against isogenic engineered strains of K. pneumoniae strains producing various beta-lactamasesa
| Cloned beta-lactamase | Host strain KPM1001 |
Host strain KPM1176 |
||||
|---|---|---|---|---|---|---|
| Strain producing beta-lactamase | MIC (μg/ml) |
Strain producing beta-lactamase | MIC (μg/ml) |
|||
| MEM | MEM + VAB | MEM | MEM + VAB | |||
| pUCP24 | KPM1116 | ≤0.06 | ≤0.06 | KPM1950 | 0.5 | 0.25 |
| KPC-2 | KPM1113 | 0.5 | ≤0.06 | KPM1931 | 64 | 2 |
| KPC-3 | KPM1049 | 0.5 | ≤0.06 | KPM2840 | 64 | 2 |
| CTX-M-15 | KPM1114 | ≤0.06 | ≤0.06 | KPM1943 | 2 | 0.5 |
| SHV-12 | KPM1115 | ≤0.06 | ≤0.06 | KPM1945 | 0.5 | 0.25 |
| CMY-2 | KPM1045 | ≤0.06 | ≤0.06 | KPM1948 | 2 | 0.5 |
| AmpC-ECL | KPM1956 | ≤0.06 | ≤0.06 | KPM1959 | 4 | 1 |
| OXA-48 | KPM1939 | ≤0.06 | ≤0.06 | KPM1941 | 64 | 64 |
Vaborbactam was used at a fixed concentration of 4 μg/ml. MEM, meropenem; VAB, vaborbactam. Based on sequence analysis (GenBank accession no. CP009208), KPM1001 (ATCC 43816) is a wild-type strain of K. pneumoniae containing full copies of acrAB, ompK35, and ompK36 and no mutations in known regulators of these genes; hence, their level of expression is the same as that in the wild type. KPM1176 is an isogenic mutant (Table S2) that contains a mutation in the gene ramR (a frameshift from amino acid 46). This mutation is responsible for overexpression of the acrAB operon and for downregulation of the gene ompK35 encoding the corresponding porin (see Table 5 and Table S5 in the supplemental material). KPM1176 also carries the loss-of-function mutation in the gene ompK36 (a frameshift from amino acid 266).
The reduction in beta-lactam MICs for KPC-producing strains by vaborbactam is greatest when vaborbactam is paired with a carbapenem rather than other classes of beta-lactams.
The effect of the vaborbactam concentration on the reduction in the MICs for several carbapenems (meropenem, biapenem, ertapenem, tebipenem, and imipenem) as well as ceftazidime, aztreonam, and cefepime against an engineered KPC-3-producing strain of K. pneumoniae KPM1271 (which has the KPM1001 and KPM1026a background and which is characterized by wild-type expression of the major efflux pump AcrAB-TolC and the outer membrane porins OmpK35 and OmpK36 [see Table 5]) was determined. The potency of vaborbactam was expressed as the minimum potentiation concentration (MPC) required to reduce the carbapenem MIC by 16-fold (MPC16) in a KPC-producing strain (KPM1271). In general, vaborbactam MPC16 values were lower when vaborbactam was tested with carbapenems than when it was tested with other beta-lactams; the vaborbactam MPC16 ranged from 0.03 to 0.06 μg/ml for all the carbapenems, with the exception of imipenem (MPC16, 0.25 μg/ml). For cefepime, aztreonam, and ceftazidime, the vaborbactam MPC16 values were 0.125 to 0.25 μg/ml (Table 3). These results indicate that vaborbactam has the most potent activity against this KPC-producing strain when it is tested in combination with most carbapenems compared with its activity when it is tested in combination with other beta-lactam antibiotics.
TABLE 5.
Effect of efflux and porins on activities of antibiotics against a panel of isogenic strains of K. pneumoniae with efflux and porin mutations
| Straina | Genotype | Effect of: |
MICb (μg/ml) |
|||
|---|---|---|---|---|---|---|
| OmpK35 | OmpK36 | AcrAB | MER | MIN | ||
| KPM1026ac | Wild type | Wild type (1) | Wild type (1) | Wild type (1) | 0.03 | 1 |
| KPM2600 | ΔompK35 | Nonfunctional | Wild type | Wild type | 0.03 | 2 |
| KPM2592 | ΔompK36 | Wild type | Nonfunctional | Wild type | 0.03 | 1 |
| KPM2040 | ompK36_2067d | Wild type | Nonfunctional | Wild type | 0.03 | 1 |
| KPM2613 | ompK36_2067 ΔompK35 | Nonfunctional | Nonfunctional | Wild type | 0.125 | 2 |
| KPM2966 | ramRe ompK36_2067 ΔompK35 | Nonfunctional | Nonfunctional | Upregulated | 0.5 | 8 |
| KPM1027f | ramRg | Downregulated (0.1) | Wild type (0.9) | Upregulated (3.1) | 0.03 | 8 |
| KPM2610 | ramR ΔompK35 | Nonfunctional | Wild type | Upregulated | 0.03 | 8 |
| KPM2658 | ramR ΔompK36 | Downregulated | Nonfunctional | Upregulated | 0.25 | 8 |
| KPM1176h | ramR ompK36_1176 | Downregulated (0.1) | Nonfunctional | Upregulated (3.9) | 0.25 | 8 |
All strains contained chromosomal SHV enzyme, encoded by blaSHV-24.
MER, meropenem; MIN, minocycline.
KPM1026a is a streptomycin-resistant mutant of wild-type strain KPM1001 (ATCC 43816). It contains a functional acrAB operon and functional ompK35 and ompK36 genes. Normalized expression of acrB, ompK35, and ompK36 in this strain was set equal to 1 (the normalized relative level of expression is shown in parentheses). The minocycline MIC for KPM1026a reflects the wild-type level of acrAB expression.
Insertion of an A reside at nucleotide 160 of ompK36, causing a frameshift from amino acid 54 of OmpK36.
A G490T substitution in ramR created TAA at amino acid 164.
KPM1027 is a derivative of KPM1026a selected on tigecycline. It has a mutation in the negative regulator gene ramR and as a result has the acrAB operon overexpressed ∼3-fold and the ompK35 gene downregulated ∼10-fold relative to their levels of expression in KPM1026a. Expression of ompK36 in KPM1027 is unchanged relative to that in KPM1026a (the normalized relative level of expression is shown in parentheses).
An 8-bp insertion in ramR causing a frameshift from amino acid 46.
KPM1176 was selected from KPM1027 on meropenem at 0.25 μg/ml. It has a mutation in the ompK36 gene that results in a frameshift in OmpK36 from amino acid 266. Similar to KPM1027, it has acrAB overexpressed and ompK35 downregulated ∼3- to 4-fold and ∼10-fold, respectively, relative to their levels of expression in KPM1026a (the normalized relative level of expression is shown in parentheses). The functional status and expression levels of acrAB, ompK35, and ompK36 in other strains were inferred from strain construction, ramR sequence analysis, and minocycline MICs.
TABLE 3.
Effect of vaborbactam concentration on MICs of antibiotics against the engineered KPC-3-producing strain of K. pneumoniae, KPM1271a
| Antibiotic | MIC (μg/ml) in the presence of the following concn of vaborbactam (μg/ml): |
MPC16 (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.015 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | ||
| Meropenem | 16 | 2 | 1 | 0.5 | 0.25 | 0.25 | 0.125 | ≤0.06 | 0.03 |
| Biapenem | 16 | 4 | 4 | 1 | 1 | 0.5 | 0.25 | 0.25 | 0.06 |
| Ertapenem | 32 | 8 | 2 | 1 | 0.5 | 0.25 | 0.25 | ≤0.06 | 0.03 |
| Tebipenem | 32 | 8 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.125 | 0.03 |
| Imipenem | 8 | 8 | 4 | 2 | 1 | 0.5 | 0.5 | 0.5 | 0.25 |
| Aztreonam | 32 | 32 | 32 | 32 | 2 | 1 | 0.25 | 0.125 | 0.125 |
| Ceftazidime | 64 | 64 | 64 | 32 | 8 | 4 | 1 | 1 | 0.25 |
| Cefepime | 4 | 4 | 2 | 0.5 | 0.125 | 0.06 | 0.03 | 0.03 | 0.125 |
MPC, minimal potentiation concentration; MPC16, the MPC of vaborbactam required to reduce the MIC by 16-fold. KPM1271 was constructed by conjugating plasmid pKpQIL, which carries the gene for KPC-3, from clinical isolate KP1074 into KPM1026a, the streptomycin-resistant mutant of wild-type strain KPM1001 (ATCC 43816 with wild-type expression of a major efflux pump AcrAB-TolC and functional porins OmpK35 and OmpK36).
The potency of vaborbactam is similar against strains producing KPC-2 or KPC-3.
The effect of the vaborbactam concentration on the meropenem MIC was determined in KPM1176 derivatives KPM1931 and KPM2840, producing KPC-2 and KPC-3, respectively (Table 4). This particular host strain with permeability defects (see above) was used to increase the sensitivity of the potentiation assay. The meropenem MICs and the concentration-response of vaborbactam inhibitory potency were comparable in these two strains, indicating that meropenem and vaborbactam have similar activities against strains producing KPC-2 or KPC-3.
TABLE 4.
Effect of vaborbactam concentration on meropenem MICs for engineered strains of K. pneumoniae producing cloned KPC-2 and KPC-3 beta-lactamasesa
| Strain | Beta-lactamase | Meropenem MIC (μg/ml) in the presence of the following concn of vaborbactam (μg/ml): |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ||
| KPM1941 | KPC-2 | 128 | 64 | 16 | 4 | 2 | 1 | 0.5 | 0.5 |
| KPM2840 | KPC-3 | 128 | 64 | 8 | 4 | 2 | 1 | 0.5 | 0.5 |
Plasmids containing cloned blaKPC-2 and blaKPC-3 were transformed into strain KPM1176, in which acrAB is upregulated and ompK35 is downregulated and which carries a loss-of-function mutation in ompK36 (see Table 5 and Table S5 in the supplemental material).
OmpK35 and OmpK36 outer membrane porins, but not efflux, are important determinants of vaborbactam uptake in K. pneumoniae.
To investigate the effect of increased efflux and various porin mutations on the microbiological potency of vaborbactam, a set of isogenic strains of K. pneumoniae with various combinations of efflux and porin mutations was constructed. The natural plasmid pKpQIL, which carries KPC-3, was introduced into each strain by conjugation using the clinical strain KP1074 (now ATCC BAA-2814) as a donor.
(i) Parent carbapenemase-negative strains and meropenem.
The MICs of meropenem and minocycline against the parent strains that lack KPC are shown in Table 5. Minocycline, a known substrate of AcrAB-TolC, was included as a control for AcrAB activity. Inactivation of ompK35 or ompK36 individually did not affect the meropenem MICs; the meropenem MICs for KPM2600 (ΔompK35) and KPM2592 (ΔompK36), or KPM2040 (ompK36_2067, with a frameshift at amino acid 54) were unchanged from the MIC for KPM1026a (the wild-type parental strain; MIC = 0.03 μg/ml). Inactivation of both ompK35 and ompK36 (KPM2613) increased the meropenem MIC 4-fold (to 0.125 μg/ml). Overexpression of acrAB due to a mutation in the negative regulator ramR did not affect the MIC of meropenem (KPM1026a versus KPM1027; Table 5 and S5). Of note, the same mutation also led to the downregulation of ompK35 (Table S5), indicating that overexpression of acrAB did not have an effect on the meropenem MIC even in combination with a decreased level of expression of ompK35. The complete inactivation of ompK35 in the strain overexpressing acrAB did not affect the MIC of meropenem (KPM2610 versus KPM1027); however, the overexpression of acrAB in the strain with both ompK35 and ompK36 inactivated increased the meropenem MIC 4-fold to 0.5 μg/ml (KPM2966 versus KPM2613). Inactivation of ompK36 in the strain with a reduced level of expression of ompK35 and overexpression of acrAB (due to the mutation in the ramR gene) increased the MIC of meropenem 8-fold to 0.25 μg/ml (KPM2658 and KPM1176 versus KPM1027). These data indicate that the overexpression of acrAB caused a 2- to 4-fold increase in the MIC of meropenem in the absence of both major porins and that the meropenem MIC could be increased up to 16-fold when double porin mutations were combined with increased efflux.
(ii) KPC-3-producing strains and meropenem.
The MICs of meropenem alone or with various concentrations of vaborbactam against isogenic KPC-producing strains are shown in Table 6. The meropenem MIC for the wild-type strain KPM1271 (in which both porins are produced and which has a normal level of acrAB), producing the KPC-3 beta-lactamase, was 16 μg/ml. Overexpression of acrAB or inactivation of ompK35 did not have any measurable effect on the MIC of meropenem (KPM1271 versus KPM1272 and KPM2601). Inactivation of ompK36 alone increased the meropenem MIC by 2-fold (KPM1271 versus KPM2599 and KPM2067); note that no such increase was seen in the absence of KPC (Table 5). Inactivation of ompK35 in the strain that did not have a functional ompK36 increased the meropenem MIC by 8-fold (from 32 to 256 μg/ml; KPM2067 versus KPM2631). Notably, overexpression of acrAB did not have any effect on the meropenem MIC even when both major porins were nonfunctional (KPM2631 versus KPM2818 or KPM2631 versus KPM2965). The kinetic parameters of meropenem efflux by AcrAB-TolC are unknown; however, if the pump is saturated at a concentration of meropenem that is lower than that required to inhibit the growth of the KPC-producing strain, the impact of efflux on the meropenem MIC is expected to be negligible.
TABLE 6.
Effects of various concentrations of vaborbactam on meropenem MICs in isogenic KPC-3-producing strains of K. pneumoniae with efflux and porin mutations
| KPC-3a-containing strain | Parent strain | Description | Meropenem MIC (μg/ml) in the presence of the following concn of vaborbactam (μg/ml): |
MPCmaxb (μg/ml) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | ||||
| Isogenic laboratory strains | ||||||||||||||||
| KPM1271 | KPM1026a | Wild type | 16 | 0.25 | 0.25 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.25 |
| KPM2601 | KPM2600 | ompK35 inactivated | 16 | 2 | 1 | 0.5 | 0.125 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 1 |
| KPM2599 | KPM2592 | ompK36 inactivated | 32 | 16 | 16 | 8 | 8 | 8 | 0.5 | 0.25 | 0.125 | 0.06 | 0.06 | 0.06 | 0.06 | 16 |
| KPM2067 | KPM2040 | ompK36 inactivated | 32 | 32 | 16 | 16 | 16 | 4 | 1 | 0.5 | 0.125 | 0.06 | 0.06 | 0.06 | 0.06 | 16 |
| KPM2631 | KPM2613 | omp35 and ompK36 inactivated | 256 | 256 | 256 | 128 | 128 | 64 | 16 | 4 | 1 | 0.5 | 0.25 | 0.25 | 0.125 | 128 |
| KPM2965 | KPM2966 | acrAB upregulated, ompK35 and ompK36 inactivated | 256 | 256 | 256 | 128 | 128 | 64 | 32 | 8 | 2 | 1 | 0.5 | 0.5 | 0.25 | 128 |
| KPM1272 | KPM1027 | acrAB upregulated, ompK35 downregulated | 16 | 8 | 2 | 2 | 0.5 | 0.25 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 | 2 |
| KPM2818 | KPM2658 | acrAB upregulated, ompK35 downregulated, ompK36 inactivated | 256 | 256 | 256 | 128 | 64 | 32 | 16 | 8 | 2 | 1 | 1 | 0.5 | 0.5 | 64 |
| Clinical strains and derivatives | ||||||||||||||||
| KP1074 | NAc | ompK35 inactivated,d OmpK36 is the same as in KP1004 but has the GD repeate | 128 | 128 | 128 | 64 | 64 | 64 | 8 | 1 | 0.5 | 0.25 | 0.125 | 0.125 | 0.125 | 32 |
| KPM2644f | KP1074 | KP1074 ΔompK36, ompK35 and ompK36 inactivated | 512 | 512 | 512 | 512 | 256 | 256 | 128 | 8 | 2 | 1 | 0.5 | 0.5 | 0.5 | 32 |
| KP1004 | NA | ompK35 inactivated, full-length ompK36 | 32 | 4 | 4 | 2 | 0.5 | 0.125 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 2 |
All strains produced KPC-3 and TEM-1, encoded by genes carried on plasmid pKpQIL. Both KPM1026a derivatives and clinical isolates also produced a chromosomal SHV enzyme, encoded by blaSHV-24 and blaSHV-11, respectively.
MPCmax, maximum potentiating concentration of the BLI required to reduce the meropenem MIC to the level seen in the parent strain that lacks KPC, corresponding to complete inhibition of KPC.
NA, not available.
A frameshift in the OmpK35 sequence at amino acid 42.
The GD repeat is a duplication of two amino acids, Gly134 and Asp135, located within the L3 internal loop and associated with reduced susceptibility to carbapenems due to constriction of the channel (29).
KPM2644 was constructed as follows. First, pKpQIL was cured from KP1074. Second, the resulting strain was used to select for an Smr mutant (on 200 μg/ml of streptomycin) to facilitate conjugation experiments. Third, ompK36 was disrupted in KPM1308, giving rise to KPM2617. Finally, plasmid pKpQIL was conjugated from KP1074 into KPM2617.
(iii) KPC-3-producing strains and meropenem-vaborbactam.
MPCmax corresponds to the concentration of vaborbactam that is required to achieve the maximal reduction of the meropenem MIC and was the parameter used to evaluate the potency of vaborbactam (maximal potentiation; Table 6). Inactivation of ompK35 alone increased the vaborbactam MPCmax 4-fold (KPM2601 versus KPM1271), while inactivation of ompK36 alone increased the vaborbactam MPCmax 64-fold. These results indicate that both porins are involved in the passage of vaborbactam across the outer membrane of K. pneumoniae, but OmpK36 plays the major role (KPM2599 and KPM2067 versus KPM1271). Inactivation of ompK35 in the strain that already lacked ompK36 (KPM2631 versus KPM2067) increased the MPCmax 4-fold, consistent with some contribution of OmpK35 to vaborbactam uptake but a contribution less significant than that of OmpK36.
Strains KPM1272 and KPM1271 were used to assess the effect of the ramR mutation. Inactivation of ramR (KPM1271) resulted in both acrAB overexpression and ompK35 downregulation. The MPCmax was 8-fold higher for KPM1272 than for KPM1271; however, the MPCmax for KPM1272 was only 2-fold higher relative to that for KPM2601, the mutant that lacked OmpK35. In addition, no effect of AcrAB overexpression on vaborbactam potency was seen in the strains that lacked (or had low levels of expression of) both OmpK35 and OmpK36 (KPM2631 versus KPM2818 and KPM2631 versus KPM2965). These data indicate that while the function of either of the major porins affects the potency of vaborbactam, AcrAB-mediated efflux has a minimal effect.
(iv) Effect of Gly134 and Asp135 duplication in OmpK36 on vaborbactam inhibition in KPC-producing strains of K. pneumoniae.
Strains of K. pneumoniae that carry a variant of OmpK36 with a duplication of two amino acids, Gly134 and Asp135 (GD repeat), are frequently reported in clinical settings (12, 13, 29, 30). We determined the MICs of meropenem alone or with various concentrations of vaborbactam against KP1074 (which expresses OmpK36 with the GD repeat) and its derivative, KPM2644, which carried a deletion of the ompK36 gene. Another clinical isolate, KP1004, was also included in this experiment. Both strain KP1074 and strain KP1004 carry the same KPC plasmid and have nonfunctional OmpK35 due to the same mutation, a frameshift at amino acid 42, and nearly identical OmpK36 amino acid sequences other than the GD repeat in the OmpK36 of KP1074.
The meropenem MIC for KP1074 (128 μg/ml) was 4-fold higher than that for KP1004 (32 μg/ml) (Table 6). While these strains are not truly isogenic, the GD repeat in KP1074 is the most likely reason for this difference. The MPCmax was 16-fold higher for KP1074 than for KP1004, implicating that the GD repeat reduced the potency of vaborbactam in this strain as well. Deletion of the ompK36 gene from KP1074 increased the MIC of meropenem 4-fold to 512 μg/ml (KPM2644 versus KP1074; Table 6); however, almost no concomitant decrease in vaborbactam potency was observed, with the MPCmax remaining unchanged. Thus, it appears that OmpK36 with a functionally constricted inner channel due to the 2-amino-acid duplication in the L3 loop still maintains some, albeit a decreased level of, meropenem uptake. Uptake is further reduced by deletion of ompK36, where vaborbactam appears to be unable to pass through the narrow channel of OmpK36 altered because of the GD duplication.
DISCUSSION
An important feature of the drug discovery program that resulted in vaborbactam was the early choice of a partner antibiotic, which allowed a specific focus on the most relevant beta-lactamase-mediated resistance mechanism. From the start of the discovery effort, we intended to combine a carbapenem antibiotic with a novel BLI that was specifically optimized for the inhibition of KPC, the most prevalent carbapenemase in the United States and many other geographic regions (31). The main focus was on the simultaneous optimization of both the kinetics of KPC inhibition and microbiological activity, i.e., enhancing the potency of carbapenems against KPC-producing strains. Since carbapenems are stable to hydrolysis by the majority of class A and class C enzymes, we focused on optimizing the potency against a specific target rather than attempting to achieve potent broad-spectrum inhibition of multiple enzymes.
KPC-2 and KPC-3 represent the most prevalent KPC variants reported worldwide (32–35). Ceftazidime hydrolysis is ∼10-fold more efficient with KPC-3 than with KPC-2 (36, 37). A recent study by Shields and colleagues (38) found that median ceftazidime-avibactam MICs were higher against KPC-3 variants than against KPC-2 variants. In contrast, the studies described here indicated that there is no difference between the KPC-2 and KPC-3 hydrolysis of meropenem. Similarly, the similar concentration-effect response for the restoration of meropenem activity by vaborbactam against engineered strains producing either KPC-2 or KPC-3 observed in this study is also consistent with the biochemistry of meropenem hydrolysis.
In panels of engineered strains expressing cloned genes for various beta-lactamases, it was demonstrated that vaborbactam is an inhibitor of various class A and class C beta-lactamases and that its potency in the restoration of the activity of beta-lactams varied depending on the specific enzyme. Vaborbactam was the most potent against KPC-producing strains and the least potent against TEM- and SHV-producing strains. Since vaborbactam is being developed in combination with meropenem, which is stable to hydrolysis by the majority of beta-lactamases, including TEM and SHV, it is its high potency against KPC and other class A carbapenemases that constitutes its most relevant attribute as a BLI. Indeed, recent studies using large collections of clinical isolates producing the KPC carbapenemase demonstrated that vaborbactam significantly increased the activity of carbapenems against these strains (31, 39, 40). Recent biochemical experiments using nitrocefin as a substrate demonstrated that in the case of KPC-2, CTX-M-15, and P99 AmpC, vaborbactam behaved as a two-step tight-binding inhibitor that forms an initial noncovalent complex with an enzyme (characterized by affinity constant K) followed by enzyme inactivation upon covalent bond formation (characterized by rate constant k2). Moreover, vaborbactam inactivated various beta-lactamases with a comparable efficiency (measured as the k2/K ratio): the inactivation efficiency was ∼0.007 μM−1 s−1 for KPC-2 and ∼0.02 μM−1 s−1 for CTX-M-15 and P99 AmpC. However, the off rate of dissociation (koff) of the vaborbactam–beta-lactamase complex varied dramatically (50- to 200-fold) for different enzymes: koff was 0.000017 s−1 (residence time [RT] = 992 min) for KPC-2, whereas koff was 0.0009 s−1 (RT = 19 min) and 0.056 s−1 (RT = 3 min) for CTX-M-15 and P99 AmpC, respectively. In the case of the SHV-12 and TEM-43 enzymes, vaborbactam exhibited a fast-on–fast-off behavior with no signs of enzyme inactivation (41). It is conceivable that the low off rate of vaborbactam for KPC plays an important role in the high potency of this compound in enhancing the activities of antibiotics against KPC-producing strains determined in microbiological experiments. By the same token, the low potency with which vaborbactam enhances the activity of antibiotics against SHV- and TEM-producing strains might be driven by its inability to form a stable inhibitory complex with these enzymes.
Vaborbactam was more potent in restoring the activity of carbapenems (with the exception of imipenem) than that of other substrates of KPC. The results of microbiological experiments are in good agreement with those of biochemical experiments that demonstrated that the Ki of KPC inhibition was ∼15-fold lower when carbapenems were used as KPC substrates than when the model cephalosporin substrate nitrocefin was used as a KPC substrate (R. Tsivkovski and O. Lomovskaya, unpublished observations). The KPC-mediated hydrolysis of imipenem was inhibited by vaborbactam with a potency ∼10-fold lower (the highest Ki) than that of other carbapenems. On a biochemical level, the lower that the ability of a beta-lactamase to inactivate a given substrate is, the easier it is to inhibit such hydrolysis. Notably, meropenem appears to be a poorer substrate for KPC than nitrocefin and imipenem: the enzymatic efficiency of meropenem hydrolysis (kcat/Km) is ∼4-fold and ∼6-fold lower than that of the hydrolysis of nitrocefin and imipenem, respectively (36, 37). These data underscore the importance of optimization of a BLI with a specific partner beta-lactam.
While the inhibitory activity of vaborbactam against class A ESBLs and class C cephalosporinases is generally less important when it is paired with a stable carbapenem, the inhibition of these beta-lactamases may be important in some non-carbapenemase-producing, carbapenem-resistant strains of Enterobacteriaceae with porin mutations (27, 28). Data obtained from studies using engineered strains indicate that vaborbactam restores the activity of meropenem against these strains. The greater potency of vaborbactam for KPC enzymes compared to that for other enzymes becomes important, in that bacterial cells rarely produce KPC alone; most frequently, a single cell simultaneously produces other beta-lactamases, in addition to KPC (38, 42). However, since it is KPC that hydrolyzes meropenem, an inhibitor with a higher affinity for KPC will mainly target this enzyme and not other beta-lactamases that hydrolyze some beta-lactams but not meropenem.
Numerous studies have demonstrated the contribution of porin mutations to the phenotype of reduced susceptibility to carbapenems in clinical isolates of Enterobacteriaceae, irrespective of the presence of a carbapenemase (11, 28, 43, 44). In addition, the major efflux pump AcrAB has been implicated in reduced carbapenem susceptibility in clinical strains of Enterobacteriaceae (45). High-level resistance to carbapenems is associated with the production of a carbapenemase in the strains that lack major porins due to mutations or downregulation of the corresponding genes and have increased efflux (10, 11, 46). The impact of porin and efflux mutations on the potency of vaborbactam and meropenem-vaborbactam against isogenic mutants of KPC-producing K. pneumoniae demonstrated that, similar to carbapenems, vaborbactam can cross the outer membrane using both the OmpK35 and OmpK36 porins; however, unlike carbapenems that can use both porins with a similar efficiency (inactivation of OmpK35 or OmpK36 individually does not affect the carbapenem MIC), OmpK36, which has a smaller channel than OmpK35, appears to play a more significant role in the passage of vaborbactam across the outer membrane than OmpK35. This conclusion was based on the finding that inactivation of either OmpK35 or Omp36 individually reduces the potency of vaborbactam. Inactivation of OmpK35 was associated with an effect much smaller than that associated with the inactivation of OmpK36, indicating that OmpK36 is the preferred porin for vaborbactam uptake. Surprisingly, the effect of the minor porin mutation was still observable in the context of the functional major porin. We hypothesize that there is some interplay between meropenem and vaborbactam uptake. When both porins are open, meropenem and vaborbactam use their preferred channels, while the closure of one porin may result in competition for passage through the single porin. An important finding was a marked reduction of the potency of vaborbactam against strains that lacked both major porins. Another finding was that a variant of OmpK36 that was predicted to have a constricted inner channel as a result of a 2-amino-acid duplication in the L3 loop and that was partially defective in meropenem uptake appeared to be completely defective in the uptake of vaborbactam. Other mutations that affect ompK36 have been reported in clinical isolates, such as the downregulation of ompK36 expression due to the insertion of IS5 in its promoter region (12); the effect of such mutations was not evaluated in this study. Unlike meropenem, vaborbactam does not appear to be a substrate of the AcrAB-TolC efflux pump, since overexpression of AcrAB had a minimal effect on vaborbactam potency, even in the absence of major porins.
This study demonstrated that established drug resistance mechanisms contribute to the potencies of meropenem and vaborbactam alone and in combination. The biggest reduction of meropenem-vaborbactam potency was observed in KPC-producing strains lacking both porins and overexpressing AcrAB. Investigation of the concentration-response for the restoration of meropenem activity by vaborbactam against various KPC-producing strains with these additional mutations indicates that with vaborbactam concentrations of ≥8 μg/ml, the meropenem MICs are ≤2 μg/ml, even for the most resistant strains containing multiple mutations. This result informed studies that were designed to identify the target in vivo exposures for both meropenem and vaborbactam required to maximize the efficacy of the combination, overcome the negative impact of preexisting mutations in the envelope genes of Gram-negative bacteria, and minimize the emergence of resistance de novo. Recently completed clinical trials demonstrate that these target exposures appear to be achievable due to the excellent safety profiles of both meropenem and vaborbactam (47, 48).
MATERIALS AND METHODS
Panels of engineered bacterial strains containing cloned beta-lactamases and various combinations of porin and efflux mutations.
The panel of engineered isogenic strains of E. coli producing individual beta-lactamases was constructed to study the profile of beta-lactamase inhibition by vaborbactam. DNA samples isolated from clinical strains producing various beta-lactamases were used as the templates for PCR with high-fidelity DNA polymerase Vent (New England BioLabs). The PCR primers contained sequences for restriction enzymes to facilitate cloning into vector plasmid pUCP24 (49) at the multiple-cloning site. After digestion with the appropriate enzymes and ligation, the ligation mix was transformed into E. coli DH5α competent cells and transformants were selected on LB agar plates containing gentamicin at 15 μg/ml. After confirmation of the correct clones by restriction digestion and sequencing, selected recombinant plasmids were introduced into E. coli ECM5497 (MG1655) and two K. pneumoniae strains, KPM1001 (ATCC 43816) and KPM1176, by transformation and selection on plates containing gentamicin at the appropriate concentrations. The sequences of the primers used for the amplification of the beta-lactamase genes are provided in Table S1 in the supplemental material. On the basis of sequence analysis (GenBank accession no. CP009208 [50]) KPM1001 is a wild-type strain containing functional genes encoding efflux pumps, such as AcrAB-TolC and the major porins OmpK35 and OmpK36. This strain does not have mutations in known regulators of efflux and porin genes; hence, expression of these genes was considered to be at the wild-type level. KPM1176 is an isogenic derivative of KPM1001 (Table S2) with acrAB overexpressed, ompK35 downregulated (Table S5) due to a mutation in the ramR gene (a frameshift at amino acid 46 of RamR), and a loss-of-function mutation in ompK36 (a frameshift at amino acid 266 of OmpK36).
The panel of engineered isogenic strains of K. pneumoniae with various combinations of porin and efflux mutations with or without KPC was used to evaluate the impact of various molecular determinants on meropenem and meropenem-vaborbactam MICs. Most of the strains are derivatives of wild-type strain KPM1026a, a streptomycin-resistant mutant of KPM1001 (ATCC 43816). All KPC-3-producing strains were constructed by conjugating the plasmid pKpQIL (51) from clinical isolate K. pneumoniae KP1074 (ATCC BAA-2814) into various isogenic derivatives of KPM1026a.
Two clinical KPC-3-producing clinical isolates, KP1074 and KP1004, were used in this study. KP1074 has a nonfunctional OmpK35 due to a frameshift mutation at amino acid 42. It also contains a defective variant of OmpK36 that has a duplication of 2 amino acids, Gly134 and Asp135 (the GD repeat). This duplication in the L3 internal loop of OmpK36 is commonly seen in sequence type 258 (ST258) isolates carrying blaKPC and is predicted to functionally constrict the inner channel of the porin (29), causing reduced susceptibility to carbapenems. KPM2644 is a derivative of KP1074 with a complete deletion of the ompK36 gene. Another clinical isolate, KP1004, similar to KP1074, carries the KPC-producing plasmid pKpQIL. It also has the same frameshift at amino acid 42 in OmpK35 found in KP1074. KP1004 and KP1074 have nearly identical OmpK36 amino acid sequences; the only difference is the absence in KP1004 of the GD repeat in OmpK36 that is found in KP1074. Therefore, OmpK36 in KP1004 is completely functional. A detailed description of all these strains is provided in Table S2.
Construction of ompK35 and ompK36 knockout strains of K. pneumoniae.
Construction of ompK35 and ompK36 knockout mutants was carried out following the method described by Datsenko and Wanner (52) with major modifications. As a selectable marker to construct knockout strains, we used resistance to rifampin and the corresponding gene arr2 since most of the K. pneumoniae strains are resistant to ampicillin or chloramphenicol, the two markers that are usually used to construct knockout strains in various organisms. The clinical isolate KPM1346, which carries the arr2 gene, was used as the template for PCR. To facilitate future removal of the arr2 gene from knockout constructs, the FRT sequence was added to each end of the arr2 gene using primers FRT-arr2-F (5′-ACAGGATCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCCAAGCAGCAAGCGCGTTAC-3′) and FRT-arr2-R (5′-CACGTCGACGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCCTAGTCTTCAATGACGTGTA-3′). BamHI and SalI restriction sites were added to facilitate cloning. The PCR product was digested with BamHI and SalI and cloned into pUC19 that had been digested with the same restriction enzymes. The correct recombinant plasmid transformed into E. coli DH5α was confirmed by DNA sequencing at Eton Biosciences (San Diego, CA). Plasmid pKD46-GM, containing bacteriophage lambda Red recombinase, was transformed into various strains to facilitate ompK35 and ompK36 knockout experiments.
To knock out the ompK35 gene, PCR products carrying the FRT-arr2 cassette flanked by 54 bp and 57 bp of sequences of the 5′ and the 3′ ends of the ompK35 coding region, respectively, were prepared by PCR using primers KP-ompK35-del-F (5′-ATGAGGGTAATAAATAATGATGAAGCGCAATATTCTGGCAGTGGTGATCCCTGCGTGAATTCGAGCTCGGTACCC-3′) and KP-ompK35-del-R (5′-TTAGAACTGGTAAACGATACCCACGGCCGCCTGGTCGTCAGTGGCGACACCAGCCGCTGACCATGATTACGCCAAGC-3′) with pUC19::FRT-arr2 as the template.
To facilitate the knockout of the ompK36 gene, the ompK36 gene coding region and 88 bp of the upstream sequence of strain KPM1026a were first cloned into pUCP24. Then, plasmid pUC19::FRT-arr2 was digested with SmaI and PstI to obtain the FRT-arr2 cassette, which was subsequently subcloned into plasmid pUCP24::ompK36 that had been digested with ScaI and PstI. The resulting plasmid, pUCP24::ompK36-FRT-arr2, had 874 bp of the ompK36 coding sequence (positions +15 to +888) deleted and replaced by the FRT-arr2 cassette. This plasmid was used as a template to generate the PCR product for ompK36 gene knockout using primers KP-ompK36-F (5′-CAGCACAATGAAATAGCCGAC-3′) and KP-ompK36-R (5′-TTAGAACTGGTAAACCAGGC-3′). This PCR product had 102 bp of the ompK36 gene 5′ region (positions −88 to +14) and the last 207 bp of the coding region (positions +904 to +1110).
The PCR products for the ompK35 and ompK36 knockouts were transformed into strains carrying plasmid pKD46-GM by electroporation using a Bio-Rad Gene Pulser apparatus (Hercules, CA). Electrocompetent cells were prepared following the manufacturer's instructions with the following modifications. After electroporation, transformants were selected on LB agar plates containing rifampin (20 mg/liter), and their sequences were confirmed by PCR using primers specific for sequences within and outside the deleted regions of the ompK35 and ompK36 genes and the primers specific for the FRT-arr2 cassette.
Conjugation experiments.
Strain KP1074 containing plasmid pKpQIL, which carries blaKPC-3, was used as a donor in conjugation experiments. pKpQIL was introduced into various derivatives of streptomycin-resistant strain KPM1026a with various combinations of efflux and porin mutations. Both donor and recipient strains were grown in LB broth at 37°C with shaking overnight. Fifty microliters of the donor culture was mixed with 50 μl of the recipient culture, followed by centrifugation for 1 min at 5,000 rpm at room temperature. After the supernatant was removed, the cells were resuspended in 50 μl of LB and spotted onto an LB agar plate without antibiotics. The plate was incubated at 37°C for 4 to 5 h to form a bacterial lawn of growth. The cells were harvested and resuspended in 1 ml of LB medium to achieve an optical density at 600 nm (OD600) of 0.2 to 0.5. Resuspended cells (0.1 ml) were plated on LB agar plates containing 1,000 μg/ml streptomycin and 8 μg/ml aztreonam to inhibit both parental strains but allow the growth of the transconjugants, which were verified by PCR to contain blaKPC-3.
Antimicrobial susceptibility testing.
Bacterial isolates were subjected to broth microdilution susceptibility testing, performed according to Clinical and Laboratory Standards Institute (CLSI) methods (53), using panels prepared in-house. A checkerboard assay conforming to the procedures described by Moody in the Clinical Microbiology Procedures Handbook (54) was used to evaluate the effects of various concentrations of vaborbactam on the MICs of various antibiotics. MPC16 and MPCmax (where MPC stands for the minimal potentiating concentration) values were used to define the potency of vaborbactam. MPC16 was defined as the concentration of vaborbactam that was required to reduce the antibiotic MIC 16-fold. MPCmax was defined as the concentration of vaborbactam that achieved the maximal effect in increasing antibiotic potency.
Meropenem was purchased from Sandoz; all other antibiotics were from Sigma-Aldrich. Vaborbactam was synthesized at The Medicines Company, San Diego, CA (lot P-232-159-2).
Determination of expression of efflux and porin genes.
Single colonies from an overnight plate were inoculated into cation-adjusted Mueller-Hinton broth and grown at 37°C until an OD600 of 0.7 was obtained. Cell pellets were collected by centrifugation, and total RNA was isolated using an Ambion RiboPure-Bacteria RNA isolation kit (Thermo Fisher, San Diego, CA). The residual DNA in the RNA samples was removed by treatment with DNase I, according to the manufacturer's instructions. Reverse transcription (RT) was performed using a TaqMan reverse transcriptase reagents kit (Thermo Fisher, San Diego, CA). A mixture of reverse primers specific for the genes to be tested, acrB, ompK35, and ompK36, and the internal control housekeeping gene, rpoB (Table S1), each at a final concentration of 0.5 μM, was used as RT primers.
The RT reaction mixture was diluted 10-fold and used in a quantitative real-time PCR (qPCR) performed on an ABI Prism 7000 sequencing system (Applied Biosystems) using SYBR Select master mix (Thermo Fisher). For these reactions, 9 μl of the diluted RT reaction mixture was used as the template and mixed with 10 μl of SYBR Select master mix (2×) and 1 μl of a qPCR primer pair mixture (Table S1) to make the final concentration of the forward and reverse primers of 0.5 μM. The qPCR was run with the following thermal cycling conditions: 55°C for 2 min, 95°C for 5 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The qPCR results (threshold cycle [CT] values) were normalized with the housekeeping gene rpoB by subtracting the CT value of the test gene from the CT value of the rpoB gene from the same RT reaction. To calculate the level of expression relative to that by strain KPM1026a, the normalized CT value of a gene in a test strain was subtracted from that of the same gene in KPM1026a, and the difference (ΔCT) was used as a logarithmic power (base 2). At least three independent RNA samples isolated from three separate cultures were used to determine the average transcript level of each strain.
Supplementary Material
ACKNOWLEDGMENTS
This work and the efforts of Olga Lomovskaya, Dongxu Sun, Debora Rubio-Aparicio, Kirk Nelson, Ruslan Tsivkovski, David C. Griffith, and Michael N. Dudley were funded in part with federal funds from the U.S. Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority (BARDA), under contract no. HHSO100201400002C with Rempex Pharmaceuticals, a wholly owned subsidiary of The Medicines Company.
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship of the manuscript, take responsibility for the integrity of the work as a whole, and have given final approval to the version to be published.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01443-17.
REFERENCES
- 1.Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem J 276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasevic AT, Canton R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 2017. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 5.Satlin MJ, Chen L, Patel G, Gomez-Simmonds A, Weston G, Kim AC, Seo SK, Rosenthal ME, Sperber SJ, Jenkins SG, Hamula CL, Uhlemann AC, Levi MH, Fries BC, Tang YW, Juretschko S, Rojtman AD, Hong T, Mathema B, Jacobs MR, Walsh TJ, Bonomo RA, Kreiswirth BN. 2017. Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother 61:e02349-16. doi: 10.1128/AAC.02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. 2017. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumbarello M, Trecarichi EM, De Rosa FG, Giannella M, Giacobbe DR, Bassetti M, Losito AR, Bartoletti M, Del Bono V, Corcione S, Maiuro G, Tedeschi S, Celani L, Cardellino CS, Spanu T, Marchese A, Ambretti S, Cauda R, Viscoli C, Viale P, Isgri S. 2015. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother 70:2133–2143. doi: 10.1093/jac/dkv086. [DOI] [PubMed] [Google Scholar]
- 8.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol 30:972–976. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 9.Alexander EL, Loutit J, Tumbarello M, Wunderink R, Felton T, Daikos G, Fusaro K, White D, Zhang S, Dudley MN. 2017. Carbapenem-resistant Enterobacteriaceae infections: results from a retrospective series and implications for the design of prospective clinical trials. Open Forum Infect Dis 4:ofx063. doi: 10.1093/ofid/ofx063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol 58:1303–1308. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clancy CJ, Chen L, Hong JH, Cheng S, Hao B, Shields RK, Farrell AN, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Mutations of the ompK36 porin gene and promoter impact responses of sequence type 258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin. Antimicrob Agents Chemother 57:5258–5265. doi: 10.1128/AAC.01069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shields RK, Nguyen MH, Potoski BA, Press EG, Chen L, Kreiswirth BN, Clarke LG, Eschenauer GA, Clancy CJ. 2015. Doripenem MICs and ompK36 porin genotypes of sequence type 258, KPC-producing Klebsiella pneumoniae may predict responses to carbapenem-colistin combination therapy among patients with bacteremia. Antimicrob Agents Chemother 59:1797–1801. doi: 10.1128/AAC.03894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries RM, Hemarajata P. 2017. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother 61:e00537-17. doi: 10.1128/AAC.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drawz SM, Bonomo RA. 2010. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D beta-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassetti M, Righi E, Viscoli C. 2008. Novel beta-lactam antibiotics and inhibitor combinations. Expert Opin Investig Drugs 17:285–296. doi: 10.1517/13543784.17.3.285. [DOI] [PubMed] [Google Scholar]
- 19.Stachyra T, Levasseur P, Pechereau MC, Girard AM, Claudon M, Miossec C, Black MT. 2009. In vitro activity of the β-lactamase inhibitor NXL104 against KPC-2 carbapenemase and Enterobacteriaceae expressing KPC carbapenemases. J Antimicrob Chemother 64:326–329. doi: 10.1093/jac/dkp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castanheira M, Mills JC, Costello SE, Jones RN, Sader HS. 2015. Ceftazidime-avibactam activity tested against Enterobacteriaceae isolates from U.S. hospitals (2011 to 2013) and characterization of beta-lactamase-producing strains. Antimicrob Agents Chemother 59:3509–3517. doi: 10.1128/AAC.00163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jonge BL, Karlowsky JA, Kazmierczak KM, Biedenbach DJ, Sahm DF, Nichols WW. 2016. In vitro susceptibility to ceftazidime-avibactam of carbapenem-nonsusceptible Enterobacteriaceae isolates collected during the INFORM Global Surveillance Study (2012 to 2014). Antimicrob Agents Chemother 60:3163–3169. doi: 10.1128/AAC.03042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shields RK, Nguyen MH, Chen L, Press EG, Potoski BA, Marini RV, Doi Y, Kreiswirth BN, Clancy CJ. 2017. Ceftazidime-avibactam is superior to other treatment regimens against carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother 61:e00883-17. doi: 10.1128/aac.00883-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krapp F, Grant JL, Sutton SH, Ozer EA, Barr VO. 2017. Treating complicated carbapenem-resistant Enterobacteriaceae infections with ceftazidime/avibactam: a retrospective study with molecular strain characterisation. Int J Antimicrob Agents 49:770–773. doi: 10.1016/j.ijantimicag.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 63:1615–1618. doi: 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/aac.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, King P, Tsivkovski R, Sun D, Sabet M, Tarazi Z, Clifton MC, Atkins K, Raymond A, Potts KT, Abendroth J, Boyer SH, Loutit JS, Morgan EE, Durso S, Dudley MN. 2015. Discovery of a cyclic boronic acid beta-lactamase inhibitor (RPX7009) with utility versus class A serine carbapenemases. J Med Chem 58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 27.Wozniak A, Villagra NA, Undabarrena A, Gallardo N, Keller N, Moraga M, Roman JC, Mora GC, Garcia P. 2012. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol 61:1270–1279. doi: 10.1099/jmm.0.045799-0. [DOI] [PubMed] [Google Scholar]
- 28.Crowley B, Benedi VJ, Domenech-Sanchez A. 2002. Expression of SHV-2 beta-lactamase and of reduced amounts of OmpK36 porin in Klebsiella pneumoniae results in increased resistance to cephalosporins and carbapenems. Antimicrob Agents Chemother 46:3679–3682. doi: 10.1128/AAC.46.11.3679-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agyekum A, Fajardo-Lubian A, Ai X, Ginn AN, Zong Z, Guo X, Turnidge J, Partridge SR, Iredell JR. 2016. Predictability of phenotype in relation to common beta-lactam resistance mechanisms in Escherichia coli and Klebsiella pneumoniae. J Clin Microbiol 54:1243–1250. doi: 10.1128/JCM.02153-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castanheira M, Huband MD, Mendes RE, Flamm RK. 2017. Meropenem-vaborbactam tested against contemporary Gram-negative isolates collected worldwide during 2014, including carbapenem-resistant, KPC-producing, multidrug-resistant, and extensively drug-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00567-17. doi: 10.1128/aac.00567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehta SC, Rice K, Palzkill T. 2015. Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog 11:e1004949. doi: 10.1371/journal.ppat.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother 49:4760–4762. doi: 10.1128/AAC.49.11.4760-4762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shields RK, Clancy CJ, Hao B, Chen L, Press EG, Iovine NM, Kreiswirth BN, Nguyen MH. 2015. Effects of Klebsiella pneumoniae carbapenemase subtypes, extended-spectrum beta-lactamases, and porin mutations on the in vitro activity of ceftazidime-avibactam against carbapenem-resistant K. pneumoniae. Antimicrob Agents Chemother 59:5793–5797. doi: 10.1128/AAC.00548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Quale J, Landman D. 2015. Activity of meropenem combined with RPX7009, a novel beta-lactamase inhibitor, against gram-negative clinical isolates in New York City. Antimicrob Agents Chemother 59:4856–4860. doi: 10.1128/AAC.00843-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castanheira M, Rhomberg PR, Flamm RK, Jones RN. 2016. Effect of the beta-lactamase inhibitor vaborbactam combined with meropenem against serine carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:5454–5458. doi: 10.1128/AAC.00711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsivkovski R, Griffith DC, Hecker SJ, Dudley MN, Lomovskaya O. 2012. Biochemical characterization of the beta-lactamase inhibitor RPX7009, abstr F-849. Abstr 52nd Intersci Conf Antimicrob Agents Chemother, San Francisco, CA American Society for Microbiology, Washington, DC. [Google Scholar]
- 42.Castanheira M, Farrell SE, Krause KM, Jones RN, Sader HS. 2014. Contemporary diversity of beta-lactamases among Enterobacteriaceae in the nine U.S. census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent beta-lactamase groups. Antimicrob Agents Chemother 58:833–838. doi: 10.1128/AAC.01896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez-Alles S, Alberti S, Alvarez D, Domenech-Sanchez A, Martinez-Martinez L, Gil J, Tomas JM, Benedi VJ. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145(Pt 3):673–679. [DOI] [PubMed] [Google Scholar]
- 44.Sho T, Muratani T, Hamasuna R, Yakushiji H, Fujimoto N, Matsumoto T. 2013. The mechanism of high-level carbapenem resistance in Klebsiella pneumoniae: underlying Ompk36-deficient strains represent a threat of emerging high-level carbapenem-resistant K. pneumoniae with IMP-1 beta-lactamase production in Japan. Microb Drug Resist 19:274–281. doi: 10.1089/mdr.2012.0248. [DOI] [PubMed] [Google Scholar]
- 45.Szabó D, Silveira F, Hujer AM, Bonomo RA, Hujer KM, Marsh JW, Bethel CR, Doi Y, Deeley K, Paterson DL. 2006. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Antimicrob Agents Chemother 50:2833–2835. doi: 10.1128/AAC.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies TA, Marie Queenan A, Morrow BJ, Shang W, Amsler K, He W, Lynch AS, Pillar C, Flamm RK. 2011. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007-09. J Antimicrob Chemother 66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 47.Griffith DC, Loutit JS, Morgan EE, Durso S, Dudley MN. 2016. Phase 1 study of the safety, tolerability, and pharmacokinetics of the beta-lactamase inhibitor vaborbactam (RPX7009) in healthy adult subjects. Antimicrob Agents Chemother 60:6326–6332. doi: 10.1128/AAC.00568-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhavnani SM, Hammel JP, Rubino CM, Tran J, Loutit JS, Griffin DC, Lomovskaya O, Dudley MN, Ambrose PG. 2017. Meropenem-vaborbactam pharmacokinetic-pharmacodynamic analyses for efficacy based on data from patients enrolled in phase 3 studies, abstr Sunday-193. Abstr ASM Microbe, New Orleans, LA American Society for Microbiology, Washington, DC. [Google Scholar]
- 49.West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 50.Broberg CA, Wu W, Cavalcoli JD, Miller VL, Bachman MA. 2014. Complete genome sequence of Klebsiella pneumoniae strain ATCC 43816 KPPR1, a rifampin-resistant mutant commonly used in animal, genetic, and molecular biology studies. Genome Announc 2(5):e00924-14. doi: 10.1128/genomeA.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clinical and Laboratory Standards Institute. 2015. M07-A10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard: ninth edition. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 54.Moody J. 2007. Synergism testing: broth microdilution checkerboard, p 12 In Garcia L. (ed), Clinical microbiology procedures handbook, 3rd ed and 2007 update ASM Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


