Abstract
The mitotic spindle is composed of dynamic microtubules and associated proteins that together direct chromosome movement during mitosis. The spindle plays a vital role in accurate chromosome segregation fidelity and is a therapeutic target in cancer. Nevertheless, the molecular mechanisms by which many spindle-associated proteins function remains unknown. The nucleolar and spindle-associated protein NUSAP1 is a microtubule-binding protein implicated in spindle stability and chromosome segregation. We show here that NUSAP1 localizes to dynamic spindle microtubules in a unique chromosome-centric pattern, in the vicinity of overlapping microtubules, during metaphase and anaphase of mitosis. Mass spectrometry-based analysis of endogenous NUSAP1 interacting proteins uncovered a cell cycle-regulated interaction between the RanBP2–RanGAP1–UBC9 SUMO E3 ligase complex and NUSAP1. Like NUSAP1 depletion, RanBP2 depletion impaired the response of cells to the microtubule poison Taxol. NUSAP1 contains a conserved SAP domain (SAF-A/B, Acinus, and PIAS). SAP domains are common among many other SUMO E3s, and are implicated in substrate recognition and ligase activity. We speculate that NUSAP1 contributes to accurate chromosome segregation by acting as a co-factor for RanBP2–RanGAP1–UBC9 during cell division.
Keywords: cell cycle, cell division, microtubule, mitosis, mitotic spindle, small ubiquitin-like modifier (SUMO), sumoylation, ubiquitin
Introduction
The accurate partitioning of chromosomes during cell division is essential for cell survival and preventing chromosome instability. The movement of chromosomes during mitosis requires the assembly and organization of a bipolar array of microtubules termed the mitotic spindle. Spindle dynamics are controlled by numerous microtubule-associated proteins, and the molecular function of many of these remains to be characterized.
Nucleolar and spindle-associated protein 1 (NUSAP1)3 is a mitotic phosphoprotein that binds microtubules and has been implicated in cell division (1–6). NUSAP1 is highly conserved among higher eukaryotes and genetic knock-out in mice is embryonic lethal due to chromosome segregation defects (4). NUSAP1 is overexpressed in numerous malignancies, and high levels correlate with poor prognosis in aggressive triple-negative breast cancer (7). A central domain in NUSAP1 directly interacts with microtubules in vitro and in vivo, and its association with the mitotic spindle is controlled by phosphorylation (1, 2, 6). NUSAP1 and has been implicated in mitotic progression, spindle formation, and stability (1–3). In addition, NUSAP1 depletion sensitized a variety of cell types to the chemotherapeutic agent Taxol, consistent with its role in spindle formation and stability (8, 9). Furthermore, studies in frog egg extracts have suggested a potential role for NUSAP1 in tethering microtubules to chromatin in a kinetochore-independent manner (1).
Mass spectrometry-based analysis of spindle-associated factors demonstrated that NUSAP1 is among a small group of proteins, which includes PRC1/Ase1 and KIF4, whose binding to microtubules increases after anaphase compared with earlier stages of mitosis (10). Consistent with this observation, NUSAP1 phosphorylation by CDK1/Cyclin B, which is active in early mitosis, displaces it from microtubules (6). Together, these studies point to a crucial role for NUSAP1 is regulating both early and late mitotic events. Importantly, they strongly suggest that there exists a pool of microtubule-free NUSAP1 in early mitosis that could contribute to its function during cell division.
We previously identified NUSAP1 as a substrate for a cell cycle-regulated, SCF-type E3 ubiquitin ligase during S/G2 phase (9). NUSAP1 is also targeted for degradation during late mitosis and in early G1 by a second E3 ligase, the anaphase promoting complex/cyclosome (APC/C) (11). In addition to its regulation by ubiquitin, NUSAP1 was also recovered in large-scale cell cycle phospho-proteomic studies (12, 13). However, the role of NUSAP1 in mitosis remains largely unknown, as does the network of proteins to which it binds during cell division. To gain mechanistic insights into how NUSAP1 regulates cell division, we applied mass spectrometry-based proteomics to identify endogenous NUSAP1-interacting proteins. This analysis identified a cell cycle-regulated interaction between NUSAP1 and a SUMO (small ubiquitin-like modifier) E3 ligase complex.
SUMO is an ubiquitin-related protein that is post-translationally appended to substrates, contributing to various aspects of signaling. SUMOylation has been linked to transcriptional activation, protein stability, and regulating protein-protein interactions (14, 15). The first described SUMO E3 ligase is composed of three proteins; Ran-binding protein 2 (RanBP2), Ran GTPase-activating protein 1 (RanGAP1), and the SUMO E2 conjugating enzyme, UBC9 (16–18). During interphase, this complex is part of the nuclear pore where it functions in Ran-mediated nuclear import and export (19, 20). However, following nuclear envelope breakdown at mitotic entry, the RanBP2 SUMO E3 ligase dissociates from the nuclear pore complex and SUMOylates proteins important for chromosome segregation (16, 21–23). The DNA decatenating enzyme TOP2A is SUMOylated at the metaphase to anaphase transition by the RanBP2 E3; SUMOylation directs TOP2A localization to centromeres, where it functions in sister chromatid disjunction (22). Failure to SUMOylate TOP2A during mitosis has been linked to severe chromosome mis-segregation (22, 24). Another RanBP2 SUMO E3 ligase target is Borealin, a member of the chromosome passenger complex (CPC), whose functions are critical to kinetochore-microtubule attachment and chromosome segregation (21, 23). The CPC is composed of Borealin, Survivin, INCENP, and Aurora B, and SUMOylation of CPC complex members is highly conserved (25). SUMOylation of the cell cycle transcription factor FoxM1 during G2/M regulates its activity (15, 26). Finally, the kinetochore-associated microtubule motor CENP-E is SUMOylated, contributing to its kinetochore localization and function (27).
The most well characterized SUMO ligases described to date are the PIAS family of SUMO E3s. PIAS proteins are a family of conserved SUMO ligases involved in various aspects of cellular physiology, including cell cycle control. PIAS proteins contain an amino-terminal SAP (SAF-A/B, Acinus, and PIAS) domain that has been implicated in both enzyme activation and substrate targeting (28). Despite the significant size of the RanBP2–RanGAP1–UBC9 complex (RanBP2 has a predicted molecular mass of 358 kDa), it lacks a recognizable SAP domain. Notably, NUSAP1 has an obvious SAP domain in its amino terminus and we propose that NUSAP1 could facilitate RanBP2 ligase function through the amino-terminal SAP domain.
Results
NUSAP1 localizes to dynamic spindle microtubules near chromatin
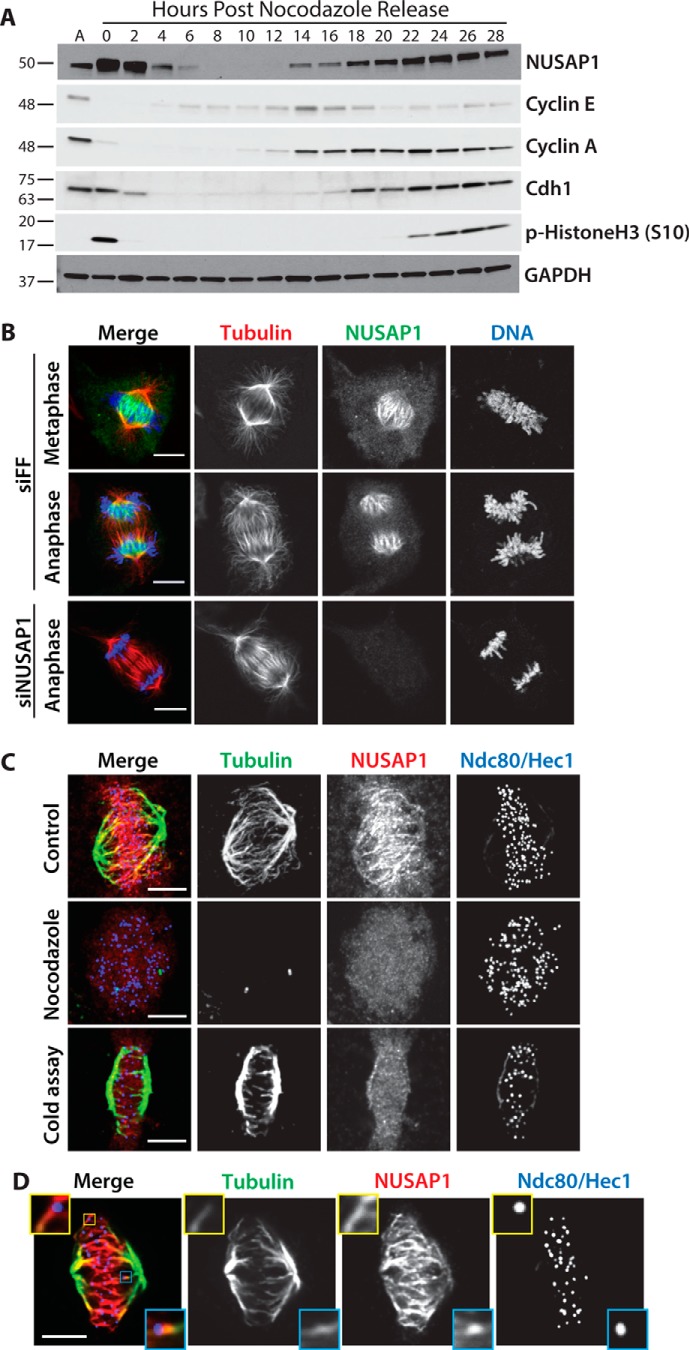
NUSAP1 is a cell cycle-regulated, microtubule-binding protein whose expression has been shown previously, by us and others, to oscillate during the cell cycle (2, 9, 29). However, experiments performed to date were done on relatively short time scales after synchronization and release, making it difficult to know if its dynamics were due to the effects of chemical synchronization. To analyze NUSAP1 protein dynamics throughout an entire cell cycle we performed immunoblots on U2OS cells synchronized using nocodazole, isolated by shake-off, and followed for 28 h after re-plating (Fig. 1A). NUSAP1 levels are elevated in mitotic cells compared with asynchronous populations, concomitant with an increase in phosphorylated histone H3 (Ser-10), a marker of mitosis. NUSAP1 levels decrease abruptly as cells enter G1 phase, consistent with degradation mediated by the APC/C. NUSAP1 levels remain low through early S phase, when Cyclin E is expressed and Cdh1 is degraded, and then begin to accumulate after the expression of Cyclin A, which marks the beginning of S phase. NUSAP1 is also targeted by another E3 ligase, the SCFCyclin-F, during S/G2 (9). Interestingly, abundance of the APC/C co-activator Cdc20, Cyclin F, and NUSAP1 are all abruptly diminished at mitotic exit, consistent with their coordinated degradation by APC/C and its other co-activator, Cdh1 (Fig. 1A and supplemental Fig. S1) (30).
Figure 1.
NUSAP1 is a cell cycle-regulated microtubule-binding protein. A, U2OS cells were synchronized by overnight treatment with nocodazole and released by mitotic shake-off. Samples were analyzed by immunoblot as cells progress through the cell cycle. B, NUSAP1 localization to the mitotic spindle analyzing by immunofluorescent imaging of mitosis in U2OS cells. (Scale bars = 10 μm.) C, NUSAP1 localization was analyzed in nocodazole-treated cells and following incubation with ice-cold buffer to destabilize non-kinetochore microtubules. (Scale bars indicate 5 μm.) D, single plane confocal imaging of NUSAP1 localization on the spindle during metaphase. Insets highlight two kinetochore-microtubule attachments. (Scale bars indicate 5 μm.)
We used high-resolution immunofluorescent (IF) imaging to interrogate the localization of NUSAP1 during mitosis, when its protein levels are at their highest. The specificity of the NUSAP1 antibody was confirmed by comparing anti-NUSAP1-stained cells treated with either control siRNA targeting firefly luciferase (FF) or oligonucleotides targeting NUSAP1. RNAi depletion of NUSAP1 completely eliminated staining, confirming antibody specificity for IF. In prometaphase, NUSAP1 staining was diffuse and localization to specific mitotic structures was not apparent (supplemental Fig. S1). Later in mitosis NUSAP1 did not localize to the whole of the mitotic spindle, like the majority of known microtubule-binding proteins in mitosis (Fig. 1B). Instead, it localizes to the central spindle with the most concentrated area of NUSAP1 being near the chromatin (Fig. 1B). Highly concentrated NUSAP1 staining in the vicinity of chromatin was visible during metaphase, anaphase, and telophase, with the bulk of NUSAP1 appearing to localize to the spindles around chromatin. NUSAP1 localization is coincident with regions of anti-parallel, overlapping microtubules in the central spindle. Notably, this chromatin-centric spindle localization is highly unique among known microtubule-binding proteins in mitosis. Interestingly, it is comparable, although not identical, to PRC1 and KIF4, which also show increased microtubule binding after anaphase and control anti-parallel microtubule assemblies in the central spindle (10, 31–33). This suggests that NUSAP1 represents a unique class of microtubule-binding protein that localize in the vicinity of inter-digitated microtubules and that tracks chromatin localization in both early and late mitosis.
The localization of a pool of NUSAP1 on spindle microtubules near chromatin prompted us to determine whether NUSAP1 localization is microtubule dependent. Prior to fixation, cells were treated with either DMSO (control) or the microtubule depolymerizing drug nocodazole. NUSAP1 localization is lost when the spindle is depolymerized by nocodazole treatment, confirming that its localization it microtubule dependent (Fig. 1C). To determine which population of microtubules NUSAP1 localizes to, we depolymerized dynamic spindle microtubules prior to fixation (Fig. 1C). Cells were cold treated prior to fixation, which leads to the destabilization of microtubules that are not stably attached to kinetochores (k-fibers). NUSAP1 localization to the spindle was lost when non-kinetochore microtubules were depolymerized, suggesting that NUSAP1 localizes to dynamic microtubules during mitosis (Fig. 1C). This observation, and the diffuse NUSAP1 staining in prometaphase cells, is consistent with the notion that NUSAP1 binds to overlapping spindle microtubules. Finally, we analyzed single focal planes of NUSAP1 and tubulin staining by confocal microscopy. We observed NUSAP1 localization along microtubules, but not at the centromere, centrosome, or kinetochore (Fig. 1D). Together, these data confirm that NUSAP1 is cell cycle regulated, and demonstrate its chromatin-centric localization to dynamic microtubules during mitosis.
Identification of NUSAP1 interacting proteins using mass spectrometry
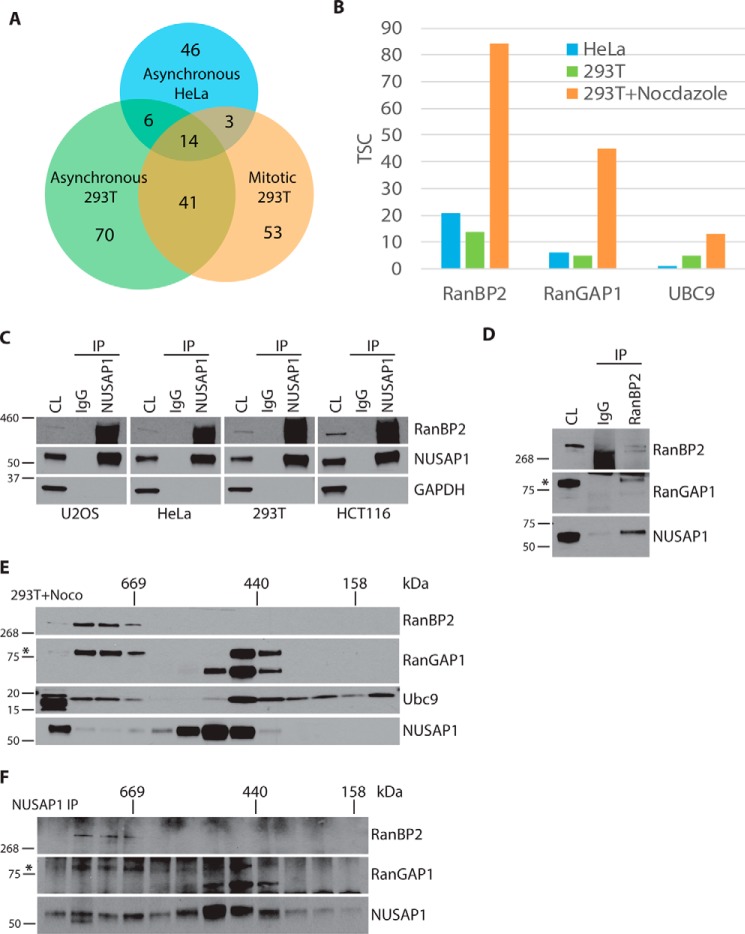
NUSAP1 has a unique mitotic localization pattern compared with known microtubule-binding proteins (Fig. 1). Because NUSAP1 has been implicated in spindle stability and chromosome segregation we were interested in the mechanism by which NUSAP1 contributes to mitotic progression. To address this question, we analyzed protein interaction partners that bind NUSAP1 using endogenous NUSAP1 immunoprecipitation (IP) followed by protein identification using mass spectrometry (MS/MS). We performed IP experiments using control IgG and endogenous NUSAP1 antibodies in multiple cell lines (HeLa and HEK-293T). In addition, because NUSAP1 levels peak during mitosis (Fig. 1A) we also performed IPs from both asynchronous and mitotic HEK-293T cells arrested using nocodazole. By performing endogenous IPs in multiple cell lines and physiological conditions we sought to identify the strongest interactors that are most likely to be physiologically relevant in controlling mitotic progression. The total spectral counts (TSCs) for each interacting protein in each pulldown experiment are reported in supplemental Table S1.
We filtered out nonspecific interactions identified in control IgG IPs, which were performed in parallel with each experiment, and removed known contaminants based on the CRAPome dataset (34). We then overlapped the remaining interactions between the three IPs to identify the highest-confidence set of NUSAP1-interacting proteins (Fig. 2A). This resulting list of 14 proteins included the known NUSAP1-interacting protein Importin-β (3).
Figure 2.
NUSAP1 interacts with the RanBP2–RanGAP1–UBC9 complex in a cell cycle-dependent manner. A, Venn diagram showing overlap of IP-MS/MS experiment results. B, TSC for each of the RanBP2–RanGAP1–UBC9 complex members determined by mass spectrometry. C, endogenous NUSAP1 IPs were performed in four different nocodazole-arrested cells and analyzed for RanBP2. D, endogenous RanBP2 IP performed in nocodazole-arrested 293T cells. E, size exclusion chromatography was performed on extracts from nocodazole-arrested 293T cells. Extracts were analyzed on a Superose 6 column. Previously tested size markers migrated in the indicated fractions. F, endogenous NUSAP1 IPs were performed using each of the gel filtration fractions from E. *, indicates SUMOylated RanGAP1.
This analysis identified all three members of the RanBP2 mitotic SUMO E3 ligase complex, which includes RanBP2, RanGAP1, and the SUMO E2-conjugating enzyme UBC9. We identified multiple RanBP2 and RanGAP1 peptides in all three experiments. Despite the fact that NUSAP1 is more abundant in mitotic cells, the IPs were saturating in that we detected a similar number of NUSAP1 TSCs between asynchronous and mitotic 293T samples (supplemental Table S1). This allowed us to compare the relative number of RanBP2, RanGAP1, and Ubc9 TSCs between asynchronous and mitotic experiments. Our data show an enrichment of all three proteins in the mitotic sample relative to asynchronous cells, indicating that their interaction is cell cycle regulated (Fig. 2B). Further supporting an interaction between NUSAP1 and RanBP2, their binding was detected in a recent, large-scale interactome study using a tagged version of NUSAP1 (35).
To confirm our IP-MS/MS findings we tested whether RanBP2 co-IPed with endogenous NUSAP1 in multiple cell lines. Importantly, isolated endogenous NUSAP1 was precipitated from nocodazole-arrested U2OS, HeLa, HEK-293T, and HCT116 cell lines which co-precipitated endogenous RanBP2 (Fig. 2C). Similarly, when we precipitated endogenous RanBP2 from nocodazole-arrested HEK-293T cells we co-precipitated endogenous NUSAP1, as well as its known interactor RanGAP1 (Fig. 2D). This interaction was also detected in Taxol-arrested cells, which prevents microtubule depolymerization, indicating that their interaction is not due to gross changes in microtubule dynamics (supplemental Fig. S2).
To further confirm these findings, we analyzed mitotic HEK-293T cell lysates using size exclusion chromatography to separate proteins and complexes based on their size and shape, followed by endogenous NUSAP1 IP. In this experiment, RanBP2, RanGAP1, and UBC9 co-migrated in a high molecular mass complex (∼1 mega dalton) (Fig. 2E, second to fourth lanes). There was a small, but detectable amount of NUSAP1 that also co-migrated with those fractions. Importantly, when we precipitated endogenous NUSAP1 from those fractions we co-precipitated both RanBP2 and RanGAP1 (Fig. 2F). Together this data strongly supports an interaction between a pool of available NUSAP1 and the RanBP2–SUMO E3 ligase complex.
Interestingly, only a subset of SUMOylated RanGAP1 co-migrated with RanBP2 based on the size exclusion chromatographic analysis. The majority of SUMOylated and unSUMOylated RanGAP1 eluted in fraction of ∼500 kDa (Fig. 2E). This demonstrates that there are RanBP2-bound and unbound pools of RanGAP1 in mitotic 293T cells and contrasts with a recent study suggesting that all of RanBP2 and RanGAP1 are complexed together in HeLa cells (21). The reason for this discrepancy is unknown, but could be cell line dependent. The peak elution of NUSAP1 partially overlapped with the peak elution of RanGAP1 that lacked RanBP2 and IPs from these fractions demonstrate that RanGAP1 and NUSAP1 interact in those fractions (Fig. 2, E and F, seventh to tenth lanes). The full composition of these different NUSAP1 complexes remains unknown.
NUSAP1 does not control RanBP2 localization during mitosis
The RanBP2 complex regulates the SUMOylation of TOP2A and Borealin, both of which have distinct mitotic localization patterns (16, 21–23). In addition, RanBP2 localizes at the kinetochore and on the spindle (36). We hypothesized that NUSAP1 could recruit the RanBP2 SUMO E3 ligase to the spindle. We performed IF, probing for RanBP2 and RanGAP1 localization in control (FF) and NUSAP1-depleted cells. We observed the previously reported RanBP2 and RanGAP1 localization patterns in control-depleted cells (37). However, in both U2OS and HeLa cells lines neither RanBP2 nor RanGAP1 localization was affected by NUSAP1 depletion (Fig. 3, A–C, and supplemental Fig. S3). Due to previous reports suggesting that SUMOylation of TOP2 regulates its centromeric localization, we also analyzed the localization of TOP2A and TOP2B on chromatin in control- and NUSAP1-depleted cells using biochemical fractionation. Similarly, we observed no change in the localization of TOP2 on chromatin in control- and NUSAP1-depleted cells (Fig. 3D). We conclude that NUSAP1 is not involved in the localization of RanBP2 and RanGAP1, nor that of the RanBP2–RanGAP1–UBC9 SUMO substrate TOP2.
Figure 3.
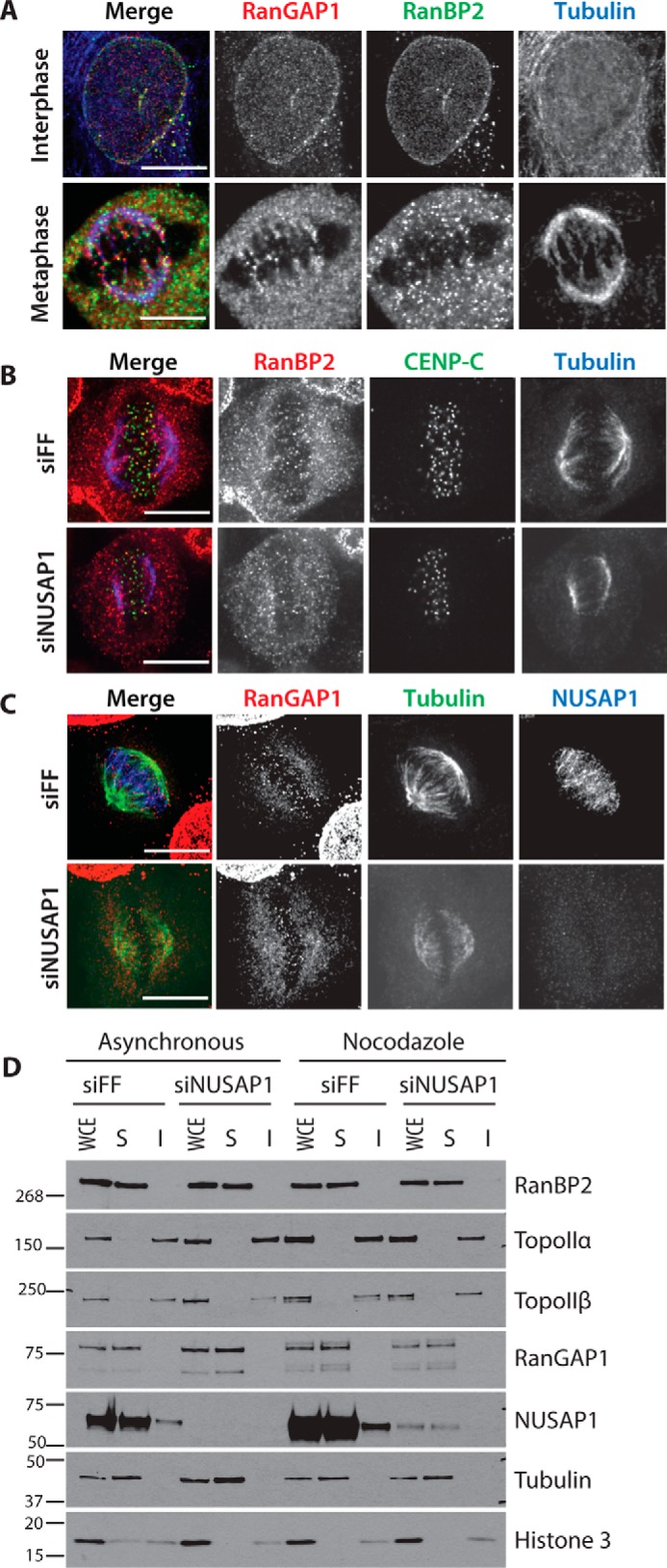
NUSAP1 depletion does not affect mitotic localization of the RanBP2–RanGAP1–UBC9 complex. A, endogenous RanBP2 and RanGAP1 localization in both interphase and metaphase HeLa cells. B, endogenous RanBP2 localization in either control or NUSAP1-depleted HeLa cells. C, endogenous RanGAP1 localization in either control or NUSAP1-depleted HeLa cells. D, chromatin fractionation in U2OS cells. Cells were transfected with either control or NUSAP1 targeting siRNA and split for overnight treatment with either DMSO or nocodazole. (WCE = whole cell lysate; S = soluble (cytoplasmic); I = insoluble (nuclear/chromatin). All scale bars indicate 10 μm.)
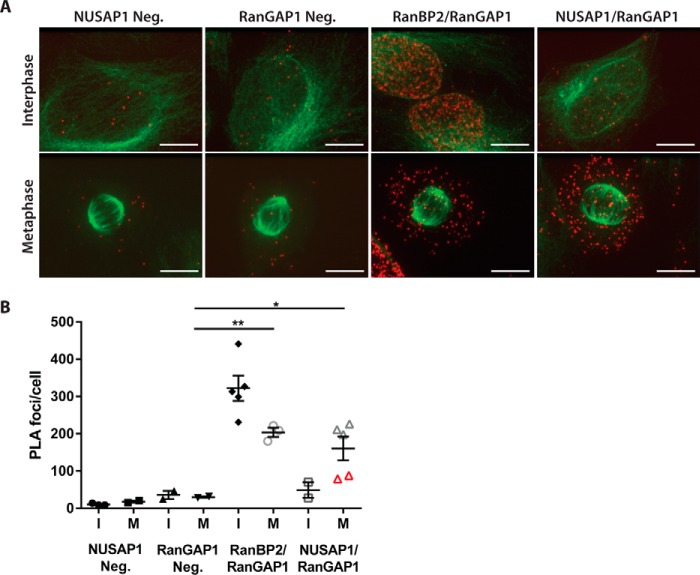
Because our IF staining was unable to distinguish clear co-localization of NUSAP1 with RanBP2 or RanGAP1 and there are soluble pools of NUSAP1, RanBP2, and RanGAP1 during mitosis, we determined where these proteins interact using a proximity ligation assay (PLA) (Fig. 4). PLA relies on the proximity of co-localizing antibodies during immune staining of fixed cells, which allows for the rolling circle amplification of a DNA probe that is detected using fluorescence hybridization. The result is a fluorescent foci at each site of interaction between the target proteins (38). Performing PLA in asynchronous cells with either NUSAP1 or RanGAP1 antibody alone produced a low background (Fig. 4A), quantified in Fig. 4B. Co-staining RanBP2 and RanGAP1 served as a positive control because they interact in both interphase and mitotic cells. Co-staining with NUSAP1 and RanGAP1 antibodies showed a strong increase in the number of foci in the cytosol of mitotic cells (Fig. 4, A and B). Intriguingly, the mitotic cells with the lowest number of foci in the NUSAP1- and RanGAP1-stained samples were in the late stages of mitosis (telophase and after; identified by red triangles in Fig. 4B). This suggests that the interaction between NUSAP1 and the RanBP2–E3 ligase complex decreases in late mitosis as the cells begin to rebuild their nuclear membranes/pores. Consistent with expression of NUSAP1 late in the cell cycle, and a cell cycle-dependent interaction between NUSAP1 and RanBP2–RanGAP1, the PLA signal was unchanged between single antibody-stained controls (NUSAP1 and RanGAP1 only) and dual antibody (combined NUSAP1/RanGAP1)-stained interphase cells. This supports the observation that NUSAP1 interacts with RanBP2–RanGAP1 in a cell cycle-dependent manner, and suggest that NUSAP1 binds RanBP2–RanGAP1 independent of the mitotic spindle, consistent with the binding observed in nocodazole-treated cells.
Figure 4.
NUSAP1 and RanBP2 interact in the cytosol of mitotic cells. A, PLA in U2OS cells using endogenous against NUSAP1, RanGAP1, RanBP2, or control IgG. Tubulin is shown in green with the PLA signal in red. (Scale bars indicate 10 μm.) B, average number of foci/cell for each PLA condition shown in A. Foci were counted using ImageJ.
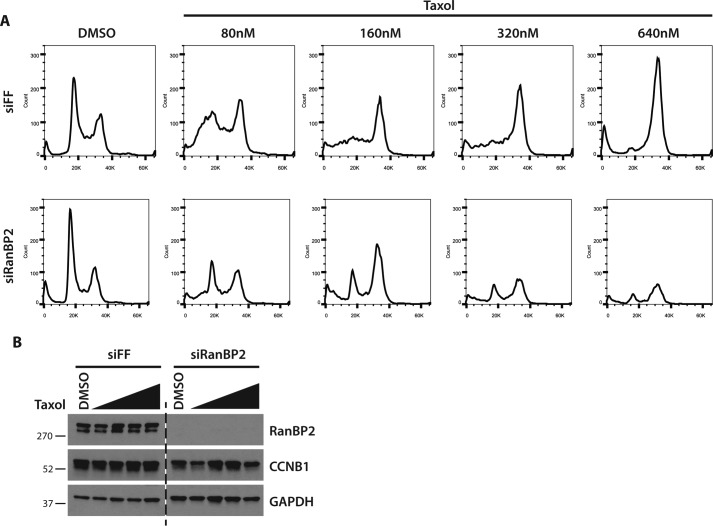
RanBP2 depletion impairs the response to Taxol
Previous reports have shown that NUSAP1 depletion sensitizes cells to spindle poisons, such as Taxol or nocodazole (9). To determine whether RanBP2 depletion would show a consistent phenotype, we depleted cells of RanBP2 using siRNA and treated them with increasing doses of Taxol overnight. RanBP2 was effectively depleted by siRNA based on immunoblot analysis (Fig. 5B). Propidium iodide staining for DNA content in control-depleted cells shows a progressive increase in G2/M phase cells in response to Taxol, indicating an increased number of cells arresting in response to spindle checkpoint activation (Fig. 5A). RanBP2-depleted cells had substantially reduced numbers of cells in the G2/M phase at all doses of Taxol tested, consistent with a defect in maintaining their mitotic arrest in response to checkpoint activation. Consistent with a slippage through mitosis, there was also a reduction in Cyclin B levels in RanBP2-depleted cells compared with controls. At higher doses of Taxol the number of surviving cells at the time of harvest was also reduced. These data are consistent with our previous studies showing that NUSAP1-depleted cells are sensitive to spindle poisons that activate the spindle checkpoint (9).
Figure 5.
RanBP2 knockdown sensitizes cells to Taxol treatment. A, U2OS cells were transfected with control of RanBP2 targeting siRNA and then treated overnight with increasing doses of Taxol. Cell cycle was analyzed by propidium iodide staining and flow cytometry. B, immunoblot analysis of cells from A.
Discussion
NUSAP1 is an important regulator of mitotic progression and chromosome segregation. NUSAP1 is essential for mouse development, and its inactivation by RNAi leads to defects in chromosome segregation (4). The NUSAP1 protein is tightly controlled post-translationally during the cell cycle. Its stability is controlled by at least two E3 ubiquitin ligases: SCFCyclin-F during S/G2 phase and APC/C in G1 (9, 11). Furthermore, NUSAP1 phosphorylation is up-regulated during cell cycle progression on upwards of 20 different residues (12, 13). Nevertheless, little is known about where NUSAP1 fits mechanistically in the mitotic spindle apparatus.
We used confocal imaging to determine the precise localization of NUSAP1 on the mitotic spindle, providing a high-resolution snapshot of NUSAP1 localization at each stage of mitosis. Interestingly, NUSAP1 exhibits a prominent, chromatin-centric localization pattern during metaphase and anaphase that is unique among microtubule-binding proteins. We demonstrate here that NUSAP1 is localized on microtubules, and that its localization is dependent on dynamic spindle microtubules, indicating a unique role for NUSAP1 in the process of cell division. The localization of NUSAP1 is most consistent with that of overlapping, inter-digitated spindle microtubules. Consistent with this, we see no significant NUSAP1 staining in prometaphase cells where the spindle poles have not yet separated. We are unaware of another microtubule-binding protein with a localization that is fully coincident with chromatin during metaphase and anaphase of mitosis. The PRC1 and KIF4 proteins show the most consistent localization with that of NUSAP1 during metaphase, but localize to the spindle mid-zone at anaphase. However, NUSAP1, PRC1, and KIF4 all showed increased microtubule binding after anaphase, suggesting a potential relationship between these factors in controlling spindle integrity (10).
To further define the role of NUSAP1 we examined endogenous binding partners using mass spectrometry. Through this analysis we identified and validated a cell cycle-regulated interaction between NUSAP1 and the RanBP2–RanGAP1–UBC9 SUMO E3 ligase. Their interaction was identified first using endogenous NUSAP1 pulldown followed by mass spectrometry and was validated by co-IP of both endogenous proteins in multiple cell lines. We were surprised not to identify a larger set of overlapping proteins between datasets, and predict that inter-cell lines differences could be explained by variances in the oncogenic repertoire of the different cell types. An interaction between NUSAP1 and RanBP2 was also detected in a large-scale study that globally mapped protein–protein interaction networks, providing further validation for their interaction (39). RanBP2–RanGAP1–UBC9 is a critical SUMO ligase involved in cell division. However, little is known about which substrates it targets, how those substrates are recognized, how its activity is regulated, and how its localization is controlled.
PIAS proteins, the most well characterized family of SUMO E3 ligases, all share a SAP domain (SAFA/B, Acinus, and PIAS protein domain) at their N terminus. Although the SAP domain of PIAS proteins has been shown to be involved in nuclear import and DNA binding, PIAS protein SAP domains also mediate substrate interactions. For example, PIAS1 interacts with its substrate C/EBP-β via its SAP domain, with deletion of its SAP domain resulting in failure to SUMOylate C/EBP-β (28). Interestingly, none of the RanBP2-associated SUMO ligase components contain an identifiable SAP domain. NUSAP1, however, has a well conserved SAP domain at its N terminus, with nearly all of the key, conserved residues found in the PIAS protein SAP domains (supplemental Fig. S4) (49). Like the PIAS proteins and other SAP domain containing proteins, the NUSAP1 SAP domain has been shown to be important for its interactions with DNA, however, this may not be its only function (40). It is unknown how RanBP2 SUMO ligase is activated and how it specifies substrates for SUMOylation. We speculate that NUSAP1 could be a regulatory subunit for the complex, mediating substrate interactions and/or complex activation, similar to the role of substrate adapters in cullin E3 ligases. Importantly, depletion of NUSAP1 using multiple siRNA reagents does not interfere with RanBP2–RanGAP1 complex assembly (supplemental Fig. S5). It is noteworthy that despite being the first discovered SUMO E3, little is known about the enzymology of the intact complex, due in large part to the size of RanBP2 (21, 41).
Despite the prominent localization of NUSAP1 during metaphase and anaphase to microtubules in the vicinity of chromatin, its binding to RanBP2–RanGAP1 is cytoplasmic. Thus, NUSAP1 could contribute to mitotic progression through multiple mechanisms: at the site overlapping microtubules on the mitotic spindle and through interactions with RanBP2 in the cytoplasm.
Recent large-scale studies have sought to identify targets of SUMOylation and have even examined cell cycle-dependent changes in SUMOylation. However, NUSAP1 has not been identified in any of these large-scale SUMO substrate screens, suggesting it is not a target for SUMOylation, despite these screens often being conducted using mitotic cells (42–45). Although this does not rule out NUSAP1 as a SUMO substrate, we currently lack evidence supporting it as target of SUMOylation and were unable to detect NUSAP1 in SUMO pulldowns.
Little is known about how SUMO E3 ligases interact with, and subsequently SUMOylate their targets and how these interactions are regulated. If NUSAP1 did mediate enzymatic activity of the RanBP2 SUMO E3 ligase, this would provide important insight into the functions of not only the RanBP2 complex, but possibly how other SUMO E3 ligases are regulated as well. Further study of the interaction between NUSAP1 and the RanBP2 SUMO E3 ligase, and possibly the SUMO pathway, could elucidate the mechanisms involved in the regulation of other SUMO E3 ligases.
Experimental procedures
Mammalian cell culture
HEK-293T, U2OS, HCT116, and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% FBS (Atlanta Biologicals) and Pen/Strep (Gibco). Nocodazole (Sigma, 487928) was used at 150 ng/ml for U2OS and 200 ng/ml for 293T. All siRNA transfections were performed using Lipofectamine RNAiMax (Thermo) following the manufacturer's protocol. Control, nonspecific siRNA-targeted firefly luciferase (siFF). Three different siRNAs against NUSAP1 were used, each at a concentration of 20 nm. The siRNA oligonucleotide sequences used in this study are detailed in supplemental Table S2.
Immunoblotting and immunoprecipitations
Samples analyzed by immunoblot were lysed in NETN (20 mm Tris-Cl, pH 8.0, 100 mm NaCl, 0.5 mm EDTA, 0.5% Nonidet P-40 (Nonidet P-40)) supplemented with 1 μg/ml of apoprotinin, 1 μg/ml of pepstatin, 10 μg/ml of leupeptin, 1 mm Na3VO4, 1 mm NaF, and 1 mm AEBSF (4-(2-aminoethyl)benzenesulfonyl fluoride). Protein concentration was estimated using the Bradford assay (Bio-Rad). Laemmli buffer was added to samples, which were then separated by SDS-PAGE gel electrophoresis using homemade or commercially available gels (Bio-Rad). Gels were transferred to nitrocellulose membranes and blotted using standard immunoblotting procedures.
NUSAP1-interacting proteins were identified using endogenous immunoprecipitation followed by tandem mass spectrometry. The mass spectrometry analysis was carried out by the UNC Hooker Proteomics Facility (described below). As a source of starting material, we used asynchronous HEK-293T and HeLa cells, or HEK-293T cells that were arrested in mitosis by overnight incubation in nocodazole. Whole cell extracts (WCE) were prepared on ice in the aforementioned NETN lysis buffer. Protein A/G-agarose beads were covalently coupled to control IgG or anti-NUSAP1 antibodies using dimethyl pimelimidate (46). WCE was clarified by centrifugation at 14,000 rpm for 10 min at 4 °C in a benchtop microcentrifuge. Clarified lysates were mixed with antibody-coated beads on a rotary mixer for 4 h at 4 °C. Samples were quickly washed three times with lysis buffer, eluted using 100 mm glycine (pH 2.5), and neutralized with Tris buffer (pH 7.5). Elutions were then digested with trypsin and analyzed by mass spectrometry (see below for details).
For the co-IP experiments in Fig. 2, cells were lysed in hypotonic lysis buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT), supplemented with 1 μg/ml of apoprotinin, 1 μg/ml of pepstatin, 10 μg/ml of leupeptin, 1 mm Na3VO4, 1 mm NaF, and 1 mm AEBSF. Protein A/G DynaBeads (Thermo) were bound to control rabbit IgG, NUSAP1, or RanBP2 antibodies overnight at 4 °C. Samples were incubated with beads for 4 h at 4 °C, which were subsequently washed three times in lysis buffer and eluted with 2× Laemmli sample buffer at 95 °C for 10 min.
Immunological reagents
Commercially available antibodies used in this study, including their use (immunoblotting, immunofluorescence, etc.), catalogue numbers, and specific dilutions are included in supplemental Table S2.
An antibody against RanBP2 was generated in-house for these studies. The DNA sequence encoding amino acids 1000–1200 was cloned into the pET28A using traditional PCR amplification to generate an amino terminally tagged hexahistidine-tagged version of the fragment. The cloning was verified by Sanger sequencing and the resulting plasmid DNA was introduced into BL21 (DE3) Escherichia coli for recombinant protein production. The expression of the His6-tagged RanBP2 fragment was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside for 22 h at 18 °C. Bacterial pellets that had been frozen and then thawed on wet ice were diluted in His6 purification buffer (20 mm Tris, pH 7.9, 500 mm NaCl, 0.5% Nonidet P-40, 5 mm imidazole, 0.5 mg/ml of lysozyme, 0.5 mm AEBSF, 1 μg/ml of apoprotinin, 1 μg/ml of pepstatin, 10 μg/ml of leupeptin, 1 mm DTT). Cells were sonicated for 5 min and lysates was centrifuged at 15,000 rpm for 30 min at 4 °C in a SS-34 fixed angle rotor. Soluble extracts were incubated in batch with nickel-nitrilotriacetic acid-agarose (Thermo) on a rotary mixer for 90 min at 4 °C. Beads were washed extensively with 20 mm Tris (pH 7.5), 500 mm NaCl, 0.5% Nonidet P-40, 30 mm imidazole and then eluted in 20 mm Tris (pH 7.5), 200 mm NaCl, 300 mm imidazole. Eluted samples were analyzed by Coomassie Blue staining, combined, and tested by Bradford. His6-RanBP21000–1200 was conjugated to keyhole limpet hemocyanin and injected into rabbits for antiserum production by Pocono Rabbit Farm & Laboratory (PRF&L, Canadensis, PA). The serum was affinity purified over a column of recombinant protein using the described protocols and dialyzed into PBS (46).
For immunoblotting, antibodies were diluted in a solution of 5% nonfat dry milk in phosphate-buffered saline, 0.05% Tween 20 (PBST). Antibodies were either incubated at room temperature for 2 h or overnight at 4 °C. Detection was performed using HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.; 1:10,000), ECL reagent (Pierce), and exposure to film.
Mass spectrometry analysis
Samples provided in solution were digested using the FASP (filter-assisted sample preparation) protocol. This includes reduction, alkylation, and digested with trypsin. The peptides were extracted, lyophilized, and resuspended in 2% acetonitrile, 98% of 0.1% formic acid. The peptides were loaded onto a 2-cm long × 360-μm outer diameter × 100-μm inner diameter microcapillary fused silica precolumn packed with Magic 5-μm C18AQ resin (Michrom Biosciences, Inc.). After sample loading, the precolumn was washed with 95% Solvent A (0.1% formic acid in water), 5% Solvent B (0.1% formic acid in acetonitrile) for 20 min at a flow rate of 2 μl/min. The pre-column was then connected to a 360-μm outer diameter × 75-μm inner diameter analytical column packed with 22 cm of 5-μm C18 resin. The peptides were eluted at a flow rate of 250 nl/min by increasing the percentage of solvent B to 40% with a Nano-Acquity HPLC solvent delivery system (Waters Corp.). The LC system was directly connected through an electrospray ionization source interfaced to an LTQ Orbitrap Velos ion trap mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was controlled by Xcalibur software and operated in the data-dependent mode in which the initial MS scan recorded the mass to charge (m/z) ratios of ions over the range 400–2000. The 10 most abundant ions were automatically selected for subsequent collision-activated dissociation. All files were searched using MASCOT (Matrix Science, version 2.3.02) via Proteome Discoverer (Thermo Fisher Scientific, version 1.3.0.339) against a recently downloaded human FASTA database. The search parameters included peptide mass tolerance of 10 ppm, and fragment ion tolerance of 0.6 mass unit. The search allowed variable modifications for methionine oxidation and carbamidomethylation of Cys.
Gel filtration chromatography
Mitotically arrested 293T cells were analyzed by gel filtration chromatography. Cells were arrested overnight in nocodazole and lysed in hypotonic buffer as described above. The cell extract was clarified via centrifugation followed by filtration through a 0.22-μm syringe filter. Protein complexes in the clarified lysate were then separated using a size exclusion column (Superose 6 10/30, GE Healthcare) that had been pre-equilibrated in hypotonic lysis buffer. During separation, 0.4-ml fractions were collected and later analyzed by immunoblot and endogenous NUSAP1 IP.
Chromatin fractionation
Cells were lysed in CSK buffer (10 mm PIPES, pH 7.0, 300 mm sucrose, 100 mm NaCl, 3 mm MgCl2, 0.1% Triton X-100) supplemented with 1 μg/ml of apoprotinin, 1 μg/ml of pepstatin, 10 μg/ml of leupeptin, 1 mm Na3VO4, 1 mm NaF, and 1 mm AEBSF. Protein concentration was determined using Bradford assay and a portion of the lysate was taken for WCE samples. Samples were then pelleted at 3,000 rpm for 5 min at 4 °C. Supernatant was saved as the soluble fraction (S). Each pellet was washed with CSK buffer on ice and pelleted. The supernatant was removed and the pellet was resuspended in Laemmli buffer diluted in CSK and boiled for 5 min before the DNA was sheared using a needle to produce the insoluble fraction (I).
Immunofluorescence imaging
Cells were plated on poly-l-lysine-coated coverslips approximately 1 day before fixation. Cells were fixed in PHEM buffer (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, 2 mm MgCl2, adjusted to pH 7.0 using KOH) + 3% paraformaldehyde for 13 min at 37 °C. Cells were washed with PHEM buffer and permeabilized using PHEM + 0.5% Nonidet P-40 for 15 min at room temperature. Cells were washed in PBS before blocking in PBS + 5% BSA. All antibodies were subsequently diluted in PBST + 5% BSA. Primary antibodies and their dilutions used were: α-NUSAP1 (1:500), α-RanGAP1 (1:100), α-RanBP2 (1:100), α-tubulin (1:200), mouse anti-HEC1 (Abcam ab3613; 1:500), guinea pig anti-CENP-C (MBL; 1:1000). Samples were incubated in primary antibody solution for 1 h at 37 °C. All fluorescent secondary antibodies (anti-mouse Alexa 594, anti-rabbit Alexa 488, anti-mouse Alexa 488, anti-guinea pig Cy5) were diluted 1:200 dilution and incubated for 1 h at 37 °C. DNA was counterstained with 1 μg/ml of Hoechst 33342 for 5 min at room temperature. All samples were mounted onto glass slides in Prolong Gold medium.
The cold stability assay was conducted as detailed in Suzuki et al. (47). Briefly, cells were treated with ice-cold media for 10 min before fixation and staining. PLA was performed using the Sigma Duolink In Situ Red Starter Kit Mouse/Rabbit (DUO92101 Sigma). Cells were plated and fixed as described above. Staining was performed following the DuoLink kit protocol, with primary antibodies against NUSAP1, RanBP2, and RanGAP1 being used at the concentrations described above. Tubulin counterstaining was performed using Alexa Fluor 488-conjugated α-tubulin at a dilution of 1:100 for 40 min at 37 °C.
For image acquisition, three-dimensional stacked images were obtained sequentially at 200 nm steps along the z axis through the cell using MetaMorph 7.8 software (Molecular Devices) and a Nikon Ti-inverted microscope equipped with the Orca-ER cooled CCD camera (Nikon) and an ×100/1.4 NA PlanApo objective (Nikon). X, Y, and Z stage movement was controlled by piezo MS2000–500 (ASI). Solid state laser (Andor) illumination at 488, 568, and 647 nm were projected through Borealis (Andor) for uniform illumination before a spinning disc confocal head (Yokogawa CSU-10, PerkinElmer Life Sciences) (48).
Flow cytometry
Cells were collected and fixed in 70% ethanol. Cells were then washed twice in 1 ml of PBS, and then resuspended in a solution of PBS containing a final concentration of 25 μg/ml of propidium iodide (Sigma) and 100 μg/ml of RNase A. Cells were sorted using a Beckman Coulter CyAn ADP. Data were analyzed using FlowJo software.
Author contributions
M. J. E. and C. A. M. conceived the experiments and analyzed data. C. A. M. was involved in all experiments. A. S. and C. A. M. performed fluorescent imaging and analysis and A. A. and C. A. M. performed cell cycle analysis. J. Y. M. and A. D. assisted with size exclusion chromatography. C. A. M. and M. J. E. prepared and wrote the manuscript and A. D. and E. D. S. provided critical feedback on the manuscript.
Supplementary Material
Acknowledgments
Special thanks to the Emanuele laboratory for critical feedback throughout this study. This work was done in part at the University of North Carolina Flow Cytometry and Proteomics Core Facilities, supported in part by Cancer Center Core Support Grant P30 CA016086 to the Lineberger Cancer Center.
This work was supported in part by start-up funds from the University Cancer Research Fund, Susan G. Komen Foundation Grant CCR14298820, the Jimmy-V Foundation, and National Institutes of Health Grant R01GM120309 (to M. J. E., C. A. M., and A. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1 and S2 and Figs. S1–S5.
- NUSAP1
- nucleolar and spindle-associated protein 1
- APC/C
- anaphase promoting complex/cyclosome
- CPC
- chromosome passenger complex
- IF
- immunofluorescent
- IP
- immunoprecipitation
- TSC
- total spectral count
- PLA
- proximity ligation assay
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride
- WCE
- whole cell extract
- PIPES
- 1,4-piperazinediethanesulfonic acid.
References
- 1. Ribbeck K., Raemaekers T., Carmeliet G., and Mattaj I. W. (2007) A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 17, 230–236 [DOI] [PubMed] [Google Scholar]
- 2. Raemaekers T., Ribbeck K., Beaudouin J., Annaert W., Van Camp M., Stockmans I., Smets N., Bouillon R., Ellenberg J., and Carmeliet G. (2003) NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 162, 1017–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ribbeck K., Groen A. C., Santarella R., Bohnsack M. T., Raemaekers T., Köcher T., Gentzel M., Görlich D., Wilm M., Carmeliet G., Mitchison T. J., Ellenberg J., Hoenger A., and Mattaj I. W. (2006) NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell 17, 2646–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vanden Bosch A., Raemaekers T., Denayer S., Torrekens S., Smets N., Moermans K., Dewerchin M., Carmeliet P., and Carmeliet G. (2010) NuSAP is essential for chromatin-induced spindle formation during early embryogenesis. J. Cell Sci. 123, 3244–3255 [DOI] [PubMed] [Google Scholar]
- 5. Xie P., Li L., Xing G., Tian C., Yin Y., He F., and Zhang L. (2011) ATM-mediated NuSAP phosphorylation induces mitotic arrest. Biochem. Biophys. Res. Commun. 404, 413–418 [DOI] [PubMed] [Google Scholar]
- 6. Chou H. Y., Wang T. H., Lee S. C., Hsu P. H., Tsai M. D., Chang C. L., and Jeng Y. M. (2011) Phosphorylation of NuSAP by Cdk1 regulates its interaction with microtubules in mitosis. Cell Cycle 10, 4083–4089 [DOI] [PubMed] [Google Scholar]
- 7. Chen L., Yang L., Qiao F., Hu X., Li S., Yao L., Yang X.-L., and Shao Z.-M. (2015) High levels of nucleolar spindle-associated protein and reduced levels of BRCA1 expression predict poor prognosis in triple-negative breast cancer. PLoS ONE 10, e0140572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okamoto A., Higo M., Shiiba M., Nakashima D., Koyama T., Miyamoto I., Kasama H., Kasamatsu A., Ogawara K., Yokoe H., Tanzawa H., and Uzawa K. (2015) Down-regulation of nucleolar and spindle-associated protein 1 (NUSAP1) expression suppresses tumor and cell proliferation and enhances anti-tumor effect of Paclitaxel in oral squamous cell carcinoma. PLoS ONE 10, e0142252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y.-N., Rush J., Hsu P. W., Yen H.-C., and Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozlü N., Monigatti F., Renard B. Y., Field C. M., Steen H., Mitchison T. J., and Steen J. J. (2010) Binding partner switching on microtubules and aurora-B in the mitosis to cytokinesis transition. Mol. Cell. Proteomics 9, 336–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song L., and Rape M. (2010) Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol. Cell 38, 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 14. Dasso M. (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Myatt S. S., Kongsema M., Man C. W., Kelly D. J., Gomes A. R., Khongkow P., Karunarathna U., Zona S., Langer J. K., Dunsby C. W., Coombes R. C., French P. M., Brosens J. J., and Lam E. W. (2014) SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 33, 4316–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pichler A., Gast A., Seeler J. S., Dejean A., and Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 17. Saitoh H., Pu R., Cavenagh M., and Dasso M. (1997) RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. U.S.A. 94, 3736–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saitoh H., Sparrow D. B., Shiomi T., Pu R. T., Nishimoto T., Mohun T. J., and Dasso M. (1998) Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8, 121–124 [DOI] [PubMed] [Google Scholar]
- 19. Mahajan R., Delphin C., Guan T., Gerace L., and Melchior F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 20. Mahajan R., Gerace L., and Melchior F. (1998) Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Werner A., Flotho A., and Melchior F. (2012) The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell 46, 287–298 [DOI] [PubMed] [Google Scholar]
- 22. Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., and van Deursen J. M. (2008) Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIα. Cell 133, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klein U. R., Haindl M., Nigg E. A., and Muller S. (2009) RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol. Biol. Cell 20, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bachant J., Alcasabas A., Blat Y., Kleckner N., and Elledge S. J. (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 25. Montpetit B., Hazbun T. R., Fields S., and Hieter P. (2006) Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J. Cell Biol. 174, 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Westman B. J., Verheggen C., Hutten S., Lam Y. W., Bertrand E., and Lamond A. I. (2010) A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol. Cell 39, 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X.-D., Goeres J., Zhang H., Yen T. J., Porter A. C., and Matunis M. J. (2008) SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y., Zhang Y.-D., Guo L., Huang H.-Y., Zhu H., Huang J.-X., Zhou S.-R., Dang Y.-J., Li X., and Tang Q.-Q. (2013) Protein inhibitor of activated STAT 1 (PIAS1) is identified as the SUMO E3 ligase of CCAAT/enhancer-binding protein β (C/EBPβ) during adipogenesis. Mol. Cell. Biol. 33, 4606–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L., Zhou Y., Sun L., Xing G., Tian C., Sun J., Zhang L., and He F. (2007) NuSAP is degraded by APC/C-Cdh1 and its overexpression results in mitotic arrest dependent of its microtubules' affinity. Cell Signal. 19, 2046–2055 [DOI] [PubMed] [Google Scholar]
- 30. Choudhury R., Bonacci T., Arceci A., Lahiri D., Mills C. A., Kernan J. L., Branigan T. B., DeCaprio J. A., Burke D. J., and Emanuele M. J. (2016) APC/C and SCF(cyclin F) constitute a reciprocal feedback circuit controlling S-phase entry. Cell Rep. 16, 3359–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu C.-K., Ozlü N., Coughlin M., Steen J. J., and Mitchison T. J. (2012) Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell 23, 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurasawa Y., Earnshaw W. C., Mochizuki Y., Dohmae N., and Todokoro K. (2004) Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kellogg E. H., Howes S., Ti S.-C., Ramírez-Aportela E., Kapoor T. M., Chacón P., and Nogales E. (2016) Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc. Natl. Acad. Sci. U.S.A. 113, 9430–9439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J.-P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Hériché J.-K., Sykora M. M., Augsburg M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., et al. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joseph J., Liu S. T., Jablonski S. A., Yen T. J., and Dasso M. (2004) The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611–617 [DOI] [PubMed] [Google Scholar]
- 37. Joseph J., Tan S.-H., Karpova T. S., McNally J. G., and Dasso M. (2002) SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 39. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verbakel W., Carmeliet G., and Engelborghs Y. (2011) SAP-like domain in nucleolar spindle associated protein mediates mitotic chromosome loading as well as interphase chromatin interaction. Biochem. Biophys. Res. Commun. 411, 732–737 [DOI] [PubMed] [Google Scholar]
- 41. Bernier-Villamor V., Sampson D. A., Matunis M. J., and Lima C. D. (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 42. Schimmel J., Eifler K., Sigurðsson J. O., Cuijpers S. A., Hendriks I. A., Verlaan-de Vries M., Kelstrup C. D., Francavilla C., Medema R. H., Olsen J. V., and Vertegaal A. C. (2014) Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 53, 1053–1066 [DOI] [PubMed] [Google Scholar]
- 43. Schou J., Kelstrup C. D., Hayward D. G., Olsen J. V., and Nilsson J. (2014) Comprehensive identification of SUMO2/3 targets and their dynamics during mitosis. PLoS ONE 9, e100692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lamoliatte F., Caron D., Durette C., Mahrouche L., Maroui M. A., Caron-Lizotte O., Bonneil E., Chelbi-Alix M. K., and Thibault P. (2014) Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5, 5409. [DOI] [PubMed] [Google Scholar]
- 45. Becker J., Barysch S. V., Karaca S., Dittner C., Hsiao H.-H., Berriel Diaz M., Herzig S., Urlaub H., and Melchior F. (2013) Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20, 525–531 [DOI] [PubMed] [Google Scholar]
- 46. Harlow E., and Lane D. (1999) Using antibodies: a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47. Suzuki A., Badger B. L., and Salmon E. D. (2015) A quantitative description of Ndc80 complex linkage to human kinetochores. Nat. Commun. 6, 8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suzuki A., Badger B. L., Wan X., DeLuca J. G., and Salmon E. D. (2014) The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev. Cell 30, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aravind L., and Koonin E. V. (2000) SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.