Abstract
Intervertebral disc degeneration (IDD) causes chronic back pain and is linked to production of proinflammatory molecules by nucleus pulposus (NP) and other disc cells. Activation of tonicity-responsive enhancer-binding protein (TonEBP)/NFAT5 by non-osmotic stimuli, including proinflammatory molecules, occurs in cells involved in immune response. However, whether inflammatory stimuli activate TonEBP in NP cells and whether TonEBP controls inflammation during IDD is unknown. We show that TNF-α, but not IL-1β or LPS, promoted nuclear enrichment of TonEBP protein. However, TNF-α-mediated activation of TonEBP did not cause induction of osmoregulatory genes. RNA sequencing showed that 8.5% of TNF-α transcriptional responses were TonEBP-dependent and identified genes regulated by both TNF-α and TonEBP. These genes were over-enriched in pathways and diseases related to inflammatory response and inhibition of matrix metalloproteases. Based on RNA-sequencing results, we further investigated regulation of novel TonEBP targets CXCL1, CXCL2, and CXCL3. TonEBP acted synergistically with TNF-α and LPS to induce CXCL1-proximal promoter activity. Interestingly, this regulation required a highly conserved NF-κB-binding site but not a predicted TonE, suggesting cross-talk between these two members of the Rel family. Finally, analysis of human NP tissue showed that TonEBP expression correlated with canonical osmoregulatory targets TauT/SLC6A6, SMIT/SLC5A3, and AR/AKR1B1, supporting in vitro findings that the inflammatory milieu during IDD does not interfere with TonEBP osmoregulation. In summary, whereas TonEBP participates in the proinflammatory response to TNF-α, therapeutic strategies targeting this transcription factor for treatment of disc disease must spare osmoprotective, prosurvival, and matrix homeostatic activities.
Keywords: chemokine, cytokine, inflammation, NF-κB (NF-KB), NFAT transcription factor, nucleus pulposus, NFAT5, TonEBP, intervertebral disc
Introduction
The transcription factor TonEBP,4 or NFAT5, is essential for the coordinated response of mammalian cells to osmotic stimuli, which includes the accumulation of non-ionic organic osmolytes and heat-shock proteins that prevent damage to cellular proteins and nucleic acids (1). Nucleus pulposus (NP) cells of the healthy intervertebral disc are subject to a hyperosmotic environment, which fluctuates with daily postural changes (2, 3). Previously, we have shown that TonEBP regulates survival of NP cells and maintains transcription of genes necessary for matrix homeostasis under hyperosmotic conditions (4–6). However, the regulation of TonEBP under inflammatory conditions and its role in disc degeneration remain unexplored.
Intervertebral disc degeneration (IDD) is an important risk factor for chronic low back and neck pain (7), which is a ubiquitous and costly pathological condition (8) that contributes heavily to disability in the United States and worldwide (9–11). During IDD, changes in both cellular behavior and extracellular matrix makeup ultimately compromise biomechanical function of the disc. NP and other disc cells produce cytokines, including tumor necrosis factor-α (TNF-α)/TNF and IL-1β, which contribute to an inflammatory environment through NF-κB- and MAPK-signaling pathways and promote production of matrix-degrading MMP and ADAMTS family members (12–16). Catabolism of the NP extracellular matrix limits its water-binding capacity, eventually leading to a stiffer matrix, decreased disc height, and altered mechanical loading (17, 18). Moreover, excessive mechanical loading (19) and matrix breakdown products (20) can also activate toll-like receptors (TLRs) expressed on NP cells.
Interestingly, independent of hyperosmolarity, TonEBP/NFAT5 expression is induced in response to inflammatory stimuli, including IFN-γ and IL-4 (21), in immune cells. Likewise, TonEBP can be induced by TLR activation through LPS (21–23), Escherichia coli (22), or Mycobacterium tuberculosis (24). Following LPS stimulation, TonEBP is required for antimicrobial response through regulation of NOS2 and other target genes (23). Moreover, in fibroblast-like synoviocytes derived from rheumatoid arthritis patients, TNF-α and IL-1β induce levels and nuclear localization of TonEBP. In these cells, TonEBP controls cell survival, proliferation, migration, and angiogenesis (25). However, whether such changes in TonEBP activation occur during disc degeneration is not known.
In this study, we investigated whether TonEBP is activated by proinflammatory stimuli in NP cells and its role in this context. TonEBP activity, but not expression, was modulated by proinflammatory stimuli in NP cells. Importantly, TNF-α-induced TonEBP selectively controls inflammatory gene expression without affecting its canonical osmoregulatory targets. Our results clearly suggest that TonEBP is critical for TNF-α-mediated expression of several molecules thought to play a key role during disc degeneration.
Results
TNF-α promotes TonEBP nuclear localization without affecting transcript levels
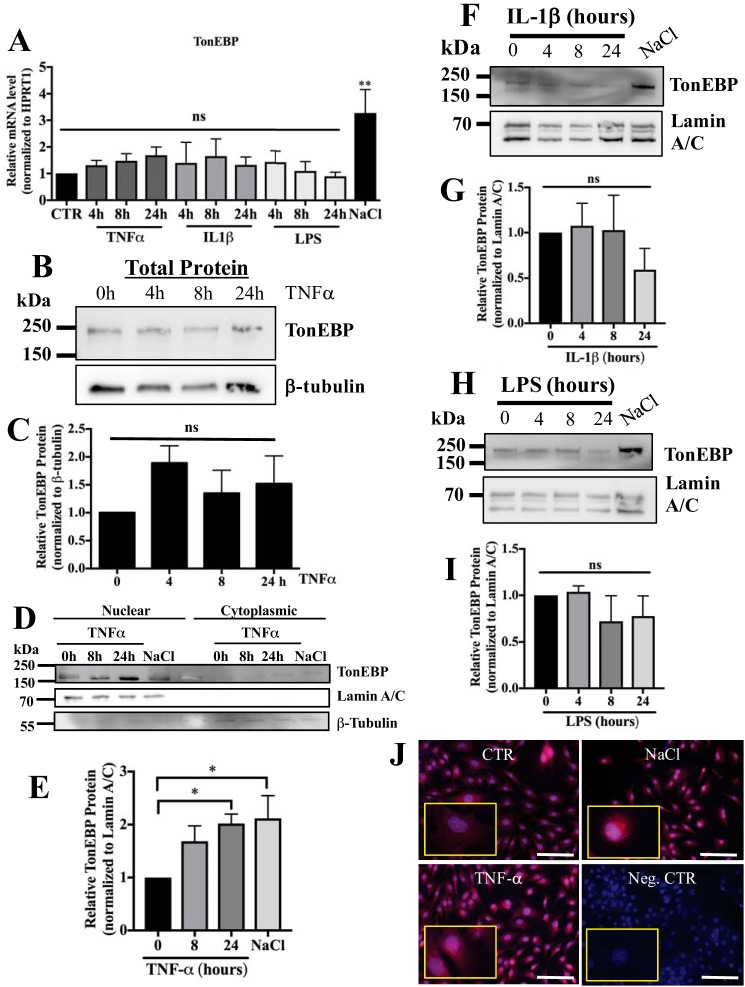
Previous studies in immune cells and rheumatoid arthritis-fibroblast-like synoviocytes have shown activation of TonEBP/NFAT5 by various inflammatory stimuli, including TNF-α. Therefore, we investigated whether TNF-α, IL-1β, and LPS promote TonEBP expression in NP cells. Interestingly, none of these proinflammatory stimuli affected TonEBP mRNA levels, although induction was seen with hyperosmolarity (Fig. 1A). We evaluated levels of total TonEBP protein following 4–24 h of stimulation with TNF-α, IL-1β, and LPS and found no significant differences with stimulation (Fig. 1, B and C). Surprisingly, only treatment with TNF-α significantly increased the nuclear pool of TonEBP protein (Fig. 1, D and E), whereas neither IL-1β (Fig. 1, F and G) nor LPS (Fig. 1, H and I) treatment had any effect on nuclear enrichment of TonEBP. This response is different from what has been reported for immune cells and fibroblast-like synoviocytes (21, 25). A similar enrichment in nuclear TonEBP was observed with immunofluorescent staining (Fig. 1J).
Figure 1.
TNF-α promotes nuclear localization of TonEBP without affecting transcript levels. A, TonEBP mRNA levels were unaffected by treatment with TNF-α, IL-1β, or LPS for 4–24 h; as expected, treatment with NaCl (110 mm) resulted in induction. B and C, levels of total cellular TonEBP did not change with TNF-α treatment for 4–24 h. D and E, nuclear and cytoplasmic fraction experiment clearly shows increased nuclear accumulation of TonEBP following TNF-α treatment for 24 h. No discernable depletion of TonEBP in cytoplasmic fraction was seen. Lamin and β-tubulin loading controls showed relatively high purity of fractions. F–I, treatment with IL-1β (F and G) and LPS (H and I) for 4–24 h did not affect the amount of nuclear TonEBP protein. J, immunofluorescent detection of TonEBP in NP cells following 24 h of treatment with NaCl or TNF-α. Inset shows high magnification image of cells. Scale bar, 100 μm. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. *, p ≤ 0.05; **, p ≤ 0.01. ns, not statistically significant. Neg. CTR, negative control.
TonEBP activation under inflammatory conditions fails to induce transcription of osmoregulatory target genes
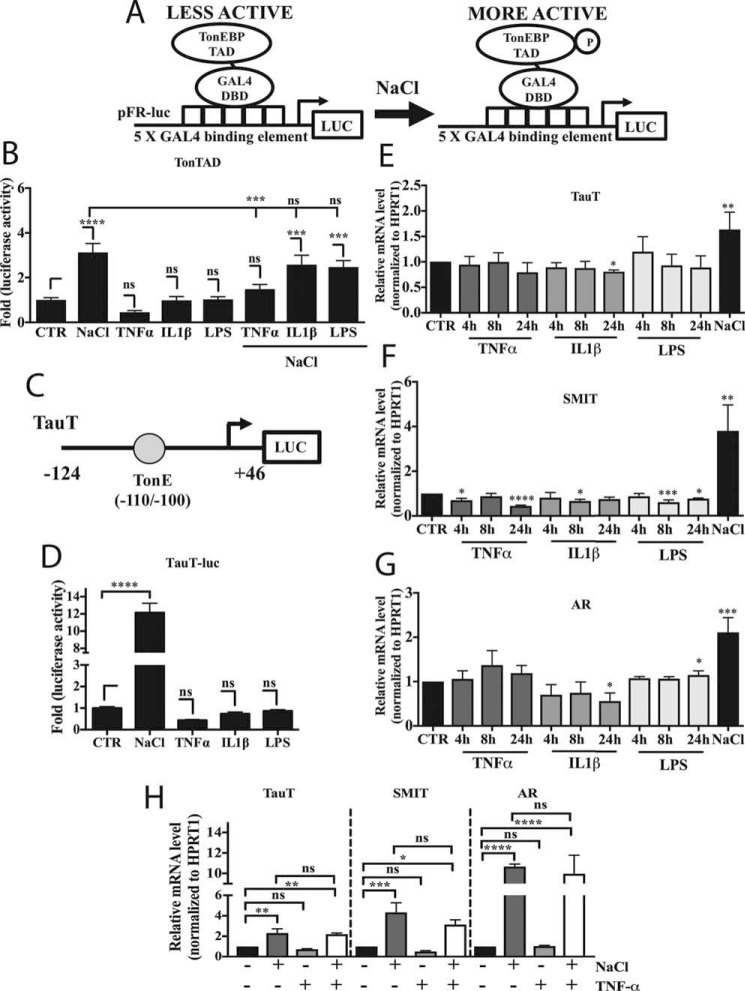
Nuclear accumulation of TonEBP is a shared feature of response to hyperosmolarity and TNF-α and an important indicator of TonEBP activation. We wanted to investigate whether TNF-α, and inflammatory stimuli such as IL-1β and LPS that do not affect nuclear translocation, can modulate the phosphorylation status of the TonEBP transactivation domain (TAD). Activity of the TonEBP-TAD (amino acids 548–1531) was assessed using a binary GAL4 system, as depicted in Fig. 2A. Interestingly, addition of TNF-α, IL-1β, or LPS had no effect on Ton-TAD activity (Fig. 2B). As expected, treatment with NaCl induced TonTAD activity. Interestingly, TAD induction by NaCl was inhibited by co-treatment with TNF-α, but not IL-1β or LPS (Fig. 2B). We then assessed the activity of the proximal TauT promoter, which contains a highly conserved TonEBP-binding site (TonE) shown to be active in NP cells (Fig. 2C) (4, 26). Similarly to TonEBP-TAD, TNF-α, IL-1β, and LPS did not affect activity, whereas NaCl strongly induced promoter activity (Fig. 2D). Because TNF-α promoted TonEBP nuclear translocation, we investigated whether mRNA levels of traditional osmoregulatory TonEBP target genes TauT/SLC6A6, SMIT/SLC5A3, and AR/AKR1B1 were affected by TNF-α as well as IL-1β and LPS. None of these target genes were significantly induced by proinflammatory stimuli; again, levels were elevated by treatment with NaCl (Fig. 2, E–G). Finally, we investigated whether co-treatment of TNF-α along with NaCl would affect inducibilty of osmoregulatory targets. TauT, SMIT, and AR were all significantly induced by treatment with 24 h of NaCl alone. Addition of TNF-α along with NaCl did not impact inducibilty of TauT, SMIT, or AR (Fig. 2H).
Figure 2.
TonEBP activation under inflammatory conditions fails to induce transcription of its canonical osmoregulatory target genes. A, schematic depicting binary TonTAD-GAL4 system used to measure TonEBP-TAD activity. B, activity of TonEBP-TAD was unaffected by treatment with TNF-α, IL-1β, or LPS alone, but it was induced by NaCl. Co-treatment with TNF-α and NaCl dampened the TAD activation by NaCl alone. C, diagram showing taurine transporter (TauT) luciferase reporter, which contains an active, highly conserved TonEBP-binding site, TonE. D, although treatment with NaCl induced activity of the TauT promoter, treatment with TNF-α, IL-1β, or LPS had no effect. E–G, TauT (E), SMIT (F), and AR (G) mRNA levels did not significantly increase with TNF-α, IL-1β, or LPS treatment for 4–24 h, whereas 8 h of NaCl treatment resulted in induction. H, co-treatment of TNF-α along with NaCl did not affect NaCl-mediated induction of TauT, SMIT, or AR. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates and, for transfection experiments, 3 technical replicates per biological replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001; ns, not statistically significant.
RNA sequencing reveals TonEBP as a critical regulator of NP cell response to TNF-α
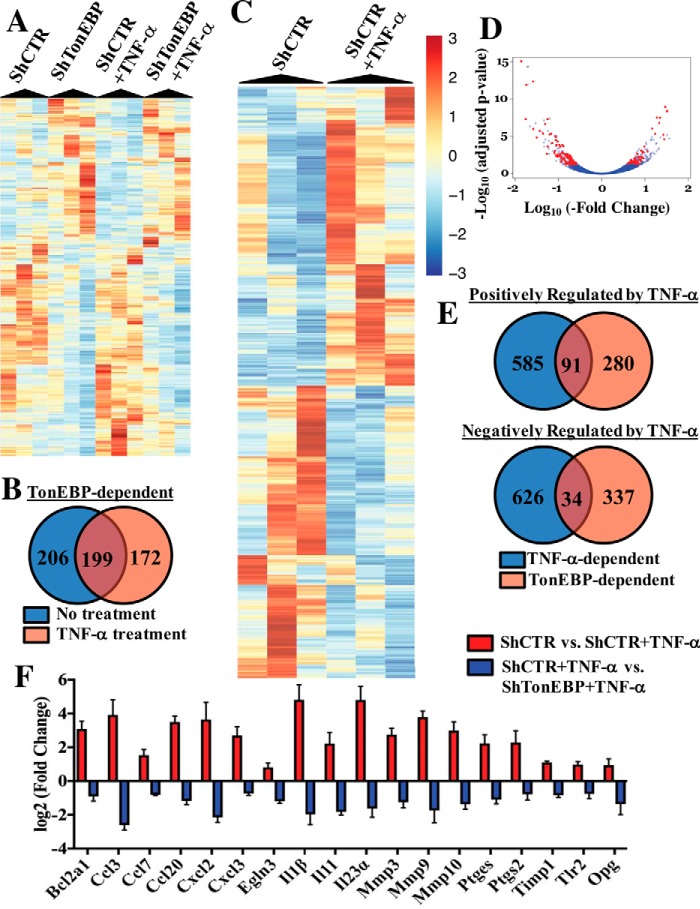
Because our results suggested that TNF-α and other inflammatory stimuli do not induce TonEBP targets concerned with cellular osmosensing, we employed an -omics approach to identify the unique transcriptional targets of TonEBP during TNF-α stimulation. We performed RNA sequencing on rat NP cells that were transduced with either lentivirally-delivered ShControl or ShTonEBP with or without TNF-α treatment for 24 h. TonEBP controlled expression of 405 genes in untreated cells and 371 genes under TNF-α stimulation (Fig. 3A). Importantly, although some TonEBP targets were shared between untreated and TNF-α-treated NP cells, a substantial number were treatment-specific (Fig. 3B). Furthermore, RNA-seq identified 1339 genes that were differentially expressed between basal and TNF-α-treated groups (Fig. 3C). We found that out of all genes differentially regulated by TNF-α, 50.6% were positively and 49.4% were negatively regulated. We then determined what proportion of the transcriptional output of TNF-α could be attributed to TonEBP in NP cells. Overall, out of all genes modulated by TNF-α, 8.5% were also regulated by TonEBP (Fig. 3D). Of the genes positively regulated by TNF-α, 12.1% (Fig. 3E) showed positive and 1.3% showed negative correlation with TonEBP. Likewise, of genes negatively regulated by TNF-α, 0.3% were positively and 4.8% were negatively correlated with TonEBP (Fig. 3E). Because TNF-α is closely linked to enhanced matrix catabolism and inflammation during disc degeneration, we decided to focus on genes that were positively regulated by both TNF-α and TonEBP and performed ingenuity pathway analysis (IPA) on this subset of genes. Several pathways important in the context of disc degeneration and metabolism emerged, including inhibition of matrix metalloproteases, IL-17-related pathways, and HIF-1α signaling (supplemental Table 1). Top diseases and disorders included inflammatory response, connective tissue disorders, and skeletal and muscular disorders (supplemental Table 2). Networks associated with this gene list included hematological system development and function, inflammatory response, and tissue morphology and cellular movement, connective tissue development and function, and tissue development (supplemental Table 3). Log2 (-fold change) values from RNA-seq results for select genes in these identified pathways is shown in Fig. 3F. These genes were significantly different between indicated experimental groups, as defined by adjusted p value < 0.05. We also validated some of the TonEBP targets shown in Fig. 3C that were most closely linked to disc degeneration using qRT-PCR and ELISA. As seen before, TonEBP and TauT levels were unaffected by TNF-α treatment but, expectedly, decreased with TonEBP silencing (Fig. 4A). Expression of genes associated with matrix catabolism such as ADAMTS4, ADAMTS5, and MMP3 were all decreased by TonEBP knockdown in the presence of TNF-α (Fig. 4B). In addition, we investigated how expression of key NP matrix molecules known to be suppressed by TNF-α were affected by TonEBP silencing. The suppressive effect of TNF-α on ACAN and COL2A1 was not changed by TonEBP knockdown (Fig. 4C). This is likely due to the fact that TonEBP has been shown to positively regulate these genes in NP and chondroprogenitor cells (4, 27).
Figure 3.
RNA sequencing reveals TonEBP as an important regulator of NP cell response to TNF-α. NP cells were transduced with either ShControl or ShTonEBP and cultured with or without TNF-α for 24 h, and RNA sequencing was performed. A, heat map depicting genes differentially expressed between shControl and shTonEBP under either basal or TNF-α-stimulated (24-h TNF-α) conditions. B, Venn diagram showing overlap between TonEBP-dependent genes in untreated versus TNF-α-treated cells. C, heat map depicting genes differentially expressed between control and TNF-α treatment groups. D, volcano plot depicting expression of transcripts that are differentially expressed between control and TNF-α treatment groups and controlled by TonEBP under TNF-α treatment. E, Venn diagrams showing overlap between genes positively or negatively regulated by TNF-α and also regulated by TonEBP. F, log2(−fold change) values for inflammation-related transcripts that were differentially expressed between control and TonEBP knockdown under TNF-α treatment; genes shown were statistically significant between the indicated experimental groups, as defined by adjusted p value <0.05. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates.
Figure 4.
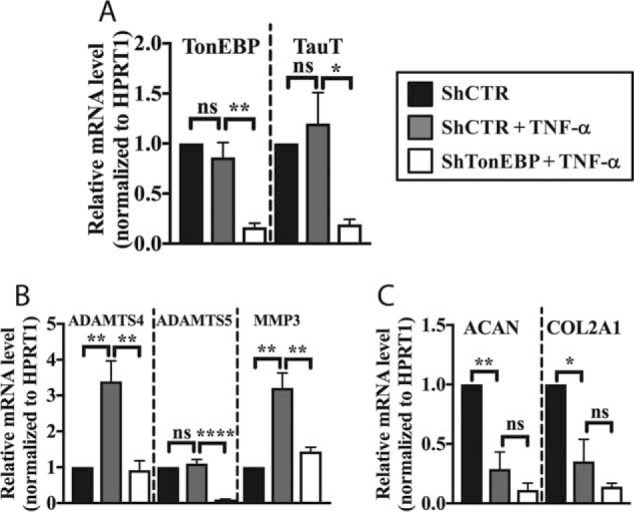
TonEBP regulates TNF-α-dependent expression of select genes linked to matrix catabolism. A, qRT-PCR confirmation of TonEBP knockdown. TonEBP and TauT mRNA levels were unaffected by TNF-α treatment in ShControl-transduced cells but decreased in cells transduced with ShTonEBP. B, TonEBP regulated transcript levels of matrix catabolic enzymes ADAMTS4, ADAMTS5, and MMP3 under TNF-α treatment. C, TNF-α treatment significantly decreases transcript levels of ACAN and COL2A1 in ShControl-transduced cells. Levels were unaffected by knockdown of TonEBP. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. *, p ≤ 0.05; **, p ≤ 0.01, ****, p ≤ 0.0001. ns, not statistically significant.
Additionally, we studied expression of several cytokines, chemokines, and regulators of the inflammatory response in TonEBP-silenced NP cells under TNF-α stimulation. TNF-α induced mRNA levels of TNF, NOS2, PHD3, and TLR2, and this induction was significantly decreased following TonEBP knockdown (Fig. 5A). Similarly, we measured mRNA and protein levels of CCL2 and IL-6 in TonEBP-silenced cells. Both genes were induced by TNF-α treatment (Fig. 5B). The transcript and protein levels were significantly suppressed by TonEBP knockdown following TNF-α treatment (Fig. 5, B–D).
Figure 5.
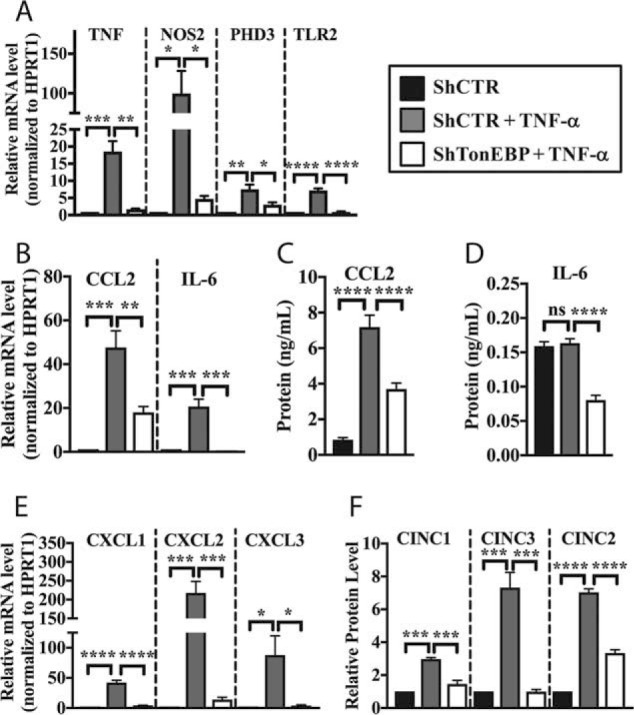
TonEBP controls TNF-α-mediated induction of key cytokines, chemokines, and inflammatory molecules in NP cells. A, TNF-α treatment induced expression of TNF, NOS2, PHD3, and TLR2 mRNAs, which was significantly inhibited by TonEBP knockdown. B and C, expression level of CCL2 and IL-6 mRNA (B) and CCL2 protein (C) were induced by TNF-α treatment; this induction was blocked by TonEBP knockdown. D, IL-6 protein levels were decreased by TonEBP knockdown under TNF-α treatment condition. E and F, CXCL1/CINC1, CXCL2/CINC3, and CXCL3/CINC2 mRNA (E) and protein (F) levels were induced by TNF-α in cell transduced with ShControl and significantly decreased by TonEBP knockdown. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. ns, not statistically significant.
TonEBP promotes TNF-α-mediated induction of CXCL1, CXCL2, and CXCL3
Our RNA-seq results suggested that TonEBP regulated expression of CXCL2 and CXCL3. Therefore, we further investigated regulation of CXCL family members by TonEBP in NP cells. Treatment with TNF-α induced mRNA levels of CXCL1, CXCL2, and CXCL3, which were significantly decreased in TonEBP-knockdown NP cells (Fig. 5E). To evaluate levels of CXCL proteins, we collected conditioned media from control and TonEBP-knockdown cells and performed immunoblots. We found that TNF-α-mediated induction of CINC1/CXCL1, CINC3/CXCL2, and CINC2/CXCL3 protein was also inhibited in TonEBP-silenced cells (Fig. 5F).
TonEBP controls activity of the CXCL1 promoter via a highly conserved NF-κB-binding site
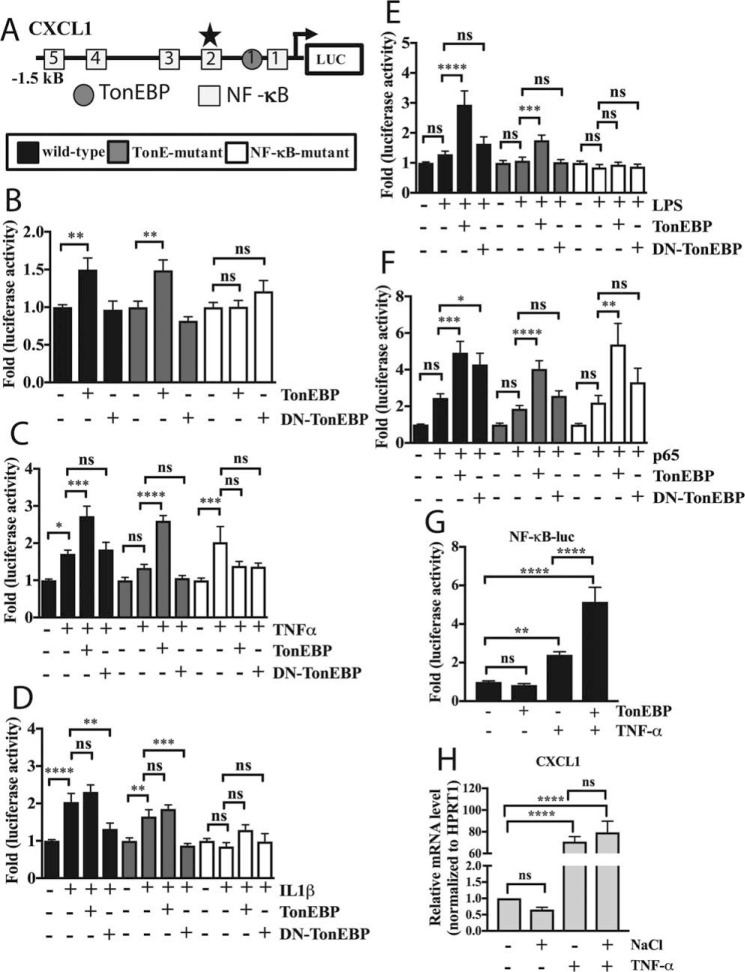
To investigate whether TonEBP acts directly on the CXCL1 promoter, we studied the proximal (1.5 kb) promoter region of the rat CXCL1 gene. Using MatInspector software, we searched for potential TonE's in the promoter, finding only a single site using a prediction score cutoff of 0.8, in addition to five NF-κB sites predicted earlier (28). To test the effects of various stimuli on the CXCL1 promoter, we measured activity of a wild-type promoter-luciferase reporter construct (Fig. 6A). Similarly, we measured activities of the construct with the predicted TonE mutation and a construct with the mutation in the most active NF-κB site, which was experimentally validated in a previous study (four other NF-κB sites are wild type) (28). To test whether TonEBP controls activity of the CXCL1 promoter and its mode of regulation, we overexpressed either wild-type (TonEBP) or dominant-negative (DN-TonEBP) TonEBP and measured activities of wild-type as well as mutant promoters. Surprisingly, although TonEBP induced activity of both wild-type and TonE-mutant CXCL1 promoters, it had no effect on the NF-κB-mutant promoter (Fig. 6B). Moreover, DN-TonEBP had no effect on any of the promoters under basal conditions (Fig. 6B). We then explored whether modulation of TonEBP would impact the responsiveness of these promoters to various inflammatory stimuli. We found that overexpression of TonEBP enhanced the inductive effect of TNF-α on the wild-type and TonE-mutant promoters. Although TNF-α increased activity of the NF-κB-mutant, TonEBP failed to further induce this activity (Fig. 6C). In contrast, TonEBP overexpression had no synergistic effect on IL1-β-mediated induction in activity of wild-type and TonE-mutant promoters. IL-1β did not affect the activity of the NF-κB mutant, and there was no modulation by co-expression of TonEBP. However, DN-TonEBP blocked IL-1β-mediated induction in wild-type and TonE-mutant promoter activity. Again, there was no effect of DN-TonEBP on the NF-κB-mutant promoter (Fig. 6D). In contrast to TNF-α and IL-1β treatment, LPS alone did not induce activity of any of the promoters. Interestingly, induction of wild-type promoter activity was seen when LPS was combined with TonEBP overexpression and to a lower extent with TonE mutation; again, there was no effect on the NF-κB-mutant (Fig. 6E). Finally, we added a small dose of p65 plasmid such that further induction of the promoter could be achieved, and we evaluated the ability of TonEBP to modulate its effects on the CXCL1 promoter. Although there was a trend of increased activity of all three promoters (wild-type, TonE-, and NF-κB-mutant) in presence of p65, it did not reach statistical significance. However, TonEBP co-expression was able to enhance activity of all promoters, including the NF-κB-mutant (Fig. 6F). We further examined the ability of TonEBP to act synergistically with NF-κB signaling by measuring activity of an NF-κB reporter in response to TNF-α treatment along with TonEBP. Although TNF-α alone induced NF-κB reporter activity, expression of TonEBP further enhanced this effect (Fig. 6G). To test whether cross-talk between osmotic response and inflammatory pathways impacted CXCL1 response to TNF-α, we measured mRNA expression following treatment with NaCl alone, TNF-α alone, or NaCl with TNF-α. We found that CXCL1 expression was unaffected by NaCl and was induced by TNF-α. Furthermore, co-treatment with NaCl and TNF-α did not impact inducibilty by TNF-α (Fig. 6H).
Figure 6.
TonEBP controls the activity of the CXCL1 promoter via a highly conserved NF-κB-binding site. A, diagram showing predicted TonEBP- and NF-κB-binding sites in the rat CXCL1 proximal promoter spanning −1.5 kb upstream of the transcription start site. Predicted TonE is depicted by gray circle and NF-κB-binding sites by open squares. NF-κB-mutant reporter contains mutation in binding site 2 (star), which has high species conservation and activity in other cell types. B, under basal conditions, TonEBP increased activities of WT and TonE-mutant promoters, whereas NF-κB-mutant was unaffected. C, WT, TonE-, and NF-κB-mutant reporters were all induced by TNF-α treatment. During TNF-α treatment, addition of TonEBP further induced activity of WT and TonE-mutant reporters only. D, WT and TonE-mutant reporters, but not NF-κB-mutant, were induced by IL-1β treatment. During IL-1β stimulation, addition of TonEBP did not affect inducibility of any of the reporters, whereas DN-TonEBP blocked induction of WT and TonE-mutant reporters. E, none of the reporters responded to LPS treatment alone. Addition of TonEBP during LPS treatment induced activity in WT and TonE-mutant reporters only. F, addition of small amount (15 ng) of p65 plasmid resulted in a trend of increased activity in all reporters, which was further enhanced by addition of TonEBP. G, expression of TonEBP further increased TNF-α-dependent induction of NF-κB reporter activity. H, treatment with TNF-α, but not NaCl, induced mRNA expression of CXCL1. TNF-α-mediated induction was unaffected by co-treatment with NaCl. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates and, for transfection experiments, 3 technical replicates per biological replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. ns, not statistically significant.
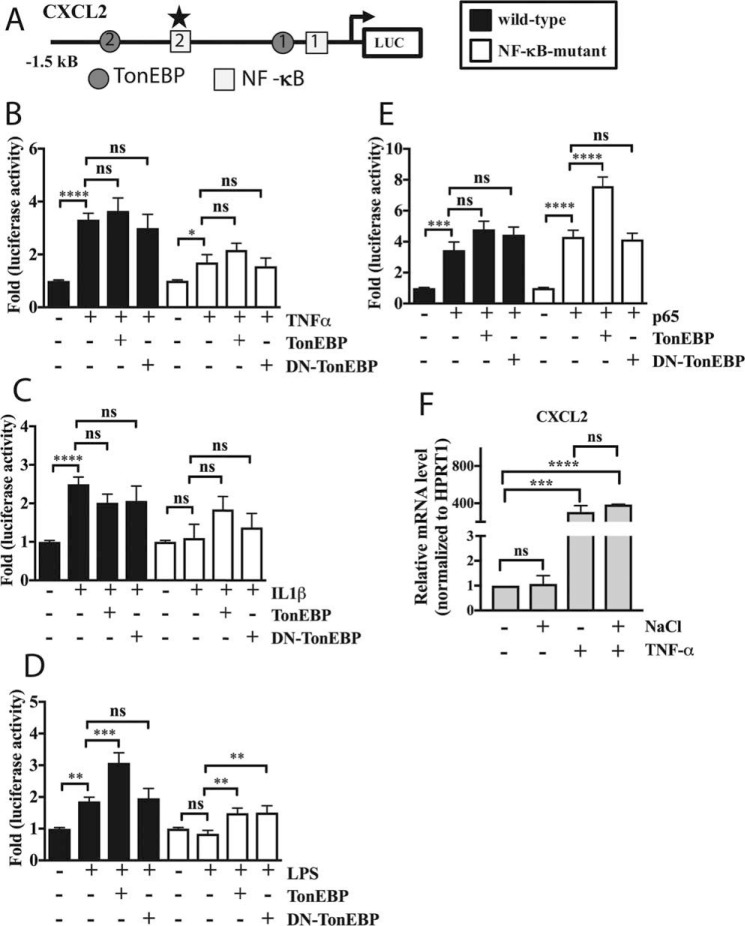
TonEBP controls the CXCL2 promoter under specific stimuli
Because CXCL1 and CXLC2 showed similar patterns of expression in response to TNF-α stimulation and TonEBP modulation, we investigated regulation of the CXCL2 promoter as well. MatInspector analysis predicted two TonEs in the 1.5-kb proximal promoter fragment (Fig. 7A) along with two previously predicted NF-κB sites, one of which is highly conserved (28). Interestingly, Multiz alignment of the rat and human genomes showed no evolutionary conservation of these TonEs, suggesting relatively low likelihood of physiological importance. We therefore chose to study only the wild-type and NF-κB-mutant (mutation of more highly conserved NF-κB site) promoters in these studies. Wild-type CXCL2 promoter showed induction in activity by TNF-α (Fig. 7B), IL-1β (Fig. 7C), and LPS (Fig. 7D). The induction by IL-1β and LPS was lost when the NF-κB site was mutated, whereas TNF-α-mediated induction was decreased by mutation, suggesting that this site was important for mediating response to inflammatory stimuli. Interestingly, although TonEBP overexpression resulted in synergistic activation of wild-type promoter only in the presence of LPS stimulation, DN-TonEBP had no effect on promoter activity in the presence of any inflammatory stimuli (Fig. 7, B–D). We then tested the effect of a small dose of p65 along with TonEBP modulation on the CXCL2 promoter activity. Surprisingly, both the wild-type and NF-κB-mutant promoters were induced by p65. Although there was a trend of TonEBP further inducing wild-type reporter activity, this was significant in the case of the NF-κB-mutant reporter (Fig. 7E), underscoring the possible contribution of TonEBP in enhancing activity of p65. DN-TonEBP did not block p65-mediated induction of either the wild-type or mutant reporter (Fig. 7E). Finally, we measured CXCL2 response to TNF-α in the presence or absence of NaCl. Similar to CXCL1, mRNA expression of CXCL2 was unaffected by NaCl, induced by TNF-α treatment, and this induction was not affected by co-treatment with NaCl (Fig. 7F). Similarly, hyperosmolarity did not affect TNF-α-dependent expression of PHD3 or TLR2; induction of MMP3 was decreased in the presence of NaCl (supplemental Fig. 1, A–C).
Figure 7.
TonEBP controls the CXCL2 promoter only under certain stimulatory conditions. A, diagram showing predicted TonEBP- and NF-κB-binding sites in the rat CXCL2 proximal promoter spanning −1.5 kb upstream of the transcription start site. TonEs are depicted by gray circles and NF-κB-binding sites by open squares. NF-κB-mutant reporter contains mutation in binding site 2 (star), which has high species conservation and activity in other cell types. B, WT and NF-κB-mutant (to a smaller extent) reporters were induced by TNF-α, whereas further addition of TonEBP or DN-TonEBP had no effect. C, WT reporter was induced by IL-1β, and further addition TonEBP or DN-TonEBP had no effect on reporter activity. D, LPS induced WT reporter activity, with further enhancement by addition of TonEBP. E, addition of small amount of p65 plasmid induced activity of both reporters. Addition of TonEBP along with p65 resulted in further induction of the NF-κB-mutant; there was a small trend of increase for the WT promoter. F, treatment with TNF-α, but not NaCl, increased CXCL2 mRNA expression. TNF-α-dependent induction was unaffected by co-treatment with NaCl. Quantitative measurements represent mean ± S.E. of ≥3 biological replicates and, for transfection experiments, 3 technical replicates per biological replicate. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. ns, not statistically significant.
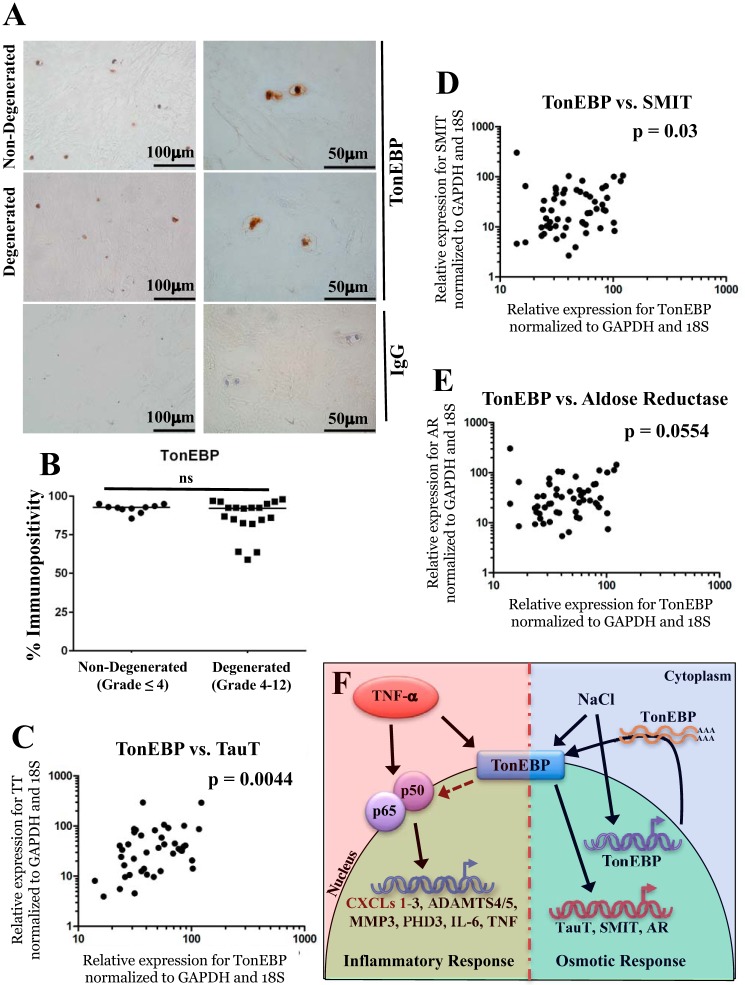
TonEBP expression is maintained during human disc degeneration and regulates osmoprotective target genes
To investigate changes in TonEBP during human disc degeneration, we measured its expression in human tissue samples. The percentage of TonEBP-immunopositive cells was unchanged between non-degenerated (grade ≤4) and degenerated (grade >4) samples (Fig. 8, A and B). Finally, we investigated the relationship between expression levels of TonEBP and its osmoregulatory targets in human tissues. Because of limited RNA quantity, we found that HPRT1 expression was not detectable in all samples. To find a suitable housekeeping gene whose expression level could be readily detected in all samples, we performed correlation analysis of HPRT1, 18S, and GAPDH expression in all human samples. Levels of 18S and GAPDH correlated well in all samples, and HPRT1 correlated with each of these in samples where HPRT1 was detectable (supplemental Fig. 2, A–C). We found strong correlations between TonEBP and TauT, SMIT, and AR in these samples (Fig. 8, C–E).
Figure 8.
TonEBP expression is correlated with levels of osmoprotective genes in human NP samples. A, immunohistochemical staining of TonEBP in non-degenerated (grades ≤4) and degenerated (grades >4) human NP samples. Isotype IgG was used as negative control for staining. B, percentage of cells immunopositive for TonEBP was not significantly different between non-degenerated and degenerated tissue samples. C–E, correlation between mRNA levels of TonEBP and osmoregulatory target genes TauT (p = 0.0044) (C), SMIT (p = 0.03) (D), and AR (p = 0.0554) (E) was seen in all tissue samples analyzed. F, schematic of proposed regulation of TonEBP by TNF-α and downstream transcriptional responses. Activation by either TNF-α or NaCl induces nuclear abundance of TonEBP, but only NaCl induces TonEBP transcript levels. Upon stimulation with TNF-α, TonEBP participates in cross-talk with NF-κB family members to promote transcription of proinflammatory genes. A, scale bar for ×40 images (left) is 100 μm, and scale bar for ×100 images (right) is 50 μm. ns, not statistically significant.
Discussion
Previous work has demonstrated that TonEBP is required for survival and function of NP cells in the osmotically challenging environment of the intervertebral disc (4–6). In the healthy NP, abundant proteoglycans and collagen afford the tissue high osmotic pressure (29). Furthermore, osmolarity of the NP is dynamic due to diurnal posture changes; water is effluxed with loading during the day and imbibed when loading is relieved at night (30–32). We have shown that, in healthy NP cells, TonEBP regulates genes involved in cell survival, matrix homeostasis, and water transport in response to hyperosmolarity (4–6, 33). However, the role of TonEBP in a degenerative disc, characterized by tissue inflammation and enhanced matrix catabolism, is unknown (16, 34). Importantly, although osmotic loads in the healthy NP reach 450–550 mosm/liter, this number declines in degenerating discs (35). Another classic feature of disc degeneration is the presence of inflammatory cytokines, including TNF-α (36) and IL-1β (37, 38), which promote matrix degradation (13, 14, 39, 40), chemokine secretion (41, 42), cellular senescence (43), and autophagy (44). The aim of this study was to determine the activity and role of TonEBP under inflammatory conditions that define the degenerative intervertebral disc milieu.
In NP cells, TNF-α treatment promoted nuclear localization of TonEBP without affecting mRNA or total protein levels. This is in contrast to osmotic activation of TonEBP, which includes a rapid mRNA induction, increase in total protein abundance, and nuclear translocation (45–47). Moreover, our result is in partial agreement with a previous report showing that TNF-α and IL-1β (25) can enhance both the total and nuclear fraction of TonEBP in other cell types. Further experiments highlighted differences between TonEBP activation by osmotic and inflammatory stimuli. Increased activity of TonEBP-TAD is a critical feature of osmotic stimulation of TonEBP activity (48). In contrast, treatment with TNF-α, IL-1β, or LPS did not increase TAD reporter activity. Importantly, genes considered to be faithful TonEBP targets were not induced by any of these inflammatory mediators in NP cells, despite evidence of increased nuclear localization of TonEBP by TNF-α. These results suggest that the mechanism of TonEBP activation differs substantially between osmotic and inflammatory conditions. It is plausible that TonEBP target selectivity may depend on TAD activation status, as has been demonstrated for other transcription factors expressed in the NP niche (49). Interestingly, previous studies suggest that cross-talk exists between osmotic and inflammatory response pathways. In RPE cells, high salt enhances IL-6 and CCL2 production in response to LPS (50). However, in human neutrophils and mononuclear cells, high salt reduces production of proinflammatory cytokines in response to LPS treatment (51). A 2013 study by Kim et al. (22) showed that both high salt and LPS induce TonEBP in macrophages via different sources of reactive oxygen species. In these cells, high salt and LPS had reciprocal effects on TonEBP target genes; pretreatment with LPS dampened the osmoprotective response to high salt, and pretreatment with NaCl decreased LPS-mediated induction of IL-6 (22). In contrast, our studies show that, whereas TNF-α influences TonEBP-TAD activation status, the effect on hyperosmolarity-dependent induction of TauT, SMIT, and AR, canonical osmoregulatory TonEBP targets, is minimal. Thus, the outcome of cross-talk between inflammatory and osmotic stimuli appears to be cell type-specific.
Despite increased nuclear TonEBP, there was lack of induction of canonical osmoregulatory TonEBP targets by TNF-α, which prompted us to perform RNA-seq to identify novel TonEBP targets in this context. Results of these experiments clearly showed that TonEBP governs pathways and functional categories related to inflammatory response, matrix degradation, tissue fibrosis, and interleukin signaling during TNF-α treatment of NP cells. We found that TonEBP controls TNF-α-dependent expression of ADAMTS4, ADAMTS5, and MMP3, matrix-degrading enzymes implicated in disc degeneration (12–14). Likewise, TonEBP controlled expression of PHD3, a recently identified regulator of NF-κB signaling in NP cells (52). In addition, TonEBP modulated levels of TNF, NOS2, TLR2, CCL2, and IL-6, molecules usually linked to tissue inflammation. These results are in agreement with previous studies in macrophages that showed TonEBP plays a role in TLR-triggered induction of NOS2, IL6, TNF, and CCL2 (23). Interestingly, we and others have shown that levels of CCL2, IL6, NOS2, and TNF are also transiently induced by hyperosmolarity in a TonEBP-dependent manner (6, 53, 54). Importantly, RNA-seq data revealed that only about 8.5% of the TNF-α-mediated response in NP cells is TonEBP-dependent. Concerning matrix homeostasis, suppression of ACAN and COL2A1 by TNF-α was not TonEBP-dependent. On the contrary, both of these critical matrix genes have been described as transcriptional targets of TonEBP under basal and hyperosmolar conditions (4, 27, 56). These results suggest that modulation of TonEBP activity may not fully reverse transcriptional changes mediated by TNF-α in NP cells.
In addition to these catabolic and inflammatory genes, our results show for the first time that TonEBP controlled TNF-α-dependent expression of CXCL1/CINC1, CXCL2/CINC3, and CXCL3/CINC2. Studies have detected expression of CXCL1, CXCL2, and CXCL3 in both healthy and degenerative NP tissues with some changes in disc infiltrated by immune cells (38). Underscoring the importance of CXCL1/CINC1, administration of anti-CINC1 antibody significantly decreased pain in a murine model of disc herniation, as measured by mechanical and thermal hyperalgesia (57). Traditionally considered chemoattractants for macrophages and neutrophils, the function and molecular regulation of CXCL1, CXCL2, and CXCL3 expression in the intact, immune privileged disc is unknown. Interestingly, NP cells express CXCR2, a receptor for both CINC1 and CINC3, and receptor expression increases with grade of degeneration (58), suggesting that these chemokines may act on NP cells in an autocrine or paracrine manner and play a role in normal tissue physiology. However, following structural damage to the NP, such as herniation, we hypothesize that high levels of these CXCL family members would drive macrophage and neutrophil migration. Our studies of the CXCL1 promoter revealed an unexpected mode of regulation; the effects of TonEBP required the presence of a highly active NF-κB-binding site (−641/−632) but not the predicted TonE (−124/−127). This result raised the question of whether TonEBP might interact with p65 at this NF-κB-binding site. Indeed, Roth et al. (59) have shown that TonEBP interacts with NF-κB-binding sites of TNF, CCL2, and IκBα in response to hyperosmolarity, presumably as part of a NF-κB–TonEBP complex. In macrophages, TonEBP plays a role in activation of LPS-induced NF-κB activity by recruiting the transcriptional co-activator p300 to the NF-κB enhanceosome (60). Likewise, we have previously shown that NF-κB activity is required for hyperosmotic induction of a subset of TonEBP targets and that NF-κB activation by hyperosmolarity depends on functional TonEBP (6). However, these studies also showed that TonEBP was not immunoprecipitated with p65 under conditions of either hypertonicity or TNF-α stimulation, suggesting either a very transient nature of interaction between these proteins or lack thereof (6). The synergistic action of TonEBP and TNF-α on the CXCL1 promoter and NF-κB prototypic reporter supports the possibility of a transient interaction between TonEBP and p65 and/or other NF-κB subunits, including p50. Interestingly, the ability of TonEBP to regulate the CXCL1 and CXCL2 promoters varied between inflammatory mediators, suggesting stimulus-specific mechanisms of regulation. Importantly, our results suggest that the highly conserved NF-κB-binding site in the CXCL2 promoter is important in mediating response to inflammatory stimuli and that the less conserved TonEs may not mediate the observed effects of TonEBP on TNF-α-mediated CXCL2 transcription. Moreover, results of p65 and TonEBP co-transfections show that TonEBP may enhance activity of p65 even on a less conserved NF-κB-binding site, such as the wild-type NF-κB site 1 in the CXCL2 NF-κB-mutant promoter. Similar to experiments that investigated the effect of TNF-α on osmoregulatory genes, we did not observe modulation of inflammatory TonEBP targets CXCL1 and CXCL2 by hyperosmolarity. There was also no effect of co-treatment with TNF-α and hyperosmolarity on TNF-α-mediated induction of CXCL1 and CXCL2 expression. These results suggest that, unlike in macrophages, in NP cells hyperosmolarity does not inhibit TonEBP action on these inflammatory target genes.
We evaluated TonEBP levels in human samples from healthy and degenerative discs. Although the number of TonEBP-positive NP cells was unaffected by degeneration, we did not quantitatively measure changes in nuclear protein level on a per cell basis because of limited amount of tissue sample. Importantly, however, strong correlation between expression levels of TonEBP and its canonical osmoregulatory targets TauT, SMIT, and AR supports in vitro findings and reflects the fact that the inflammatory milieu of disc degeneration does not interfere with the ability of TonEBP to act on its osmoregulatory targets.
Together, our results show that TNF-α activates TonEBP by promoting its nuclear localization to regulate expression of a subset of the TNF-α-induced inflammatory response in NP cells (schematic, Fig. 8F). These results invite the possibility that targeted TonEBP inhibition might be a novel therapeutic strategy to treat disc degeneration. Indeed, TonEBP heterozygosity partially protects mice from complete Freund's adjuvant-induced arthritis (61). However, it is important to consider the pro-survival, matrix homeostatic, and other physiological activities of TonEBP in the NP when developing such a therapy.
Experimental procedures
Isolation and treatment of NP cells
Rat NP cells were isolated using a method described previously (62). Collection of animal tissues for cell isolation was approved by the IACUC at Thomas Jefferson University. Cells were maintained in DMEM with 10% FBS and antibiotics. For hypertonic culture, 110 mm NaCl was added to medium; for TNF-α treatment, 50 ng/ml was added to medium; for IL-1β treatment, 10 ng/ml was added to medium; for LPS treatment, 2 ng/ml was added to medium.
Human tissue collection and grading
Human lumbar intervertebral disc (IVD) tissue was obtained either at surgery or post-mortem (PM) examination with informed consent of the patient or relatives (Sheffield Research Ethics Committee no. 09/H1308/70). Four PM IVDs were recovered from three donors. They consisted of intact IVDs within the complete motion segment from which the IVDs were removed. Sixty surgical IVD tissue samples were obtained from patients undergoing microdiscectomy procedures for the treatment of low back pain and root pain caused by prolapse of the IVD. NP tissue was divided into two: one-half was fixed in 10% neutral buffered formalin and processed for histological and immunohistochemical examination, and the remaining tissue was used for RNA isolation. H&E-stained sections were used to score the degree of morphological degeneration. Briefly, tissue sections were scored for the presence of cell clusters, the presence of fissures, loss of demarcation and loss of hematoxophilia. A score of 0–4 indicates a histologically normal (non-degenerate) IVD, and a grade of 4.1–12 (>4) indicates evidence of degeneration. Gene-expression study samples were classified as non-degenerate (≤4) and degenerate (>4) based on histological examination. Grading was performed independently by two researchers, and grades were averaged.
Plasmids and reagents
Luciferase reporter plasmids were provided by the following: TauT-luc, Ito et al. (26); WT CXCL1-luc, WT CXCL2-luc, NF-κB Mut CXCL1-luc, and NF-κB Mut CXCL2-luc, Collier and co-workers (28); Ton-TAD, pFR-luc, Burg and co-workers (48); and NF-κB-luc, Fu and Taubman (63). psPAX2 (12260), pMD2G (12259), and FLAG-p65 (20012) (55) were from Addgene. As transfection control, pRL-TK (Promega) was used. Lentiviral ShTonEBP (TRCN0000020019) and control ShRNA pLKO.1 were from Sigma.
Lentiviral studies
HEK-293T cells in 10-cm plates (1.3 × 106 cells/plate) were transfected with 9 μg of either lentiviral ShCTR (pLKO.1) or ShTonEBP plasmids, 6 μg of psPAX2, 3 μg of pMD2.G. After 16 h, the media were removed and replaced with DMEM with 5% FBS. Lentiviral particles were harvested at 48 and 60 h post-transfection and concentrated using PEG solution. NP cells were transduced with media containing viral particles and 8 μg/ml Polybrene. Cells and conditioned medium were collected 5 days post-transduction.
RNA sequencing
Illumina TruSeq-stranded total RNA sample preparation with Ribo-Zero was used to prepare the library. Libraries were chemically denatured and applied to an Illumina HiSeq version 4 single-read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR cluster kit version 4-cBot. After transfer of the flow cell to an Illumina HiSeq 2500, a 50-cycle single-read sequence run was performed (HiSeq SBS kit version 4). For data analysis, Rn5 Ensembl annotations (Build 75) were downloaded and converted to genePred format. Reads were aligned to the transcriptome reference index using Novoalign (version 2.08.01), allowing up to 50 alignments for each read. Read counts were generated using USeq's DefinedRegionDifferentialSeq application and used in DESeq2 to measure the differential expression between each condition. Because of inherent variability between sets arising from independent primary cell isolations, sample preparation batch was accounted for during data normalization. Data have been deposited to the GEO Database (accession no. GSE97496). For IPA, differentially expressed gene lists were used as input to identify enriched related pathways, diseases, and networks.
Real-time qRT-PCR
For in vitro assays, total DNA-free RNA was extracted from NP cells using RNeasy mini columns (Qiagen), and cDNA was made using EcoDry premix (Clontech). For human tissue samples, extracted RNA was subjected to treatment with DNase (Qiagen, Crawley, UK) and purified using Qiagen MinElute Cleanup kit prior to cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (Bioline, London, UK) and random hexamers (Invitrogen). Gene expression of TonEBP was determined in 42 IVD patient samples (2 post-mortem and 40 surgical) via real-time PCR analysis using predesigned, 6-carboxyfluorescein-labeled TaqMan Gene Expression Assays (Applied Biosystems). 10 histologically non-degenerated samples (mean histological grade, 2.7 (range 1–4); mean age, 36 (range 20–45)) and 32 histologically degenerated samples (mean histological grade, 8.2 (range 4.8–12); mean age, 40.9 (range 25–66)) were used for this component of the study.
Western blotting
Cells were placed on ice following treatment and washed with ice-cold PBS. Buffers included 1× protease inhibitor mixture (Roche Applied Science), NaF (4 mm), Na3VO4 (20 mm), NaCl (150 mm), β-glycerophosphate (50 mm), and DTT (0.2 mm). Nuclear proteins were isolated using the CellLytic NuCLEAR extraction kit (Sigma). Proteins were resolved on 8% SDS-polyacrylamide gels and transferred by electroblotting to PVDF membranes (Bio-Rad). Membranes were blocked with 5% nonfat dry milk in TBST and incubated overnight at 4 °C in blocking buffer with rabbit anti-TonEBP (1:1000, Abcam, catalogue no. ab3446, lot GR303959-2), rabbit anti-lamin A/C (1:1000, Cell Signaling, catalogue no. 2032), or mouse anti-β-tubulin antibody (1:2000, DSHB, catalogue no. E-7). Specificity of the TonEBP antibody has been demonstrated previously (6). Immunolabeling was detected with ECL reagent. Chemiluminescence was detected using Digital Imaging System ImageQuant LAS-400 (GE Healthcare). Densitometric analysis was performed (ImageQuant) by first normalizing protein-of-interest levels to the housekeeping protein (β-tubulin) and then normalizing to the experimental control group.
Immunohistochemical analysis
Expression of TonEBP was confirmed and localized via immunohistochemistry in 30 IVDs (2 PM and 28 surgical samples) (supplemental Table 4). 10 histologically non-degenerate samples (mean histological grade, 3.4 (range 2–4); mean age, 35.9 (range 21–46)) and 20 histologically degenerate samples (mean histological grade, 7.7 (range 5–11); mean age, 44.1 (range 18–74)) were used. 4-μm tissue sections were dewaxed and rehydrated, and endogenous peroxidases were blocked. Following heat antigen retrieval, sections were blocked in goat serum and incubated overnight at 4 °C with rabbit polyclonal antibody against human TonEBP (1:100; Abcam, catalogue no. ab3446, lot GR223014-6). Rabbit polyclonal IgG (Abcam, catalogue no. ab27478, lot GR166285-4) was used as a negative control. After washing, sections were incubated with biotinylated goat anti-rabbit antibody (1:500; Abcam, catalogue no. ab97049, lot GR137025-3). Binding was detected using VECTASTAIN Elite ABC HRP reagent (Vector Laboratories, Peterborough, UK) and 3,3′-diaminobenzidine tetrahydrochloride solution (Sigma). Sections were counterstained with Mayer's hematoxylin (Leica Microsystems, Milton Keynes, UK), dehydrated, cleared, and mounted in Pertex (Leica Microsystems). Images were captured using an Olympus BX60 microscope, QImaging Micropublisher 5.0 RTV camera, and QCapture Pro version 8.0 software (Media Cybernetics, Marlow, UK) with Olympic UPlan F1 ×40 or ×100 objective. A total of 200 NP cells were counted in each section and the number of immunopositive cells expressed as a percentage of total count.
Immunofluorescence
NP cells were plated on glass coverslips. After incubation with various stimuli, cells were fixed and permeabilized with ice-cold methanol for 15 min, blocked with PBS containing 5% normal goat serum and 0.3% Triton X-100 for 1 h at room temperature, and incubated with anti-TonEBP (1:150, Abcam) at 4 °C overnight. Cells were washed and incubated with Alexa-Fluor-594-conjugated anti-rabbit antibody (1:800, Jackson ImmunoResearch). After washing, cells were mounted with DAPI-containing mountant (Invitrogen) and visualized using a fluorescence microscope (Zeiss Axio Imager A2 and imaged with Axiocam mRm camera and Plan Apochromat ×63 objective).
ELISA and dot blot
Conditioned medium was filtered (0.45 μm) and supplemented with 1× protease inhibitor mixture (Roche Applied Science). ELISA was performed using Mini ELISA kits for CCL2 and IL-6 (PeproTech). CINC1/CXCL1, CINC3/CXCL2, and CINC2/CXCL3 were detected using the RayBio Rat Cytokine Antibody Array 2 (Ray Biotech Inc., Norcross, GA) following the manufacturer's instructions. Membranes were blocked, incubated with conditioned medium, and incubated with biotin-conjugated primary antibody mixture and then HRP-conjugated secondary antibody. Chemiluminescence was detected using Digital Imaging System ImageQuant LAS-400 (GE Healthcare).
Bioinformatic analysis of promoters and TonE prediction
Rat CXCL1 and CXCL2 promoter sequences were downloaded from the UCSC Genome Table Browser. MatInspector (Genomatix) was used to identify predicted TonEBP-binding sites (TonE), with score cutoff of 0.8. Ensembl browser was used for Multiz alignments of TonE predicted in the rat promoter against human genome.
Transfections and Dual-LuciferaseTM assay
Cells were transferred to 48-well plates (2 × 104 cells/well) 1 day prior to transfection. To measure effects of stimuli on TauT-luc activity, cells were transfected with 250 ng of TauT promoter reporter and 250 ng of pRL-TK plasmid and cultured in control, hypertonic, TNF-α, IL-1β, or LPS treatment conditions. For gain- and loss-of-function studies, FLAG-TonEBP, FLAG-DN-TonEBP, or backbone plasmid (150 ng) was co-transfected with CXCL1, CXCL2, or NF-κB reporter and pRLTK. For experiments with p65 overexpression, 15 ng of FLAG-p65 or backbone was added along with reporter plasmid, FLAG-TonEBP or FLAG-DN-TonEBP, and pRLTK. In all experiments, plasmids were premixed with the transfection reagent, Lipofectamine 2000 (Invitrogen). 48 h after transfection, cells were harvested, and firefly and Renilla luciferase activities were measured using the Dual-LuciferaseTM reporter assay (Promega) and a luminometer (Tecan Infinite M200).
Site-directed mutagenesis
Site-directed mutagenesis of the rat CXCL1 promoter was performed according to the manufacturer's protocol, using the Q5 site-directed mutagenesis kit (New England Biolabs). Primers used for CXCL1 promoter mutants are (mutated TonE underlined): forward, 5′-CCGGTTGTGGTTCCACACCCTGTG-3′; reverse, 5′-AAAGGACATCGCTCCTCC-3′. Mutations were verified by sequencing (Applied Biosystems 3730 DNA sequencer).
Statistics
For quantitative measurements, results are presented as the mean ± S.E. Differences between groups were assessed by analysis of variance and Student's t test using GraphPad Prism software. p values <0.05 were considered statistically significant. Human expression data were non-parametric; thus, Kruskal-Wallis test with post hoc analysis by Conover-Inman test was used to determine significance between 2ΔCT values and percentage immunopositivity between groups. Spearman's rank correlation was used to determine correlations between gene and expression for different targets.
Author contributions
Z. I. J., J. W. S., C. L. L., I. M. S., and M. V. R. conceived the study. Z. I. J. conducted the experiments, analyzed the data, and wrote the manuscript. A. C. D. conducted the experiments, analyzed the data, and wrote the manuscript. J. W. S. conducted the experiments, analyzed the data, and wrote the manuscript. C. L. L. conducted the experiments, analyzed the data, interpreted the results, and wrote the manuscript. I. M. S. designed the study, wrote the manuscript, and secured funding. M. V. R. designed the experiments, the interpreted results, secured funding, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank surgeons Ashley Cole, Neil Chiverton, Anthony Michael, and Lee Breakwell for the human tissue samples.
This work was supported in part by National Institutes of Health Grants AR055655 and AR064733 (to M. V. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S4 and Figs. S1 and S2.
- TonEBP
- tonicity-responsive enhancer-binding protein
- IDD
- intervertebral disc degeneration
- NP
- nucleus pulposus
- TLR
- toll-like receptor
- PM
- post mortem
- TAD
- transactivation domain
- RNA-seq
- RNA-seq
- DN
- dominant-negative
- IPA
- ingenuity pathway analysis
- qRT
- quantitative RT
- IVD
- intervertebral disc.
References
- 1. Burg M. B., Ferraris J. D., and Dmitrieva N. I. (2007) Cellular response to hyperosmotic stresses. Physiol. Rev. 87, 1441–1474 [DOI] [PubMed] [Google Scholar]
- 2. Adams M. A., and Hutton W. C. (1983) The effect of posture on the fluid content of lumbar intervertebral discs. Spine 8, 665–671 [DOI] [PubMed] [Google Scholar]
- 3. Nazari J., Pope M. H., and Graveling R. A. (2015) Feasibility of magnetic resonance imaging (MRI) in obtaining nucleus pulposus (NP) water content with changing postures. Magn. Reson. Imaging 33, 459–464 [DOI] [PubMed] [Google Scholar]
- 4. Tsai T. T., Danielson K. G., Guttapalli A., Oguz E., Albert T. J., Shapiro I. M., and Risbud M. V (2006) TonEBP/OREBP is a regulator of nucleus pulposus cell function and survival in the intervertebral disc. J. Biol. Chem. 281, 25416–25424 [DOI] [PubMed] [Google Scholar]
- 5. Hiyama A., Gajghate S., Sakai D., Mochida J., Shapiro I. M., and Risbud M. V (2009) Activation of TonEBP by calcium controls β1,3-glucuronosyltransferase-I expression, a key regulator of glycosaminoglycan synthesis in cells of the intervertebral disc. J. Biol. Chem. 284, 9824–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson Z. I., Shapiro I. M., and Risbud M. V. (2016) RNA sequencing reveals a role of TonEBP transcription factor in regulation of proinflammatory genes in response to hyperosmolarity in healthy nucleus pulposus cells. J. Biol. Chem. 291, 26686–26697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoy D., Bain C., Williams G., March L., Brooks P., Blyth F., Woolf A., Vos T., and Buchbinder R. (2012) A systematic review of the global prevalence of low back pain. Arthritis Rheum. 64, 2028–2037 [DOI] [PubMed] [Google Scholar]
- 8. Katz J. N. (2006) Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J. Bone Joint Surg. Am. 88, 21–24 [DOI] [PubMed] [Google Scholar]
- 9. Murray C. J., Atkinson C., Bhalla K., Birbeck G., Burstein R., Chou D., Dellavalle R., Danaei G., Ezzati M., Fahimi A., Flaxman D., Foreman Gabriel S., Gakidou E., Kassebaum N., et al. (2013) The State of US Health, 1990–2010. JAMA 310, 591–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Livshits G., Popham M., Malkin I., Sambrook P. N., Macgregor A. J., Spector T., and Williams F. M. (2011) Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women: the UK Twin Spine Study. Ann. Rheum. Dis. 70, 1740–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stewart W. F., Ricci J. A., Chee E., Morganstein D., and Lipton R. (2003) Lost productive time and cost due to common pain conditions in the U.S. workforce. JAMA 290, 2443–2454 [DOI] [PubMed] [Google Scholar]
- 12. Wang J., Markova D., Anderson D. G., Zheng Z., Shapiro I. M., and Risbud M. V. (2011) TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J. Biol. Chem. 286, 39738–39749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Wang H., Yang H., Li J., Cai Q., Shapiro I. M., and Risbud M. V. (2014) Tumor necrosis factor-α- and interleukin-1β-dependent matrix metalloproteinase-3 expression in nucleus pulposus cells requires cooperative signaling via syndecan 4 and mitogen-activated protein kinase-NF-κB axis. Am. J. Pathol. 184, 2560–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tian Y., Yuan W., Fujita N., Wang J., Wang H., Shapiro I. M., and Risbud M. V. (2013) Inflammatory cytokines associated with degenerative disc disease control aggrecanase-1 (ADAMTS-4) expression in nucleus pulposus cells through MAPK and NF-κB. Am. J. Pathol. 182, 2310–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Binch A. L., Shapiro I. M., and Risbud M. V. (2016) Syndecan-4 in intervertebral disc and cartilage: saint or synner? Matrix Biol. 52, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson Z. I., Schoepflin Z. R., Choi H., Shapiro I. M., and Risbud M. V. (2015) Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur. Cell Mater. 30, 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng H., Danfelter M., Strömqvist B., and Heinegård D. (2006) Extracellular matrix in disc degeneration. J. Bone Joint Surg. 88, 25–29 [DOI] [PubMed] [Google Scholar]
- 18. Kumaresan S., Yoganandan N., Pintar F. A., Macias M., and Cusick J. F. (2000) Morphology of young and old cervical spine intervertebral disc tissues. Biomed. Sci. Instrum. 36, 141–146 [PubMed] [Google Scholar]
- 19. Gawri R., Rosenzweig D. H., Krock E., Ouellet J. A., Stone L. S., Quinn T. M., and Haglund L. (2014) High mechanical strain of primary intervertebral disc cells promotes secretion of inflammatory factors associated with disc degeneration and pain. Arthritis Res. Ther. 16, R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quero L., Klawitter M., Schmaus A., Rothley M., Sleeman J., Tiaden A. N., Klasen J., Boos N., Hottiger M. O., Wuertz K., and Richards P. J. (2013) Hyaluronic acid fragments enhance the inflammatory and catabolic response in human intervertebral disc cells through modulation of toll-like receptor 2 signalling pathways. Arthritis Res. Ther. 15, R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeong G. R., Im S.-K., Bae Y.-H., Park E. S., Jin B. K., Kwon H. M., Lee B.-J., Bu Y., Hur E.-M., and Lee B. D. (2016) Inflammatory signals induce the expression of tonicity-responsive enhancer binding protein (TonEBP) in microglia. J. Neuroimmunol. 295, 21–29 [DOI] [PubMed] [Google Scholar]
- 22. Kim N. H., Hong B.-K., Choi S. Y., Moo Kwon H., Cho C.-S., Yi E. C., and Kim W. U. (2013) Reactive oxygen species regulate context-dependent inhibition of NFAT5 target genes. Exp. Mol. Med. 45, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buxadé M., Lunazzi G., Minguillón J., Iborra S., Berga-Bolaños R., Del Val M., Aramburu J., and López-Rodríguez C. (2012) Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 209, 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ranjbar S., Jasenosky L. D., Chow N., and Goldfeld A. E. (2012) Regulation of Mycobacterium tuberculosis-dependent HIV-1 transcription reveals a new role for NFAT5 in the toll-like receptor pathway. PLoS Pathog. 8, e1002620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoon H. J., You S., Yoo S.-A., Kim N. H., Kwon H. M., Yoon C. H., Cho C.-S., Hwang D., and Kim W. U. (2011) NF-AT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum. 63, 1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T., Fujio Y., Hirata M., Takatani T., Matsuda T., Muraoka S., Takahashi K., and Azuma J. (2004) Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem. J. 382, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caron M. M., van der Windt A. E., Emans P. J., van Rhijn L. W., Jahr H., and Welting T. J. (2013) Osmolarity determines the in vitro chondrogenic differentiation capacity of progenitor cells via nuclear factor of activated T-cells 5. Bone 53, 94–102 [DOI] [PubMed] [Google Scholar]
- 28. Burke S. J., Lu D., Sparer T. E., Masi T., Goff M. R., Karlstad M. D., and Collier J. J. (2014) NF-κB and STAT1 control CXCL1 and CXCL2 gene transcription. Am. J. Physiol. Endocrinol. Metab. 306, E131–E149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Urban J. P., and McMullin J. F. (1985) Swelling pressure of the inervertebral disc: influence of proteoglycan and collagen contents. Biorheology 22, 145–157 [DOI] [PubMed] [Google Scholar]
- 30. Paajanen H., Lehto I., Alanen A., Erkintalo M., and Komu M. (1994) Diurnal fluid changes of lumbar discs measured indirectly by magnetic resonance imaging. J. Orthop. Res. 12, 509–514 [DOI] [PubMed] [Google Scholar]
- 31. Boos N., Wallin A., Gbedegbegnon T., Aebi M., and Boesch C. (1993) Quantitative MR imaging of lumbar intervertebral disks and vertebral bodies: influence of diurnal water content variations. Radiology 188, 351–354 [DOI] [PubMed] [Google Scholar]
- 32. Nachemson A., and Morris J. M. (1964) In vivo measurements of intradiscal pressure. Discometry, a method for the determination of pressure in the lower lumbar discs. J. Bone Joint Surg. Am. 46, 1077–1092 [PubMed] [Google Scholar]
- 33. Johnson Z. I., Shapiro I. M., and Risbud M. V. (2014) Extracellular osmolarity regulates matrix homeostasis in the intervertebral disc and articular cartilage: evolving role of TonEBP. Matrix Biol. 40, 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Risbud M. V., and Shapiro I. M. (2014) Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat. Rev. Rheumatol. 10, 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urban J. P. (2002) The role of the physicochemical environment in determining disc cell behaviour. Biochem. Soc. Trans. 30, 858–864 [DOI] [PubMed] [Google Scholar]
- 36. Bachmeier B. E., Nerlich A. G., Weiler C., Paesold G., Jochum M., and Boos N. (2007) Analysis of tissue distribution of TNF-α, TNF-α-receptors, and the activating TNF-α-converting enzyme suggests activation of the TNF- system in the aging intervertebral disc. Ann. N.Y. Acad. Sci. 1096, 44–54 [DOI] [PubMed] [Google Scholar]
- 37. Le Maitre C. L., Freemont A. J., and Hoyland J. A. (2005) The role of interleukin-1 in the pathogenesis of human intervertebral disc degeneration. Arthritis Res. Ther. 7, R732–R745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phillips K. L., Chiverton N., Michael A. L., Cole A. A., Breakwell L. M., Haddock G., Bunning R. A., Cross A. K., and Le Maitre C. L. (2013) The cytokine and chemokine expression profile of nucleus pulposus cells: implications for degeneration and regeneration of the intervertebral disc. Arthritis Res. Ther. 15, R213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Séguin C. A., Pilliar R. M., Roughley P. J., and Kandel R. A. (2005) Tumor necrosis factor-α modulates matrix production and catabolism in nucleus pulposus tissue. Spine 30, 1940–1948 [DOI] [PubMed] [Google Scholar]
- 40. Shen B., Melrose J., Ghosh P., and Taylor F. (2003) Induction of matrix metalloproteinase-2 and -3 activity in ovine nucleus pulposus cells grown in three-dimensional agarose gel culture by interleukin-1β: a potential pathway of disc degeneration. Eur. Spine J. 12, 66–75 [DOI] [PubMed] [Google Scholar]
- 41. Kepler C. K., Markova D. Z., Hilibrand A. S., Vaccaro A. R., Risbud M. V., Albert T. J., and Anderson D. G. (2013) Substance P stimulates production of inflammatory cytokines in human disc cells. Spine 38, E1291–E1299 [DOI] [PubMed] [Google Scholar]
- 42. Kepler C. K., Markova D. Z., Dibra F., Yadla S., Vaccaro A. R., Risbud M. V., Albert T. J., and Anderson D. G. (2013) Expression and relationship of proinflammatory chemokine RANTES/CCL5 and cytokine IL-1β in painful human intervertebral discs. Spine 38, 873–880 [DOI] [PubMed] [Google Scholar]
- 43. Purmessur D., Walter B. A., Roughley P. J., Laudier D. M., Hecht A. C., and Iatridis J. (2013) A role for TNFα in intervertebral disc degeneration: a non-recoverable catabolic shift. Biochem. Biophys. Res. Commun. 433, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen C., Yan J., Jiang L. S., and Dai L. Y. (2011) Autophagy in rat annulus fibrosus cells: evidence and possible implications. Arthritis Res. Ther. 13, R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miyakawa H., Woo S. K., Dahl S. C., Handler J. S., and Kwon H. M. (1999) Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc. Natl. Acad. Sci. U.S.A. 96, 2538–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ko B. C., Turck C. W., Lee K. W., Yang Y., and Chung S. S. (2000) Purification, identification, and characterization of an osmotic response element binding protein. Biochem. Biophys. Res. Commun. 270, 52–61 [DOI] [PubMed] [Google Scholar]
- 47. Woo S. K., Dahl S. C., Handler J. S., and Kwon H. M. (2000) Bidirectional regulation of tonicity-responsive enhancer binding protein in response to changes in tonicity. Am. J. Physiol. Renal Physiol. 278, F1006–F1012 [DOI] [PubMed] [Google Scholar]
- 48. Ferraris J. D., Williams C. K., Persaud P., Zhang Z., Chen Y., and Burg M. B. (2002) Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration. Proc. Natl. Acad. Sci. U.S.A. 99, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dayan F., Roux D., Brahimi-Horn M. C., Pouyssegur J., and Mazure N. M. (2006) The oxygen sensor factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bifunctional transcriptional character of hypoxia-inducible factor-1. Cancer Res. 66, 3688–3698 [DOI] [PubMed] [Google Scholar]
- 50. Zhang D., Wang C., Cao S., Ye Z., Deng B., Kijlstra A., and Yang P. (2015) High-salt enhances the inflammatory response by retina pigment epithelium cells following lipopolysaccharide stimulation. Mediators Inflamm. 2015, 197521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hatanaka E., Shimomi F. M., Curi R., and Campa A. (2007) Sodium chloride inhibits cytokine production by lipopolysaccharide-stimulated human neutrophils and mononuclear cells. Shock 27, 32–35 [DOI] [PubMed] [Google Scholar]
- 52. Fujita N., Markova D., Anderson D. G., Chiba K., Toyama Y., Shapiro I. M., and Risbud M. V (2012) Expression of prolyl hydroxylases (PHDs) is selectively controlled by HIF-1 and HIF-2 proteins in nucleus pulposus cells of the intervertebral disc: distinct roles of PHD2 and PHD3 proteins in controlling HIF-1α activity in hypoxia. J. Biol. Chem. 287, 16975–16986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kojima R., Taniguchi H., Tsuzuki A., Nakamura K., Sakakura Y., and Ito M. (2010) Hypertonicity-induced expression of monocyte chemoattractant protein-1 through a novel cis-acting element and MAPK signaling pathways. J. Immunol. 184, 5253–5262 [DOI] [PubMed] [Google Scholar]
- 54. Ueno M., Shen W. J., Patel S., Greenberg A. S., Azhar S., and Kraemer F. B. (2013) Fat-specific protein 27 modulates nuclear factor of activated T cells 5 and the cellular response to stress. J. Lipid Res. 54, 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sanjabi S., Williams K. J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., and Smale S. T. (2005) A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 19, 2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van der Windt A. E., Haak E., Das R. H., Kops N., Welting T. J., Caron M. M., van Til N. P., Verhaar J. A., Weinans H., and Jahr H. (2010) Physiological tonicity improves human chondrogenic marker expression through nuclear factor of activated T-cells 5 in vitro. Arthritis Res. Ther. 12, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Souza Grava A. L., Ferrari L. F., and Defino H. L. (2012) Cytokine inhibition and time-related influence of inflammatory stimuli on the hyperalgesia induced by the nucleus pulposus. Eur. Spine J. 21, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phillips K. L., Cullen K., Chiverton N., Michael A. L., Cole A. A., Breakwell L. M., Haddock G., Bunning R. A., Cross A. K., and Le Maitre C. L. (2015) Potential roles of cytokines and chemokines in human intervertebral disc degeneration: interleukin-1 is a master regulator of catabolic processes. Osteoarthr. Cartil. 23, 1165–1177 [DOI] [PubMed] [Google Scholar]
- 59. Roth I., Leroy V., Kwon H. M., Martin P. Y., Féraille E., and Hasler U. (2010) Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-κB activity. Mol. Biol. Cell 21, 3459–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee H. H., Sanada S., An S. M., Ye B. J., Lee J. H., Seo Y. K., Lee C., Lee-Kwon W., Küper C., Neuhofer W., Choi S. Y., and Kwon H. M. (2016) LPS-induced NFκB enhanceosome requires TonEBP/NFAT5 without DNA binding. Sci. Rep. 6, 24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim N. H., Choi S., Han E. J., Hong B. K., Choi S. Y., Kwon H. M., Hwang S. Y., Cho C. S., and Kim W.-U. (2014) The xanthine oxidase-NFAT5 pathway regulates macrophage activation and TLR-induced inflammatory arthritis. Eur. J. Immunol. 44, 2721–2736 [DOI] [PubMed] [Google Scholar]
- 62. Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., and Shapiro I. M. (2006) Nucleus pulposus cells express HIF-1α under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem. 98, 152–159 [DOI] [PubMed] [Google Scholar]
- 63. Fu J., and Taubman M. B. (2010) Prolyl hydroxylase EGLN3 regulates skeletal myoblast differentiation through an NF-κB-dependent pathway. J. Biol. Chem. 285, 8927–8935 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.